Abstract
Rapid identification of Mycobacterium avium complex (MAC) is possible by use of AccuProbe (Gen-Probe, San Diego, Calif.). To evaluate the reliability of the MAC AccuProbe for testing 7H9 cultures inoculated with broth from MGIT cultures positive for acid-fast bacilli or growth on a solid medium, we compared probe results to results obtained by sequencing a portion of the 16S rRNA gene. Isolates were sequenced if the MAC probe result was <100,000 relative light units (RLU) or if it was ≥100,000 RLU and the colony morphology was not classic or there were two colony types. For the 1,389 cultures tested in phase 1, conducted to evaluate cutoff values for the MAC probe in testing of 7H9 cultures inoculated with broth from MGIT cultures, the sensitivity and specificity of the MAC AccuProbe were 97.7% and 88.8%, respectively, according to the manufacturer's interpretive criteria (≥30,000 RLU is positive). If the cutoff for a positive result were 80,000 RLU, the specificity would be 100% and the sensitivity 92.3%. Of the 344 isolates in phase 2, which was conducted to confirm the 80,000-RLU cutoff for a positive result and therefore included only isolates with a MAC probe result of ≤100,000 RLU, 13 of 16 with results of ≥30,000 but <80,000 RLU were identified as mycobacteria other than MAC, including five Mycobacterium tuberculosis complex isolates. These data support the use of 80,000 RLU as the cutoff for a positive result in testing of 7H9 broth cultures with the MAC AccuProbe.
In the early to mid-1990s, the use of rapid methods for growth and identification of mycobacteria was stressed, primarily to ensure timely diagnosis of tuberculosis, although time to identification of nontuberculous mycobacteria was impacted as well (7). To provide rapid diagnosis, laboratories must use a broth system for mycobacterial growth and test aliquots of broth from positive cultures by using a rapid identification method. Of the methods that provide rapid identification, AccuProbes (Gen-Probe, Inc., San Diego, Calif.) are most commonly used.
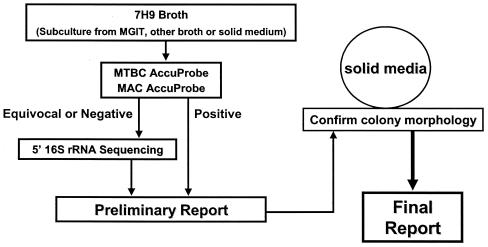
The algorithm implemented at ARUP in August 2000 for mycobacterial culture and identification (Fig. 1) involved using the Mycobacterium Growth Indicator Tube (MGIT 960; Becton Dickinson, Sparks, Md.) for growth and detection, subculturing broth from signal-positive cultures containing acid-fast bacilli (AFB) to Middlebrook 7H9 broth, and testing the 7H9 broth for Mycobacterium tuberculosis complex (MTBC) and M. avium complex (MAC) using AccuProbes. Additionally, identification was performed on isolates sent to ARUP on a solid medium or in a broth culture (e.g., positive BACTEC 12B, ESP, or BacT/Alert bottles) which were subsequently subcultured to 7H9 broth. With the MTBC probe, virtually all results were either >500,000 or <20,000 relative light units (RLU); a result between 30,000 and 100,000 RLU was rare. With the MAC probe, most results were similar to those described for the MTBC probe. However, for occasional cultures, results were between 30,000 and 100,000 RLU, and subcultures from some of these, which were presumed to contain MAC based on the manufacturer's interpretive criteria, grew colonies on a solid medium that were inconsistent with MAC. This observation prompted the investigation described here.
FIG. 1.
Algorithm for identification of Mycobacterium species from 7H9 broth cultures.
The goal of phase 1 of this study was to evaluate appropriate cutoff values for the MAC AccuProbe in testing of 7H9 broth that had been inoculated with growth from an AFB-positive culture in a liquid or on a solid medium. In phase 2 the cutoff for a positive probe result was confirmed.
(Portions of this study were presented at the 103rd General Meeting of the American Society for Microbiology, Washington, D.C., 2003 [abstr. C-226]).
Specimens from sites contaminated with normal flora were decontaminated prior to inoculation by using N-acetyl-l-cysteine-2% NaOH (5). For each specimen, 0.5 ml was inoculated into a MGIT tube, and 0.2 ml was inoculated onto a selective Lowenstein-Jensen (LJ) flask (Hardy Diagnostics, Santa Maria, Calif.). Grossly bloody specimens were inoculated to two LJ flasks only and therefore were not included in the study. MGIT cultures were placed in the MGIT 960 instrument, where they were continuously monitored for growth for 6 weeks. LJ flasks were incubated at 37°C under 5 to 10% CO2 and examined weekly for 7 weeks. When an MGIT culture was flagged positive, a smear of the broth was stained with the Kinyoun stain. If AFB only were present, aliquots of the broth were subcultured to 7H9 broth (two tubes) and Middlebrook 7H11 and sheep blood agar plates, all of which were incubated at 37°C. Sheep blood agar plates were examined at 24 and 48 h. If bacteria were seen in the Kinyoun-stained smear or grew on the sheep blood agar plate, broth from the MGIT was subcultured to a selective 7H11 flask, and the remainder of the broth was re-decontaminated and cultured to 7H11 agar. Cultures (in broth or on a solid medium) sent for mycobacterial identification were treated as described for positive MGIT cultures.
Tests to identify mycobacteria were performed on cultures containing AFB only, based on the smear result and lack of bacterial growth on sheep blood agar. The first step in the identification process was to examine the sheep blood agar plates for rapidly growing mycobacteria. If they were present, sequencing was performed as described below. If rapidly growing mycobacteria were not recognized by 48 h, cultures were tested for MAC and MTBC as follows. When the density of growth in a 7H9 broth culture was approximately equal to a 1.0 McFarland standard, the AccuProbe procedure was performed according to the directions in the package insert. The positive control was either M. avium (ATCC 25177) or M. intracellulare (ATCC 13950). The negative control was M. tuberculosis (ATCC 27294 or ATCC 25177). Temperatures for all incubations were checked daily prior to and during the performance of the assay. Interpretive criteria recommended by the manufacturer are as follows: ≥30,000 RLU, positive; <20,000 RLU, negative; 20,000 to 29,999 RLU, repeat. If the value was in the same range upon repeat testing, the isolate was subcultured to a solid medium to verify its purity.
If only the MTBC probe was positive (≥30,000 RLU), a preliminary report of MTBC was issued and appropriate susceptibility testing was performed. A final report was issued when colonies consistent with MTBC were visible on either the original LJ medium or the 7H11 subculture. These cultures are not discussed further. In both phases of the study, when the probe results were <30,000 RLU for MTBC and <100,000 RLU for MAC, identification was determined by partial 16S rRNA gene sequencing. Probe tests for these cultures, including those with AccuProbe results between 20,000 and 29,999 RLU (the manufacturer's recommended repeat range), were not repeated. Sequencing was performed as described previously (1, 2). Sequences were assembled and edited using MicroSeq software, version 1.4.3 (Applied Biosystems, Foster City, Calif.). Each consensus sequence was compared to the MicroSeq 500 bacterial database, version 500-0125, and, if necessary, to the Ribosomal Differentiation of Medical Microorganisms (RIDOM) database (3) (www.ridom-rdna.de), which contains several sequevars of M. intracellulare and M. avium. If the degree of relatedness between the sequenced organism and the most closely related sequence in the databases was 99.6 to 100%, a preliminary report with that identification was issued, pending growth on Middlebrook agar. A final report was issued if the colony morphology, pigmentation, and growth rate were consistent with the species identified. If the degree of relatedness was <99.6%, a descriptive report indicating that sequencing did not provide an identification to the species level was issued. The degree of relatedness was determined by (i) our internal validation, our accumulated 5-year experience, and data analysis based on hundreds of strains added to our library and (ii) consulting the RIDOM website (http://www.ridom.de/doc/Ridom_16S_rDNA_tour.pdf; slide 25), which uses statistical analysis to determine cutoff values. We chose an error probability of 0.01%, which correlated to a similarity of 99.5%. We have added more species to our database, so we arbitrarily chose a more stringent cutoff of 99.6%.
16S rRNA gene sequencing also was performed in phase 1 of the study if the MAC probe result was ≥100,000 and a client requested identification to the species level or if, when growth was visible on the 7H11 subculture, there were two colony types or the colony morphology and/or pigmentation was inconsistent with MAC. When there were two or more colony types, a single colony of each morphology was transferred to 7H9 broth and 7H11 agar. When the density of growth in each broth culture was equal to a 1.0 McFarland standard, sequencing was performed as described above.
Using colonies from a solid medium, isolates from positive cultures were stored at −70°C for 6 to 12 months. In phase 2 of the study, if the sequencing identification was other than M. avium or M. intracellulare and the MAC probe result was 30,000 to 100,000 RLU, AccuProbe testing was performed using colonies subcultured to 7H11 agar from the frozen isolate, if it was available.
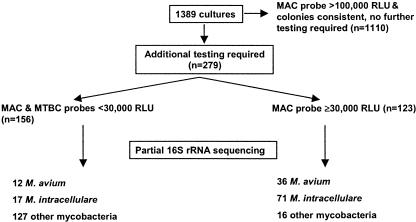
Phase 1 of the study included 1,389 cultures, of which 1,110 had MAC probe results of >100,000 RLU and typical colony morphology on solid media. No additional testing was performed on these 1,110 cultures, for which the MAC probe result was considered a true positive. MTBC and MAC probes and 16S rRNA gene sequencing were performed on the remaining 279 broth cultures (results are summarized in Fig. 2). Both probe results were <30,000 RLU for 156 of these cultures. 16S rRNA gene sequencing identified 29 (18.6%) of the 156 probe-negative isolates as either M. avium (n = 12) or M. intracellulare (n = 17) (Table 1); the rest were other mycobacterial species. For 40 cultures identified by sequencing as mycobacteria other than MAC, the MAC probe result was 20,000 to 29,999 RLU (data not shown).
FIG. 2.
Summary of results of testing performed in phase 1 of this study.
TABLE 1.
AccuProbe results for cultures identified as M. avium or M. intracellulare by 16S rRNA gene sequencing
| RLU value | No. of cultures with the indicated RLU value in:
|
|||
|---|---|---|---|---|
| Phase 1
|
Phase 2a
|
|||
| M. avium | M. intracellulare | M. avium | M. intracellulare | |
| <5,000 | 0 | 2 | 0 | 0 |
| 5,000-9,999 | 2 | 2 | 0 | 0 |
| 10,000-19,999 | 2 | 5 | 0 | 1 |
| 20,000-29,999 | 8 | 8 | 0 | 1 |
| 30,000-49,999 | 8 | 17 | 0 | 0 |
| 50,000-79,999 | 12 | 30 | 0 | 3 |
| 80,000-99,999 | 2 | 2 | 0 | 0 |
| ≥100,000 | 14 | 22 | NA | NA |
NA, not applicable (phase 2 included only isolates with RLU values of <100,000).
One hundred twenty-three of the 279 cultures had a MAC probe result of ≥30,000 RLU (positive according to the product insert). Of these 123 cultures, 36 were identified by sequencing as M. avium and 71 as M. intracellulare (Table 1). For the other 16 (13.0%) cultures, all of which had a MAC probe result of <80,000 RLU, sequencing results (Table 2) were as follows: 1 M. terrae complex, 1 MTBC, 1 M. kansasii, and 1 M. interjectum culture, 2 M. lentiflavum and 2 M. che-lonae/M. abscessus cultures, 5 M. gordonae cultures, and 3 cultures of rapidly growing mycobacteria that could not be identified to the species level.
TABLE 2.
Mycobacteria other than M. avium or M. intracellulare with MAC AccuProbe results of ≥30,000 RLU
| Species | Phase 1
|
Phase 2
|
||
|---|---|---|---|---|
| No. of cultures | RLU value(s) | No. of cultures | RLU value(s) | |
| M. gordonae | 5 | 33,000, 34,000, 36,000, 40,000, 48,000 | 6 | 32,000, 36,000, 36,000, 48,000, 60,000, 72,000 |
| MTBC | 1 | 72,000 | 5 | 34,000, 34,500, 37,000, 41,000, 46,000 |
| M. chelonae/M. abscessus | 2 | 32,000, 53,000 | 0 | NAa |
| M. kansasii | 1 | 39,000 | 0 | NA |
| M. lentiflavum | 2 | 72,000, 76,000 | 0 | NA |
| M. interjectum | 1 | 30,000 | 0 | NA |
| M. fortuitum complex | 0 | NA | 1 | 30,000 |
| M. simiae | 0 | NA | 1 | 69,000 |
| M. terrae complex | 1 | 31,000 | 0 | NA |
| Mycobacterium sp., unable to identify to species level | 3 | 30,000, 31,000, 53,000 | 0 | NA |
NA, not applicable.
For the 1,389 cultures tested in phase 1, the sensitivity and specificity of the MAC AccuProbe were 97.7% and 88.8%, respectively, using the manufacturer's recommended interpretive criteria (≥30,000 RLU is positive) and considering the probe result to be a true positive for the 1,110 cultures not tested by sequencing (discussed above). If the cutoff value required for a positive result were raised to 80,000 RLU, the specificity of the MAC probe would increase to 100% and its sensitivity would decrease to 92.3%.
Phase 2 of the study, conducted to confirm our suggested cutoff, included only those cultures with a MAC probe result of <100,000 RLU; 16S rRNA gene sequencing was performed on all of these. For 328 of the 344 cultures tested with the MAC probe, the result was <30,000 RLU; sequencing identified 2 of them as M. intracellulare (Table 1) and the rest as mycobacteria other than MAC. The MAC probe result was ≥30,000 RLU for 16 cultures, of which 3 were identified as M. intracellulare by sequencing (Table 1). The other 13 cultures were identified as M. simiae (n = 1), M. fortuitum complex (n = 1), MTBC (n = 5), or M. gordonae (n = 6) (Table 2). Of the latter 13, 10 frozen isolates were available for testing colonies from solid media with the MAC probe, and for all 10, results were <10,000 RLU.
For the 587 cultures with a MAC AccuProbe result of <30,000 RLU in both phases of the study, the sensitivity and specificity of the probe compared with sequencing were 78.0% and 93.5%, respectively. As in the analysis for phase 1 only, the specificity would increase to 100% if the cutoff for a positive probe result were raised to 80,000 RLU.
As supported by other investigators (4, 6), repeat probe testing on colonies from solid media when the original result from a liquid medium is <30,000 RLU often detects additional isolates of MAC. Reisner et al. (6) reported a sensitivity of 78.5% for the MAC probe when testing positive BACTEC 12B cultures. When testing positive BacT/Alert cultures, Louro et al. reported a sensitivity of 82.3% (4). In both of the latter studies, colonies from subcultures of the probe-negative broth samples were MAC probe positive. Although repeating the probe on solid media is likely more cost-effective, especially for laboratories where the less common nontuberculous mycobacteria are rarely encountered, this procedure adds several days or weeks to the turnaround time. Therefore, to facilitate rapid reporting, we routinely sequence cultures from 7H9 broth if the initial probe yields a result of <80,000 RLU.
To our knowledge, ours is the first study to show false-positive results when testing AFB-positive broth cultures with the MAC AccuProbe. Of the 482 broth cultures containing mycobacteria other than MAC in both phases of our evaluation, MAC probe results for 29 (6%) were ≥30,000 but <80,000 RLU. In all cases, solid media were critically examined for colonies resembling MAC, and none were found. Additionally, if susceptibility testing was performed, results were consistent with the species. The most disconcerting aspect of these false-positive results was that six (20.7%) occurred with cultures that contained only MTBC (Table 2). An incorrect report of MAC in these cases could likely delay treatment and initiation of a contact investigation or cause discontinuation of therapy, if it had already been started. Theoretically, both actions could result in increased spread of tuberculosis.
The cause of the false-positive results with the MAC probe is not clear. They did not occur consistently, and all available isolates gave clearly negative results when colonies from subcultures on a solid medium were tested with the MAC probe. Furthermore, colony morphology was consistent with sequencing results. None of the broth cultures were bloody, which theoretically could cause a low-level false-positive result. None of the cultures contained M. saskatchewanense, a recently described slowly growing scotochromogen that is AccuProbe positive for MAC (10). We also reviewed partial 16S rRNA gene sequences from this study that were 99.6 to 99.9% similar to those of MAC and found none to match the sequences of the newer species once considered members of the MAC, M. palustre (8) and M. chimaera (9). Because rapid reporting of results is important for our clients, sequencing was performed on cultures yielding a MAC probe result between 30,000 and 80,000 RLU; AccuProbe testing was not repeated. Although technical error as a cause of the false-positive result cannot be unequivocally excluded, all positive and negative controls gave the expected results and equipment was functioning properly.
The results of this study support the use of 80,000 RLU as the cutoff for a positive MAC AccuProbe result in testing of 7H9 cultures inoculated with growth from an AFB-positive culture in broth (e.g., MGIT, ESP, BACTEC 12B, BacT/Alert) or on a solid medium. If the laboratory has the resources, testing of AFB-positive, MAC probe-negative (<80,000-RLU) broth cultures by a different identification method—e.g., DNA sequencing or high-pressure liquid chromatography—provides the most rapid and accurate results. Alternatively, a laboratory could wait for growth on a solid medium and perform probe testing directly on colonies consistent with MAC; isolates with negative results would require further evaluation. Our results also show that occasionally, broth cultures containing either M. avium or M. intracellulare give results of <20,000 RLU with the MAC probe. Therefore, a preliminary report should not exclude the possibility that MAC is present.
Acknowledgments
This study was supported by the ARUP Institute for Clinical and Experimental Pathology.
We thank Andrea Stetler for secretarial assistance.
REFERENCES
- 1.Cloud, J. L., P. S. Conville, A. Croft, D. Harmsen, F. G. Witebsky, and K. C. Carroll. 2004. Evaluation of partial 16S ribosomal DNA sequencing for identification of Nocardia species by using the MicroSeq 500 system with an expanded database. J. Clin. Microbiol. 42:578-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cloud, J. L., H. Neal, R. Rosenberry, C. Y. Turenne, M. Jama, D. R. Hillyard, and K. C. Carroll. 2002. Identification of Mycobacterium spp. by using a commercial 16S ribosomal DNA sequencing kit and additional sequencing libraries. J. Clin. Microbiol. 40:400-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harmsen, D., J. Rothganger, M. Frosch, and J. Albert. 2002. RIDOM: Ribosomal Differentiation of Medical Micro-organisms Database. Nucleic Acids Res. 30:416-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Louro, A. P. S., K. B. Waites, E. Georgescu, and W. H. Benjamin, Jr. 2001. Direct identification of Mycobacterium avium complex and Mycobacterium gordonae from MB/BacT bottles using AccuProbe. J. Clin. Microbiol. 39:570-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfyffer, G. E., B. A. Brown-Elliot, and R. J. Wallace, Jr. 2003. Mycobacterium: general characteristics, isolation, and staining procedures, p. 532-559. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed., vol. 1. American Society for Microbiology, Washington, D.C.
- 6.Reisner, B., A. Gatson, and G. Woods. 1994. Use of Gen-Probe AccuProbes to identify Mycobacterium avium complex, Mycobacterium tuberculosis complex, Mycobacterium kansasii, and Mycobacterium gordonae directly from BACTEC TB broth cultures. J. Clin. Microbiol. 32:2995-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tenover, F. C., J. T. Crawford, R. E. Huebner, L. J. Geiter, C. R. Horsburgh, Jr., and R. C. Good. 1993. The resurgence of tuberculosis: is your laboratory ready? J. Clin. Microbiol. 31:767-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torkko, P., S. Suomalainen, E. Iivanainen, E. Tortoli, M. Suutari, J. Seppanen, L. Paulin, and M. L. Katila. 2002. Mycobacterium palustre sp. nov., a potentially pathogenic, slowly growing mycobacterium isolated from clinical and veterinary specimens and from Finnish stream waters. Int. J. Syst. Evol. Microbiol. 52:1519-1525. [DOI] [PubMed] [Google Scholar]
- 9.Tortoli, E., L. Rindi, M. J. Garcia, P. Chiaradonna, R. Dei, C. Garzelli, R. M. Kroppenstedt, N. Lari, R. Mattei, A. Mariottini, G. Mazzarelli, M. I. Murcia, A. Nanetti, P. Piccoli, and C. Scarparo. 2004. Proposal to elevate the genetic variant MAC-A, included in the Mycobacterium avium complex, to species rank as Mycobacterium chimaera sp. nov. Int. J. Syst. Evol. Microbiol. 54:1277-1285. [DOI] [PubMed] [Google Scholar]
- 10.Turenne, C. Y., L. Thibert, K. Williams, T. V. Burdz, V. J. Cook, J. N. Wolfe, D. W. Cockcroft, and A. Kabani. 2004. Mycobacterium saskatchewanense sp. nov., a novel slowly growing scotochromogenic species from human clinical isolates related to Mycobacterium interjectum and Accuprobe-positive for Mycobacterium avium complex. Int. J. Syst. Evol. Microbiol. 54:659-667. [DOI] [PubMed] [Google Scholar]