Abstract
The occurrence of an adverse drug event (ADE) has become a serious social concern of public health. Early detection of ADEs can lower the risk of drug safety as well as the expense of the drug. While post-market spontaneous reports of ADEs remain a cornerstone of pharmacovigilance, most existing signal detection algorithms rely on substantial accumulated data, limiting their applicability to early ADE detection when reports are scarce. To address this issue, we propose a label propagation model for generating enhanced drug safety signals using multiple drug features. We first construct multiple drug similarity networks using a range of drug features. We then calculate initial drug safety signals using conventional signal detection algorithms. These original signals are subsequently propagated across each drug similarity network to obtain enhanced drug safety signals. We evaluate our proposed model using two common signal detection algorithms on data from the FDA Adverse Event Reporting System (FAERS). Results demonstrate that enhanced drug safety signals with pre-clinical information outperform the standard safety signal detection algorithms on early ADE detection. In addition, we systematically evaluate the performance of different drug similarities against different types of ADEs. Furthermore, we have developed a web interface (http://drug-drug-sim.aimedlab.net/) to display our multiple drug similarity scores, facilitating access to this valuable resource for drug safety monitoring.
1. Introduction
Adverse drug events (ADEs) refer to unexpected and harmful reactions that occur when medications are used as intended. ADEs have been a persistent challenge in the medical and healthcare communities due to their significant medical and financial burden on patients, and in rare cases, can result in death [1], [2]. ADEs become the fourth leading cause of death in the United States, exceeding serious diseases such as pulmonary disease, diabetes and AIDS etc [3]. Each year, over two million major ADEs occur in patients, leading to approximately 100,000 deaths [2]. As a consequence, ADE detection is crucial for drug safety. Early detection of potential ADEs associated with drug candidates during the initial clinical phases of drug development can mitigate risks for patients and reduce hospital costs.
However, pre-marketing clinical studies often fail to fully detect adverse events due to their limited cohort sizes, short durations, and lack of patient diversity [4], [5]. As a result, most ADEs are revealed only after the drugs have been marketed. In contrast, post-marketing monitoring, with its larger sample sizes and longer observation periods, allows for a more comprehensive discovery of ADEs.
Spontaneous reporting systems (SRSs) [6] are regulatory agencies-developed mechanisms for monitoring drug safety throughout the post-marketing period. SRSs are gathered from a range of sources, including healthcare professionals, governmental agencies, pharmaceutical firms, medical literature, and direct patient contact. With an abundance of valuable data, SRSs are critical in delivering early warnings about suspected ADEs. A number of signal detection techniques have been proposed for detecting drug safety signals in SRSs. The most widely used methods are Proportional Reporting Rate (PRR) [7] and Reporting Odds Ratio (ROR) [8], [9], both of which are based on the ADEs' most frequent statistical analysis. Bayesian approaches are also popular in signal detection files, the most common methods are Bayesian Confidence Propagation Neural Network (BCPNN) [10] and Multi-item Gamma Poisson Shrinker (MGPS) [11]. Drug safety signals (also called signal scores), calculated using these signal detection techniques, are surrogate measures of statistical relationships between drug-ADE pairs, with higher scores indicating stronger associations [6].
Recently, researchers have explored a variety of strategies to improve the detection of ADEs. Most existing methods are designed to generate signals and/or re-rank original signals for drugs with sufficient reports in SRS. For example, Vilar et al. [12], [13], [14], [15] predict the adverse effects of pharmacological compounds based on their chemical structure and additional biological features. To address this, one recent study [16] is proposed to generate safety signals for newly approved drugs with few or no safety reports in SRS. Specifically, they develop a label propagation framework that enhances drug safety signals by combining drug chemical structures with data from the FDA Adverse Event Reporting System (FAERS), allowing for the early identification of potential ADEs for newly approved drugs compared to conventional signal detection methods. However, their study primarily focuses on a single source of drug features, which may limit the scalability and comprehensiveness of ADE detection.
To date, a variety of pre-clinical drug property data has been accumulated and is readily accessible. Given that this data from various sources contains a diverse range of properties, it is crucial to comprehensively evaluate different drug features (including chemical, biological, and phenotypic characteristics) for ADE detection. Thus, in this paper, we leverage these multiple drug features to enhance ADE detection. Our approach consists of three main steps. First, we compute the original FAERS-based drug safety signals using conventional signal detection algorithms. Next, we construct drug-drug similarity networks based on a range of drug characteristics, including target sequences, ATC codes, chemical structures, and GO annotations. Finally, we apply a label propagation operation on each similarity network separately to obtain enhanced drug safety signals from the original signal scores.
Overall, our contributions are as follows:
-
•
We propose a label propagation framework utilizing multiple drug similarity networks to generate enhanced drug safety signals for the early detection of ADEs.
-
•
We compare different drug similarity networks for enhancing drug safety signal generation and demonstrate that the proposed framework outperforms existing safety signal detection methods.
-
•
Furthermore, we group ADEs into their respective MedDRA System Organ Classes (SOCs) and compare the performance of the proposed framework on the top 10 most frequent SOCs.
-
•
We perform a case study on two drug-ADE pairs to demonstrate the efficacy of different similarities on different ADE mechanisms.
-
•
We have also developed a website to display the drug similarity scores, providing a valuable resource for drug safety monitoring.
2. Overall framework
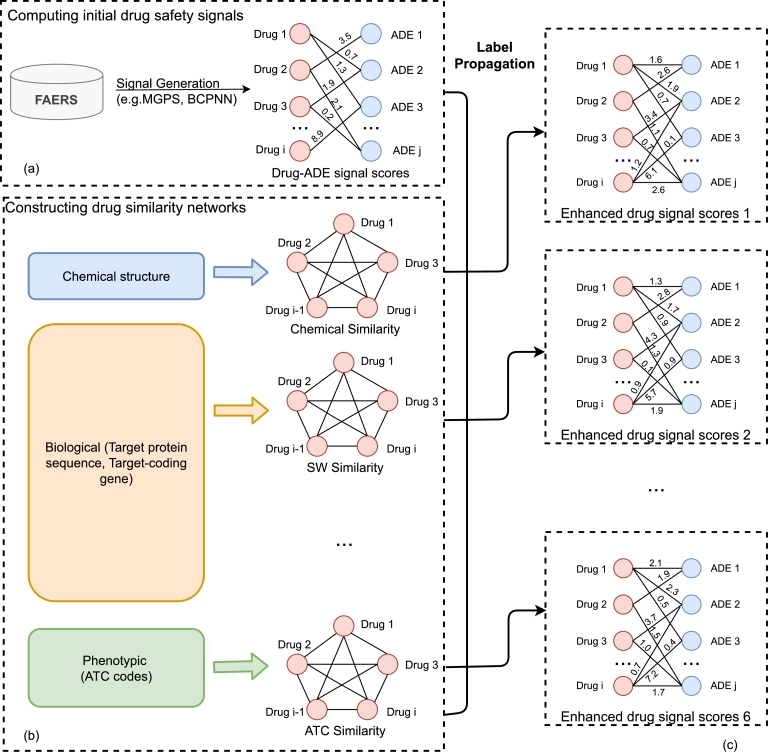
Fig. 1 shows the overall framework of this project. The entire workflow is divided into 3 parts. First, the original signal scores for Drug-ADE pairs from the cumulative FAERS dataset are calculated using the MGPS and BCPNN algorithms. Then we created the 6 unique drug similarity networks using various characteristics of the drugs. Finally, the enhanced drug safety signal scores are generated by combining the original scores and similarity networks using a label propagation method.
Figure 1.
Overall framework of label propagation using multiple similarity networks. The method includes three parts: (a) Original drug safety signal score computation, (b) Multiple similarity network construction, and (c) Enhanced drug safety signal score generation with similarity networks.
3. Results
3.1. Dataset
FAERS database. Because the terms used in the FAERS database [17] are determined by the reporter, inaccurate descriptions may be included, we used the standardized FAERS data from 2004 to 2022 [18]. They curated and standardized the FAERS database entries using the preferred term (PT) from the Medical Dictionary for Regulatory Activity (MedDRA) [19]. We extracted 2872 individual drugs and 20,772 ADEs, resulting in a total of 4,207,448 unique drug-ADE combinations. Fig. 2 displays a histogram depicting the number of drugs associated with each ADE. This histogram is divided into 50 bins of size ≈48 which indicates ≈9200 ADEs drugs are associated with 1 to 48 drugs, ≈2600 ADEs are associated with 49 to 96 drugs with subsequent bins representing higher drug counts. The vertical dashed line shows the mean of the distribution that indicates on average 203 drugs are associated with each ADE. Similarly, the histogram in Fig. 3 shows the number of ADEs associated with each drug. In this figure the there are also 50 bins each covering ≈246 ADEs. Here, ≈1000 drugs are associated with 1 to 246 ADEs, ≈350 drugs are associated with 246 to 492 ADEs and so on. the dashed lines shows on average 1465 ADEs are associated with each drug. Both of these histograms show a highly right-skewed distribution, which represents that a small number of drugs are linked to a large amount of ADEs and vice-versa.
Figure 2.
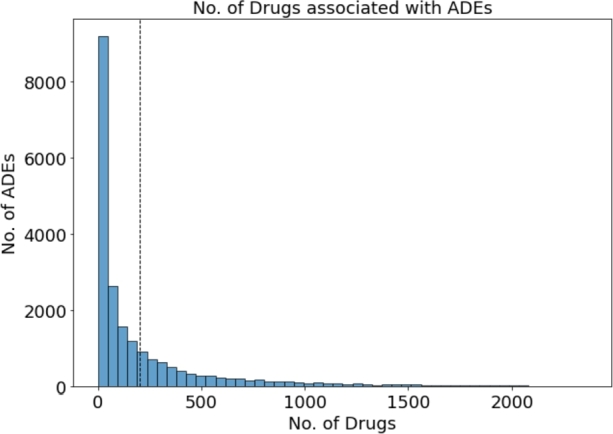
Frequency of no. of drugs associated with ADEs.
Figure 3.
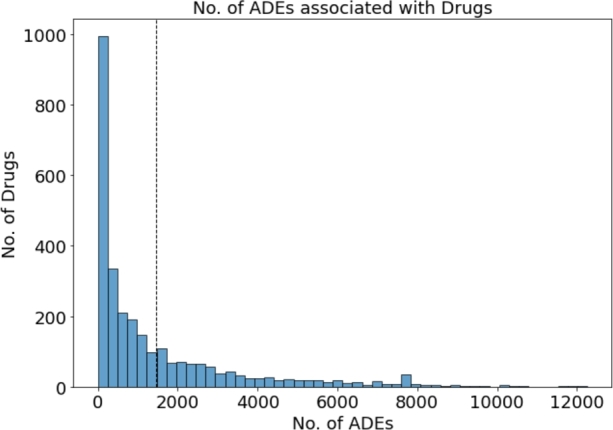
Frequency of no. of ADEs associated with drugs.
Drugbank database. Drugbank [20] is an open and free drug database that contains a great deal of information on drugs (e.g. target, chemical properties, pharmacology, toxicology). Each drug in this database has its own unique identifier, Drugbank_ID. SMILES annotation is used to represent the chemical structure. The uniprot_ID of the target is retrieved from the Uniprot database. To measure similarities, the chemical structure, sequence of targets, Go words for targets, and ATC code were used as features of the medications.
SIDER database. The side effects were collected from the Side Effect Resource (SIDER) database [21]. The SIDER database currently contains 4251 adverse reactions to 1344 drugs, totaling 152,277 drug-ADE pairs. A total of 643 drugs with all the characteristics (chemical structure, ATC code, sequence, Go terms) were mapped from the Drugbank database to the SIDER database. Both the Preferred Term (PT) and Lowest Level Term (LLT) of MedDRA are used to record ADEs in SIDER. We only selected the Preferred Term(PT) as our ADEs for evaluation.
3.2. Experiment setup
As positive controls, we used known drug-ADE pairs from the SIDER data set and unknown drug-ADE pairs as negative controls. Due to a scarcity of positive samples, we create evaluation datasets that contain the drugs present in both the drug similarity matrix and SIDER, and the top 1000 most frequent ADEs from FAERS data. As the six proposed drug similarity matrices have different numbers of drugs, we generate separate evaluation datasets for each one of them. We evaluate our proposed framework against two baselines (MGPS, BCPNN). We chose MGPS and BCPNN as our baseline because they are not affected by the sampling variance issue and are considered more reliable by the FDA [22]. As evaluation can be viewed as a binary classification job in this work, we used the Area Under the Curve (AUC), the Area Under the Reaction-Recall Curve (AUPR), and the F1 score. The AUC score represents the true positive rate (TPR) and false positive rate (FPR). The TPR and FPR can be computed as follows:
| (1) |
AUPR can also be calculated using the precision and recall scores, which illustrates the trade-off between precision and recall at various decision thresholds. Precision may be defined as the probability that the output safety signal score is accurate. The possibility of true safety signals being assessed as outputs may be determined using recall. The definition of precision and recall is shown in Eq. (2).
| (2) |
The F1 score is the harmonic mean of the precision and recall:
| (3) |
Furthermore, we divide the datasets into validation (80%) and test (20%) sets. The validation set is used to calculate the optimal value of the model parameter for the Label-Propagation framework. Then we run the best performing model on the test set and compare its result with results obtained by the baseline methods on the same test set.
3.3. Performance assessment
Performance among all ADEs. To evaluate the proposed method, we first compared it to two baseline reference methods (MGPS, and BCPNN) on all-year data (from 2004 to 2022). As shown in Table 1, Table 2, BCPNN outperforms MGPS in terms of both AUC and AUPR. On AUC, AUPR, and F1 scores, our strategy surpasses the corresponding baseline methods. Overall, our method can improve AUC score by about , and AUPR score by about compared with all the baseline methods. The results show that drug-drug similarity helps enhance safety signals, as similar drugs may cause the same adverse effects. The method incorporates information from similar drugs and improves the original drug safety signal.
Table 1.
Comparison between different label propagation framework and corresponding baseline methods using MGPS signal scores on all years reports.
| Score | Baseline | LP-ATC | LP-Chem | LP-Seq | LP-GO_BP | LP-GO_CC | LP-GO_MF |
|---|---|---|---|---|---|---|---|
| AUC | 0.743 | 0.753 | 0.759 | 0.759 | 0.752 | 0.756 | 0.755 |
| AUPR | 0.226 | 0.231 | 0.236 | 0.242 | 0.243 | 0.246 | 0.246 |
| F1 | 0.319 | 0.322 | 0.330 | 0.335 | 0.335 | 0.339 | 0.339 |
Table 2.
Comparison between different label propagation framework and corresponding baseline methods using BCPNN signal scores on all years reports.
| Score | Baseline | LP-ATC | LP-Chem | LP-Seq | LP-GO_BP | LP-GO_CC | LP-GO_MF |
|---|---|---|---|---|---|---|---|
| AUC | 0.743 | 0.763 | 0.786 | 0.784 | 0.764 | 0.776 | 0.775 |
| AUPR | 0.224 | 0.253 | 0.280 | 0.293 | 0.263 | 0.290 | 0.289 |
| F1 | 0.319 | 0.332 | 0.371 | 0.378 | 0.350 | 0.378 | 0.378 |
Performance among different similarity networks. The results shown in Table 1, demonstrate that all the similarity network has similar results with MGPS signal scores on cumulated all-year data. However, there is more difference among the similarity network results with BCPNN data (Table 2). Here, LP-Chem achieves the highest AUC of 0.759 and 0.786 using MGPS and BCPNN signal scores respectively. LP-Seq also achieves a comparable AUC of 0.759 and 0.784 with the corresponding signal scores.
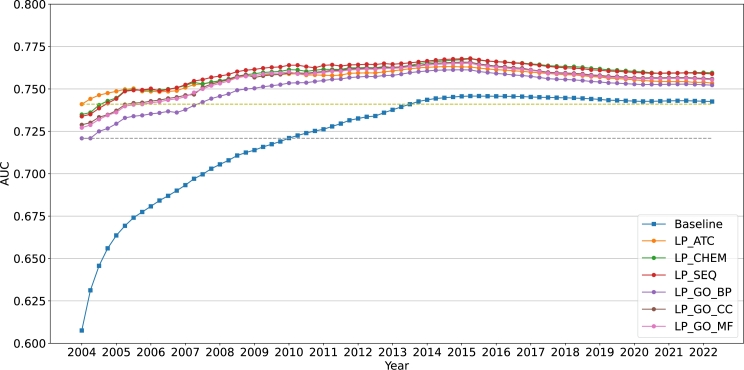
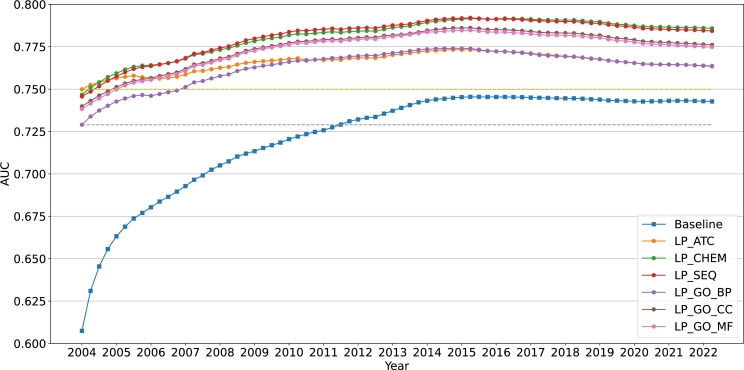
Additionally, we presented the annual change curve for MGPS and BCPNN using the AUC scores (shown in Figure 4, Figure 5). These graphs show that in general, preclinical drug similarities enhance ADE detection - as early as 10 years. The dashed reference lines show how long it takes the baseline to reach the AUC scores obtained by our enhanced safety scores in 2004 Q1, which is the first quarter in our FAERS dataset. The bottom dashed line compares the baseline to the worst performing LP score whereas the top dashed line compares it to the best performing LP-score. In the case of MGPS, the baseline reaches the bottom reference line in 6 years and the top reference line in almost 10 years. In the case of BCPNN, it takes the baseline over 7 years to attain similar performance as the lower reference line, and it never achieves as high AUC as the upper reference line.
Figure 4.
Comparison of the different methods based on MGPS on yearly cumulative reports.
Figure 5.
Comparison of the different methods based on BCPNN on yearly cumulative reports.
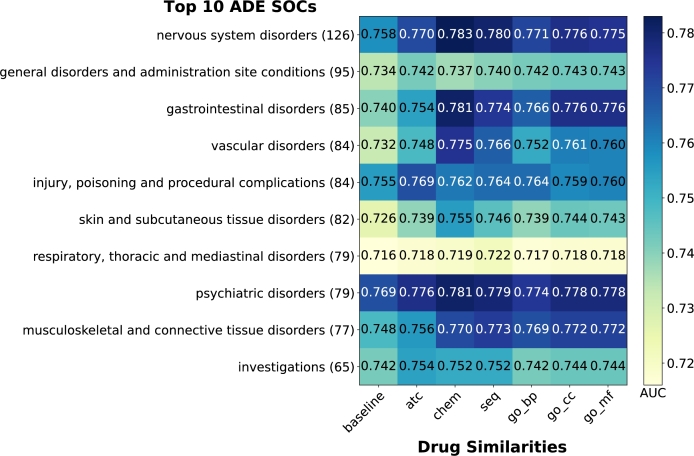
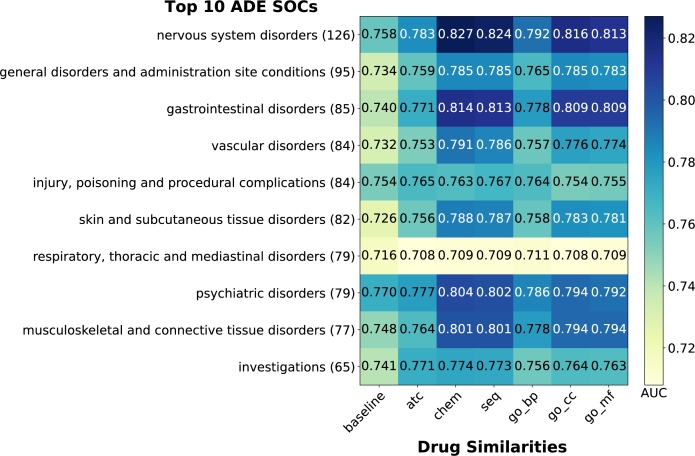
Performance among top 10 most frequent MedDRA SOC classes. The MedDRA vocabulary is divided into 27 non-mutually exclusive System Organ Classes (SOCs). These SOCs are further divided into High Level Group Term (HLGT), then into High Level Term (HLT), then Preferred Term (PT), and lastly Low Level Term (LLT). To investigate the performance of our proposed model, we grouped PT ADEs in our result into their corresponding SOCs and calculated AUC for the top 10 most frequent SOCs based on cumulated FAERS data.
The heatmaps in Fig. 6 and Fig. 7 demonstrate that all LP scores outperform baselines in both MGPS and BCPNN methods, with one exception: the SOC “Respiratory, Thoracic and Mediastinal disorders” using BCPNN. For MGPS, for MGPS, the SOC “Nervous System Disorders” achieves the highest AUC of 0.783 using LP-Chem. LP-Chem performs best for 6 out of 10 SOCs, while LP-ATC performs best for 3 SOCs. BCPNN shows similar patterns to MGPS, albeit with higher overall AUC scores. The SOC “Nervous System Disorders” again achieves the highest AUC of 0.827 using LP-Chem. LP-Chem performs best for 8 out of 10 SOCs, while LP-Seq performs best for 1 out of 10 SOCs.
Figure 6.
Comparison of AUC achieved using the different methods based on MGPS on top 10 most frequent MedDRA SOCs. (#) at the end of SOC names in the y-axes denotes the number of unique MedDRA PTs from our filtered SIDER dataset mapped to each SOC.
Figure 7.
Comparison of AUC achieved using the different methods based on BCPNN on top 10 most frequent MedDRA SOCs. (#) at the end of SOC names in the y-axes denotes the number of unique MedDRA PTs from our filtered SIDER dataset mapped to each SOC.
These results highlight the consistent superiority of LP-Chem across both methods. “Respiratory, Thoracic and Mediastinal disorders” achieves the lowest AUC among the SOCs for all LP and baseline scores in both methods.
3.4. Website
We designed a website (http://drug-drug-sim.aimedlab.net/) to display our drug similarity scores obtained using the methods described in the methodologies section.
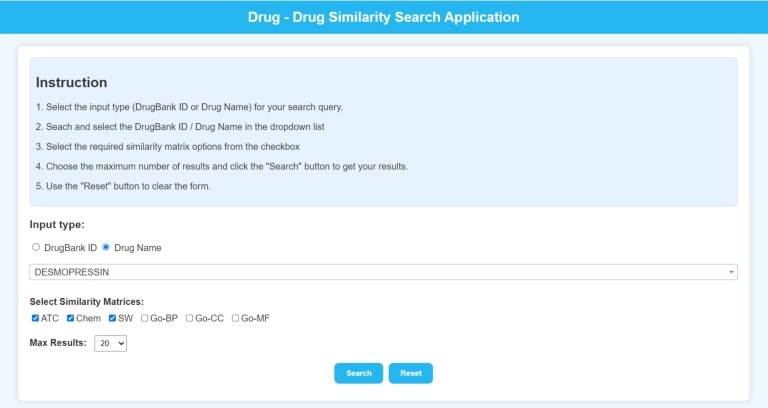
The home page, shown in Fig. 8, presents a user-friendly interface for initiating a drug search. Users can choose to search by either DrugBank ID (DB ID) or Drug Name. After that, users can select the drug of choice in the searchable drop-down list which dynamically updates based on the user input. The search results can be further refined by selecting required similarity matrices from the 6 options, i.e., ATC, Chem, Seq, Go-BP, Go-CC, and Go-MF. Users also have the option to specify the maximum number of results they wish to see.
Figure 8.
Landing page of the website.
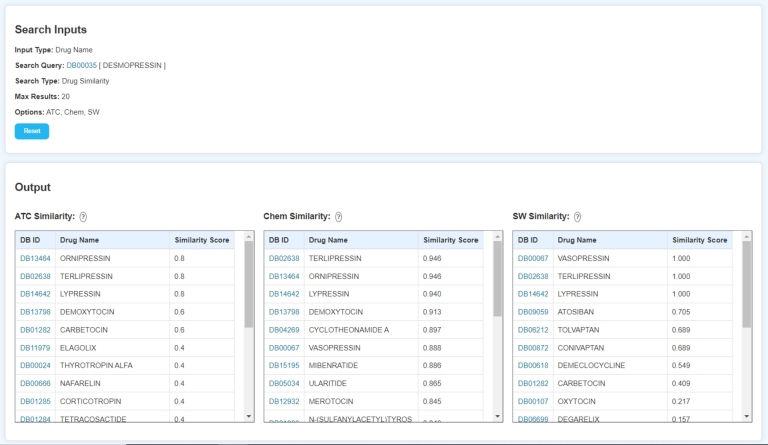
Fig. 9 displays the result page of the website, which provides a comprehensive overview of the search results. In this example, a search was performed for the drug “DESMOPRESSIN” using the Drug Name as the input type. The search utilized the ATC, Chem, and Seq similarity matrices, with a maximum of 20 results requested. The results are presented in a clear, tabular format representing the selected similarity matrices. Each table is divided into 3 columns that displays the DB ID, Drug Name, and a corresponding Similarity Score. The drugs in the similarity tables are sorted in descending order of their similarity scores. Every DB ID in the result page provides a hyperlink to the corresponding page for that drug on the DrugBank website. Additionally, the question mark icon beside the similarity names provide the formula used to calculate those similarities.
Figure 9.
Result page of the website.
4. Discussion
In this paper, we explore a label propagation based on an approach for early detection of adverse effect caused by drugs using drug similarities and post-market spontaneous adverse effect reports. We calculate six drug similarity metrics using various drug characteristics such as chemical structure, target protein sequence, target coding gene and phenotype. Additionally, we generate baseline drug safety signal scores from the FAERS quarterly reports using MGPS and BCPNN. After that, we employ the proposed label propagation framework to generate the enhanced drug safety signal scores using the drug similarities and baseline similarity scores. Lastly, we compare the performance of our enhanced drug safety signal scores with the baseline. We use SIDER side effects database as our positive controls. As demonstrated in the previous section our proposed method outperforms the baseline scores.
Statistical significance of model performance To demonstrate that the differences between the results of the proposed label propagation methods and the baseline methods are statistically significant, we conduct a paired t-test on the AUC scores of the models and calculate the p-value. Table 3 shows the p-value of the six label propagation models with different drug similarities against their respective baselines. We consider 0.05 to be our threshold for this case. As all the p-values in Table 3 is less than the threshold improvement in performance achieved by the label propagation models are statistically significant.
Table 3.
P-values for AUC comparisons between label propagation models against baseline methods.
| Model | MGPS | BCPNN |
|---|---|---|
| LP-ATC | 6.945 × 10−17 | 5.299 × 10−23 |
| LP-Chem | 1.125 × 10−20 | 4.576 × 10−38 |
| LP-Seq | 4.466 × 10−21 | 1.979 × 10−38 |
| LP-GO_BP | 1.111 × 10−17 | 1.185 × 10−26 |
| LP-GO_CC | 6.039 × 10−20 | 9.025 × 10−35 |
| LP-GO_MF | 9.950 × 10−20 | 3.861 × 10−34 |
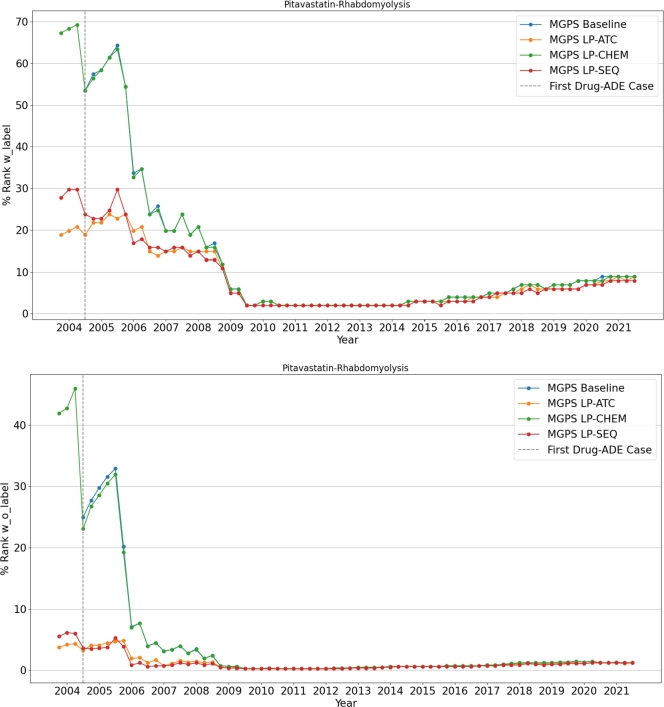
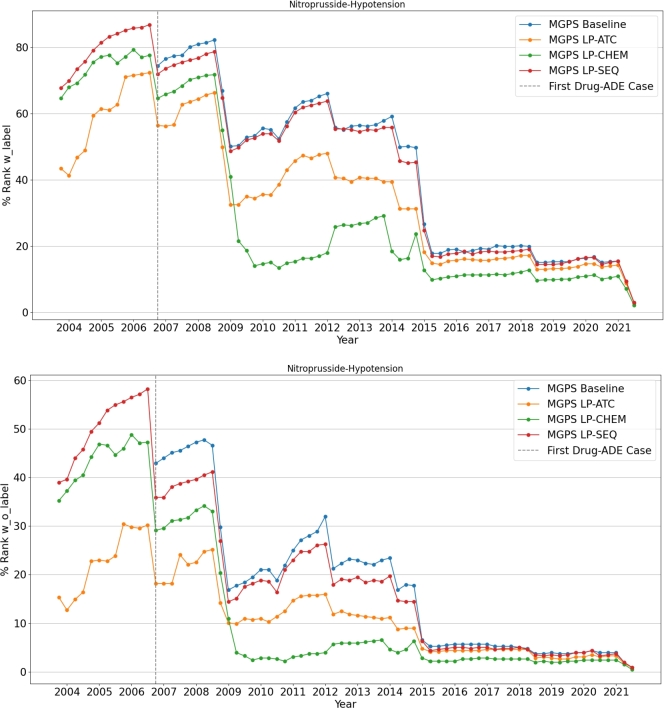
Case study We would like to discuss two drug-ADE pairs documented by drug labels: pitavastatin - rhabdomyolysis (Fig. 10) and nitroprusside - hypotension (Fig. 11). The mechanism of pitavastatin-induced rhabdomyolysis remains unclear and may involve complex mechanisms such as mitochondrial dysfunction or autoimmunity [23],[24]. Nitroprusside can naturally trigger hypotension, as the drug acts as a blood pressure lowering agent [25]. In our analysis, we identify protein sequence-based similarity has the best performance on prioritizing pitavastatin - rhabdomyolysis and chemical structure-based similarity has the best performance on prioritizing nitroprusside - hypotension. Given these observations, we hypothesize that an ADE with complex mechanisms could be better characterized by protein sequence-based similarity, and an ADE related to the mechanism of action of the corresponding drug could be better characterized by chemical similarity. Further studies are warranted to investigate these hypotheses.
Figure 10.
Quarterly change in the rank of Pitavastatin-Rhabdomyolysis Drug-ADE pair.
Figure 11.
Quarterly change in the rank of Nitroprusside-Hypotension Drug-ADE pair.
The quarter by quarter change in the ranks of these two drug-ADE pairs among all drugs from SIDER with and without labels are shown in Fig. 10 and Fig. 11. To calculate the rank, all drugs are divided into two categories based on their ground truth labels in relation to the target ADE. If a drug and target ADE pair exists in SIDER, the drug is classified as “with label”; otherwise, it is classified as “without label”. The “rank with label” for a drug-target ADE pair in a given quarter is determined by its position in a descending list of signal scores for all “with label” drug-target ADE pairs present in the FAERS data for that quarter. Similarly, the “rank without label” for a drug-target ADE pair in a given quarter is determined by its position in a descending list of signal scores for all “without label” drug-target ADE pairs present in the FAERS data for that quarter. These ranks are then normalized into a percentage by dividing it by the total number of drugs in their respective lists and multiplying by one hundred. The vertical dashed lines in the figures indicate the quarter when the first case of the specified drug-ADE pair was reported.
5. Methodology
5.1. Drug similarity network construction
We implement several drug similarity networks with different types of biological entities. For drug features, we use molecular structure and ATC code to calculate drug similarity. For drug-related target features, sequence and Go terms are utilized to measure the drug similarity.
Chemical structure similarity. We extract chemical structure information from the DrugBank database and the chemical structure similarity is constructed based on Pubchem fingerprint [26]. PubChem defines 881 chemical substructures, so an element can describe a drug in 881 dimensions, with each substructure having a value of 1 or 0, indicating its presence or absence. The chemical similarity of a drug-drug pair is calculated by the Jaccard Score using the vector form of chemical fingerprints.
| (4) |
where A and B signify two distinct drug profiles. In our work, we compute the chemical structure similarity among all the drugs with small molecules from the Drugbank database. Then we got a matrix of size .
ATC code-based similarity. ATC codes organize drugs hierarchically by organ or system on which they work, therapeutic impact, and chemical properties. All FDA-approved drugs have ATC codes that may be derived from the public database Drugbank. In this paper, we define the kth level ATC code based similarity of drug a and drug b as
| (5) |
where represents all ATC codes from 1st to kth level. Then the similarity score is defined as follows:
| (6) |
where n represents the five levels of ATC codes and ranges from 1 to 5. In this paper, we totally computed similarities among 3224 drugs.
Sequence based similarity. We extract all target proteins for each drug from the Drugbank database. The similarity of drug targeted protein sets A and B for a given pair of drugs is calculated as follows:
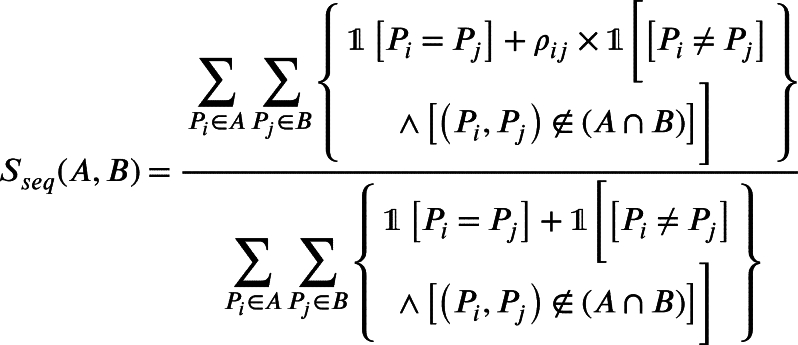 |
(7) |
Where and are the protein sequences from set A and B respectively, is the similarity between and and denotes the indicator function. ρ is calculated using R Biostrings package [27]. We got the similarity network of size .
Go-term based similarity. All pharmacological target-coding genes have their Gene Ontology (GO) annotation [28]. Biological processes (BP), molecular function (MF), and cellular component (CC) are three categories of empirically proven or literature-driven evidence that we employ. Semantic comparison of GO annotations enables comparisons of genes and gene products. In this paper, we use an R package, named GOSemSim [29], to compute the similarity between two drug-targeted genes.
If A and B are two sets of genes targeted by a drug pair and , respectively. The GO similarity of the drug pair is determined by Equation (8). Here is the similarity between genes and which is calculated using GOSemSim. For the three types, we calculate the similarity of 700 drugs.
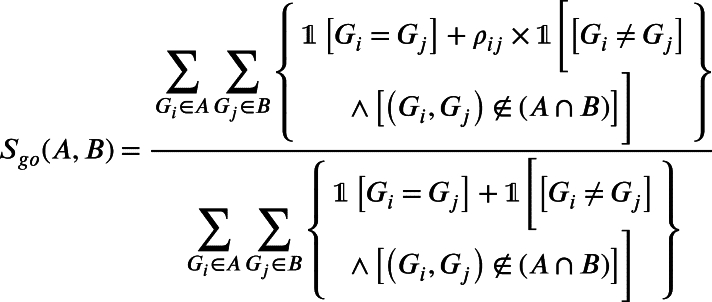 |
(8) |
5.2. Regenerate drug safety signal scores
Inspired by the label propagation method [30], the original signals are propagated on each drug similarity network. We use MGPS and BCPNN as baseline methods for generating the original safety signals.
The initial label for the drug is the kth row of the original signal scores matrix S, . Then, we used the Bregmanian Bi-Stochastication algorithm to normalize the drug similarity matrix A, ensuring that both the row and column sums are equal to one.
As with the initial label propagation based on the graph's weighted edges, the second step recursively propagates labels from nodes with labels to nodes without labels using W. During each iteration, each node's label information is updated by absorbing labels from neighbors with a probability (γ) and maintaining prior labels with a probability . From step to step t, the revised label information for a drug node i can be used as follows:
| (9) |
where denotes the tth iteration's updated label information for drug node i.
After t iteration, for all nodes, the label information is denoted as:
| (10) |
Apparently, , the spectral radius , and . Therefore, we get the updated label information for drugs, which is denoted as:
| (11) |
Note that I is an identity matrix, and S is the matrix for initial label information. Actually, the coverage solution can also be written as follows:
| (12) |
where signifies the matrix's trace and denotes the matrix's Frobenius norm. The first term is a smoothness term, which indicates consistency throughout the intrinsic network, resulting in a smooth change in the enhanced signal score for each reference drug-ADE pair. This is consistent with our hypothesis that similar drugs have similar adverse drug reactions (ADEs). The second term is a fitting term that stipulates that the increased signal scores must be near to their initial values. The parameter γ is used to arbitrate between these two contradictory terms. Due to the fact that the formula for Y is convex, we can derive the global optimal solution by fixing the first derivative of J with respect to Y to 0, retaining .
As for the parameter γ, we run the label propagation with γ from . The final model is built with γ that yields the maximum AUC score. For a detailed comparison of performance for all γ values, refer to Table A1 and Table A2.
Code availability
The source code for this paper can be downloaded from the GitHub repository at: https://github.com/BiswajitPadhi99/ADE-prediction-using-Drug-Similarities
Additional information
Correspondence and requests for materials should be addressed to PeZ and PiZ.
CRediT authorship contribution statement
Biswajit Padhi: Investigation, Methodology, Software, Validation, Visualization, Writing – original draft. Ruoqi Liu: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Supervision, Writing – original draft, Project administration, Writing – review & editing. Yuedi Yang: Data curation, Investigation, Methodology. Xueqiao Peng: Data curation, Writing – original draft. Lang Li: Conceptualization, Funding acquisition, Investigation, Writing – review & editing. Pengyue Zhang: Conceptualization, Formal analysis, Funding acquisition, Supervision, Writing – review & editing. Ping Zhang: Conceptualization, Data curation, Formal analysis, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded in part by the National Institutes of Health (NIH) under award number R01GM141279. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Supplementary material related to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e39728.
Contributor Information
Pengyue Zhang, Email: zhangpe@iu.edu.
Ping Zhang, Email: zhang.10631@osu.edu.
Appendix A. Supplementary material
The following is the Supplementary material related to this article.
Comparison of results for different values of model parameter (gamma). To generate the label propagated signal scores we use a probability parameter γ as outlined in equation 12, which is discussed in detail in subsection 5.2. We calculate the AUC of the label propagation methods for γ ranging from 0.1 to 0.9, in increments of 0.1. We perform this step separately for all six drug similarity matrices to find the optimal value of γ that achieves the maximum AUC score for each matrix.
Data availability
The quarterly drug-ADE reports used in this work was downloaded from the FAERS website. The side effects data was collected from the SIDER website. Additional drug information was extracted from DrugBank. The six drug similarity matrices used in this paper are available at: https://zenodo.org/records/13270611. We performed further data processing to filter and prepare these datasets for our use.
References
- 1.Eicher T., et al. Metabolomics and multi-omics integration: a survey of computational methods and resources. Metabolites. 2020;10:202. doi: 10.3390/metabo10050202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lazarou J., Pomeranz B.H., Corey P.N. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA. 1998;279:1200–1205. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- 3.Giacomini K.M., et al. When good drugs go bad. Nature. 2007;446:975–977. doi: 10.1038/446975a. [DOI] [PubMed] [Google Scholar]
- 4.Rosen A.C., et al. Impact of dermatologic adverse events on quality of life in 283 cancer patients: a questionnaire study in a dermatology referral clinic. Am. J. Clin. Dermatol. 2013;14:327–333. doi: 10.1007/s40257-013-0021-0. [DOI] [PubMed] [Google Scholar]
- 5.Sultana J., Cutroneo P., Trifirò G. Clinical and economic burden of adverse drug reactions. J. Pharmacol Pharmacother. 2013;4:S73. doi: 10.4103/0976-500X.120957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harpaz R., et al. Performance of pharmacovigilance signal-detection algorithms for the fda adverse event reporting system. Clin. Pharmacol. Ther. 2013;93:539–546. doi: 10.1038/clpt.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans S.J., Waller P.C., Davis S. Use of proportional reporting ratios (prrs) for signal generation from spontaneous adverse drug reaction reports. Pharmacoepidemiol. Drug Saf. 2001;10:483–486. doi: 10.1002/pds.677. [DOI] [PubMed] [Google Scholar]
- 8.Rothman K.J., Lanes S., Sacks S.T. The reporting odds ratio and its advantages over the proportional reporting ratio. Pharmacoepidemiol. Drug Saf. 2004;13:519–523. doi: 10.1002/pds.1001. [DOI] [PubMed] [Google Scholar]
- 9.Waller P., Van Puijenbroek E., Egberts A., Evans S. The reporting odds ratio versus the proportional reporting ratio: ‘deuce’. Pharmacoepidemiol. Drug Saf. 2004;13:525–526. doi: 10.1002/pds.1002. [DOI] [PubMed] [Google Scholar]
- 10.Bate A., et al. A Bayesian neural network method for adverse drug reaction signal generation. Eur. J. Clin. Pharmacol. 1998;54:315–321. doi: 10.1007/s002280050466. [DOI] [PubMed] [Google Scholar]
- 11.DuMouchel W. Bayesian data mining in large frequency tables, with an application to the fda spontaneous reporting system. Am. Stat. 1999;53:177–190. [Google Scholar]
- 12.Vilar S., et al. Facilitating adverse drug event detection in pharmacovigilance databases using molecular structure similarity: application to rhabdomyolysis. J. Am. Med. Inform. Assoc. 2011;18:i73–i80. doi: 10.1136/amiajnl-2011-000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vilar S., Harpaz R., Santana L., Uriarte E., Friedman C. Enhancing adverse drug event detection in electronic health records using molecular structure similarity: application to pancreatitis. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0041471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vilar S., et al. Similarity-based modeling applied to signal detection in pharmacovigilance. CPT: Pharmacometr. Syst. Pharmacol. 2014;3:1–9. doi: 10.1038/psp.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vilar S., Tatonetti N.P., Hripcsak G. 3d pharmacophoric similarity improves multi adverse drug event identification in pharmacovigilance. Sci. Rep. 2015;5:1–9. doi: 10.1038/srep08809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu R., Zhang P. Towards early detection of adverse drug reactions: combining pre-clinical drug structures and post-market safety reports. BMC Med. Inform. Decis. Mak. 2019;19:1–9. doi: 10.1186/s12911-019-0999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.https://open.fda.gov/data/faers/ Fda's adverse event reporting system (faers), [EB/OL]
- 18.Banda J.M., et al. A curated and standardized adverse drug event resource to accelerate drug safety research. Sci. Data. 2016;3:1–11. doi: 10.1038/sdata.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown E.G., Wood L., Wood S. The medical dictionary for regulatory activities (meddra) Drug Safety. 1999;20:109–117. doi: 10.2165/00002018-199920020-00002. [DOI] [PubMed] [Google Scholar]
- 20.Wishart D.S., et al. Drugbank 5.0: a major update to the drugbank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn M., Campillos M., Letunic I., Jensen L.J., Bork P. A side effect resource to capture phenotypic effects of drugs. Mol. Syst. Biol. 2010;6:343. doi: 10.1038/msb.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao C., Li Y., Baytas I.M., Zhou J., Wang F. An mcem framework for drug safety signal detection and combination from heterogeneous real world evidence. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-19979-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Safitri N., Alaina M.F., Pitaloka D.A.E., Abdulah R. A narrative review of statin-induced rhabdomyolysis: molecular mechanism, risk factors, and management. Drug Healthc. Patient Saf. 2021;13:211–219. doi: 10.2147/DHPS.S333738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouitbir J., Sanvee G.M., Panajatovic M.V., Singh F., Krähenbühl S. Mechanisms of statin-associated skeletal muscle-associated symptoms. Pharmacol. Res. 2020;154 doi: 10.1016/j.phrs.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 25.Hottinger D.G., Beebe D.S., Kozhimannil T., Prielipp R.C., Belani K.G. Sodium nitroprusside in 2014: a clinical concepts review. J. Anaesthesiol. Clin. Pharmacol. 2014;30:462–471. doi: 10.4103/0970-9185.142799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.ftp://ftp.ncbi.nlm.nih.gov/pubchem/specifications/pubchem_fingerprints.txt Pubchem substructure fingerprint v1.3, [EB/OL]
- 27.Pagès H., Aboyoun P., Gentleman R., DebRoy S. Biostrings: efficient manipulation of biological strings. 2024. https://bioconductor.org/packages/Biostrings R package version 2.72.1.
- 28.Consortium T.G.O., et al. The Gene Ontology knowledgebase in 2023. Genetics. 2023;224 doi: 10.1093/genetics/iyad031. https://academic.oup.com/genetics/article-pdf/224/1/iyad031/51074934/iyad031.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu G., et al. Gosemsim: an r package for measuring semantic similarity among go terms and gene products. Bioinformatics. 2010;26:976–978. doi: 10.1093/bioinformatics/btq064. [DOI] [PubMed] [Google Scholar]
- 30.Zhang P., Wang F., Hu J., Sorrentino R. Label propagation prediction of drug-drug interactions based on clinical side effects. Sci. Rep. 2015;5:1–10. doi: 10.1038/srep12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of results for different values of model parameter (gamma). To generate the label propagated signal scores we use a probability parameter γ as outlined in equation 12, which is discussed in detail in subsection 5.2. We calculate the AUC of the label propagation methods for γ ranging from 0.1 to 0.9, in increments of 0.1. We perform this step separately for all six drug similarity matrices to find the optimal value of γ that achieves the maximum AUC score for each matrix.
Data Availability Statement
The quarterly drug-ADE reports used in this work was downloaded from the FAERS website. The side effects data was collected from the SIDER website. Additional drug information was extracted from DrugBank. The six drug similarity matrices used in this paper are available at: https://zenodo.org/records/13270611. We performed further data processing to filter and prepare these datasets for our use.