Abstract
A field isolate of Actinobacillus pleuropneumoniae, the causative agent of porcine fibrinohemorrhagic necrotizing pleuropneumonia, was sent to the diagnostic laboratory for serotyping. The isolate presented a clear reaction, with both polyclonal antibodies against serotype 1 and monoclonal antibodies against the capsular polysaccharide of serotype 1. It also exhibited a PCR profile of Apx toxins expected for serotype 1. The isolate, however, failed to react with monoclonal antibodies against the O-antigen of serotype 1 lipopolysaccharide (LPS), suggesting a rough phenotype. The lipid A-core region of the isolate migrated faster than the corresponding region of the serotype 1 reference strain S4074 by Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis, suggesting the presence of a truncated core. Sugar analysis and mass spectrometry analysis of the O-deacylated LPS from the field isolate were consistent with the absence of O-antigen and truncation of the outer core compared to the wild-type reference strain. Experimental infection of pigs confirmed the virulence of the isolate. This is the first report of an isolate of A. pleuropneumoniae serotype 1 with a truncated outer core and a rough LPS phenotype. Veterinary diagnostic laboratories should be vigilant, since infections caused by such an isolate will not be detected by serological tests based on LPS O-antigen.
Actinobacillus pleuropneumoniae is the etiologic agent of porcine pleuropneumonia, a highly contagious respiratory disease with major economic implications for the swine industry worldwide (27). Twelve serotypes of NAD-dependent A. pleuropneumoniae have been recognized based on capsular polysaccharide (CPS) and lipopolysaccharide (LPS) antigens (19), and an additional serotype has recently been proposed by Blackall et al. (2). Two additional serotypes of NAD-independent A. pleuropneumoniae have been reported (20). In North America, serotypes 1, 5, and 7 are the most prevalent serotypes recovered from diseased animals (4).
Infection by A. pleuropneumoniae is a multifactorial process governed by many virulence factors acting alone or in concert and by host susceptibility. Several virulence factors, such as CPS, LPS, outer membrane proteins, and Apx toxins, have already been identified (3, 5, 10). Studies by our group have focused on surface polysaccharides, namely, CPS and LPS, of A. pleuropneumoniae (4, 12). We have previously shown that the LPS molecule plays an important role in adherence of the bacterium to porcine respiratory tract cells and mucus (11). LPS molecules are a major constituent of outer membranes of gram-negative bacteria. They consist of a polysaccharide and a lipid moiety. The polysaccharide part is composed of a core region, which is an oligosaccharide containing 3-deoxy-d-manno-2-octulosonic acid (Kdo), and the O antigen, a polysaccharide chain consisting of repeated units. We have generated rough LPS mutants as well as core LPS mutants by using transposon mutagenesis and showed that the core region of LPS seems to play an important role in the adherence of A. pleuropneumoniae to porcine respiratory tract cells (7, 14, 23).
A field isolate of A. pleuropneumoniae (FMV 91-6514) originating from a clinical case of swine pleuropneumonia and recovered from lungs was sent to the diagnostic laboratory of the Veterinary College of Université de Montréal for serotyping. The isolate presented a clear reaction with both polyclonal antibodies against serotype 1 (18) and monoclonal antibodies against the CPS of serotype 1 (15). It also exhibited a PCR profile of the Apx toxins (positive for ApxI, ApxII, and ApxIV) expected for serotype 1 (6). The isolate, however, failed to react with monoclonal antibodies against the O-antigen of serotype 1 (16). The aim of the present study was to characterize further this atypical isolate of A. pleuropneumoniae serotype 1 at both phenotypic and genotypic levels.
Bacterial strains and growth conditions.
A. pleuropneumoniae field isolate FMV 91-6514 and reference strain S4074 (ATCC 27088) were grown on brain heart infusion agar plates (Difco Laboratories, Detroit, MI) supplemented with 15 μg/ml of NAD. Plates were incubated at 37°C in 5% CO2 for 18 to 24 h.
SDS-PAGE, Tricine-SDS-PAGE, and immunoblotting.
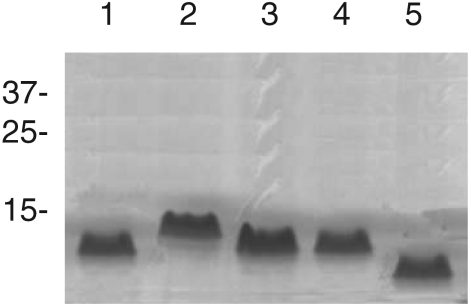
LPS profiles with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Tricine-SDS-PAGE were determined using proteinase K-digested whole-cell lysates and silver staining (23). LPSs were also transferred to nitrocellulose membranes for immunoblotting. A whole-cell lysate of strain FMV 91-6514 digested with proteinase K was run on SDS-PAGE. No high-molecular-weight bands corresponding to long O-chains were observed after staining with silver nitrate (data not shown). Furthermore, no reaction was observed with a monoclonal antibody 5.1G8F10, which is specific for the O-antigen of serotype 1 (16), suggesting the presence of a rough LPS phenotype (absence of O-antigen) (Fig. 1). Proteinase K-digested whole-cell lysate of strain FMV 91-6514 was also run on a Tricine-SDS-PAGE, which gives a better resolution of the low-molecular-weight lipid A-core region of LPS than SDS-PAGE does (Fig. 2). The lipid A-core region of strain FMV 91-6514 migrated faster than the corresponding region of the reference strain S4074, indicating the presence of a truncated core.
FIG. 1.
Immunoblot of whole-cell, proteinase K-treated preparations of A. pleuropneumoniae reference strain S4074 (lane 1) and strain FMV 91-6514 (lane 2). The immunoblot was probed with a monoclonal antibody against A. pleuropneumoniae serotype 1 O-antigen. Molecular mass markers (in kilodaltons) are indicated on the left.
FIG. 2.
Silver-stained Tricine-SDS-PAGE profiles of whole-cell, proteinase K-treated preparations of A. pleuropneumoniae reference strain S4074 (lane 2), strain FMV 91-6514 (lane 3), and strain FMV 91-6514 recovered from lungs of experimentally infected animals (lane 4). For comparison, LPS from Salmonella enterica serovar Typhimurium TV119 (Ra mutant, lane 1) and SL1181 (Re mutant, lane 5) are shown. Molecular mass markers (in kilodaltons) are indicated on the left.
Detection of LPS biosynthesis genes by PCR.
Genes known to be involved in the biosynthesis of the O-antigen (open reading frame 6 [ORF6] to ORF18 [this study and reference 14]) or the core oligosaccharide (ORFcg1 and ORFcg3 [7, 14], rfaE [21], galU [23], and rfaD and waaF [this study]) of A. pleuropneumoniae serotype 1 were amplified by PCR (Table 1) as previously described (14). The A. pleuropneumoniae serotype 1 reference strain S4074 served as the control. Although PCRs were negative with strain FMV 91-6514 for a gene (ORF15) involved in the biosynthesis of the O-antigen (14) as well as a heptosyltransferase (waaF) and an UTP-d-glucose-1-phosphate uridylyltransferase (galU) (23) involved in the biosynthesis of the LPS core, Southern blot hybridization (14) revealed that these genes were still present in strain FMV 91-6514.
TABLE 1.
Primers used for PCR amplification of A. pleuropneumoniae serotype 1 genes involved in LPS biosynthesis
| Primer | Sequence | Reference |
|---|---|---|
| Sense primer | ||
| FORF6 | TGGTGCAGGTTTTATTGG | This study; 14 |
| FORF7 | TATTATTCTTGCGGGCGG | This study; 14 |
| FORF8 | CTTAGCAGTAGATCGTGA | This study; 14 |
| FORF9 | AGTGTTTGGTGATGAGCG | This study; 14 |
| FORF10 | TCCGAATTTTGCAGTGTA | This study; 14 |
| FORF11 | TAATGTAACAGTCCGCTT | This study; 14 |
| FORF12 | AACGCTTCTTCCTATGCA | 14 |
| FORF13 | CACCTGATGAATTTGCTC | This study; 14 |
| FORF14 | CACCTGATGAATTTGCTC | This study; 14 |
| FORF15 | TGTGATCAAGGTAGTGGT | This study; 14 |
| FORF16 | GGATTTTACCGGTAGTGG | 14 |
| FORF17 | AGGATTATCTTGGCAGGA | 14 |
| FORF18 | TATCCACTTATCGTTAGG | 14 |
| FgalU | CTTAAGGGAAGAAAAACTATC | 23 |
| Fcg1a | CTTTAGTAATGGGTGGGG | 7 |
| Fcg3b | GAGAGTGCTTTAAACGGT | 7 |
| FrfaDc | CCTTCGGCTACGGTTTTA | This study |
| FrfaE | GTGCCACCAACCGTATTT | 21 |
| FwaaFd | GTCGGCGATATGATGATG | This study |
| Antisense primer | ||
| RORF6 | TAATACTCGACTCCACCA | This study; 14 |
| RORF7 | CGCCATCGGTTTTGCTAA | This study; 14 |
| RORF8 | CTGGTCGTTTTGCTGGTG | This study; 14 |
| RORF9 | GCTCGGCTCACCATTAAG | This study; 14 |
| RORF10 | AGTAAATCTATGGCAGTA | This study; 14 |
| RORF11 | CTTTGACAGAGCTCCCTT | This study; 14 |
| RORF12 | ACTTGGTATAGATCCGTG | 14 |
| RORF13 | AATACCTTCCTTGCACAC | This study; 14 |
| RORF14 | CATAATGGTTCCTGTTGG | This study; 14 |
| RORF15 | CATAATGGTTCCTGTTGG | This study; 14 |
| RORF16 | GAGATACTCCATCCGATT | 14 |
| RORF17 | CCATCTAGGTAATTTCTC | 14 |
| RORF18 | CCTTCTCGGATCCTTAAT | 14 |
| RgaIU | GTCGACTATGCACCTTGTAA | 23 |
| Rcgla | CACTCCTTACTCACTTCA | 7 |
| Rcg3b | GATCATTCACACTCTGC | 7 |
| RrfaDc | GGCGGCTTTGGTATGATC | This study |
| RrfaE | ACACTTTAACTTCGCCGC | 21 |
| RwaaFd | GATGATAGCCTTCGGC | This study |
Fcgl (forward) and Rcgl (reverse) primers are designed from the ORF of mutant CGI (AF143904) that has homology with WaaB of serovar Typhimurium (AF0263861) (7).
Fcg3 (forward) and Rcg3 (reverse) primers are designed from the ORF of mutant CG3 and CG5 (AF143905) that has homology with LbgB of Haemophilus ducreyi (U58147) (7).
FrfaD (forward) and RrfaD (reverse) primers are designed from A. pleuropneumoniae S4074 (ZP_00135423) that has homology with RfaD of serovar Typhimurium LT2.
FwaaF (forward) and RwaaF (reverse) primers are designed from A. pleuropneumoniae S4074 (ZP_00135584) that has homology with WaaF of serovar Typhimurium LT2.
Sugar analysis and mass spectrometry analysis of O-deacylated LPS (LPS-OH).
Strain FMV 91-6514 was grown on a chocolate agar plate overnight at 37°C. Cells were scraped off and resuspended in a 2% aqueous phenol solution for 4 h. The cells were pelleted and washed with H2O three times. The cell pellet was then checked for viability before the continuation of the LPS-OH isolation. The cell pellet was dissolved in H2O and lyophilized. The lyophilized material was dissolved in H2O, and a 0.05 volume of a proteinase K solution (1 mg in 4 ml) was added and incubated at 37°C for 5 h, inactivated at 65°C for 10 min, and lyophilized. The lyophilized material was dissolved in 20 mM NH4Ac buffer (pH 7.4), a 0.05 volume of RNase solution (1 mg in 10 ml of 20 mM NH4Ac) and a 0.05 volume of DNase solution (2 mg in 10 ml of 20 mM NH4Ac) were added, and the mixture was incubated at 37°C for 4 h and lyophilized. The lyophilized material was treated with anhydrous hydrazine with stirring at 37°C for 1 h to prepare LPS-OH following precipitation with ice-cold acetone. The final pellet of LPS-OH was suspended in H2O and lyophilized. Sugars were determined as their alditol acetate derivatives (24) by gas-liquid chromatography-mass spectrometry (GLC-MS) as described previously (26). All mass spectrometry experiments were performed as described previously (26).
Sugar analysis was carried out on LPS-OH from strain FMV 91-6514, which consists of a somewhat crude preparation, having not been subjected to a thorough LPS isolation protocol. This analysis revealed galactose (Gal) and N-acetylglucosamine (GlcNAc), the sugars of the capsular polysaccharide of serotype 1 strains, as major constituents with minor amounts of glucose (Glc), l-glycero-d-manno-heptose (LD-Hep) and d-glycero-d-manno-heptose (DD-Hep), with sugars from the core oligosaccharide also being identified. No rhamnose, the major O-antigen sugar from serotype 1 strains, was identified in this analysis, consistent with the failure to react with monoclonal antibodies against the O-antigen of serotype 1 and SDS-PAGE data, which suggested the absence of O-antigen in this strain.
LPS-OH prepared from plate-grown cells was also analyzed by capillary electrophoresis-MS (Table 2), which revealed a mass profile that was similar to that observed for serotype 1 reference strain S4074 (25) but with the major species now being a triply charged ion of m/z 889.3, which would correspond to loss of a HexNAc residue compared to the most prominent species (m/z 957.0) from strain S4074 LPS-OH. These data are therefore consistent with the truncated behavior of the strain FMV 91-6514 LPS on SDS-PAGE, which had an increased mobility compared to the wild-type reference strain, suggesting a truncated LPS phenotype. Sugar analysis and MS analysis of the LPS-OH derived from strain FMV 91-6514 were therefore consistent with the absence of O-antigen from this strain and suggested that the core oligosaccharide was truncated compared to the wild-type serotype 1 reference strain S4074 missing the terminal open chain GalNAc residue. Interestingly, we have identified isogenic mutants of the reference strain S4074 with a truncated outer core that still contain the O-antigen (unpublished data); it is therefore unlikely that the absence of the terminal HexNAc residue is the reason no O-antigen is expressed in strain FMV 91-6514.
TABLE 2.
Negative ion capillary electrophoresis-electrospray-MS data and proposed compositions of O-deacylated LPS from A. pleuropneumoniae strain FMV 91-6514a
| Proposed composition | Observed ions (m/z)
|
Molecular mass (Da)
|
||
|---|---|---|---|---|
| (M-2H)2− | (M-3H)3− | Observed | Calculated | |
| 4Hex, 4Hep, Kdo, P, lipid A-OH | 1,334.8 | 889.3 | 2,671.3 | 2,669.4 |
| 4Hex, 4Hep, Kdo, P, PEtn, lipid A-OH | 1,396.3 | 930.3 | 2,794.3 | 2,792.5 |
PEtn, phosphoethanolamine. Average mass units were used for calculations of molecular weights based on proposed composition as follows: Hex, 162.15; Hep, 192.17; Kdo, 220.18; P, 79.98, PEtn, 123.05. O-deacylated lipid A (Lipid A-OH) is 952.00.
Experimental infection.
The research facilities were thoroughly cleaned and disinfected prior to pig placement. The trial took place in controlled rooms with concrete floors. The virulence of strain FMV 91-6514 in 40 day-old pigs originating from a herd free of all serotypes of A. pleuropneumoniae was evaluated, as determined by routine serology during the last 3 years. All experimental procedures were conducted in accordance with the guidelines of the Canadian Council on Animal Care. A total of 19 pigs were used in the present study. Three of the pigs were randomly selected and infected intranasally with a 6-h culture of strain FMV 91-6514 (107 CFU/pig) to become seeder pigs (17). Individual pigs were observed daily for clinical signs of illness. Within the following days, all pigs showed clinical signs typical of porcine pleuropneumonia, including dyspnea, coughing, fever, and prostration, which persisted for 3 to 4 days for most pigs. Interestingly, the isolate was so virulent that it killed two of the seeder pigs within 48 h and five contact pigs within 10 days postinfection. Lung tissue samples were then recovered and cultured on blood agar plates with a streak of Staphylococcus aureus. Suspicious colonies were further identified and serotyped as described previously (18). Lung lesions and bacteriological examination of necrotic lung tissue confirmed that the animals died from acute pleuropneumonia. In addition, four pigs found to be heavily affected were removed and euthanized. At necropsy, they also presented typical lesions of porcine pleuropneumonia, and pure cultures of A. pleuropneumoniae serotype 1 were obtained from lung tissue samples. Interestingly, the LPS profile of the strains recovered from lungs of experimentally infected animals were similar to the strain FMV 91-6514 used for challenge (Fig. 2, lanes 3 and 4).
The eight animals that recovered from infection were kept for 14 weeks in order to evaluate their serological response. Two different enzyme-linked immunosorbent assays (ELISAs) were used, the long-chain LPS (LC-LPS) ELISA, a highly specific and sensitive test routinely used in Canada as a diagnostic test, and an ELISA using a saline extract of boiled formalinized whole cells (8). The LC-LPS antigen has also been adapted by Danish diagnostic laboratories (1, 9, 13). Both methodologies were performed with strain FMV 91-6514. In addition, all sera were tested with the LC-LPS ELISA, using the antigen from the reference strain S4074. The LC-LPS antigens are composed mainly of the O-LC polysaccharides, whereas the SBE contains also proteins, neutral sugars, hexosamines, and Kdo (22). The LC-LPS antigens obtained from strain FMV 91-6514 will not contain O-long chain polysaccharides because of its rough LPS profile. Both ELISAs were carried out as previously described (8). As expected, sera from convalescent animals did not show any increase by either LC-LPS ELISA (mean optical densities were <0.1 for both preinfection and 14-week postinfection sera). On the other hand, the SBE-ELISA was able to detect a clear increase of antibody titers (optical densities of 0.05 and 0.37, respectively). This result indicates that animals could not develop antibodies against the O-chain fraction of the LPS of this specific strain.
To the best of our knowledge, this is the first report of an isolate of A. pleuropneumoniae serotype 1 with a truncated outer core and a rough LPS phenotype. Veterinary diagnostic laboratories should be vigilant, since infections caused by such an isolate will not be detected by serological tests based on LPS O-antigen. It is important to note that the SBE-ELISA cannot be used in routine conditions, due to the presence of cross-reactions among different serotypes of A. pleuropneumoniae (8). Although we do not know at present the importance of such strains of A. pleuropneumoniae serotype 1 and whether similar strains can also be found in other serotypes, most field cases of infection due to A. pleuropneumoniae are easily detected by the LC-LPS, indicating a probable low prevalence of these atypical strains.
Acknowledgments
This work was supported by grants from Natural Sciences and Engineering Research Council of Canada (Research Networks 225155 and DGPIN3428 to M.J.), from Fonds québécois de la recherche sur la nature et les technologies (2002-ER-71900), and from Elanco, Division of Eli Lilly and Co.
We thank Marie Schneider for technical help and Lisa Morrison for mass spectrometry.
REFERENCES
- 1.Andresen, L. O., J. Klausen, K. Barfod, and V. Sorensen. 2002. Detection of antibodies to Actinobacillus pleuropneumoniae serotype 12 in pig serum using a blocking enzyme-linked immunosorbent assay. Vet. Microbiol. 89:61-67. [DOI] [PubMed] [Google Scholar]
- 2.Blackall, P. J., H. L. Klaasen, H. Van Den Bosch, P. Kuhnert, and J. Frey. 2002. Proposal of a new serovar of Actinobacillus pleuropneumoniae: serovar 15. Vet. Microbiol. 84:47-52. [DOI] [PubMed] [Google Scholar]
- 3.Bossé, J. T., H. Janson, B. J. Sheehan, A. J. Beddek, A. N. Rycroft, J. Simon Kroll, and P. R. Langford. 2002. Actinobacillus pleuropneumoniae: pathobiology and pathogenesis of infection. Microbes Infect. 4:225-235. [DOI] [PubMed] [Google Scholar]
- 4.Dubreuil, J. D., M. Jacques, K. R. Mittal, and M. Gottschalk. 2000. Actinobacillus pleuropneumoniae surface polysaccharides: their role in diagnosis and immunogenicity. Anim. Health Res. Rev. 1:73-93. [DOI] [PubMed] [Google Scholar]
- 5.Frey, J. 1995. Virulence in Actinobacillus pleuropneumoniae and RTX toxins. Trends Microbiol. 3:257-261. [DOI] [PubMed] [Google Scholar]
- 6.Frey, J. 2003. Detection, identification, and subtyping of Actinobacillus pleuropneumoniae, p. 87-95. In K. Sachse and J. Frey (ed.), Methods in molecular biology, vol. 216. PCR detection of microbial pathogens: methods and protocols. Humana Press, Inc., Totowa, N.J. [DOI] [PubMed] [Google Scholar]
- 7.Galarneau, C., S. Rioux, and M. Jacques. 2000. Core oligosaccharide mutants of Actinobacillus pleuropneumoniae serotype 1 obtained by mini-Tn10 mutagenesis. Pathogenesis 1:253-264. [Google Scholar]
- 8.Gottschalk, M., E. Altman, N. Charland, F. De Lasalle, and J. D. Dubreuil. 1994. Evaluation of a saline boiled extract, capsular polysaccharides and long-chain lipopolysaccharides of Actinobacillus pleuropneumoniae serotype 1 as antigens for the serodiagnosis of swine pleuropneumonia. Vet. Microbiol. 42:91-104. [DOI] [PubMed] [Google Scholar]
- 9.Grondahl-Hansen, J., K. Barfod, J. Klausen, L. O. Andresen, P. M. Heegaard, and V. Sorensen. 2003. Development and evaluation of a mixed long-chain lipopolysaccharide based ELISA for serological surveillance of infection with Actinobacillus pleuropneumoniae serotypes 2, 6 and 12 in pig herds. Vet. Microbiol. 96:41-51. [DOI] [PubMed] [Google Scholar]
- 10.Haesebrouck, F., K. Chiers, I. Van Overbeke, and R. Ducatelle. 1997. Actinobacillus pleuropneumoniae infections in pigs: the role of virulence factors in pathogenesis and protection. Vet. Microbiol. 58:239-249. [DOI] [PubMed] [Google Scholar]
- 11.Jacques, M. 1996. Role of lipo-oligosaccharides and lipopolysaccharides in bacterial adherence. Trends Microbiol. 4:408-410. [DOI] [PubMed] [Google Scholar]
- 12.Jacques, M. 2004. Surface polysaccharides and iron uptake systems of Actinobacillus pleuropneumoniae. Can. J. Vet. Res. 68:81-85. [PMC free article] [PubMed] [Google Scholar]
- 13.Klausen, J., L. O. Andresen, K. Barfod, J., and V. Sorensen. 2002. Evaluation of an enzyme-linked immunosorbent assay for serological surveillance of infection with Actinobacillus pleuropneumoniae serotype 5 in pig herds. Vet. Microbiol. 88:223-232. [DOI] [PubMed] [Google Scholar]
- 14.Labrie, J., S. Rioux, M. M. Wade, F. R. Champlin, S. C. Holman, W. W. Wilson, C. Savoye, M. Kobisch, M. Sirois, C. Galarneau, and M. Jacques. 2002. Identification of genes involved in biosynthesis of Actinobacillus pleuropneumoniae serotype 1 O-antigen and biological properties of rough mutants. J. Endotoxin Res. 8:27-38. [PubMed] [Google Scholar]
- 15.Lacouture, S., K. R. Mittal, M. Jacques, and M. Gottschalk. 1997. Serotyping Actinobacillus pleuropneumoniae by the use of monoclonal antibodies. J. Vet. Diagn. Investig. 9:337-341. [DOI] [PubMed] [Google Scholar]
- 16.Lairini, K., E. Stenbaek, S. Lacouture, and M. Gottschalk. 1995. Production and characterization of monoclonal antibodies against Actinobacillus pleuropneumoniae serotype 1. Vet. Microbiol. 46:369-381. [DOI] [PubMed] [Google Scholar]
- 17.Lechtenberg, K. F., T. R. Shryock, and G. Moore. 1994. Characterization of an Actinobacillus pleuropneumoniae seeder pig challenge-exposure model. Am. J. Vet. Res. 55:1703-1709. [PubMed] [Google Scholar]
- 18.Mittal, K. R., R. Higgins, and S. Larivière. 1987. An evaluation of agglutination and coagglutination techniques for serotyping of Haemophilus pleuropneumoniae isolates. Am. J. Vet. Res. 48:219-226. [PubMed] [Google Scholar]
- 19.Nielsen, R. 1986. Serological characterization of Actinobacillus pleuropneumoniae strains and proposal of a new serotype: serotype 12. Acta Vet. Scand. 27:453-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen, R., L. O. Andresen, T. Plambeck, J. P. Nielsen, L. T. Krarup, and S. E. Jorsal. 1997. Serological characterization of Actinobacillus pleuropneumoniae biotype 2 strains isolated from pigs in two Danish herds. Vet. Microbiol. 54:35-46. [DOI] [PubMed] [Google Scholar]
- 21.Provost, M., J. Harel, J. Labrie, M. Sirois, and M. Jacques. 2003. Identification, cloning and characterization of rfaE of Actinobacillus pleuropneumoniae serotype 1, a gene involved in lipopolysaccharide inner-core biosynthesis. FEMS Microbiol. Lett. 223:7-14. [DOI] [PubMed] [Google Scholar]
- 22.Radacovici, S., R. Lallier, S. Larivière, and J. D. Dubreuil. 1992. Biochemical characterization of an antigenic saline extract of Actinobacillus pleuropneumoniae serotype 5 and identification of a serotype-specific antigen for ELISA serodiagnosis. Vet. Microbiol. 30:369-385. [DOI] [PubMed] [Google Scholar]
- 23.Rioux, S., C. Galarneau, J. Harel, J. Frey, J. Nicolet, M. Kobisch, J. D. Dubreuil, and M. Jacques. 1999. Isolation and characterization of mini-Tn10 lipopolysaccharide mutants of Actinobacillus pleuropneumoniae serotype 1. Can. J. Microbiol. 45:1017-1026. [DOI] [PubMed] [Google Scholar]
- 24.Sawardeker, D. G., J. H. Sloneker, and A. Jeanes. 1965. Quantitative determination of monosaccharides as their alditol acetates by gas liquid chromatography. Anal. Chem. 37:1602-1604. [Google Scholar]
- 25.St. Michael, F., J.-R. Brisson, S. Larocque, M. Monteiro, J. Li, M. Jacques, M. B. Perry, and A. D. Cox. 2004. Structural analysis of the lipopolysaccharide derived core oligosaccharide of Actinobacillus pleuropneumoniae serotypes 1, 2, 5a and the genome strain 5b. Carbohydr. Res. 339:1973-1984. [DOI] [PubMed] [Google Scholar]
- 26.St. Michael, F., J. Li, E. Vinogradov, S. Larocque, M. Harper, and A. D. Cox. 2005. Structural analysis of the lipopolysaccharide of Pasteurella multocida VP161: identification of both Kdo-P and Kdo-Kdo species in the lipopolysaccharide. Carbohydr. Res. 340:59-68. [DOI] [PubMed] [Google Scholar]
- 27.Taylor, D. J. 1999. Actinobacillus pleuropneumoniae, p. 343-354. In B. E. Straw, S. D'Allaire, W. I. Mengeling, and D. J. Taylor (ed.), Diseases of Swine, 8th ed. Iowa State University Press, Ames, Iowa.