Abstract
Plasmid Rms149, the archetype of Pseudomonas plasmid incompatibility group IncP-6, was identified in Pseudomonas aeruginosa as an agent conferring resistance to streptomycin, sulfanilamide, gentamicin, and carbenicillin in 1975. It has been classed as a broad-host-range plasmid due to its ability to replicate in both Escherichia coli (where it is designated IncG) and Pseudomonas species, although both species are γ-proteobacteria. To provide reference information on this Inc group, we have determined the complete sequence of Rms149 and found that, although the genome comprises 57,121 bp, it is essentially a small mobilizable plasmid carrying multiple mobile elements, which make up 79% (>45 kb) of its genome. A replicon has been identified which encodes a single polypeptide with moderate identity to other replication proteins. The region encoding this protein can replicate in Pseudomonas putida and E. coli. This sequence is directly downstream of a putative partitioning region highly similar to that of pRA2. A functional IncQ-type mobilization region is also present. Thus, the backbone appears to be a novel combination of modules already identified in other plasmid systems. Analysis of the segments that fall outside this core of stable inheritance and transfer functions show that this plasmid has been subject to multiple insertion events and that the plasmid appears to carry a considerable load of DNA that no longer should be phenotypically advantageous. The plasmid therefore functions not just as a vehicle for spread of selective traits but also as a store for DNA that is not currently under selection.
The key features of a plasmid are the ability to replicate autonomously and to be maintained in a cell lineage without a high rate of segregational loss. An optional but frequently encountered property is the ability to transfer or to be mobilized from one bacterium to another. From these properties it follows that genes that become associated with a plasmid may spread rapidly from one genetic background to another. Some plasmids appear to consist of nothing more than functions that confer the above core abilities, and these have been termed cryptic plasmids.
To succeed as such a selfish element, a plasmid must be stable and confer minimal burden on its host, or at least overcome competitive losses by horizontal transfer (42). Conversely, a plasmid carrying genes that promote the growth of its host, relative to competitor bacteria, will benefit by increased propagation. However, since maintenance of a plasmid is generally found to place a metabolic or phenotypic load on the cell, universally useful genes will be selectively reacquired by the chromosome over evolutionary time. Therefore, successful plasmids typically carry payload genes, favorable in some environments but not others (12), or genes with a transient plasmid association that are spreading through a microbial community. In passing through different strains or species, plasmids are exposed to different genetic contexts and provide the opportunity for combinations of genes to form new patterns of selective benefit. Thus, plasmids are key genetic elements in the diversity and adaptability of bacteria.
The genus Pseudomonas consists of a range of species that are found in diverse habitats. Many plasmids, conferring a range of phenotypes of both clinical and environmental importance, have been identified in members of this genus (43). To characterize the genomes of Pseudomonas species, it is therefore important to include a knowledge of the plasmid content of different strains of the species as well as the plasmids to which the species has access. Rapid classification of plasmids depends on the existence of DNA sequence information that can be exploited through hybridization or PCR to establish the relationship to previously studied plasmids (11). A key requirement is to establish the nature of the replication functions, since these will indicate the lineage and possible linked properties of the plasmid. While some Pseudomonas plasmids have been studied in detail (43, 45), little is known about many others, and consequently, existing probes have often been singularly unsuccessful in classifying new Pseudomonas plasmids from strain collections (26). A major purpose of our current work is to provide information to help rectify this defect. We have therefore set out to sequence the archetypes of the various Pseudomonas plasmid groups (7, 24).
Plasmid Rms149 was discovered in a clinical strain, Pseudomonas aeruginosa Ps142, from Frankfurt in Germany (37); it confers multiple antibiotic resistances. It has since been shown to replicate in Escherichia coli after mobilization by an IncP-1 plasmid (21, 22). Although tested against many plasmids, it remains the sole member of its designated incompatibility group (24). This may simply reflect the absence of intense work to classify plasmids by incompatibility testing and the absence of DNA sequence information for rapid screening. In this paper we report the complete sequence of Rms149. This should allow us to test for the distribution of this replicon.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
For E. coli K-12 the strains used were as follows: DH5α [F− endA1 hsdR17(rK− mK+) supE44 thi-1 recA1 gyrA96 relA1 deoR Δ(lacZYA-argF)U169 φ80lacZΔM15 λ− phoA] (19) and HB101 [F− Δ(gpt-proA)62 leuB6 supE44 hsdS20(rB− mB−) ara-14 galK2 lacY1 Δ(mcrC-mrr) rpsL20 (Str) xyl-5 mtl-1 recA13] (8). For E. coli C, the strain used was: C2110 (polA1 his rha P2S), obtained from D. R. Helinski, University of California, San Diego. For Pseudomonas putida the strain used was KT2440 (hsdR1 hsdM+) (2). For P. aeruginosa the strain used was PU21 (FP− ilvB112 leu-1 strr-1 rifr) (23). Plasmids used or constructed during this work are described in Table 1.
TABLE 1.
Plasmids used in this study for functional analysis.
| Plasmid | Relevant featuresa | Reference or source |
|---|---|---|
| Rms149 | IncP-6, Cbr Gmr Smr Spr Sur Mob+ | 37 |
| pUB307 | IncP-1, deletion derivative of RP1, Kmr Tcr Tra+ | 4 |
| pGEM-T Easy | Cloning vector for PCR products, received linearized Pnr | Promega |
| pCR-4Blunt-TOPO | Cloning vector for shotgun sequencing, received linearized, Kmr Pnr | Invitrogen |
| pAH149-1 | Clone of Rms149 bases 54514 to 56668 (repA) in pCR-4Blunt-TOPO, Kmr Pnr Rep+ | This work |
| pAH149-2 | Clone of Rms149 bases 55315 to 349 (parABC) in pCR-4Blunt-TOPO, Kmr Pnr Rep− | This work |
| pAH149-3 | Clone of Rms149 bases 55923 to 753 (parABC) in pCR-4Blunt-TOPO, Kmr Pnr Rep− | This work |
| pAH149-4 | Clone of Rms149 bases 53432 to 55556 (′repA) in pCR-4Blunt-TOPO, Kmr Pnr Rep− | This work |
| pAH149-5 | Clone of Rms149 bases 53922 to 56224 (repA) in pCR-4Blunt-TOPO, insert orientation is opposite pAH149-1 to 4, Kmr Pnr Rep+ | This work |
| pAH149-6 | Clone of Rms149 bases 4961 to 7198 [intI1 aac(3)-I] in pCR-4Blunt-TOPO, Gmr Kmr Pnr | This work |
| pAH149-7 | Clone of Rms149 bases 4125 to 6601 [intI1′ aac(3)-I aadA5] in pCR-4Blunt-TOPO, Gmr Kmr Pnr Smr Spr | This work |
| pAH149-12 | Derivative of pAH149-1 and pAH149-2 carrying Rms149 bases 54514 to 349, Kmr Pnr | This work |
| pAH149-13 | Derivative of pAH149-1 and pAH149-3 carrying Rms149 bases 54514 to 753, Kmr Pnr | This work |
| pAH149-1r | Deletion derivative of pAH149-1 without vector replicon or penicillin resistance, Kmr | This work |
| pAH149-12r | Deletion derivative of pAH149-12 without vector replicon or penicillin resistance, Kmr | This work |
| pAH149-13r | Deletion derivative of pAH149-13 without vector replicon or penicillin resistance, Kmr | This work |
| pGEM-149mob | PCR product of mobilisation region, bases 30665 to 35361 cloned into pGEM-T Easy, Pnr Mob+ | This work |
Antibiotic resistances: Cb, carbenicillin; Gm, gentamicin; Km, kanamycin; Pn, penicillin; Sm, streptomycin; Sp, spectinomycin; Su, sulfonamide. Other phenotypic traits: Mob+, mobilizable; Tra+, conjugative. For pAH149-1 to 5, Rep+ and Rep− indicate ability to replicate in P. putida from a replicon on the Rms149 insert.
In general standard microbiological techniques were used for growth and manipulation of bacteria. The standard medium was LB. Blood plates used were 40 g/liter tryptone soy agar with 50 ml/liter human blood. Minimal plates were 1.5% agar with M9 salts (6 g/liter NaH2PO4, 3 g/liter KH2PO4, 1 g/liter NH4Cl, 0.5 g/liter NaCl) supplemented with 2 g/liter glucose, 0.1 μM CaCl2, 1 mM MgSO4 and 5 mg/liter thiamine. Antibiotics used at standard concentrations for work with E. coli were as follows: kanamycin, 50 μg/ml; penicillin, 150 μg/ml (broth) and 300 μg/ml (agar); and streptomycin, 30 μg/ml.
DNA isolation, manipulation, and sequencing.
Large-scale purification of plasmid DNA from P. aeruginosa strain PU21 carrying Rms149 was performed using a slightly modified Birnboim and Doly technique followed by cesium chloride gradient purification from chromosomal DNA (39). Crude, small-scale purification of plasmid DNA during plasmid construction work used the modified Birnboim and Doly preparation only. The purified DNA was sheared and fragments were cloned using the TOPO shotgun subcloning kit from Invitrogen.
DNA preparation for sequencing used the QIAGEN MagAttract 96 miniprep system or in some cases Promega Wizard plus SV miniprep kits. Clones were initially sequenced using universal primers from both insert ends on an ABI 3700 sequencer. Finishing reads used custom primers on clones or PCR products. A few PCR products for sequencing or further study were cloned using the Promega pGEM-T Easy vector system.
The sequence was produced and assembled using the Phred/Phrap/Consed package (13, 14, 17, 18). Analysis and annotation of the sequence used Artemis and ACT (36) and WU-BLAST2 (16). Analysis of codon usage used the Pseudomonas aeruginosa PAO1 table (accessed from http://www.kazusa.or.jp/codon/).
Functional assays.
Conjugative transfer was performed by mixing loops picked from selective plates of donor and recipient onto L agar plates with parental control patches spread singly alongside. After incubation overnight, a loop of the mating mixture and parental strains were streaked to single colonies on individual and combined selection plates for the desired plasmid and recipient bacterium.
Estimations of plasmid loss rates were performed as described previously (5). Transformants were restreaked onto selective agar before growth overnight with selection and then diluted into nonselective L-broth. Viable cell counts were made using selective and nonselective agar both before and after growth. The number of generations is given by G = log2(CFU per unit volume after growth/CFU per unit volume before growth). If the proportions of cells carrying plasmid before and after growth are given by P0 and PE, then the apparent loss rate (as the percentage of plasmid-free daughter-cells per division) is L = (1 − (PE/P0)1/G) × 100.
Nucleotide sequence accession number.
The nucleotide sequence of Rms149 has been submitted to the EMBL database (accession number AJ877225).
RESULTS AND DISCUSSION
Overview.
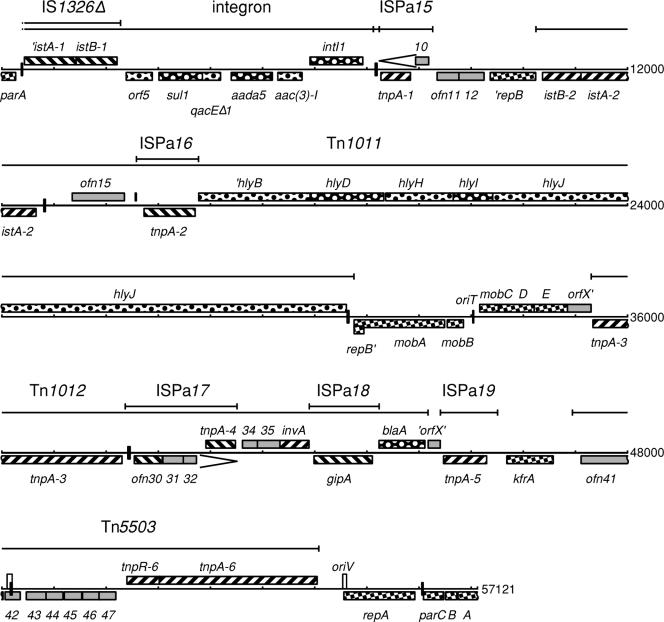
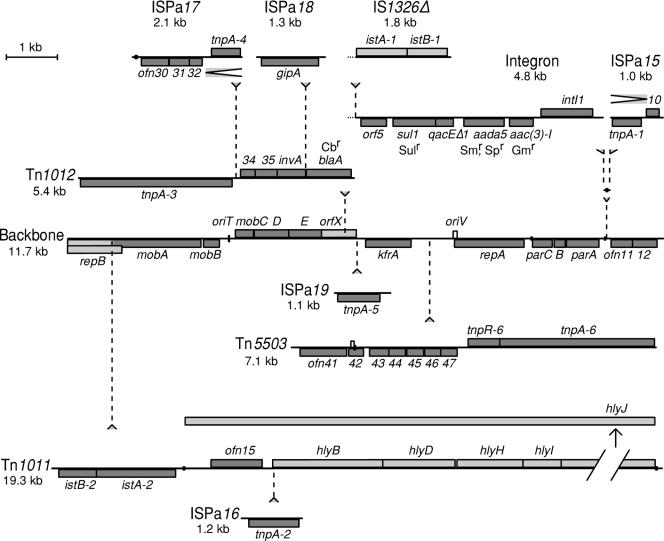
The 57,121-base-pair sequence of Rms149 was determined by sequencing a combination of cloned random fragments and PCR products. The entire range is covered by more than one clone or PCR and sequenced on both strands. Consed reports 0.0015 expected errors for this sequence. The average GC content is 59.5%, but using a 500-bp window, the content varies from 39 to 75%. The sequence was recursively analyzed for biologically relevant structures. Table 2 lists the 53 protein coding sequences identified, of which 10 appear to be inactivated by internal or upstream deletions or mobile element insertion. A map showing the locations of these coding regions is presented in Fig. 1. The functions of each open reading frame (ORF) were predicted by BLAST search of the EMBL databases: the products of 34 ORFs showed a predicted function by similarity to known proteins. Of these, five are predicted to be involved in replication and stable inheritance, six in transfer/mobilization, 14 in transposition, and nine in phenotypic determinants such as antibiotic resistance or virulence. The mosaic structure of Rms149 we have determined is shown in Fig. 2.
TABLE 2.
Biologically relevant reading frames
| Gene name | Coordinates and directiona | Size (no. of residues) | GC (%) | Most closely related gene product (EMBL accession no. of sequence)b | Product identity (%) | Proposed function |
|---|---|---|---|---|---|---|
| istA-1 | 434-1438 | 334 | 54 | IstA of IS1326, e.g., on Tn21 (AF071413) | 100 | Transposition, 5′ truncated, presumed inactive |
| istB-1 | 1425-2210 | 261 | 50 | IstB of IS1326, e.g., on Tn21 (AF071413) | 100 | Transposition, presumed inactive |
| orf5 | 2386-2886 c | 166 | 65 | Orf5 in 3′ conserved region of integron, e.g., In2 on Tn21 (AF071413) | 100 | Not known; conserved in integrons |
| sul1 | 3014-3853 c | 279 | 62 | Sul1 in 3′ conserved region of integron, e.g., In2 on Tn21 (AF071413) | 100 | Sulfonamide resistance, conserved in integrons |
| qacEΔ1 | 2847-4194 c | 115 | 50 | QacEΔ1 in 3′ conserved region of integron, e.g., In2 on Tn21 (AF071413) | 100 | Minimal resistance to quaternary ammonium compounds |
| aadA5 | 4400-5188 c | 262 | 58 | AadA5, integron cassette (e.g., AF137361) | 100 | Streptomycin and spectinomycin resistance |
| aac(3)-I | 5293-5757 c | 154 | 55 | Aac(3)-I, integron cassette (AF318077) | 95 | Gentamicin resistance |
| intI1 | 5909-6922 | 337 | 61 | IntI1 in 5′ conserved region of integron, e.g., In2 on Tn21 (AF071413) | 100 | Gene cassette integration |
| tnpA-1 | 7270-7836 c | 188 | 67 | DNA recombinase of composite transposon on pFBAOT6 (CR376602)c | 100 | Transposition |
| ofn10 | 7935-8186 | 83 | 63 | Unannotated open reading frame on pFBAOT6 (CR376602) | 100 | Unknown |
| ofn11 | 8347-8769 c | 140 | 63 | No significant protein matches | Unknown | |
| ofn12 | 8766-9248 c | 160 | 56 | 72% DNA identity to region of Xylella fastidiosa (AE003901) | - | Unknown |
| repB | 9366-10238, 30756-30944 c | 353 | 65 | C-terminus of polyprotein of pRAS3.2 (AY043299); equivalent to RepB in other IncQ-like plasmids | 97 | Primase of IncQ replicon; not required by Rms149, interrupted by Tn1011 |
| istB-2 | 10370-11110 c | 246 | 57 | IstB on pB4 (AJ431260) | 52 | Transposition |
| istA-2 | 11107-12657 c | 516 | 59 | IstA on pB4 (AJ431260) | 45 | Transposition |
| ofn15 | 13350-14351 | 333 | 58 | No significant matches | Unknown | |
| tnpA-2 | 14727-15707 c | 326 | 60 | Transposases, e.g., TnpA3 of ISPre2 on pCAR-1 (AB088420) | 95 | Transposition |
| hlyB | 14559-14579, 15776-17914 | 719 | 61 | Hemolysin export, ABC transporter protein of Photorhabdus luminescens subsp. laumondii (BX571861) | 80 | Toxin secretion ABC transporter protein-ISPa16 insertion, presumed inactive |
| hlyD | 17911-19326 | 471 | 63 | Hemolysin export, membrane fusion protein of Photorhabdus luminescens subsp. laumondii (BX571861) | 64 | Toxin secretion, presumed inactive |
| hlyH | 19362-20669 | 435 | 56 | Probable glycosyltransferase of Chromobacterium violaceum (AE016911) | 62 | Possible glycosyltransferase, presumed inactive |
| hlyI | 20656-21429 | 257 | 43 | No significant matches | Toxin pathway related? presumed inactive | |
| hlyJ | 21414-30608 | 3064 | 58 | Putative hemolysin, Ralstonia solanacearum (AL646058) (after removing annotated IS) | 41 | Putative hemolysin, presumed inactive |
| mobA | 30687-32492 c | 601 | 61 | N-terminus of polyprotein of pRAS3.2 (AY043299); equivalent to MobA in other IncQ-like plasmids | 87 | Mobilization presumed functional; reads in to Tn1011 |
| mobB | 32542-32853 c | 103 | 58 | MobB, IncQ-like plasmids, e.g. pTF-FC2 (M57717) (unannotated in this plasmid) | 92 | Mobilization |
| mobC | 33165-33521 | 118 | 55 | MobC, IncQ-like plasmids, e.g., pRAS3.2 (AY043299) | 98 | Mobilization |
| mobD | 33534-34217 | 227 | 53 | MobD, IncQ-like plasmids, e.g., pRAS3.2 (AY043299) | 99 | Mobilization |
| mobE | 34210-34851 | 213 | 60 | MobE, IncQ-like plasmids, e.g., pTF-FC2 (M57717) | 93 | Mobilization |
| orfX | 34848-35300, 44177-44408 | 228 | 55 | OrfX, IncQ-like plasmids, e.g., pRAS3.2 (AY043299) (unannotated in this plasmid) | 97 | Mobilization?, possibly inactive due to Tn1012 insertion |
| tnpA-3 | 35336-38305 c | 989 | 65 | Transposase of Tn3 family transposon on pFBAOT6 (CR376602) | 95 | Transposition |
| ofn30 | 38541-39077 c | 178 | 63 | Transposase-like protein of apparent insertion into mobile genome island TNCP23 on plasmid pKLC102 from P. aeruginosa (AY257539) | 100 | Transposition? |
| ofn31 | 39089-39484 c | 131 | 62 | Hypothetical protein of apparent insertion into mobile genome island TNCP23 on plasmid pKLC102 from P. aeruginosa (AY257539) | 100 | Unknown |
| ofn32 | 39481-39732 c | 83 | 62 | Hypothetical protein of apparent insertion into mobile genome island TNCP23 on plasmid pKLC102 from P. aeruginosa (AY257539) | 100 | Unknown |
| tnpA-4 | 39914-40480 | 188 | 67 | Recombinase of apparent insertion into mobile genome island TNCP23 on plasmid pKLC102 from P. aeruginosa (AY257539)c | 100 | Transposition |
| ofn34 | 40615-40905 | 96 | 63 | Putative nucleotidyltransferase of Tn3 family transposon on pFBAOT6 (CR376602) | 98 | Unknown |
| ofn35 | 40902-41339 | 145 | 61 | Hypothetical protein of Tn3 family transposon on pFBAOT6 (CR376602) (C-terminus affected by insertion/deletion, majority is 92% identical) | 87 | Unknown |
| invA | 41333-41890 | 185 | 63 | Recombinase of Tn3 family transposon on pFBAOT6 (CR376602) | 95 | Transposition |
| gipA | 41987-43105 c | 372 | 55 | Peyer's patch-specific virulence factor of lysogenic phage Gifsy-1, Salmonella enterica (AF246666) | 72 | Transposition |
| blaA | 43233-44114 | 293 | 60 | β-Lactamase, Aeromonas hydrophila (U14748) | 58 | Carbenicillin resistance |
| tnpA-5 | 44471-45298 c | 275 | 64 | Hypothetical exonuclease in bacteriophage 186 (U32222) | 45 | Transposition (putative) |
| kfrA | 45684-46574 c | 296 | 73 | KfrA of IncP-1 plasmids, e.g., pB4 (AJ431260) | 38 | Replication/partition associated |
| ofn41 | 47109-48023 c | 304 | 62 | C-terminal (major) part of hypothetical proteins, e.g., of Pseudomonas aeruginosa (AE004827) | 58 | Unknown |
| ofn42 | 48062-48352 c | 96 | 60 | N-terminal (minor) part of hypothetical proteins, e.g., of Pseudomonas aeruginosa (AE004827) | 41 | Unknown |
| ofn43 | 48470-48850 c | 126 | 64 | Conserved hypothetical protein of Tn3 family transposon on pFBAOT6 (CR376602)d | 84 | Unknown |
| ofn44 | 48847-49173 c | 108 | 63 | Conserved hypothetical protein of Tn3 family transposon on pFBAOT6 (CR376602)d | 94 | Unknown |
| ofn45 | 49197-49532 c | 111 | 59 | Putative transposase repressor of unmarked transposon on plasmid pND6-1 (AY208917) | 100 | Unknown |
| ofn46 | 49547-49882 c | 111 | 58 | Hypothetical protein of unmarked transposon on plasmid pND6-1 (AY208917) | 100 | Unknown |
| ofn47 | 49863-50186 c | 107 | 59 | Hypothetical protein of unmarked transposon on plasmid pND6-1 (AY208917) | 100 | Unknown |
| tnpR-6 | 50399-51034 | 211 | 64 | Resolvase of unmarked transposon on plasmid pND6-1 (AY208917) | 100 | Transposition |
| tnpA-6 | 51018-54047 | 1009 | 61 | Transposase of unmarked transposon on plasmid pND6-1 (AY208917) | 100 | Transposition |
| repA | 54559-55920 c | 453 | 66 | Putative replication protein of broad-host-range IncU plasmid pFBAOT6 (CR376602) | 71 | Replication |
| parC | 56086-56472 c | 128 | 54 | Hypothetical protein of pRA2, Pseudomonas alcaligenes (U88088) | 98 | Partitioning |
| parB | 56509-56727 c | 73 | 61 | Partitioning protein ParB of pRA2, Pseudomonas alcaligenes (U88088) | 100 | Partitioning |
| parA | 56749-57121, 1-266 c | 211 | 64 | Partitioning protein ParA of pRA2, Pseudomonas alcaligenes (U88088) | 100 | Partitioning |
c indicates the ORF is present on the complementary strand.
Where similar strength matches are found in multiple genera, organism names are not given.
TnpA-1 and TnpA-4 are identical. Different matches are given to represent the different contexts.
The DNA sequence covering ofn43 and ofn44 is highly conserved in an unmarked transposon on plasmid pND6-1, but these reading frames are not annotated in favor of an open reading frame in the opposite direction.
FIG. 1.
Linear map of Rms149. Bars above the map indicate the extent of mobile elements. Genes are shown above (sense strand) or below (complementary strand) the DNA line and filled according to known or putative function. Genes involved with plasmid functions are patterned with alternating squares, and phenotypic load genes are dotted. Genes involved with transposition are striped, with the direction being reversed for secondary insertions. Open reading frames of unknown function are shown in grey. Stem-loops are shown as black boxes over the DNA. The series of repeats in ofn42 is shown as a white box. The repeat between tnpA-1 and tnpA-4 is shown by chevrons.
FIG. 2.
Map of Rms149, showing insertions into the plasmid backbone. DNA is represented by black horizontal lines, with identified open reading frames as grey boxes above (sense strand) or below (complementary strand). Light grey boxes indicate apparently nonfunctional genes. Insertions of mobile elements are indicated by vertical dotted lines leading to the element. Stem-loops are shown as black boxes centered on the DNA. The tandem repeats in Tn5503 and the putative oriV are shown as white boxes. Terminal repeats of mobile elements are not shown. The deletion from inside IS1326 is indicated by a dotted DNA strand. The gene hlyJ has been shown as an outgroup because of its size. The large repeat between ISPa15 and ISPa17 is shown by chevrons.
Replication and partition region.
The putative genes for partitioning of Rms149 were identified by high sequence similarity (98% identity) to a 1.6-kb region of the plasmid pRA2, encoding an operon of three genes, parABC. In pRA2, the parAB segment confers segregational stability on low-copy-number plasmids (27). In Rms149 there is an additional gene downstream of these par genes. This encodes a product with weak similarity to several hypothetical replication proteins and strong similarity (71% identity) to the putative Rep protein of the recently sequenced IncU plasmid pFBAOT6 from Aeromonas caviae (34). In pRA2 this gene is absent and the region is bordered by a transposon downstream of parC.
To determine whether the region containing this gene is indeed capable of replication, several shotgun library clones were tested for their ability to transform P. putida strain KT2440, in which vector replicon is nonfunctional. It was found that clones carrying the downstream gene (and some fraction of parC) produced colonies, while control clones carrying parABC or other segments of Rms149 did not. Although small-scale plasmid DNA preparation did not yield enough DNA to visualize by agarose gel electrophoresis, the extracted DNA could be used to generate further transformants with competent bacteria. This confirms that free plasmid DNA was present even if the copy number was very low. We named this gene repA.
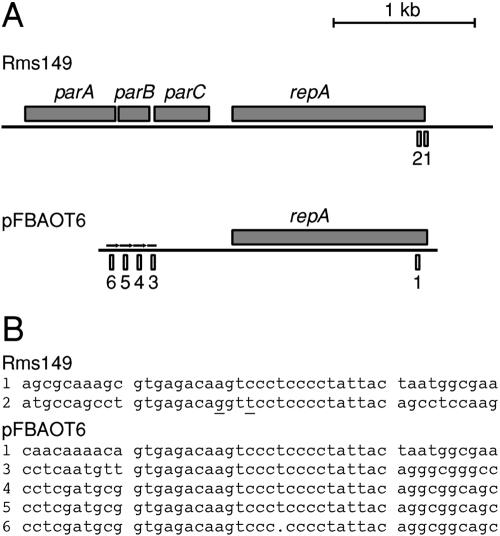
Since both orientations of the repA insert within the vector were found to allow growth in KT2440, it seems that replication ability is independent of transcription from the vector, and it is therefore likely that a promoter exists upstream of repA. We were unable to identify any close matches to promoter consensus boxes in this sequence, although we did find a putative promoter as a weak match. We did not find an obvious candidate for a replication origin. However, overlapping the 3′ end of the repA protein coding sequence are two repeats of the sequence GTGAGACA(G/A)GT(T/C)CCTCCCCTATTAC, which may be part of the origin of replication. In addition, although there is only one copy of the repeat at the 3′ end of the homologous gene in pFBAOT6 (34), there are three copies separated by 73-bp spacers 548 bases upstream of the start codon, followed by a copy with a single-base-pair deletion (Fig. 3).
FIG. 3.
Comparison of proposed replicons of Rms149 and pFBAOT6. A. Comparison of putative replicon structure. Sense-strand protein-coding regions are indicated by grey boxes above the black DNA line. Repeats putatively involved with replication are represented by numbered boxes below the DNA line. Repeats previously identified in pFBAOT6 (34) are shown as black arrows above the DNA line. The region of Rms149 shown is flanked by the truncated integron on the left and Tn5503 on the right. The region of pFBAOT6 shown is flanked by Tn1721 on the left and a further series of repeats on the right. B. Alignment of repeats proposed to be involved in replication. Ten surrounding bases are shown for each repeat. Repeat 2 of Rms149 has two base mismatches compared to the other repeats (underlined), while repeat 6 of pFBAOT6 has a single base deletion.
One reason for the very poor yield of plasmid DNA from clones with just repA in P. putida may be a high rate of segregational instability due to low copy number and lack of a partitioning system. Using clones from the random shotgun library digested with BstAPI and BstXI (which cut once in the insert and once in the vector, respectively), it was possible to construct plasmids carrying both parABC and repA. Surprisingly, the addition of the parABC region did not appear to improve the yield of plasmid DNA.
Hedges and Jacoby (21) reported that Rms149 could be mobilized to E. coli, implying that it was also capable of replication in this species. We wished to verify this property for the rep region identified. The shotgun library clones can replicate in E. coli under the control of the high-copy-number vector (pMB1) replicon, so we attempted to introduce these plasmids by transformation into C2110, a polA mutant strain of E. coli C in which the pMB1 replicon is known not to function. All attempts with plasmids carrying repA or parABC-repA of Rms149 proved negative. We concurrently transformed E. coli DH5α with samples of the same DNA and transformed the competent E. coli C2110 with a control plasmid. All these control transformations produced colonies. This suggests that this minimal IncP-6 replicon is either not functional in E. coli or is unable to function without DNA polymerase I.
To distinguish between the above possibilities, we deleted the pMB1 replicon from repA and parABC-repA plasmids in vitro and transformed P. putida, producing kanamycin-resistant but penicillin-sensitive clones. When plasmid DNA from these were used to transform E. coli DH5α, colonies were obtained. Plasmid DNA prepared from these transformants yielded faint bands with the expected restriction patterns. The mini replicon plasmids that we constructed could not be introduced into E. coli C2110. These results strongly suggest that the IncP-6 replicon depends on DNA polymerase I for replication.
Plasmid stability.
To determine whether the parABC region is important for stabilization of the replicon, we compared the stability of plasmids carrying only repA with that of two plasmids carrying parABC and repA. Of the two plasmids with par regions, one carries all the sequences that appear to be involved with partitioning, while the other one has the parABC coding region and the consensus promoter boxes but lacks one of three ParB regulatory sites identified in pRA2 (27). In E. coli, all three plasmids appeared to be stable, with 100% retention after approximately 20 generations of growth in nonselective medium. In P. putida, however, all three plasmids appeared very unstable, with a very low proportion of cells carrying the plasmid even after preliminary growth in selective broth. All had loss rates of 22 to 25% per generation over the 20-generation experiment. This suggests that there may be additional factors encoded by the plasmid that normally are involved in plasmid replication or stability.
It may be that the regions we have cloned are not the full sequence required for efficient replication or partitioning in some species. Downstream of repA, a transposon has inserted (Tn5503). However, beyond this insertion there appears to be a further region of plasmid backbone, encoding a predicted protein with significant identity and similar general characteristics to KfrA encoded by IncP-1 plasmids, particularly a predicted high α-helical content and heptad repeats characteristic of coiled-coil proteins (10). Such genes are found close to replication or partition regions of plasmids from many incompatibility groups (1). Upstream of the par region, beyond an integron insertion, are two orphan genes. Either or both of these regions may be necessary for reliable replication or full stability in Pseudomonas species, or alternatively the IncP-6 replicon may not be fully functional in the P. putida strain we used.
Mobilization.
A 5.6-kb region similar to the mobilization cassette of the IncQ-like plasmids pTF-FC2 (35) and pRAS3 (29) is present within Rms149. This places it in the MOBP family of mobilization regions (15). Transposons have inserted close to both ends of the region, truncating the mobA and repB gene products at one end and an unnamed open reading frame at the other. Although Hedges and Jacoby reported mobilization of Rms149 (21), attempts here using the IncP-1-mobilizing plasmids pUB307 and R751 failed to reproduce this. As an alternative way to test this property, the putative mob region between the two transposons was amplified by PCR and ligated into pGEM-T Easy to produce pGEM-149mob. This plasmid could be mobilized from E. coli DH5α to E. coli HB101 using pUB307, an IncP-1α plasmid lacking Tn1 and not conferring penicillin resistance. No mobilization was obtained with the negative control, a nonmobilizing pGEM derivative.
Insertions into the putative plasmid backbone and phenotypic determinants.
Rms149 carries several mobile elements, as is apparent from the number of transposition-related genes (Fig. 1 and Table 2). Figure 2 presents the genetic map redrawn so as to make clear how the backbone components and the transposons and insertion sequences are integrated. The central core of the plasmid consists of the replication, stable inheritance, and mobilization functions. Into this are inserted three major transposons, a partial, composite transposon carrying an integron, and one putative insertion sequence. A further four insertion sequences are inserted into these transposons. These insertions into the backbone are described below, starting with the integron, which provides the majority of the known phenotypic determinants.
In2-related truncated integron.
This integron is very similar to In2, a widespread element (9) originally discovered in Tn21 (32), and the related In0 from pVS1 (6). It contains the insertion sequence IS1326, which is also present in In0 and In2. However, a deletion has removed the integron's transposition gene-proximal terminus and part of this insertion sequence, which probably leaves both elements immobile.
The integron expression operon retains the conserved downstream-end genes qacEΔ1, sul1, and orf5. In addition, the first two integrated gene cassettes are highly related to genes known to confer gentamicin resistance (EMBL accession no. AF347074) and streptomycin-spectinomycin resistance (38). We have confirmed the resistances conferred by these two genes in E. coli, using shotgun-cloned DNA segments expressed from the integron promoter, by streaking to single colonies on selective media.
ISPa15.
This 1.0-kb element lies adjacent to the In2-related integron and is separated from it by a 114-bp segment. The integron terminal repeats are similar to ISPa15 termini: the remaining integron repeat in Rms149 matches 23 of 25 bp, in highly related integrons, the other one is identical. It therefore seems likely that ISPa15 would be capable of mobilizing a complete integron as a composite transposon. This would be consistent with the absence of direct repeats of insertion for ISPa15.
The 114-bp fragment between ISPa15 and the integron has several perfect matches in the EMBL nucleotide databases. Where neighboring sequence is available, these matching sequences themselves appear to be between an In2 family integron on one side and an ISPa15-related insertion sequence on the other. In particular, plasmid pFBAOT6 (34) appears to have a complete composite transposon, comprising the structure described above of an In2-like integron, a 114-bp spacer, and an ISPa15-like element, and in addition a different insertion sequence element on the other side of the integron. This opposing element has the same terminal repeats as ISPa15, and the whole structure is flanked by 5-bp direct repeats, indicating that it has moved as a unit. Therefore, we propose that in Rms149, a structure including at least ISPa15 and the integron previously formed a composite transposon.
On Rms149, a 715-bp global inverted repeat of 99% identity is a match of the majority of ISPa15 to ISPa17. A second small, divergently expressed open reading frame exists in the nonduplicated portion of ISPa15.
Tn1011.
This transposon is the largest element (19.3 kb) in the Rms149 mosaic and is inserted in the mobA-repB region, presumably inactivating repB but leaving the Mob function of mobA intact. Its transposition appears to be directed by a two-component transposase. At the termini there are multiple adjacent, degenerate repeats (four on the left, three on the right). The repeats are extended by a few bases at the outside ends to uniquely define the termini. Outside these there are direct repeats of 6 base pairs.
Tn1011 carries an operon of five genes, hlyBDHIJ, which we interpret to encode a pathway for the production and secretion of a large, repetitive hemolysin-like protein (HlyJ). The protein products of the first two genes, HlyB and HlyD, exhibit 80 and 64% sequence identity, respectively, across their entire length to known proteins involved in hemolysin export, while the fifth, HlyJ, shows a lower but still significant similarity (41% identity) to hemolysin proteins. The third encoded protein, HlyH, may be involved in glycosylation of one of the other proteins, as it is similar to several probable glycosyltransferases. The closest match found (62% identity) is expressed from an operon between the hlyBD and hlyJ-like genes. We could find no related proteins for the fourth encoded protein, HlyI.
The putative hly operon appears to have been rendered inactive by the simple insertion of the IS5 family insertion sequence ISPa16, just after the start of hlyB. This insertion has created a direct repeat of the 4-base-pair target site CTAG, typical of elements in this family. P. aeruginosa PU21(Rms149) did not produce halos when streaked onto blood plates, which would have been expected if it were producing active hemolysin (data not shown).
The GC content of the end of hlyH and hlyI and the start of hlyJ is relatively low, averaging 43%, so this region may have been integrated from another source, or alternatively the high AT content may have a structural purpose. Because of the low G+C content there will be many stop codons on a random basis unless counterselected, as in a protein-coding sequence. However, it was striking that the extra stop codons in this region were predominantly in one of the two alternative reading frames over both hlyI and the start of hlyJ. The expected stop codon frequency given the GC content is 5.7% for hlyI and 5.5% for the first 1,500 bases of hlyJ. The plus-one reading frame over both regions matches quite well to this (5.0% for hlyI, 5.6% for hlyJ). However, the plus-two reading frames have two to three times more stop codons than expected (13.2% for hlyI, 14.8% for hlyJ). We do not have an explanation for this.
Tn1012.
This transposon consists of a gene encoding a transposase divergently transcribed from an operon of three or four ORFs, including an invertase and a probable β-lactamase gene. It is also disrupted by two probable insertion sequences, one between the two transcriptional units and the other inserted just upstream of the β-lactamase. However, since the plasmid confers carbenicillin resistance and this is the only such gene, we conclude that the bla gene is still functional. The termini of the transposon are flanked by 5-bp direct repeats, suggesting that Tn1012 was acquired as a simple insertion into orfX without loss of information.
The first of the two insertions in Tn1012, ISPa17, appears to encode a transposase divergently from an operon of three genes ending with a second transposase-like gene. This structure is similar to that of Tn1012 prior to ISPa17 insertion. ISPa17 has a perfect, full-length bounded match in the sequence of transposon TNCP23 (of plasmid pKLC102 from P. aeruginosa) (25). In TNCP23, unlike in Rms149, it does not have direct repeats beyond its termini, which is presumably why it was not identified as an autonomous unit.
The second element disrupting Tn1012, ISPa18, encodes a single protein most similar to GipA, a transposase-like protein from the lysogenic phage Gifsy-1. While in the bacteriophage this gene is reported to confer virulence (40), it is flanked by matching palindromic repeats which are similar to those of ISPa18. Further, this protein shows similarity to the transposase of the IS605 family. This family does not have inverted repeats at its termini and does not generate direct repeats on insertion. Therefore, we identified the ends of this insertion sequence by comparing the alignment of several related sequences identified in a BLAST search.
Putative ISPa19.
We have identified what we believe may be a new kind of insertion sequence element. This would contain one ORF (tnpA-5) and be bordered by three 20-bp imperfect repeats at each end, the terminal repeats being extended by 9 bp at their outside ends. At the presumed promoter-proximal end, the repeats are separated, while at the other they are adjacent.
TnpA-5 has reasonable similarity to several putative exonucleases. There are no direct repeats bordering this sequence. However, the homology of Rms149 to pRAS3.2 stops at the insertion sequence border. This suggests that a deletion involving two insertion sequence copies in direct repeat may have occurred.
Tn5503.
This Tn3 family transposon shows 99% identity to a transposon on pND6-1 (31) and 97% to Tn5501 (30), a plasmid-carried transposon of P. putida. However, after a partial terminal inverted repeat-like sequence, this transposon encodes an additional two genes followed by a full-length terminal repeat. This extended structure is similar in nature to that described for Tn5502 (30). Outside the terminal repeats are 5-bp direct repeats, suggesting that these represent the ends of an element that was inserted as a single unit. The main part of the transposon encodes a two-component transposase system divergently from an operon of five small genes. Expression from these may continue into the extender region.
Taken together, the two open reading frames of the extra sequence show similarity to almost the whole of a single ORF in the P. aeruginosa PAO1 genome. In the first of the two Rms149 ORFs, there is a region which is not present in the P. aeruginosa PAO1 sequence. It consists of a 16-bp-stem perfect stem-loop that overlaps the first in a series of repeats by 6 bp. The repeats consist of two 9-bp repeats of sequence CCCAGAGAG, followed by seven 7-bp repeats of sequence CCCAGAG. One possibility is that this region may be involved in programmed genetic variation; a mutation deleting one (or adding two) 7-bp repeats from the current set of seven would create a fusion protein of ofn42 and ofn41.
Relationship of Rms149 sequences to Tn21.
Rms149 has two regions of similarity to Tn21, a transposon that carries an integron between the transposition and mercury resistance cassettes (32). The similar regions are the integron (excluding two captured antibiotic resistance genes) and the transposase and terminus of Tn1012. Adjacent to these is a long inverted repeat from the related insertion sequences ISPa15 and ISPa17. Therefore, a recombination event could rearrange these sequences into a conformation similar to that of Tn21. Conceivably, this process may have proceeded “in reverse”; a Tn21-like transposon could have been broken up. That this is not the case is clear from the direct repeats on either side of ISPa17 and Tn1012 itself, as well of the identification of an alternative, composite transposon carrying an integron on pFBAOT6. Indeed, the existence of this composite transposon in forms both with IS1326 (Rms149) and without (pFBAOT6) suggests that the insertion sequence insertion occurred in this context. Subsequent transfer of the IS1326-carrying integron has yielded a family of integrons, including In0, In2, and In5, in a variety of locations (9).
Propagation and descent of Rms149.
Given the large number of apparent insertion sequence function knockouts in this sequence, we were concerned that the plasmid may have evolved during subculture by removal of functions advantageous in the clinical context. However, Hedges and Jacoby (21) presented an EcoRI digest pattern for Rms149 which consists of four visible bands representing fragments with estimated sizes of 16, 12.6, 6, and 3 kb (after conversion from MDa). Within the degree of accuracy that is possible from the molecular weight markers available at the time and the relationship between size and mobility, this matches the predicted pattern from our DNA sequence. Were any of the detected mobile elements to have inserted after the initial restriction mapping, this would either modify the restriction pattern or require a larger error in estimation of fragment sizes. Similarly, the 3′ conserved fragment of the integron is 100% identical to that of In2, as far as the truncation point, and the integron of pFBAOT6 up to the IS1326 insertion. If this conservation were to continue to the end of the integron (9) or the end of a composite transposon, as in pFBAOT6, then the restriction map would be clearly different prior to the deletion.
However, we cannot exclude the loss of DNA mediated by a direct duplication and resolution of the putative element ISPa19. Indeed, the neighboring mobilization region's similarity terminates at one border of the element, suggesting that just such a deletion has occurred. The restriction fragment in the digestion pattern of Hedges and Jacoby (21) that we interpreted as covering this region equates to 12.6 kb, while we determined a size of 11.7 kb. The removal of up to around 1.7 kb by ISPa19 duplication and deletion by homologous recombination would increase the agreement between the two restriction patterns.
Conclusions.
Plasmids can be divided into two general groups: small, high-copy-number plasmids and larger, low-copy-number plasmids. Large plasmids are often self-transmissible, encoding a system that enables their horizontal transmission to other bacteria. Small plasmids, on the other hand, are often mobilizable, parasitizing a conjugative plasmid to transfer if one is present in the cell. From the size of Rms149, we expected it to fall into the former category, but the sequence reveals it to have characteristics of both smaller mobilizable plasmids and larger low-copy-number plasmids.
The largest group of backbone genes (mobA, mobB, mobC, mobD, and mobE) are those related to the IncQ superfamily of plasmids, but rather than providing both replication and mobilization, they only appear to provide mobilization functions. This is significant because IncQ-like plasmids use a sequential single-strand displacement mechanism of replication, where the DNA strands are synthesized separately. This may result in structural instability when plasmid size grows too large (33), and would thus not be compatible with expansion to 57 kb. Also, as the IncQ family replicon is optimized for high copy number, it does not include a partitioning system, so the low copy number necessary to prevent a plasmid of this size from being a significant burden on its host would result in segregational instability (3). It thus makes sense that the plasmid carries a different replicon, linked to widely distributed active partitioning genes.
Accumulating studies on the backbone functions of plasmids suggest that generally, once maintenance functions have been acquired by a plasmid, there is optimization of clustering and/or coregulation of the genes (41). In the case of Rms149, the mob block comes from a long lineage, while the rep-par region, although showing suitable integration, is composed of segments that are also found with other partners, suggesting relatively recent mixing and matching of these functions. Unsurprisingly, the relationship between the mob and rep-par systems does not reveal any evidence of optimization of organization or regulation. Plasmid pRA2, carrying the closest relative of the Rms149 par region, has a different rep system and distantly related mob and kfrA, although some of the linear organization of the plasmid is similar (28). Plasmid pFBAOT6, carrying the closest relative of the rep region, again has a distantly related kfrA gene and a different par and mob/tra system (34). It may be significant that although Rms149 has a novel combination of backbone functions, almost all are related to other known systems rather than representing novel replication, transfer, or partitioning genes. This is consistent with the view that a limited number of different plasmid module types are available for construction of new plasmids.
While we have emphasized that Rms149 carries a large amount of genetic baggage, this just represents a rather extreme version of a general tendency for plasmids to consist of a high proportion of mobile elements of one sort or another. We have previously noted how the multiple genetic layers of a plasmid can be uncovered by careful analysis of the DNA sequences present, as for example in our study of the TOL plasmid pWW0 (20). Our dissection of Rms149 illustrates this very clearly, although other plasmids have been found to have an even higher proportion, with 53% of identified genes encoding transposition functions (44). Apart from the antibiotic resistance determinants, the potential phenotypic determinants in Rms149 are probably nonfunctional. The putative secretory pathway on the large transposon Tn1011 takes up 15 kb but is apparently rendered inactive by insertion sequence ISPa16. While we cannot exclude the possibility that this insertion occurred during laboratory culture prior to the restriction mapping, it may be that it is a sign of rapid evolution in the clinical context, with conferred phenotypes switching rapidly between advantageous and detrimental. These lessons from Rms149 should help us to add to the general rules that apply to plasmid genome evolution, an important part of bacterial genomics.
Acknowledgments
This work was supported by a project grant from the Wellcome Trust (063083). The DNA sequencing was carried out in the University of Birmingham Functional Genomics Laboratory, funded by BBSRC grant JI6/F13209.
We thank Grazyna Jagura-Burdzy for training in the method for randomly sheared fragment library construction. The visit to the Institute of Biochemistry and Biophysics was partially funded by an EC Centre of Excellence grant to IBB-PAS, Warsaw. Poland.
REFERENCES
- 1.Adamczyk, M., and G. Jagura-Burdzy. 2003. Spread and survival of promiscuous IncP-1 plasmids. Acta Biochim. Pol. 50:425-453. [PubMed] [Google Scholar]
- 2.Bagdasarian, M., R. Lurz, B. Rückert, F. C. H. Franklin, M. M. Bagdasarian, J. Frey, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237-247. [DOI] [PubMed] [Google Scholar]
- 3.Becker, E. C., and Meyer, R. J. 1997. Acquisition of resistance genes by the IncQ plasmid R1162 is limited by its high copy number and lack of a partitioning mechanism. J. Bacteriol. 179:5947-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett, P. M., J. Grinsted, and M. H. Richmond. 1977. Transposition of TnA does not generate deletions. Mol. Gen. Genet. 154:205-211. [DOI] [PubMed] [Google Scholar]
- 5.Bignell, C. R., A. S. Haines, D. Khare, and C. M. Thomas. 1999. Effect of growth rate and incC mutation on symmetric plasmid distribution by the IncP-1 partitioning apparatus. Mol. Microbiol. 34:205-216. [DOI] [PubMed] [Google Scholar]
- 6.Bissonnette, L., and P. H. Roy. 1992. Characterization of In0 of Pseudomonas aeruginosa plasmid pVS1, an ancestor of integrons of multiresistance plasmids and transposons of gram-negative bacteria. J. Bacteriol. 174:1248-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boronin, A. M. 1992. Diversity of Pseudomonas plasmids: to what extent? FEMS Microbiol. Lett. 100:461-468. [DOI] [PubMed] [Google Scholar]
- 8.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in E. coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 9.Brown, H. J., H. W. Stokes, and R. M. Hall. 1996. The integrons In0, In2, and In5 are defective transposon derivatives. J. Bacteriol. 178:4429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Combet, C., C. Blanchet, C. Geourjon, and G. Deléage. 2000. NPS@: Network protein sequence analysis. Trends Biochem. Sci. 25:147-150. [DOI] [PubMed] [Google Scholar]
- 11.Couturier, M., F. Bex, P. L. Berquist, and W. Maas. 1988. Identification and classification of bacterial plasmids. Microbiol. Rev. 52:375-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eberhard, W. G. 1989. Why do bacterial plasmids carry some genes and not others? Plasmid 21:167-174. [DOI] [PubMed] [Google Scholar]
- 13.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 14.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Research. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 15.Francia, M. V., A. Varsaki, M. P. Garcillán-Barcia, A. Latorre, C. Drainas, and F. de la Cruz. 2004. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 28:79-100. [DOI] [PubMed] [Google Scholar]
- 16.Gish, W. 2004. http://blast.wustl.edu.
- 17.Gordon, D., C. Abajian, and P. Green. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 18.Gordon, D., C. Desmarais, and P. Green. 2001. Automated Finishing with Autofinish. Genome Res. 11:614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grant, S. G. N., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greated, A., L. Lambertsen, P. A. Williams, and C. M. Thomas. 2002. Complete sequence of the IncP-9 TOL plasmid pWW0 from Pseudomonas putida. Environ. Microbiol. 4:856-871. [DOI] [PubMed] [Google Scholar]
- 21.Hedges, R. W., and G. A. Jacoby. 1980. Compatibility and molecular properties of plasmid Rms149 in Pseudomonas aeruginosa and E. coli. Plasmid 3:1-6. [DOI] [PubMed] [Google Scholar]
- 22.Hedges, R. W., and M. Matthew. 1979. Acquisition by Escherichia coli of plasmid-bourne β-lactamases normally confined to Pseudomonas spp. Plasmid 2:269-278. [DOI] [PubMed] [Google Scholar]
- 23.Jacoby, G. A. 1974. Properties of R plasmids determining gentamycin resistance by acetylation in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 6:239-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacoby, G. A. 1986. Resistance plasmids of Pseudomonas, p. 265-293. In J. R. Sokatch (ed.), The bacteria, vol. X. Academic Press, New York, N.Y.
- 25.Klockgether, J., O. Reva, K. Larbig, and B. Tümmler. 2004. Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa C. J. Bacteriol. 186:518-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi, N., and M. J. Bailey. 1994. Plasmids Isolated from the sugar-beet phyllosphere show little or no homology to molecular probes currently available for plasmid typing. Microbiology UK 140:289-296. [DOI] [PubMed] [Google Scholar]
- 27.Kwong, S. M., C. C. Yeo, and C. L. Poh. 2001. Molecular analysis of the pRA2 partitioning region: ParB autoregulates parAB transcription and forms a nucleoprotein complex with the plasmid partition site, parS. Mol. Microbiol. 40:621-633. [DOI] [PubMed] [Google Scholar]
- 28.Kwong, S. M., C. C. Yeo, A. Suwanto, and C. L. Poh. 2000. Characterization of the endogenous plasmid from Pseudomonas alcaligenes NCIB 9867: DNA sequence and mechanism of transfer. J. Bacteriol. 182:81-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.L'Abée-Lund, T. M., and H. Sørum. 2002. A global non-conjugative Tet C plasmid, pRAS3, from Aeromonas salmonicida. Plasmid 47:172-181. [DOI] [PubMed] [Google Scholar]
- 30.Lauf, U., C. Müller, and H. Herrmann. 1998. The transposable elements resident on the plasmids of Pseudomonas putida strain H, Tn5501 and Tn5502, are cryptic transposons of the Tn3 family. Mol. Gen. Genet. 259:674-678. [DOI] [PubMed] [Google Scholar]
- 31.Li, W., J. Shi, X. Wang, Y. Han, W. Tong, L. Ma, B. Liu, and B. Cai. 2004. Complete nucleotide sequence and organization of the naphthalene catabolic plasmid pND6-1 from Pseudomonas sp. strain ND6. Gene 336:231-240. [DOI] [PubMed] [Google Scholar]
- 32.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rawlings, D. E., and E. Tietze. 2001. Comparative biology of IncQ and IncQ-like plasmids. Microbiol. Mol. Biol. Rev. 65:481-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhodes, G., J. Parkhill, C. Bird, K. Ambrose, M. Jones, G. Huys, J. Swings, R. W. Pickup. 2004. The complete nucleotide sequence of the conjugative tetracycline resistance plasmid pFBAOT6, a member of a group of IncU plasmids with global ubiquity. Appl. Environ. Microbiol. 70:7497-7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohrer, J., and D. E. Rawlings. 1992. Sequence analysis and characterization of the mobilization region of a broad-host-range plasmid, pTF-FC2, isolated from Thiobacillus ferrooxidans. J. Bacteriol. 174:6230-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M.-A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualisation and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 37.Sagai, H., V. Krcmery, K. Hasuda, S. Iyobe, H. Knothe, and S. Mitsuhashi. 1975. R factor-mediated resistance to aminoglycoside antibiotics in Pseudomonas aeruginosa. Jpn. J. Microbiol. 19:427-432. [DOI] [PubMed] [Google Scholar]
- 38.Sandvang, D. 1999. Novel streptomycin and spectinomycin resistance gene as a gene cassette within a class 1 integron isolated from Escherichia coli. Antimicrob. Agents Chemother. 43:3036-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, C. A., and C. M. Thomas. 1983. Deletion mapping of kil and kor functions in the trfA and trfB regions of broad host range plasmid RK2. Mol. Gen. Genet. 190:245-254. [DOI] [PubMed] [Google Scholar]
- 40.Stanley, T. L., C. D. Ellermeier, and J. M. Slauch. 2000. Tissue-specific gene expression identifies a gene in the lysogenic phage Gifsy-1 that affects Salmonella enterica serovar Typhimurium survival in Peyer's patches. J. Bacteriol. 182:4406-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas, C. M. 2000. Paradigms of plasmid organization. Mol. Microbiol. 37:485-491. [DOI] [PubMed] [Google Scholar]
- 42.Thomas, C. M. 2004. Evolution and population genetics of bacterial plasmids, p. 509-528. In B. E. Funnell and G. J. Phillips (ed.), Plasmid biology. American Society for Microbiology, Washington, D.C.
- 43.Thomas, C. M., and A. S. Haines. 2004. Plasmids of the genus Pseudomonas, p. 197-231. In J.-L. Ramos (ed.), Pseudomonas, vol. 1. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 44.Venkatesan, M. M., M. B. Goldberg, D. J. Rose, E. J. Grotbeck, V. Burland, and F. R. Blattner. 2001. Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infect. Immun. 69:3271-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams, P. A., R. M. Jones, and G. Zylstra. 2004. Genomics of catabolic plasmids. p. 165-195. In J-L. Ramos (ed.), Pseudomonas, vol. 1. Kluwer Academic/Plenum Publishers, New York, NY.