Abstract
Crop production currently relies on the widespread use of agrochemicals to ensure food security. This practice is considered unsustainable, yet has no viable alternative at present. The plant microbiota can fulfil various functions for its host, some of which could be the basis for developing sustainable protection and fertilization strategies for plants without relying on chemicals. To harness such functions, a detailed understanding of plant‒microbe and microbe‒microbe interactions is necessary. Among interactions within the plant microbiota, those between bacteria are the most common ones; they are not only of ecological importance but also essential for maintaining the health and productivity of the host plants. This review focuses on recent literature in this field and highlights various consequences of bacteria‒bacteria interactions under different agricultural settings. In addition, the molecular and genetic backgrounds of bacteria that facilitate such interactions are emphasized. Representative examples of commonly found bacterial metabolites with bioactive properties, as well as their modes of action, are given. Integrating our understanding of various binary interactions into complex models that encompass the entire microbiota will benefit future developments in agriculture and beyond, which could be further facilitated by artificial intelligence-based technologies.
Keywords: Plant microbiome, Bacteria‒bacteria interaction, Plant pathogen, Crop production, Molecular interaction
Abstract
化学农药防治是当前农业生产中防治作物病虫害最主要的措施,对粮食安全生产起到至关重要的作用。化学农药的不合理使用会导致有害生物抗药性和环境污染等问题,因此利用有益农业微生物资源控制作物有害生物被认为是替代化学防治的可持续举措之一。植物微生物群落对宿主植物的生长发育等具有重要作用,其中部分微生物具有开发为微生物农药和肥料的潜力。全面解析微生物群落中细菌之间的相互作用及其生态功能,对合理利用微生物菌落的功能来维持植物的健康和生产力至关重要。本综述重点关注微生物群落中细菌-细菌之间互作机理及其在不同环境下对作物发育与健康的影响,并强调如何通过调控这种互作改善作物生长环境,以及小分子物质和信号调控通路在细菌-细菌相互作用中的关键作用。本文列举了具有生物活性的细菌代谢产物介导细菌-细菌互作的典型案例及其互作机制。未来需要充分利用人工智能等先进技术,将微生物群落中各种二元互作方式整合到整个微生物组的复杂模型中,以进一步了解微生物群落中各组分间的互作机理,为更好地利用农业有益微生物资源解决作物病虫害等问题提供解决方案。
Keywords: 植物微生物组, 细菌-细菌互作, 植物病原物, 农作物生产, 分子机理
1. Introduction
Plants live in close associations with diverse microbial communities, collectively termed as plant microbiota. Recent technological advances, especially the emergence of various omics-based techniques, have allowed us to better understand the implications of a range of functions of these microbiota that are essential for their host ( Berg et al., 2020). Among other roles, plant microbiomes can support the health of their hosts in various ways including the modulation of immunity, specific metabolic pathways, and direct pathogen antibiosis ( Singh et al., 2015; Chen et al., 2018; Finkel et al., 2020; Matsumoto et al., 2021; Xu et al., 2022; Zhou et al., 2022; Yang et al., 2023). A detailed understanding of mechanisms involved in the recruitment of beneficial microorganisms by plants is of outstanding importance at times of increasing environmental concerns due to the prevalence of unsustainable agricultural practices ( Munir et al., 2022). Recently, microorganisms were described that can confer holistic disease resistance to plants, which become susceptible to distinct pathogens in their absence. These have been termed as “soterobionts” and highlighted as a highly valuable resource for plant protection ( Cernava and Berg, 2022). The discovery of further such microorganisms currently poses a significant challenge, as most of the knowledge about bacteria–bacteria/fungi and plant–bacteria/fungi interaction mechanisms has been obtained through reductionist methods, such as pure cultures ( Wang and Cernava, 2023). Furthermore, it remains largely unclear which biotic or abiotic factors are determinative for the establishment of soterobionts and whether they can be supported by other microorganisms from the same community.
Specifically, bacteria–bacteria interactions are of high relevance due to the predominance of prokaryotes in most microbial communities. Different mechanisms of cell–cell interactions have been described for bacteria; however, these are mostly based on a low number of general interaction principles ( Venturi and Bez, 2021). Computational approaches facilitated by high-throughput sequencing have unraveled a high diversity of bacteria that are often found as major constituents of plant microbiomes. A recently published review by Lyng and Kovács (2023) specifically addresses bacteria–bacteria interactions between different species of Bacillus and Pseudomonas, shedding light on their interplay at the taxonomic and molecular levels. The authors highlight the manifold interactions in which these two common constituents of the plant microbiota engage. Given the overall diversity and dynamics within bacterial communities, it is becoming evident that the number of potential binary interactions therein is enormous, which cannot be captured yet holistically despite the technological advances. Here, we compile recent insights into mechanisms related to bacteria–bacteria interactions and highlight their roles in enhancing or compromising plant health and productivity. Studies focusing on bacterial interactions are essential to understand how a unique set of microbiota is established in different hosts, influences health or disease, and provides the basis for the development of new applications, such as synthetic communities (SynComs) or microbiome engineering ( Su et al., 2024).
2. Modes of bacteria‒bacteria interactions in plants
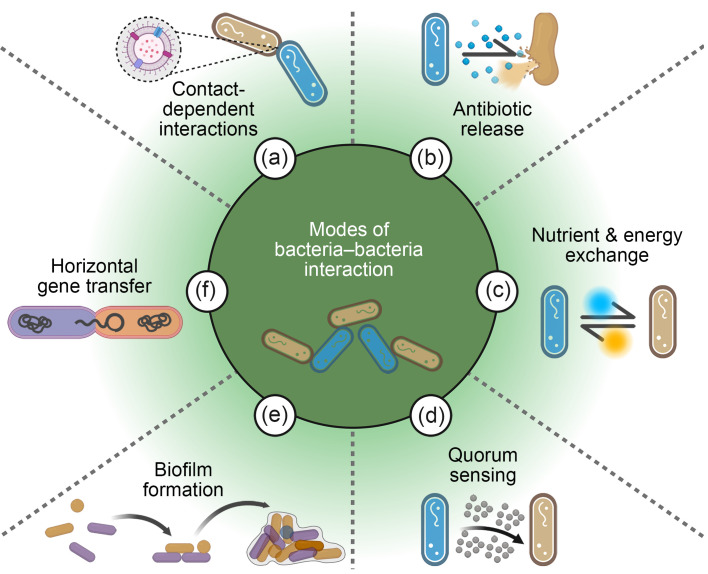
The interspecific interactions between bacteria, which can be both positive and negative, control the establishment and/or proliferation of beneficial bacteria and potential pathogens in plants. Current research on the mechanisms of bacteria–bacteria interactions in plants mainly focuses on the following aspects: (1) contact-dependent interactions; (2) chemical substances; (3) nutrient and energy exchange; (4) quorum sensing (QS) systems; (5) biofilm formation; (6) horizontal gene transfer (HGT) ( Fig. 1).
Fig. 1. Modes of bacteria‒bacteria interactions. (a) Bacteria can deliver toxic proteins or DNA into cells through secretion systems that rely on cell-to-cell contact. (b) By releasing antibiotics into their surrounding environment, certain bacteria can inhibit others. (c) Nutrient exchange between different bacterial species can result in mutual benefits for both. (d) By means of quorum sensing (QS), bacteria are able to regulate gene expression, enabling the detection of signaling molecules within microbial communities. (e) In environments containing antibiotics, the development of biofilms assists bacteria in better adapting to their surroundings. (f) Bacterial DNA can be transferred to other bacteria via horizontal gene transfer, facilitating genetic diversity and adaptation.
2.1. Cell-to-cell contact-dependent interactions
Bacteria employ two main strategies to counter invaders: the release of small molecules with antimicrobial activity into the surrounding environment and the delivery of toxic effector proteins into nearby competitors through specialized secretion systems. The latter includes type IV secretion system (T4SS), type VI secretion system (T6SS), and type VII secretion system (T7SS), which allow different types of cell-to-cell contact. The T4SS is a complex multi-protein structure capable of transporting DNA, effector proteins, and protein-DNA complexes either into the extracellular environment or directly into eukaryotic or prokaryotic target cells. This system facilitates bacterial conjugation transfer, DNA release and uptake, and effector protein secretion, thereby enhancing bacterial adaptability and survival capabilities. For example, Pseudomonas putida IsoF defends tomato plants against Ralstonia solanacearum using a type IVB secretion system (T4BSS) to deliver toxic effectors to competing bacteria in a contact-dependent manner. This strategy is supported by the kib gene cluster in a rare genomic island, likely acquired through HGT, enabling P. putida IsoF to eliminate bacterial competitors and penetrate biofilms for environmental persistence ( Purtschert-Montenegro et al., 2022). The T6SS is one of the most widely researched secretion systems, predominantly occurring in Proteobacteria and Bacteroidetes ( Galán and Waksman, 2018). The T6SS, similar to the tail spike of T4 phage, pierces bacterial cells to deliver genetic material ( Ho et al., 2014). The beneficial plant bacterium P. putida can kill other bacteria by secreting the toxic effector Tke2 via its K1-T6SS secretion system ( Bernal et al., 2017). The T7SS was first discovered in the pathogen Mycobacterium tuberculosis and is primarily found in Gram-positive bacteria. Bacilli subtilis can secrete the LXG antimicrobial effector YxiD through T7SS to antagonize other bacteria ( Tassinari et al., 2022). Many studies have reported on the roles of bacterial secretion systems in interactions between beneficial bacteria and pathogens. In addition, research into these secretion systems provides new insights into the pathogenic mechanisms of plant pathogens, offering possibilities for developing novel plant protection strategies. However, the mechanisms of bacterial secretion systems are extremely complex, involving multiple molecular and cellular interactions; a variety of advanced techniques are required to decipher the underlying mechanisms, such as molecular biology assays, proteomics, and microscopy technologies.
2.2. Interactions via chemical substances
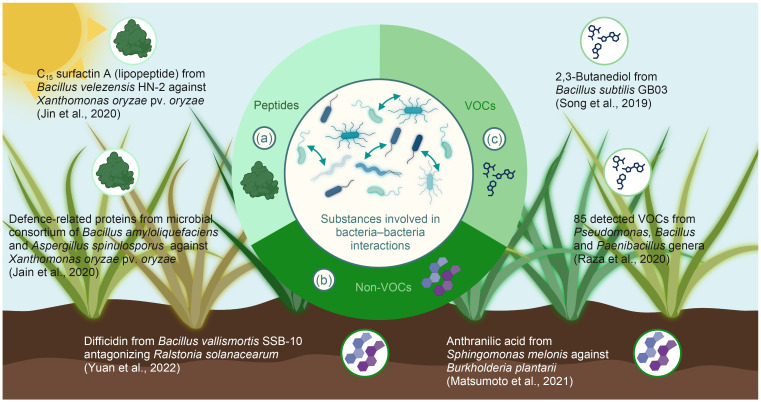
A wide range of chemical substances have been described from bacteria–bacteria interactions, with a single one having different effects depending on the target cell. Based on their size and structure, these substances can be classified into three main groups: peptides, non-volatile organic compounds (non-VOCs), and VOCs ( Fig. 2). Peptides, especially lipopeptides from interacting bacteria, can play an important role in plant health and productivity. Bacillus species can synthesize a diverse set of antimicrobial peptides. Iturin, bacillomycin, bacilysin, fengycin, surfactin, ericin, subtilin, mersacidin, subtilosin, and mycosubtilin are some examples reported in the literature ( Chung et al., 2008; Özcengiz and Öğülür, 2015; Nakkeeran et al., 2020). A few of these have been classified as lipopeptide antibiotics ( Tsuge et al., 2005). These biomolecules comprise a lipid tail linked to a cyclic or short linear oligopeptide and can contribute to plant health by antagonizing pathogens ( Raaijmakers et al., 2010). The surfactant properties of putisolvin I and II produced by a plant root-associated Pseudomonas isolate render the molecule capable of destroying biofilms formed by other bacteria ( Kuiper et al., 2004). Such compounds are not only relevant for agricultural applications but can also be used in biotechnology ( Kuiper et al., 2004).
Fig. 2. Schematic illustration showing the three different substance groups commonly involved in bacteria‒bacteria interactions: (a) peptides (e.g., lipopeptides, such as surfactins), (b) non-volatile organic compounds (non-VOCs) (e.g., anthranilic acid, pyrrole-derivates, and fatty acids), and (c) VOCs (e.g., 2,3-butandiol).
During the past few years, various lipopeptides were identified as promising agents for biological control applications. In Arabidopsis roots, the inhibition of Pseudomonas syringae was shown to depend on the concentration of surfactin produced by Bacillus spp. ( Bais et al., 2004). Moreover, C15 surfactin A, produced by Bacillus velezensis HN-2, displayed antibacterial activity against Xanthomonas oryzae pv. oryzae ( Xoo) and effectively prevented it from infecting rice at low concentrations. Thorough assessments based on scanning electron and transmission electron microscopy showed that this compound causes significant damage to the cell wall structure of Xoo ( Jin et al., 2020). In parallel, C15 surfactin A exposure upregulated the expression of the plant genes phenylalanine ammonia-lyase ( Pal), profilin-1A ( Pr1a), and catalase-A ( CatA). C15 surfactin A could therefore be employed as a biocontrol agent due to its efficacy in inhibiting the growth of Xoo and effectively inducing rice resistance by triggering a hypersensitive reaction ( Jin et al., 2020). Additionally, Bacillus amyloliquefaciens in rice plants was identified as a promising source of new peptides with antibiotic function against plant pathogens ( Jain et al., 2020). García-Bayona and Comstock (2018) published a review focused on bacterial antagonism mediated by peptides and proteins produced by members of host-associated microbial communities. These compounds play important roles in ecosystem defense, pathogen invasion, and spatial segregation.
An exhaustive review focusing on siderophores was published by Ferreira et al. (2019). Siderophores are low-weight molecules (500‒1500 Da), possessing strong affinity and selectivity towards the binding of Fe(III) ( Ahmed and Holmström, 2014). More than 500 siderophores have been reported in the literature. Among these, at least 270 have been structurally characterized ( Barry and Challis, 2009). Despite this great variety, siderophores share some common moieties such as the negatively charged oxygen atoms for binding Fe(Ⅲ). The following bacterial genera were described to produce such molecules: Azotobacter ( Baars et al., 2016; McRose et al., 2017; Romero-Perdomo et al., 2017), Azospirillum ( Banik et al., 2016), Bacillus ( Kesaulya et al., 2018; Pourbabaee et al., 2018), Dickeya ( Sandy and Butler, 2011), Klebsiella ( Zhang et al., 2017; Bailey et al., 2018), Nocardia ( Hoshino et al., 2011), Pantoea ( Burbank et al., 2015; Soutar and Stavrinides, 2018), Paenibacillus ( Liu et al., 2020), Pseudomonas ( Baune et al., 2017; Deori et al., 2018; Pourbabaee et al., 2018), Serratia ( Gerc et al., 2014), and Streptomyces ( Schütze et al., 2015; Gáll et al., 2016; Goudjal et al., 2016). These molecules are often involved in bacteria–bacteria interactions and therefore of high importance for the development of biological control agents.
Comparative analyses of experimental data have shown that volatile metabolites have a greater contribution to microbial interactions than non-volatile ones ( Kanchiswamy et al., 2015). VOCs are a broad group of lipophilic compounds with high vapour pressure, low boiling point, and typically 100‒500 Da molecular weight ( Schulz-Bohm et al., 2018; Raza et al., 2020). Bacterial VOCs have been reported to affect different organisms, such as bacteria, fungi, plants, and animals. Among them, many have inhibitory effects towards plant pests and pathogens ( Garbeva and Weisskopf, 2020). Over the last 15 years, the significance of VOCs as mediators of mutualistic interactions between plant-associated bacteria and their hosts has become evi‑dent. Garbeva and Weisskopf (2020) summarized the current knowledge concerning bacterial VOC-mediated plant protection against biotic and abiotic stresses in a detailed review. They also highlighted the scientific gaps that need to be answered to better understand molecular mechanisms, such as factors underlying the biosynthesis of bacterial volatiles, the complex regulation of their emission in natural communities, the perception of bacterial volatiles by plants, and their modes of actions ( Garbeva and Weisskopf, 2020). Song et al. (2019) investigated the plant protection efficiency of 2,3-butanediol using filter paper soaked with this substance or a culture of B. subtilis GB03. In both cases, continuous exposure to an atmosphere enriched in volatiles led to a decrease in the leaf spot symptoms of cucumber. Raza et al. (2020) demonstrated in a model plant system that bacterial community richness and composition are important for VOC production, pathogen suppression, and plant growth-promotion. VOC-producing bacterial communities with different richness levels were assembled, varying from 1 to 12 strains and with three soil-dwelling genera ( Paenibacillus, Bacillus, and Pseudomonas). Their VOC production correlated positively with the suppression of pathogens and plant growth promotion. All bacteria produced a diverse set of VOCs. However, the pathogen suppression effect reached its maximum at intermediate community richness levels. Based on the results, they proposed that both the number of interacting bacterial species and the structure of the rhizosphere microbiome affect the balance between VOC-mediated microbe‒pathogen and microbe‒plant interactions, regulating plant disease occurrence in agricultural ecosystems ( Raza et al., 2020).
Revealing the chemical communication between bacteria provides valuable insights into understanding how a certain plant microbiome is shaped. The current research on bacterial communication mainly focuses on culturable bacteria; however, these represent only a small part of the entire microbiota. Future research requires a more diverse set of methods to better describe chemical interactions in complex communities.
2.3. Nutrient and energy exchange
Antagonistic interactions are commonly observed in natural microbial communities. However, in recent years, it was increasingly observed that bacteria also engage in a wide range of cooperative behaviours. For instance, they use substantial resources to produce metabolic by-products for utilization by other bacteria, thereby assisting them. In most cases, cooperating bacteria are related, and by aiding their kin, they can increase the chances of their own genes being indirectly propagated ( D'Souza et al., 2018). However, associations between non-related bacteria have also been observed. The rhizobacterium Bacillus cereus UW85 stimulates the growth of bacteria from the Cytophaga- Flavobacterium group (Bacteroidetes) in the soybean rhizosphere. The growth-promoting mechanism likely involves bacterial cell wall components, because peptidoglycan isolated from B. cereus cultures stimulated the growth of Flavobacterium johnsoniae in vitro. In another study, B. velezensis SQR9 was able to induce the enrichment of Pseudomonas stutzeri in the cucumber rhizosphere. These two micro‑organisms formed robust biofilms both in vitro and on the surface of plant roots. Metabolomic and transcriptomic analyses showed that SQR9 facilitates the growth of P. stutzeri through metabolic cross-feeding ( Sun et al., 2022).
2.4. Quorum sensing systems
QS, a critical determinant of microbial interactions, enables cells in a microbial community to detect signaling molecules, leading to a cascade of reactions and regulating gene expression. In this way, QS affects nutrient absorption, material transport, and energy metabolism, and orchestrates microbial interactions that define the community structure, facilitating swift adaptation to changing environmental conditions ( Zeng et al., 2023). QS systems differ between Gram-negative and Gram-positive bacteria. In the former, signal molecules are typically acyl-homoserine lactone (AHL) or cis-11-methyl-2-dodecenoic acid (also known as diffusible signal factor (DSF)), mainly found in genera like Pseudomonas and Xanthomonas. Conversely, in Gram-positive bacteria, these molecules are generally peptides ( Lazdunski et al., 2004; Danhorn and Fuqua, 2007). Notably, in species such as B. subtilis and Streptococcus pneumoniae, these peptide signals play a crucial role in triggering sporulation and the development of competence.
Pathogens can utilize QS molecules to regulate the expression of virulence factors. For instance, the plant pathogen Pantoea stewartii regulates its biofilm formation and colonization abilities through the EsaI/ EsaR QS system, leading to its pathogenicity. QS mutants of P. stewartii are unable to disperse and migrate in the vascular system, thereby reducing disease severity ( Koutsoudis et al., 2006). Some bacteria possess the ability to release compounds that disrupt the production and dispersal of autoinducers, hence meddling with the QS systems of specific microbial species. This interference hampers microbial communication, diminishing the levels of harmful factor expression, and thus modulates bacterial growth and reproduction mechanisms. This process is identified as QS quenching, with the involved compounds referred to as QS inhibitors. The Lysobacter enzymogenes OH11 strain, which lacks AHL synthesis capability, can utilize the T4SS to inject the effector protein Le1288 (LqqE1) into the cytoplasm of another soil bacterium, Pseudomonas fluorescens 2P24 (2P24), which possesses an AHL communication system. Unlike typical AHL quenching proteins, LqqE1 does not degrade AHL but inhibits AHL synthesis by directly binding to the AHL synthase PcoI in 2P24 ( Liao et al., 2023). With more in-depth research into QS mechanisms, we will better understand bacterial communication and develop new strategies to control bacterial plant diseases.
2.5. Biofilm formation
Biofilms, often harboring complex microbial communities, represent a collective behaviour of bacteria adapting to specific and often extreme surface envir‑onments. It is well-known that the formation of biofilms involves various collective behaviours and depends on the complex regulation of QS. In the development of biofilms, synergistic and antagonistic interactions both exist between bacteria. Pseudoalteromonas sp. MEBiC 03485 boosts the extracellular polymeric substance (EPS) production of Porphyridium cruentum UTEX 161 ( Han et al., 2020). This expansion in EPS productivity occurs because of specific chemical compounds discharged by bacteria. Using rheology and microscopy-based approaches, Yannarell et al. (2019) recently explored a dual-species biofilm consisting of B. subtilis and Pantoea agglomerans. Several features of the mixed-community biofilm appeared to be distinct from those identified in monocultures, highlighting the importance of characterizing multispecies consortia for a better understanding of natur‑ally occurring bacterial interactions. The formation of biofilms can also serve as a physical barrier to help bacteria adapt to the presence of other bacteria. Molina-Santiago et al. (2019) explored the interaction between B. subtilis 3610 and Pseudomonas chlororaphis PCL1606, and revealed that the extracellular matrix of B. subtilis protects colonies from P. chlororaphis. B. subtilis is a soil-dwelling bacterium engaging in mutualistic interactions with plants, frequently used as a model of biofilm formation ( Beauregard et al., 2013). The absence of extracellular matrix from B. subtilis resulted in its increased fluidity and loss of colony structure. The P. chlororaphis T6SS was activated upon contact with B. subtilis, stimulating sporulation mediated by histidine kinases KinA and KinB. Molina-Santiago et al. (2019) also demonstrated the importance of the extracellular matrix and the P. chlororaphis T6SS in modulating the coexistence of these two species on melon leaves and seeds. Microorganisms in biofilms engage in complex interactions, involving both known and obscure mechanisms. Although our understanding of these interactions has significantly deepened, there are still unresolved questions about biofilm maintenance and stability, creating space for new discoveries.
2.6. Horizontal gene transfer
Bacteria acquire new DNA through HGT, enabling them to adapt to constantly changing environments. This exogenous DNA may contain elements that can expand the bacterial habitat range, alter the relationship between bacteria and their hosts, or provide advantages in competition with other organisms. DNA can be transferred in multiple ways, with conjugation, transposition, and transformation being the most studied mechanisms. However, additional mechanisms are gradually being discovered, such as acquisition through outer membrane vesicles and transfer via virus-like particles ( Brito, 2021). By combining theoretical deduction and numerical simulations, researchers have found that HGT unexpectedly exceeds the scale of biodiversity predicted by classical models. HGT is achieved by facilitating dynamic neutrality among competing species, thus permitting the coexistence of multiple competitive species ( Zhu et al., 2024).
HGT within microbial communities aids bacterial adaptation to diverse environmental stimuli. This process is critically important in the context of the spread of antibiotic resistance genes, a phenomenon further amplified by the formation of biofilms. As improved detection methods and innovative theoretical and computational approaches are increasingly being implemented in relevant studies, our understanding of bacterial HGT has significantly widened. In the future, in-depth sequencing of metagenomes will allow the recovery of complete genomes to reveal a multitude of co-existing species, including those that are unculturable in the laboratory. These data will greatly enhance our knowledge of the complex microbial dynamics facilitated by HGT.
3. Agricultural relevance of bacteria‒ bacteria interactions
3.1. Harnessing bacteria‒ bacteria interactions to enhance plant health and disease resistance
Plant health is often compromised by resistant phytopathogens that cannot be controlled by conventional strategies. While the most prevalent diseases are caused by fungi, phytopathogenic bacteria also account for substantial yield losses ( Yin et al., 2011; Elshafie et al., 2012; Weise et al., 2012; Molina-Santiago et al., 2019; Purtschert-Montenegro et al., 2022). Alternatives to conventional agrochemical-based protection strategies can emerge from observations of specific bacteria–bacteria interactions. Specifically, beneficial bacteria can engage in interactions that are detrimental for their pathogenic counterparts, thus protecting different plant species from the seedling stage onwards ( Venturi and Bez, 2021). A conceptual framework on this phenomenon was provided by Hassani et al. (2018), illustrating that interactions within the microbiota are critical for the establishment and maintenance of host health. During the past decades, manifold bacteria–bacteria interactions were regarded as crucial to maintain plant health under laboratory, field, or greenhouse conditions. Members of the bacterial genera Pseudomonas, Bacillus, Rhizobium, Pantoea, Azospirillum, and Azotobacter have been considered as highly promising for agricultural applications ( Ferreira et al., 2019). Among these, Pseudomonas spp. and Bacillus spp. are the most frequently described plant-beneficial bacteria. The ability of beneficial bacteria to protect plants against pathogens ( Combes-Meynet et al., 2011; Zeriouh et al., 2011; Bernal et al., 2017; Gu et al., 2017; Molina-Santiago et al., 2019; Purtschert-Montenegro et al., 2022) and to promote their growth ( Combes-Meynet et al., 2011; Roca et al., 2013; Lally et al., 2017) has been widely acknowledged. A representative set of interaction types as well as their consequences for host plant health is provided in Table 1. Typical plant characteristics that can be improved by bacteria–bacteria interactions are schematically visualized in Fig. 3a. In general, many natural products formed by Pseudomonas-like soil-dwelling microorganisms are a consistent source of potential antimicrobial metabolites and biopesticides. Yang et al. (2023) successfully isolated a Pseudomonas mosselii strain from the rice rhizosphere that specifically inhibited the growth of plant-pathogenic Xanthomonas species. The bioactive compound produced by P. mosselii was identified as pseudoiodinine. An approximately 20-fold increase in the production of pseudoiodinine was achieved by optimizing the growth medium, via overexpressing the biosynthetic operon and removing the CsrA (a carbon storage regulator)-binding sites. P. mosselii as well as purified pseudoiodinine inhibited the pathogen without affecting the rice host, suggesting the applicability of this compound in controlling certain diseases ( Yang et al., 2023).
Table 1.
Examples of different bacteria‒ bacteria interactions and their consequences for the health and productivity of plants
| Bacteria–bacteria interaction | Description of the interaction | Category | Consequence on the plant | Reference |
|---|---|---|---|---|
| Pseudomonas mosselii 923 against Xanthomonas species | Xanthomonas species are inhibited by pseudoiodinine from P. mosselii 923 | Negative, amensalism | Rice plants are protected against bacterial and fungal pathogens | Yang et al., 2023 |
| Pseudomonas putida IsoF against Ralstonia solanacearum | Delivery of toxic effector to R. solanacearum via type IVB secretion system (T4BSS) | Negative, amensalism | Tomato plants are protected against the pathogen R. solanacearum | Purtschert-Montenegro et al., 2022 |
| Aeromicrobium Leaf245 against Nocardioides Leaf374 | Nocardioides is lysed by an antimicrobial peptidase from Aeromicrobium | Negative, amensalism | Potential growth-promotion and disease-suppression | Schäfer et al., 2022 |
| Sphingomonas melonis against Burkholderia plantarii | S. melonis produces anthranilic acid that interferes with the sigma factor RpoS of B. plantarii, likely causing impairment of upstream cascades required for virulence factor biosynthesis | Negative, amensalism | Rice becomes disease-resistant against B. plantarii | Matsumoto et al., 2021 |
| Bacillus velezensis HN-2 against Xoo | C15 surfactin A produced by B. velezensis HN-2 shows antibacterial activity against Xoo | Negative, amensalism | Xoo infection in rice is inhibited | Jin et al., 2020 |
| Bacilli subtilis 3610 against Pseudomonas chlororaphis PCL1606 | B. subtilis protects itself with extracellular matrix from P. chlororaphis | Negative, amensalism | Establishment of mutualistic interactions with B. subtilis | Molina-Santiago et al., 2019 |
| Pseudomonas fluorescens F113 and endophytic microbial consortium ( P. fluorescens L111, L228, and L321; Pseudomonas sp. L117, L130, L132, and Rt03; Serratia sp. R324 and S120; Enterobacter sp. R232) | Plant-associated bacteria with PGP properties | Positive, mutualism | Significant increment in crop height and pod biomass | Lally et al., 2017 |
| B. subtilis UMAF6614 and UMAF6639 against Xanthomonas campestris pv. cucurbitae and Pectobacterium carotovorum subsp. carotovorum | Antibiotics (iturin A and bacillomycin) from Bacillus have cytotoxic effect on phytopathogens | Negative, amensalism | Control of the bacterial leaf spot and soft rot pathogens of cucurbits | Zeriouh et al., 2011 |
| P. fluorescens F113 and Azospirillum brasilense Sp245-Rif | DAPG-producing Pseudomonas enhances Azospirillum's cell motility, biofilm formation, and production of PBH and auxin | Positive, mutualism | Enhanced phytostimulation effects | Combes-Meynet et al., 2011 |
Xoo: Xanthomonas oryzae pv. oryzae; PGP: plant growth promoting; DAPG: 2,4-diacetylphloroglucinol; PBH: poly-β-hydroxybutyrate.
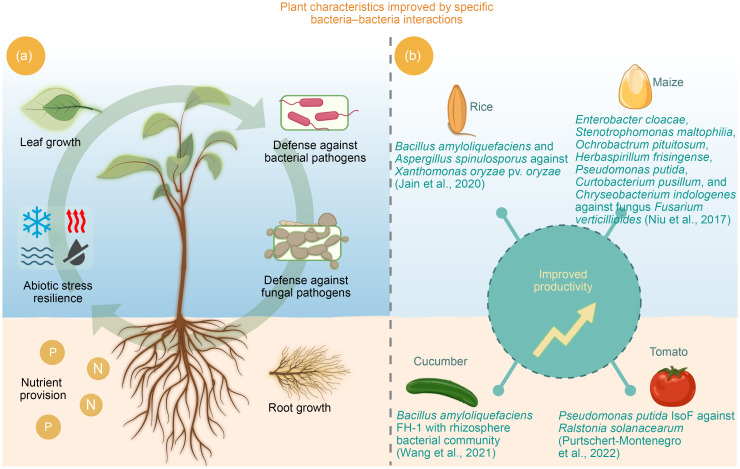
Fig. 3. Schematic representation of various scenarios where plant fitness and/or productivity are improved by specific bacteria‒bacteria interactions. (a) Such interactions can result in improved root and leaf growth, defense against fungal and bacterial pathogens, abiotic stress resilience, and nutrient provision. (b) The productivity of different plants (e.g., rice, maize, tomato, and cucumber) can also be enhanced by different bacteria‒bacteria interactions.
A study on the core bacterial endophytes of rice revealed 14 prevailing amplicon sequence variants ( Zhang et al., 2022). Especially, Pantoea and Xanthomonas were identified as potentially vertically transmitted taxa. They exhibited cellulase activity and were found to produce indole-3-acetic acid. Five Pantoea strains showed moderate antagonism against major pathogens of rice diseases like Magnaporthe oryzae and Xoo. These data provide an important basis for harnessing functions of naturally occurring members of the plant microbiota for crop protection. Matsumoto et al. (2021) observed that rice plants of the same genotype under the same pathogen pressure can be differentiated into disease-resistant and susceptible phenotypes. After the identification of a seed-endophytic bacterium ( Sphingomonas melonis) as the resistance-conferring agent against the seed-borne pathogen Burkholderia plantarii, the integration of high-throughput data, gene mutagenesis, and molecular interaction assays was performed, facilitating the discovery of the mode of action. S. melonis, accumulated and transmitted across generations in disease-resistant rice seeds, confers resistance to disease-susceptible phenotypes producing anthranilic acid. This compound interferes with the sigma factor RpoS of rice-pathogenic B. plantarii, likely causing the impairment of upstream cascades required for virulence factor biosynthesis ( Matsumoto et al., 2021). In another study focusing on rice plants, Xoo was found to be antagonized by B. velezensis HN-2 ( Jin et al., 2020). This study highlighted the importance of identifying core endophytes, vertically transmitted microbiota, and network hubs, which can be crucial for the development of an effective strategy for the targeted manipulation of microbiomes towards improving plant health.
Currently, the use of numerous biological control agents is hindered by various factors. These include the poor survival rate and colonization capacity of the introduced biocontrol strains, along with a reduction in the expression of essential antagonistic traits in field application. Thus, it is critical to comprehensively understand the dynamic interactions among pathogens, antagonistic microorganisms, and the environment to formulate effective biological control strategies.
3.2. Plant fitness and productivity improved by bacteria‒bacteria interactions
Heat, drought, flood, and cold weather are among the most frequent abiotic stresses faced by agricultural plants, reducing their productivity. Due to climate change, the impact and intensity of these stresses have gradually increased during recent decades ( Chaudhry and Sidhu, 2022). Many microorganisms can produce secondary and primary metabolites, which help plants mitigate these stresses ( Biró et al., 2006; Rajkumar et al., 2013; Garcia et al., 2022; Munir et al., 2022). When bacteria interact within microbiomes, the outcome can coincide with a better productivity of the host plant, improved growth, or higher fitness ( Yuan et al., 2016; Niu et al., 2017; Hawkes et al., 2021; Wang et al., 2021). Various studies have shown that the productivity of rice, maize, tomato, cucumber, and other agricultural plants can be enhanced by bacteria–bacteria interactions ( Fig. 3b). A recent review by Munir et al. (2022) summarized the impacts of abiotic stressors on agricultural productivity and the role of beneficial microorganisms in alleviating negative effects. Targeted inoculation with beneficial microbes can also reduce the effects of different biotic stresses, enhancing plant fitness ( Wei et al., 2015; Niu et al., 2017; Ansari and Ahmad, 2019; Jain et al., 2020; Tan et al., 2021; Wang et al., 2021). To this end, Liu et al. (2020) proposed the "DefenseBiome" concept, providing a framework for the design and construction of beneficial microbial synthetic communities to improve the fundamental understanding of plant–microbial interactions and the development of related products for agriculture.
Jain et al. (2020) evaluated the defense network provided by beneficial microorganisms in the rice rhizosphere and observed reduced plant mortality as well as an increase in dry biomass and chlorophyll content in the presence of this network. Another study focused on the beneficial functions of P. fluorescens FAP2 and Bacillus licheniformis B642, two well-characterized plant-growth-promoting rhizobacteria (PGPR) strains ( Ansari and Ahmad, 2019). Rhizosphere and rhizoplane colonization of wheat seedlings by both isolates resulted in the production of indole acetic acid (IAA), siderophores, ammonia, and phosphate solubilization, as well as some biofilm-related functions (e.g., production of exopolysaccharides, alginate, and cell surface hydrophobicity). The effects of co-inoculation revealed the significant enhancement of vegetative growth, photosynthetic parameters (e.g., chlorophyll content, transpiration rate, internal CO2 concentration, stomatal conductance, and net photosynthetic rate), and leaf water potential, in comparison with single inoculation and the uninoculated control. Combes-Meynet et al. (2011) have shown that the Pseudomonas-produced secondary metabolite 2,4-diacetylphloroglucinol (DAPG) can act as a signal molecule for PGPR, enhancing their phytostimulation effects. In wheat, DAPG-producing P. fluorescens F113 can enhance cell motility, biofilm formation, and the production of poly-β-hydroxybutyrate and auxin of Azospirillum brasilense Sp245-Rif (Sp245-Rif). A differential fluorescence induction promoter-trapping approach was used to confirm the high expression of Sp245-Rif gene in the presence of DAPG. Compared with its phl-negative mutant, the expression of four genes ( nirK, nifX- nifB, ppdC, and flgE) was upregulated in the presence of P. fluorescens F113 ( Combes-Meynet et al., 2011). In a different study, Tan et al. (2021) demonstrated that the fitness of Lemna minor is modified due to interactions between the microbiome and P. fluorescens. The microbiome promoted the rapid evolution of P. fluorescens, which reciprocally altered the microbiome, and these eco-evolutionary dynamics subsequently modified the fitness of the host. The results suggested that the microbial effects probably occur through changes in the production of the plant growth hormone auxin by the microbiome ( Tan et al., 2021). Wang et al. (2021) aimed to explore the ecological roles of B. amyloliquefaciens FH-1 (FH) on rhizosphere properties, cucumber seedlings, and the bacterial community. FH was shown to promote cucumber seedling growth, reduce the rhizosphere bacterial diversity, increase Proteobacteria abundance, and decrease Acidobacteria abundance. The weight, height, and length of cucumber seedlings were significantly correlated with 18 inhibited taxa and the enrichment of Nannocystaceae. FH might therefore promote cucumber seedling growth via the enrichment of Nannocystaceae and the inhibition of various taxa belonging to Acidobacteria, Actinobacteria, Chloroflexi, Plantctomycetes, and Verrucomicrobia ( Wang et al., 2021).
In order to study bacterial dynamics under soil nitrogen limitation, Garcia et al. (2022) recently applied selection pressure for the microbiome-driven enhancement of Brassica rapa biomass. The selection pressure enhanced the seed yield of B. rapa and the utilization efficiency of nitrogen. Among specific bacterial phyla such as Proteobacteria and Bacteroidetes, distinct interactions emerged in response to selection, and without selection, the bacterial communities showed less complex interactions. These results suggested that group-level bacterial synergies can modify microbiome functions, thereby affecting the growth of the host plant under soil nitrogen limitation ( Garcia et al., 2022). Zhang et al. (2022) found that the stem xylem selectively recruits highly conserved microbes dominated by γ-Proteobacteria, and assessed the assembly and functions of maize microbiomes across soil types, climate zones, and genotypes. The authors revealed that the proportion of bacterial taxa carrying the nitrogenase gene ( nifH) in stem xylem was higher than that in the root and leaf endosphere. They constructed synthetic communities consisting of two core diazotrophs and two helpers. By utilizing green fluorescent protein ( gfp)-tagged strains and 15N isotopes, it was demonstrated that these strains contributed to biological nitrogen fixation in maize stems ( Zhang et al., 2022). In a different study related to abiotic stress, Mueller et al. (2021) developed a method to select for rhizosphere microbiomes that confer tolerance to the model grass Brachypodium distachyon under sodium or aluminum salt stress. The proposed selection generated microbiomes that were able to enhance plant fitness. Yuan et al. (2016) hypothesized that the high halo-tolerance of the seepweed Suaeda salsa is due to a specialized belowground microbiome interplay. To test this hypothesis, they performed a phylogenetic trait-based analysis. Rhizospheric and endophytic bacteria associated with S. salsa were shown to contribute to salt stress acclimatization, nutrient solubilization, and competitive root colonization ( Yuan et al., 2016). Plant growth was shown to increase linearly with the inoculation of different species, with Klebsiella pneumoniae and Brevibacillus agri playing important roles in improving fitness in monocultures ( Singh et al., 2015).
Phytoremediation can serve as a sustainable and effective technology for cadmium remediation, especially in agricultural soil. To this end, the rhizosphere microbiome can promote the growth and cadmium accumulation in hyperaccumulator plants. A recent study by Fan et al. (2022) used accumulating and non-accumulating ecotypes of Sedum alfredii as model plants to investigate the assembly and influence of the rhizosphere microbiome on plant growth under high- cadmium conditions. Their results showed distinct root microbiomes assembled in both ecotypes of S. alfredii. The bacterial genera Sphingomonas, Lysobacter, Afipia, Hydrogenispora, Luedemannella, Adhaeribacter, and Allorhizobium- Neorhizobium- Pararhizobium- Rhizobium were enriched in cadmium-contaminated soils. The rhizosphere microbiome, assembled in the accumulating ecotype, contributed to plant fitness via nitrogen fixation, phosphorus solubilization, siderophore metabolism, and IAA synthesis, as well as cadmium uptake ( Fan et al., 2022).
3.3. Understanding bacteria‒bacteria interactions to guide the design of synthetic communities
Synthetic microbial communities are composed of two or more defined microorganisms. Within such communities, members can engage in complex communication via exchanging, detecting, and responding to signaling molecules. They can perform different tasks in this setting, thereby achieving complex functions that a single microbial strain cannot accomplish. In natural systems, microbial communities play important roles in various environments, which often inspires the construction of synthetic microbial communities. Niu et al. (2017) constructed a synthetic bacterial community consisting of seven bacterial species: Enterobacter cloacae, Stenotrophomonas maltophilia, Ochrobactrum pituitosum, Herbaspirillum frisingense, P. putida, Curtobacterium pusillum, and Chryseobacterium indologenes. These represent three of the four most dominant bacterial phyla typically found on maize roots. The findings suggested that E. cloacae plays a key role in this model ecosystem; this synthetic community synergistically inhibited the phytopathogenic fungus Fusarium verticillioides in planta and in vitro ( Niu et al., 2017).
By selectively removing or replacing individual species to test their impact on synthetic microbial communities, insights into microbial interactions can be obtained. Schäfer et al. (2022) recently studied bacterial interactions in the phyllosphere microbiota. They screened microbial interactions in the model plant Arabidopsis by individually adding 200 endogenous strains to a synthetic community composed of 15 bacterial strains, and tracked changes in microbial community composition after colonization. It was found that 90% of the bacterial interactions were negative, and strains that were phylogenetically more related had more consistent effects on the synthetic community ( Schäfer et al., 2022). Finkel et al. (2020) used a bacterial synthetic community consisting of 185 isolates for inoculation experiments. They constructed the synthetic community based on four classified modules of co-occurring bacteria and identified interactions between microorganisms that determine the root phenotype. The interactions were shown to mainly involve the bacterial genus Variovorax, which reversed the inhibition of root growth induced by a wide diversity of other bacteria. Furthermore, Variovorax was shown to manipulate the levels of plant hormones, balancing the effects of the synthetic community on root growth.
Combining knowledge from synthetic biology and ecology can inform the construction of synthetic microbial communities. However, in natural environments, even a simple microbial ecosystem typically contains at least ten interwoven species, which can yield combinations of hundreds or even thousands of interactions ( Li et al., 2022). Thus far, unravelling and studying the vast number of interactions in microbial ecosystems has been a challenging task, partly due to the lack of feasible and manageable tools to decipher species interactions in complex communities. However, a detailed understanding of such interactions will be required for the design of more reliable synthetic communities applicable for plant protection.
4. Concluding remarks and perspectives
The potential of bacteria–bacteria interactions to improve agricultural practices, especially the reduction of pesticide use, has been explored intensively during recent years. Many of these findings could be translated into sustainable applications during the next decade. However, viable solutions will require a deeper understanding of soil and plant microbiomes as well as key interactions therein. The development of biological control applications can profit from microbiome analyses at various stages ( Cernava, 2021). Such analyses can support the identification of synergistic as well as antagonistic interactions between biological control agents and members of naturally occurring bacterial communities. Future developments will also need to rely on the efficient production of living microorganisms, especially bacteria, for agricultural use. For this purpose, the industrial-scale fermentation of mixed bacterial communities could provide viable means not only to reduce production costs but also to increase their efficacy under field conditions ( Rändler-Kleine et al., 2020). To identify microorganisms that are suitable for such approaches, the large-scale profiling of various microbiomes is necessary to identify prevailing synergistic interactions between bacteria under distinct environmental conditions.
The advent of artificial intelligence (AI) technologies has allowed us to significantly enhance our understanding of complex microbial interactions. AI algorithms are capable of processing and analyzing large volumes of biological data, including genomics, transcriptomics, and proteomics, supporting the identification of patterns and correlations in microbial interactions. Moreover, machine-learning models can be used to predict the behavior of microbes under various environmental conditions and how they interact with each other. Specifically developed models can forecast microbial responses to environmental changes based on historical data. Overall, AI provides a vital basis for a range of powerful tools and methods that can accelerate discovery processes and enhance the precision and efficiency of research towards a comprehensive understanding of microbial interactions ( Hernández Medina et al., 2022).
A deeper knowledge of bacteria–bacteria interactions will also facilitate the development of microbiome engineering approaches. In the past, the modulatory effect of microbiome was often observed when plants were inoculated with various beneficial bacteria ( Berg et al., 2021). It is highly probable that we will able to engineer plant and soil microbiomes with great precision in the near future not only to increase sustainability in agriculture but also to boost plant health and productivity to a level unachievable by applying conventional agrochemicals.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (No. 2022YFD1400100), the Natural Science Foundation of Zhejiang Province (No. LZ23C140004), the National Natural Science Foundation of China (No. 32172356), and the China Agriculture Research System (No. CARS-3-1-15).
Author contributions
Yun CHEN and Tomislav CERNAVA contributed to the manuscript’s conceptualization, review, and editing. Zhonghua MA contributed to the manuscript’s editing. Giovanni Davide BARONE and Yaqi ZHOU compiled the data and wrote the manuscript. Hongkai WANG and Sunde XU contributed to the figure and table design. All authors have read and approved the final manuscript.
Compliance with ethics guidelines
Zhonghua MA and Yun CHEN are an Editorial Board Member and a Young Scientist Committee Member, respectively, for Journal of Zhejiang University-SCIENCE B (Biomedicine & Biotechnology), and were not involved in the editorial review or the decision to publish this review. Giovanni Davide BARONE, Yaqi ZHOU, Hongkai WANG, Sunde XU, Zhonghua MA, Tomislav CERNAVA, and Yun CHEN declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- Ahmed E, Holmström SJM, 2014. Siderophores in environmental research: roles and applications. Microb Biotechnol, 7(3): 196- 208. 10.1111/1751-7915.12117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari FA, Ahmad I, 2019. Fluorescent Pseudomonas -FAP2 and Bacillus licheniformis interact positively in biofilm mode enhancing plant growth and photosynthetic attributes. Sci Rep, 9: 4547. 10.1038/s41598-019-40864-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baars O, Zhang XN, Morel FMM, et al. , 2016. The siderophore metabolome of Azotobacter vinelandii . Appl Environ Microbiol, 82(1): 27- 39. 10.1128/aem.03160-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey DC, Alexander E, Rice MR, et al. , 2018. Structural and functional delineation of aerobactin biosynthesis in hypervirulent Klebsiella pneumoniae . J Biol Chem, 293(20): 7841- 7852. 10.1074/jbc.RA118.002798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais HP, Fall R, Vivanco JM, 2004. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol, 134(1): 307- 319. 10.1104/pp.103.028712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banik A, Mukhopadhaya SK, Dangar TK, 2016. Characterization of N2-fixing plant growth promoting endophytic and epiphytic bacterial community of Indian cultivated and wild rice ( Oryza spp. ) genotypes. Planta, 243(3): 799- 812. 10.1007/s00425-015-2444-8 [DOI] [PubMed] [Google Scholar]
- Barry SM, Challis GL, 2009. Recent advances in siderophore biosynthesis. Curr Opin Chem Biol, 13(2): 205- 215. 10.1016/j.cbpa.2009.03.008 [DOI] [PubMed] [Google Scholar]
- Baune M, Qi YL, Scholz K, et al. , 2017. Structural characterization of pyoverdines produced by Pseudomonas putida KT2440 and Pseudomonas taiwanensis VLB120. BioMetals, 30(4): 589- 597. 10.1007/s10534-017-0029-7 [DOI] [PubMed] [Google Scholar]
- Beauregard PB, Chai YR, Vlamakis H, et al. , 2013. Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci USA, 110(17): E1621- E1630. 10.1073/pnas.1218984110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G, Rybakova D, Fischer D, et al. , 2020. Microbiome definition re-visited: old concepts and new challenges. Microbiome, 8: 103. 10.1186/s40168-020-00875-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G, Kusstatscher P, Abdelfattah A, et al. , 2021. Microbiome modulation—toward a better understanding of plant microbiome response to microbial inoculants. Front Microbiol, 12: 650610. 10.3389/fmicb.2021.650610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal P, Allsopp LP, Filloux A, et al. , 2017. The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J, 11(4): 972- 987. 10.1038/ismej.2016.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biró B, Köves-Péchy K, Tsimilli-Michael M, et al. , 2006. Role of beneficial microsymbionts on the plant performance and plant fitness. In: Mukerji KG, Manoharachary C, Singh J (Eds. ), Microbial Activity in the Rhizosphere. Springer, Berlin, Heidelberg, p. 265- 296. 10.1007/3-540-29420-1_14 [DOI] [Google Scholar]
- Brito IL, 2021. Examining horizontal gene transfer in microbial communities. Nat Rev Microbiol, 19(7): 442- 453. 10.1038/s41579-021-00534-7 [DOI] [PubMed] [Google Scholar]
- Burbank L, Mohammadi M, Roper MC, 2015. Siderophore-mediated iron acquisition influences motility and is required for full virulence of the xylem-dwelling bacterial phytopathogen Pantoea stewartii subsp. stewartii. Appl Environ Microbiol, 81(1): 139- 148. 10.1128/aem.02503-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernava T, 2021. How microbiome studies could further improve biological control. Biol Control, 160: 104669. 10.1016/j.biocontrol.2021.104669 [DOI] [Google Scholar]
- Cernava T, Berg G, 2022. The emergence of disease-preventing bacteria within the plant microbiota. Environ Microbiol, 24(8): 3259- 3263. 10.1111/1462-2920.15896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry S, Sidhu GPS, 2022. Climate change regulated abiotic stress mechanisms in plants: a comprehensive review. Plant Cell Rep, 41(1): 1- 31. 10.1007/s00299-021-02759-5 [DOI] [PubMed] [Google Scholar]
- Chen Y, Wang J, Yang N, et al. , 2018. Wheat microbiome bacteria can reduce virulence of a plant pathogenic fungus by altering histone acetylation. Nat Commun, 9: 3429. 10.1038/s41467-018-05683-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Kong H, Buyer JS, et al. , 2008. Isolation and partial characterization of Bacillus subtilis ME488 for suppression of soilborne pathogens of cucumber and pepper. Appl Microbiol Biotechnol, 80(1): 115- 123. 10.1007/s00253-008-1520-4 [DOI] [PubMed] [Google Scholar]
- Combes-Meynet E, Pothier JF, Moënne-Loccoz Y, et al. , 2011. The Pseudomonas secondary metabolite 2, 4-diacetylphloroglucinol is a signal inducing rhizoplane expression of Azospirillum genes involved in plant-growth promotion. Mol Plant-Microbe Int, 24(2): 271- 284. 10.1094/MPMI-07-10-0148 [DOI] [PubMed] [Google Scholar]
- Danhorn T, Fuqua C, 2007. Biofilm formation by plant-associated bacteria. Annu Rev Microbiol, 61: 401- 422. 10.1146/annurev.micro.61.080706.093316 [DOI] [PubMed] [Google Scholar]
- Deori M, Jayamohan NS, Kumudini BS, 2018. Production, characterization and iron binding affinity of hydroxamate siderophores from rhizosphere associated fluorescent Pseudomonas. J Plant Prot Res, 58(1): 36- 43. 10.24425/119116 [DOI] [Google Scholar]
- D'Souza G, Shitut S, Preussger D, et al. , 2018. Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat Prod Rep, 35(5): 455- 488. 10.1039/c8np00009c [DOI] [PubMed] [Google Scholar]
- Elshafie HS, Camele I, Racioppi R, et al. , 2012. In vitro antifungal activity of Burkholderia gladioli pv. agaricicola against some phytopathogenic fungi. Int J Mol Sci, 13(12): 16291- 16302. 10.3390/ijms131216291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan WJ, Deng JM, Shao L, et al. , 2022. The rhizosphere microbiome improves the adaptive capabilities of plants under high soil cadmium conditions. Front Plant Sci, 13: 914103. 10.3389/fpls.2022.914103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira CMH, Soares HMVM, Soares EV, 2019. Promising bacterial genera for agricultural practices: an insight on plant growth-promoting properties and microbial safety aspects. Sci Total Environ, 682: 779- 799. 10.1016/j.scitotenv.2019.04.225 [DOI] [PubMed] [Google Scholar]
- Finkel OM, Salas-González I, Castrillo G, et al. , 2020. A single bacterial genus maintains root growth in a complex microbiome. Nature, 587(7832): 103- 108. 10.1038/s41586-020-2778-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán JE, Waksman G, 2018. Protein-injection machines in bacteria. Cell, 172(6): 1306- 1318. 10.1016/j.cell.2018.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gáll T, Lehoczki G, Gyémánt G, et al. , 2016. Optimization of desferrioxamine E production by Streptomyces parvulus . Acta Microbiol Immunol Hung, 63(4): 475- 489. 10.1556/030.63.2016.029 [DOI] [PubMed] [Google Scholar]
- Garbeva P, Weisskopf L, 2020. Airborne medicine: bacterial volatiles and their influence on plant health. New Phytol, 226(1): 32- 43. 10.1111/nph.16282 [DOI] [PubMed] [Google Scholar]
- Garcia J, Gannett M, Wei LP, et al. , 2022. Selection pressure on the rhizosphere microbiome can alter nitrogen use efficiency and seed yield in Brassica rapa . Commun Biol, 5: 959. 10.1038/s42003-022-03860-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Bayona L, Comstock LE, 2018. Bacterial antagonism in host-associated microbial communities. Science, 361(6408): eaat2456. 10.1126/science.aat2456 [DOI] [PubMed] [Google Scholar]
- Gerc AJ, Stanley-Wall NR, Coulthurst SJ, 2014. Role of the phosphopantetheinyltransferase enzyme, PswP, in the biosynthesis of antimicrobial secondary metabolites by Serratia marcescens Db10. Microbiology, 160(8): 1609- 1617. 10.1099/mic.0.078576-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudjal Y, Zamoum M, Meklat A, et al. , 2016. Plant-growth-promoting potential of endosymbiotic actinobacteria isolated from sand truffles ( Terfezia leonis Tul. ) of the Algerian Sahara. Ann Microbiol, 66(1): 91- 100. 10.1007/s13213-015-1085-2 [DOI] [Google Scholar]
- Gu Q, Yang Y, Yuan QM, et al. , 2017. Bacillomycin D produced by Bacillus amyloliquefaciens is involved in the antagonistic interaction with the plant-pathogenic fungus Fusarium graminearum . Appl Environ Microbiol, 83(19): e01075-17. 10.1128/aem.01075-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SI, Jeon MS, Heo YM, et al. , 2020. Effect of Pseudoalteromonas sp. MEBiC 03485 on biomass production and sulfated polysaccharide biosynthesis in Porphyridium cruentum UTEX 161. Bioresour Technol, 302: 122791. 10.1016/j.biortech.2020.122791 [DOI] [PubMed] [Google Scholar]
- Hassani MA, Durán P, Hacquard S, 2018. Microbial interactions within the plant holobiont. Microbiome, 6: 58. 10.1186/s40168-018-0445-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes CV, Kjøller R, Raaijmakers JM, et al. , 2021. Extension of plant phenotypes by the foliar microbiome. Annu Rev Plant Biol, 72: 823- 846. 10.1146/annurev-arplant-080620-114342 [DOI] [PubMed] [Google Scholar]
- Hernández Medina R, Kutuzova S, Nielsen KN, et al. , 2022. Machine learning and deep learning applications in microbiome research. ISME Commun, 2(1): 98. 10.1038/s43705-022-00182-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho BT, Dong TG, Mekalanos JJ, 2014. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe, 15(1): 9- 21. 10.1016/j.chom.2013.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino Y, Chiba K, Ishino K, et al. , 2011. Identification of nocobactin NA biosynthetic gene clusters in Nocardia farcinica . J Bacteriol, 193(2): 441- 448. 10.1128/jb.00897-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Chatterjee A, Das S, 2020. Synergistic consortium of beneficial microorganisms in rice rhizosphere promotes host defense to blight-causing Xanthomonas oryzae pv. oryzae. Planta, 252(6): 106. 10.1007/s00425-020-03515-x [DOI] [PubMed] [Google Scholar]
- Jin PF, Wang Y, Tan Z, et al. , 2020. Antibacterial activity and rice-induced resistance, mediated by C15surfactin A, in controlling rice disease caused by Xanthomonas oryzae pv. oryzae. Pestic Biochem Physiol, 169: 104669. 10.1016/j.pestbp.2020.104669 [DOI] [PubMed] [Google Scholar]
- Kanchiswamy CN, Malnoy M, Maffei ME, 2015. Chemical diversity of microbial volatiles and their potential for plant growth and productivity. Front Plant Sci, 6: 151. 10.3389/fpls.2015.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesaulya H, Hasinu JV, Tuhumury GNC, 2018. Potential of Bacillus spp produces siderophores insuppressing thewilt disease of banana plants. IOP Conf Ser Earth Environ Sci, 102: 012016. 10.1088/1755-1315/102/1/012016 [DOI] [Google Scholar]
- Koutsoudis MD, Tsaltas D, Minogue TD, et al. , 2006. Quorum-sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii . Proc Natl Acad Sci USA, 103(15): 5983- 5988. 10.1073/pnas.0509860103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper I, Lagendijk EL, Pickford R, et al. , 2004. Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Mol Microbiol, 51(1): 97- 113. 10.1046/j.1365-2958.2003.03751.x [DOI] [PubMed] [Google Scholar]
- Lally RD, Galbally P, Moreira AS, et al. , 2017. Application of endophytic Pseudomonas fluorescens and a bacterial consortium to Brassica napus can increase plant height and biomass under greenhouse and field conditions. Front Plant Sci, 8: 2193. 10.3389/fpls.2017.02193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazdunski AM, Ventre I, Sturgis JN, 2004. Regulatory circuits and communication in Gram-negative bacteria. Nat Rev Microbiol, 2(7): 581- 592. 10.1038/nrmicro924 [DOI] [PubMed] [Google Scholar]
- Li SY, Xiao J, Sun TZ, et al. , 2022. Synthetic microbial consortia with programmable ecological interactions. Methods Ecol Evol, 13(7): 1608- 1621. 10.1111/2041-210x.13894 [DOI] [Google Scholar]
- Liao JX, Li ZH, Xiong D, et al. , 2023. Quorum quenching by a type IVA secretion system effector. ISME J, 17(10): 1564- 1577. 10.1038/s41396-023-01457-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HW, Brettell LE, Qiu ZG, et al. , 2020. Microbiome-mediated stress resistance in plants. Trends Plant Sci, 25(8): 733- 743. 10.1016/j.tplants.2020.03.014 [DOI] [PubMed] [Google Scholar]
- Lyng M, Kovács ÁT, 2023. Frenemies of the soil: Bacillus and Pseudomonas interspecies interactions. Trends Microbiol, 31(8): 845- 857. 10.1016/j.tim.2023.02.003 [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Fan XY, Wang Y, et al. , 2021. Bacterial seed endophyte shapes disease resistance in rice. Nat Plants, 7(1): 60- 72. 10.1038/s41477-020-00826-5 [DOI] [PubMed] [Google Scholar]
- McRose DL, Baars O, Morel FMM, et al. , 2017. Siderophore production in Azotobacter vinelandii in response to Fe-, Mo- and V-limitation. Environ Microbiol, 19(9): 3595- 3605. 10.1111/1462-2920.13857 [DOI] [PubMed] [Google Scholar]
- Molina-Santiago C, Pearson JR, Navarro Y, et al. , 2019. The extracellular matrix protects Bacillus subtilis colonies from Pseudomonas invasion and modulates plant co-colonization. Nat Commun, 10: 1919. 10.1038/s41467-019-09944-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller UG, Juenger TE, Kardish MR, et al. , 2021. Artificial selection on microbiomes to breed microbiomes that confer salt tolerance to plants. mSystems, 6(6): e0112521. 10.1128/mSystems.01125-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir N, Hanif M, Abideen Z, et al. , 2022. Mechanisms and strategies of plant microbiome interactions to mitigate abiotic stresses. Agronomy, 12(9): 2069. 10.3390/agronomy12092069 [DOI] [Google Scholar]
- Nakkeeran S, Surya T, Vinodkumar S, 2020. Antifungal potential of plant growth promoting Bacillus species against blossom blight of rose. J Plant Growth Regul, 39(1): 99- 111. 10.1007/s00344-019-09966-1 [DOI] [Google Scholar]
- Niu B, Paulson JN, Zheng XQ, et al. , 2017. Simplified and representative bacterial community of maize roots. Proc Natl Acad Sci USA, 114(12): E2450- E2459. 10.1073/pnas.1616148114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcengiz G, Öğülür İ, 2015. Biochemistry, genetics and regulation of bacilysin biosynthesis and its significance more than an antibiotic. New Biotechnol, 32(6): 612- 619. 10.1016/j.nbt.2015.01.006 [DOI] [PubMed] [Google Scholar]
- Pourbabaee AA, Shoaibi F, Emami S, et al. , 2018. The potential contribution of siderophore producing bacteria on growth and Fe ion concentration of sunflower ( Helianthus annuus L. ) under water stress. J Plant Nutr, 41(5): 619- 626. 10.1080/01904167.2017.1406112 [DOI] [Google Scholar]
- Purtschert-Montenegro G, Cárcamo-Oyarce G, Pinto-Carbó M, et al. , 2022. Pseudomonas putida mediates bacterial killing, biofilm invasion and biocontrol with a type IVB secretion system. Nat Microbiol, 7(10): 1547- 1557. 10.1038/s41564-022-01209-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JM, de Bruijn I, Nybroe O, et al. , 2010. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol Rev, 34(6): 1037- 1062. 10.1111/j.1574-6976.2010.00221.x [DOI] [PubMed] [Google Scholar]
- Rajkumar M, Prasad MNV, Swaminathan S, et al. , 2013. Climate change driven plant‒metal‒microbe interactions. Environ Int, 53: 74- 86. 10.1016/j.envint.2012.12.009 [DOI] [PubMed] [Google Scholar]
- Rändler-Kleine M, Wolfgang A, Dietel K, et al. , 2020. How microbiome approaches can assist industrial development of biological control products. In: Gao YL, Hokkanen HMT, Menzler-Hokkanen I (Eds.), Integrative Biological Control. Springer, Cham, p. 201- 215. 10.1007/978-3-030-44838-7_13 [DOI] [Google Scholar]
- Raza W, Wang JN, Jousset A, et al. , 2020. Bacterial community richness shifts the balance between volatile organic compound-mediated microbe‒pathogen and microbe‒plant interactions. Proc Roy Soc B Biol Sci, 287(1925): 20200403. 10.1098/rspb.2020.0403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca A, Pizarro-Tobías P, Udaondo Z, et al. , 2013. Analysis of the plant growth-promoting properties encoded by the genome of the rhizobacterium Pseudomonas putida BIRD-1. Environ Microbiol, 15(3): 780- 794. 10.1111/1462-2920.12037 [DOI] [PubMed] [Google Scholar]
- Romero-Perdomo F, Abril J, Camelo M, et al. , 2017. Azotobacter chroococcum as a potentially useful bacterial biofertilizer for cotton ( Gossypium hirsutum): effect in reducing N fertilization. Rev Argent Microbiol, 49(4): 377- 383. 10.1016/j.ram.2017.04.006 [DOI] [PubMed] [Google Scholar]
- Sandy M, Butler A, 2011. Chrysobactin siderophores produced by Dickeya chrysanthemi EC16. J Nat Prod, 74(5): 1207- 1212. 10.1021/np200126z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer M, Vogel CM, Bortfeld-Miller M, et al. , 2022. Mapping phyllosphere microbiota interactions in planta to establish genotype‒phenotype relationships. Nat Microbiol, 7(6): 856- 867. 10.1038/s41564-022-01132-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz-Bohm K, Gerards S, Hundscheid M, et al. , 2018. Calling from distance: attraction of soil bacteria by plant root volatiles. ISME J, 12(5): 1252- 1262. 10.1038/s41396-017-0035-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütze E, Ahmed E, Voit A, et al. , 2015. Siderophore production by streptomycetes—stability and alteration of ferrihydroxamates in heavy metal-contaminated soil. Environ Sci Pollut Res, 22(24): 19376- 19383. 10.1007/s11356-014-3842-3 [DOI] [PubMed] [Google Scholar]
- Singh M, Awasthi A, Soni SK, et al. , 2015. Complementarity among plant growth promoting traits in rhizospheric bacterial communities promotes plant growth. Sci Rep, 5: 15500. 10.1038/srep15500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song GC, Riu M, Ryu CM, 2019. Beyond the two compartments Petri-dish: optimising growth promotion and induced resistance in cucumber exposed to gaseous bacterial volatiles in a miniature greenhouse system. Plant Methods, 15: 9. 10.1186/s13007-019-0395-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutar CD, Stavrinides J, 2018. The evolution of three siderophore biosynthetic clusters in environmental and host-associating strains of Pantoea . Mol Genet Genomics, 293(6): 1453- 1467. 10.1007/s00438-018-1477-7 [DOI] [PubMed] [Google Scholar]
- Su P, Kang HX, Peng QZ, et al. , 2024. Microbiome homeostasis on rice leaves is regulated by a precursor molecule of lignin biosynthesis. Nat Commun, 15: 23. 10.1038/s41467-023-44335-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XL, Xu ZH, Xie JY, et al. , 2022. Bacillus velezensis stimulates resident rhizosphere Pseudomonas stutzeri for plant health through metabolic interactions. ISME J, 16(3): 774- 787. 10.1038/s41396-021-01125-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JQ, Kerstetter JE, Turcotte MM, 2021. Eco-evolutionary interaction between microbiome presence and rapid biofilm evolution determines plant host fitness. Nat Ecol Evol, 5(5): 670- 676. 10.1038/s41559-021-01406-2 [DOI] [PubMed] [Google Scholar]
- Tassinari M, Doan T, Bellinzoni M, et al. , 2022. The antibacterial type VII secretion system of Bacillus subtilis: structure and interactions of the pseudokinase YukC/EssB. mBio, 13(5): e0013422. 10.1128/mbio.00134-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge K, Inoue S, Ano T, et al. , 2005. Horizontal transfer of iturin A operon, itu, to Bacillus subtilis 168 and conversion into an iturin A producer. Antimicrob Agents Chemother, 49(11): 4641- 4648. 10.1128/aac.49.11.4641-4648.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi V, Bez C, 2021. A call to arms for cell‒cell interactions between bacteria in the plant microbiome. Trends Plant Sci, 26(11): 1126- 1132. 10.1016/j.tplants.2021.07.007 [DOI] [PubMed] [Google Scholar]
- Wang JJ, Xu S, Yang R, et al. , 2021. Bacillus amyloliquefaciens FH-1 significantly affects cucumber seedlings and the rhizosphere bacterial community but not soil. Sci Rep, 11: 12055. 10.1038/s41598-021-91399-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MC, Cernava T, 2023. Soterobionts: disease-preventing microorganisms and proposed strategies to facilitate their discovery. Curr Opin Microbiol, 75: 102349. 10.1016/j.mib.2023.102349 [DOI] [PubMed] [Google Scholar]
- Wei Z, Yang TJ, Friman VP, et al. , 2015. Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat Commun, 6: 8413. 10.1038/ncomms9413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise T, Kai M, Gummesson A, et al. , 2012. Volatile organic compounds produced by the phytopathogenic bacterium Xanthomonas campestris pv. vesicatoria 85-10. Beilstein J Org Chem, 8: 579- 596. 10.3762/bjoc.8.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SD, Liu YX, Cernava T, et al. , 2022. Fusarium fruiting body microbiome member Pantoea agglomerans inhibits fungal pathogenesis by targeting lipid rafts. Nat Microbiol, 7(6): 831- 843. 10.1038/s41564-022-01131-x [DOI] [PubMed] [Google Scholar]
- Yang RH, Shi Q, Huang TT, et al. , 2023. The natural pyrazolotriazine pseudoiodinine from Pseudomonas mosselii 923 inhibits plant bacterial and fungal pathogens. Nat Commun, 14: 734. 10.1038/s41467-023-36433-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannarell SM, Grandchamp GM, Chen SY, et al. , 2019. A dual-species biofilm with emergent mechanical and protective properties. J Bacteriol, 201(18): e00670-18. 10.1128/jb.00670-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin XT, Xu LN, Xu L, et al. , 2011. Evaluation of the efficacy of endophytic Bacillus amyloliquefaciens against Botryosphaeria dothidea and other phytopathogenic microorganisms. Afr J Microbiol Res, 5(4): 340- 345. 10.5897/AJMR10.679 [DOI] [Google Scholar]
- Yuan WF, Ruan S, Qi GF, et al. , 2022. Plant growth-promoting and antibacterial activities of cultivable bacteria alive in tobacco field against Ralstonia solanacearum . Environ Microbiol, 24(3): 1411- 1429. 10.1111/1462-2920.15868 [DOI] [PubMed] [Google Scholar]
- Yuan ZL, Druzhinina IS, Labbé J, et al. , 2016. Specialized microbiome of a halophyte and its role in helping non-host plants to withstand salinity. Sci Rep, 6: 32467. 10.1038/srep32467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng XY, Zou YM, Zheng J, et al. , 2023. Quorum sensing-mediated microbial interactions: mechanisms, applications, challenges and perspectives. Microbiol Res, 273: 127414. 10.1016/j.micres.2023.127414 [DOI] [PubMed] [Google Scholar]
- Zeriouh H, Romero D, García-Gutiérrez L, et al. , 2011. The iturin-like lipopeptides are essential components in the biological control arsenal of Bacillus subtilis against bacterial diseases of cucurbits. Mol Plant Microbe Interact, 24(12): 1540- 1552. 10.1094/MPMI-06-11-0162 [DOI] [PubMed] [Google Scholar]
- Zhang WL, Zhang Y, Wang XX, et al. , 2017. Siderophores in clinical isolates of Klebsiella pneumoniae promote ciprofloxacin resistance by inhibiting the oxidative stress. Biochem Biophys Res Commun, 491(3): 855- 861. 10.1016/j.bbrc.2017.04.108 [DOI] [PubMed] [Google Scholar]
- Zhang XX, Ma YN, Wang X, et al. , 2022. Dynamics of rice microbiomes reveal core vertically transmitted seed endophytes. Microbiome, 10: 216. 10.1186/s40168-022-01422-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YQ, Wang HK, Xu SD, et al. , 2022. Bacterial‒fungal interactions under agricultural settings: from physical to chemical interactions. Stress Biol, 2: 22. 10.1007/s44154-022-00046-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SB, Hong JK, Wang T, 2024. Horizontal gene transfer is predicted to overcome the diversity limit of competing microbial species. Nat Commun, 15: 800. 10.1038/s41467-024-45154-w [DOI] [PMC free article] [PubMed] [Google Scholar]