Abstract
Background
The emergence of drug-resistant Tuberculosis (TB) has made treatment challenging. Although fluoroquinolones (FQs) are used as key drugs in the treatment of multidrug-resistant tuberculosis (MDR-TB), the problem of FQs resistance is becoming increasingly serious. Rifampicin (RIF) resistance is considered a risk factor for FQs resistance. The objective of this study was to investigate the impact of RIF and isoniazid (INH) resistance on the FQs resistance in vitro experiment.
Methods
FQs resistant strains were selected in vitro from RIF and/or INH resistant Mycobacterium smegmatis (M.sm). The sequencing of the gyrA gene, and the minimum inhibitory concentration (MIC) of FQs (ciprofloxacin, levofloxacin, moxifloxacin and gatifloxacin) were performed for FQs-resistant strains.
Results
A total of 222 FQs-resistant M.sm strains were selected, all of which had the gyrA mutation. Seven gyrA mutations were detected, with mutations at loci 90 and 94 being the most common. There were no differences in FQs resistance developed from RIF and/or INH resistant M.sm. There was a significant difference in the MIC of the gyrA mutant types to FQs. The highest resistance to FQs was observed in the Gly88Cys mutant strains. M.sm with the identical gyrA mutation showed the highest resistance to ciprofloxacin and relatively low resistance to gatifloxacin and moxifloxacin.
Conclusions
In this study, we found no evidence that RIF and/or INH resistance directly affects FQs resistance in M.sm in vitro experiments. Resistance profiles of different gryA mutations to the four FQs drugs were also presented. These findings provide a more comprehensive understanding of FQs resistance.
Introduction
Tuberculosis (TB) is one of the three major infectious diseases threatening global public health, and is caused by infection with Mycobacterium tuberculosis (M.tb) [1]. The WHO reports that 10.6 million people worldwide had TB in 2022, an increase of 300,000 compared to 2021. Of these, 7.5 million were newly diagnosed with TB, the highest peak in 28 years of global TB surveillance[2, 3]. Approximately 500,000 multidrug-resistant/rifampicin-resistant TB (MDR/RR-TB) cases are reported each year, and drug-resistant is a major cause of TB deaths [4]. The cure rate for MDR/RR-TB patients is only 63%, posing a serious threat to public health [5–8].
Fluoroquinolones (FQs) are important drugs in the treatment of MDR/RR-TB [9]. FQs are commonly used in the treatment of drug-resistant TB because of their efficacy, oral administration, relatively few side effects and low cost compared with other second-line injectable drugs [10]. FQs can inhibit bacterial DNA gyrase and disrupt genome replication to exert bactericidal activity. Bacteria develop resistance to FQs through mutations in the gyrA gene, which encodes a DNA gyrase [11, 12]. In recent years, FQs resistant TB has become increasingly common in clinics, posing a challenge to treating the disease. Treatment success rates for pre-XDR (resistant to one of the FQs while fulfil the definition of MDR/RR-TB) and XDR (resistant to bedaquiline or linezolid while fulfil the definition of pre-XDR) TB are only 30~40% [5, 13–15].
Clinical data show that the drug resistance profile may influence the risk of other drug resistance in TB. Compared to susceptible TB, rifampicin (RIF) monoresistant patients and MDR patients had a 6.3-fold and 13.8-fold risk of FQs resistance, respectively [16, 17]. Therefore, to guide the use of FQs in the clinic, it is essential to clarify whether the drug-resistant background influences the development of resistance to FQs. Due to the slow growth and biosafety concerns of M.tb, Mycobacterium smegmatis (M.sm) is often used as a model organism to study M.tb drug resistance [18–22]. Therefore, we used M.sm to investigate whether RIF and/or isoniazid (INH) resistance influences the development of FQs resistance.
Materials and methods
RIF-R, INH-R and MDR strains
To investigate the effect of different drug-resistant M.sm on FQ resistance, the RIF-resistant (RIF-R), INH-resistant (INH-R), MDR (both RIF and INH resistant), and drug-susceptible (both RIF and INH susceptible) MC2-155 strains were used, which were rpoB gene S531L mutant (MSRIF-R), katG gene G280D mutant (MSINH-R), rpoB gene S531L and katG gene G280D double mutant (MSMDR), and wild type (MSS), respectively.
In vitro selection of FQs-resistant mutants
We isolated spontaneous mutant colonies using 2× minimal inhibitory concentration (MIC) [23] of levofloxacin (0.5 μg/mL) with the Luria-Delbrück fluctuation assay [24]. For the initial preparation of the cell suspension, the optical density (OD) at 600 nm was used to measure the CFU, as a bacterial solution with an OD600 equal to 1.0 corresponds to approximately 2×108 CFU/ml [25]. For each strain, an approximately 2.3 × 105 CFU were added to 90 ml of Middlebrook 7H9 liquid medium containing 10% OADC (7H9-OADC), mixed well, and divided into 20 culture tubes. Each tube contains 4ml of cell suspension, approximately 104 CFU. These parallel cultures were then incubated at 37°C until the OD600 reached 1.0 [26]. Among twenty parallel cultures for each strain, 15 were used for isolating spontaneous FQ-resistant mutant colonies and 5 for counting CFU. Spontaneous FQ-resistant mutant colonies were selected on Middlebrook 7H11 solid medium with 10% OADC (7H11-OADC) and 1.0 μg/mL levofloxacin (2× MIC). Serially diluted cell suspensions were added to drug-free 7H11-OADC for CFU counting. Mutation rates and 95% confidence intervals (CI) were calculated by the online tools of FluCalc, which was able to accurately estimate mutation rates using the fluctuation assay [27].
DNA extraction and gyrA gene sequencing
Bacterial genomic DNA was extracted by boiling method [28, 29]. The gyrA-1R (5’-CGACGAACTGTTCTCCATCC-3’) and gyrA-1F (5’-CCGGTCTTGTACGTGTCCTC-3’) primers were used to amplify the 869 bp fragment of the gyrA gene [13]. The obtained PCR products were sent for Sanger sequencing to identify the FQs-resistant mutations.
Minimal inhibitory concentration (MIC) assay for FQs
MICs were determined in 7H9-OADC by the resazurin microtiter assay (REMA) using a twofold FQ drugs dilution [30, 31]. Ciprofloxacin was tested at a range of 0.25 to 128 μg/mL, while levofloxacin at a range of 0.0156 to 8 μg/mL, moxifloxacin at a range of 0.0625 to 32 μg/mL and gatifloxacin at a range of 0.0625 to 32 μg/mL. The MIC is interpreted as the minimum drug concentration that prevents the color changing from blue to purple or pink.
Statistical analysis
The chi-squared test was used to compare the mutations between the groups, while the one-way ANOVA method and the t-test were used to compare the MIC. All analyses were performed with SPSS version 29.0 and results with a p-value less than 0.05 were considered statistically significant.
Results
Selection of FQs-resistant clones
A total of 609 FQs-resistant clones were selected by the Luria-Delbrück fluctuation assay. The number of FQs-resistant clones from MSS, MSINH-R, MSRIF-R and MSMDR were 96, 349, 88 and 76, respectively. The frequency of FQs resistance was 2.93×10−10 (95% CI: 1.67–4.42×10−10) for MSS, 5.28×10−10 (95% CI: 3.17–7.77×10−10) for MSINH-R, 8.11×10−10 (95% CI: 4.33–12.72×10−10) for MSRIF-R and 5.09×10−10 (95% CI: 2.56–8.23×10−10) for MSMDR. Of the 609 FQs-resistant clones, 222 were randomly selected for gyrA sequencing and FQs MICs testing, of which 66 clones were from MSS, 66 clones from MSINH-R, 41 clones from MSRIF-R and 49 clones from MSMDR.
FQs-resistant mutations
Each sequenced clone harbored a mutation in the gyrA gene. There were seven types of mutation, located at loci 88, 90, 91 and 94 of the gyrA gene (Table 1). Mutations at loci 90 and 94 were the most common, accounting for 54.1% (120/222) and 30.6% (68/222) of all samples, respectively. Chi-squared test showed a significant difference in the mutation profile between MSS and MSINH-R (p = 0.004), and between MSINH-R and MSRIF-R (p = 0.002) (S1 Table). The mutation profile comparisons between MSS and MSINH-R, and between MSINH-R and MSRIF-R were performed independently. Compared to MSINH-R, the MSS strain had a significantly higher proportion of Gly88Cys and Asp94Tyr mutations, but a significantly lower proportion of Asp94Gly mutation. And MSINH-R strain had a significantly higher proportion of Ala90Val mutation, but a significantly lower proportion of Ser921Pro mutation than MSRIF-R strain.
Table 1. gyrA mutation profile of M.sm strains.
| Mutation | MSS | MSINH-R | MSRIF-R | MSMDR |
|---|---|---|---|---|
| n = 66 (%) | n = 66 (%) | n = 41 (%) | n = 49 (%) | |
| Gly88Cys | 11 (16.7) | 2 (3.0) | 5 (12.2) | 3 (6.1) |
| Ala90Val | 36 (54.5) | 40 (60.6) | 14 (34.1) | 30 (61.2) |
| Ser91Pro | 3 (4.5) | 1 (1.5) | 9 (22.0) | 0 (0.0) |
| Asp94His | 0 (0.0) | 1 (1.5) | 1 (2.4) | 0 (0.0) |
| Asp94Tyr | 6 (9.1) | 0 (0.0) | 0 (0.0) | 2 (4.1) |
| Asp94Gly | 10 (15.2) | 21 (31.8) | 12 (29.3) | 14 (28.6) |
| Asp94Ala | 0 (0.0) | 1 (1.5) | 0 (0.0) | 0 (0.0) |
Abbreviations: Gly, Glycine; Cys, cysteine; Ala, Alanine; Val, Valine; Ser, Serine; Pro, Proline; Asp, Aspartic acid; His, Histidine; Tyr, Threonine.
FQs MIC of M.sm strains
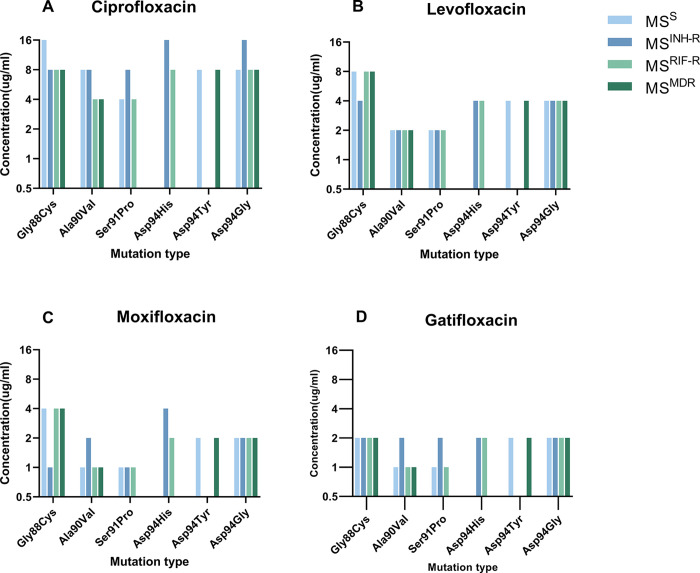
In order to ascertain whether drug-resistance backgrounds influence FQs resistance, this study analyzed the MIC of FQs in MSS, MSINH-R, MSRIF-R, and MSMDR with identical gyrA mutations. The MICs of ciprofloxacin, levofloxacin, moxifloxacin and gatifloxacin for strains with the same mutation were found to be generally consistent, with MIC differences observed primarily within a range of one titer (Fig 1). The results indicated that resistance to RIF and/or INH did not influence the level of FQs resistance. Consequently, the MIC results of different strains with the identical mutation were combined and subjected to further analysis. (S2 and S3 Tables).
Fig 1. The FQs MIC among M.sm strains.
(A) Ciprofloxacin, (B) Levofloxacin, (C) Moxifloxacin, (D) Gatifloxacin. No clone with gyrA Ser91Pro mutation was selected form MSMDR, no clone with gyrA Asp94His mutation was selected form MSS and MSMDR, and no clone with gyrA Asp94Tyr mutation was selected form MSINH-R and MSRIF-R.
FQs MIC of gyrA mutations
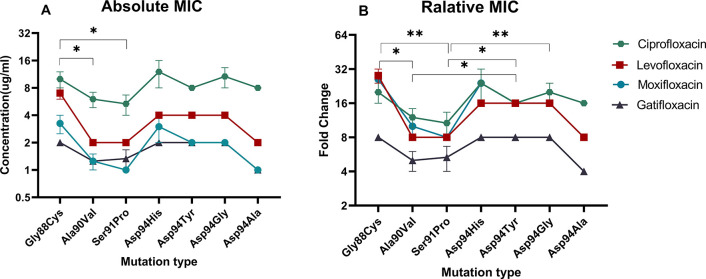
The gyrA mutation increased the MIC of ciprofloxacin from 0.5μg/mL to 4.0~16.0 μg/mL, levofloxacin from 0.25μg/mL to 2.0~8.0 μg/mL, moxifloxacin from 0.125μg/mL to 1.0~4.0 μg/mL, and gatifloxacin from 0.25μg/mL to 1.0~2.0 μg/mL (Fig 1). An analysis of the FQs MICs for different mutations revealed that the MIC of the Gly88Cys mutation was significantly higher than that of the Ala90Val (p = 0.015) and Ser91Pro (p = 0.016) mutations (Fig 2A). This suggests that the Gly88Cys mutation may confer higher resistance to FQs, while Ala90Val and Ser91Pro confer lower. No significant difference was observed in the MICs between the remaining mutations.
Fig 2.
The FQs absolute MIC (A) and relative MIC (B) of gryA mutations. * indicates a p-value of less than 0.05, ** indicates a p-value of less than 0.01.
Differences in MIC of four FQ drugs
In general, the mutant clones exhibited variability in their MICs for the four FQ drugs. The mutant clones had the highest resistance to ciprofloxacin (4.0~16.0 μg/mL), followed by levofloxacin (2.0~8.0μg/mL), and the lowest resistance to moxifloxacin (1.0~4.0 μg/mL) and gatifloxacin (1.0~2.0 μg/mL) (Fig 1). With the exception of no difference being found in MICs between moxifloxacin and gatifloxacin, significant differences were observed in the comparisons of MICs for all the drugs (p<0.001) (S4 Table)
The relative changes in the FQs MIC of the mutant strains relative to the wild-type strain were further analyzed. Resistance mutations resulted in the smallest mean increase in minimum inhibitory concentration (MIC) for gatifloxacin, with a mean increase of 6.8-fold (interquartile range: 4–8), whereas levofloxacin, ciprofloxacin, and moxifloxacin showed a larger mean increase in MIC, with a mean increase of 16-fold (IQR: 8–16) (Fig 2B). The relative changes in MIC of gatifloxacin were significantly lower than those of levofloxacin, ciprofloxacin, and moxifloxacin (p<0.001) (S5 Table). Among all the mutation types, Ala90Val and Ser91Pro showed lower relative changes than Gly88Cys, Asp94Tyr and Asp94Gly (Fig 2B).
Discussion
In this study, a total of 222 FQs-resistant M.sm strains were selected in vitro from RIF and/or INH resistant M.sm, all of which had the gyrA mutation. The MICs of FQs developed from M.sm strains resistant to RIF and/or INH were found to be no significantly different. Among seven gyrA mutations were detected, the highest resistance to FQs was observed in the Gly88Cys mutant strains. M.sm with the identical gyrA mutation showed the highest resistance to ciprofloxacin and relatively low resistance to gatifloxacin and moxifloxacin. The findings will contribute to a more comprehensive understanding of drug resistance in FQs.
The gyrA gene encodes the α-subunit of bacterial DNA gyrase, which is the target of FQ drugs [32, 33]. FQ resistance is the consequence of the drug-resistant mutations in the gyrA gene [34, 35]. A total of seven FQ-resistant mutations at four codon sites have been observed in this study. All of these mutations have been reported in clinical FQ-resistant M.tb strains [36–38]. We also found more than one mutation type in codon 94 conferring FQ resistance, which is consistent with clinical M.tb strains [36, 39]. These results suggest that M.sm can be used as a model for the study of FQ resistance in M.tb.
Clinical studies have demonstrated that the prevalence of FQs resistance in MDR/RR-TB cases is considerably higher than in susceptible cases, indicating that INH/RIF resistance may contribute to the risk of FQs resistance [40, 41]. Nevertheless, our study did not identify a significant difference in the frequency of FQs resistance among RIF and/or INH-resistant M.sm. This suggests that it is not drug resistance profiles but other factors that increase the risk of FQ resistance. One potential explanation is that FQs are primarily used for the treatment of drug-resistant TB [9], whereas patients with susceptible TB have limited opportunity to use FQs. Consequently, the overall FQs resistance rate is higher in drug-resistant TB than in susceptible TB [41]. Studies have shown that subinhibitory concentrations of drugs induce bacteria to generate reactive oxygen species (ROS) that increase the mutation rate [42], thereby increasing the risk of drug resistance. Because the in vivo environment is complex and drug concentrations in tissues vary spatially and temporally [43, 44], it is possible that subinhibitory concentrations may exist in patients and increase the rate of drug resistance mutations. Compared to susceptible strains, drug-resistant strains have more opportunities to be in a subinhibitory environment, which could potentially favour the development of drug resistance.
Not only patient factors, but also bacterial factors other than drug resistance may promote resistance to FQs. Bacteria can prevent drugs from entering into cells to reduce its susceptibility, and can also excrete drugs out of bacteria through efflux pumps to increase the ability of drug resistance, which involving many genes, such as pe11, mymA, virS, cpnT, mmpL3, bacA, efpA [45]. These mechanisms that affect drug susceptibility are usually not specific to a single drug, but affect multiple drugs [46]. Drug-resistant strain can also acquire compensatory mutation that partially or fully restore the deficiency caused by the drug resistance conferring mutations [47]. A secondary in the rpoA, ropB and rpoC genes was a common compensatory mutation to restore the deficiency caused by RIF resistance [48], may have a potential contribution to the development of FQs resistance. In addition, genetic background may have an effect on susceptibility to FQs, as the MIC of lineage 3 strains was lower than that of lineages 4 [49]. Whether these bacterial factors contribute to the promotion of FQs resistance requires further investigation.
FQs are frequently used to treat MDR-TB patients as an important component of the WHO-recommended short-course treatment regimen for TB [50, 51]. FQs were initially discovered as broad-spectrum antibiotics in 1962, and four generations of FQ drugs have been developed to date [52]. Our study involved three generations of FQs, including second-generation (ciprofloxacin), third-generation (levofloxacin), and fourth-generation (moxifloxacin and gatifloxacin) [53]. Strains with the same gyrA mutation differed in their resistance to different generations of FQs. In general, the more recent generation of FQs exhibited a lower MIC. The MIC of ciprofloxacin for the Gly88Cys mutant strain was 16 μg/mL, while that of gatifloxacin was 2 μg/mL, representing an 8-fold difference. This phenomenon makes it possible for patients to achieve the same therapeutic effect and reduce the risk of side effects by taking lower doses of new-generation FQs. It is also possible to treat low-level drug-resistant TB by using high-dose new-generation FQs.
This study had several limitations. Although, M.sm is often used as a model organism to study M.tb drug resistance [18–22], there are genetic and physiological differences between them. The results of this study require further validation using both M.tb reference and clinical strains with different drug resistance profiles. Secondly, the diversity and complexity of drug resistance mechanisms in clinical M.tb strains, which may have evolved under different selective pressures in vivo [54], were not considered in this study. Thirdly, this study did not assess the effect of other bacterial factors on the development of resistance to FQs, in particular compensatory mutations, which are common in clinical RIF-resistant strains. These factors need to be studied in order to draw more comprehensive conclusions.
In summary, this study assesses the impact of RIF and/or INH-resistant profile on the development of FQs resistance in M.sm. We found that there was no significant direct effect of RIF and/or INH resistance profile on FQs resistance in M.sm in vitro experiments. This indicates that there are other reasons for the higher FQs-resistant rate in drug-resistant TB. Furthermore, the distribution of MICs between generations of FQs and gyrA mutations was presented. These findings provide a more comprehensive understanding of FQs resistance.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by Shenzhen Science and Technology Program (No. JCYJ20230807153604010) awarded to PX, Shenzhen High-level Hospital Construction Fund (No. G2022157) to Peng Xu, Shenzhen Clinical Research Center for Tuberculosis (No. 20210617141509001) awarded to PX, National College Students' innovation and entrepreneurship training program (202310661058) awarded to QZ and NP, and Students' innovation and entrepreneurship training program of Guizhou Province (S202310661110) awarded to QZ and NP. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Singh R, Dwivedi SP, Gaharwar US, Meena R, Rajamani P, Prasad T. Recent updates on drug resistance in Mycobacterium tuberculosis. J Appl Microbiol. 2020;128(6):1547–67. Epub 20191029. doi: 10.1111/jam.14478 . [DOI] [PubMed] [Google Scholar]
- 2.Bagcchi S. WHO’s Global Tuberculosis Report 2022. Lancet Microbe. 2023;4(1):e20. Epub 20221212. doi: 10.1016/S2666-5247(22)00359-7 . [DOI] [PubMed] [Google Scholar]
- 3.Organization WH. Global tuberculosis report 2023 2023. Available from: https://www.who.int/zh. [Google Scholar]
- 4.Organization WH. WHO consolidated guidelines on tuberculosis. Module 1: prevention–tuberculosis preventive treatment 2020. Available from: https://www.who.int/zh. [PubMed] [Google Scholar]
- 5.Dheda K, Gumbo T, Maartens G, Dooley KE, McNerney R, Murray M, et al. The epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant, extensively drug-resistant, and incurable tuberculosis. Lancet Respir Med. 2017. Epub 20170315. doi: 10.1016/S2213-2600(17)30079-6 . [DOI] [PubMed] [Google Scholar]
- 6.Lange C, Dheda K, Chesov D, Mandalakas AM, Udwadia Z, Horsburgh CR, Jr. Management of drug-resistant tuberculosis. Lancet. 2019;394(10202):953–66. doi: 10.1016/s0140-6736(19)31882-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lange C, Chesov D, Heyckendorf J, Leung CC, Udwadia Z, Dheda K. Drug-resistant tuberculosis: An update on disease burden, diagnosis and treatment. Respirology. 2018;23(7):656–73. Epub 20180411. doi: 10.1111/resp.13304 . [DOI] [PubMed] [Google Scholar]
- 8.Garcia PK, Annamalai T, Wang W, Bell RS, Le D, Martin Pancorbo P, et al. Mechanism and resistance for antimycobacterial activity of a fluoroquinophenoxazine compound. PLOS ONE. 2019;14(2):e0207733. doi: 10.1371/journal.pone.0207733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanino E, Granozzi B, Akkerman OW, Munoz-Torrico M, Palmieri F, Seaworth B, et al. Update of drug-resistant tuberculosis treatment guidelines: A turning point. Int J Infect Dis. 2023;130 Suppl 1:S12–s5. Epub 20230312. doi: 10.1016/j.ijid.2023.03.013 . [DOI] [PubMed] [Google Scholar]
- 10.Lan Z, Ahmad N, Baghaei P, Barkane L, Benedetti A, Brode SK, et al. Drug-associated adverse events in the treatment of multidrug-resistant tuberculosis: an individual patient data meta-analysis. Lancet Respir Med. 2020;8(4):383–94. Epub 20200317. doi: 10.1016/S2213-2600(20)30047-3 ; PubMed Central PMCID: PMC7384398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aldred KJ, Blower TR, Kerns RJ, Berger JM, Osheroff N. Fluoroquinolone interactions with Mycobacterium tuberculosis gyrase: Enhancing drug activity against wild-type and resistant gyrase. Proc Natl Acad Sci U S A. 2016;113(7):E839–46. Epub 20160120. doi: 10.1073/pnas.1525055113 ; PubMed Central PMCID: PMC4763725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spencer AC, Panda SS. DNA Gyrase as a Target for Quinolones. Biomedicines. 2023;11(2). Epub 20230127. doi: 10.3390/biomedicines11020371 ; PubMed Central PMCID: PMC9953508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo T, Yuan J, Peng X, Yang G, Mi Y, Sun C, et al. Double mutation in DNA gyrase confers moxifloxacin resistance and decreased fitness of Mycobacterium smegmatis. J Antimicrob Chemother. 2017;72(7):1893–900. doi: 10.1093/jac/dkx110 . [DOI] [PubMed] [Google Scholar]
- 14.Kabir S, Tahir Z, Mukhtar N, Sohail M, Saqalein M, Rehman A. Fluoroquinolone resistance and mutational profile of gyrA in pulmonary MDR tuberculosis patients. BMC Pulm Med. 2020;20(1):138. Epub 20200511. doi: 10.1186/s12890-020-1172-4 ; PubMed Central PMCID: PMC7216623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tritar F, Daghfous H, Ben Saad S, Slim-Saidi L. [Management of multidrug-resistant tuberculosis]. Rev Pneumol Clin. 2015;71(2–3):130–9. Epub 20140819. doi: 10.1016/j.pneumo.2014.05.001 . [DOI] [PubMed] [Google Scholar]
- 16.Xu P, Li X, Zhao M, Gui X, DeRiemer K, Gagneux S, et al. Prevalence of fluoroquinolone resistance among tuberculosis patients in Shanghai, China. Antimicrob Agents Chemother. 2009;53(7):3170–2. Epub 20090413. doi: 10.1128/AAC.00177-09 ; PubMed Central PMCID: PMC2704692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang JY, Lee LN, Lai HC, Wang SK, Jan IS, Yu CJ, et al. Fluoroquinolone resistance in Mycobacterium tuberculosis isolates: associated genetic mutations and relationship to antimicrobial exposure. J Antimicrob Chemother. 2007;59(5):860–5. Epub 20070405. doi: 10.1093/jac/dkm061 . [DOI] [PubMed] [Google Scholar]
- 18.Borrell S, Teo Y, Giardina F, Streicher EM, Klopper M, Feldmann J, et al. Epistasis between antibiotic resistance mutations drives the evolution of extensively drug-resistant tuberculosis. Evol Med Public Health. 2013;2013(1):65–74. Epub 20130308. doi: 10.1093/emph/eot003 ; PubMed Central PMCID: PMC3868377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danilchanka O, Pavlenok M, Niederweis M. Role of porins for uptake of antibiotics by Mycobacterium smegmatis. Antimicrob Agents Chemother. 2008;52(9):3127–34. Epub 20080616. doi: 10.1128/AAC.00239-08 ; PubMed Central PMCID: PMC2533485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida M, Nakata N, Miyamoto Y, Fukano H, Ato M, Hoshino Y. A rapid and non-pathogenic assay for association of Mycobacterium tuberculosis gyrBA mutations and fluoroquinolone resistance using recombinant Mycobacterium smegmatis. FEMS Microbiol Lett. 2018;365(23). doi: 10.1093/femsle/fny266 . [DOI] [PubMed] [Google Scholar]
- 21.Lelovic N, Mitachi K, Yang J, Lemieux MR, Ji Y, Kurosu M. Application of Mycobacterium smegmatis as a surrogate to evaluate drug leads against Mycobacterium tuberculosis. J Antibiot (Tokyo). 2020;73(11):780–9. Epub 20200529. doi: 10.1038/s41429-020-0320-7 ; PubMed Central PMCID: PMC7554168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim KH, An DR, Yoon HJ, Yang JK, Suh SW. Structure of Mycobacterium smegmatis Eis in complex with paromomycin. Acta Crystallogr F Struct Biol Commun. 2014;70(Pt 9):1173–9. Epub 20140829. doi: 10.1107/S2053230X14017385 ; PubMed Central PMCID: PMC4157414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillespie SH, Basu S, Dickens AL, O’Sullivan DM, McHugh TD. Effect of subinhibitory concentrations of ciprofloxacin on Mycobacterium fortuitum mutation rates. J Antimicrob Chemother. 2005;56(2):344–8. Epub 20050614. doi: 10.1093/jac/dki191 . [DOI] [PubMed] [Google Scholar]
- 24.Sebastian J, Swaminath S, Nair RR, Jakkala K, Pradhan A, Ajitkumar P. De Novo Emergence of Genetically Resistant Mutants of Mycobacterium tuberculosis from the Persistence Phase Cells Formed against Antituberculosis Drugs In Vitro. Antimicrob Agents Chemother. 2017;61(2). Epub 20170124. doi: 10.1128/AAC.01343-16 ; PubMed Central PMCID: PMC5278719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang H, Yu D, Hong Y, Zhou X, Li C, Sun B. The LuxR family regulator Rv0195 modulates Mycobacterium tuberculosis dormancy and virulence. Tuberculosis (Edinb). 2013;93(4):425–31. Epub 20130511. doi: 10.1016/j.tube.2013.04.005 . [DOI] [PubMed] [Google Scholar]
- 26.Yan S, Xu M, Wang R, Li Q, Yu Z, Xie J. Overexpression of Rv2788 increases mycobacterium stresses survival. Microbiol Res. 2017;195:51–9. Epub 20161117. doi: 10.1016/j.micres.2016.11.007 . [DOI] [PubMed] [Google Scholar]
- 27.Radchenko EA, McGinty RJ, Aksenova AY, Neil AJ, Mirkin SM. Quantitative Analysis of the Rates for Repeat-Mediated Genome Instability in a Yeast Experimental System. Methods Mol Biol. 2018;1672:421–38. doi: 10.1007/978-1-4939-7306-4_29 ; PubMed Central PMCID: PMC5741187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Afghani B, Stutman HR. Polymerase chain reaction for diagnosis of M. tuberculosis: comparison of simple boiling and a conventional method for DNA extraction. Biochem Mol Med. 1996;57(1):14–8. doi: 10.1006/bmme.1996.0003 . [DOI] [PubMed] [Google Scholar]
- 29.Hosseinali Z, Mohammadshahi J, Teimourpour A, Habibzadeh S, Esmaelizad M, Arzanlou M, et al. Molecular identification of multiple drug resistance (MDR) strain of Mycobacterium tuberculosis. Mol Biol Rep. 2023;50(12):10271–5. Epub 20231116. doi: 10.1007/s11033-023-08867-7 . [DOI] [PubMed] [Google Scholar]
- 30.Palomino JC, Martin A, Camacho M, Guerra H, Swings J, Portaels F. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2002;46(8):2720–2. doi: 10.1128/AAC.46.8.2720-2722.2002 ; PubMed Central PMCID: PMC127336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rivoire N, Ravololonandriana P, Rasolonavalona T, Martin A, Portaels F, Ramarokoto H, et al. Evaluation of the resazurin assay for the detection of multidrug-resistant Mycobacterium tuberculosis in Madagascar. Int J Tuberc Lung Dis. 2007;11(6):683–8. . [PubMed] [Google Scholar]
- 32.Dighe SN, Collet TA. Recent advances in DNA gyrase-targeted antimicrobial agents. Eur J Med Chem. 2020;199:112326. Epub 20200428. doi: 10.1016/j.ejmech.2020.112326 . [DOI] [PubMed] [Google Scholar]
- 33.Mayer C, Takiff H. The Molecular Genetics of Fluoroquinolone Resistance in Mycobacterium tuberculosis. Microbiol Spectr. 2014;2(4):Mgm2-0009-2013. doi: 10.1128/microbiolspec.MGM2-0009-2013 . [DOI] [PubMed] [Google Scholar]
- 34.Chaiyachat P, Chaiprasert A, Nonghanphithak D, Smithtikarn S, Kamolwat P, Pungrassami P, et al. Whole-genome analysis of drug-resistant Mycobacterium tuberculosis reveals novel mutations associated with fluoroquinolone resistance. Int J Antimicrob Agents. 2021;58(3):106385. Epub 20210620. doi: 10.1016/j.ijantimicag.2021.106385 . [DOI] [PubMed] [Google Scholar]
- 35.Maruri F, Guo Y, Blackman A, van der Heijden YF, Rebeiro PF, Sterling TR. Resistance-Conferring Mutations on Whole-Genome Sequencing of Fluoroquinolone-resistant and -Susceptible Mycobacterium tuberculosis Isolates: A Proposed Threshold for Identifying Resistance. Clin Infect Dis. 2021;72(11):1910–8. doi: 10.1093/cid/ciaa496 ; PubMed Central PMCID: PMC8315129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Gao X, Luo T, Wu J, Sun G, Liu Q, et al. Association of gyrA/B mutations and resistance levels to fluoroquinolones in clinical isolates of Mycobacterium tuberculosis. Emerg Microbes Infect. 2014;3(3):e19. Epub 20140312. doi: 10.1038/emi.2014.21 ; PubMed Central PMCID: PMC3974338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uddin MKM, Ather MF, Nasrin R, Rahman T, Islam A, Rahman SMM, et al. Correlation of gyr Mutations with the Minimum Inhibitory Concentrations of Fluoroquinolones among Multidrug-Resistant Mycobacterium tuberculosis Isolates in Bangladesh. Pathogens. 2021;10(11). Epub 20211102. doi: 10.3390/pathogens10111422 ; PubMed Central PMCID: PMC8623510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chien JY, Chiu WY, Chien ST, Chiang CJ, Yu CJ, Hsueh PR. Mutations in gyrA and gyrB among Fluoroquinolone- and Multidrug-Resistant Mycobacterium tuberculosis Isolates. Antimicrob Agents Chemother. 2016;60(4):2090–6. Epub 20160325. doi: 10.1128/AAC.01049-15 ; PubMed Central PMCID: PMC4808166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malik S, Willby M, Sikes D, Tsodikov OV, Posey JE. New insights into fluoroquinolone resistance in Mycobacterium tuberculosis: functional genetic analysis of gyrA and gyrB mutations. PLoS One. 2012;7(6):e39754. Epub 20120628. doi: 10.1371/journal.pone.0039754 ; PubMed Central PMCID: PMC3386181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaidi SMA, Haseeb A, Habib SS, Malik A, Khowaja S, SaifUllah N, et al. Emergence of fluoroquinolone resistance among drug resistant tuberculosis patients at a tertiary care facility in Karachi, Pakistan. BMC Res Notes. 2017;10(1):313. Epub 20170725. doi: 10.1186/s13104-017-2633-6 ; PubMed Central PMCID: PMC5526320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ali S, Khan MT, Khan AS, Abbas Q, Irfan M. Fluoroquinolone Resistance Among Isolates of Mycobacterium tuberculosis in Khyber Pakhtunkhwa, Pakistan. Microb Drug Resist. 2021;27(6):786–91. Epub 20201030. doi: 10.1089/mdr.2020.0118 . [DOI] [PubMed] [Google Scholar]
- 42.Bhattacharya G, Dey D, Das S, Banerjee A. Exposure to sub-inhibitory concentrations of gentamicin, ciprofloxacin and cefotaxime induces multidrug resistance and reactive oxygen species generation in meticillin-sensitive Staphylococcus aureus. J Med Microbiol. 2017;66(6):762–9. Epub 20170609. doi: 10.1099/jmm.0.000492 . [DOI] [PubMed] [Google Scholar]
- 43.Turnidge J. Pharmacokinetics and pharmacodynamics of fluoroquinolones. Drugs. 1999;58 Suppl 2:29–36. doi: 10.2165/00003495-199958002-00006 . [DOI] [PubMed] [Google Scholar]
- 44.Ji B, Lounis N, Maslo C, Truffot-Pernot C, Bonnafous P, Grosset J. In vitro and in vivo activities of moxifloxacin and clinafloxacin against Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1998;42(8):2066–9. doi: 10.1128/AAC.42.8.2066 ; PubMed Central PMCID: PMC105862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu G, Liu H, Jia X, Wang X, Xu P. Mechanisms and detection methods of Mycobacterium tuberculosis rifampicin resistance: The phenomenon of drug resistance is complex. Tuberculosis. 2021;128:102083. Epub 2021/05/12. doi: 10.1016/j.tube.2021.102083 . [DOI] [PubMed] [Google Scholar]
- 46.Gygli SM, Borrell S, Trauner A, Gagneux S. Antimicrobial resistance in Mycobacterium tuberculosis: mechanistic and evolutionary perspectives. FEMS Microbiol Rev. 2017;41(3):354–73. doi: 10.1093/femsre/fux011 . [DOI] [PubMed] [Google Scholar]
- 47.Alame Emane AK, Guo X, Takiff HE, Liu S. Drug resistance, fitness and compensatory mutations in Mycobacterium tuberculosis. Tuberculosis (Edinb). 2021;129:102091. Epub 20210521. doi: 10.1016/j.tube.2021.102091 . [DOI] [PubMed] [Google Scholar]
- 48.Brunner VM, Fowler PW. Compensatory mutations are associated with increased in vitro growth in resistant clinical samples of Mycobacterium tuberculosis. Microb Genom. 2024;10(2). doi: 10.1099/mgen.0.001187 ; PubMed Central PMCID: PMC10926696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Consortium CR. Quantitative measurement of antibiotic resistance in Mycobacterium tuberculosis reveals genetic determinants of resistance and susceptibility in a target gene approach. Nature Communications. 2024;15(1):488. Epub 20240112. doi: 10.1038/s41467-023-44325-5 ; PubMed Central PMCID: PMC10786857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Falzon D, Schünemann HJ, Harausz E, González-Angulo L, Lienhardt C, Jaramillo E, et al. World Health Organization treatment guidelines for drug-resistant tuberculosis, 2016 update. Eur Respir J. 2017;49(3). Epub 20170322. doi: 10.1183/13993003.02308-2016 ; PubMed Central PMCID: PMC5399349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mirzayev F, Viney K, Linh NN, Gonzalez-Angulo L, Gegia M, Jaramillo E, et al. World Health Organization recommendations on the treatment of drug-resistant tuberculosis, 2020 update. Eur Respir J. 2021;57(6). Epub 20210604. doi: 10.1183/13993003.03300-2020 ; PubMed Central PMCID: PMC8176349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahashi H, Hayakawa I, Akimoto T. [The history of the development and changes of quinolone antibacterial agents]. Yakushigaku Zasshi. 2003;38(2):161–79. . [PubMed] [Google Scholar]
- 53.King DE, Malone R, Lilley SH. New classification and update on the quinolone antibiotics. Am Fam Physician. 2000;61(9):2741–8. . [PubMed] [Google Scholar]
- 54.Castro RAD, Borrell S, Gagneux S. The within-host evolution of antimicrobial resistance in Mycobacterium tuberculosis. FEMS Microbiol Rev. 2021;45(4). doi: 10.1093/femsre/fuaa071 ; PubMed Central PMCID: PMC8371278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.