Abstract
Multiple-locus variable-number tandem-repeat analysis (MLVA), a new PCR-based method of typing Staphylococcus aureus, was compared to pulsed-field gel electrophoresis (PFGE), spa typing, and multilocus sequence typing (MLST) on a group of 59 S. aureus (mostly methicillin-resistant) clinical isolates. The aim of the study was to establish possible criteria of clustering MLVA patterns and to check concordance levels between the results produced by MLVA and the three other typing methods. As in our earlier study, MLVA turned out to have discriminatory power similar to that of PFGE. Comparison of data obtained by the two approaches allowed us to propose a 70% or ca. 80% cutoff value of the similarity between two MLVA patterns, depending on a cutoff level applied to interpret the PFGE results, 75% or ca. 90%, respectively. The cutoff values corresponded to the difference of up to six or four bands, respectively, among maximum 14 bands in total produced by two isolates in the analysis. The MLVA clusters matched well those obtained by PFGE, and they were also consistent in general with clusters generated by spa typing and MLST, these latter methods characterized lower resolution. Our results suggest that MLVA may be reliable in shorter-term S. aureus epidemiological studies, including analyses of outbreaks and hospital-to-hospital strain transmission events. Well-known advantages of typing methods based on PCR (low cost, short time, and easiness of performance) make MLVA a method that may be useful in a variety of laboratories, including those performing routine microbiological analyses within medical centers.
Staphylococcus aureus, especially methicillin-resistant S. aureus (MRSA), is one of the most important bacterial pathogens in humans responsible for the constantly increasing number of nosocomial and community-acquired infections. Therefore, adequate and precise typing of S. aureus isolates, which allows monitoring of local outbreaks and wider-scale dissemination of specific dangerous clones, is of great concern. Similar to other microorganisms, S. aureus typing has been recently dominated by molecular biology techniques based on variation analysis of DNA sequences in bacterial isolates. Different molecular methods refer to different genome characteristics that may change independently of each other, and this greatly affects their essential parameters, mainly discriminatory power. This is also a reason why sometimes relationships among isolates that are inferred by one typing method do not correspond with those obtained by another method. The choice of a proper typing approach is crucial in various kinds of epidemiological studies: for example, outbreak analyses require methods with high discriminatory power, whereas those with lower resolution potential are suitable for long-term evolutionary studies.
Pulsed-field gel electrophoresis (PFGE) has been considered to be the “gold standard” in typing of a variety of bacteria, including S. aureus. Being highly discriminatory, it is an excellent tool to analyze outbreaks and center-to-center strain transmission events (3, 21, 22, 26), but it has also been used successfully in large-scale S. aureus (mostly MRSA) epidemiological investigations (6, 15, 16). Such studies have been further developed by application of multilocus sequence typing (MLST), which measures sequence variation at seven housekeeping loci (8). MLST characterized lower resolution and, as a DNA sequencing-based method, gives full reproducibility of results between laboratories. The recently introduced spa typing method analyzes sequence polymorphism at a single locus, and with its discriminatory power, between those of PFGE and MLST (2, 6, 11, 19, 29), it seems to be useful not only in macroevolution but also in smaller-scale studies (14). Unfortunately, all of these methods are technically demanding, expensive, and time-consuming and, in the case of PFGE, interlaboratory comparisons of results are difficult (16, 17, 18).
Rapid, easy, and relatively inexpensive typing techniques are those that are based on the PCR method. Several of these have been used in S. aureus outbreak analyses; however, all have important limitations, such as low reproducibility or limited discriminatory power (4, 7, 11, 31). It is possible that these shortfalls will be eliminated with new S. aureus (or MRSA)-specific methods, such as triplex PCR for spa and coa genes, the hypervariable region adjacent to mecA (28, 32), or the restriction profile analysis of the repetitive element STAR (20). Recently, multiplex PCR referred to as MLVA (25) has been developed in our laboratory. It analyzes the variation in number of repeats in seven individual genes (sspA, spa, sdrC, sdrD, sdrE, clfA, and clfB) and in the initial study was found to be comparable in discriminatory power with PFGE. The aim of the present study was to determine the congruence between the isolate or clonal groupings recognized by MLVA, and PFGE, spa typing, and MLST and to propose possible MLVA clustering criteria based on comparison of MLVA and PFGE DNA banding patterns.
MATERIALS AND METHODS
Bacterial strains.
A group of 56 MRSA and 2 methicillin-susceptible S. aureus (MSSA) isolates was selected from a collection of ca. 500 S. aureus isolates from 1992 to 2001, deposited at the National Institute of Public Health in Warsaw. Strain selection was based on preliminary PFGE data and was intended to include isolates with various degrees of genetic relatedness. Isolates were derived from a variety of human infections. Their geographic origins included mainly Poland but there were also isolates from several other European countries. A reference S. aureus strain NCTC 8325/0 (MSSA) was included in the study.
PFGE.
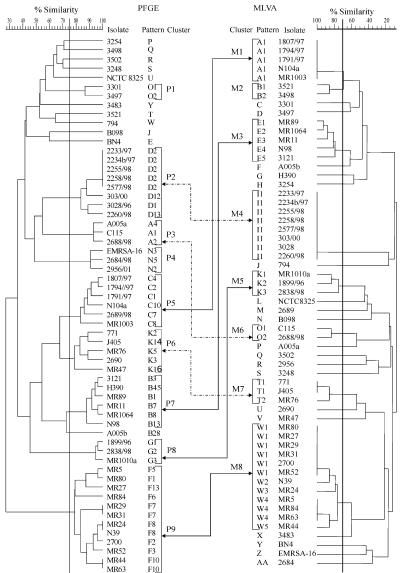
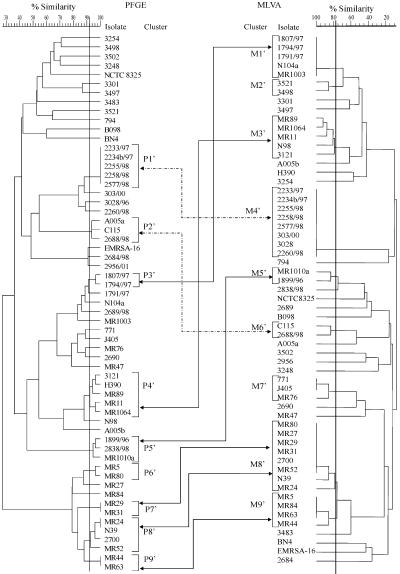
Preparation of genomic DNA of the isolates, followed by SmaI (MBI Fermentas, Vilnius, Lithuania) digestion, and separation in a CHEF DR II apparatus (Bio-Rad, Hercules, Calif.) was performed as described previously (5). SmaI PFGE patterns were saved in the TIF format, exported to the Molecular Analyst database (Bio-Rad), and analyzed by using the Dice coefficient-UPGMA (unweighted pair-group method with arithmetic averages). A dendrogram was generated to examine relatedness of PFGE profiles for all study isolates, and cutoff levels of 75 and 92% were applied to this dendrogram (Fig. 1 and 2, respectively). With the 75% cutoff, isolates differing by up to six DNA fragments were clustered together, whereas the 92% cutoff corresponded to the difference of up to three bands within a cluster.
FIG. 1.
PFGE (left) and MLVA (right) dendrograms of the study isolates generated by the UPGMA algorithm. Isolate clusters were delineated with a 75 and 70% similarity cutoff values for PFGE and MLVA, respectively, as indicated by vertical lines. Double arrows connect the corresponding clusters discerned by both methods (different arrow forms are used only for clarity of visualization).
FIG. 2.
PFGE (left) and MLVA (right) dendrograms of the study isolates. Isolate clusters were delineated with a 92 and 77% similarity cutoff values for PFGE and MLVA, respectively, as indicated by vertical lines. Double arrows connect the corresponding clusters discerned by both methods (different arrow forms are used only for clarity of visualization).
Preparation of total DNA for PCR.
Total DNA of the isolates was purified by using the Genomic DNA Prep Plus kit (A&A Biotechnology, Gdynia, Poland) as previously described (25). The purified DNA concentration was estimated with a spectrophotometer (CE3021; Cecil Instruments, Cambridge, United Kingdom), and stock solutions were diluted to a concentration of 5 ng/μl. A total of 5 ng of DNA were then used in each PCR.
MLVA typing.
MLVA typing was performed as described previously (25). Gel images were exported as TIF files for further analysis by using the Molecular Analyst database. Construction of a dendrogram of banding patterns by using UPGMA was performed by using the Dice coefficient.
spa typing.
Amplification of the spa gene X region was performed as described previously (27), and amplicons were then sequenced by using an ABI 377 sequencer (Applied Biosystems, Foster City, Calif.). The spa types were determined with the Ridom SpaServer (12). The spa types with identical or similar repeat profiles were grouped into clusters according to the method of Koreen et al. (14). Any two spa types that differed in the number of repeats but contained many identical repeats in common or showed a single deletion of the internal spa sequence were classified into the same cluster.
MLST.
MLST was performed according to protocol described by Enright et al. (8). Sequences of each locus were submitted to the Internet database (www.mlst.net), and resulting allelic profiles were assigned to particular sequence types (ST) for each isolate. START software (13) was used to classify different STs into clusters or clonal complexes (CCs) of phylogenetic relationships. Such clusters were composed of two or more isolates of the same STs or STs which differed at a single locus (single-locus variants) or two loci (double-locus variants) (10).
Calculation of concordance.
Intermethod concordance was calculated as the maximum percentage proportion of isolates grouped together into unique patterns/profiles or clusters by two methods compared (24).
RESULTS
PFGE.
The 59 S. aureus isolates produced 52 PFGE patterns. Using a cutoff similarity value of 75%, 48 of the isolates were classified into nine clusters designated from P1 to P9, while 11 isolates had separate positions in the dendrogram (Fig. 1). The clusters almost perfectly matched PFGE types of the isolates that had been discerned in an earlier study aimed at description of the clonal structure of MRSA in Poland (original designations of the types O, D, A, N, C, K, B, G, and F, respectively), in which a much wider collection of isolates had been used (J. Krzyszton-Russjan, J. Empel, T. Leski, M. Gniadkowski, and W. Hryniewicz, Abstr. 11th International Symposium on Staphylococci and Staphylococcal Infections, abstr. ME-15, 2004). Isolates belonging to the same cluster or PFGE type fulfilled the criteria by Tenover et al. (30). The only exception was isolate A005b, which in the present study was excluded from cluster P7, corresponding to the original PFGE type B. The reason for the latter was that for the present study only a sample of isolates of this type were selected and did not include those that had linked this isolate by pattern similarity with the remaining isolates of the cluster.
When the cutoff of 92% was applied, nine PFGE clusters comprising 28 isolates were distinguished and designated from P1′ to P9′ (Fig. 2). Of the clusters identified with the 75% cutoff, only two clusters, P3 and P8, remained unchanged as clusters P2′ and P5′, respectively. Clusters P2, P5, and P7 were reduced by three, four, and one isolate, respectively, into clusters P1′, P3′, and P4′. Cluster P9 was split into four smaller ones—P6′, P7′, P8′, and P9′—and two unique isolates. Finally, all isolates of clusters P1, P4, and P6 were classified as nonrelated by PFGE with the raised cutoff value (Fig. 2).
spa typing.
Our analysis yielded 20 spa types among 59 S. aureus isolates, including six new types (t386 to t391) (Fig. 3). With the use of the criteria by Koreen et al. (14), isolates with similar spa repeat profiles were grouped into four clusters designated S1 to S4. Only a single isolate, isolate BN4 (t159), was classified outside any cluster.
FIG. 3.
Comparison of PFGE and MLVA results (cutoffs 75 and 70%, respectively) with those obtained by MLST and spa typing. The order in which isolates are listed from top to bottom is based on the MLST dendrogram (left side of the figure). Gaps within spa profiles indicate possible deletion/insertion events in the spa locus and were introduced in order to optimize the alignment of the profiles. In PFGE and MLST columns only cluster designations were used; the unique PFGE and MLVA types were not included for clarity.
MLST.
MLST analysis identified 14 distinct allelic profiles or STs among the isolates, including one, designated ST501 (isolate 3301) (Fig. 3), that has not been described previously in the database at http://saureus.mlst.net/. Ten STs were represented by more than one isolate. Six clusters of closely related STs were distinguished, and these corresponded to S. aureus clonal complexes CC1, CC5, CC8, CC30, and CC45, with a cluster of three isolates belonging to ST80 (1, 9, 10). Only one isolate, isolate 794 of ST15, could not be assigned to any of the clusters.
MLVA results and their comparison with the PFGE data. (i) Criteria for defining MLVA clusters.
MLVA produced 40 different DNA banding patterns among 59 S. aureus isolates. In order to establish criteria for clustering the patterns into similarity groups, the MLVA typing results were compared to those obtained by PFGE. With the PFGE similarity cutoff of 75%, a comparable set of MLVA clusters was observed when the cutoff value between two MLVA patterns was set up at the level of 70% (Fig. 1). Isolates belonging to the same cluster differed by up to six bands, and isolates classified into different clusters differed by more than six bands. A total of eight MLVA clusters that comprised 40 isolates were distinguished. Almost all clusters delineated in the PFGE dendrogram had corresponding clusters in the MLVA dendrogram. PFGE clusters P2, P8, and P9 were identical to the MLVA clusters M4, M5, and M8, respectively (Fig. 1). A very good correlation was also observed between PFGE clusters P3, P5, P6, and P7 and MLVA clusters M6, M1, M7, and M3, respectively. The only difference was that MLVA groups were smaller by one or two isolates than their PFGE counterparts; however, none of the “missing” isolates was clustered by MLVA together with any other isolates (isolate A005a, PFGE cluster P3; 2689/98, PFGE cluster P5; 2690 and MR47, PFGE cluster P6; and H390, PFGE cluster P7). Isolates in PFGE clusters P1 (isolates 3301 and 3497) and P4 (isolates EMRSA-16, 2684/98, and 2956/01) were all classified as unique isolates by MLVA analysis. The only isolates defined as similar by MLVA and nonrelated by PFGE were two isolates of the MLVA cluster M2 (isolates 3521 and 3498) (Fig. 1).
For the nine PFGE clusters defined by the 92% cutoff, a corresponding set of nine MLVA clusters, M1′ to M9′, was distinguished when the MLVA cutoff level was raised to 77% (Fig. 2). These clusters comprised 39 isolates, and isolates within a single cluster differed from each other by up to four bands. Clusters M1′, M2′, M3′, M4′, M6′, and M7′ were identical to those from the MLVA analysis with the 70% cutoff. The original cluster M5 was reduced by one isolate into M5′, and cluster M8 was divided into two smaller ones, M8′ and M9′. All but one PFGE clusters correlated with particular MLVA clusters; however, none of these were identical to its MLVA counterpart. PFGE clusters P1′, P2′, P3′, P4′, P5′, and P9′ corresponded to MLVA clusters M4′, M6′, M1′, M3′, M5′, and M9′, respectively, and were usually smaller by one to three isolates than the MLVA ones. Isolates from clusters P7′ and P8′ were classified together by MLVA into cluster M8′. The only exception was PFGE cluster P6′, which had no a corresponding MLVA cluster, and its two isolates were split between MLVA clusters M8′ and M9′ (Fig. 2). On the other hand, isolates of the MLVA clusters M2′ and M7′ were defined as nonrelated by PFGE with the 92% cutoff.
We also searched for a level of variability in PFGE patterns that would always allow classifying any two isolates to corresponding clusters by using MLVA and PFGE methods. Any two isolates with either indistinguishable PFGE patterns (the cutoff 92% clusters P1′, P7′, and P9′, and isolates MR24 and N39 of cluster P8′) or differing by a single band (cluster P3′) were always assigned to a single, separate MLVA cluster. These differed by no more than four bands within the MLVA clusters.
(ii) Comparison of MLVA with spa and MLST clusters.
In all cases, S. aureus isolates of a given MLVA cluster, defined by the 70% cutoff value, were grouped together within the same spa cluster and MLST clonal complex or cluster (Fig. 3). Moreover, such isolates were usually indistinguishable from each other by spa typing and MLST. Exceptions included isolates 3121 (cluster M3) and N39 (cluster M8) that differed in spa types from other isolates in their clusters, and isolates MR1003 (cluster M1), and 2260/98, 3028 and 303/00 (cluster M4) that varied with respect to their STs.
Of the four clusters revealed by spa typing, almost each one was split by MLVA into several clusters and singular isolates. The one exception was the spa cluster S4 with 13 isolates, 12 of which corresponded to the MLVA cluster M8 and one isolate (isolate 3483) was sorted as unique. Similar observations were made when the six MLST clusters were compared to MLVA clusters. The CC5 complex, corresponding to the spa cluster S4, matched precisely the MLVA cluster M8 and the single isolate 3483. Isolate 3483 slightly differed from remaining isolates of the group in spa typing and MLST. The only other MLST clusters that overlapped with MLVA (and spa) clusters were those occurring less often among isolates analyzed: cluster ST80 (corresponding to M5 and S3) and CC45 (M6 and the spa subcluster S2′). CC30 was split completely into unique isolates by MLVA, as was the case of CC1 and all other typing methods used in the present study.
Concordance between methods.
The concordance values between the typing methods compared are listed in Table 1. The highest levels of correlation were found between the results produced on one hand by MLVA and PFGE (72.9%), and, on the other, by spa typing and MLST (72.9%). Of the remaining combinations of method pairs, a higher correlation level was observed only in the case of MLVA and spa typing (62.7%).
TABLE 1.
Correlation between four typing methods for S. aureus
| Typing method | % Correlation between patterns/profiles (correlation between clusters)
|
|||
|---|---|---|---|---|
| PFGE | MLST | MLVA | spa typing | |
| PFGE | - | - | - | - |
| MLST | 32.2 (49.1) | - | - | - |
| MLVA | 72.9 (64.4) (39.0)a | 44.1 (42.4) | - | - |
| spa typing | 42.4 (49.1) | 72.9 (66.1) | 62.7 (47.5) | - |
-, Concordance value referring to the comparison of PFGE and MLVA clusters, in which similarity cutoff levels of 92% and 77% were used in the two methods, respectively. In all other cases, comparisons were done with a PFGE cutoff of 75% and an MLVA cutoff of 70%.
DISCUSSION
In this study we compared the new S. aureus typing method, MLVA (25), with PFGE, MLST, and spa typing on a group of 59 nosocomial S. aureus, mostly MRSA isolates. In contrast to other approaches it is rapid, inexpensive, and easy to use, which is a general characteristic of PCR-based typing techniques. DNA banding patterns generated by MLVA are simple and easy to interpret. Moreover, together with spa typing and MLST, MLVA offers the possibility of unambiguous interlaboratory comparisons of results.
In general, typing techniques with high discriminatory power have a better level of concordance between themselves than with those of lower resolution potential. By analogy, methods with low discriminatory power correlate better with each other than with highly discriminatory techniques. According to this rule we have observed a high level of congruence between PFGE and MLVA on one hand and spa typing and MLST on the other. Comparability of the resolution potential between MLVA and PFGE has already been demonstrated in our earlier study (25). What is interesting, however, is that the level of concordance between typing techniques can vary and depends on the collection of isolates used for investigation. With a highly diverse collection of isolates, one may observe a good correlation even between typing techniques which differ greatly in discriminatory power. In this way we can explain the difference in the concordance level between PFGE and MLST obtained in the present study (32.2%) and in the analysis performed by Grundmann et al. (67%) (11). In these two studies two different types of S. aureus collections were used. Whereas our collection included only nosocomial and mostly related MRSA isolates, Grundmann et al. (11) studied a collection of mainly MSSA isolates circulating in the community.
The concordance level between MLVA and PFGE was remarkably high. The comparison of the results obtained by both methods was performed at two similarity cutoff levels in the PFGE analysis: 75 and 92%. With the PFGE cutoff of 75%, which referred to the interpretation criteria by Tenover et al. (30), isolates that differed by no more than six bands in MLVA (MLVA cutoff of 70%) could be regarded as related in PFGE and therefore classified into the same MLVA cluster. MLVA differentiated all isolates of different PFGE clusters, and almost all clusters distinguished with one of the methods had their counterparts in the second analysis. Most differences observed in PFGE and MLVA clustering were not accidental but could be explained by other observations. Sporadic isolates of PFGE clusters P1, P3, P4, P5, P6, and P7 that were segregated as unique by MLVA differed slightly from each other or from remaining isolates of their clusters also in spa types (isolates 2689/98, 2690, and A005a), STs (isolates 3301 and 3497), or both (isolates 2684/98 and EMRSA-16). On the other hand, two isolates of the MLVA cluster M2 (isolates 3521 and 3498), which were separated from each other by PFGE, turned out to be the same spa type and ST. The PFGE interpretation criteria by Tenover et al. (30) allow for identifying broader isolate clusters that have been useful in larger-scale epidemiological or phylogenetic studies (6, 15, 16). However, short-term epidemiological studies aimed mostly at outbreak investigations usually require more stringent criteria, especially in the case of highly clonal organisms, such as S. aureus.
Using the PFGE cutoff of 92%, which reflected up to a three-band difference between two PFGE patterns, it was possible to raise the MLVA cutoff level to 77% in order to obtain the most stringent criteria in MLVA interpretation. Such MLVA clusters grouped DNA patterns that differed by up to four bands from each other. Under these conditions, almost each of the PFGE clusters had a corresponding MLVA cluster, with the only exception of the PFGE cluster P6′, the two isolates of which were split into MLVA clusters M8′ and M9′. With the MLVA cutoff of 70%, these two clusters were combined into a single, broad M8, and it is possible that with more diverse isolates of this phylogenetic lineage included in the analysis, clusters M8′ and M9′ would form a single one also under the stringent conditions. The even better results were obtained when the MLVA cutoff 77% clusters were assigned to PFGE clusters that grouped isolates of up to one band difference only. In this case, all PFGE clusters had their MLVA counterparts. All of the observations discussed above suggest that MLVA, with its resolution power and clustering capability comparable to PFGE, may be a good tool for the same applications, especially in outbreak analyses and monitoring shorter-time interhospital spread of S. aureus strains.
Lower levels of correlation between MLVA (cutoff, 70%) and spa typing and MLST at the cluster level could be attributed to obvious differences in discriminatory power between these methods; however, isolates of a specific MLVA type were usually of the same spa type and ST. Exceptional cases of variation (isolates 3121 and N39 in spa and isolates MR1003, 2260/98, 3028, and 303/00 in MLST) were still confined to the same spa cluster (S2 or S4) or MLST clonal complex or cluster (CC8) and were either due to a single deletion/insertion event within the spa locus or due to changes in only one or two of the loci analyzed by MLST. The correlation between MLVA and spa typing at the DNA banding pattern level was especially noteworthy. In order to show dynamics of clone circulation, MLVA and spa typing may be combined, which would decrease the time and cost of typing procedures and allow for interlaboratory comparisons of results. Since all spa types corresponded to specific MLVA patterns, it may be proposed that representatives of MLVA types by spa gene may be routinely typed by using partial sequencing. On the other hand, because spa typing results correlated very well with the MLST data, the representatives of spa types should be good candidates for MLST analysis.
The good agreement observed between MLVA and PFGE on one hand and spa typing and MLST on the other indicates a low level of genetic recombination in S. aureus. Its clonal population structure arises mostly from point mutations (10), which explains the presence of a phylogenetic signal within the S. aureus population (14). However, long-term evolution may be also influenced by chromosomal replacements that complicate the view of the relationships between distantly related isolates. Clusters revealed by spa typing and MLST have not always corresponded in our study. The largest MLST clonal complex CC8 was also the most diverse with six different STs, including ST8 and ST239. This cluster was split into two spa clusters, S1 and S2 (with ST8 and ST239 isolates, respectively), of which cluster S2 was also identified in another MLST group, CC30 (with ST30). Isolates of ST8 and ST239 were considered closely related because they were single-locus variants of each other, differing only in the arcC locus. However, analysis of Robinson and Enright, which included spa and seven surface protein-encoding genes sas, revealed that a large (∼557-kb) chromosomal fragment containing all of these genes and arcC was in ST239 identical to that of ST30 and not that of ST8. Therefore, a chromosomal replacement between the ST30 parent and the ST8 parent which resulted in the ST239 mosaic chromosome (23) was most likely responsible for the observed incongruence. The second case was found for the spa cluster S3 and the MLST complex CC1 and cluster ST80. The CC1 isolate 3502 was indistinguishable from isolate BN4 by MLST but different by spa type. On the other hand, its spa type t386 was closely related to the type t044, which is characteristic for three ST80 isolates (cluster S3). There are no detailed data on genome structure of these clones; therefore, we suggest that a chromosomal replacement encompassing the spa locus, followed by its mutational changes, could have also occurred in the evolution of S. aureus CC1 (ST1) clone.
In summary, the main objective of the present study was to compare MLVA with other approaches of typing of S. aureus clinical isolates. Its discriminatory power was found to be lower but comparable with PFGE; moreover, both of these methods were highly concordant in terms of discerning clusters of related and probably related isolates. Therefore, we conclude that MLVA may appear to be useful and reliable in shorter-term epidemiological investigations of S. aureus. The obvious advantage of this method is that it is PCR based, which makes it available for many laboratories, including hospital-based ones. Its good correlation with methods of lower resolution, such as spa typing, suggests that MLVA may also be useful in indicating clonal representatives for larger-scale analyses performed at the Reference Center level. However, this method needs further validation against other typing methods, especially PFGE, with use of other collections of S. aureus isolates. Studies focused on MSSA isolates would also be of interest.
Acknowledgments
This study was supported by two grants (grants 2 P04A 001 27 and 2P05D 081 26) from the Polish Committee for Scientific Research (KBN).
REFERENCES
- 1.Aires de Sousa, M., C. Batzavali, I. Spiliopoulou, I. Santos Sanches, M. I. Crisóstomo, and H. de Lencastre. 2003. Two international methicillin-resistant Staphylococcus aureus clones endemic in a university hospital in Patras, Greece. J. Clin. Microbiol. 41:2027-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aires de Sousa, M., and H. de Lencastre. 2004. Bridges from hospitals to the laboratory: genetic portraits of methicillin-resistant Staphylococcus aureus clones. FEMS Immunol. Med. Microbiol. 40:101-111. [DOI] [PubMed] [Google Scholar]
- 3.Bannerman, T. L., G. A. Hancock, F. C. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiou, C. S., H. L. Wei, and L. C. Yang. 2000. Comparison of pulsed-field gel electrophoresis and coagulase gene restriction profile analysis techniques in the molecular typing of Staphylococcus aureus. J. Clin. Microbiol. 38:2186-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung, M., H. de Lencastre, P. Matthews, A. Tomasz, I. Adamsson, M. Aires de Sousa, T. Camou, C. Cocuzza, A. Corso, I. Couto, A. Dominguez, M. Gniadkowski, R. Goering, A. Gomes, K. Kikuchi, A. Marchese, R. Mato, O. Melter, D. Oliveira, R. Palacio, R. Sa-Leao, I. Santos Sanches, J. H. Song, P. T. Tassios, and P. Villari. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6:189-198. [DOI] [PubMed] [Google Scholar]
- 6.Crisostomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 98:9865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deplano, A., A. Schuermans, J. Van Eldere, W. Witte, H. Meugnier, J. Etienne, H. Grundmann, D. Jonas, G. T. Noordhoek, J. Dijkstra, A. van Belkum, W. van Leeuwen, P. T. Tassios, N. J. Legakis, A. van der Zee, A. Bergmans, D. S. Blanc, F. C. Tenover, B. C. Cookson, G. O'Neil, M. J. Struelens, et al. 2000. Multicenter evaluation of epidemiological typing of methicillin-resistant Staphylococcus aureus strains by repetitive-element PCR analysis. J. Clin. Microbiol. 38:3527-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright, M. C., D. A. Robinson, R. Randle, E. J. Feil, G. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grundmann, H., S. Hori, M. C. Enright, C. Webster, A. Tami, E. J. Feil, and T. Pitt. 2002. Determining the genetic structure of the natural population of Staphylococcus aureus: a comparison of multilocus sequence typing with pulsed-field gel electrophoresis, randomly amplified polymorphic DNA analysis, and phage typing. J. Clin. Microbiol. 40:4544-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harmsen, D., H. Claus, W. Witte, J. Rothganger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 14.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mato, R., F. Campanile, S. Stefani, M. I. Crisostomo, M. Santagati, S. I. Sanches, and H. de Lencastre. 2004. Clonal types and multidrug resistance patterns of methicillin-resistant Staphylococcus aureus (MRSA) recovered in Italy during the 1990s. Mikrob. Drug Resist. 10:106-113. [DOI] [PubMed] [Google Scholar]
- 16.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulvey, M. R., L. Chui, J. Ismail, L. Louie, C. Murphy, N. Chang, and M. Alfa. 2001. Development of a Canadian standardized protocol for subtyping methicillin-resistant Staphylococcus aureus using pulsed-field gel electrophoresis. J. Clin. Microbiol. 39:3481-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 20.Quelle, L. S., A. Corso, M. Galas, and D. O. Sordelli. 2003. STAR gene restriction profile analysis in epidemiological typing of methicillin-resistant Staphylococcus aureus: description of the new method and comparison with other polymerase chain reaction (PCR)-based methods. Diagn. Microbiol. Infect. Dis. 47:455-464. [DOI] [PubMed] [Google Scholar]
- 21.Roberts, R. B., A. de Lencastre, W. Eisner, E. P. Severina, B. Shopsin, B. N. Kreiswirth, A. Tomasz, et al. 1998. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. J. Infect. Dis. 178:164-171. [DOI] [PubMed] [Google Scholar]
- 22.Roberts, R. B., A. M. Tennenberg, W. Eisner, J. Hargrave, L. M. Drusin, R. Yurt, and B. N. Kreiswirth. 1998. Outbreak in a New York City teaching hospital burn center caused by the Iberian epidemic clone of MRSA. Microb. Drug Resist. 4:175-183. [DOI] [PubMed] [Google Scholar]
- 23.Robinson, D. A., and M. C. Enright. 2004. Evolution of Staphylococcus aureus by large chromosomal replacements. J. Bacteriol. 186:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson, D. A., S. K. Hollingshead, J. M. Musser, A. J. Parkinson, D. E. Briles, and M. J. Crain. 1998. The IS1167 insertion sequence is a phylogenetically informative marker among isolates of serotype 6B Streptococcus pneumoniae. J. Mol. Evol. 47:222-229. [DOI] [PubMed] [Google Scholar]
- 25.Sabat, A., J. Krzyszton-Russjan, W. Strzalka, R. Filipek, K. Kosowska, W. Hryniewicz, J. Travis, and J. Potempa. 2003. New method for typing Staphylococcus aureus strains: multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J. Clin. Microbiol. 41:1801-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlichting, C., C. Branger, J. M. Fournier, W. Witte, A. Boutonnier, C. Wolz, P. Goullet, and G. Doring. 1993. Typing of Staphylococcus aureus by pulsed-field gel electrophoresis, zymotyping, capsular typing, and phage typing: resolution of clonal relationships. J. Clin. Microbiol. 31:227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stranden, A., R. Frei, and A. F. Widmer. 2003. Molecular typing of methicillin-resistant Staphylococcus aureus: can PCR replace pulsed-field gel electrophoresis? J. Clin. Microbiol. 41:3181-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang, Y. W., M. G. Waddington, D. H. Smith, J. M. Manahan, P. C. Kohner, L. M. Highsmith, H. Li, F. R. Cockerill III, R. L. Thompson, S. O. Montgomery, and D. H. Persing. 2000. Comparison of protein A gene sequencing with pulsed-field gel electrophoresis and epidemiologic data for molecular typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 38:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelson, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Belkum, A., J. Kluytmans, W. van Leeuwen, R. Bax, W. Quint, E. Peters, A. Fluit, C. Vandenbroucke-Grauls, A. van den Brule, H. Koeleman, W. Melchers, J. Meis, A. Elaichouni, M. Vaneechoutte, F. Moenes, N. Maes, M. Struelens, F. Tenover, and H. Verbrugh. 1995. Multicenter evaluation of arbitrarily primed PCR for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 33:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wichelhaus, T. A., K. P. Hunfeld, B. Boddinghaus, P. Kraiczy, V. Schafer, and V. Brade. 2001. Rapid molecular typing of methicillin-resistant Staphylococcus aureus by PCR-RFLP. Infect. Control Hosp. Epidemiol. 22:294-298. [DOI] [PubMed] [Google Scholar]