Abstract
Hendra virus (HeV) and Nipah virus (NiV) belong to the genus Henipavirus of the family Paramyxoviridae and are unique in that they exhibit a broad species tropism and cause fatal disease in both animals and humans. They infect cells through a pH-independent membrane fusion process mediated by their fusion and attachment glycoproteins. Previously, we demonstrated identical cell fusion tropisms for HeV and NiV and the protease-sensitive nature of their unknown cell receptor and identified a human cell line (HeLa-USU) that was nonpermissive for fusion and virus infection. Here, a microarray analysis was performed on the HeLa-USU cells, permissive HeLa-CCL2 cells, and two other permissive human cell lines. From this analysis, we identified a list of genes encoding known and predicted plasma membrane surface-expressed proteins that were highly expressed in all permissive cells and absent from the HeLa-USU cells and rank-ordered them based on their relative levels. Available expression vectors containing the first 10 genes were obtained and individually transfected into HeLa-USU cells. One clone, encoding human ephrin-B2 (EFNB2), was found capable of rendering HeLa-USU cells permissive for HeV- and NiV-mediated cell fusion as well as infection by live virus. A soluble recombinant EFNB2 could potently block fusion and infection and bind soluble recombinant HeV and NiV attachment glycoproteins with high affinity. Together, these data indicate that EFNB2 serves as a functional receptor for both HeV and NiV. The highly conserved nature of EFNB2 in humans and animals is consistent with the broad tropism exhibited by these emerging zoonotic viruses.
The broad species tropisms and the ability to cause fatal disease in both animals and humans have distinguished Hendra virus (HeV) and Nipah virus (NiV) from all other known paramyxoviruses (1). These viruses can be amplified and cause disease in large animals and be transmitted to humans where infection is manifested as a severe respiratory illness and/or febrile encephalitis, and they are now classified as biological safety level-4 agents. HeV appeared first in eastern Australia in 1994 and was transmitted to humans from infected horses; NiV first appeared in 1998-1999 in peninsular Malaysia and was predominantly passed from infected pigs to humans, but several other animal species also became infected (2, 3). In recent years, both HeV and NiV have continued to reemerge; there were two NiV outbreaks in early 2004 in Bangladesh and another in January of 2005, 74 human cases in all, and HeV reappeared in northern Australia in late 2004 with two cases of fatal infection in horses and one nonfatal human case (4, 5). The recent NiV outbreaks were characterized by a higher incidence of acute respiratory distress syndrome, person-to-person transmission, higher case fatality rates near 75%, and no direct link to infected livestock or domestic animals (6-9). Although ribavirin therapy may be of clinical benefit, there are presently no vaccines or approved therapeutics for NiV or HeV infection (10).
HeV and NiV possess two principal glycoproteins in their envelope membrane required for virion attachment to and fusion with the membrane of an appropriate host cell (11). Depending on the biological properties of the virus, attachment proteins are either the hemagglutinin-neuraminidase, the hemagglutinin, or, as is the case for HeV and NiV, the attachment glycoprotein (G), which lacks hemagglutinating and neuraminidase activities (12). Several paramyxovirus attachment proteins have been shown to be tetrameric, comprised of a dimer of homodimers (13-16). After receptor binding, the fusion glycoprotein (F), an oligomeric homotrimer, facilitates the fusion of virion and host cell membranes (11, 17). Among the paramyxoviruses, measles virus was the first one shown to use a cell surface protein as a receptor, and its hemagglutinin glycoprotein binds to CD46 (18-20). In addition, measles virus field isolates and vaccine strains can use signaling lymphocyte activation molecule as a receptor (CD150) (21), and signaling lymphocyte activation molecule is also used by canine distemper virus (22). Previously, we characterized the glycoproteins from HeV and NiV (23-25). These studies demonstrated a broad species tropism similar to that observed in natural (2, 3) and experimental (26-30) infections. We also provided evidence that the virus receptor was a surface-expressed protein and identified nonpermissive cell lines of human and animal origins, including a HeLa cell line derivative (HeLa-USU). In contrast, the original HeLa-CCL2 cell line from the American Type Culture Collection is permissive for HeV and NiV (23, 24).
A cytogenetic analysis of both the HeLa-USU and HeLa-CCL2 cell lines confirmed the derivation of the former from HeLa-CCL2. A microarray examination of expressed genes in the HeLa-USU, HeLa-CCL2, and two additional permissive human cell lines was performed. From this analysis a panel of candidate receptor proteins was assembled and tested for their ability to confer cell fusion permissiveness to HeLa-USU cells. A single clone encoding human ephrin-B2 (EFNB2) could render HeLa-USU cells permissive for both HeV and NiV cell fusion and virus infection. A recombinant mouse EFNB2 protein could block HeV and NiV cell fusion and live virus infection and could bind and coprecipitate G from both viruses. Taken together, these data indicate that EFNB2 serves as a receptor for both HeV and NiV. These findings will aid our understanding of the henipavirus-host relationships and provide insight into the mechanisms of host-cell infection and pathogenesis.
Experimental Procedures
Cells and Culture Conditions. HeLa-USU cells were provided by Anthony Maurelli, Uniformed Services University. HeLa-CCL2 was from the American Type Culture Collection (ATCC CCL 2). The human glioblastoma U373 cell line and the human head and neck carcinoma PCI-13 cell line have been described (15). Cells were maintained as described (see Supporting Text, which is published as supporting information on the PNAS web site).
Cytogenetic Analysis. HeLa-USU and HeLa-CCL2 cell lines were subjected to cytogenetic analysis (Applied Genetics Laboratories, Melbourne, FL). Five karyotypes from HeLa-CCL2 and HeLa-USU cells showed both lines to be of human origin. Of the four described markers characteristic of the HeLa cell line (ATCC CCL-2), four markers were present in the HeLa-CCL2 cells but only two, M1 and M2, were present in HeLa-USU cells.
Viruses. HeV and NiV F- and G-encoding vaccinia viruses have been described (23, 24). Vaccinia viruses, WR, vTF7-3 (T7 RNA polymerase), and vCB21R-LacZ (Escherichia coli lacZ linked to T7) have been described (25, 31). HeV and NiV stock viruses [titers 1 × 108 tissue culture 50% infective dose (TCID50) per ml and 3 × 107 TCID50 per ml, respectively] were prepared as described (32, 33).
Microarray Analysis. HeLa-USU, HeLa-CCL2, U373, and PCI-13 cells (2 × 106 cells) were pelleted, frozen at -80°C and prepared for microarray analysis (Microarray Core Facility, State University of New York Upstate Medical University, Syracuse). Affymetrix (Santa Clara, CA) GeneChip sample preparation and data acquisition were performed by Frank Middleton and Karen Gentile (State University of New York Upstate Medical University) (see Supporting Text). Data mining was as follows: data were initially filtered based on the detection parameters given by the software, and genes were eliminated if they were present in HeLa-USU cells or absent in HeLa-CCL2, U373, or PCI-13 cells. Next, the data were filtered based on the change in signal intensity over HeLa-USU cells, and genes were eliminated if they showed a signal intensity change of decrease, marginal decrease, or no change, and genes that were not associated with or expressed in the plasma membrane were also eliminated. A cut-off of accepted negative signals for the HeLa-USU group was three times the background expression (≈120); genes were ranked in descending order according to the signal strength of PCI-13 cells. Only proteins with a calculated molecular mass in the range of 20 to 90 kDa were included based on previous observations with virus-overlay blotting (12).
Expression Plasmids, Antibodies, and Reagents. Plasmids containing human EFNB2 (GenBank accession no. NM_004093), human transmembrane 4 superfamily member 6 (GenBank accession no. NM_003270), and human G protein-coupled receptor 160 (Gen-Bank accession no. NM_014373) were obtained from Origene (Rockville, MD). Plasmids containing human activated leukocyte cell adhesion molecule (GenBank accession no. NM_001627), and human transmembrane 6 superfamily member 1 (GenBank accession no. NM_023003) were obtained from Genecopoeia (German-town, MD). Plasmids containing human coagulation factor II (thrombin) receptor-like 1 (GenBank accession no. BC002464), human endothelial differentiation G protein-coupled receptor 2 (GenBank accession no. AY322546), human histamine receptor H1 (GenBank accession no. D28481), human endothelin receptor type A (GenBank accession no. AY275462), and human adenosine receptor A2B (GenBank accession no. NM_000676) were obtained from the University of Missouri-Rolla cDNA Resource Center. Recombinant, soluble mouse EFNB2/Fc and goat anti-mouse-EFNB2 polyclonal antibody (Pab) came from R & D Systems, and rabbit anti-human-EFNB2 was from Santa Cruz Biotechnology. The peptide NiV-FC2 has been described (23, 33).
Cell Fusion Assays. Fusion between HeV or NiV F and G-expressing cells (effector cells) and target cells was measured by reporter gene assay as described (23-25). Expression of HeV or NiV F and G, with vCB21R-LacZ (34) was performed in HeLa-USU cells. Target cells were prepared by infection with vTF7-3 (35). Candidate receptors were screened by transfection of the expression plasmids with Lipofectamine 2000 (Invitrogen) in HeLa-USU target cells. For inhibition by antibodies or soluble EFNB2/Fc, serial dilutions were made and added to effector cells 20 min before addition of target cells. Assays were performed as described (25).
Virus Infection Assays. HeLa-USU (5 × 104), HeLa-CCL2 cells (4 × 104), and Vero cells (4 × 104) were plated into eight-well Lab-Tek II chamber slides (Nalge Nunc) and incubated overnight at 37°C in 5% CO2. The HeLa-USU cells were transfected with expression plasmids for human EFNB2 or an irrelevant gene (SH of J virus) (36) for 6 h at 37°C in 5% CO2 with 0.5 μg of DNA and 0.6 μg of Lipofectamine 2000 (Invitrogen) in 50 μl of DMEM-2.5, and the media were replaced with 300 μl of DMEM-10 and incubated overnight at 37°C in 5% CO2. HeLa cells were infected with of 8 × 103 TCID50 HeV or 4 × 103 TCID50 NiV in 200 μl of DMEM 2.5. Vero cells were infected with 4 × 103 TCID50 of HeV or 2 × 103 TCID50 NiV in 200 μl of DMEM 2.5. Infections were conducted in a biological safety level-4 laboratory. For inhibition, either 20 or 2 μg of soluble EFNB2/Fc was mixed with either HeV or NiV in 200 μl in duplicate and incubated for 30 min before addition to cells. Virus-infected cells were incubated overnight and methanol-fixed. Infection was analyzed by immunofluoresence using antiserum to henipavirus phosphoprotein as described (15, 33).
Binding of EFNB2 to HeV and NiV G. For ELISA, a 96-well plate was coated with 150 ng of EFNB2/Fc or either HeV or NiV soluble G (sG) (15) in coating buffer 16 h at 4°C and washed three times with PBS/0.05% Tween-20 (PBST). All reagents were diluted in 5% skim milk/PBST; all incubations were 45 min at 37°C followed by washing. Plates were blocked with 5% skim milk/PBST. One hundred fifty nanograms of HeV or NiV sG was added to wells coated with EFNB2/Fc, followed by rabbit anti-HeV sG (15) (1:500). Two hundred nanograms of EFNB2/Fc, anti-HeV (1:250), anti-NiV (1:250), or anti-Tioman virus (1:250) antisera was added to wells coated with HeV or NiV sG. Protein G-horseradish peroxidase conjugate (Pierce) (1:5,000) was added to HeV or NiV sG-coated wells, and goat anti-rabbit-horseradish peroxidase (Amresco, Solon, OH) (1:2,000) was added to EFNB2/Fc-coated wells. ELISA was developed with a TMB substrate (Sigma). For coprecipitations, metabolically labeled myc epitope-tagged HeV and NiV G expressed by recombinant vaccinia viruses was used with 100 μCi of [35S]-met-cys/ml (Amersham Pharmacia) and cells were lysed as described (15, 24). Lysates were mixed with 10 μg of EFNB2/Fc or 25 μl of a 2 mg/ml 9E10 mAb ascite (15) and incubated for 2 h at 25°C. Complexes were precipitated with Protein G-Sepharose and washed twice with lysis buffer and once with 0.1% sodium deoxycholate and 0.1% SDS-containing lysis buffer. Proteins were separated by 10% SDS/PAGE and visualized by autoradiography. The EFNB2/Fc-G interaction was analyzed by surface plasmon resonance technology using a BIACORE 1000 with purified proteins (Biacore, Piscataway, NJ) as described (37). sG was covalently immobilized onto a sensor chip (CM5) by using carbodiamide coupling. A control reference surface was prepared for nonspecific binding and refractive index changes. For kinetics of interaction, concentrations of EFNB2/Fc (150, 50, 25, 10, 5, and 1 nM) were injected at a flow rate of 30 μl/min in running buffer (10 mM phosphate/150 mM NaCl/3 mM EDTA/0.005% P-20, pH 7.4), and regeneration of the chip was performed by using 15 μl of 10 mM glycine·HCl, pH 2.0. Specific binding was obtained by subtracting the background signal from the reference surface, and kinetic parameters were determined by using biaevaluation 3.2 software.
Results
Microarray Analysis of Permissive and Nonpermissive Human Cell Lines. Previously, we identified human and nonhuman cell lines that were nonpermissive for NiV- and HeV-mediated fusion, while the majority of cell types and cell lines were fusogenic to varying levels (15, 23, 24). We performed a microarray analysis on mRNA from four human cell lines to rapidly screen for genes that were differentially expressed in fusogenic versus nonfusogenic cells. Using HeLa-USU cells as the nonfusogenic cell control, we compared the signals of >55,000 human mRNA sequences in this cell line with signals from three different fusogenic cell lines, HeLa-CCL2, PCI-13, and U373, by using an Affymetrix human GeneChip microarray. The data were then analyzed for resident plasma membrane proteins expressed on all of the fusogenic cells, but absent in the HeLa-USU cells; and only those proteins with calculated molecular masses within the range of 20 to 90 kDa were tabulated based on prior speculations (12). A group of 21 candidate receptor-encoding genes was compiled (Table 1, which is published as supporting information on the PNAS web site). Ten genes that were expressed at elevated levels in the most fusogenic PCI-13 cells (15) were obtained from commercial sources and tested for their ability to rescue cell fusion when transfected into the HeLa-USU cells.
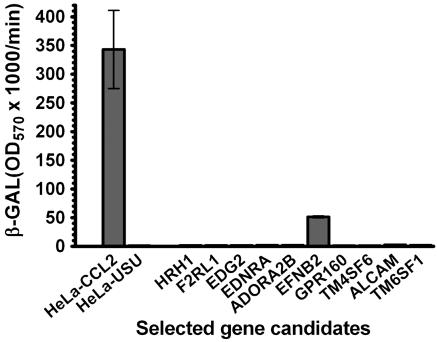
EFNB2 Expression Confers HeV and NiV Fusion Permissiveness. Ten potential receptor-encoding plasmids were used to transfect HeLa-USU cells, each assigned to a separate well in two six-well plates. HeLa-CCL2 cells were used as a positive control for fusion. Because of their greater fusogenic nature only effector cells expressing HeV F and G were chosen for the initial screen. When the target cells were used in the cell fusion assay, only cells transfected with the EFNB2 plasmid (human EFNB2) were rendered fusion competent (Fig. 1).
Fig. 1.
EFBN2 expression confers HeV fusion permissiveness to HeLa-USU cells. HeLa-CCL2 cells were used as the positive control, and mock-transfected HeLa-USU cells were used as the negative control for cell fusion. Each plasmid (1 μg) was transfected into a single well containing 106 HeLa-USU cells. The cell fusion assay was carried out as detailed in Experimental Procedures. Fusion was obtained only with the EFNB2 gene (human EFNB2). The transfection efficiency was ≈10%, monitored by GFP expression.
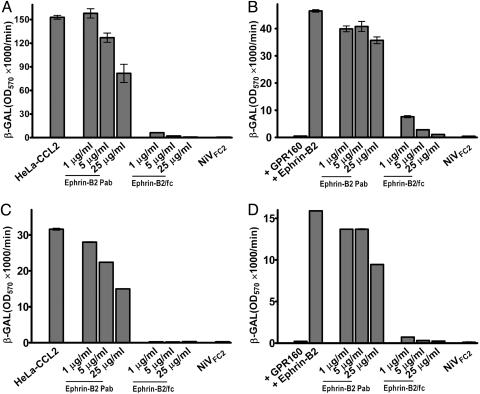
To confirm EFNB2 as a receptor for HeV and to determine whether it also serves as a receptor for NiV, a mouse-derived EFNB2/Fc protein and goat anti-mouse EFNB2 Pab were tested for their ability to block fusion. The ectodomain of murine and human EFNB2 share 98% amino acid homology. HeV and NiV cell fusion reactions, using HeLa-CCL2 cells and HeLa-USU cells transfected to express human EFNB2 as target cells, were conducted with and without the addition of 1, 5, or 25 μg/ml EFNB2/Fc or anti-EFNB2 Pab. As expected, human EFNB2 transfection also rendered the HeLa-USU cells permissive for NiV cell fusion (Fig. 2). EFNB2/Fc could completely inhibit cell fusion with the human EFNB2-transfected HeLa-USU cells and the HeLa-CCL2 cells (Fig. 2). The anti-mouse-EFNB2 Pab exhibited only low inhibitory activity at 25 μg/ml, suggesting that it did not contain significant levels of cross-reactive antibodies capable of blocking the human EFNB2 and G interaction. We also tested another rabbit anti-human-EFNB2 antisera raised against a peptide, and it showed only marginally better inhibitory activity (Fig. 6, which is published as supporting information on the PNAS web site). Normal rabbit sera had no inhibitory effect (data not shown) (23, 24). The specificity of the fusion reactions was confirmed by blocking with a peptide fusion inhibitor (NiV-FC2), corresponding to the HR-2 of NiV F, which also blocks NiV and HeV infection (Fig. 2) (15, 24, 33).
Fig. 2.
Soluble EFNB2 blocks HeV and NiV fusion in human EFNB2-transfected HeLa-USU cells and HeLa-CCL2 cells. The cells were prepared for fusion as described in Experimental Procedures. (A) HeV-mediated cell fusion with HeLa-CCL2 cells in the presence of anti-EFNB2 Pab or EFNB2/Fc. (B) HeV-mediated cell fusion with HeLa-USU cells expressing human EFNB2 in the presence of anti-EFNB2 Pab or EFNB2/Fc. (C) NiV-mediated cell fusion with HeLa-CCL2 cells in the presence of anti-EFNB2 Pab or EFNB2/Fc. (D) NiV-mediated cell fusion with HeLa-USU cells expressing human EFNB2 in the presence of anti-EFNB2 Pab or EFNB2/Fc. NiV-FC2 (1 μM) was used as a specific inhibitor for NiV- and HeV-mediated fusion in all instances. HeLa-USU cells transfected with G protein-coupled receptor 160 (GPR160) were used as the negative control in B and D.
EFNB2 Interacts with HeV and NiV G. The interaction of EFNB2 with HeV and NiV G was examined by ELISA using purified, soluble, oligomeric HeV and NiV G (sG) (15). Both HeV and NiV sG and EFNB2/Fc could be specifically and reciprocally captured by ELISA (Fig. 3 A and B). To confirm the interaction of EFNB2 with G, lysates containing metabolically labeled HeV and NiV G were prepared and subjected to coprecipitation with EFNB2/Fc. Both Gs were efficiently coprecipitated by EFNB2/Fc (Fig. 3C). No other coprecipitated proteins were observed when EFNB2/Fc was used with a control lysate (WR) and EFNB2/Fc did not coprecipitate HeV and NiV F (data not shown). Binding of the EFNB2/Fc and G was also measured by Biacore surface plasmon resonance. Under the conditions used here the binding was very strong with an equilibrium dissociation constant equal to 1.0 nM and with association and dissociation rate constants typical for high-affinity protein-protein interactions: 1.3 × 105 M-1·s-1 and 1.4 × 10-4 s-1, respectively (Fig. 3D). We also found that the EFNB2/Fc-G interaction could be inhibited by two human mAbs (m101 and m102) we recently identified as potent neutralizers of HeV and NiV (Z. Zhu, A.S.D., K.N.B., G.C., K.A.B., V.C., B.A.M., Y. R. Feng, A. Choudhary, M. Y. Zhang, Y. Feng, L.-F.W., X. Xiao, B. T. Eaton, C.C.B., and D.S.D., unpublished work), confirming the specificity of EFNB2 binding to G (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 3.
Binding of EFNB2 to HeV and NiV G. Two ELISA formats were conducted as detailed in Experimental Procedures. (A) EFNB2/Fc was used as the capture antigen for HeV and NiV sG, detected with anti-sG antisera. (B) HeV and NiV sG were used as capture antigens and detected with either anti-HeV or anti-NiV antisera or EFNB2/Fc. Anti-Tioman virus (TIV) was used as negative control. (C) HeV and NiV coprecipitations. Cell lysates containing myc-tagged metabolically labeled G were precipitated with 9E10 mAb (lanes 1-3) or EFNB2/Fc (lanes 4-6) and protein G (15). Precipitates were separated by SDS/PAGE, and visualized by autoradiography. WR, control vaccimia virus. (D) Interaction between EFNB2/Fc and sG by surface plasmon resonance using a BIACORE 1000. Two surfaces where sG was associated at two different concentrations in two independent experiments were used for test of reproducibility. In all of the experiments EFNB2 was stripped with 10 mM glycine·HCl (pH 2.0), and regeneration did not affect its binding at the same concentration in the next cycle. The arrow indicates the beginning of the dissociation phase.
EFNB2 Expression Confers Susceptibility to HeV and NiV Infection. Viral glycoprotein-mediated cell fusion assays are well known to be a valid surrogate assay for virus infection (38) and are ideal for studying biological safety level-4 agents (25). Nevertheless, confirmation of the use of EFNB2 by infectious HeV and NiV was required. Here, live HeV and NiV infection assays were conducted with Vero cells, which are the standard for assaying henipaviruses and virus propagation, and with human EFNB2-transfected HeLa-USU cells and HeLa-CCL2 cells. In the first experiment, EFNB2/Fc was used to block the infection of Vero cells by HeV and NiV with an assay that relied on immunological detection of henipavirus phosphoprotein in productively infected cells (15, 33, 39). As shown in Fig. 4, for both HeV and NiV, 100 μg/ml of EFNB2/Fc was capable of completely preventing infection, and at 10 μg/ml infection foci were barely detectable. Next, infection assays with both viruses were conducted with human EFNB2-transfected HeLa-USU cells and HeLa-CCL2 cells (Fig. 5). The transfection of human EFNB2 into the HeLa-USU cells conferred infection by both HeV and NiV, and this infection was completely blocked by EFNB2/Fc. Transfection of an irrelevant gene (SH of J virus) had no effect. Infection of HeLa-CCL2 cells is shown for comparison.
Fig. 4.
Inhibition of HeV (Left) and NiV (Right) by EFNB2/Fc. Vero cells were infected with either HeV or NiV as described in Experimental Procedures. For inhibition, HeV and NiV were preincubated with either 100 or 10 μg/ml EFNB2/Fc for 30 min before addition to the cells. All infections were incubated for 24 h, fixed, and immunofluorescently stained for phosphoprotein before digital microscopy. Images were obtained at an original magnification of ×40.
Fig. 5.
HeV (Right) and NiV (Left) infection of EFNB2-transfected HeLa-USU cells. (A) HeLa-USU cells transfected and expressing an irrelevant gene (SH of J virus). (B) HeLa-USU cells expressing human EFNB2. (C) HeLa-USU cells expressing human EFNB2 viruses preincubated with 10 μg/ml EFNB2/Fc before infection. (D) HeLa-CCL2 cells. Infections were conducted as described in Experimental Procedures. After 24 h, cells were fixed and immunofluorescently stained for phosphoprotein before digital microscopy. Images were obtained at an original magnification of ×40.
Discussion and Conclusion
Paramyxoviruses have a class I, pH-independent, fusion mechanism and generally require an attachment glycoprotein along with F for fusion (38, 40-43), and a key feature is that receptor engagement is central to initiating the process. Whereas most well characterized paramyxoviruses use sialic acid moieties as receptors, only measles virus and canine distemper virus have been shown to use proteins as receptors: CD46 and signaling lymphocyte activation molecule (18, 19, 21, 22). Earlier studies on HeV and NiV revealed that each possessed a G (44-46), and it was speculated that like certain morbilliviruses, they also used a surface protein as a receptor. Our studies on the HeV and NiV glycoproteins demonstrated that their putative receptor was protease-sensitive and each virus possessed a broad cellular tropism (23, 24). We also identified a HeLa cell line derivative to be completely resistant in supporting HeV and NiV cell fusion (23). This HeLa-USU cell line is used as the F- and G-expressing effector cell in fusion studies, circumventing aberrant cell fusion between effector cells (15, 24, 25). Subsequently, we noted that HeLa-CCL2 cells from the American Type Culture Collection were permissive for HeV and NiV.
In the present study we conducted a comparative microarray analysis of the expressed genes present in the HeLa-USU and HeLa-CCL2 cells. We also included two additional human cell lines that were highly permissive for HeV and NiV infection (15). From an analysis of >55,000 mRNA sequences, a list of 21 candidate receptors was generated (Table 1). Among the first group of 10 cDNA clones, human EFNB2 was found to confer cell fusion permissiveness to HeLa-USU cells. EFNB2 is a member of the family of transmembrane-anchored ligands of the ephrin receptor tyrosine kinase family, the largest subgroup of receptor tyrosine kinases known (47, 48), and EFNB proteins are type I membrane glycoproteins. Collectively known as ephrin proteins, the ephrin receptors and their ephrin ligands make up an important group of bidirectional signaling molecules. The ephrin proteins are known to participate in many instances of cell-cell interactions, including those of vascular endothelial cells, and are modulators of cell migration in remodeling events, especially in the nervous system. EFNB2 expression is seen in most human tissues to varying degrees and appears highest in lung and lowest in lymphocytes (49). Indeed, we have observed that primary human peripheral blood mononuclear cells and human macrophages exhibit near-negative cell fusion activities (23, 24), which would be consistent with an absence or very low level of EFNB2 expression. Analysis of the ephrin genes in the cell lines we examined indicated only EFNB2 was absent from HeLa-USU cells and present in all three fusogenic cell lines (Table 2, which is published as supporting information on the PNAS web site). Whether other molecules unrelated to EFNB2 or its family members can serve as functional receptors for HeV or NiV remains possible, and whether EFNB2 expression renders all types of otherwise henipavirus nonpermissive cells permissive remains to be determined.
The identification of a henipavirus receptor will provide the opportunity for more detailed studies on the mechanisms underlying the paramyxovirus fusion process in general. Typically, viruses that use protein receptors to trigger their membrane fusion process display a high-affinity interaction between their glycoprotein and receptor (38). In agreement with such a mechanism, we observed that a EFNB2/Fc protein could potently inhibit fusion and infection, efficiently coprecipitate G, and interact with G with very high affinity. The use of soluble EFNB2 should also facilitate studies on the HeV and NiV F and G interaction and how their association is modulated during receptor engagement. Paramyxovirus attachment proteins are type II membrane proteins and consist of a stalk and globular head structure described as a six-bladed propeller (50) and both dimeric and/or tetrameric configurations exist (14-16, 51-54). Also, the crystal structure of an EFNB2 ectodomain has been shown to be dimeric (55). It will be of interest to determine the nature of the stoichiometry that exists between EFNB2 and G oligomers. The identification of EFNB2 as a receptor for the henipaviruses also suggests that they infect cells through areas known as lipid rafts, a recurring theme in virus entry (56-59), as the ephrin proteins are well known to be localized in these membrane microdomains (60).
Serological surveillance and virus isolation studies have indicated that HeV and NiV reside naturally in several species of flying foxes in the genus Pteropus (61). In light of the highly conserved nature of EFNB2 seen between the mouse and human proteins, we speculate that significant homology will also exist in the EFNB2 homologue from flying foxes. It will be of interest to determine whether EFNB2 serves as the virus receptor in these natural animal hosts. Other questions that now arise are why HeV and NiV use this receptor and whether it plays a role in pathogenesis. Certain relationships seem to be in place; both viruses can cause systemic infections and can infect the lung where EFNB2 is highly expressed, causing severe respiratory disease; both can also invade the central nervous system where EFNB2 expression is observed. Human NiV infection in particular is a systemic endothelial infection where again EFNB2 expression is observed. The identification of a receptor used by NiV and HeV will help facilitate vaccine and therapeutic strategies to treat infection as well as aid our understanding of their pathogenic features in human and animal hosts.
Supplementary Material
Acknowledgments
We thank Dr. Frank Middleton and Karen Gentile for their help and consultation in the microarray analysis; Yan-Ru Feng (Uniformed Services University) and Lemin Wang (Uniformed Services University) for viruses and cells; Jennifer McEachern for assistance in the biological safety level-4 laboratory at the Australian Animal Health Laboratory; Zhongyu Zhu (National Cancer Institute) for providing Fab m101 and Fab m102; and Dimple Khetawat, Jared R. Patch, and Xiaodong Xiao for helpful discussions. This study was supported by the National Institute of Allergy and Infectious Diseases Intramural Biodefense Program (D.S.D.) and National Institutes of Health Grant AI057168 (to C.C.B.).
Abbreviations: EFNB2, ephrin-B2; F, fusion glycoprotein; G, attachment glycoprotein; sG, soluble G; Pab, polyclonal antibody; HeV, Hendra virus; NiV, Nipah virus; TCID50, tissue culture 50% infective dose.
References
- 1.Eaton, B. T. (2001) Microbes Infect. 3, 277-278. [DOI] [PubMed] [Google Scholar]
- 2.Chua, K. B. (2003) J. Clin. Virol. 26, 265-275. [DOI] [PubMed] [Google Scholar]
- 3.Tan, C. T. & Wong, K. T. (2003) Ann. Acad. Med. Singapore 32, 112-117. [PubMed] [Google Scholar]
- 4.Anonymous (2005) Commun. Dis. Rep. Wkly. 15, 6. [Google Scholar]
- 5.Anonymous (2004) ProMed-mail, the International Society for Infectious Disease (December 14, 2004, archive no. 20041214.3307). Available at www.promedmail.org. Accessed December 17, 2004.
- 6.Enserink, M. (2004) Science 303, 1121. [DOI] [PubMed] [Google Scholar]
- 7.Anonymous (2004) Wkly. Epidemiol. Rec. 79, 168-171.15132054 [Google Scholar]
- 8.Butler, D. (2004) Nature 429, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anonymous (2004) Health Sci. Bull. 2, 5-9. [Google Scholar]
- 10.Chong, H. T., Kamarulzaman, A., Tan, C. T., Goh, K. J., Thayaparan, T., Kunjapan, S. R., Chew, N. K., Chua, K. B. & Lam, S. K. (2001) Ann. Neurol. 49, 810-813. [DOI] [PubMed] [Google Scholar]
- 11.Lamb, R. A. & Kolakofsky, D. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott-Williams & Wilkins, Philadelphia), pp. 1305-1340.
- 12.Eaton, B. T., Wright, P. J., Wang, L. F., Sergeyev, O., Michalski, W. P., Bossart, K. N. & Broder, C. C. (2004) Arch. Virol., Suppl., 122-31. [PubMed]
- 13.Markwell, M. A. & Fox, C. F. (1980) J. Virol. 33, 152-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrison, T. G. (1988) Virus Res. 10, 113-135. [DOI] [PubMed] [Google Scholar]
- 15.Bossart, K. N., Crameri, G., Dimitrov, A. S., Mungall, B. A., Feng, Y. R., Patch, J. R., Choudhary, A., Wang, L. F., Eaton, B. T. & Broder, C. C. (2005) J. Virol. 79, 6690-6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crennell, S., Takimoto, T., Portner, A. & Taylor, G. (2000) Nat. Struct. Biol 7, 1068-1074. [DOI] [PubMed] [Google Scholar]
- 17.Baker, K. A., Dutch, R. E., Lamb, R. A. & Jardetzky, T. S. (1999) Mol. Cell 3, 309-319. [DOI] [PubMed] [Google Scholar]
- 18.Dorig, R. E., Marcil, A., Chopra, A. & Richardson, C. D. (1993) Cell 75, 295-305. [DOI] [PubMed] [Google Scholar]
- 19.Naniche, D., Varior-Krishnan, G., Cervoni, F., Wild, T. F., Rossi, B., Rabourdin-Combe, C. & Gerlier, D. (1993) J. Virol. 67, 6025-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nussbaum, O., Broder, C. C., Moss, B., Stern, L. B., Rozenblatt, S. & Berger, E. A. (1995) J. Virol. 69, 3341-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tatsuo, H., Ono, N., Tanaka, K. & Yanagi, Y. (2000) Nature 406, 893-897. [DOI] [PubMed] [Google Scholar]
- 22.Tatsuo, H., Ono, N. & Yanagi, Y. (2001) J. Virol. 75, 5842-5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bossart, K. N., Wang, L. F., Eaton, B. T. & Broder, C. C. (2001) Virology 290, 121-135. [DOI] [PubMed] [Google Scholar]
- 24.Bossart, K. N., Wang, L. F., Flora, M. N., Chua, K. B., Lam, S. K., Eaton, B. T. & Broder, C. C. (2002) J. Virol. 76, 11186-11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bossart, K. N. & Broder, C. C. (2004) Methods Mol. Biol. 269, 309-332. [DOI] [PubMed] [Google Scholar]
- 26.Westbury, H. A., Hooper, P. T., Brouwer, S. L. & Selleck, P. W. (1996) Aust. Vet. J. 74, 132-134. [DOI] [PubMed] [Google Scholar]
- 27.Williamson, M. M., Hooper, P. T., Selleck, P. W., Gleeson, L. J., Daniels, P. W., Westbury, H. A. & Murray, P. K. (1998) Aust. Vet. J. 76, 813-818. [DOI] [PubMed] [Google Scholar]
- 28.Middleton, D. J., Westbury, H. A., Morrissy, C. J., van der Heide, B. M., Russell, G. M., Braun, M. A. & Hyatt, A. D. (2002) J. Comp. Pathol. 126, 124-136. [DOI] [PubMed] [Google Scholar]
- 29.Wong, K. T., Grosjean, I., Brisson, C., Blanquier, B., Fevre-Montange, M., Bernard, A., Loth, P., Georges-Courbot, M. C., Chevallier, M., Akaoka, H., et al. (2003) Am. J. Pathol. 163, 2127-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hooper, P. T., Westbury, H. A. & Russell, G. M. (1997) Vet. Pathol. 34, 323-329. [DOI] [PubMed] [Google Scholar]
- 31.Nussbaum, O., Broder, C. C. & Berger, E. A. (1994) J. Virol. 68, 5411-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, L. F., Michalski, W. P., Yu, M., Pritchard, L. I., Crameri, G., Shiell, B. & Eaton, B. T. (1998) J. Virol. 72, 1482-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bossart, K. N., Mungall, B. A., Crameri, G., Wang, L. F., Eaton, B. T. & Broder, C. C. (2005) Virol. J., in press. [DOI] [PMC free article] [PubMed]
- 34.Feng, Y., Broder, C. C., Kennedy, P. E. & Berger, E. A. (1996) Science 272, 872-877. [DOI] [PubMed] [Google Scholar]
- 35.Alexander, W. A., Moss, B. & Fuerst, T. R. (1992) J. Virol. 66, 2934-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jack, P. J. M., Boyle, D. B., Eaton, B. T. & Wang, L. F. (2005) J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 37.Zhang, M. Y., Xiao, X., Sidorov, I. A., Choudhry, V., Cham, F., Zhang, P. F., Bouma, P., Zwick, M., Choudhary, A., Montefiori, D. C., et al. (2004) J. Virol. 78, 9233-9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dimitrov, D. S. (2000) Cell 101, 697-702. [DOI] [PubMed] [Google Scholar]
- 39.Michalski, W. P., Crameri, G., Wang, L., Shiell, B. J. & Eaton, B. (2000) Virus Res. 69, 83-93. [DOI] [PubMed] [Google Scholar]
- 40.Weissenhorn, W., Dessen, A., Calder, L. J., Harrison, S. C., Skehel, J. J. & Wiley, D. C. (1999) Mol. Membr. Biol. 16, 3-9. [DOI] [PubMed] [Google Scholar]
- 41.Russell, C. J., Jardetzky, T. S. & Lamb, R. A. (2001) EMBO J. 20, 4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eckert, D. M. & Kim, P. S. (2001) Annu. Rev. Biochem. 70, 777-810. [DOI] [PubMed] [Google Scholar]
- 43.Lamb, R. A. (1993) Virology 197, 1-11. [DOI] [PubMed] [Google Scholar]
- 44.Murray, K., Selleck, P., Hooper, P., Hyatt, A., Gould, A., Gleeson, L., Westbury, H., Hiley, L., Selvey, L., Rodwell, B., et al. (1995) Science 268, 94-97. [DOI] [PubMed] [Google Scholar]
- 45.Yu, M., Hansson, E., Langedijk, J. P., Eaton, B. T. & Wang, L. F. (1998) Virology 251, 227-233. [DOI] [PubMed] [Google Scholar]
- 46.Harcourt, B. H., Tamin, A., Ksiazek, T. G., Rollin, P. E., Anderson, L. J., Bellini, W. J. & Rota, P. A. (2000) Virology 271, 334-349. [DOI] [PubMed] [Google Scholar]
- 47.Poliakov, A., Cotrina, M. & Wilkinson, D. G. (2004) Dev. Cell 7, 465-480. [DOI] [PubMed] [Google Scholar]
- 48.Pasquale, E. B. (2004) Nat. Neurosci. 7, 417-418. [DOI] [PubMed] [Google Scholar]
- 49.Hafner, C., Schmitz, G., Meyer, S., Bataille, F., Hau, P., Langmann, T., Dietmaier, W., Landthaler, M. & Vogt, T. (2004) Clin. Chem. 50, 490-499. [DOI] [PubMed] [Google Scholar]
- 50.Langedijk, J. P., Daus, F. J. & van Oirschot, J. T. (1997) J. Virol. 71, 6155-6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Russell, R., Paterson, R. G. & Lamb, R. A. (1994) Virology 199, 160-168. [DOI] [PubMed] [Google Scholar]
- 52.Plemper, R. K., Hammond, A. L. & Cattaneo, R. (2000) J. Virol. 74, 6485-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lawrence, M. C., Borg, N. A., Streltsov, V. A., Pilling, P. A., Epa, V. C., Varghese, J. N., McKimm-Breschkin, J. L. & Colman, P. M. (2004) J. Mol. Biol. 335, 1343-1357. [DOI] [PubMed] [Google Scholar]
- 54.Sheehan, J. P., Iorio, R. M., Syddall, R. J., Glickman, R. L. & Bratt, M. A. (1987) Virology 161, 603-606. [DOI] [PubMed] [Google Scholar]
- 55.Toth, J., Cutforth, T., Gelinas, A. D., Bethoney, K. A., Bard, J. & Harrison, C. J. (2001) Dev. Cell 1, 83-92. [DOI] [PubMed] [Google Scholar]
- 56.Nayak, D. P. & Hui, E. K. (2004) Subcell. Biochem. 37, 443-491. [DOI] [PubMed] [Google Scholar]
- 57.Rawat, S. S., Viard, M., Gallo, S. A., Rein, A., Blumenthal, R. & Puri, A. (2003) Mol. Membr. Biol. 20, 243-254. [DOI] [PubMed] [Google Scholar]
- 58.Phogat, S. K. & Dimitrov, D. S. (2003) Biochim. Biophys. Acta 1614, 85-88. [DOI] [PubMed] [Google Scholar]
- 59.Chazal, N. & Gerlier, D. (2003) Microbiol. Mol. Biol. Rev. 67, 226-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murai, K. K. & Pasquale, E. B. (2004) Neuroscientist 10, 304-314. [DOI] [PubMed] [Google Scholar]
- 61.Field, H., Young, P., Yob, J. M., Mills, J., Hall, L. & Mackenzie, J. (2001) Microbes Infect. 3, 307-314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.