Abstract
A critical part of the hepatitis B virus (HBV) life cycle is the packaging of the pregenomic RNA (pgRNA) into nucleocapsids. While this process is known to involve several viral elements, much less is known about the identities and roles of host proteins in this process. To better understand the role of host proteins, we isolated pgRNA and characterized its protein interactome in cells expressing either packaging-competent or packaging-incompetent HBV genomes. We identified over 250 host proteins preferentially associated with pgRNA from the packaging-competent version of the virus. These included proteins already known to support capsid formation, enhance viral gene expression, catalyze nucleocapsid dephosphorylation, and bind to the viral genome, demonstrating the ability of the approach to effectively reveal functionally significant host-virus interactors. Three of these host proteins, AURKA, YTHDF2, and ATR, were selected for follow-up analysis. RNA immunoprecipitation qPCR (RIP-qPCR) confirmed pgRNA-protein association in cells, and siRNA knockdown of the proteins showed decreased encapsidation efficiency. This study provides a template for the use of comparative RNA-protein interactome analysis in conjunction with virus engineering to reveal functionally significant host-virus interactions.
Keywords: RNA-protein interactions, host-pathogen interactions, hepatitis B virus, viral packaging, RNA binding proteins, viral proteomics
Introduction
Hepatitis B virus (HBV) infects over 290 million people globally and increases the risk of cirrhosis of the liver and hepatocellular carcinoma for those chronically infected.1 While there is a vaccine to protect against infection, once infected there is no cure.2 The HBV virus is an enveloped, reverse-transcribing DNA virus with a small 3.2 kilobase genome. Upon hepatocyte infection, virions fuse with the cellular membrane to reveal the nucleocapsid which delivers the relaxed-circular DNA (rcDNA) into the nucleus. The rcDNA is converted by host enzymes into the covalently closed circular DNA (cccDNA).3 The cccDNA serves as the transcription template for several subgenomic RNAs (sgRNAs), and the full-length RNA genome, pregenomic RNA (pgRNA). Both pgRNA and the sgRNAs serve as mRNAs for the translation of viral proteins. Additionally, pgRNA is the form of the genome packaged into the viral nucleocapsids.4
The specific packaging of pgRNA is an important part of the viral life cycle and involves several viral elements: the ε structure of pgRNA, the multifunctional viral polymerase (P), and the viral core protein (Cp). The ε structure is a cis-acting encapsidation element folded into a stem loop found at both the 5’ and 3’ ends of pgRNA. However, only the ε loop on the 5’ end is used for genome packaging.5, 6 The 5’ ε element interacts with P and this pgRNA-P complex is then packaged into the nucleocapsid composed of 120 Cp dimers. In the nucleocapsid, pgRNA is reverse transcribed into single stranded DNA which then serves as the template for plus strand synthesis to generate the double stranded DNA genome.7–10 Previous work has highlighted the role of host factors in the viral genome packaging process.11–13 However, further studies are needed to better understand the identities and functions these host proteins may be playing.
One strategy to further investigate the role of host proteins in viral RNA packaging is RNA-protein interaction analysis such as hybridization purification of RNA-protein complexes followed by mass spectrometry (HyPR-MS).14–16 In HyPR-MS, cells are cross-linked with formaldehyde to stabilize RNA-protein complexes, and the complexes are then specifically captured by hybridization to biotin-conjugated complementary oligonucleotides. The interacting proteins are then identified and quantified by liquid chromatography tandem mass spectrometry (LC-MS/MS). We applied this strategy to define and compare the protein interactomes of packaging-competent pgRNAs and packaging-incompetent pgRNAs. We identified over 250 host proteins specifically associated with the packaging-competent HBV pgRNA. Through RNA immunoprecipitation qPCR (RIP-qPCR), we were able to validate packaging-specific interactions with pgRNA for three host proteins, AURKA, YTHDF2, and ATR. Gene-specific siRNA knockdowns showed decreases in the encapsidation efficiency of HBV pgRNA for all three proteins. This study is the first in-depth characterization of proteins binding specifically to packaging-competent pgRNA and provides new insights into the role of host proteins in the encapsidation of the viral genome in HBV.
Materials and Methods
Molecular Clones
The HBV molecular clones used in this study were derived from the ayw HBV strain (GenBank accession no. V01460.1). The packaging-competent plasmid, pLJ144 (henceforth LJ144), contains the complete HBV genome.17–19 The packaging-incompetent plasmid pEL43 (henceforth EL43) is defective for pgRNA packaging, pEL43 encodes a deletion of the upstream segment of the epsilon (ε) sequence.5 Additionally, it is also defective in P protein expression due to a premature stop codon in the P ORF’s thirteenth codon, changing the CTG to TAG.20, 21 Although not relevant to this study that focused on viral post-transcriptional stages, neither plasmid expresses the envelope proteins due to two mutations: (i) the start codon of the S open reading frame (ORF) was changed from the ATG to ACG (T154C) and (ii) a premature termination codon TAA was introduced into the sixth codon of the S protein ORF (C169A).22 For both plasmids, pgRNA is under transcriptional control of the CMV-IE promoter.
Cell lines and DNA transfections
The human liver carcinoma cell line Huh7 was cultured in Dulbecco’s modified Eagle essential minimal medium nutrient mixture with Ham’s F-12 medium (DMEMF12; Gibco) supplemented to a final concentration of 10% with heat-inactivated fetal bovine serum (FBS) and 1% penicillin-streptomycin-L-glutamine (PSG) solution (both from Sigma). All transfections were performed using Lipofectamine 3000 (Invitrogen) according to manufacturer’s protocol. On the day of transfection, cells were 60% to 80% confluent. The total mass of DNA transfected per condition was 1μg/3.3 cm2 cell culture growth area. Transfection mixes were added directly to the cell culture medium in each well, with the transfection medium replaced with fresh, prewarmed complete medium at 6 hours post transfection. At 72 hours post transfection, cells were washed three times with PBS and cross-linked by suspending in 1% formaldehyde at room temperature for 10 min. The formaldehyde was quenched by adding 100mM Tris-HCl and incubating at room temperature for 10 minutes. Cells were then scraped off the dish into 1.5ml tubes, pelleted by centrifugation, and washed twice with PBS before being stored frozen at −80°C.
Cell lysis, hybridization, capture, and release of target RNA
The lysis, hybridization, capture, and release protocols are adapted from the HyPR-MS protocol in Henke et al., summarized here to include any changes. Each capture replicate was approximately 5×107 cells and 4 capture replicates were performed for each plasmid condition. Cells were lysed and sonicated as previously described.23 Lysates were incubated with pgRNA capture oligonucleotides (COs) and scrambled oligonucleotides (SOs) at 37°C for three hours with gentle nutation (Supplementary Table 1). Sera-Mag Streptavidin Coated Magnetic Speedbeads (Fisher Scientific) were washed twice at RT and added to the lysate. The samples were rocked for one hour before bead collection and lysate removal. Beads were then washed and resuspended in release buffer with release oligonucleotides (ROs) (Supplementary Table 1). The beads were gently rocked at RT for one hour to release RNA-protein complexes, then the supernatant was isolated and stored at 4 °C prior to RT-qPCR and mass spectrometric analyses. The release process was first done with pgRNA ROs and then scrambled ROs.
RNA extraction, reverse transcription, and qPCR analysis
A 2% aliquot of each release sample was incubated overnight at 37°C with 1mg/mL Proteinase K (Sigma), 4mM CaCl2, and 0.2% LiDS. The RNA was then extracted from each sample using TRIzol Reagent (Invitrogen) per the manufacturer’s protocol and precipitated in 75% ethanol with 1μL of GlycoBlue coprecipitant (ThermoFisher) at −20°C overnight. The RNA was pelleted by centrifugation at 20,000 g at 4°C for 15 min, washed twice with 75% ethanol, centrifuged at 20,000 g at 4°C for 15 min, then resuspended in 30μL of nuclease free water (Invitrogen). An aliquot of 10μL of the purified RNA was reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) per the manufacturer’s protocol. The 20μL reverse transcription product was diluted with 60μL of nuclease free water (Invitrogen) and analyzed using sequence-specific qPCR primers and probes (Supplementary Table 1; Integrated DNA Technologies) and Light Cycler 480 Probes Master Mix (Roche) for relative quantitation of RNA on a CFX96 Touch real-time PCR detection system (Bio-Rad).
Protein purification, trypsin digestion, and mass spectrometry analysis of peptides
Proteins from the remainder of each capture sample were purified and digested into peptides with trypsin using an adapted version of eFASP, as previously described.23, 24 For removal of salts from the sample, an OMIX C18 solid-phase extraction pipette tip (Agilent) was used according to manufacturer’s instructions. The samples were then dried in a SpeedVac and reconstituted in 95:5 H2O: acetonitrile (ACN), 0.1% formic acid. The samples were analyzed using an HPLC-ESI-MS/MS system consisting of a high-performance liquid chromatography (nanoAcquity, Waters) set in line with an electrospray ionization (ESI) Orbitrap mass spectrometer (QE-HF orbitrap, ThermoFisher). A 100μm id × 365μm od fused silica capillary micro-column packed with 20cm of 1.7μm diameter, 130 Å pore size, C18 beads (Waters BEH), and an emitter tip pulled to approximately 1μm using a laser puller (Sutter Instruments) was used for HPLC separation of peptides. Peptides were loaded on-column with 2% acetonitrile in 0.1% formic acid at a flowrate of 400nL/min for 30 min. Peptides were then eluted at a flowrate of 300nL/min over 150 min with a gradient from 2% to 30% acetonitrile, in 0.1% formic acid. Full-mass profile scans were performed in the orbitrap between 375 and 1500 m/z at a resolution of 120,000, followed by MS/MS collision induced dissociation scans of the 10 highest intensity parent ions at 35% relative collision energy and 1500 resolution, with a mass range starting at 100 m/z. Dynamic exclusion was enabled with a repeat count of one over a duration of 30 seconds.
Mass spectrometry data analysis
Mass spectral files were analyzed with the free and open-source search software program MetaMorpheus and the reviewed Swiss-Prot human XML (canonical) database.25 For HBV, the Hepatitis B virus genotype C subtype ayr (isolate Human/Japan/Okamoto/-) database was downloaded from UniProt and amino acid differences between this strain and the plasmids were made manually. Samples were searched allowing for a fragment ion mass tolerance of 20 ppm and cysteine carbamidomethylation (static) and methionine oxidation (variable). A 5% FDR for both peptides and proteins was applied. Up to two missed cleavages per peptide were allowed for protein identification and quantitation. To determine the differential protein interactomes of the RNAs, pairwise comparisons were made between protein abundances from each pgRNA interactome and to protein abundances from the scrambled control using a Student’s T-test (no multiple testing correction) in Perseus software.26 Proteins that met a p-value and fold-abundance threshold (p < 0.05, fold-change >2 or <−2) in the pairwise comparisons were then evaluated for Gene Ontology enrichment (using the human genome as the background gene set) and protein-protein interactors using STRING.27 For each capture, individual protein intensities were normalized to total protein intensity.
siRNA transfections
Approximately 1×106 cells were plated in 60mm dishes and incubated for 24 hr. All siRNA transfections were performed using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocol. On each plate, cells were transfected with 120pmol of siRNA (Supplementary Table 1). Transfection mixes were added directly to the cell culture medium in each well, with the transfection medium replaced with fresh, prewarmed complete medium at 4 hours post transfection. 48 hours after the first siRNA transfection, the cells were again transfected this time with 120pmol of siRNA, 6ug of the LJ144 plasmid, and 1.2ug of the GFP plasmid. Transfection mixes were added directly to the cell culture medium in each well, with the transfection medium replaced with fresh, prewarmed complete medium at 4 hours post transfection. 72 hours post transfection, cells were washed three times with PBS and stored at −80°C.
Capsid blot analysis
Cells were lysed with buffer composed of 0.2% NP-40, 50mM Tris-HCl, and 1mM EDTA, pH 8.0, for 15 min at 37°C. Lysed cells were centrifuged at 14000 rpm for 3min at 4°C to pellet the nuclei for removal. Cytoplasmic lysates (supernatants) containing intact capsids were adjusted to 2mM CaCl2. To digest plasmid DNA in samples, 44U of micrococcal nuclease (New England Biolabs) was added to each sample and incubated at 37°C for 2h. The capsids were then analyzed by native gel electrophoresis. Briefly, cytoplasmic lysates were electrophoresed through a 1% Tris-acetate-EDTA (TAE) gel, then transferred by passive upward capillary action in 1x TNE buffer (10mM Tris, 150mM NaCl, 1mM EDTA, pH 8.0). The gel was transferred to two membranes: a PVDF membrane, and a Hybond-N+ membrane. The PVDF membrane was immunoassayed to detect native core proteins and the Hybond-N+ membrane was probed with 32P end-labeled oligonucleotides to detect encapsidated positive strand HBV nucleic acids (genome coordinates ayw 2007- to 2498-).
RNA immunoprecipitation
Approximately 1×107 cells were lysed with 20mM Tris-HCl (pH 7.5), 100mM LiCl, 5mM Mg2Cl, 0.5% NP-40, Halt Protease Inhibitor (ThermoFisher), and RNasin Recombinant Ribonuclease Inhibitor (Promega). Cells were lysed on ice for 10 minutes with periodic vortexing followed by sonication on a Model 505 Sonic Dismembrator (Fisherbrand) for 10 sec on and 10 sec off for three cycles, keeping the samples on ice between cycles. The lysate was centrifuged (16000g at 4°C, 5 min) and the supernatant was quantified with a BCA protein assay (ThermoFisher). Between 500 and 1000ug of protein was used for each immunoprecipitation (IP) replicate and with 1% of the input used for control. For each IP, the total volume of lysate was brought up to 200μL with lysis buffer and 1ug of antibody was added to each. All IPs and input controls were incubated for 2 hours at 4°C, rocking. During this incubation, 25μl per IP of protein A magnetic beads (Pierce) were washed 3 times in lysis buffer and resuspended in the original volume. Beads were added and the tubes were incubated for another 1 hour at 4°C with rocking. After incubation, beads were washed twice with lysis buffer and resuspended. Template DNA was removed by adding 10 units of DNase I and incubating samples for 15 minutes at 37°C. Proteins were removed by adding 1mg/mL Proteinase K (Sigma) and 4mM CaCl2 and incubating for 1 hour at 37°C.
Results and Discussion
Purification of pgRNA-protein complexes from cross-linked cells
Hybridization purification of RNA-protein complexes followed by mass spectrometry (HyPR-MS) has been used to characterize the protein interactomes of viral RNAs, mRNAs, and long noncoding RNAs from a variety of cell lines and tissues.14, 15, 28, 29 We employed HyPR-MS here to isolate Hepatitis B Virus (HBV) pgRNAs and identify proteins that specifically associate with pgRNA during viral genome packaging, a critical part of the viral life cycle where host proteins are known to play a role. To do this, we isolated pgRNA from cells expressing one of two distinct viral constructs, LJ144 or EL43. LJ144 enables wildtype pgRNA packaging by expressing the epsilon (ε) sequence and the viral proteins Cp and P that function to package pgRNAs into nucleocapsids in cis to the pgRNA genome. LJ144 has been shown in previous work to serve as a reliable model for studying nucleocapsid formation and viral genome replication.20, 21 The second plasmid, EL43, is a packaging-deficient construct. EL43 expresses Cp; however, it has the ε sequence omitted and a premature stop codon that prevents P expression. This results in a pgRNA that is not competent for packaging which yields “empty” capsids formed by Cp (Figure 1A).5, 20, 21 We hypothesized that proteins that bind to pgRNA during aspects of the viral life cycle unrelated to packaging, which includes proteins mediating functions such as transcription, nuclear export, and translation, should be present in the pgRNA interactomes from both EL43 and LJ144. However, those specifically associated with pgRNA packaging (e.g., bound to epsilon or recruited by P) should be specific to the interactome of LJ144.
Figure 1. Purification of HBV pgRNA.
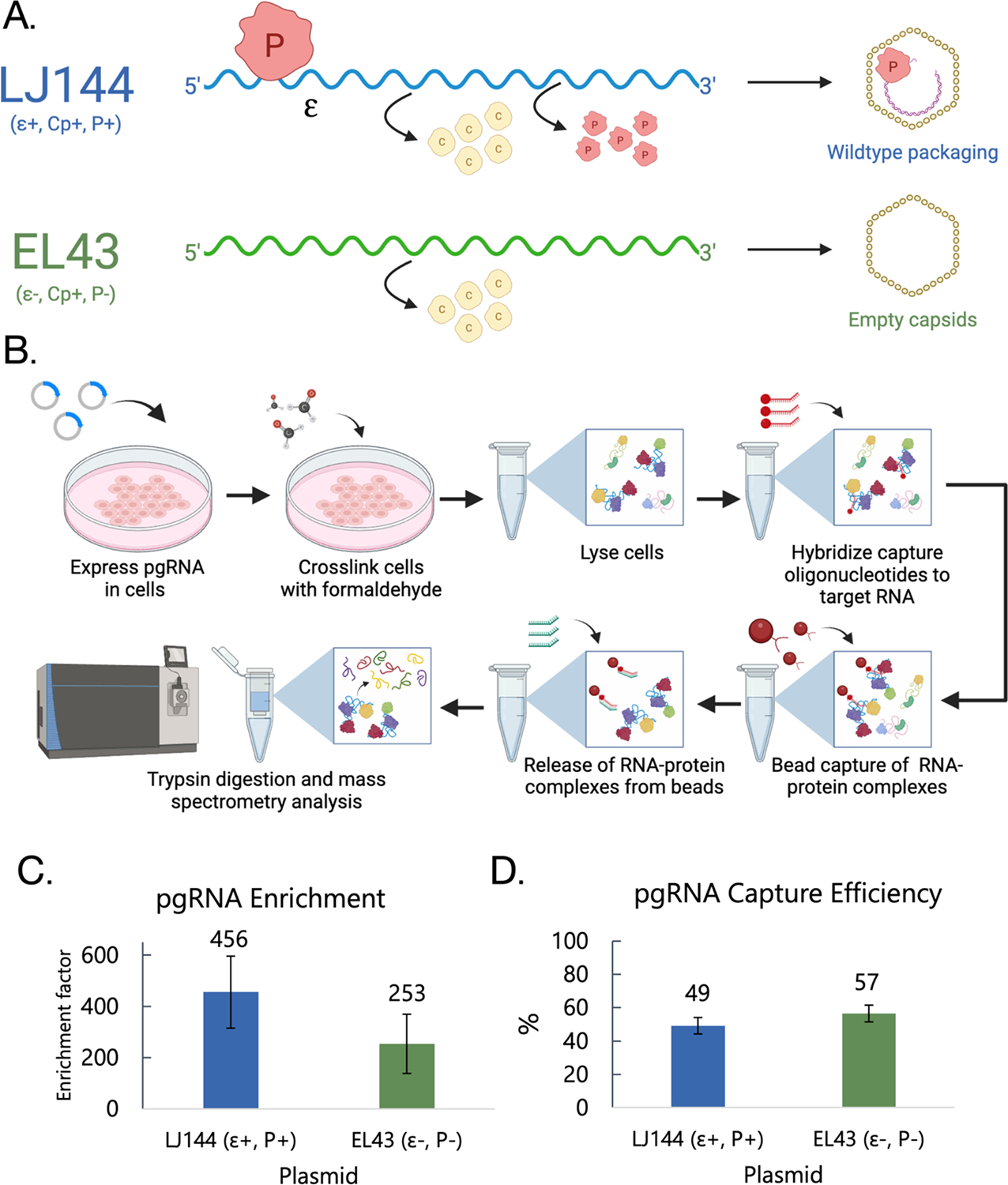
(A) Outline of HBV pgRNA constructs (LJ144 and EL43) including translated proteins and packaging phenotype. (B) Overview of HyPR-MS procedure. (C) pgRNA enrichment was calculated by ΔΔCt analysis comparing the differences in pgRNA and GAPDH Ct values in capture and input samples. (D) Capture efficiency was determined by calculating % pgRNA depletion from the input lysate. Error bars in (C) and (D) represent the standard error of the mean for the 4 capture replicates.
Specific capture of pgRNA was accomplished by using specific capture oligonucleotides (CO) designed to be complementary to a segment of the pgRNA that does not overlap with any of the viral subgenomic RNAs (Supplementary Table 1).18 Two different COs were used to maximize capture efficiency and a scrambled CO with a similar melting temperature and GC content to that of the target COs was designed to serve as a control for nonspecific protein binding. Four replicates were performed each using 5×107 Huh7 cells for each of the two viral constructs (LJ144 and EL43). Lysates were incubated with both pgRNA and scrambled COs and then released sequentially, pgRNA COs first then scrambled (Figure 1B). After RNA capture, a 2% aliquot of the sample was used to evaluate RNA capture specificity and efficiency using RT-qPCR assays specific to the unique pgRNA region (Supplementary Table 1). pgRNA capture efficiencies averaged 49% and 57% for LJ144 and EL43 transfected cells, respectively, and pgRNA was enriched by a factor of 456 and 253 compared to the scrambled control for LJ144 and EL43, respectively (Figure 1C and 1D; Supplementary Table 2). These data were consistent with previous HyPR-MS experiments and demonstrated the high degree of specificity necessary for mass spectrometry and comparative protein analysis.15, 16, 28
Characterization of the pgRNA-protein interactome
The remaining capture material was prepared for mass spectrometry analysis of proteins by detergent removal and digestion of proteins into peptides. Peptides were then analyzed using LC-MS/MS. Protein intensities for each replicate were normalized to total protein intensity to account for differences in cell pellet size, transfection efficiency, capture efficiency and sample loss (Supplementary Tables 3 and 4). We were first interested in identifying the overall protein interactome of packaging-competent pgRNA from LJ144-expressing cells. To look broadly, the pgRNA interactome was compared to the scrambled control (Figure 2A. Next, to specifically identify proteins that were involved in pgRNA packaging and maturation, we compared the LJ144 pgRNA interactome to the pgRNA interaction from EL43-expressing cells (Figure 2A). Student’s T-tests were performed to determine the proteins with significant changes between conditions. Proteins with a p-value < 0.05 and a fold-change > 2 were considered statistically significantly different (Figure 2A). Based on these criteria, 247 proteins were identified as differing significantly between LJ144 pgRNA and the scrambled control, and 268 proteins identified as differing between LJ144 and EL43 pgRNA conditions with 35 proteins common between the two (Supplementary Table 3; Figures S3, S4, S5, and S6). Comparisons to both scrambled control and EL43 pgRNA showed the viral protein P as a statistically significant interactor elevated in LJ144 pgRNA. It has been well established that P interacting with the ε structure near the 5’ end of pgRNA initiates packaging of the pgRNA-P complex into capsids and reverse transcription.7, 9 10 Therefore, the presence of P was expected and provides further evidence that the capture and analysis were successful.
Figure 2. Enriched pgRNA-associated proteins and GO analysis.
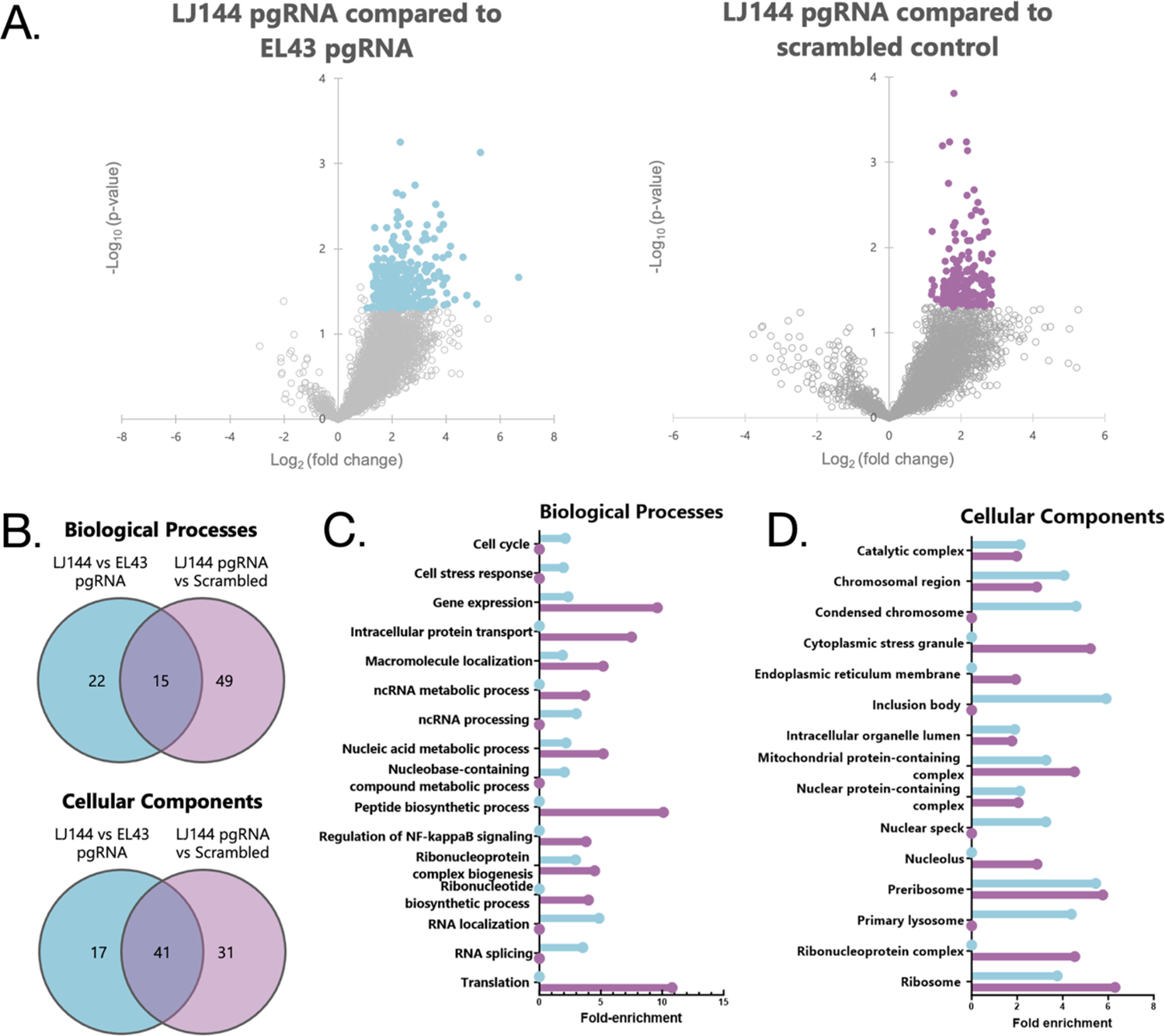
For all parts of the figure purple indicates comparison between pgRNA from LJ144-expressing cells and the scrambled control while blue indicates comparisons between pgRNA from LJ144-expressing cells and pgRNA from EL43-expressing cells. (A) Volcano plots displaying the distribution of all identified proteins with the significance level (negative log10 p-value) plotted against the relative protein abundance (log2 fold change). In both plots, a positive log2 fold change indicates a greater abundance in the pgRNA capture from LJ144 transfected cells. Significantly changing proteins (p < 0.05, fold-change >2 or <−2) are highlighted. (B) Venn diagrams displaying overlap in gene ontology (GO) analysis terms identified as statistically significant. GO enrichment analysis for (C) biological processes and (D) cellular components.
Gene ontology analysis and literature review
Gene ontology (GO) term enrichment analysis was used to evaluate pgRNA-associated proteins from LJ144-expressing cells as compared to the scrambled control and pgRNA from EL43-expressing cells. Terms were categorized as either being involved in certain biological processes or relating to specific cellular components. In general, there was substantial overlap between the enriched terms of the two interactomes. There were 15 exact matches of the 86 biological processes terms and 41 of the 89 cellular components terms (Figure 2B; Supplementary Table 4). There were also many terms that were not identical but were a part of the same broad categories (ribonucleoprotein complexes, RNA processing, etc.) Major differences included translation, NF-κB signaling, and cytoplasmic stress granules which were only found in the scrambled control comparison, and RNA localization, which was only found in the comparison to pgRNA from EL43 transfected cells (Figures 2C and D).
Comprehensive literature research of the proteins enriched in LJ144 pgRNA in either comparison found that over 60 of the proteins have previously been identified as effectors of the HBV life cycle (Table 1; Supplementary Table 5).30–61 Some interactors, such as HSP90AB1 and LIG1, are required for essential aspects of the viral life cycles: reverse transcription and cccDNA conversion, respectively.3, 62 Other interactors, such as HNRNPK and AURKA, are associated with increased viral replication efficiency.63, 64 We also identified antiviral factors including Z3CHAV1 and SACM1L.65, 66 Two of the proteins, EIF4E and PPP1CA, are known to be packaged into capsids.67, 68 The identification of these known important host proteins provides further support indicating an effective characterization of the pgRNA interactome.
Table 1.
Abbreviated list of proteins enriched in pgRNA from LJ144 that were previously found to interact with HBV.
| Protein Accession | Gene | Known interactions with HBV | Comparison Identified in |
|---|---|---|---|
| HOYFL3 | AICDA | Forms an RNP complex with viral RNA and viral proteins; deamidates both viral RNA and ssDNA71 | LJ144 pgRNA vs SCR |
| Q13315 | ATM | HBV replication is tightly connected to ATM signaling69 | LJ144 pgRNA vs EL43 pgRNA |
| Q13535 | ATR | HBV replication is tightly connected to ATR signaling69 | LJ144 pgRNA vs EL43 pgRNA |
| O14965 | AURKA | Enhances viral replication independent of kinase activity63 | LJ144 pgRNA vs EL43 pgRNA |
| P06730 | EIF4E | Interacts with P protein and is packaged into nucleocapsids67 | LJ144 pgRNA vs EL43 pgRNA |
| P61978 | HNRNPK | Binds to HBV genome and increases replication efficiency64 | LJ144 pgRNA vs SCR |
| P08238 | HSP90AB1 | Required for reverse transcriptase activity62; facilitates capsid formation through Cp interaction72 | LJ144 pgRNA vs SCR |
| P18858 | LIG1 | Required for minus strand repair in the conversion of rcDNA to cccDNA3 | LJ144 pgRNA vs EL43 pgRNA |
| Q9UBU9 | NXF1 | Involved in Cp and pgRNA export from nucleus73 | LJ144 pgRNA vs EL43 pgRNA |
| O15294 | OGT | Regulates HBV replication74 | LJ144 pgRNA vs EL43 pgRNA |
| P62136 | PPP1CA | Catalyzes core protein dephosphorylation and is packaged in capsids68; inhibited by HbX75 | LJ144 pgRNA vs SCR |
| Q9NTJ5 | SACM1L | Suppresses HBV replication65 | LJ144 pgRNA vs EL43 pgRNA |
| A6NHR9 | SMCHD1 | Recruited to cccDNA and induce associated histone PTMs76 | LJ144 pgRNA vs SCR |
| Q9Y5A9 | YTHDF2 | m6A reader previously identified as binding to HBV transcripts70 | Both |
| Q7Z2W4 | ZC3HAV1 | Restriction factor that degrades HBV RNA66 | LJ144 pgRNA vs SCR |
This list also served as a tool to help select proteins for follow-up analysis. Three proteins were selected for further experiments: ATR, AURKA, and YTHDF2. ATR is a serine/threonine kinase and DNA damage sensor. Knockdown of ATR is associated with reduction in HBV DNA and RNA levels while activation of ATR is associated with increased RNA transcription and higher levels of DNA both within the cell and secreted from the cell. However, the exact mechanisms of this process are unclear.69 AURKA is a cell cycle-regulated kinase, and its inhibition has been found to reduce levels of core protein, viral capsid, viral DNA, and viral RNA. This was true for both WT and catalytically dead AURKA, indicating that its effect on viral replication is independent of kinase activity.63 Similar to ATR, the specific mechanism of this effect remains to be understood. The final protein selected was YTHDF2 which specifically recognizes and binds to N6-methyladenosine (m6A) containing RNAs regulating their stability. m6A modifications have been found on the ε structure of pgRNA and have been hypothesized to regulate pgRNA packaging.11 YTHDF2 specifically has been shown to associate with HBV RNA transcripts via RNA immunoprecipitation analysis.70
RIP-qPCR validation of interactions
To orthogonally validate the interaction between pgRNA and the three proteins selected for follow-up (ATR, AURKA, and YTHDF2), the protein-centric method RIP-qPCR was used. In contrast to the HyPR-MS strategy of isolating an RNA of interest and identifying interacting proteins, in RIP-qPCR immunoaffinity was used to purify the protein of interest and associated pgRNA was detected via qPCR. The amount of pgRNA detected was compared to the amount present in an input aliquot. The pgRNA associated with the immunoprecipitated proteins was calculated as a percent of the total pgRNA. This experiment was done for both LJ144- and EL43-expressing cells and the percent capture was compared between the two as a means of confirming the specificity of the pgRNA-protein interactions. For LJ144-expressing cells, the fractions of pgRNA associated with AURKA, YTHDF2, and ATR, respectively, were 2%, 2.1%, and 3.7% (Figure 3A). In contrast, in the EL43 transfected cells, the fractions of pgRNA associated with AURKA, YTHDF2, and ATR, respectively, were 0.7%, 1%, and 1.1% of total pgRNA (Figure 3B; Supplementary Table 6). Overall, target protein associated-pgRNA levels were between two and four-fold higher in LJ144-expressing cells compared to EL43-expressing cells for all three immunoprecipitations (Figure 3C). This ratio should be independent of the antibody efficiency. While the fold-changes in RIP-qPCR were of lower magnitude than what was found in the HyPR-MS analysis (2- to 4-fold compared to 10- to 15-fold; Supplementary Table 3), the agreement between these complementary analyses provides further evidence that these interactions are specific to the packaging competent version of the virus.
Figure 3. RIP-qPCR assay results.
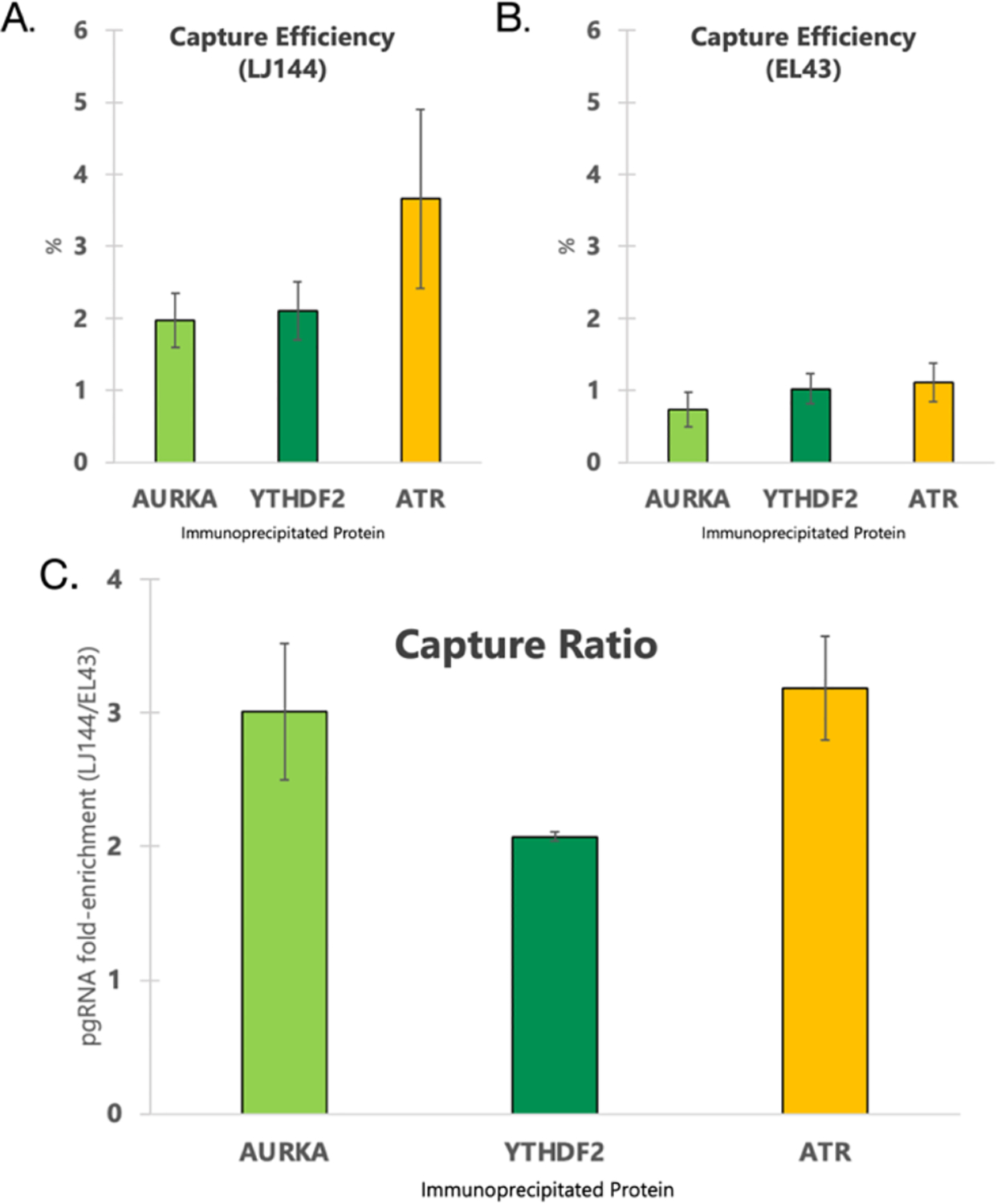
For (A) and (B) Capture efficiency was calculated by comparing the Ct of pgRNA in the immunoprecipitated sample to the Ct of pgRNA in a 1% input sample assuming 100% qPCR assay efficiency. (C) Fold-enrichment was calculated by dividing the capture efficiency of pgRNA from LJ144 transfected cells by the capture efficiency of pgRNA from EL43 transfected cells. Comparisons were made between immunoprecipitations performed side by side. Error bars for (A), (B), and (C) represent standard error of the mean.
siRNA knockdown and functional assays
To investigate possible functional roles for these proteins in the packaging of pgRNA, siRNA knockdowns of the three host proteins were done followed by analysis of encapsidation efficiency. For each target protein, two knockdowns were performed with separate siRNA assays each targeting a different region of the RNA to control for off-target effects. The knockdowns were compared to cells transfected with a nontargeting (NT) siRNA control nonhomologous to the transcriptome. Cells were transfected with the siRNA twice, co-transfecting with LJ144 plasmid the second time. After 72-hours of LJ144 plasmid transfection, cytoplasmic lysates were obtained from the cells and intact capsids were analyzed via native gel electrophoresis. The capsids were transferred to two membranes by passive upward capillary transfer where the first membrane bound proteins and the second bound nucleic acids. Bound Cp was detected by antibody binding, and bound nucleic acids were detected by hybridization with 32P-labeled probes complementary to HBV (+) strand nucleic acid (genome coordinates ayw 2007- to 2498-) (Figures 4A, S1, and S2). Encapsidation efficiency was calculated by comparing the amount of HBV nucleic acid detected to the amount of Cp detected in the blots. This ratio of HBV nucleic acid to capsids in the NT control was set to 100% encapsidation efficiency and the encapsidation efficiencies of the knockdown samples were calculated as percentages relative to the NT control. Knockdowns for each of the three proteins resulted in statistically significant reductions in encapsidation efficiency for each of the two siRNA assays with p-values ranging between .01 and .05 (Figure 4B; Supplementary Table 7). Reductions in encapsidation efficiencies ranged from 39% to 81% consistent with all three proteins playing a role in the genome packaging process. Having two independent siRNA assays for each protein both have statistically significant reductions in encapsidation efficiency clearly demonstrates a correlation even if knockouts may be incomplete. While this work provides strong evidence that these proteins are playing a role in viral genome encapsidation, further experiments are required to better elucidate their specific roles. CRISPR-mediated knockouts could provide a more robust way to evaluate the roles of the proteins upon packaging. RIP-seq analysis could also be helpful, by enabling the determination of where on the pgRNA these interactions are occurring. Additionally, the engineering of catalytically dead versions of the kinases could help to establish if the loss of packaging efficiency is related to phosphorylation, or instead to a different function of the proteins. Future experiments of this type could provide useful insight into the mechanisms involved in HBV packaging and help to determine if these RNA-binding proteins might serve as useful drug targets.
Figure 4. Effects of siRNA knockdowns on packaging efficiency.
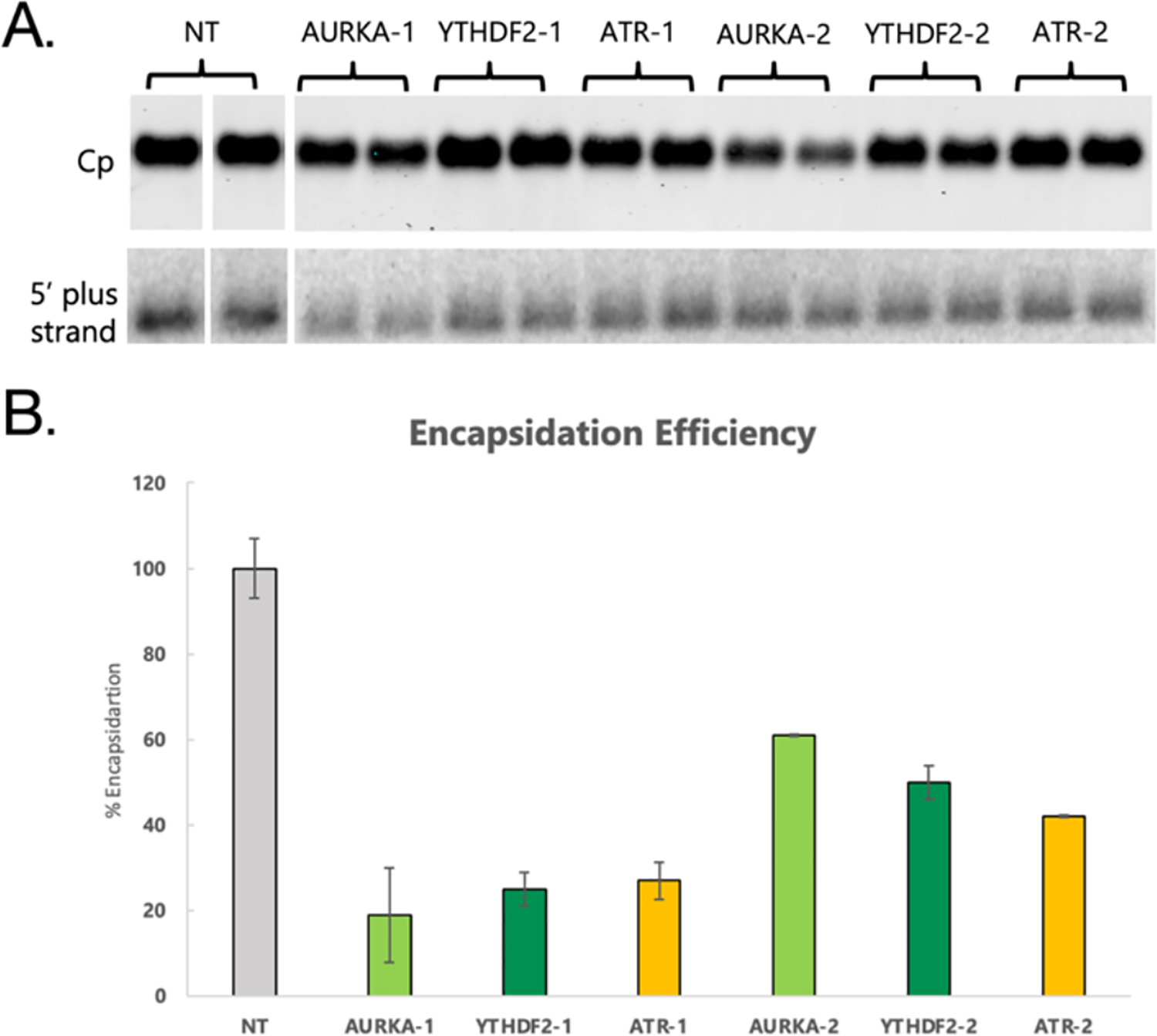
(A) Protein and nucleic acid blots from HBV capsids separated by native gel electrophoresis and transferred to membranes. Top shows detection of Cp and bottom shows detection of HBV (+) strand nucleic acid. (B) Encapsidation efficiency after siRNA knockdowns defined as the ratio of HBV nucleic acid to Cp in the capsid blots. The encapsidation efficiency of cells transfected with a NT siRNA control assay was set to 100% and all other encapsidation efficiency measurements were normalized to that value. Error bars represent standard error of the mean.
Conclusion
In this study HyPR-MS was used to identify the protein interactome of HBV pregenomic RNA (pgRNA) in cells expressing either packaging-competent or packaging-incompetent pgRNAs, to identify host factors hypothesized to be involved in the viral genome packaging process. We thoroughly characterized the protein interactomes of the two pgRNAs and used the differences observed to identify over 250 host proteins involved in pgRNA packaging. These included host factors previously shown to play important roles in the HBV viral life cycle. We selected three proteins for follow-up analysis, AURKA, ATR, and YTHDF2, which were confirmed to be packaging-specific pgRNA interactors using RIP-qPCR. siRNA knockdown of these three proteins showed statistically significant decreases in pgRNA encapsidation. These results underscore the effectiveness of our strategy at revealing functionally significant host proteins playing important roles in viral genome packaging. More broadly, the findings show the efficacy of using differential HyPR-MS to discover host-pathogen interactions occurring during specific steps of the viral life cycle. By directed engineering of selected regions of the viral genome, it is possible to dissect viral replication pathways into manageable subsystems and reveal which host proteins are important for each. This strategy is applicable to a broad range of viruses, enabling the identification of essential host proteins, providing insight into the mechanisms of pathogenesis, and revealing potential novel therapeutic targets.
Supplementary Material
Acknowledgements
The authors would like to acknowledge Dr. Daniel D. Loeb for his comments and discussions related to this project.
Funding
This study was supported by the National Institutes of Health (T32GM008349 to ITW, T32AI078985 to SR, T32HG002760 to RK, P01CA022443 to NMS, R01CA193481 to LMS).
Footnotes
Supplemental Data
Supplementary Table 1 presents all qPCR assay sequences, siRNA assay information, capture/release oligonucleotide sequences and radiolabeled probe sets. Supplementary Table 2 presents raw data from qPCR assays evaluating RNA capture. Supplementary Table 3 presents proteomics data and results from Student’s t-tests. Supplementary Table 4 presents gene ontology analysis results. Supplementary Table 5 presents a comprehensive literature research of the proteins enriched in HyPR-MS captures. Supplementary Table 6 presents raw qPCR data from RIP-qPCR experiment. Supplementary Table 7 presents raw data from encapsidation blot quantification. Supplementary File 1 contains all Supplementary Figures. Supplementary Figure 1 shows the total number of protein groups identified in each of the samples. Supplementary Figure 2 shows the median intensity of proteins before and after imputation from each sample. Supplementary Figure 3 shows a heatmap of the differentially expressed proteins and Supplementary Figure 4 shows enriched protein overlap. Supplementary Figures 5 and 6 show uncropped capsid blots and uncropped nucleic acid blots, respectively.
Data Availability
The mass spectrometry proteomics data have been deposited to MassIVE with the following identifier MSV000094286.
References
- (1).Easterbrook P; Luhmann N; Newman M; Walsh N; Lesi O; Doherty M New WHO guidance for country validation of viral hepatitis B and C elimination. Lancet Gastroenterol Hepatol 2021, 6 (10), 778–780. DOI: 10.1016/S2468-1253(21)00267-3. [DOI] [PubMed] [Google Scholar]
- (2).Pattyn J; Hendrickx G; Vorsters A; Van Damme P Hepatitis B Vaccines. J Infect Dis 2021, 224 (12 Suppl 2), S343–S351. DOI: 10.1093/infdis/jiaa668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Wei L; Ploss A Hepatitis B virus cccDNA is formed through distinct repair processes of each strand. Nat Commun 2021, 12 (1), 1591. DOI: 10.1038/s41467-021-21850-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ganem D; Varmus HE The molecular biology of the hepatitis B viruses. Annu Rev Biochem 1987, 56, 651–693. DOI: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- (5).Junker-Niepmann M; Bartenschlager R; Schaller H A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J 1990, 9 (10), 3389–3396. DOI: 10.1002/j.1460-2075.1990.tb07540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Knaus T; Nassal M The encapsidation signal on the hepatitis B virus RNA pregenome forms a stem-loop structure that is critical for its function. Nucleic Acids Res 1993, 21 (17), 3967–3975. DOI: 10.1093/nar/21.17.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hirsch RC; Lavine JE; Chang LJ; Varmus HE; Ganem D Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as wel as for reverse transcription. Nature 1990, 344 (6266), 552–555. DOI: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- (8).Porterfield JZ; Dhason MS; Loeb DD; Nassal M; Stray SJ; Zlotnick A Full-length hepatitis B virus core protein packages viral and heterologous RNA with similarly high levels of cooperativity. J Virol 2010, 84 (14), 7174–7184. DOI: 10.1128/JVI.00586-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Bartenschlager R; Schaller H Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J 1992, 11 (9), 3413–3420. DOI: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Pollack JR; Ganem D Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J Virol 1994, 68 (9), 5579–5587. DOI: 10.1128/JVI.68.9.5579-5587.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kim GW; Moon JS; Siddiqui A N6-methyladenosine modification of the 5’ epsilon structure of the HBV pregenome RNA regulates its encapsidation by the viral core protein. Proc Natl Acad Sci U S A 2022, 119 (7). DOI: 10.1073/pnas.2120485119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Hu J; Flores D; Toft D; Wang X; Nguyen D Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function. J Virol 2004, 78 (23), 13122–13131. DOI: 10.1128/JVI.78.23.13122-13131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Yao Y; Yang B; Chen Y; Wang H; Hu X; Zhou Y; Gao X; Lu M; Niu J; Wen Z; et al. RNA-Binding Motif Protein 24 (RBM24) Is Involved in Pregenomic RNA Packaging by Mediating Interaction between Hepatitis B Virus Polymerase and the Epsilon Element. J Virol 2019, 93 (6). DOI: 10.1128/JVI.02161-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Knoener R; Evans E 3rd; Becker JT; Scalf M; Benner B; Sherer NM; Smith LM Identification of host proteins differentially associated with HIV-1 RNA splice variants. Elife 2021, 10. DOI: 10.7554/eLife.62470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Knoener RA; Becker JT; Scalf M; Sherer NM; Smith LM Elucidating the in vivo interactome of HIV-1 RNA by hybridization capture and mass spectrometry. Sci Rep 2017, 7 (1), 16965. DOI: 10.1038/s41598-017-16793-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Whitworth IT; Knoener RA; Puray-Chavez M; Halfmann P; Romero S; Baddouh M; Scalf M; Kawaoka Y; Kutluay SB; Smith LM; Sherer NM Defining Distinct RNA-Protein Interactomes of SARS-CoV-2 Genomic and Subgenomic RNAs. J Proteome Res 2024, 23 (1), 149–160. DOI: 10.1021/acs.jproteome.3c00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Haines KM; Loeb DD The sequence of the RNA primer and the DNA template influence the initiation of plus-strand DNA synthesis in hepatitis B virus. J Mol Biol 2007, 370 (3), 471–480. DOI: 10.1016/j.jmb.2007.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Abraham TM; Lewellyn EB; Haines KM; Loeb DD Characterization of the contribution of spliced RNAs of hepatitis B virus to DNA synthesis in transfected cultures of Huh7 and HepG2 cells. Virology 2008, 379 (1), 30–37. DOI: 10.1016/j.virol.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Nassal M The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol 1992, 66 (7), 4107–4116. DOI: 10.1128/JVI.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Lewellyn EB; Loeb DD The arginine clusters of the carboxy-terminal domain of the core protein of hepatitis B virus make pleiotropic contributions to genome replication. J Virol 2011, 85 (3), 1298–1309. DOI: 10.1128/JVI.01957-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Lewellyn EB; Loeb DD Serine phosphoacceptor sites within the core protein of hepatitis B virus contribute to genome replication pleiotropically. PLoS One 2011, 6 (2), e17202. DOI: 10.1371/journal.pone.0017202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Liu N; Ji L; Maguire ML; Loeb DD cis-Acting sequences that contribute to the synthesis of relaxed-circular DNA of human hepatitis B virus. J Virol 2004, 78 (2), 642–649. DOI: 10.1128/jvi.78.2.642-649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Henke KB; Miller RM; Knoener RA; Scalf M; Spiniello M; Smith LM Identifying Protein Interactomes of Target RNAs Using HyPR-MS. Methods Mol Biol 2022, 2404, 219–244. DOI: 10.1007/978-1-0716-1851-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Erde J; Loo RR; Loo JA Enhanced FASP (eFASP) to increase proteome coverage and sample recovery for quantitative proteomic experiments. J Proteome Res 2014, 13 (4), 1885–1895. DOI: 10.1021/pr4010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Solntsev SK; Shortreed MR; Frey BL; Smith LM Enhanced Global Post-translational Modification Discovery with MetaMorpheus. J Proteome Res 2018, 17 (5), 1844–1851. DOI: 10.1021/acs.jproteome.7b00873. [DOI] [PubMed] [Google Scholar]
- (26).Tyanova S; Temu T; Sinitcyn P; Carlson A; Hein MY; Geiger T; Mann M; Cox J The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 2016, 13 (9), 731–740. DOI: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- (27).Szklarczyk D; Gable AL; Lyon D; Junge A; Wyder S; Huerta-Cepas J; Simonovic M; Doncheva NT; Morris JH; Bork P; et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019, 47 (D1), D607–D613. DOI: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Spiniello M; Knoener RA; Steinbrink MI; Yang B; Cesnik AJ; Buxton KE; Scalf M; Jarrard DF; Smith LM HyPR-MS for Multiplexed Discovery of MALAT1, NEAT1, and NORAD lnctiNA Protein Interactomes. J Proteome Res 2018, 17 (9), 3022–3038. DOI: 10.1021/acs.jproteome.8b00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Whitworth IT; Henke KB; Yang B; Scalf M; Frey BL; Jarrard DF; Smith LM Elucidating the RNA-Protein Interactomes of Target RNAs in Tissue. Anal Chem 2023, 95 (18), 7087–7092. DOI: 10.1021/acs.analchem.2c05635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Van Damme E; Vanhove J; Severyn B; Verschueren L; Pauwels F The Hepatitis B Virus Interactome: A Comprehensive Overview. Front Microbiol 2021, 12, 724877. DOI: 10.3389/fmicb.2021.724877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Zhang Y; Liu J; Liu H; He Y; Yi R; Niu Y; Chen T; Yang Q; Zhao Y Comparative study of the different activities of hepatitis B virus whole-X protein and HBx in hepatocarcinogenesis by proteomics and bioinformatics analysis. Arch Virol 2015, 160 (7), 1645–1656. DOI: 10.1007/s00705-015-2421-3. [DOI] [PubMed] [Google Scholar]
- (32).Li G; Ma L; He S; Luo R; Wang B; Zhang W; Song Y; Liao Z; Ke W; Xiang Q; et al. WTAP-mediated m(6)A modification of lncRNA NORAD promotes intervertebral disc degeneration. Nat Commun 2022, 13 (1), 1469. DOI: 10.1038/s41467-022-28990-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Herrscher C; Pastor F; Burlaud-Gaillard J; Dumans A; Seigneuret F; Moreau A; Patient R; Eymieux S; de Rocquigny H; Hourioux C; et al. Hepatitis B virus entry into HepG2-NTCP cells requires clathrin-mediated endocytosis. Cell Microbiol 2020, 22 (8), e13205. DOI: 10.1111/cmi.13205. [DOI] [PubMed] [Google Scholar]
- (34).Zhu C; Song H; Xu F; Yi W; Liu F; Liu X Hepatitis B virus inhibits the expression of complement C3 and C4, in vitro and in vivo. Oncol Lett 2018, 15 (5), 7459–7463. DOI: 10.3892/ol.2018.8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Nishitsuji H; Ujino S; Shimizu Y; Harada K; Zhang J; Sugiyama M; Mizokami M; Shimotohno K Novel reporter system to monitor early stages of the hepatitis B virus life cycle. Cancer Sci 2015, 106 (11), 1616–1624. DOI: 10.1111/cas.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Xu Z; Zhai L; Yi T; Gao H; Fan F; Li Y; Wang Y; Li N; Xing X; Su N; et al. Hepatitis B virus X induces inflammation and cancer in mice liver through dysregulation of cytoskeletal remodeling and lipid metabolism. Oncotarget 2016, 7 (43), 70559–70574. DOI: 10.18632/oncotarget.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Lim Z; Mohd-Ismail NKB; Png E; Sze CW; Lin Q; Hong W; Lim SG; Tan YJ; Gunaratne J Phosphoproteomics Unravel HBV Triggered Rewiring of Host Phosphosignaling Events. Int J Mol Sci 2022, 23 (9). DOI: 10.3390/ijms23095127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Chen JY; Chen WN; Liu LL; Lin WS; Jiao BY; Wu YL; Lin JY; Lin X Hepatitis B spliced protein (HBSP) generated by a spliced hepatitis B virus RNA participates in abnormality of fibrin formation and functions by binding to fibrinogen gamma chain. J Med Virol 2010, 82 (12), 2019–2026. DOI: 10.1002/jmv.21918. [DOI] [PubMed] [Google Scholar]
- (39).Minor MM; Slagle BL Hepatitis B virus HBx protein interactions with the ubiquitin proteasome system. Viruses 2014, 6 (11), 4683–4702. DOI: 10.3390/v6114683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Genera M; Quioc-Salomon B; Nourisson A; Colcombet-Cazenave B; Haouz A; Mechaly A; Matondo M; Duchateau M; Konig A; Windisch MP; et al. Molecular basis of the interaction of the human tyrosine phosphatase PTPN3 with the hepatitis B virus core protein. Sci Rep 2021, 11 (1), 944. DOI: 10.1038/s41598-020-79580-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Wei XF; Fan SY; Wang YW; Li S; Long SY; Gan CY; Li J; Sun YX; Guo L; Wang PY; et al. Identification of STAU1 as a regulator of HBV replication by TurboID-based proximity labeling. iScience 2022, 25 (6), 104416. DOI: 10.1016/j.isci.2022.104416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Taha TY; Anirudhan V; Limothai U; Loeb DD; Petukhov PA; McLachlan A Modulation of hepatitis B virus pregenomic RNA stability and splicing by histone deacetylase 5 enhances viral biosynthesis. PLoS Pathog 2020, 16 (8), e1008802. DOI: 10.1371/journal.ppat.1008802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Kann M; Sodeik B; Vlachou A; Gerlich WH; Helenius A Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J Cell Biol 1999, 145 (1), 45–55. DOI: 10.1083/jcb.145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Xi R; Kadur Lakshminarasimha Murthy P; Tung KL; Guy CD; Wan J; Li F; Wang Z; Li X; Varanko A; Rakhilin N; et al. SENP3-mediated host defense response contains HBV replication and restores protein synthesis. PLoS One 2019, 14 (1), e0209179. DOI: 10.1371/journal.pone.0209179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Kim K; Kim KH; Cheong J Hepatitis B virus X protein impairs hepatic insulin signaling through degradation of IRS1 and induction of SOCS3. PLoS One 2010, 5 (3), e8649. DOI: 10.1371/journal.pone.0008649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Chou SF; Tsai ML; Huang JY; Chang YS; Shih C The Dual Role of an ESCRT-0 Component HGS in HBV Transcription and Naked Capsid Secretion. PLoS Pathog 2015, 11 (10), e1005123. DOI: 10.1371/journal.ppat.1005123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Zhi-ming L; Yu-lian J; Zhao-lei F; Chun-xiao W; Zhen-fang D; Bing-chang Z; Yue-ran Z Polymorphisms of killer cell immunoglobulin-like receptor gene: possible association with susceptibility to or clearance of hepatitis B virus infection in Chinese Han population. Croat Med J 2007, 48 (6), 800–806. DOI: 10.3325/cmj.2007.6.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Nakanishi A; Okumura H; Hashita T; Yamashita A; Nishimura Y; Watanabe C; Kamimura S; Hayashi S; Murakami S; Ito K; et al. Ivermectin Inhibits HBV Entry into the Nucleus by Suppressing KPNA2. Viruses 2022, 14 (11). DOI: 10.3390/v14112468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Du K; Ohsaki E; Wada M; Ueda K Identification of the Interaction between Minichromosome Maintenance Proteins and the Core Protein of Hepatitis B Virus. Curr Issues Mol Biol 2023, 45 (1), 752–764. DOI: 10.3390/cimb45010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Zhang Y; Lu W; Chen X; Cao Y; Yang Z A Bioinformatic Analysis of Correlations between Polymeric Immunoglobulin Receptor (PIGR) and Liver Fibrosis Progression. Biomed Res Int 2021, 2021, 5541780. DOI: 10.1155/2021/5541780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Chavalit T; Nimsamer P; Sirivassanametha K; Anuntakarun S; Saengchoowong S; Tangkijvanich P; Payungporn S Hepatitis B Virus-Encoded MicroRNA (HBV-miR-3) Regulates Host Gene PPM1A Related to Hepatocellular Carcinoma. Microrna 2020, 9 (3), 232–239. DOI: 10.2174/2211536608666191104105334. [DOI] [PubMed] [Google Scholar]
- (52).Zeyen L; Doring T; SReler JT; Prange R Hepatitis B subviral envelope parRcles use the COPII machinery for intracellular transport via selective exploitation of Sec24A and Sec23B. Cell Microbiol 2020, 22 (6), e13181. DOI: 10.1111/cmi.13181. [DOI] [PubMed] [Google Scholar]
- (53).Miyakawa K; Jeremiah SS; Ogawa M; Nishi M; Ohnishi M; Ryo A Crosstalk between the innate immune system and selective autophagy in hepatitis B virus infection. Autophagy 2022, 18 (8), 2006–2007. DOI: 10.1080/15548627.2022.2059747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Hofmann S; Plank V; Groitl P; Skvorc N; Hofmann K; Luther J; Ko C; Zimmerman P; Bruss V; Stadler D; et al. SUMO Modification of Hepatitis B Virus Core Mediates Nuclear Entry, Promyelocytic Leukemia Nuclear Body Association, and Efficient Formation of Covalently Closed Circular DNA. MicrobiolSpectr 2023, 11 (3), e0044623. DOI: 10.1128/spectrum.00446-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Yuan S; Liao G; Zhang M; Zhu Y; Wang K; Xiao W; Jia C; Dong M; Sun N; Walch A; et al. Translatomic profiling reveals novel self-restricting virus-host interactions during HBV infection. J Hepatol 2021, 75 (1), 74–85. DOI: 10.1016/j.jhep.2021.02.009. [DOI] [PubMed] [Google Scholar]
- (56).Makokha GN; Abe-Chayama H; Chowdhury S; Hayes CN; Tsuge M; Yoshima T; Ishida Y; Zhang Y; Uchida T; Tateno C; et al. Regulation of the Hepatitis B virus replication and gene expression by the multi-functional protein TARDBP. Sci Rep 2019, 9 (1), 8462. DOI: 10.1038/s41598-019-44934-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Fan H; Zhang H; Pascuzzi PE; Andrisani O Hepatitis B virus X protein induces EpCAM expression via active DNA demethylation directed by RelA in complex with EZH2 and TET2. Oncogene 2016, 35 (6), 715–726. DOI: 10.1038/onc.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Gad SA; Sugiyama M; Tsuge M; Wakae K; Fukano K; Oshima M; Sureau C; Watanabe N; Kato T; Murayama A; et al. The kinesin KIF4 mediates HBV/HDV entry through the regulation of surface NTCP localization and can be targeted by RXR agonists in vitro. PLoS Pathog 2022, 18 (3), e1009983. DOI: 10.1371/journal.ppat.1009983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Waris G; Huh KW; Siddiqui A Mitochondrially associated hepatitis B virus X protein constitutively activates transcription factors STAT-3 and NF-kappa B via oxidative stress. Mol Cell Biol 2001, 21 (22), 7721–7730. DOI: 10.1128/MCB.21.22.7721-7730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Iwamoto M; Cai D; Sugiyama M; Suzuki R; Aizaki H; Ryo A; Ohtani N; Tanaka Y; Mizokami M; Wakita T; et al. Functional association of cellular microtubules with viral capsid assembly supports efficient hepatitis B virus replication. Sci Rep 2017, 7 (1), 10620. DOI: 10.1038/s41598-017-11015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).You H; Zhang N; Yu T; Ma L; Li Q; Wang X; Yuan D; Kong D; Liu X; Hu W; et al. Hepatitis B virus X protein promotes MAN1B1 expression by enhancing stability of GRP78 via TRIM25 to facilitate hepatocarcinogenesis. Br J Cancer 2023, 128 (6), 992–1004. DOI: 10.1038/s41416-022-02115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Hu J; Seeger C Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc Natl Acad Sci U S A 1996, 93 (3), 1060–1064. DOI: 10.1073/pnas.93.3.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Jeong GU; Ahn BY Aurora kinase A promotes hepatitis B virus replication and expression. Antiviral Res 2019, 170, 104572. DOI: 10.1016/j.antiviral.2019.104572. [DOI] [PubMed] [Google Scholar]
- (64).Ng LF; Chan M; Chan SH; Cheng PC; Leung EH; Chen WN; Ren EC Host heterogeneous ribonucleoprotein K (hnRNP K) as a potential target to suppress hepatitis B virus replication. PLoS Med 2005, 2 (7), e163. DOI: 10.1371/journal.pmed.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Zheng J; Deng Y; Wei Z; Zou H; Wen X; Cai J; Zhang S; Jia B; Lu M; Lu K; Lin Y Lipid phosphatase SAC1 suppresses hepatitis B virus replication through promoting autophagic degradation of virions. Antiviral Res 2023, 213, 105601. DOI: 10.1016/j.antiviral.2023.105601. [DOI] [PubMed] [Google Scholar]
- (66).Mao R; Nie H; Cai D; Zhang J; Liu H; Yan R; Cuconati A; Block TM; Guo JT; Guo H Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein. PLoS Pathog 2013, 9 (7), e1003494. DOI: 10.1371/journal.ppat.1003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Kim S; Wang H; Ryu WS Incorporation of eukaryotic translation initiation factor eIF4E into viral nucleocapsids via interaction with hepatitis B virus polymerase. J Virol 2010, 84 (1), 52–58. DOI: 10.1128/JVI.01232-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Hu Z; Ban H; Zheng H; Liu M; Chang J; Guo JT Protein phosphatase 1 catalyzes HBV core protein dephosphorylation and is co-packaged with viral pregenomic RNA into nucleocapsids. PLoS Pathog 2020, 16 (7), e1008669. DOI: 10.1371/journal.ppat.1008669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Kostyusheva A; Brezgin S; Bayurova E; Gordeychuk I; Isaguliants M; Goptar I; Urusov F; Nikiforova A; Volchkova E; Kostyushev D; Chulanov V ATM and ATR Expression Potentiates HBV Replication and Contributes to Reactivation of HBV Infection upon DNA Damage. Viruses 2019, 11 (11). DOI: 10.3390/v11110997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Imam H; Khan M; Gokhale NS; McIntyre ABR; Kim GW; Jang JY; Kim SJ; Mason CE; Horner SM; Siddiqui A N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc Natl Acad Sci U S A 2018, 115 (35), 8829–8834. DOI: 10.1073/pnas.1808319115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Liang G; Kitamura K; Wang Z; Liu G; Chowdhury S; Fu W; Koura M; Wakae K; Honjo T; Muramatsu M RNA editing of hepatitis B virus transcripts by activation-induced cytidine deaminase. Proc Natl Acad Sci U S A 2013, 110 (6), 2246–2251. DOI: 10.1073/pnas.1221921110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Shim HY; Quan X; Yi YS; Jung G Heat shock protein 90 facilitates formation of the HBV capsid via interacting with the HBV core protein dimers. Virology 2011, 410 (1), 161–169. DOI: 10.1016/j.virol.2010.11.005. [DOI] [PubMed] [Google Scholar]
- (73).Yang CC; Huang EY; Li HC; Su PY; Shih C Nuclear export of human hepatitis B virus core protein and pregenomic RNA depends on the cellular NXF1-p15 machinery. PLoS One 2014, 9 (10), e106683. DOI: 10.1371/journal.pone.0106683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Wang X; Lin Y; Liu S; Zhu Y; Lu K; Broering R; Lu M O-GlcNAcylation modulates HBV replication through regulating cellular autophagy at multiple levels. FASEB J 2020, 34 (11), 14473–14489. DOI: 10.1096/f.202001168RR. [DOI] [PubMed] [Google Scholar]
- (75).Cougot D; Allemand E; Riviere L; Benhenda S; Duroure K; Levillayer F; Muchardt C; Buendia MA; Neuveut C Inhibition of PP1 phosphatase activity by HBx: a mechanism for the activation of hepatitis B virus transcription. Sci Signal 2012, 5 (205), ra1. DOI: 10.1126/scisignal.2001906. [DOI] [PubMed] [Google Scholar]
- (76).Cheng J; Zhao Q; Zhou Y; Tang L; Sheraz M; Chang J; Guo JT Interferon Alpha Induces Multiple Cellular Proteins That Coordinately Suppress Hepadnaviral Covalently Closed Circular DNA Transcription. J Virol 2020, 94 (17). DOI: 10.1128/JVI.00442-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to MassIVE with the following identifier MSV000094286.


