Abstract
Background
Intraductal papillary mucinous neoplasms (IPMNs) are precursors to pancreatic cancer, but not all IPMNs progress to cancer. The objective of this study was to identify the germline genetic variants associated with IPMN clinical progression by conducting the first genome‐wide association study (GWAS) and computing a polygenic hazard score (PHS) in 338 patients with IPMN.
Methods
The study population was divided into two subsets, and a Cox analysis adjusted for sex, age, cyst size at diagnosis, and the top 10 principal components was performed. A PHS was calculated using the genotypes of common variants associated with IPMN progression identified.
Results
Eight loci with significant associations (p < 5 × 10−8) were identified, and the most significant was 7q21.11‐rs117620617 (hazard ratio, 16.35; 95% confidence interval, 6.93–38.60; p = 1.80 × 10−10). All variants were associated with inflammatory processes, suggesting that alleles that predispose to an inflammatory prone phenotype may promote progression. The PHS indicated a statistically significant association (hazard ratio, 18.05; 95% confidence interval, 7.96–45.80; p = 6.18 × 10−11) with IPMN progression among individuals who had the highest number of effect alleles (fourth quartile) compared with those who had the lowest number (first quartile).
Conclusions
The current results study advance the understanding of individual predisposition to IPMN progression and underscore the potential use of genetics in the stratification of patients who have IPMN.
Keywords: genome‐wide association study (GWAS), intraductal papillary mucinous neoplasms, pancreatic cancer, polygenic score, progression
Short abstract
This genome‐wide association study identifies eight loci associated with the progression of intraductal papillary mucinous neoplasms toward malignancy. The findings provide valuable insights into the genetic predisposition for intraductal papillary mucinous neoplasm progression, highlighting the role of inflammatory processes.
INTRODUCTION
Intraductal papillary mucinous neoplasms (IPMNs), the most common pancreatic premalignant lesions, are usually diagnosed incidentally in asymptomatic individuals. 1 Because IPMNs may evolve into pancreatic ductal adenocarcinoma (PDAC), lifelong surveillance is frequently recommended for patients who are fit for resection. 2 , 3 Current guidelines identify worrisome features (WFs) and high‐risk stigmata (HRSs) as clinical markers of IPMN progression and, in some cases, as an indication for surgery. 2 , 3 However, there is no consensus in follow‐up protocols in terms of duration and check‐up intervals; therefore, it is of the utmost importance to discover biomarkers useful to stratify the patients by their risk of clinical progression, with the long‐term goal of being able to predict who will eventually develop PDAC.
The investigation of germline genetic variants is potentially of great interest since single‐nucleotide variants (SNVs) are associated with the risk of PDAC 4 , 5 , 6 , 7 , 8 and have been proposed to be involved in IPMN malignant transformation. 9 , 10 , 11 , 12 For example, Giaccherini and colleagues identified an SNV involved in telomere maintenance that was associated with IPMN clinical progression. 9 Another example is a study conducted by Gentiluomo and colleagues in which two PDAC risk SNVs (GRP reference SNV number 1517037 [rs1517037] and PVT1 rs10094872) displayed suggestive associations with IPMN progression. 11 In addition, the role of the genetic variability of the ABO locus, has been investigated in relation to IPMN progression. Several authors have demonstrated that patients in the ABO_AA blood group have a higher risk of IPMN progression compared with those in the O blood group, whereas others reported no association. 12 , 13 , 14
In the last decade, genome‐wide association studies (GWAS) have been very successful in discovering genotype–phenotype associations, including statistically significant associations with the risk of developing PDAC. 15 , 16 , 17 , 18 , 19 , 20 However, to our knowledge, a GWAS has never been attempted with the aim of understanding the genetic background of IPMN evolution into malignancy. With these premises, we have performed the first GWAS and computed a polygenic hazard score (PHS) to identify new genetic determinants of IPMN clinical evolution.
MATERIALS AND METHODS
Study participants and study design
In total, 338 patients with IPMN who were undergoing surveillance were retrospectively enrolled (from 2006 to 2023) by the San Raffaele Research Hospital, Milan, Italy (n = 266), and the Ospedale Cisanello‐University of Pisa, Pisa, Italy (n = 72). Among these 338 patients, 319 presented with a diagnosis of branch‐duct IPMN, 17 presented with a mixed‐type, and two presented with a main‐duct IPMN. The diagnoses were formulated by magnetic resonance imaging, magnetic resonance cholangiopancreatography, and/or endoscopic ultrasonography, according to the 2017 International Association of Pancreatology guidelines (Table 1). 3
TABLE 1.
Description of the study population.
| No. of patients (%) | ||||||
|---|---|---|---|---|---|---|
| Set 1, n = 279 | Set 2, n = 59 | |||||
| With progression | Without progression | p a | With progression | Without progression | p a | |
| Age, median [IQR], years | 68 [61.0–73.5] | 66 [60.0–72.0] | 63.5 [60.8–69.3] | 66 [56–73.5] | ||
| Follow‐up: Median [IQR], months | 67.2 [35.5–105.4] | 58.4 [31.4–87.6] | 41.9 [18.5–85.4] | 40.2 [13.8–80.7] | ||
| Sex | .351 | .197 | ||||
| Men | 37 | 66 | 3 | 12 | ||
| Women | 53 | 123 | 17 | 27 | ||
| BMI, kg/m2 | .848 | .815 | ||||
| <18.0 | 3 | 4 | 2 | 0 | ||
| 18.0–25.9 | 47 | 121 | 11 | 18 | ||
| 26.0–29.9 | 23 | 30 | 7 | 16 | ||
| ≥30 | 11 | 16 | 0 | 3 | ||
| Not available | 6 | 8 | 0 | 2 | ||
| Smoking | .899 | .651 | ||||
| No, never | 59 | 124 | 14 | 25 | ||
| Yes | 32 | 65 | 6 | 14 | ||
| Alcohol consumption | .606 | .795 | ||||
| No, never | 53 | 106 | 13 | 24 | ||
| Yes | 37 | 83 | 7 | 15 | ||
| Type 2 diabetes | .069 | .080 | ||||
| No | 75 | 172 | 15 | 36 | ||
| Yes | 15 | 17 | 5 | 3 | ||
| Cyst size at diagnosis, mm | 8.92e−06 | .448 | ||||
| <15.0 | 36 | 126 | 10 | 21 | ||
| 15.1–29.9 | 54 | 63 | 10 | 18 | ||
| Family history of pancreatic cancer | .421 | .198 | ||||
| No | 79 | 172 | 19 | 32 | ||
| Yes | 11 | 17 | 1 | 7 | ||
Abbreviations: BMI, body mass index; IQR, interquartile range.
aThe p value of the variable distribution between the groups of patients with and without clinical progression.
Individuals were required to be aged 18 years or older and to be willing to provide blood samples for research purposes. A follow‐up period of at least 12 months was also necessary for inclusion. None of the 338 patients presented with WFs or HRSs at the time of IPMN diagnosis.
The primary outcome of the study was the appearance of WFs or HRSs, whichever occurred first, as a proxy of clinical progression. According to the International Association of Pancreatology guidelines, WFs were defined as: (1) acute pancreatitis; (2) main pancreatic duct (MPD) size between 5 and 9 mm; (3) an abrupt change in greatest MPD dimension with distal pancreatic atrophy; (4) enhancement of the mural nodule <5 mm; (5) cyst size >3 cm; (6) cyst growth rate >5 mm in 2 years; (7) thickened, enhanced cyst walls; (8) lymphadenopathy; and (9) an increased serum level of carbohydrate antigen 19‐9. HRSs were defined as: (1) enhancing of the mural nodule ≥5 mm, (2) MPD ≥10 mm, and (3) obstructive jaundice caused by the IPMN. 3
Considering the capricious nature of association studies and to mitigate the occurrence of type 1 error, the study population was divided into two groups. The sample division was random (although it balanced the ratio of individuals with and without progression), including 80% of patients in the first set (n = 279; 90 with clinical progression and 189 without) and 20% in the second set (n = 59; 20 with clinical progression and 39 without). From this point onward, the first set is referred to as S1, and the second set is referred to as S2.
Genotyping
DNA was extracted from peripheral whole blood collected at the time of IPMN diagnosis or during patient follow‐up. Genotyping was performed using the Infinium Global Screening Array‐24, version 3.0 BeadChip (Illumina, Inc.). Raw genotypes were used to identify and exclude individuals with sex mismatches (discrepancy between the self‐declared sex and the sex determined by the genotyping), a call rate <0.95, or cryptic relatedness (p̂ > 0.2, where p̂ indicates the proportion of the genome shared identical‐by‐descent between pairs of individuals). In addition, polymorphisms that had a minor allele frequency (MAF) <0.01 or evidence of violating Hardy–Weinberg equilibrium (p < 5 × 10−5) were excluded. The quality controls (QCs) were performed using PLINK software (www.cog‐genomics.org/plink/1.9/, version released 11 December 2023). 21
Principal component analysis (PCA) was used to assess population stratification and exclude outliers. The individuals from phase 3 of the 1000 Genomes Project (International Genome Sample Resource) were used as the reference population, and PCA was performed with PLINK software. Samples that did not show European ancestry were excluded.
FTER the genotyping and QCs, the data set was imputed using the Michigan Imputation Server and Minimac4 (https://imputationserver.sph.umich.edu; data access May 2024). The Haplotype Reference Consortium (HRC version r1.1; Wellcome Sanger Institute) was used as a reference panel. After imputation, only polymorphisms with an imputation quality (INFO) score > 0.7 were included in the analyses. PCA was conducted using PLINK software on the data set that passed postimputation QC analyses using independent polymorphisms (linkage disequilibrium [LD]; coeffeient of determination [r2] < 0.8). PCA was also conducted separately for each set of patients. After the association analysis, the lambda inflation factor was calculated.
ABO blood group evaluation
The ABO blood group was suggested by using genotype data from specific variants: rs505922 for the O and non‐O groups, rs8176746 for the B group, and rs8176747 for the A group, as detailed elsewhere. 22
Statistical analyses
To evaluate the association between SNVs, ABO blood groups, and clinical IPMN progression, a multivariable Cox regression analysis adjusted for age, sex, and the first 10 principal components was performed by computing hazard ratios (HRs) and 95% confidence intervals (95% CIs). The exposome variables (body mass index, smoking status, alcohol consumption, type 2 diabetes, cyst size at diagnosis, and family history of pancreatic cancer) were also tested by using a univariable model, and those that resulted in an association with progression were added as adjustment factors in a sensitivity analysis model. The Cox regression analyses were conducted in R‐Studio using the package gwasurvivr (The R Foundation). The model used the time intervals between the initial diagnosis of IPMN (expressed in months) and the onset of the first WF or HRS to assess the likelihood of progression. Three individual Cox analyses were conducted: one for S1, one for S2, and, finally, one for all study participants (global phase). SNVs with p values < .05 in the S1 and S2 sets and SNVs with p < 5 × 10−8 in the global phase that had the effect allele direction (HR, <1.00 or >1.00) consistent between all three analyses were considered to be associated with IPMN progression.
Kaplan–Meier curves were used to assess differences in progression based on genotype using the R packages survival and ggsurvplot.
Power calculation
Power calculation was performed using the R package survSNP. 23 For power calculation, α was set at .05. Considering a population size of 338 patients and 110 events of clinical progression, the study had statistical power >80% to observe statistically significant associations with an HR of 4.80 for variants with a frequency of 1% and an HR of 2.20 for variants with a frequency of 5%.
SNVs and gene annotation
Multiple bioinformatic tools were used to understand the possible function of the SNVs. To evaluate whether they were associated with tissue‐specific gene expression (expression quantitative trait loci [QTL]) or alternative splicing of premessenger RNA (splicing QTL), the Genotype‐Tissue Expression portal and QTLbase (Mulinlab; http://www.mulinlab.org/qtlbase; access June 2024) were used. The National Human Genome Research Institute‐European Bioinformations Institute GWAS Catalog (https://www.ebi.ac.uk/gwas/; access June 2024) and the Open Targets Genetics tool (Wellcome Sanger Institute [https://genetics.opentargets.org/; access June 2024]; see Table S1) were used to assess the association of the statistically significant SNVs with other human traits. Unlike the GWAS Catalog, the Open Targets Genetics tool reports the summary statistics of all studies conducted in the FinnGen and UK‐Biobank projects and further compiles all summary statistics deposited in the GWAS Catalog using p < .005. LDtrait (National Cancer Institute, National Institutes of Health; see Table S2) was used to test the associations with additional traits of the SNVs in LD with the polymorphisms associated with IPMN progression. The RegulomeDB portal was used to obtain a score predicting the potential regulatory function of the SNVs. 24 The HaploReg v4.2 web tool (Broad Institute, Massachusetts Institute of Technology) was used to explore annotations of the noncoding genome at SNVs on haplotype blocks based on LD. The Combined Annotation Dependent Depletion (CADD) score was used (University of Washington; https://cadd.gs.washington.edu/) to assess the potential deleteriousness of nucleotide substitutions.
Polygenic hazard score
The PHS was computed for all patients in the study. The SNVs selected to compute the PHS were chosen by including only SNVs with an MAF >5%, as recommended for a study population with <1000 participants, and with a p value < 5 × 10−7.in the global analysis (see Table S3). 25 The list of SNVs was obtained by adopting a shrinkage strategy, which entailed the selection of independent SNVs (LD threshold, r2 < 0.2) prioritizing the variant with the lowest p value in each LD block. The data from the analyses adjusted by sex, age, and the top 10 principal components were used. The PHS was calculated for each patient by summing the number of alleles that increased the probability of clinical progression. A value of 0, 1, or 2 was assigned to each SNV, representing homozygotes without effect alleles (value = 0), heterozygotes (value = 1), and homozygotes with two effect alleles (value = 2). To obtain a reference value that represented the general population, a PHS with the same SNVs was computed using genotyping data from European participants in the 1000 Genomes Project. Individuals of British and Finnish descent were excluded to reduce the possible effect of population stratification. The final data set for the reference population was comprised of 313 individuals. By using quartiles derived from the reference population, IPMNs were divided into four groups. Patients with the lowest allele counts associated with progression were grouped in the first quartile, and those with the highest number of effect alleles were grouped in the fourth quartile.
To assess the level of the association of the PHS with the probability of IPMN clinical progression, a generalized linear model (GLM) analysis was performed using the group within the first quartile of PHS as the reference group and adjusting for sex, age, and cyst size. The predictiveness of the PHS is indicated by the area under the receiver operating characteristic curve (AUC) and the Nagelkerke R‐squared value (or pseudo‐R–squared). The results of PHS analyses were validated by permutation analysis using one million iterations.
GLM and permutations were performed using R studio with the packages stats and predictmeans. The AUC was computed using the pROC package, and the Nagelkerke R‐squared value was estimated using the fmsb package.
The study obtained internal review board approval from both enrolling centers: San Raffaele Research Hospital (protocol number 133/2016 [for recording clinical information]; protocol BIOGASTRO 2011, amendment November 11, 2017 [for blood sample biobanking and for biomarker investigation]) and Cisanello Hospital‐University of Pisa (the ethical committee was the Comitato Etico Regionale per la Sperimentazione Clinica della Toscana sezione Area Vasta Nord Ovest [protocol code IPMN‐PDAC transition]). The patients/participants provided written informed consent to participate in this study.
RESULTS
Development of worrisome features or high‐risk stigmata
For all 338 patients included in the study, follow‐up data were recorded for a median follow‐up of 60.2 months, with an interquartile range (IQR) of 29.3–90.1 months. During this period, 110 patients developed at least one WF, one HRS, or both. The median time to develop either WFs or HRSs was 36.8 months (IQR, 16.34–70.32 months). In detail, 22 patients developed at least one HRS, and 109 developed at least one WF. The number of WFs and HRSs observed during follow‐up are reported in Table S4.
Exposome variables
None of the variables showed a statistically significant association with IPMN progression with the exception of cyst size at diagnosis (Table 1); therefore, cyst size at diagnosis was used as adjustment factor in the sensitivity analysis.
Genome‐wide analysis results
All patients and 10,904,148 genetic SNVs passed postimputation QCs and were analyzed. The lambda inflation factors were 0.92 for the patients in S1, 0.94 for those in S2, and 0.95 for all patients when adjusted for sex, age, and the top 10 principal components. When further adjusted for cyst size, the factors were 0.99, 1.10, and 1.04, respectively. The results of the GWAS analysis revealed five loci in the analysis without cyst size as an adjustment factor (2p14‐rs115567527, 4p15.1‐rs10517223, 7q21.11‐rs117620617, 11q14.3‐rs145104689, and 15q22.2‐rs113897592) and four in the analysis with cyst size (1q41‐rs116276774, 1q42.12‐rs112528149, 7q21.11‐rs117620617, and 13q31.3‐rs77789312,). Notably, 7q21.11‐rs117620617 was associated in both analyses with almost superimposable HRs and p values. SNVs 4p15.1‐rs10517223 and 15q22.2‐rs113897592 are common variants with MAFs >5%, whereas the others are low‐frequency SNVs with MAFs within 1% and 5%. The results of the association analysis are reported in Table 2. The associations of the SNVs with other traits are reported in Figure S1. A circular Manhattan plot of results from the S1 set, the S2 set, and the global phase has been used to visualize the results (Figure 1). Kaplan–Meier curves indicate a clear effect of the genotypes on progression in patients with IPMN (see Figure S1).
TABLE 2.
Polymorphisms associated with clinical progression in intraductal papillary mucinous neoplasm.
| CHR | POS | A0 | A1 | rsID | Model | Set 1, n = 279; 90 events | Set 2, n = 59; 20 events | Global, n = 338; 110 events a | MAF, % | CADD b | Overlapped gene | Nearest gene, upstream‐downstream | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR (95%CI) | p | HR (95% CI) | p | HR (95%CI) | p | Study | ALFA | |||||||||
| 1q41 | 216477597 | C | G | rs116276774 | u | 9.13 (2.77–30.03) | 2.73e−4 | 5.78 (2.28–14.66) | 2.17e−4 | 4.40 (2.43–7.97) | 1.01e−6 | 1.0 | 3.0 | 0.972 | USH2A | |
| a | 8.55 (2.57–28.41) | 4.62e−4 | 6.93 (2.48–19.41) | 2.27e−4 | 5.71 (3.13–10.42) | 1.43e−8 | Intron variant | |||||||||
| 1q42.12 | 225029458 | A | G | rs112528149 | u | 3.74 (1.92–7.29) | 1.05e−4 | 8.84 (1.62–48.20) | 1.17e−2 | 4.41 (2.45–7.96) | 8.31e−7 | 3.0 | 4.0 | 3.227 | None | RP11‐449J1.1 (68,415 bp) |
| a | 4.84 (2.43–9.61) | 6.85e−6 | 10.04 (1.68–59.94) | 1.14e−2 | 5.50 (3.00–10.07) | 3.28e−8 | DNAH14 (54,506 bp) | |||||||||
| 2p14 | 65725624 | C | G | rs115567527 | u | 9.39 (4.16–21.2) | 7.01e−8 | 13.79 (1.65–114.87) | 1.53e−2 | 8.14 (3.86–17.15) | 3.50e−8 | 2.0 | 2.0 | 9.935 | LOC105374781 | |
| a | 7.82 (3.26–18.71) | 3.92e−6 | 17.95 (1.88–171.22) | 1.21e−2 | 8.25 (3.83–17.8) | 7.29e−8 | Intron variant | |||||||||
| 4p15.1 | 31154618 | A | G | rs10517223 | u | 2.44 (1.73–3.45) | 4.13e−7 | 5.44 (1.21–24.44) | 2.71e−2 | 2.48 (1.8–3.42) | 2.96e–8 | 12.0 | 11.0 | 6.110 | None | PCDH7 (6,196bp) |
| a | 2.38 (1.65–3.42) | 3.22e−6 | 5.51 (1.15–26.48) | 3.29e−2 | 2.36 (1.69–3.29) | 4.04e–7 | RP11‐617I14.1 (18,148 bp) | |||||||||
| 7q21.11 | 79203535 | T | C | rs117620617 | u | 11.52 (4.42–29.98) | 5.56e−7 | 101.34 (4.08–2518.18) | 4.84e−3 | 13.15 (5.69–30.39) | 1.67e–9 | 1.0 | 1.0 | 4.417 | None | AC091813.2 (29,366 bp) |
| a | 13.92 (5.19–37.33) | 1.69e−7 | 120.32 (4.47–3238.91) | 4.36e−3 | 16.35 (6.93–38.60) | 1.80e–10 | RNA5SP234 (79,890 bp) | |||||||||
| 11q14.3 | 92196765 | C | T | rs145104689 | u | 10.16 (3.00–34.37) | 1.93e−4 | 28.05 (4.46–176.38) | 3.79e−4 | 13.39 (5.37–33.4) | 2.64e–8 | 1.0 | 2.0 | 0.621 | FAT3 | |
| a | 7.55 (2.21–25.74) | 1.24e−3 | 31.62 (4.77–209.66) | 3.45e−4 | 12.00 (4.83–29.82) | 8.94e–8 | Intron variant | |||||||||
| 13q31.3 | 92820315 | T | C | rs77789312 | u | 11.76 (4.70–29.42) | 1.39e−7 | 11.86 (1.04–135.44) | 4.65e−2 | 8.01 (3.46–18.52) | 1.15e−6 | 1.0 | 2.0 | 0.929 | GPC5 | |
| a | 14.11 (5.60–35.60) | 2.05e−8 | 16.30 (1.23–215.59) | 3.41e−2 | 11.24 (4.83–26.14) | 1.94e−8 | Intron variant | |||||||||
| 15q22.2 | 62751925 | G | A | rs113897592 | u | 3.28 (2.02–5.34) | 1.69e−6 | 5.39 (1.50–19.4) | 9.85e−3 | 3.52 (2.26–5.48) | 2.73e−8 | 6.0 | 9.0 | 0.026 | TLN2 | |
| a | 2.54 (1.54–4.19) | 2.75e−4 | 6.06 (1.6–22.86) | 7.88e−3 | 2.98 (1.89–4.69) | 2.63e−6 | Intron variant | |||||||||
Abbreviations: a, adjusted model; A, adenine; A0, minor and effect allele; A1, nonminor allele; ALFA, Allele Frequency Aggregator (National Library of Medicine, National Center for Biotechnology Information); bp, base pairs; C, cytosine; CADD, Combined Annotation‐Dependent Depletion; CHR, chromosome; CI, confidence interval; G, guanine; GRCh37, Genome Reference Consortium Human Build 37; HR, hazard ratio; MAF, minor allele frequencies (measured in the study population and reported by the ALFA Project; POS, genomic position; rsID, reference single‐nucleotide polymorphism identification number; T, thymine; u, unadjusted model.
aResult: Divided by set 1, set 2, and global phases with report HRs.
bCADD scores were PHRED‐scaled (a quality score) by expressing the rank in order of magnitude terms rather than the precise rank itself; e.g., reference genome single‐nucleotide variants with CADD scores at the 10th percentile were assigned to CADD‐10, those with scores in the top 1% were assigned to CADD‐20, those with scores in the top 0.1% were assigned to CADD‐30, etc.; for each variant, the results of the analyses reported are adjusted by (model a) sex, age, cyst size, and the top 10 principal components; and by (model u) sex, age, and the top 10 principal components.
FIGURE 1.
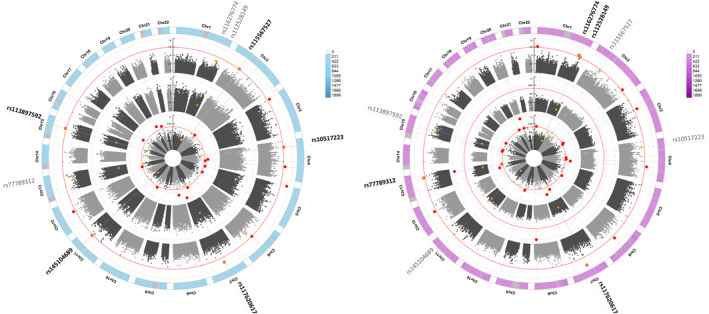
Circular Manhattan plots. Light blue indicates graphs of the Cox analyses conducted adjusting for sex, age, and the top 10 principal components; purple, graphs of the Cox analyses conducted adjusting for sex, age, the top 10 principal components, and cyst size at the time of diagnosis. In centrifuge order, starting from the center, graphs illustrate: analyses conducted on the first data set of patients (S1), analyses conducted on the second data set of patients (S2), and the outermost circle represents the analyses conducted on all study participants. The rs numbers of the eight identified SNVs are reported outside the outer circle, with SNVs that meet the p value threshold in the respective analyses indicated in bold. Red dots indicate SNVs with a p value < 5e−8 in the respective analysis. Yellow dots represent the eight identified SNVs, and yellow dots with a red border indicate the identified SNVs for analyses in which the p value threshold of 5e−8 was reached. Chr indicates chromosome; rs, reference single‐nucleotide variant; SNV, single‐nucleotide variant.
Functional annotation
1q41‐rs116276774
The cytosine (C) allele is associated with clinical progression, with an HR of 4.40 (95% CI, 2.43–9.97; p = 1.01 × 10−6) in the analyses not adjusted for cyst size and an HR of 5.71 (95% CI, 3.13–10.42; p = 1.43 × 10−8) in the analyses adjusted for cyst size. 1q41‐rs116276774 is an intron variant of the Usher syndrome 2A gene. This gene encodes a protein that contains laminin epidermal growth factor motifs, a pentaxin domain, and many fibronectin type III motifs involved in hearing and vision. According to data from Open Targets Genetics, 1q41‐rs116276774 is associated with 45 different traits. Interestingly, several of these traits are IPMN progression risk factors, such as type 1 diabetes and chronic inflammation; galectin‐3, interleukin‐1 receptor type 2, and interleukin‐6 receptor subunit alpha levels in plasma; and juvenile arthritis.
1q42.12‐rs112528149
The adenine (A) allele exhibited an HR of 4.41 (95% CI, 2.45–7.96; p = 8.31 × 10⁻7) in the unadjusted analyses and an HR of 5.50 (95% CI, 3.00–10.07; p = 3.28 × 10−8) in the analyses adjusted for cyst size. 1q42.12‐rs112528149 is an intergenic region, between the RP11‐449j1 gene (68,415 base pairs [bp]) and the DNAH14 gene (54,506 bp). 1q42.12‐rs112528149 is reported as an expression QTL and a splicing QTL for DNAH14 in cultured fibroblasts cells and in the testis. 1q42.12‐rs112528149 is associated with inflammatory and immune response traits, such as chemokine ligand 28 (CCL28) levels and psoriasis.
2p14‐rs115567527
In the analyses not adjusted for cyst size, the C allele had an HR of 8.14 (95% CI, 3.86–17.15; p = 3.50 × 10−8) and, adjusting for cyst size, it had an HR of 8.25 (95% CI, 3.83–17.80; p = 7.29 × 10−8). Notably, the C allele of 2p14‐rs115567527 was observed in 10 of 110 patients who had clinical progression and only in four of 228 patients without clinical progression. In addition, in patients who manifested clinical progression, 2p14‐rs115567527‐C had a frequency (MAF, 4.5%) that was more than double that of the general European population in the 1000 Genome Project (MAF, 2%). 2p14‐rs115567527 is located in intron 2 of the LOC105374781 long noncoding RNA gene. RegulomeDB indicated a rank of 3a, which indicates the possible binding of transcription factors and a chromatin accessibility peak.
4p15.1‐rs10517223
The A allele is associated with clinical progression, with an HR of 2.48 (95% CI, 1.80–3.42; p = 2.96 × 10−8) in the analyses not adjusted and an HR of 2.36 (95% CI, 1.69–3.29; p = 4.04 × 10−7) in the analyses adjusted for cyst size. It is situated in an intergenic region with protocadherin 7 (PCDH7) as the nearest coding gene (432,627 bp). 4p15.1‐rs10517223 is associated with body mass traits, such as appendicular lean mass, and allergies, including those to drugs, medicaments, and biologic substances. According to the data obtained by LDtrait, 4p15.1‐rs10517223 is in a block of LD (D′ > 0.80) with other variants associated with smoking behaviors and body mass index.
7q21.11‐rs117620617
For this SNV, the thymine (T) allele had the highest effect size observed. T allele carriers have a greater probability of clinical progression compared with C allele carriers, with an HR of 13.15 (95% CI, 5.69–30.39; p = 1.67 × 10−9) in the analyses not adjusted for cyst size and 16.35 (95% CI, 6.93–38.60; p = 1.80 × 10−10) in the analyses adjusted for cyst size. This SNV is situated in an intergenic region between two genes, AC091813.2 (29,366 bp) and RNA5SP234 (79,890 bp). The nearest coding gene is the membrane‐associated guanylate kinase WW and PDZ domain‐containing 2 gene, which is 120,552 bp away from 7q21.11‐rs117620617. 7q21.11‐rs117620617 is associated with CCL19 and signaling lymphocytic activation molecule 1 (SLAMF1) levels, both of which are involved in immunoregulatory and inflammatory processes.
11q14.3‐rs145104689
In the analyses not adjusted for cyst size, the C allele was associated with clinical progression, with an HR of 13.39 (95% CI, 5.37–33.40; p = 2.64 × 10−8) and a slightly less statistically significant result with an HR of 12.00 (95% CI, 4.83–29.82; p = 8.94 × 10−8) in the analyses adjusted for cyst size. It is an intron variant of the FAT atypical cadherin 3 (FAT3) gene that codes for a protein involved in cell–cell adhesion. 11q14.3‐rs145104689 is linked to allergic or anaphylactic drug reactions.
13q31.3‐rs77789312
The T allele is associated with an HR of 8.01 (95% CI, 3.46–18.52; p = 1.15 × 10−6) in the analyses not adjusted for cyst size and an HR of 11.24 (95% CI, 4.83–26.14; p = 1.94 × 10−8) in the analyses adjusted for cyst size. It is an intron variant of the glypican proteoglycan 5 gene. This gene codes for a protein that, according to GeneCard data, may play a role in controlling cell division and regulating growth. 13q31.3‐rs77789312 is linked to the white blood cell count and asthma, markers of immune response status and inflammatory processes.
15q22.2‐rs113897592
In unadjusted analyses, the guanine (G) allele was associated with clinical progression, with an HR of 3.52 (95% CI, 2.26–5.48; p = 2.73 × 10−8), and the HR was 2.98 (95% CI, 1.89–4.69; p = 2.63 × 10−6) in analyses adjusted for cyst size. 15q22.2‐rs113897592 is associated with body mass traits, such as arm fat percentage and body fat percentage, and with interleukin‐1 receptor type 2 levels. 15q22.2‐rs113897592 is partially in LD (D′ = 0.98; r2 = 0.17) with rs17205757, which is associated with abdominal adipose tissue volumes.
Association with the risk of developing PDAC
None of the eight identified SNVs have been associated with the risk of developing PDAC based on the results of GWAS conducted across multiple populations. 15 , 16 , 18 , 19 , 20 , 26 , 27 , 28 , 29 , 30 , 31
Association between ABO blood group and progression
No statistically significant association was observed between carriers of blood type O and carriers of other blood type groups (Table 3) in either analysis.
TABLE 3.
Blood group analyses.
| Analysis | Model [Link] , [Link] | HR | SE | Z score | pr(>|z|) |
|---|---|---|---|---|---|
| A group vs. O group | u | 0.882 | 0.207 | −0.606 | .545 |
| A group vs. O group | a | 0.821 | 0.209 | −0.942 | .346 |
| AB group vs. O group | u | 1.220 | 0.413 | 0.482 | .630 |
| AB group vs. O group | a | 0.970 | 0.413 | −0.074 | .941 |
| B group vs. O group | u | 0.482 | 0.433 | −1.684 | .092 |
| B group vs. O group | a | 0.490 | 0.434 | −1.647 | .100 |
| Non‐O groups vs. O group | u | 0.836 | 0.194 | −0.925 | .355 |
| Non‐O groups vs. O group | a | 0.777 | 0.195 | −1.293 | .196 |
Abbreviations: a, adjusted model; HR, hazard ratio; pr(>|z|), p value of the test for whether the coefficient point estimate is significantly different from 0; SE standard error; u, unadjusted model.
aModel a was adjusted by sex, age, cyst size, and the top 10 principal components.
bModel u was adjusted by sex, age, and the top 10 principal components.
Polygenic hazard score
The PHS demonstrated a statistically significant association (HR, 18.05; 95% CI, 7.96–45.80; p = 6.18 × 10−11) with IPMN progression when comparing individuals who had the highest number of effect alleles (fourth quartile) versus those who had the lowest (first quartile). The p values for PHS were validated using 1 million permutations (Figure 2). The AUC of the model was 0.77, and the estimated pseudo–R‐squared value was 0.27, indicating that PHS accounted for 27% of the variability in the likelihood of progression in this data set. Less than 10% of patients within the first PHS quartile showed progression before the 130th month, whereas 50% of patients in the fourth quartile manifested clinical progression before the 50th month (Figure 2, Kaplan–Meier curve). Interestingly, the frequency of effect alleles among the simulated general population was higher than that observed in patients who had IPMN without clinical progression (p = .0048), whereas it was lower than that in thosed who had IPMN with clinical progression (p = 3.20 × 10−8). The SNVs included in the PHS are listed in Table S3.
FIGURE 2.
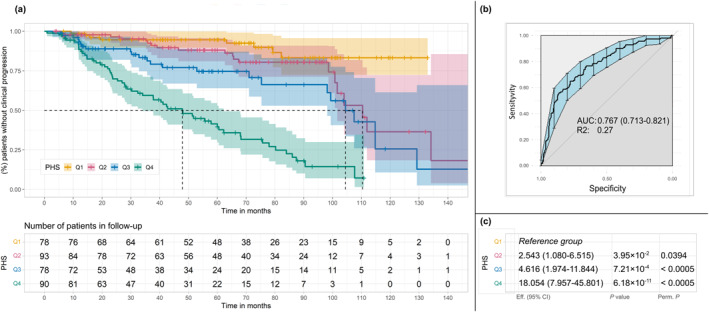
Results of PHS analyses. The PHS was calculated by summing the number of effect alleles and was divided into quartiles based on the distribution of the score calculated in a reference population (1000 Genome Project; International Genome Sample Resource). In the first group (Q1) are patients with a lower number of effect alleles and, in each rank, are those with an increased number of effect alleles. (A) Kaplan–Meier curves of PHS. The table below the Kaplan–Meier curves represents the number of patients who were still undergoing follow‐up who had not yet manifested clinical progression. (B) The AUC of PHS analyses. (C) Results of the analyses adjusted by sex and age to evaluate the PHS, comparing each quartile with the first quartile. AUC indicates area under the receiver operating characteristic curve; CI, confidence interval; PHS, polygenic hazard score; Q, quartile; R2, Nagelkerke R‐squared (coefficient of determination) value; rs, reference single‐nucleotide polymorphism. Eff. (effect) refers to the hazard ratios derived from the Cox model for each quartile relative to Q1. P.perm: p‐value calculated using permutation testing, based on one million random permutations of the dataset to assess the significance of the observed results under the null hypothesis.
DISCUSSION
The current study, which is the first GWAS on IPMN clinical progression, identified eight SNVs that reached the conventional GWAS threshold of p < 5 × 10⁻⁸. Specifically, these were 2p14‐rs115567527, 4p15.1‐rs10517223, 11q14.3‐rs145104689, and 15q22.2‐rs113897592 in the crude analysis and 1q41‐rs116276774, 1q42.12‐rs112528149, and 13q31.3‐rs77789312 when also adjusting for cyst size. In addition, 7q21.11‐rs117620617 was significant in both analyses. However, all SNVs had an association with the p value was close to p < 5 × 10⁻⁸ in both analyses, with consistent HRs and CIs, as reported in Table 2. This consistency suggests that the small change in p values reflects statistical fluctuations caused by the samples size and does not reflect real biologic differences.
The most intriguing finding is represented by 7q21.11‐rs117620617, which met all p‐value thresholds in both models (adjusted and unadjusted for cyst size), as mentioned above. This variant is associated with plasma levels of the proinflammatory chemokine CCL19, a protein that may induce antitumor immune responses and, conversely, can sustain an inflammatory condition that, modifying the microenvironment, could promote carcinogenesis. 32 , 33 7q21.11‐rs117620617 is also associated with plasma levels of the SLAMF1 receptor, which is a crucial protein involved in negative regulation of the CD40 signaling pathway, positive regulation of the MAPK cascade, and the activation of B and T lymphocytes, highlighting its crucial role in the immune response. 34 The role of SLAM proteins in tumor immunity has been reported in several cancers, including PDAC. 34 Interestingly, all of the other SNVs are also associated with various inflammatory processes and/or immune responses. For example, 1q41‐rs116276774 is associated with interleukin‐6 receptor subunit alpha and interleukin‐1 receptor type 2 levels, both of which are involved in the recruitment of tumor‐associated macrophages. In addition, 1q41‐rs116276774 is associated with blood levels of CCL28, a cytokine that plays a role in both innate and adaptive immune responses. Moreover, 13q31.3‐rs77789312 is associated with white blood cell and lymphocyte counts, 2p14‐rs115567527 is associated with the granulocytes ratio, 15q22.2‐rs113897592 is associated with interleukin‐1 receptor type 2 levels, 4p15.1‐rs10517223 is associated with a personal history of drug allergies, and, finally, 11q14.3‐rs145104689 is associated with the percentage of neutrophils among white blood cells, which is reported to be a predictive marker for invasive malignancy in IPMN. 35
Inflammatory processes play a significant role in IPMN development. 36 Arima and colleagues reported a notable decrease in the neutrophil‐to‐lymphocyte ratio among patients who had IPMN compared with healthy volunteers, 37 whereas Ohno and colleagues observed a higher concentration of neutrophils in patients who had malignant IPMN compared with those who had nonmalignant IPMN. 38 Tumor‐associated neutrophils and cytokines, such as TNFα and IL‐1ß, have been linked to the progression of IPMN toward malignancy. 39 , 40 Roth and colleagues observed that macrophages (CD68‐positive) accumulate already at early stages of IPMN and increase in number during progression. 41
All this evidence points toward the involvement of genetic variants in IPMN progression through their effect on chronic inflammation, suggesting that alleles that predispose to an inflammatory‐prone phenotype may promote progression. However, because all of the SNVs were located in intergenic or intronic regions, our current knowledge, which is largely based on silico analyses and on annotation databases, is not enough to identify a precise link between the association of the variants and their function. When interpreting SNV analysis results, it is important to consider that the magnitude of the HRs is likely overestimated because of the limited sample size. We also analyzed the cumulative effect of the SNVs using a polygenic hazards score, and we observed a dramatic difference in the progression rate between individuals in the fourth and the first quartiles of the distribution of the effect alleles (HR, 18.05; 95% CI, 7.96–45.80; p = 6.18 × 10−11). In addition, 50% of patients with IPMNs in the fourth quartile develop clinical progression in approximately 4 years, whereas only 10% of those in the lowest quartile develop progression after 10 years of follow‐up. Finally, the division of the PHS into quartiles based on the distribution of the score in the reference population clearly demonstrated an enrichment of effect alleles in patients with IPMN who had clinical progression (p = 3.20 × 10−8) and a significant decrease in those without clinical progression (p = .0048). We are aware of the inflation bias caused by the possible overfitting of the data. However, although the results must be approached with caution, to our knowledge, they represent the first exploratory use of a PHS in IPMN stratification based on genetic background.
A possible limitation of the study is that, although WFs and HRSs are helpful surrogate markers for indicating clinical progression, they may not necessarily be indicative of carcinogenesis. However, a recent study demonstrated that patients with IPMN are more likely to develop malignancy if they develop novel or additional WFs or HRSs during surveillance. 42
The current study has allowed the identification of eight novel germline genetic variants, marking a substantial leap forward in the comprehension of this complex process. The results of the PHS hold promise for personalized patient care, even if they require validation and refining.
Future studies are warranted to validate these findings and to identify additional germline variants associated with clinical progression. Such efforts will enhance the precision of predictive tools like PHS, which offer noninvasive assessments based on genetic variants from DNA extracted from peripheral blood samples. Moreover, integrating somatic variants identified from tissue biopsies or cyst fluid could further refine the PHS, enabling more accurate and personalized predictions of IPMN progression.
Conclusions
In conclusion, the current findings underscore the role of genetics in IPMN progression, offering a potential avenue for identifying individuals predisposed to malignancy. This highlights the potential future integration of genetics into clinical practice, paving the way for more personalized and effective IPMN management.
AUTHOR CONTRIBUTIONS
Manuel Gentiluomo: Writing–original draft, writing–review and editing, data curation, formal analysis, investigation, and methodology. Chiara Corradi: Investigation, writing–review and editing, formal analysis, and data curation. Laura Apadula: Data curation and writing–review and editing. Annalisa Comandatore: Writing–review and editing and data curation. Gaetano Lauri: Writing–review and editing and data curation. Gemma Rossi: Writing–review and editing and data curation. Giulia Peduzzi: Writing–review and editing. Stefano Crippa: Writing–review and editing and data curation. Cosmeri Rizzato: Writing–review and editing. Massimo Falconi: Writing–review and editing and data curation. Paolo Giorgio Arcidiacono: Writing–review and editing and data curation. Luca Morelli: Writing–review and editing and data curation. Gabriele Capurso: Conceptualization, funding acquisition, writing–review and editing, resources, and data curation. Daniele Campa: Conceptualization, funding acquisition, writing–original draft, writing–review and editing, methodology, project administration, supervision, resources, and data curation.
CONFLICT OF INTEREST STATEMENT
Gabriele Capurso reports personal/consulting fees from Amgen, Boston Scientific Corporation, and Viatris outside the submitted work. The remaining authors disclosed no conflicts of interest.
Supporting information
Supplementary Material
Figure S1
Table S1
ACKNOWLEDGMENTS
The research that led to these results received funding from La Fondazione Italiana per la Recerca sul Cancero under IG 2019‐ID 23672 project (principal inestigator, Campa Daniele), and IG 2021‐ID 26201 project (principal inestigator, Capurso Gabriele). Open access publishing facilitated by Universita degli Studi di Pisa, as part of the Wiley ‐ CRUI‐CARE agreement.
Gentiluomo M, Corradi C, Apadula L, et al. A genome‐wide association study identifies eight loci associated with intraductal papillary mucinous neoplasm progression toward malignancy. Cancer. 2025;e35678. doi: 10.1002/cncr.35678
The first two authors contributed equally to this article as first co‐authors.
The last two authors contributed equally to this article as last co‐authors.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in the GWAS Catalog under study accession number GCST90435438.
REFERENCES
- 1. Crippa S, Arcidiacono PG, De Cobelli F, Falconi M. Review of the diagnosis and management of intraductal papillary mucinous neoplasms. United European Gastroenterol J. 2020;8(3):249‐255. doi: 10.1177/2050640619894767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Del Chiaro M, Besselink MG, Scholten L, et al. European evidence‐based guidelines on pancreatic cystic neoplasms. Gut. 2018;67(5):789‐804. doi: 10.1136/gutjnl-2018-316027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tanaka M, Fernández‐Del Castillo C, Kamisawa T, et al. Revisions of international consensus Fukuoka guidelines for the management of IPMN of the pancreas. Pancreatology. 2017;17(5):738‐753. doi: 10.1016/j.pan.2017.07.007 [DOI] [PubMed] [Google Scholar]
- 4. Campa D, Pastore M, Gentiluomo M, et al. Functional single nucleotide polymorphisms within the cyclin‐dependent kinase inhibitor 2A/2B region affect pancreatic cancer risk. Oncotarget. 2016;7(35):57011‐57020. doi: 10.18632/oncotarget.10935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gentiluomo M, Canzian F, Nicolini A, Gemignani F, Landi S, Campa D. Germline genetic variability in pancreatic cancer risk and prognosis. Semin Cancer Biol. 2022;79:105‐131. doi: 10.1016/j.semcancer.2020.08.003 [DOI] [PubMed] [Google Scholar]
- 6. Campa D, Matarazzi M, Greenhalf W, et al. Genetic determinants of telomere length and risk of pancreatic cancer: a PANDoRA study. Int J Cancer. 2018;144(6):1275‐1283. doi: 10.1002/ijc.31928 [DOI] [PubMed] [Google Scholar]
- 7. Galeotti AA, Gentiluomo M, Rizzato C, et al. Polygenic and multifactorial scores for pancreatic ductal adenocarcinoma risk prediction Med Genet. 2021;58(6):369‐377. doi: 10.1136/jmedgenet-2020-106961 [DOI] [PubMed] [Google Scholar]
- 8. Nodari Y, Gentiluomo M, Mohelnikova‐Duchonova B, et al. Genetic and non‐genetic risk factors for early‐onset pancreatic cancer. Dig Liver Dis. 2023;55(10):1417‐1425. doi: 10.1016/j.dld.2023.02.023 [DOI] [PubMed] [Google Scholar]
- 9. Giaccherini M, Gentiluomo M, Arcidiacono PG, et al. A polymorphic variant in telomere maintenance is associated with worrisome features and high‐risk stigmata development in IPMNs. Carcinogenesis. 2022;43(8):728‐735. doi: 10.1093/carcin/bgac051 [DOI] [PubMed] [Google Scholar]
- 10. Campa D, Felici A, Corradi C, et al. Long or short? Telomere length and pancreatic cancer and its precursor lesions, a narrative review. Mutagenesis. Published online November 17, 2023. doi: 10.1093/mutage/gead034 [DOI] [PubMed] [Google Scholar]
- 11. Gentiluomo M, Corradi C, Arcidiacono PG, et al. Role of pancreatic ductal adenocarcinoma risk factors in intraductal papillary mucinous neoplasm progression. Front Oncol. 2023;13:1172606. doi: 10.3389/fonc.2023.1172606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Capurso G, Crippa S, Vanella G, et al. Factors associated with the risk of progression of low‐risk branch‐duct intraductal papillary mucinous neoplasms. JAMA Netw Open. 2020;3(11):e2022933. doi: 10.1001/jamanetworkopen.2020.22933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Poruk KE, Griffin J, Makary MA, et al. Blood type as a predictor of high‐grade dysplasia and associated malignancy in patients with intraductal papillary mucinous neoplasms. J Gastrointest Surg. 2019;23(3):477‐483. doi: 10.1007/s11605-018-3795-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zelga P, Hernández‐Barco YG, Qadan M, et al. ABO blood group distribution and risk of malignancy in patients undergoing resection for intraductal papillary mucinous neoplasm (IPMN). Pancreatology. 2022;22(2):264‐269. doi: 10.1016/j.pan.2021.12.012 [DOI] [PubMed] [Google Scholar]
- 15. Zhang M, Wang Z, Obazee O, et al. Three new pancreatic cancer susceptibility signals identified on chromosomes 1q32.1, 5p15.33 and 8q24.21. Oncotarget. 2016;7(41):66328‐66343. doi: 10.18632/oncotarget.11041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wolpin BM, Rizzato C, Kraft P, et al. Genome‐wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat Genet. 2014;46(9):994‐1000. doi: 10.1038/ng.3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klein AP, Wolpin BM, Risch HA, et al. Genome‐wide meta‐analysis identifies five new susceptibility loci for pancreatic cancer. Nat Commun. 2018;9(1):556. doi: 10.1038/s41467-018-02942-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Amundadottir L, Kraft P, Stolzenberg‐Solomon RZ, et al. Genome‐wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41(9):986‐990. doi: 10.1038/ng.429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Petersen GM, Amundadottir L, Fuchs CS, et al. A genome‐wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42(3):224‐228. doi: 10.1038/ng.522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Childs EJ, Mocci E, Campa D, et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat Genet. 2015;47(8):911‐916. doi: 10.1038/ng.3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chang CC, Chow CC , Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ. Second‐generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yamamoto F, Clausen H, White T, Marken J, Hakomori S. Molecular genetic basis of the histo‐blood group ABO system. Nature. 1990;345(6272):229‐233. doi: 10.1038/345229a0 [DOI] [PubMed] [Google Scholar]
- 23. Owzar K, Li Z, Cox N, Jung SH. Power and sample size calculations for SNP association studies with censored time‐to‐event outcomes. Genet Epidemiol. 2012;36(6):538‐548. doi: 10.1002/gepi.21645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dong S, Zhao N, Spragins E, et al. Annotating and prioritizing human non‐coding variants with RegulomeDB v.2. Nat Genet. 2023;55(5):724‐726. doi: 10.1038/s41588-023-01365-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Choi SW, Mak TSH, O’Reilly PF. Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc. 2020;15(9):2759‐2772. doi: 10.1038/s41596-020-0353-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klein AP, Wolpin BM, Risch HA, et al. Genome‐wide meta‐analysis identifies five new susceptibility loci for pancreatic cancer. Nat Commun. 2018;9(1):556. doi: 10.1038/s41467-018-02942-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang J, Tian J, Zhu Y, et al. Exome‐wide analysis identifies three low‐frequency missense variants associated with pancreatic cancer risk in Chinese populations. Nat Commun. 2018;9(1):3688. doi: 10.1038/s41467-018-06136-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakatochi M, Lin Y, Ito H, et al. Prediction model for pancreatic cancer risk in the general Japanese population. PLoS One. 2018;13(9):e0203386. doi: 10.1371/journal.pone.0203386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Low SK, Kuchiba A, Zembutsu H, et al. Genome‐wide association study of pancreatic cancer in Japanese population. PLoS One. 2010;5(7):e11824. doi: 10.1371/journal.pone.0011824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu C, Miao X, Huang L, et al. Genome‐wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations. Nat Genet. 2011;44(1):62‐66. doi: 10.1038/ng.1020 [DOI] [PubMed] [Google Scholar]
- 31. Ozçelik H, Schmocker B, Di Nicola N, et al. Germline BRCA2 6174delT mutations in Ashkenazi Jewish pancreatic cancer patients. Nat Genet. 1997;16(1):17‐18. doi: 10.1038/ng0597-17 [DOI] [PubMed] [Google Scholar]
- 32. Gowhari Shabgah A, Al‐Obaidi ZMJ, Sulaiman Rahman H, et al. Does CCL19 act as a double‐edged sword in cancer development? Clin Exp Immunol. 2022;207(2):164‐175. doi: 10.1093/cei/uxab039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Crusz SM, Balkwill FR. Inflammation and cancer: advances and new agents. Nat Rev Clin Oncol. 2015;12(10):584‐596. doi: 10.1038/nrclinonc.2015.105 [DOI] [PubMed] [Google Scholar]
- 34. Farhangnia P, Ghomi SM, Mollazadehghomi S, Nickho H, Akbarpour M, Delbandi AA. SLAM‐family receptors come of age as a potential molecular target in cancer immunotherapy. Front Immunol. 2023;14:1174138. doi: 10.3389/fimmu.2023.1174138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gemenetzis G, Bagante F, Griffin JF, et al. Neutrophil‐to‐lymphocyte ratio is a predictive marker for invasive malignancy in intraductal papillary mucinous neoplasms of the pancreas. Ann Surg. 2017;266(2):339‐345. doi: 10.1097/sla.0000000000001988 [DOI] [PubMed] [Google Scholar]
- 36. Shockley KE, To B, Chen W, Lozanski G, Cruz‐Monserrate Z, Krishna SG. The role of genetic, metabolic, inflammatory, and immunologic mediators in the progression of intraductal papillary mucinous neoplasms to pancreatic adenocarcinoma. Cancers (Basel). 2023;15(6):1722. doi: 10.3390/cancers15061722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arima K, Okabe H, Hashimoto D, et al. The diagnostic role of the neutrophil‐to‐lymphocyte ratio in predicting pancreatic ductal adenocarcinoma in patients with pancreatic diseases. Int J Clin Oncol. 2016;21:940‐945. doi: 10.1007/s10147-016-0975-z [DOI] [PubMed] [Google Scholar]
- 38. Ohno R, Kawamoto R, Kanamoto M, et al. Neutrophil to lymphocyte ratio is a predictive factor of malignant potential for intraductal papillary mucinous neoplasms of the pancreas. Biomark Insights. 2019;14:117727191985150. doi: 10.1177/1177271919851505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sadot E, Basturk O, Klimstra DS, et al. Tumor‐associated neutrophils and malignant progression in intraductal papillary mucinous neoplasms: an opportunity for identification of high‐risk disease. Ann Surg. 2015;262(6):1102‐1107. doi: 10.1097/sla.0000000000001044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Simpson RE, Yip‐Schneider MT, Flick KF, Wu H, Colgate CL, Schmidt CM. Pancreatic fluid interleukin‐1β complements prostaglandin E2 and serum carbohydrate antigen 19‐9 in prediction of intraductal papillary mucinous neoplasm dysplasia. Pancreas. 2019;48(8):1026‐1031. doi: 10.1097/mpa.0000000000001377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roth S, Zamzow K, Gaida MM, et al. Evolution of the immune landscape during progression of pancreatic intraductal papillary mucinous neoplasms to invasive cancer. EBioMedicine. 2020;54:102714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marchegiani G, Pollini T, Andrianello S, et al. Progression vs cyst stability of branch‐duct intraductal papillary mucinous neoplasms after observation and surgery. JAMA Surg. 2021;156(7):654‐661. doi: 10.1001/jamasurg.2021.1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Figure S1
Table S1
Data Availability Statement
The data that support the findings of this study are openly available in the GWAS Catalog under study accession number GCST90435438.


