Abstract
Methicillin-resistant Staphylococcus aureus (MRSA), regarded as a tenacious pathogen in the hospital, has recently become increasingly prevalent as a community pathogen. We evaluated the prevalence and characteristics of methicillin-resistant staphylococci in the Japanese community by testing nasal samples of 818 children of five day care centers and two kindergartens in three districts. We found that methicillin-resistant staphylococci are already prevalent among healthy children. Among 818 children, 35 children (4.3%) carried MRSA and 231 children (28.2%) carried methicillin-resistant coagulase-negative staphylococci (MRC-NS). The types of staphylococcal cassette chromosome mec (SCCmec) found among 44 MRSA isolates were as follows: type IIa, 11 isolates; type IIb, 19 isolates; and type IV, 14 isolates. The type IIb SCCmec element was a new SCCmec element found in this study. Eleven (25%) strains which belonged to clonal complex 5 (CC5) carried type IIa SCCmec, and they produced type 2 coagulase and toxic shock syndrome toxin 1. They were indistinguishable from health care-associated MRSA (H-MRSA) strains in Japan, represented by strain N315. On the other hand, 33 (75%) strains, most of which belonged to CC78 or CC91, carried small SCCmec elements, such as type IIb or type IV, and they produced type 1 or type 3 coagulase and exfoliative toxin. The data indicated that MRSA clones distinct from H-MRSA have disseminated in healthy children. The fact that MRC-NS strains were prevalent in the community suggested that they might serve as a reservoir for the SCCmec element carried by MRSA strains disseminated in the community.
Since the discovery of the first clinical isolates in 1960, methicillin-resistant Staphylococcus aureus (MRSA) has remained a major hospital pathogen throughout the world (19). However, recent reports suggest that it became increasingly prevalent in the community as well since the 1990s (6, 8, 13, 31). Now, the MRSA strains designated community-acquired or community-associated MRSA (C-MRSA) are increasingly found in healthy individuals without conventional risk factors for MRSA colonization (2, 11, 14, 26, 33).
MRSA strains carry methicillin resistance gene mecA, encoded by a mobile genetic element designated staphylococcal cassette chromosome mec (SCCmec) (15, 22). We define the type of SCCmec by the combination of the type of ccr gene complex, composed of cassette chromosome recombinase genes and the surrounding open reading frames (ORFs), and the class of the mec gene complex, composed of the mecA gene and its surrounding ORFs. A total of five allelic types have been identified in SCCmec elements (16, 17, 21). Three types of SCCmec elements (type I, type II, and type III) are carried mostly by health care-associated MRSA (H-MRSA) strains throughout the world (9, 16). On the other hand, novel types of SCCmec elements (type IV and type V) have been widely disseminated among C-MRSA strains (7, 17, 25, 29). The type IV and type V SCCmec elements are characterized by their small sizes (21 to 28 kbp) and lack of resistance genes, other than mecA (17, 25, 29).
MRSA clones are defined by the type of SCCmec element and the genotype of the methicillin-susceptible S. aureus chromosome in which the SCCmec element is integrated (12). We have shown that the C-MRSA strains isolated in Australia and the United States were derived from more diverse S. aureus clones than H-MRSA strains by determination of the types of the SCCmec elements and the types of their chromosomes by multilocus sequence typing (MLST) (29). C-MRSA strains grew faster than H-MRSA strains and carried virulence genes, such as Panton-Valentine leucocidin (PVL) genes (1, 29, 37).
This study was undertaken to investigate the prevalence of MRSA strains and methicillin-resistant coagulase-negative staphylococci (MRC-NS) among healthy Japanese children. In addition, we describe the characteristic features of MRSA strains distributed in the Japanese community.
MATERIALS AND METHODS
Isolation of methicillin-resistant staphylococci from nasal swabs of healthy children.
To establish the prevalence of methicillin-resistant staphylococci in the community, we have isolated staphylococci from nasal swabs of healthy children from five day care centers and two kindergartens in three different districts: Miyagi, Kyoto, and Saga. To understand the colonization of methicillin-resistant staphylococci, we obtained samples from children in Miyagi twice, with an 8-month interval. In the first sampling, in July 2001, 362 children were sampled; and in the second sampling, in March 2002, 292 children were sampled. Among the 292 children sampled in the second sampling, 236 children who had been sampled in the first test were resampled. In Kyoto and Saga, we sampled 150 and 250 children, respectively. The children who were absent on the day of investigation and the children who did not receive parental consent were not included in this study. A total of 818 children were tested.
Samples were obtained from both nares of the children by using a sterile dry-cotton swab (Medical Wire & Equipment Co., Ltd., Corsham, United Kingdom) and were inoculated directly onto mannitol-salt agar (Eiken Chemical Co., Ltd., Tokyo, Japan), with or without 10 mg/liter of ceftizoxime (Fujisawa Pharmaceutical Co., Ltd., Osaka, Japan), and incubated at 37°C for 48 h. Yellow colonies that grew on the agar plates were tested for the production of clumping factor and protein A by using a Staphylo LA test kit (Denka Seiken Co., Ltd., Niigata, Japan) to distinguish S. aureus from other species. The species of the strains that showed negative reactions in the Staphylo LA test were determined by using an identification kit (StaphyoGram; Wako Pure Chemical Industries, Ltd., Osaka, Japan).
Identification of exotoxin genes and SCCmec elements.
Chromosomal DNAs were extracted from cells that had been cultured overnight by the phenol-chloroform extraction methods described previously (29). Detection of the mecA gene and typing of the SCCmec elements were carried out by PCR, as described previously (16, 29). PCR amplification was performed in a 50-μl reaction mixture composed of 2 U of Ex Taq (Takara Shuzo Co., Ltd., Kyoto, Japan), 10 pmol of each primer, 0.2 mM deoxynucleoside triphosphate mixture, 10 ng of chromosomal DNA, 1× reaction buffer (Takara Shuzo Co., Ltd.), and H2O. Thermal cycling was set at 30 cycles (30 s for denaturation at 94°C, 1 min for annealing at 50°C, and 2 min for elongation at 72°C) and was performed with a Gene Amp PCR system 9600 (Perkin-Elmer, Wellesley, Mass.). Localization of five exotoxin genes, the toxic shock syndrome toxin 1 gene (tst-1), the exfoliative toxin A gene (eta), the exfoliative toxin B gene (etb), the exfoliative toxin D gene (etd), and the Panton-Valentine leucocidin genes (lukS and lukF); and all MRSA isolates were examined by PCR with the sets of primers listed in Table 1. The S. aureus strains used as references for the five exotoxins were N315 for tst-1, ZM for eta, N20 for etb, TY114 for etd, and MW2 for lukS and lukF. S. aureus ZM and N20 were kindly provided by Y. Yoshizawa, Jikei University, Tokyo, Japan. S. aureus TY114 was kindly provided by M. Sugai, Department of Microbiology, Hiroshima University, Hiroshima, Japan (39-41).
TABLE 1.
Primers used for SCCmec typing and toxin gene detection
| Primer group and gene(s) or gene allele detected | Primer name | Nucleotide sequence (5′→3′) | Reference or source |
|---|---|---|---|
| mecA | mA1 | TGCTATCCACCCTCAAACAGG | 29 |
| mA2 | AACGTTGTAACCACCCCAAGA | 29 | |
| ccr gene complex type | |||
| ccrB | βc | ATTGCCTTGATAATAGCCITCTa | 16 |
| ccrA1 | α1 | AACCTATATCATCAATCAGTACGT | 16 |
| ccrA2 | α2 | TAAAGGCATCAATGCACAAACACT | 16 |
| ccrA3 | α3 | AGCTCAAAAGCAAGCAATAGAAT | 16 |
| ccrC | γ1 | AGCCCAATTTTGATGGTTATTGA | This study |
| γ2 | TGGAGAAGTACTCGTTACAATGT | This study | |
| mec gene complex class | |||
| mecI-mecRI (class A) | mI4 | CAAGTGAATTGAAACCGCCT | 29 |
| mcR3 | GTCTCCACGTTAATTCCATT | 21 | |
| IS1272-mecA (class B) | IS5 | AACGCCACTCATAACATATGGAA | 29 |
| mA6 | TATACCAAACCCGACAAC | 21 | |
| IS431mecL-mecA (class C) | mA2 | AACGTTGTAACCACCCCAAGA | 21 |
| IS2 | TGAGGTTATTCAGATATTTCGATGT | 21 | |
| Primers for subtyping | |||
| IIa | 2a1 | ATGTCAGAGCTTTCTAACTTAGTCA | This study |
| 2a2 | TGAAAATGAAAGCCGTGCCG | This study | |
| IIb | 2b1 | AGCAATTTTTTCTCCTTCTGCTA | This study |
| 2b2 | TTATTAGATCAAGAGCCAAGTG | This study | |
| IVa | 4a1 | TTTGAATGCCCTCCATGAATAAAAT | 29 |
| 4a2 | AGAAAAGATAGAAGTTCGAAAGA | 29 | |
| IVb | 4b1 | AGTACATTTTATCTTTGCGTA | 29 |
| 4b2 | AGTCATCTTCAATATCGAGAAAGTA | 29 | |
| IVc | 4c1 | TCTATTCAATCGTTCTCGTATTT | This study |
| 4c2 | TCGTTGTCATTTAATTCTGAACT | This study | |
| IVd | 4d1 | TTTGAGAGTCCGTCATTATTTCTT | This study |
| 4d2 | AGAATGTGGTTATAAGATAGCTA | This study | |
| Exotoxic genes | |||
| tst-1 | TSST-1A | TGATATGTGGATCCGTCAT | This study |
| TSST-1B | AAACACAGATGGCAGCAT | This study | |
| eta | ET-1 | CTATTTACTGTAGGAGCTAG | 39 |
| ET-2 | ATTTATTTGATGCTCTCTAT | 39 | |
| etb | ET-3 | ATACACACATTACGGATAAT | 40 |
| ET-4 | CAAAGTGTCTCCAAAAGTAT | 40 | |
| etd | ET-14 | AACTATCATGTATCAAGG | 41 |
| ET-15 | CAGAATTTCCCGACTCAG | 41 | |
| lukS and lukF | PVLup | AAGACTATTAGCTGCAACATTGTC | 29 |
| PVLdn | AATCTATCTGTTTAGCTCATAGGA | 29 |
I, inosine.
Coagulase isotyping.
The coagulase isotypes of MRSA strains were determined by the coagulation inhibition test for coagulation by using commercially available rabbit antisera specific to each of the eight isotypes of staphylocoagulases (Denka Seiken Co., Ltd., Tokyo, Japan), as described previously (29). Briefly, S. aureus cells were cultured overnight in brain heart infusion broth (Becton Dickinson Co., Ltd., Paramus, N.J.), and the supernatant was collected by centrifugation. The 0.1-ml aliquot of the supernatant, which was diluted appropriately with diluent (2.0% polypeptone, 1.0% sodium citrate, 0.1% sodium azide), was distributed into nine tubes, and 0.1 ml of anticoagulase type I to VIII sera was added to tubes 1 to 8. In the ninth tube, 0.1 ml of 5% normal rabbit serum in diluent was added as a control. Then, the tubes were incubated at 37°C for 1 h. After the incubation, 0.2 ml of diluted rabbit plasma was added to each tube, followed by incubation at 37°C for at least 1 h. The coagulation of the plasma was judged by visual inspection after 1, 2, 4, 6, and 24 h. The type of antiserum that inhibited the coagulation alone was regarded as the type of staphylocoagulase produced by the strain.
PFGE.
Chromosomal DNAs of the MRSA strains were digested with SmaI and were separated by pulsed-field gel electrophoresis (PFGE) with a Gene Path system (CHEF-DR, PULS WAVE.760; Bio-Rad, Hercules, Calif.). The settings for PFGE were as follows: initial switch time, 5.0 s; final switch time, 40.0 s; included angle, 120°; current, 200 V; and run time, 22 h. The buffer temperature was maintained at 14°C. The correlations of the banding patterns were analyzed with BioNumerics software (version 2.5; Applied Maths, Kortrijk, Belgium). A similarity index was determined for each pair of strains by using the Dice coefficient with 0.5% band tolerance. Clustering correlation coefficients were calculated by using the unweighted pair group method of arithmetic averages. Isolates were considered “potentially genetically related” if their macrorestriction DNA patterns differed by less than seven bands (35).
MLST.
The MLSTs of the MRSA strains representing each pulsotype were determined according to the method described previously (10). Sequencing reactions were performed with fluorescent dideoxy chain termination chemistry by using a Big Dye Terminator (version 1.1) cycle sequencing kit (Applied Biosystems). DNA sequencing was performed with an ABI Prism 3100 genetic analyzer (Applied Biosystems). The nucleotide sequences of each of the seven genes were assigned allele numbers by comparing them with those of the extant alleles listed on the MLST website (http://www.mlst.net). The sequence type (ST) was determined according to the pattern of the combination of the seven alleles, and the clonal complex (CC) was defined by the BURST (based upon related sequence types) program by accessing the MLST website.
Antimicrobial susceptibility testing.
The minimal growth-inhibitory concentrations of isolates against nine antibiotics (MICs) were determined by the agar dilution method recommended by CLSI (formerly the National Committee for Clinical Laboratory Standards) (28). The antibiotics tested were as follows: oxacillin and gentamicin, Sigma Chemical Co., Ltd., St. Louis, Mo.; ceftizoxime, Fujisawa Pharmaceutical Co., Ltd.; imipenem, Banyu Pharmaceutical Co., Ltd., Tokyo, Japan; erythromycin and tetracycline, Wako Pure Chemical Industries, Ltd.; tobramycin, Shionogi Co., Ltd., Osaka, Japan; kanamycin, Meiji Seika Kaisha, Ltd., Tokyo, Japan; and norfloxacin, Kyorin Pharmaceutical Co., Ltd., Tokyo, Japan.
Nucleotide sequence accession number.
The nucleotide sequence of the region containing the junkyard 1 (J1) region and the ccr gene complex of SCCmec type IIb of strain JCSC3063 has been deposited in the DDBJ/EMBL/GenBank database under accession no. AB127982.
RESULTS
Carriage of MRSA and MRC-NS in healthy children.
We took nasal smear samples from healthy children in five day care centers and two kindergartens in three prefectures of Japan, Miyagi, Kyoto, and Saga, from north to south, respectively. To elucidate the stability of MRSA carriage by individuals, the Miyagi study was done twice with overlapping subjects: the first study was done in July 2001 and the second one was done in March 2002. The number of children who participated was 818 in total: 418 from the first and second studies in Miyagi, 150 from Kyoto, and 250 from Saga (Table 2). Table 2 summarizes the results of the surveillance for MRSA and MRC-NS among the children tested. Of the 818 children, 231 (28.2%) children carried S. aureus, and 35 (4.3%) children carried MRSA. The ratio of MRSA strains among the S. aureus isolates was 15.1%. The percentages of MRSA carriers among the children in the four studies were 5.2% (19 of 362) in the first Miyagi study, 3.7% (11 of 292) in the second Miyagi study, 4.0% (6 of 150) in Kyoto, and 3.2% (8 of 250) in Saga. Interestingly, 231 (28.2%) children carried MRC-NS. The rates of carriage of MRC-NS in each study were 39.7% (144 of 362) in the first Miyagi study, 24.0% (70 of 292) in the second Miyagi study, 19.3% (29 of 150) in Kyoto, and 15.2% (38 of 250) in Saga.
TABLE 2.
Carriage of methicillin-resistant staphylococci by healthy Japanese children
| District or group | No. (%) of children
|
|||
|---|---|---|---|---|
| Total tested | With S. aureus | With MRSA | With MRC-NS | |
| Miyagi, first study | 362 | 133 (36.7) | 19 (5.2) | 144 (39.7) |
| Miyagi,a second study | 56 | 12 (21.4) | 2 (3.6) | 20 (35.7) |
| Children sampled a second time in Miyagi | 236 | 56 (23.7) | 9 (3.8) | 50 (21.1) |
| Kyoto | 150 | 28 (18.7) | 6 (4.0) | 29 (19.3) |
| Saga | 250 | 58 (23.2) | 8 (3.2) | 38 (15.2) |
| Totalb | 818 | 231 (28.2) | 35 (4.3) | 231 (28.2) |
In the second test, a total of 292 children were tested. Among them, 56 children were sampled only in the second test, whereas 236 children were sampled twice, in both the first and the second tests. The numbers of children carrying S. aureus, MRSA, or MRC-NS among the 236 resampled children are also shown.
To eliminate overlapping of the data, we excluded the children from the second sampling when calculating the total number of tested children. Therefore, the total numbers of children are the cumulative sums of the numbers of children participating in the first and second tests in Miyagi and the tests in Kyoto and Saga.
A new SCCmec subtype (type IIb) identified in MRSA strains from healthy children.
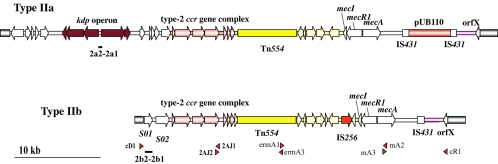
To determine the clonal compositions of the MRSA strains carried by healthy children, we analyzed 44 MRSA strains isolated in this study. Among the 44 MRSA strains, 30 strains carried the type II SCCmec element and 14 strains carried the type IV SCCmec element. Among the 30 type II SCCmec strains, 11 strains carried the kdp operon and were judged to carry the same type IIa SCCmec element as strain N315. Since the subtype of the type II SCCmec elements of the other 19 strains seemed to be new, we determined the nucleotide sequence from the J1 region to the mec gene complex of a type II SCCmec element from strain JCSC3063 (SD036-1). Figure 1 illustrates the comparison of structures between a new subtype of SCCmec designated type IIb SCCmec and the type IIa SCCmec element that we reported on previously (18, 23). The J1 region of the type IIb SCCmec element in JCSC3063 (SD036-1) was shorter than that in the type IIa SCCmec element and contained two open reading frames which had no similarity to those found in the latter element. Since the element did not carry plasmid pUB110, which encodes bleomycin and tobramycin resistance, its size was estimated to be approximately 29 kb, which was confirmed by the long-range PCR analysis of the entire element (Fig. 1). The remaining 19 strains were judged to carry the type IIb SCCmec element by PCR with a set of primers constructed to be specific for J1-region DNAs of type IIb, as shown in Fig. 1.
FIG. 1.
Structural comparison of type IIa and type IIb SCCmec elements. The structures of the type IIa and type IIb SCCmec elements are illustrated based on the nucleotide sequences deposited in the DDBJ/EMBL/GenBank databases under accession nos. D86934 and AB127982, respectively. The entire SCCmec regions of the type IIb SCCmec elements were amplified by PCR with four sets of primers, indicated with arrowheads. The four sets of primers are as follows: cR1 and mA3, mA2 and ermA1 (5′-TGAAACAATTTGTAACTATTGA-3′), ermA3 (5′-TGGGTAAACCGTGAATATCGTGT-3′) and 2AJ1 (5′-ATTAGCCGATTTGGTAATTGAA-3′), and 2AJ2 (5′-TCGTACTTTGACGTAAATAGCCT-3′) and cD1 (5′-TAGTAAAGACTGTGAAATCTCATAT-3′). The nucleotide sequences of primers cR1, mA2, and mA3 were reported previously (25, 29). Type II SCCmec is defined as an SCCmec element which typically possesses a class A mec gene complex combined with a type 2 ccr gene complex. The element was further subtyped according to the differences in the nucleotide sequences of the junkyard 1 regions. The novel subtype of type II SCCmec element, type IIb, carried Tn554, which encodes macrolide-lincomycin-spectinomycin resistance, but did not carry pUB110, which encodes aminoglycoside resistance. Although IS256 was inserted upstream of the mecI gene, we are not sure whether or not it is happenstance that it was inserted in the element. The locations of the two sets of primers used for the identification of the type IIa SCCmec-specific J1 region and the type IIb SCCmec-specific J1 region are shown in bars. Two primers, primers 2a1 and 2a2, were constructed on the basis of the sequence of the kdp operon; and two primers, primers 2b1and 2b2, were constructed on the basis of the sequence of open reading frame S02.
Among the 14 strains that carried the type IV SCCmec element, 5 of them carried the type IVa SCCmec element and the remaining 9 strains carried an unknown subtype of the type IV SCCmec element so far tested with sets of primers used to identify four subtypes. There was a distinct geographical distribution of the SCCmec types of the MRSA strains carried by healthy children. In the Kyoto and Saga surveys, 12 of 14 MRSA isolates carried the type IV SCCmec element, whereas in the Miyagi study 28 of 30 MRSA strains carried the type II SCCmec element (Table 3).
TABLE 3.
Types of SCCmec elements carried by 44 MRSA strains isolated from 42 healthy childrena
| District or group | No. of MRSA strains tested | No. of strains with the following type of SCCmec element:
|
|||
|---|---|---|---|---|---|
| IIa | IIb | IVa | IVn | ||
| Miyagi, first | 19 | 7 | 11 | 0 | 1 |
| Miyagi,a second | 2 | 1 | 1 | 0 | 0 |
| Children sampled a second time in Miyagi | 9 | 1 | 7 | 1 | 0 |
| Kyoto | 6 | 1 | 0 | 2 | 3 |
| Saga | 8 | 1 | 0 | 2 | 5 |
| Total | 44 | 11 | 19 | 5 | 9 |
Among the 236 children sampled twice, 2 children continuously carried MRSA strains, and in the second test 7 children carried newly acquired MRSA strains. Therefore, 44 MRSA strains were isolated from 42 children.
MRSA clones distinct from H-MRSA were carried by healthy children.
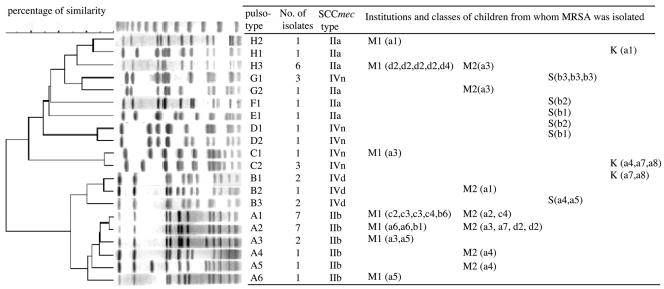
We determined the pulsotypes of all MRSA isolates. A total of 20 pulsotypes were identified among the 44 strains, and they were classified into eight major groups, groups A to H, by the mutual correlations of their banding patterns (Fig. 2). All of the 19 MRSA strains that were isolated in Miyagi and that carried the type IIb SCCmec element belonged to pulsotype A. Strains that carried the type IIa SCCmec element belonged to pulsotypes E, F, G, and H, whereas type IV SCCmec strains belonged to four pulsotypes, B, C, D, and G, indicating that their genetic backgrounds are very divergent. Furthermore, the multilocus STs of 20 MRSA strains representing each pulsotype were determined. Six STs (ST5, ST8, ST78, ST89, ST90, and ST91) were identified, and they were classified into four clonal complexes (CC5, CC8, CC78, and CC91).
FIG. 2.
Dendrogram of PFGE banding patterns of 44 MRSA isolates. A total of 44 MRSA strains were classified into eight pulsotypes. Pulsotypes of a representative strain of each type were compared by using the BioNumerics software program. The institutions and classes of children from whom MRSA strains were isolated are also indicated. The institution (kindergarten or day care center) to which a child belonged is indicated by the following abbreviations: M1, a child in Miyagi tested the first time; M2, a child in Miyagi tested the second time; S, a child in Saga; K, a child in Kyoto. Seven institutions have been subdivided according to the classes that they have. Their classes are indicated in parentheses by abbreviations. The names of the institutions are shown with letters, and the classes are indicated with numerals. Four institutions in Miyagi had multiple classes. They are indicated as a (a1 to a7), b (b1 to b7), c (c1 to c6), and d (d1 to d4). Two institutions in Saga and an institution in Kyoto are also indicated as a (a1 to a7) and b (b1 to b6) and a (a1 to a8), respectively.
Table 4 shows the characteristic features of the 44 MRSA strains and the susceptibilities of the strains to various antibiotics.
TABLE 4.
Characterization of MRSA strains isolated from healthy children
| Pulso- type | No. of isolates | Districta | coab | SCCmec type | Presence of exotoxin gene(s)c
|
Strain chosen | MLST
|
MIC (mg/liter)d
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| et allele | tst-1 | lukS and lukF | ST | Allelic profile | CC | CZX | OXA | IMP | TET | NOR | ERY | GEN | TOB | KAN | ||||||
| A1 | 7 | M | 1 | 2A2(IIb) | b | − | − | SD205-1 | 89 | 1-26-28-18-18-33-50 | 91 | >128 | 4 | 0.13 | 0.5 | 2 | >128 | >128 | >128 | >128 |
| A2 | 7 | M | 1 | 2A2(IIb) | b | − | − | SD036-1 | 89 | 1-26-28-18-18-33-50 | 91 | >128 | 32 | 0.5 | 0.5 | 1 | >128 | 64 | 32 | >128 |
| A3 | 2 | M | 1 | 2A2(IIb) | b | − | − | SD084-1 | 89 | 1-26-28-18-18-33-50 | 91 | 64 | 1 | <0.06 | 0.5 | 2 | >128 | >128 | 32 | >128 |
| A4 | 1 | M | 1 | 2A2(IIb) | b | − | − | SD2:063-3 | 89 | 1-26-28-18-18-33-50 | 91 | >128 | 64 | 1 | 0.5 | 4 | >128 | >128 | >128 | >128 |
| A5 | 1 | M | 1 | 2A2(IIb) | b | − | − | SD2:050-2 | 89 | 1-26-28-18-18-33-50 | 91 | >128 | 8 | 0.25 | 0.5 | 4 | >128 | >128 | 32 | >128 |
| A6 | 1 | M | 1 | 2A2(IIb) | − | − | − | SD007-1 | 89 | 1-26-28-18-18-33-50 | 91 | >128 | 64 | 0.5 | 0.5 | 2 | >128 | 32 | >128 | >128 |
| B3 | 2 | S | 1 | 2B1(IVa) | − | − | − | SG25 | 89 | 1-26-28-18-18-33-50 | 91 | 64 | 4 | 0.25 | 0.2 | 2 | 0.5 | 64 | 64 | >128 |
| B2 | 1 | M | 1 | 2B1(IVa) | b | − | − | SD2:176-3 | 91 | 1-26-28-18-18-54-50 | 91 | >128 | 16 | 0.13 | 0.5 | 4 | 0.5 | 64 | >128 | >128 |
| B1 | 2 | K | 1 | 2B1(IVa) | b | − | − | W12 | 91 | 1-26-28-18-18-54-50 | 91 | >128 | 16 | 0.13 | 0.25 | 1 | >128 | 32 | 128 | >128 |
| E1 | 1 | S | 2 | 2A1(IIa) | − | + | − | SG144-1 | 5 | 1-4-1-4-12-1-10 | 5 | >128 | >128 | 64 | 64 | >128 | >128 | 64 | 128 | >128 |
| F1 | 1 | M | 2 | 2A1(IIa) | − | + | − | SD183-1 | 5 | 1-4-1-4-12-1-10 | 5 | >128 | >128 | 32 | 64 | 64 | >128 | 32 | >128 | >128 |
| G2 | 1 | M | 2 | 2A1(IIa) | − | + | − | SD2:083-2 | 5 | 1-4-1-4-12-1-10 | 5 | >128 | >128 | 32 | 0.5 | >128 | >128 | 2 | >128 | >128 |
| H1 | 1 | K | 2 | 2A1(IIa) | − | + | − | W143 | 5 | 1-4-1-4-12-1-10 | 5 | >128 | 32 | 0.5 | 0.25 | 1 | >128 | 32 | 32 | >128 |
| H3 | 6 | M | 2 | 2A1(IIa) | − | + | − | SD370-1 | 5 | 1-4-1-4-12-1-10 | 5 | >128 | >128 | 32 | 8 | 64 | >128 | 32 | >128 | >128 |
| H2 | 1 | M | 2 | 2A1(IIa) | − | + | − | SD172-1 | 90 | 1-4-1-4-42-1-10 | 5 | >128 | >128 | 32 | 64 | 32 | >128 | 0.5 | >128 | >128 |
| G1 | 3 | S | 2 | 2Bn(IVnt) | − | − | − | SG167-2 | 5 | 1-4-1-4-12-1-10 | 5 | 128 | 2 | 0.13 | 0.5 | 2 | 128 | 0.5 | 0.25 | 4 |
| C1 | 1 | M | 3 | 2Bn(IVnt) | − | − | − | SD179-1 | 88 | 22-1-14-23-12-4-31 | 78 | >128 | 64 | 0.5 | 0.5 | 2 | >128 | 32 | 16 | >128 |
| C2 | 3 | K | 3 | 2Bn(IVnt) | a | − | − | W17 | 88 | 22-1-14-23-12-4-31 | 78 | >128 | 4 | 0.25 | 0.25 | 0.5 | >128 | 32 | 8 | >128 |
| D1 | 1 | S | 3 | 2Bn(IVnt) | − | + | − | SG208 | 8 | 3-3-1-1-4-4-3 | 8 | >128 | 8 | 0.25 | 0.5 | 1 | 0.25 | 0.25 | 128 | 128 |
| D2 | 1 | S | 3 | 2Bn(IVnt) | − | + | − | SG143-1 | 8 | 3-3-1-1-4-4-3 | 8 | >128 | 32 | 0.5 | 1 | 0.5 | 0.5 | 0.5 | >128 | >128 |
The districts where the strains were isolated are designated as follows; M, Miyagi Prefecture; K, Kyoto Prefecture; S, Saga Prefecture.
coa, coagulase type.
The strains were tested for the carriage of exotoxin genes, eta, etb, etd, tst-1, and lukS and lukF.
CZX, ceftizoxime; OXA, oxacillin; IMP, imipenem; TET, tetracycline; NOR, norfloxacin; ERY, erythromycin; GEN, gentamicin; TOB, tobramycin; KAN, kanamycin. Boldface indicates resistance.
nt, nontypeable.
All 11 strains that carried the type IIa SCCmec element produced type 2 coagulase and carried the toxic shock syndrome toxin 1 (tst-1) gene. They belonged to CC5, although their pulsotypes were not identical and they showed a multidrug resistance phenotype. The characteristic features of the strains were indistinguishable from those of the H-MRSA strains in Japan represented by strain N315 (23). On the other hand, 19 strains carrying the type IIb SCCmec element and 14 strains carrying the type IV SCCmec element differed greatly from the H-MRSA strains. The strains carrying the type IIb SCCmec element produced type 1 coagulase, and 12 of 19 strains carried exfoliative toxin B genes. All of them belonged to pulsotype A, and all six strains whose MLSTs were tested belonged to CC91. The strains carrying the type IV SCCmec element produced type 1 coagulase, and three of the five strains carried exfoliative toxin B genes. Although all three strains tested belonged to the same clonal complex (CC91) as the strains carrying the type IIb SCCmec element, their pulsotypes were different. They belonged to pulsotype B. On the other hand, the strains carrying the type IV SCCmec element of unknown subtype produced type 2 or type 3 coagulase. Two of them carried the tst-1 gene, and the other three carried the exfoliative toxin A gene. Their chromosomal genetic backgrounds were judged to be divergent; at the least they covered three pulsotypes, pulsotypes C, D, and G, and three clonal complexes, CC5, CC8, and CC78.
The oxacillin MICs of the 33 strains carrying either the type IIb or the type IV SCCmec element were rather low compared with those of type IIa strains. Furthermore, they were susceptible to imipenem, tetracycline, and norfloxacin. Those features of the MRSA strains isolated from healthy children differed greatly from those of the highly methicillin-resistant and multiple-drug-resistant MRSA strains disseminated in Japanese hospitals.
Mode of dissemination and colonization of MRSA strains in children.
We noticed that some children attending the same institutions or different institutions carried the same MRSA clone (Fig. 2). In Miyagi, SCCmec type IIb and pulsotype A1 strains were isolated from seven children in three different facilities, SCCmec type IIb and pulsotype A2 strains were isolated from seven children in three different facilities, and SCCmec type IIb and pulsotype A3 strains were isolated from two children who attended different classes (Fig. 2). Furthermore, SCCmec type IIa and pulsotype H3 strains were isolated from six children. Interestingly, four of the six children belonged to the same class in the same facility. In Saga, SCCmec type IVn and pulsotype G strains were isolated from three children in the same class; and also in Kyoto, SCCmec type IVn and pulsotype C2 strains and SCCmec type IVn and pulsotype B1 strains were isolated from three and two children in the same facility, respectively. In these cases, the data clearly showed that the dissemination of MRSA strains occurred in children attending a kindergarten or a day care center.
In the two studies conducted in Miyagi, we successfully collected consecutive nasal swab samples from 236 children. Among them were 13 children who were MRSA positive in the first survey. Of these 13 children, only 2 were positive for MRSA in the second test, and 7 MRSA-positive children were newly identified in the second survey. When we compared the pulsotypes of four MRSA strains isolated from the two children in the first and the second studies, only two MRSA strains isolated from a child (SD084-1 in the first study and SD084-15 in the second study) belonged to the same pulsotype (pulsotype A), with one band difference. It was also noted that the MRSA strain isolated from one of the two consistent carriers changed to a coagulase isotype 1-producing MRSA isolate with a type IIb SCCmec element from a coagulase isotype 2-producing MRSA isolate with a type IIa SCCmec element.
Our data suggested that the carriage of MRSA was a temporary phenomenon in most of the cases, with only a small number of persistent carrier cases.
Questionnaire survey.
Prior to the collection of nasal swab samples, the parents were asked whether their children had taken antibiotics on the test day or in the past 1 month (in Kyoto and Saga) or in the past 3 months (in Miyagi) and whether their children had seen a clinician(s). The rates of response to the questionnaires were 91.4% in Miyagi (first), 93.5% in Miyagi (second), 100% in Kyoto, and 100% in Saga. A total of 19.3% of children had seen a clinician, 9.6% of the children had taken antibiotics on the test day, 34.3% of the children (in Kyoto and Saga) had taken antibiotics in the past 1 month, and 56.1% of the children (in Miyagi) had taken antibiotics in the past 3 months. However, no significant differences in the answers to the questionnaires were observed between the carriers and the noncarriers of MRSA.
DISCUSSION
Dissemination of methicillin-resistant staphylococci (MRS) in the community.
Recent reports have indicated an increasing incidence of community-acquired MRSA infection, including increasing numbers of isolates from patients without the usual risk factors associated with MRSA infection or colonization (2, 8, 14, 26, 27, 31, 32).
In this study, we investigated the prevalence of MRS among healthy children. The prevalence of MRSA nasal carriage in children was 4.3%. This ratio was higher than those (0.2 to 2.2%) reported in previous surveys of healthy children and outpatients (14, 33, 36). Furthermore, we identified MRC-NS from 28.2% of the children in seven institutions across three districts. Although the prevalence of methicillin-resistant staphylococci among healthy children was high, no apparent correlation was found between the carriage of MRS and antibiotic use or medical examination at the individual level. However, the questionnaire survey clearly showed that the population itself was a high-risk one, in that more than 34.3% and 56.1% of the children had taken antibiotics in the last 1 month or the last 3 months before the tests, respectively. The medication seemed to have been prescribed by physicians on the occasion of the children's visits for common colds. In addition, determination of the genotypes of the MRSA strains showed that endemic strains were disseminating among children, presumably through close physical contact among the children in the same institution.
Forty-three percent of the MRSA strains carried the type IIb SCCmec element, which is rarely found among the strains in Japanese hospitals; and 32% of the MRSA strains carried the type IV SCCmec element, which is mostly found in community-acquired MRSA isolates. These data suggest that children are acquiring and disseminating MRSA clones which are not associated with hospital MRSA strains, although some minor populations do temporarily carry a typical H-MRSA strain (type IIa SCCmec and CC5). H-MRSA were seen in the first Miyagi study, and most of them disappeared in the second Miyagi study (Table 3). Colonization with MRSA did not continue for a long period; instead, it seemed to circulate among children. This is because 11 of 13 previously MRSA-positive children did not carry MRSA in the second test; instead, 7 previously MRSA-negative children newly acquired the same MRSA strains as those found among MRSA-positive children in the first test. Outbreaks of C-MRSA have been reported among some members of sport teams and in jails, where sweating and a greater chance of direct physical contact are expected (3-5, 24, 30, 38). Day care centers are the same in this regard. Children play in close physical contact, especially in the summertime, with tens of children sharing a small pool in the garden. The first Miyagi study was done during the hot season, which might have been the reason why the MRSA carriage rate was more pronounced than in the other investigations done in the colder seasons. There is a report that Japanese physicians prescribe antibiotics to children under 6 years old for as many as 90% of the cases of common colds (20). The high frequency of antibiotic use and the high chance of physical contact may be reasons why MRSA and MRC-NS prevail in Japanese day care centers and kindergartens.
Characteristic features of MRSA and MRC-NS isolates.
Until now, five types of SCCmec elements have been reported. Four subtypes of the type IV SCCmec element are known. SCCmec typing has become an important tool in MRSA epidemiology. We have reported previously that well-defined C-MRSA and nonmultiresistant oxacillin-resistant Staphylococcus aureus strains carry the type IV or the type V SCCmec element, whereas H-MRSA strains carry the type I, II, and III SCCmec elements. Enright et al. (9) have reported that the H-MRSA strains in the world belong to five major clones. In this study, we found that a type IIb SCCmec element of a new subtype was carried by 63.3% of MRSA strains in Miyagi, and 83.3% and 87.5% of MRSA strains isolated in Saga and in Kyoto carried the type IV SCCmec element. Both elements were rather short and did not carry many antibiotic resistance-conferring genes on the element, although Tn554-encoding macrolide-lincomycin-spectinomycin resistance was carried by the type IIb element. Thus, the rule for community-acquired SCCmec of a small size and carriage of smaller numbers of resistance genes still applies to the new type IIb element. Evidently, the kinds of antibiotic selective pressure for day care centers and kindergartens are limited compared to those for hospitals. Oral cephalosporins and macrolides constitute more than 90% of the antibiotics used for the treatment of Japanese children (20). Thus, only the carriage of Tn554 and the mecA gene is required for Japanese C-MRSA strains so that they can survive in the community.
Small SCCmec elements may be more prone to transfer from strain to strain, presumably via phage-associated transduction. We proposed the hypothesis based on the extreme heterogeneity of the C-MRSA genotypes (29). Here, again, we observed diversity in the MRSA strains in the community. CC91 is a novel chromosome type, and two MRSA clones of SCCmec type IIb and CC91 and SCCmec type IVa and CC91 are novel MRSA clones transformed by the introduction of the community-type SCCmec. Further study to determine nontypeable subtypes of type IV SCCmec elements is ongoing and may add to the repertoire of the highly efficient transformer SCCmec.
Highly virulent C-MRSA strains carrying PVL genes are known to prevail in the world. No PVL-positive strains were identified in the present study. However, the possibility that some C-MRSA strains carrying PVL are distributed in the Japanese community cannot be excluded, since in the early 1980s we found that many type IV SCCmec strains carry PVL genes. In fact, in 2002 we found a type IV SCCmec strain carrying PVL from an outpatient who visited our facility with a deep skin abscess (X. X. Ma, unpublished data). However, the MRSA strains found in the 1980s declined, and a new MRSA represented by strain N315 appeared in the 1990s (34). Thus, we believe that PVL-positive strains are not widely disseminated in Japan at this moment.
MRC-NS in the community.
We found that MRC-NS strains are widely disseminated in the Japanese community and investigated the characteristics of those MRC-NS strains. Most MRC-NS strains carried the type IV SCCmec element (K. Kuwahara-Arai, unpublished data). The data might suggest that MRC-NS in the community may serve as a reservoir for type IV SCCmec elements, which are widely disseminated among the C-MRSA strains in the world. A larger number of surveys in the community is warranted to explore from where and how the SCCmec element enters the S. aureus chromosome.
Acknowledgments
We thank Zetty Maztura, T. J. Tengku, and Bruce E. Allen for their kind help with the preparation of the manuscript.
This work was supported by a Grant-in-Aid for 21st Century COE Research and a Grant-in-Aid for Scientific Research on Priority Areas (grant 13226114) from the Ministry of Education, Science, Sports, Culture and Technology of Japan.
REFERENCES
- 1.Centers for Disease Control. 1981. Community-acquired methicillin-resistant Staphylococcus aureus infections—Michigan. Morb. Mortal. Wkly. Rep. 30:185-187. [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997-1999. JAMA 282:1123-1125. [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2003. Methicillin-resistant Staphylococcus aureus infections among competitive sports participants—Colorado, Indiana, Pennsylvania, and Los Angeles County, 2000-2003. Morb. Mortal. Wkly. Rep. 52:793-795. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2003. Methicillin-resistant Staphylococcus aureus infections in correctional facilities—Georgia, California, and Texas, 2001-2003. Morb. Mortal. Wkly. Rep. 52:992-996. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2001. Methicillin-resistant Staphylococcus aureus skin or soft tissue infections in a state prison—Mississippi, 2000. Morb. Mortal. Wkly. Rep. 50:919-922. [PubMed] [Google Scholar]
- 6.Chambers, H. F. 2001. The changing epidemiology of Staphylococcus aureus. Emerg. Infect. Dis. 7:178-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daum, R. S., T. Ito, K. Hiramatsu, F. Hussain, K. Mongkolrattanothai, M. Jamklang, and S. Boyle-Vavra. 2002. A novel methicillin-resistance cassette in community-acquired methicillin-resistant Staphylococcus aureus isolates of diverse genetic backgrounds. J. Infect. Dis. 186:1344-1347. [DOI] [PubMed] [Google Scholar]
- 8.Eady, E. A., and J. H. Cove. 2003. Staphylococcal resistance revisited: community-acquired methicillin resistant Staphylococcus aureus—an emerging problem for the management of skin and soft tissue infections. Curr. Opin. Infect. Dis. 16:103-124. [DOI] [PubMed] [Google Scholar]
- 9.Enright, M. C., D. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herold, B. C., L. C. Immergluck, M. C. Maranan, D. S. Lauderdale, R. E. Gaskin, S. Boyle-Vavra, C. D. Leitch, and R. S. Daum. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593-598. [DOI] [PubMed] [Google Scholar]
- 12.Hiramatsu, K., N. Kondo, and T. Ito. 1996. Genetic basis for molecular epidemiology of MRSA. J. Infect. Chemother. 2:117-129. [DOI] [PubMed] [Google Scholar]
- 13.Hiramatsu, K., K. Okuma, X. X. Ma, M. Yamamoto, S. Hori, and M. Kapi. 2002. New trends in Staphylococcus aureus infections: glycopeptide resistance in hospital and methicillin resistance in the community. Curr. Opin. Infect. Dis. 15:407-413. [DOI] [PubMed] [Google Scholar]
- 14.Hussain, F. M., S. Boyle-Vavra, and R. S. Daum. 2001. Community-acquired methicillin-resistant Staphylococcus aureus colonization in healthy children attending an outpatient pediatric clinic. Pediatr. Infect. Dis. J. 20:763-767. [DOI] [PubMed] [Google Scholar]
- 15.Ito, T., K. Okuma, X. X. Ma, H. Yuzawa, and K. Hiramatsu. 2003. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist. Update 6:41-52. [DOI] [PubMed] [Google Scholar]
- 16.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, T., Y. Katayama, and K. Hiramatsu. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 43:1449-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jevons, M. P. 1961. “Celbenin”-resistant staphylococci. Br. Med. J. 124:124-125. [Google Scholar]
- 20.Kaji, S., and K. Hayashida. 1987. Prophylactic antifacterial chemotherapy in cold syndrome. Sogorinsyo 36:2251-2252. [Google Scholar]
- 21.Katayama, Y., T. Ito, and K. Hiramatsu. 2001. Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 45:1955-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcal cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 24.Lindenmayer, J. M., S. Schoenfeld, R. O'Grady, and J. K. Carney. 1998. Methicillin-resistant Staphylococcus aureus in a high school wrestling team and the surrounding community. Arch. Intern. Med. 158:895-899. [DOI] [PubMed] [Google Scholar]
- 25.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maguire, G. P., A. D. Arthur, P. J. Boustead, B. Dwyer, and B. J. Currie. 1998. Clinical experience and outcomes of community-acquired and nosocomial methicillin-resistant Staphylococcus aureus in a northern Australian hospital. J. Hosp. Infect. 38:273-281. [DOI] [PubMed] [Google Scholar]
- 27.Naimi, T. S., K. H. LeDell, K. Como-Sabetti, S. M. Borchardt, D. J. Boxrud, J. Etienne, S. K. Johnson, F. Vandenesch, S. Fridkin, C. O'Boyle, R. N. Danila, and R. Lynfield. 2003. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 290:2976-2984. [DOI] [PubMed] [Google Scholar]
- 28.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for growth aerobically, 5th ed. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 29.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan, E. S., B. A. Diep, H. A. Carleton, E. D. Charlebois, G. F. Sensabaugh, B. L. Haller, and F. Perdreau-Remington. 2003. Increasing prevalence of methicillin-resistant Staphylococcus aureus infection in California jails. Clin. Infect. Dis. 37:1384-1388. [DOI] [PubMed] [Google Scholar]
- 31.Salgado, C. D., B. M. Farr, and D. P. Calfee. 2003. Community-acquired methicillin-resistant Staphylococcus aureus: a meta-analysis of prevalence and risk factors. Clin. Infect. Dis. 36:131-139. [DOI] [PubMed] [Google Scholar]
- 32.Shopsin, B., B. Mathema, J. Martinez, E. Ha, M. L. Campo, A. Fierman, K. Krasinski, J. Kornblum, P. Alcabes, M. Waddington, M. Riehman, and B. N. Kreiswirth. 2000. Prevalence of methicillin-resistant and methicillin-susceptible Staphylococcus aureus in the community. J. Infect. Dis. 182:359-362. [DOI] [PubMed] [Google Scholar]
- 33.Suggs, A. H., M. C. Maranan, A. Boyle-Vavra, and R. S. Daum. 1999. Methicillin-resistant and borderline methicillin-resistant asymptomatic Staphylococcus aureus colonization in children without identifiable risk factors. Pediatr. Infect. Dis. J. 18:410-414. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka, T., K. Okuzumi, A. Iwamoto, and K. Hiramatsu. 1995. A retrospective study on methicillin-resistant Staphylococcus aureus clinical strains in Tokyo University Hospital. J. Infect. Chemother. 1:40-49. [Google Scholar]
- 35.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murry, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torano, G., D. Quinones, I. Hernandez, T. Hernandez, I. Tamargo, and S. Borroto. 2001. Nasal carriers of methicillin-resistant Staphylococcus aureus among Cuban children attending day-care centers. Enferm. Infecc. Microbiol. Clin. 19:367-370. (In Spanish.) [DOI] [PubMed] [Google Scholar]
- 37.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, J., S. Barth, M. Richardson, K. Corson, and J. Mader. 2003. An outbreak of methicillin-resistant Staphylococcus aureus cutaneous infection in a saturation diving facility. Undersea Hyperb. Med. 30:277-284. [PubMed] [Google Scholar]
- 39.Yamaguchi, T., T. Hayashi, H. Takami, K. Nakasone, M. Ohnishi, K. Nakayama, S. Yamada, H. Komatsuzawa, and M. Sugai. 2000. Phage conversion of exfoliative toxin A production in Staphylococcus aureus. Mol. Microbiol. 38:694-705. [DOI] [PubMed] [Google Scholar]
- 40.Yamaguchi, T., T. Hayashi, H. Takami, M. Ohnishi, T. Murata, K. Nakayama, K. Asakawa, M. Ohara, H. Komatsuzawa, and M. Sugai. 2001. Complete nucleotide sequence of a Staphylococcus aureus exfoliative toxin B plasmid and identification of a novel ADP-ribosyltransferase, EDIN-C. Infect. Immun. 69:7760-7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamaguchi, T., K. Nishifuji, M. Sasaki, Y. Fudaba, M. Aepfelbacher, T. Takata, M. Ohara, H. Komatsuzawa, M. Amagai, and M. Sugai. 2002. Identification of the Staphylococcus aureus etd pathogenicity island which encodes a novel exfoliative toxin, ETD, and EDIN-B. Infect. Immun. 70:5835-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]