Abstract
Adenoviruses (AdV) can cause life-threatening infections in immunosuppressed patients. Reliable diagnostic tests are therefore of paramount importance. Apparently, any of the six AdV species (A to F), currently comprising 51 different serotypes, can play a clinically important role in patients with impaired immune response. It is imperative therefore that diagnostic assays cover the entire spectrum of these viruses. We have sequenced presumably conserved regions of the adenoviral genome in all AdV serotypes. Based on the complete sequence information of the hexon gene, we were able to develop a two-reaction real-time PCR assay covering all human adenoviruses with equally high specificity and sensitivity. The detection systems were tested using reference strains for all 51 serotypes and >1,000 clinical samples derived from peripheral blood and stool specimens from pediatric patients after allogeneic stem cell transplantation. The two-reaction assay presented permits highly specific detection and quantification of adenoviral DNA of any serotype. From the perspective of routine clinical diagnosis, the assay represents an important improvement over existing approaches by providing a sensitive and economic technique for early detection and monitoring of adenoviral infections.
Adenoviruses (AdV) are pathogens causing serious infections in immunosuppressed patients, particularly in human immunodeficiency virus-positive individuals and in allogeneic bone marrow transplant recipients (4, 5, 8, 10, 11, 29, 31, 32). To date, 51 different human AdV serotypes have been identified (8, 35). They are divided into six major subgroups (subgenera or species A to F) on the basis of their oncogenic, hemagglutinating, morphological, and DNA sequence properties (3, 13, 16). Any adenovirus species may cause life-threatening infections (4, 13, 26, 23). In most clinical situations involving adenovirus infection, species identification of an AdV isolate is as informative as a finer identification by serotype (16).
Infections can cause localized disease such as enteritis, upper respiratory tract infection, encephalitis, or cystitis (19). However, AdV infections in immunocompromised patients tend to become invasive, and disseminated disease is associated with very high mortality rates (7, 9, 13, 17, 20, 24, 28). Currently available treatment options, which include antiviral agents such as cidofovir and ribavirin, immunomodulation, and, more recently, cytotoxic T cells, have been reported to work successfully at least in a proportion of patients, thus emphasizing the importance of adequate diagnostic techniques permitting timely detection and monitoring of the course of infection (18, 25, 33).
Earlier diagnostic approaches to AdV detection relied mainly on serological tests and cell culture (1, 2, 6, 14, 15). In immunosuppressed patients, however, the use of serological tests is limited due to the impaired immune response, and evaluation of positive cultures is a relatively slow method (28). The recent introduction of PCR-based assays has opened new ways to rapid, specific, and highly sensitive AdV detection (9, 12, 17, 19, 20, 28, 27, 30, 35). Many of these diagnostic approaches, however, do not effectively cover all AdV types or use lower-stringency conditions to permit detection of the genetically highly diverse adenoviruses. We have recently introduced a five-reaction real-time PCR assay permitting reliable detection and quantification of all 51 currently known human AdV serotypes (23). However, the relatively high cost and labor associated with this assay have been a major impediment to its use in the routine diagnostic setting. In view of the clinical importance of adenovirus screening in immunocompromised patients, it was desirable to develop a more economic diagnostic test. Based on the AdV sequence information generated in our laboratory, we have established a two-reaction real-time PCR (RQ-PCR) assay, relying on the use of primers and probes targeting the hexon or the fiber gene and permitting quantitative analysis of the entire spectrum of known human adenoviruses at a sensitivity and specificity that compare favorably to the earlier five-reaction assay. The new two-reaction test system presented herein provides a rapid and cost-effective approach to adenovirus detection in the routine clinical setting.
MATERIALS AND METHODS
Clinical samples.
Forty-five consecutive pediatric patients undergoing allogeneic stem cell transplantation at St. Anna Children's Hospital, Vienna, Austria, between May 2003 and September 2004 were screened by the two-reaction PCR technique for the presence of latent AdV infection in peripheral blood (PB) leukocytes prior to transplantation. Subsequently, prospective virus screening of PB and stool was performed at short intervals until immunologic reconstitution, as previously described (23). In AdV-positive samples, virus load was determined by real-time PCR as outlined below. In total, >1,000 clinical samples were screened by the technique described.
Isolation of DNA.
Cell culture supernatants derived from reference strains of all 51 serotypes (kindly provided by H. Niesters, Department of Virology, University of Rotterdam, The Netherlands, and A. Heim, Department of Virology, Medical University of Hannover, Germany) were lysed, and DNA was extracted using the QIAmp DNA Blood Mini kit (QIAGEN, Hilden, Germany). The DNA content was quantified by fluorometric analysis using the PicoGreen dsDNA Quantitation kit (Molecular Probes Inc., Eugene, OR) and the Fluorescence Spectrophotometer F-2500 (HITACHI, Japan).
For AdV analysis in PB, virus DNA was usually isolated from whole-blood samples using the QIAmp Blood DNA Mini kit. For the assessment of latent versus active infection, the same kit and extraction protocol were used for the isolation of viral DNA from white blood cells and serum or plasma, respectively. For the isolation of DNA from stool samples, the QIAmp DNA Stool Mini kit was used (QIAGEN, Hilden, Germany) according to the manufacturer's recommendations. The positive controls employed to document successful DNA isolation were previously described in detail (23, 34).
Real-time quantitative PCR.
Primers and probes for the two-reaction panadenovirus detection system were placed within conserved regions of the hexon and the fiber genes, according to sequences available in part from public databases (National Center for Biotechnology Information) and, for the most part, on the basis of new hexon gene sequence data generated in our laboratory. The primer and probe design was based on the alignment of previously published hexon and fiber gene sequences of serotypes 2, 3, 4, 5, 7, 11, 12, 16, 21, 34, 35, 40, 41, and 48 (serotypes and accession numbers for the hexon gene are AdV2, AJ293905; AdV3, X76549; AdV4, X84646; AdV5, J01966; AdV7, AF515814; AdV12, NC_001460; AdV16, X74662; AdV21, AY008279; AdV34, AB052911; AdV35, AB052912; AdV40, X51782; AdV41, D13781; AdV48, U20821; and for the fiber gene, serotype AdV40 and accession number L19443) and on the sequence data of the remaining 37 AdV serotypes determined in our laboratory (data not shown).
Prior to experimental testing, the primer and probe sequences were controlled by standard nucleotide-nucleotide BLAST (National Center for Biotechnology Information) searches for the absence of homology with any other relevant organism. All primers and probes included in the two-reaction assay (Table 1) were designed with the help of Primer Express software (Applied Biosystems, Foster City, CA) to work under identical cycling conditions.
TABLE 1.
Sequences and concentrations of primers and probes included in the two- reaction real-time PCR assaya
| Reaction and primer or probe | 5′ Labeling | Oligonucleotide sequence (5′-3′) | 3′ Labeling | Nucleotide positions | Concn (μM) |
|---|---|---|---|---|---|
| Reaction 1 | |||||
| AdV ACF forward I | GGK CTG GTG CAA TTC GCC | 78-96 | 0.3 | ||
| AdV ACF probe I | FAM | CCA CGG ACA CCT ACT TCA CCC TGG G | TAMRA | 101-125 | 0.2 |
| AdV ACF reverse I | CAC GGG CAC AAA ACG CA | 197-213 | 0.3 | ||
| AdV ACF forward II | ACC TGG GCC AAA ACC TTC TC | 2659-2679 | 0.3 | ||
| AdV ACF probe II | FAM | AAC TCC GCC CAC GCG CTA GA | TAMRA | 2686-2705 | 0.2 |
| AdV ACF reverse II | CGT CCA TGG GAT CCA CCT C | 2716-2734 | 0.9 | ||
| AdV ACF forward III | CCC GTG TTT GAC AAC GAA GG | 1324-1343 | 0.3 | ||
| AdV ACF probe III | FAM | ATC GAC AAG GAC AGT CTG CCA ACA CTA ACG | TAMRA | 1373-1402 | 0.2 |
| AdV ACF reverse III | TTA GAG CTA GGC ATA AAT TCT ACA GCA | 1410-1436 | 0.3 | ||
| Reaction 2 | |||||
| AdV BDE forward | ACA TGC ACA TCG CCG G | 35-50 | 0.4 | ||
| AdV BDE probe I | FAM | CGG GTC TGG TGC AGT | NFQ | 77-91 | 0.2 |
| AdV BDE probe II | FAM | CGG GTT TGG TGC AGT | NFQ | 77-91 | 0.2 |
| AdV BDE reverse | CGG TCS GTG GTC ACA TC | 163-179 | 0.4 |
A mixture of primers and probes for the detection of all adenovirus serotypes belonging to the species A, C, and F and B, D, and E, respectively, was used. The nucleotide positions are derived from the hexon gene, except for the primers and probe AdV ACF III, which are derived from the fiber-2 gene. NFQ, nonfluorescent quencher (not specified by the manufacturer [Applied Biosystems]).
PCRs were set up in a total volume of 25 μl, including 12.5 μl TaqMan Universal Master Mix (Eurogentec, Seraing, Belgium) and 6 μl template DNA. For reaction 1, covering AdV species A, C, and F, TaqMan probes were used, which were labeled with 6-carboxyfluorescein at the 5′ end and with carboxy tetramethyl-rhodamine as a quencher at the 3′ end (Eurogentec, Seraing, Belgium). For reaction 2, covering AdV species B, D, and E, minor groove binding probes labeled with 6-carboxyfluorescein at the 5′ end and with a nonfluorescent quencher at the 3′ end (Applied Biosystems, Foster City, CA,) were employed (Table 1). Amplifications were carried out using the ABI Prism 7700 or 7900 (Applied Biosystems, Foster City, CA) for a total of 50 cycles. After an initial denaturation step for 10 min at 95°C, each cycle consisted of denaturation for 15 s at 95°C and annealing and primer extension for 60 s at 60°C. Strict precautions were undertaken to prevent contamination of PCRs with exogenous products as previously described (21). To reduce the risk of false-positive results due to contamination with PCR products, dTTP was partially replaced by dUTP in the reaction master mixture and a dUTP glycosylase step was performed prior to each PCR, as described previously (34). Each DNA sample was analyzed in duplicate, and multiple negative controls were included in each assay. DNA isolates derived from different AdV serotypes served as positive controls. In largely cell-free clinical samples such as plasma, serum, cerebrospinal fluid, or stool, an additional control for efficient DNA extraction and the presence of inhibitors of amplification was the seal herpesvirus PHHV (kindly provided by H. Niesters, Department of Virology, University of Rotterdam, The Netherlands). The PHHV stock solutions were quantified by fluorometric measurement of extracted viral DNA and subsequent calculation of viral particles based on the size of the virus genome. Defined concentrations of PHHV were spiked into each sample prior to isolation of DNA. Successful extraction of viral DNA and absence of inhibitory effects were documented by real-time PCR quantification of PHHV copy number, as described previously (34). The presence of spiked control virus had no effect on the sensitivity of AdV detection in clinical samples. In cell-rich clinical samples, such as peripheral blood or tissue biopsy material, a human single-copy housekeeping gene (beta2-microglobulin, designated B2-MG) (22, 34) was tested in parallel by RQ-PCR as an additional amplification control.
Clinical samples that tested AdV positive were controlled by an independent assay based on the species-specific RQ-PCR described earlier (23), to verify the results. For the quantification of virus load, external standard curves were established using serial dilutions of fluorometrically quantified virus DNA preparations corresponding to defined virus particle equivalents. Viral DNA was derived from reference strains of serotypes 2, 4, 7, 18, 24, and 40, as representatives of all six AdV species. The standard curves were very similar, permitting linear virus quantification over the entire range tested, and corresponded well with amplification curves obtained from all other serotypes. For the two-reaction PCR assay presented, we therefore selected two standard curves, one representative of the species A, C, and F (reaction 1) and B, D, and E (reaction 2), respectively, using serotypes 2 and 7, as specified in Results. For assessment of virus copies per cell, a single-copy gene (B2-MG) was quantified in parallel by real-time PCR (33). When investigating largely cell-free liquids such as plasma, quantitative results were expressed as the number of virus copies per milliliter. In stool samples, the virus copies were calculated per gram of material, and intracellular virus copies were indicated per 1E + 6 cells.
RESULTS AND DISCUSSION
Two-reaction panadenovirus PCR detection system (patent pending). Specificity of the assay.
The analysis of DNA sequences of the hexon gene in the entire panel of currently known human adenoviruses revealed a certain level of similarity between serotypes belonging to the AdV species A, C, and F (ACF) and B, D, and E (BDE), respectively. The degree of homology within the two groups of AdV species permitted the establishment of a single RQ-PCR covering all serotypes of the species A, C, and F (reaction 1) and a second reaction facilitating the detection of all serotypes of the species B, D, and E (reaction 2). The disparity of DNA sequences between the two groups precluded the design of a single PCR covering all 51 serotypes with optimal specificity. As indicated in Table 1, PCR-1 included three primer sets and three TaqMan probes, and reaction 2 comprised a single primer set and two minor groove binding probes.
The strategy of combining a mixture of primers and probes in a single reaction was based on earlier results indicating that the presence of mismatches with the targeted sequences may result in decreased sensitivity of detection for certain AdV serotypes. Conversely, lower-stringency conditions, mainly including a decreased temperature of the annealing step that was implemented in attempts to ensure adequate sensitivity despite the presence of mismatches, may compromise the specificity of the assay (unpublished observations). The RQ-PCR assay presented was carried out under high-stringency conditions with an annealing temperature of 60°C, thus ensuring high specificity. For individual serotypes there was only marginal cross-reactivity between the ACF and the BDE detection systems: serotype 40 belonging to species F showed minimal cross-reactivity with the BDE detection system, and the serotypes 42, 44, and 45 belonging to species D showed borderline cross-reactivity with the ACF detection system. However, the difference in Ct values between the appropriate and the nonappropriate detection systems obtained for these serotypes was in the range of 20 cycles, reflecting a >5-log-greater amplification efficiency of the correct RQ-PCR. Hence, the observed level of cross-reactivity was rather of theoretical interest and had no relevance for clinical diagnosis.
Additionally, >200 patient specimens were tested in parallel by the two-reaction RQ-PCR presented and the species-specific five-reaction RQ-PCR published previously (23). There was complete concordance between the results obtained, both for the AdV-negative (n = 144) and -positive (n = 56) findings. Under the stringent assay conditions used, no false positive results were observed in >1,000 clinical specimens investigated by the two-reaction assay, the majority of which reproducibly tested negative, as documented by Ct values of 50.
Sensitivity of the assay.
The lower detection limit of the diagnostic test described was assessed by the analysis of dilution series prepared using fluorometrically quantified viral DNA derived from reference strains of all 51 serotypes. The assay has been tested for comparable amplification efficiency and sensitivity in different types of clinical specimens by spiking experiments with defined preparations of control virus. In largely cell-free specimens, it was possible to reliably detect the equivalent of 1E + 2 virus particles per milliliter or gram of the sample investigated (Fig. 1). For reproducible quantification of virus load, however, the presence of ≥1E + 3 particles per milliliter or gram were necessary. When analyzing cell material, sensitivity of the assays permitted detection and quantification of virus copies at a level of 10 particles per 1E + 6 cells. These results are equivalent to the five-reaction adenovirus assay we have developed previously (23) and are in line with other real-time PCR virus detection assays established in our laboratory (34).
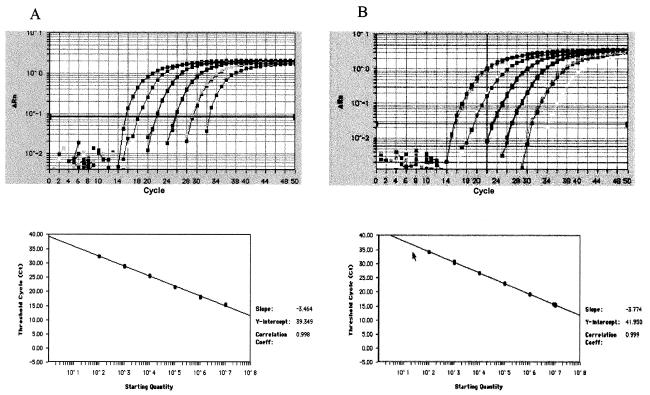
FIG. 1.
Amplification plots and standard curves. (A) Serial dilution of adenoviral DNA (species A, serotype 2) ranging from 102 to 107 virus particles per reaction. The analysis was done in duplicate using the ACF RQ-PCR system. (B) Serial dilution of adenoviral DNA (species B, serotype 7) ranging from 102 to 107 virus particles per reaction. The analysis was done in duplicate using the BDE system. The amplification plots displayed were concordant with those established using serial dilutions of serotypes representative of other AdV species. The amplification plots generated on the basis of the above serial dilutions therefore served as external standard curves for the ACF and BDE RQ-PCR detection systems, respectively.
Quantification of virus load.
Virus particles in clinical samples were quantified according to standard curves established with reference strains of AdV serotypes 2 and 7, representative of species ACF and BDE, respectively (Fig. 1), as outlined in Materials and Methods. The two-reaction assay presented permitted analysis of virus copy numbers across a range of >7 logs. In the current series of >1,000 peripheral blood and stool samples derived from 45 pediatric patients after allogeneic stem cell transplantation, AdV positivity was detected in 11 (25%) of the individuals tested. The majority of AdV-positive patients had merely a localized intestinal infection, as revealed by repeatedly positive stool samples. In total, 115 specimens were AdV positive, including 94 and 21 positive test results in stool and peripheral blood samples, respectively. These findings are in accordance with earlier data, which indicated that the intestinal tract is the most common site of AdV infection during the posttransplant period. Our long-term observations revealed intestinal AdV infections in approximately one-third of all patients undergoing allogeneic stem cell transplantation at our center, whereas invasive infections, defined by virus detection in peripheral blood samples, were seen in <10% of transplant recipients (reference 23 and unpublished results). All samples that tested AdV positive by the two-reaction assay were retested by the species-specific RQ-PCR method described previously (23) to verify the results. The AdV species identified included the subgenera A (one patient), C (nine patients), and D (one patient). There was complete concordance between the results obtained by the two methods, both qualitatively and quantitatively. The AdV levels in patients who displayed self-limiting intestinal infection were below 5E + 5 virus copies per gram of stool, without showing increasing viral load in serial samples. Similar to earlier findings, three patients in the present study, who had higher virus levels in stool samples and showed rapid proliferation kinetics, experienced an invasive infection, as documented by subsequent detection of the virus in peripheral blood samples. This observation preceded the onset of clinical symptoms of disseminated AdV disease by more than a week in one of the patients, in agreement with the findings described earlier in a cohort of pediatric transplant recipients (23). This patient died from disseminated adenovirus disease, despite preemptive antiviral treatment. Two other patients with invasive AdV infection responded to preemptive antiviral therapy, which resulted in clearance of the virus from peripheral blood and in a dramatic decrease of viral load in serial stool samples.
Concluding comments.
The two-reaction panadenovirus RQ-PCR assay described facilitates highly sensitive and specific detection of all currently known human serotypes of the virus and permits quantitative assessment of viral load in a variety of clinical specimens. The assay can be completed within 4 h from sample collection to the report of results and has proven to be very well suited for high-throughput clinical screening. The performance of this assay is comparable to the species-specific RQ-PCR test established previously in our laboratory (23). The latter test, however, was too cost and labor intensive to permit its wide application in the clinical setting. Hence, the main advantages of the two-reaction assay presented include substantially (∼60%) lower costs with regard to reagents and labor, while the broad specificity and sensitivity have been maintained. The test permits rapid quantitative detection of any adenovirus serotype, which is an important prerequisite for timely onset of appropriate treatment. Wide application of the assay in clinical virus screening may therefore contribute to an improvement of the outcome of adenovirus infections in immunocompromised patients.
Acknowledgments
This work has been supported by a grant from the Jubiläumsfonds of the National Bank of Austria (grant no. 11168).
REFERENCES
- 1.Akalu, A., W. Seidel, H. Liebermann, U. Bauer, and L. Dohner. 1998. Rapid identification of subgenera of human adenovirus by serological and PCR assays. J. Virol. Methods 71:187-196. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin, A., H. Kingman, M. Darville, A. B. Foot, D. Grier, J. M. Cornish, N. Goulden, A. Oakhill, D. H. Pamphilon, C. G. Steward, and D. I. Marks. 2000. Outcome and clinical course of 100 patients with adenovirus infection following bone marrow transplantation. Bone Marrow Transplant. 26:1333-1338. [DOI] [PubMed] [Google Scholar]
- 3.Benkö, M, B. Harrach, and W. C. Russell. 1999. Family Adenoviridae. Mastadenovirus, p. 227-238. In M. H. V. Van Regenmortel, C. M. Fauquet, D. K. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: seventh report of the International Committee on Taxonomy of Viruses. Academic Press, New York, N.Y.
- 4.Blanke, C., C. Clark, E. R. Broun, G. Tricot, I. Cunningham, K. Cornetta, A. Hedderman, and R. Hromas. 1995. Evolving pathogens in allogeneic bone marrow transplantation: increased fatal adenoviral infections. Am. J. Med. 99:326-328. [DOI] [PubMed] [Google Scholar]
- 5.Bruno, B., R. A. Zager, M. J. Boeckh, T. A. Gooley, D. H. Myerson, M. L. Huang, and R. C. Hackman. 2004. Adenovirus nephritis in hematopoietic stem-cell transplantation. Transplantation 77:1049-1057. [DOI] [PubMed] [Google Scholar]
- 6.Carrigan, D. R. 1997. Adenovirus infections in immunocompromised patients. Am. J. Med. 102:71-74. [DOI] [PubMed] [Google Scholar]
- 7.Chakrabarti, S., K. E. Collingham, C. D. Fegan, D. Pillay, and D. W. Milligan. 2000. Adenovirus infections following haematopoietic cell transplantation: is there a role for adoptive immunotherapy? Bone Marrow Transplant. 26:305-307. [DOI] [PubMed] [Google Scholar]
- 8.De Jong, J. C., A. G. Wermenbol, M. W. Verweij-Uijterwaal, K. W. Slaterus, P. Wertheim-Van Dillen, G. J. Van Doornum, S. H. Khoo, and J. C. Hierholzer. 1999. Adenoviruses from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotypes Ad50 and Ad51 of species B1 and D, respectively. J. Clin. Microbiol. 37:3940-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Echavarria, M., M. Forman, M. J. van Tol, J. M. Vossen, P. Charache, and A. C. Kroes. 2001. Prediction of severe disseminated adenovirus infection by serum PCR. Lancet. 358:384-385. [DOI] [PubMed] [Google Scholar]
- 10.Flomenberg, P., V. Piaskowski, J. Harb, A. Segura, and J. T. Casper. 1996. Spontaneous, persistent infection of a B-cell lymphoma with adenovirus. J. Med. Virol. 48:267-272. [DOI] [PubMed] [Google Scholar]
- 11.Hale, G. A., H. E. Heslop, R. A. Krance, M. A. Brenner, D. Jayawardene, D. K. Srivastava, and C. C. Patrick. 1999. Adenovirus infection after pediatric bone marrow transplantation. Bone Marrow Transplant. 23:277-282. [DOI] [PubMed] [Google Scholar]
- 12.Heim, A., C. Ebnet, G. Harste, and P. Pring-Akerblom. 2003. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol. 70:228-239. [DOI] [PubMed] [Google Scholar]
- 13.Hierholzer, J. C. 1992. Adenoviruses in the immunocompromised host. Clin. Microbiol. Rev. 5:262-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman, J. A., A. J. Shah, L. A. Ross, and N. Kapoor. 2001. Adenoviral infections and a prospective trial of cidofovir in pediatric hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 7:388-394. [DOI] [PubMed] [Google Scholar]
- 15.Howard, D. S., G. L. Phillips, I. I., D. E. Reece, R. K. Munn, J. Henslee-Downey, M. Pittard, M. Barker, and C. Pomeroy. 1999. Adenovirus infections in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 29:1494-1501. [DOI] [PubMed] [Google Scholar]
- 16.Kidd, A. H., M. Jonsson, D. Garwicz, A. E. Kajon, A. G. Wermenbol, M. W. Verweij, and J. C. De Jong. 1996. Rapid subgenus identification of human adenovirus isolates by a general PCR. J. Clin. Microbiol. 34:622-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lankester, A. C., M. J. van Tol, E. C. Claas, J. M. Vossen, and A. C. Kroes. 2002. Quantification of adenovirus DNA in plasma for management of infection in stem cell graft recipients. Clin. Infect. Dis. 34:864-867. [DOI] [PubMed] [Google Scholar]
- 18.Lankester, A. C., B. Heemskerk, E. C. Claas, M. W. Schilham, M. F. Beersma, R. G. Bredius, M. J. van Tol, and A. C. Kroes. 2004. Effect of ribavirin on the plasma viral DNA load in patients with disseminating adenovirus infection. Clin. Infect. Dis. 38:1521-1525. [DOI] [PubMed] [Google Scholar]
- 19.La Rosa, A. M., R. E. Champlin, N. Mirza, J. Gajewski, S. Giralt, K. V. Rolston, I. Raad, K. Jacobson, D. Kontoyiannis, L. Elting, and E. Whimbey. 2001. Adenovirus infections in adult recipients of blood and marrow transplants. Clin. Infect. Dis. 32:871-876. [DOI] [PubMed] [Google Scholar]
- 20.Legrand, F., D. Berrebi, N. Houhou, F. Freymuth, A. Faye, M. Duval, J. F. Mougenot, M. Peuchmaur, and E. Vilmer. 2001. Early diagnosis of adenovirus infection and treatment with cidofovir after bone marrow transplantation in children. Bone Marrow Transplant. 27:621-626. [DOI] [PubMed] [Google Scholar]
- 21.Lion, T., A. Gaiger, T. Henn, E. Horth, O. A. Haas, K. Geissler, and H. Gadner. 1995. Use of quantitative polymerase chain reaction to monitor residual disease in chronic myelogenous leukemia during treatment with interferon. Leukemia 9:1353-1360. [PubMed] [Google Scholar]
- 22.Lion, T. 2001. Current recommendations for positive controls in RT-PCR assays. Leukemia 15:1033-1037. [DOI] [PubMed] [Google Scholar]
- 23.Lion, T., R. Baumgartinger, F. Watzinger, S. Matthes-Martin, M. Suda, S. Preuner, B. Futterknecht, A. Lawitschka, C. Peters, U. Potschger, and H. Gadner. 2003. Molecular monitoring of adenovirus in peripheral blood after allogeneic bone marrow transplantation permits early diagnosis of disseminated disease. Blood 102:1114-1120. [DOI] [PubMed] [Google Scholar]
- 24.Ljungman, P. 1997. Respiratory virus infections in bone marrow transplant recipients: the European perspective. Am. J. Med. 102:44-47. [DOI] [PubMed] [Google Scholar]
- 25.Ljungman, P. 2004. Treatment of adenovirus infections in the immunocompromised host. Eur. J. Clin. Microbiol. Infect. Dis. 2:583-588. [DOI] [PubMed] [Google Scholar]
- 26.Morris, D. J., G. Corbitt, A. S. Bailey, M. Newbould, E. Smith, S. Picton, and R. F. Stevens. 1993. Fatal disseminated adenovirus type 2 infection following bone marrow transplantation for Hurler's syndrome: a primary infection. J. Infect. 26:181-184. [DOI] [PubMed] [Google Scholar]
- 27.Pring-Akerblom, P., F. E. Trijssenaar, T. Adrian, and H. Hoyer. 1999. Multiplex polymerase chain reaction for subgenus-specific detection of human adenoviruses in clinical samples. J. Med. Virol. 58:87-92. [PubMed] [Google Scholar]
- 28.Runde, V., S. Ross, R. Trenschel, E. Lagemann, O. Basu, K. Renzing-Kohler, U. W. Schaefer, M. Roggendorf, and E. Holler. 2001. Adenoviral infection after allogeneic stem cell transplantation (SCT): report on 130 patients from a single SCT unit involved in a prospective multi center surveillance study. Bone Marrow Transplant. 28:51-57. [DOI] [PubMed] [Google Scholar]
- 29.Schilham, M. W., E. C. Claas, W. van Zaane, B. Heemskerk, J. M. Vossen, A. C. Lankester, R. E. Toes, M. Echavarria, A. C. Kroes, and M. J. van Tol. 2002. High levels of adenovirus DNA in serum correlate with fatal outcome of adenovirus infection in children after allogeneic stem-cell transplantation. Clin. Infect. Dis. 35:526-532. [DOI] [PubMed] [Google Scholar]
- 30.Seidemann, K., A. Heim, E. D. Pfister, H. Koditz, A. Beilken, A. Sander, M. Melter, K. W. Sykora, M. Sasse, and A. Wessel. 2004. Monitoring of adenovirus infection in pediatric transplant recipients by quantitative PCR: report of six cases and review of the literature. Am. J. Transplant. 4:2102-2108. [DOI] [PubMed] [Google Scholar]
- 31.Shields, A. F., R. C. Hackmann, K. H. Fife, L. Corey, and J. D. Meyers. 1985. Adenovirus infections in patients undergoing bone-marrow transplantation. N. Engl. J. Med. 312:529-533. [DOI] [PubMed] [Google Scholar]
- 32.Teramura, T., M. Naya, T. Yoshihara, G. Kanoh, A. Morimoto, and S. Imashuku. 2004. Adenoviral infection in hematopoietic stem cell transplantation: early diagnosis with quantitative detection of the viral genome in serum and urine. Bone Marrow Transplant. 33:87-92. [DOI] [PubMed] [Google Scholar]
- 33.Watzinger, F., and T. Lion. 1998. Multiplex PCR for quality control of template RNA/cDNA in RT-PCR assays. Leukemia 12:1984-1986. [DOI] [PubMed] [Google Scholar]
- 34.Watzinger, F., M. Suda, S. Preuner, R. Baumgartinger, K. Ebner, L. Baskova, H. G. M. Niesters, A. Lawitschka, and T. Lion. 2004. Real-time quantitative PCR assays for the detection and monitoring of pathogenic human viruses in immunosuppressed pediatric patients. J. Clin. Microbiol. 42:5189-5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu, W., M. C. McDonough, and D. D. Erdman. 2000. Species-specific identification of human adenoviruses by a multiplex PCR assay. J. Clin. Microbiol. 38:4114-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]