Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is a major nosocomial pathogen in India, and up to 70% methicillin resistance has been reported from hospitals in various parts of India. Hospitals use phenotyping for the most part, and molecular genotyping is not done. Here we report on the genotyping of 82 single-patient isolates from two hospitals in Bangalore, South India, for the first time. Most of the strains possessed type III or IIIA staphylococcal cassette chromosome (SCCmec) cassettes, and we did not detect strains with type I, IA, or II cassettes. Most isolates also contained the type III cassette chromosome recombinase (ccr) AB region. Multilocus sequence typing (MLST) and staphylococcal protein A (spa) typing of a selected number of isolates have been carried out. Although most isolates that were chosen for MLST and spa typing had the same patterns, they were quite diverse in their pulsed-field gel electrophoresis (PFGE) patterns. PFGE, MLST, and spa typing of the Indian strains revealed that they are related to the previously described Hungarian and Brazilian clones.
Methicillin-resistant Staphylococcus aureus (MRSA) is an important pathogen causing pyogenic, disseminated, and toxin-mediated infections (7, 18, 20). MRSA bacteremia is associated with significantly higher mortality than is known for methicillin-susceptible S. aureus bacteremia (6). Genotyping data from large international studies have shown that a few clones of MRSA are responsible for the spread of the disease in various parts of the world (4, 8, 18). Methicillin resistance among S. aureus isolates has reached phenomenal proportions in Indian hospitals, with some cities reporting that up to 70% of the strains are resistant to methicillin (2). About 40 to 50% of S. aureus strains isolated from the burn and trauma wards in hospitals in and around Bangalore, India, are resistant (13). For the present study, clinical isolates have been collected from two major hospitals in the city of Bangalore. Many of these MRSA strains are multidrug resistant, and they are characterized only phenotypically at present. The discriminatory power of most of the phenotypic methods is restricted and ambiguous (10, 21). Molecular typing methods have in the last few years paved the way for sophisticated techniques to track the source and transmission route of bacterial pathogens in hospital outbreaks and have also helped in establishing epidemiological investigations comparing strains across continents (1, 4, 23). Pulsed-field gel electrophoresis (PFGE) has been shown to be highly discriminatory in analyzing hospital outbreaks and tracking genetic changes which occur in a relatively short time, while multilocus sequence typing (MLST) is more suitable for studying long-term genetic variations (5, 8, 16, 24). The aim of this study was to characterize the Indian isolates by PFGE, MLST, and spa typing techniques, which would aid in controlling hospital outbreaks, epidemiological studies, and comparison with international strains.
MATERIALS AND METHODS
Hospitals.
St. John's Medical College (SJ) is a tertiary-care teaching hospital. Manipal Hospital (M) is a multi-superspecialty tertiary-care hospital with 650 beds. Both hospitals report about 40 to 50% methicillin resistance among their S. aureus isolates.
Samples.
Isolates were grown from culturing pus, urine, sputum, and blood, and a few were grown from culturing miscellaneous sites such as tracheal aspirates, at SJ and M. The isolates were inoculated into peptone water or semisolid nutrient agar deeps, sealed, and sent to us.
Bacterial strains.
Forty-five clinical isolates were obtained from SJ and 37 from M during the period of April 2003 to May 2004. S. aureus strains NCTC 8325, HUSA 304 (Hungarian), and HSJ 216 (Brazilian) were the kind gift of Herminia De Lencastre, Rockefeller University, New York, N.Y. Strain BB 255 was the kind gift of Brigitte Berger-Bächi, University of Zurich. DNA samples from S. aureus isolates possessing staphylococcal cassette chromosome (SCCmec) cassette types I, II, and III were the kind gift of T. Ito and K. Hiramatsu, Juntendo University, Tokyo, Japan.
Growth.
The strains were inoculated on brain heart infusion (BHI) agar (Himedia Laboratories, Mumbai, India) and grown for 24 h at 37°C. Typical staphylococcal colonies were examined under the microscope by Gram staining. A few strains which were contaminated were reinoculated on mannitol salt agar (Himedia) to inhibit the growth of nonstaphylococcal organisms, and colonies were picked and grown on BHI agar. Strains were characterized by catalase, coagulase, and DNase tests by established procedures for S. aureus (3).
Antibiotic susceptibility testing was performed by Kirby-Bauer disk diffusion according to the guidelines recommended by the NCCLS (17) on Mueller-Hinton agar plates at 37°C, using antibiotic disks containing penicillin, gentamicin, erythromycin, tetracycline, methicillin, and vancomycin (HiMedia). The MIC of oxacillin was determined by the broth dilution method in Mueller-Hinton broth after 24 h of incubation at 37°C in microtiter plates.
Preparation of chromosomal DNA.
Cells from an overnight culture in BHI broth collected by centrifugation were suspended in lysis buffer (phosphate-buffered saline containing 0.5% sodium dodecyl sulfate and 100 μg/ml proteinase K). The cell suspension was incubated at 37°C for 1 h, and an equal volume of phenol:chloroform (1:1) mixture was added to the cell suspension and vortexed. The samples were centrifuged, and the aqueous phase was transferred to a fresh tube. The DNA was precipitated by the addition of 30 μl of 3 M sodium acetate and 3 volumes of cold 99% ethanol. The DNA pellet was washed twice with cold 99% alcohol, air dried, and suspended in 500 μl of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8]).
Multiplex PCR.
The multiplex PCR was performed according to the procedure of Oliveira et al. (19). The presence of the mecA (gene coding for penicillin binding protein 2A) and femA (factor essential for methicillin resistance) genes was used as internal controls for detection of MRSA, and the genes were detected by PCR using the forward primer 5′-ACTGCTATCCACCCTCAAAC-3′ and the reverse primer 5′-CTGGTGAAGTTGTAATCTGG-3′ for mecA and the forward primer 5′-AAAAAAGCACATAACAAGCG-3′ and the reverse primer 5′-GATAAAGAAGAAACCAGCAG-3′ for femA in all the strains. The sizes of the amplified products for mecA and femA were 169 and 133 base pairs, respectively. The conditions for PCR were as described by Mehrotra et al. (15).
The remaining primers for detecting type I, II and III SCCmec cassettes; the conditions for PCR; and the sizes of the amplified products were as described by Oliviera et al. (19).
PCR for typing ccrAB.
PCR was performed for detection of the types of recombinase systems of the various strains with 5′-ATTGCCTTGATAATAGCCITC T-3′ (forward) and 5′-AGCTCAAAAGCAAGCAATAGAAT-3′ (reverse) as cassette chromosome recombinase (ccrAB) primers under the conditions described by Hanssen et al. (9).
MLST and spa typing.
MLST was performed as previously described by Enright et al. (8). Internal fragments of the seven housekeeping genes for carbamate kinase (arcC), shikimate dehydrogenase (aroE), glycerol kinase (glpF), guanylate kinase (gmK), phosphate acetyltransferase (pta), triosephosphate isomerase (tpi), and acetyl coenzyme A acetyltransferase (yqiL) were amplified by PCR with the specified primers, and the PCR products were purified (QiaQuick PCR purification kit; QIAGEN GmbH, Germany) and sequenced using an ABI Prism 377 DNA sequencer. Consensus sequences were assembled from both orientations. spa typing was performed according to the procedure of Shopsin et al. (22) using the forward primer 5′-GAACAACGTAACGGCTTCATCC-3′ and reverse primer 5′-CAGCAGTAGTGCCGTTTG-3′, and consensus sequences were assembled from both forward and reverse sequences (12, 22).
PFGE.
PFGE was done as previously described by McDougal et al. (14). Briefly, a bacterial pellet obtained from centrifuging 200 μl of an overnight-grown culture from a single colony was processed for lysostaphin treatment, and agarose plugs were prepared by mixing the culture with PFGE-grade agarose (Bio-Rad Laboratories, Inc., Richmond, California). The plugs were digested with EC lysis buffer for 4 h, washed with TE buffer, and then digested with SmaI restriction enzyme (Promega Corporation, Madison, Wis.) for 3 h. The restriction fragments were separated on a 1.5% gel (Bio-Rad PFGE agarose) with an initial switch time of 5 s and a final switch time of 35 s, a voltage of 6 V/cm, an included angle of 120°, and a running time of 21 h, using the CHEF-DRIII device (Bio-Rad). After electrophoresis, gels were stained using ethidium bromide, rinsed in water, and photographed under UV light with the GelDoc system (Bio-Rad).
Dendrogram.
The NCTC 8325 pattern on PFGE was used as a standard with assigned molecular weights according to Tenover et al. (25), and the dendrogram based on the similarities was derived from the unweighted pair group method using arithmetic averages (UPGMA) and Dice coefficients with Quantity One software (Bio-Rad).
RESULTS AND DISCUSSION
All isolates from both hospitals were multidrug resistant and were resistant to all the five antibiotics tested (penicillin G, methicillin, erythromycin, gentamicin, and tetracycline), but all were sensitive to vancomycin. The MIC of oxacillin varied from 6 to >800 μg/ml for isolates from SJ and from 200 to >800 μg/ml for isolates from M. The MICs for the majority of isolates from both hospitals fell into the range of 200 to 400 μg/ml.
Table 1 depicts the origins of the isolates from both hospitals. Eighty percent of isolates from SJ were cultured from pus, while only 43% of isolates from M were from pus and 35% of isolates were from tracheal culture. A small percentage of cultures from both hospitals came from sputum, urine, blood, and drainage fluid.
TABLE 1.
Origins of the isolates from the two hospitals
| Hospital | No. of isolates from:
|
|||||
|---|---|---|---|---|---|---|
| Pus | Urine | Blood | Sputum | Miscellaneousa | Drainage fluidb | |
| SJ | 36 | 1 | 0 | 4 | 4 | 0 |
| M | 16 | 3 | 2 | 1 | 13 | 2 |
Tracheal culture, fluid from pulmonary artery sheath tip, and suction tip.
Fluid from the wound site (carcinoma of the breast).
SCCmec typing of all of the strains is presented in Table 2, along with the PFGE patterns. All 82 isolates had femA by PCR, and 7 isolates gave no amplification with the mecA primers used in this study. In the multiplex PCR for determining the SCCmec type, out of the total of 82 strains from SJ and M, 49 had SCCmec type III (209-, 243-, 303-, and 414-bp PCR products present in the multiplex PCR) and 26 had SCCmec type IIIA (209-, 243-, and 414-bp products present and 303-bp band absent). The only difference between type III and IIIA is the absence of plasmid pT181 from IIIA (absence of the 303-bp product) (1). DNA from P2300 generated only the 414- and 243-bp products; P3035, P2478, and P3122 generated only the 303-bp product. These four strains from SJ amplified partial cassettes and were nontypeable. Three isolates from M did not amplify any of the primers that were used. In all, there were seven strains which did not amplify the mecA primers, and these might have different SCCmec cassettes. As none of the isolates were typed as SCCmec type I, IA, or II, to reconfirm that the majority of the isolates were type III or IIIA, a PCR was performed for detection of type III ccrAB with β2 and α4 primers. All the isolates generated a 1,600-bp product indicating the presence of type III ccrAB, except for the four mecA-negative nontypeable strains from SJ, which gave no amplification with the primers. These strains will be further investigated, as only part of the type III SCCmec cassette was amplified with the primers used in the multiplex PCR. Ito et al. (11) have suggested that the staphylococcal cassette chromosome could exist independently of the antibiotic resistance genes and may serve as a general genetic information exchange system in staphylococci. The four mecA-negative isolates from SJ did not have the type III recombinase system, which is unlike the findings of Hanssen et al. (9), who reported the existence of three types of ccrAB genes in the absence of mec genes in coagulase-negative staphylococci. Whether the nontypeable strains have a truncated or a different recombinase system needs to be studied.
TABLE 2.
Phenotypic and genotypic characteristics of MRSA isolates from two hospitals in Bangalore, South India
| PFGE type | No. of isolates | Antimicrobial resistancea | SCCmec type (no. of isolates) |
|---|---|---|---|
| A1 | 17 | P, M, E, G, T | III (16), NAb (1) |
| A2 | 7 | P, M, E, G, T | III |
| A3 | 8 | P, M, E, G, T | III |
| A4 | 2 | P, M, E, G, T | III |
| B1 | 7 | P, M, E, G, T | III (2), IIIA (5), III (1) |
| B2 | 1 | P, M, E, G, T | |
| B3 | 1 | P, M, E, G, T | IIIA |
| B4 | 4 | P, M, E, G, T | III (3), IIIA (1) |
| B5 | 2 | P, M, E, G, T | IIIA (1), NA (1) |
| B6 | 1 | P, M, E, G, T | III |
| B7 | 1 | P, M, E, G, T | IIIA |
| B8 | 1 | P, M, E, G, T | III |
| B9 | 1 | P, M, E, G, T | IIIA |
| B10 | 1 | P, M, E, G, T | NA |
| B11 | 1 | P, M, E, G, T | IIIA |
| C1 | 8 | P, M, E, G, T | III (2), IIIA (6) |
| C2 | 5 | P, M, E, G, T | III (1), IIIA (3), NTc (1) |
| C3 | 1 | P, M, E, G, T | III |
| C4 | 3 | P, M, E, G, T | III (1), IIIA (2) |
| C5 | 1 | P, M, E, G, T | IIIA |
| C6 | 1 | P, M, E, G, T | IIIA |
| C7 | 1 | P, M, E, G, T | IIIA |
| C8 | 4 | P, M, E, G, T | III (3), NT(1) |
| D1 | 2 | P, M, E, G, T | NT (2) |
P, penicillin; M, methicillin; E, erythromycin; G, gentamicin; T, tetracycline.
NA, not amplified with the set of primers used.
NT, nontypeable (not belonging to SCCmec type I, II, III, or IIIA).
The 82 isolates from SJ and M were grouped into three major PFGE patterns, A, B, and C. The majority of samples from SJ and M had PFGE patterns A1, A2, A3, B1, and C1. Patterns B2 to B11 had 14 isolates altogether from both hospitals, although B6, B7, B8, B9, and B11 were present only in isolates from SJ, represented by one strain per pattern. The largest diversity was in PFGE pattern B subsets, followed by pattern C. There were 21 isolates from SJ with patterns C1 to C8, of which 12 isolates belonged to type IIIA SCCmec, and only 2 from M belonged to type IIIA. Among the four nontypeable strains of SJ, two belonged to pattern D, which was quite different from the three other patterns, and one each of the nontypeable strains belonged to patterns C2 and C8, respectively.
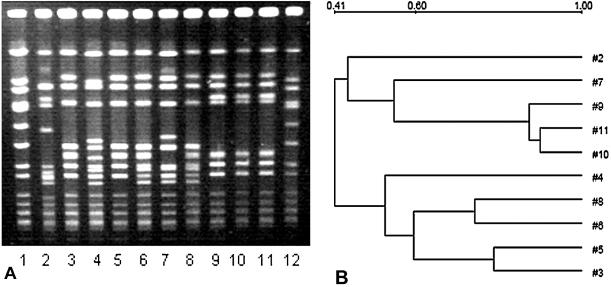
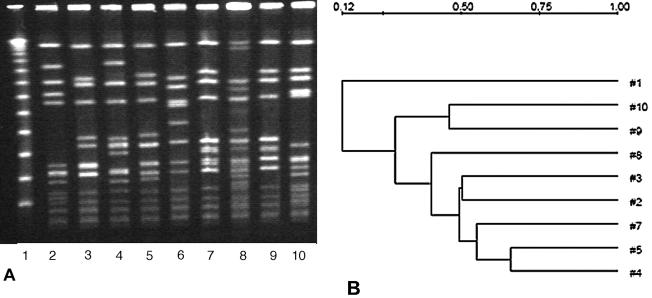
Recently, Aires de Sousa et al. (1) reported the recovery of predominantly type III and IIIA isolates from two hospitals in Taiwan and China which were related to the Hungarian and the Brazilian epidemic clones. PFGE was performed with Hungarian strain HUSA304 (SCCmec type III) and Brazilian strain HSJ216 (type IIIA; pT181 absent) along with samples from SJ and M, as shown in Fig. 1A and 2A. Figure 1A represents the PFGE patterns of SmaI macrorestriction fragments of selected strains from SJ and M, and Fig. 2A shows patterns with the Hungarian and the Brazilian clones along with some strains from SJ and M. Figure 1B reveals the relatedness between the SJ and M isolates, and Fig. 2B shows the relatedness of isolates from SJ and M to the Hungarian and the Brazilian clones. Patterns A1 and C1 of the clinical isolates differ from those of the Hungarian clone HUSA 304 and Brazilian clone HSJ 216 by two and three bands, respectively. Pattern A1 differs from B1 by only two bands. A1 and C1 are the predominant PFGE patterns among all the isolates. Aires de Sousa et al. have shown that the PFGE patterns of other Brazilian clones, i.e., HU 25, BRA 5, and BRA 101, have a high degree of similarity to those of HSJ 216. Similarly, the other Hungarian clones HU 101, HUSA 88, and HUSA 176 are closely related to HUSA 304 (1).
FIG. 1.
(A) PFGE patterns of SmaI digests of MRSA isolates from the two hospitals. Lanes 1 and 12, NCTC 8325; lanes 2 to 5, SJ isolates; lanes 6 to 11, M isolates. (B) Dendrogram based on the similarities derived from the UPGMA and Dice coefficients, using Quantity One software. The scale at the top represents similarity.
FIG. 2.
(A) PFGE patterns of SmaI digests of MRSA isolates from the two hospitals. Lane 6, NCTC 8325; lane 10, HUSA 304 (Hungarian clone); lane 2, HSJ 216 (Brazilian clone); lanes 4, 5, and 9, SJ isolates; lanes 3, 7, and 8, M isolates; lane 1, lambda molecular weight marker. (B) Dendrogram based on the similarities derived from the UPGMA and Dice coefficients, using Quantity One software. The scale at the top represents similarity.
Selected strains from SJ and M were processed for MLST and spa typing, and the data are presented in Table 3. Two isolates each from SJ and M belonged to sequence type (ST) 239, and one isolate from M was ST 241, which has yqiL allele 30 and is a single-locus variant of yqiL 3 (MLST database). STs 239 and 241 have also been detected in Thailand (7), as well as in China and Taiwan (1). As MLST involves analyzing 14 sequences for each strain and is expensive, spa typing was done for 30 isolates, 15 from SJ and 15 from M. Twenty-eight isolates had the same spa type (WGKAOMQ) by the Kreisworth nomenclature (12), and two isolates, P2300 and P3400 from SJ, had different repeat sequences (XKAOMQ and XKAQKAOMQ, respectively). The common sequence KAOMQ is considered the signature spa sequence for type III and IIIA isolates. Strain P2300 also has the common sequence KAOMQ, although it has only part of the type III cassette. The MIC for this strain also is very low (0.78 μg/ml), and further studies are under way. The MLST, ST, and spa typing results for the SJ and M strains are also in concordance with similar patterns for the Hungarian and the Brazilian clones, where ST 239 and spa type WGKAOMQ predominate (1).
TABLE 3.
MICs, SCCmec typing, MLST, and spa typing for selected strains from SJ and M
| No. of isolatesa | Oxacillin MIC (μg/ml) | SCCmec type (no. of isolates) | spa typec | MLSTd | ST |
|---|---|---|---|---|---|
| 3 | 6-50b | IIIA | WGKAOMQ | ||
| 20 | 200-800 | III (11), IIIA (5), NTe (1), NAf (3) | WGKAOMQ | ||
| 2 | 200 | IIIA | WGKAOMQ | 2-3-1-1-4-4-3 | 239 |
| III | WGKAOMQ | 2-3-1-1-4-4-30 | 241 | ||
| 1 | 0.78 | NT | XKAOMQ | ||
| 1 | 25 | IIIA | WGKAOMQ | 2-3-1-1-4-4-3 | 239 |
| 1 | 100 | IIIA | XKAQKAOMQ | ||
| 1 | 400 | III | WGKAOMQ | 2-3-1-1-4-4-3 | 239 |
| 1 | >800 | III | WGKAOMQ | 2-3-1-1-4-4-3 | 239 |
| HUSA 304 | III | WGKAOMQ | 2-3-1-1-4-4-3 | 239 | |
| HSJ 216 | IIIA | WGKAOMQ | 2-3-1-1-4-4-3 | 239 |
HUSA 304 (ATCC BAA-39) is the Hungarian clone, and HSJ 216 (ATCC BAA-43) is the Brazilian clone.
Range of MICs obtained for different isolates.
DNA sequence of the repeat region of protein A gene.
Allelic profile based on the DNA sequences of seven housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi, and yqiL).
NT, nontypeable.
NA, not amplified with the set of primers used.
Both MLST and spa typing suggested a very limited divergence between the isolates included in this study, but PFGE generated a more diverse pattern. This may be explained by PFGE reflecting chromosomal rearrangements that may be more common than mutations changing the genes analyzed for MLST and spa typing. It may be an indication that these isolates have acquired the resistance genes on different occasions, perhaps from a common reservoir of commensal bacteria.
Indian S. aureus strains had not been genotyped until now, except for a report by Hanssen et al. (9), who performed PFGE for two Indian strains and reported that they belong to ccr type 3. Among the 82 strains from two hospitals in Bangalore that we have genotyped, 75 isolates were type III or IIIA. Although the majority of samples from SJ were isolated from pus, there was diversity in the PFGE patterns, and types III and IIIA were distributed equally. There was also a more varied range of MICs with SJ samples. Strains from M were cultured from a variety of sources, with pus and miscellaneous sources comprising 43% and 38%, respectively, and yet type III (84%) was the predominant SCCmec type among these isolates. SJ, being a teaching hospital, perhaps treats a more ethnically and socioeconomically diverse group of people, while M is a private hospital with a restricted clientale. However, PFGE, MLST, and spa typing data indicate that most of the strains obtained from these two hospitals are genetically related and are clonally similar to the Hungarian and the Brazilian epidemic strains. Although we have looked at a limited number of strains from two hospitals in Bangalore, it is clear that isolates related to the Brazilian and Hungarian epidemic clones have made their appearance in India. We are collecting MRSA isolates from various parts of India to genotype them and to follow the patterns and trends to help us in long-term epidemiological studies.
Acknowledgments
We acknowledge the help of Savita Nagaraj from St. John's Hospital and H. B. Beena and Mary Rajammal from Manipal Hospital with the collection of samples. We are grateful to G. Padmanaban for critically reviewing the manuscript.
This work was supported mainly by funds from the Dorabji Tata Trust and partially with a planning grant from the Swedish Research Link program of the Swedish Agency for Research Cooperation with Developing Countries (SIDA/SAREC) and the Swedish Research Council.
REFERENCES
- 1.Aires de Sousa, M., M. I. Crisostomo, I Santos Sanches, J. S. Wu, J. Fuzhong, A. Tomasz, and H. de Lencastre. 2003. Frequent recovery of a single clonal type of multidrug-resistant Staphylococcus aureus from patients in two hospitals in Taiwan and China. J. Clin. Microbiol. 41:159-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anupurba, S., M. R. Sen, G. Nath, B. M. Sharma, A. K. Gulati, and T. M. Mohapatra. 2003. Prevalence of methicillin resistant Staphylococcus aureus in a tertiary referral hospital in eastern Uttar Pradesh. Indian J. Med. Microbiol. 21:49-51. [PubMed] [Google Scholar]
- 3.Baird, D. 1996. Staphylococcus: cluster-forming gram positive cocci, p. 245-261. In J. G. Collee, A. G. Fraser, B. P. Marmion, and A. Simmons (ed.), Practical medical microbiology. Churchill Livingstone, New York, N.Y.
- 4.Chung, M., G. Dickenson, H. de Lencastre, and A. Tomasz. 2004. International clones of methicillin resistant Staphylococcus aureus in two hospitals in Miami, Florida. J. Clin. Microbiol. 42:542-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cookson, B. D., P. Aparicio, A. Deplano, M. Struelens, R. Goering, and R. Marples. 1996. Inter-centre comparision of pulsed-field gel electrophoresis for the typing of methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 44:179-184. [DOI] [PubMed] [Google Scholar]
- 6.Cosgrove, S. E., G. Sakoulas, E. N. Perencevich, M. J. Schwaber, A. W. Karchmer, and Y. Carmeli. 2003. Comparison of mortality associated with methicillin resistant and methicillin susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin. Infect. Dis. 36:53-59. [DOI] [PubMed] [Google Scholar]
- 7.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enright, M. C., N. P. J. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanssen, A., G. Kjeldsen, and J. U. E. Sollid. 2004. Local variants of staphylococcal cassette chromosome mec in sporadic methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci: evidence of horizontal gene transfer? Antimicrob. Agents Chemother. 48:285-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ip, M., D. J. Lyon, F. Chio, M. C. Enright, and A. F. Cheng. 2003. Characterization of isolates of methicillin-resistant Staphylococcus aureus from Hong Kong by phage typing, pulsed-field gel electrophoresis, and fluorescent amplified-fragment length polymorphism analysis. J. Clin. Microbiol. 41:4980-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koreen, L., S. V. Ramaswamy, E. A. Gravis, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krishnan, P. U., K. Miles, and N. Shetty. 2002. Detection of methicillin and mupirocin resistance in Staphylococcus aureus isolates using conventional and molecular methods: a descriptive study from a burns unit with high prevalence of MRSA. J. Clin. Pathol. 55:745-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mehrotra, M., G. Wang, and W. M. Johnson. 2000. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J. Clin. Microbiol. 38:1032-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murchan, S., M. E. Kaufmann, A. Deplano, Raf de Ryck, M. Stuelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. V. Leeuwen, A. V. Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility testing for bacteria that grow aerobically, 5th ed. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 18.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 19.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson, D. A., and M. C. Enright. 2003. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:3926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlichting, C., C. Branger, Jean-Michel Fournier, W. Witte, A. Boutonnier, C. Wolz, P. Goullet, and G. Goring. 1993. Typing of Staphylococcus aureus by pulsed-field gel electrophoresis, zymotyping, capsular typing, and phage typing: resolution of clonal relationships. J. Clin. Microbiol. 31:227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Straden, A., R. Frei, and A. F. Windmer. 2003. Molecular typing of methicillin-resistant Staphylococcus aureus: can PCR replace pulsed-field gel electrophoresis? J. Clin. Microbiol. 41:3181-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang, Y.-W., M. G. Waddington, D. H. Smith, J. M. Manahan, P. C. Kohner, L. M. Highsmith, H. Li, F. R. Cockerll III, R. L. Thompson, S. O. Montgomery, and D. H. Persing. 2000. Comparision of protein A gene sequencing with pulsed-field gel electrophoresis and epidemiologic data for molecular typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 38:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]