Abstract
The genome sequence of the hyperthermophilic bacterium Thermotoga maritima MSB8 presents evidence for lateral gene transfer events between bacterial and archaeal species. To estimate the extent of genomic diversity across the order Thermotogales, a comparative genomic hybridization study was initiated to compare nine Thermotoga strains to the sequenced T. maritima MSB8. Many differences could be associated with substrate utilization patterns, which are most likely a reflection of the environmental niche that these individual species occupy. A detailed analysis of some of the predicted variable regions demonstrates many examples of the deletion/insertion of complete cassettes of genes and of gene rearrangements and insertions of DNA within genes, with the C or N terminus being retained. Although the mechanism for gene transfer in this lineage remains to be elucidated, this analysis suggests possible associations with repetitive elements and highlights the possible benefits of rampant genetic exchange to these species.
The genome of the hyperthermophilic bacterium Thermotoga maritima strain MSB8 (4) was sequenced in 1999 (9). Comparative genome analysis of the single circular chromosome (1.86 Mbp; 46% G+C) with the genomes of other completely sequenced microbial species presented evidence for lateral gene transfer (LGT), with approximately 24% of the open reading frames (ORFs) having their best matches to genes from archaeal species. Many of these genes were also of atypical composition and were often found in clusters (termed “archaeal islands”), which had a conservation of gene order with the archaeal species that was the closest apparent relative. Additional analysis of the T. maritima genome showed the presence of clustered regularly interspaced short palindromic repeats (CRISPR) in eight distinct loci on the chromosome. CRISPR have a remarkable structure that consists of a 30-bp repeat element interspersed with a variable and nonrepetitive 39- to 40-bp sequence called the “spacer.” They are thought to increase in size by duplicating the repeat sequences and adding at least one new spacer by a mechanism that is still not known; the origin of the variable spacer sequences also remains elusive. These CRISPR elements have been identified in a broad range of microbial species, such as Salmonella enterica serovar Typhimurium, Streptococcus pyogenes, Mycobacterium tuberculosis and Campylobacter jejuni (5, 18). The unique structure of CRISPR and their association with a group of conserved genes (called CAS genes, for CRISPR-associated sequences), which are potentially involved in DNA recombination and repair, provide additional clues for an active role of CRISPR elements in the mobilization of DNA.
Subsequent to the completion of the T. maritima strain MSB8 genome sequence, Nesbo and coworkers presented a series of studies that investigated potential LGT events in the Thermotoga lineage (14-16). In the first of these studies, the patterns of acquisition of two “archaeal-like” genes in the order Thermotogales were investigated, and the results lent additional support to the movement of the predicted “archaeal-like” genes across the domains. Suppressive subtractive hybridization (SSH) (1) was subsequently used to compare the genome of the sequenced strain MSB8 to Thermotoga sp. strain RQ2 (99.7% identity in the small-subunit rRNA sequence), which was isolated from the geothermally heated sea floor in Ribiera Quente, the Azores. This SSH study allowed for a partial identification of strain-specific sequences and resulted in a subset of sequences comprising approximately 48 kb of strain RQ2-specific DNA. Based on this finding, it was estimated that 20% of the strain RQ2 genome was not present in the genome of strain MSB8. Most recently, Nesbo and coworkers have screened lambda libraries that were created from strain RQ2 DNA for five regions that are absent from the MSB8 genome (14). Among the gene clusters found to be unique to strain RQ2 were an archaeal-type ATPase, a rhamnose biosynthesis operon, and an arabinosidase island.
With the advent of whole-genome sequencing, many new examples of gene transfer events between archaea and bacteria have come to the forefront (2, 3). However, although it is now evident that there is a high level of genetic exchange in the Thermotoga lineage, the study of LGT “is still in its adolescence” (7, 8), and the mechanism(s) of the exchange, its direction, and the degree to which it occurs are still not known. In an attempt to gain further insight into gene transfer in the Thermotoga lineage, we initiated a comparative genome hybridization (CGH) study with the reference-sequenced MSB8 genome against nine strains of Thermotoga (including strain RQ2) that have been isolated from different locations throughout the world.
MATERIALS AND METHODS
Strains.
All genomic DNA that was used in this study was provided by Karl Stetter and Robert Huber from the University of Regensburg, Germany. The name, location of origin, and optimal growth temperature of the strains that were included in the analysis are presented in Table 1. To test for the closeness of the strains to each other, the complete 16S rDNA gene for all of the strains was PCR amplified and sequenced as previously described (11, 13). Phylogenetic analyses on the 16S rDNA sequences were performed using the PHYLIP phylogeny inference package version 3.5. Pairwise evolutionary distances were computed from sequences aligned by ClustalX using the PHYLIP program, DNADIST, and a Kimura model for substitution. Stability associated with treeing orders was evaluated by using the programs SEQBOOT, DNADIST, NEIGHBOR, and CONSENSE within the PHYLIP program. One hundred bootstrap trees were generated for each data set.
TABLE 1.
Thermotoga strains used in the comparative genomic hybridization study
| Strain | Habitat | Literature name | Optimal growth temp (°C) |
|---|---|---|---|
| MSB8 | Geothermal heated seafloor, Vulcano Island, Italy | T. maritima | 55-90 |
| VMA1/L2B | Vulcano Island, Italy | 90 | |
| LA4 | Shore of Lac Abbc, Djibouti | T. neapolitana LA4 | 82 |
| LA10 | Shore of Lac Abbe, Djibouti | T. neapolitana LA10 | 87 |
| NS-E | Shallow submarine hot spring, Naples, Italy | T. neapolitana NS-E | 55-90 |
| NE2x/L8B | Naples, Italy | 90 | |
| NE7/L9B | Naples, Italy | 90 | |
| S1/L12B | Naples, Italy | 90 | |
| RQ2 | Geothermal heated seafloor, Ribeira Quente, the Azores | Thermotoga sp. strain RQ2 | 76-82 |
| RQ7 | Geothermal heated seafloor, Ribeira Quente, the Azores | Thermotoga sp. strain RQ7 | 76-82 |
| PB1platt | Oil field at Prudhoe Bay, Alaska | 90 |
Microarray procedure.
The microarray procedure used in this study is as previously described at http://www.tigr.org/microarray/Vanco_Paper/ (supplemental material for reference 8a). Briefly, 1,865 unique PCR products representing 99.4% of the T. maritima MSB8 genome were printed in duplicate onto UltraGAPS slides (Corning Life Sciences, Acton, MA) by means of a Molecular Dynamics Generation III array spotter (Amersham Biosciences, Piscataway, NJ). Genomic DNA was labeled by indirect coupling to Cy3 or Cy5 dyes as described at http://www.tigr.org/microarray/Vanco_Paper/. The quality and specific activity of the probes were confirmed by a spectrophotometric scan from 200 nm to 700 nm. Typically, 8 μg of aminoallyl DNA was obtained from 1 μg of genomic DNA template, with 40 to 50 pmol of dye nucleotide incorporated per μg of aminoallyl DNA produced.
Tagged image file format images of the hybridized arrays were analyzed using TIGR Spotfinder software (http://www.tigr.org/software/), the data set was normalized by applying the linear regression algorithm of the MIDAS software (http://www.tigr.org/software/), and values were then averaged to determine the final ratio (R = MSB8/test strain) reported for each ORF. For each comparison, at least two flip-dye experiments (four hybridizations) were performed. Statistical analysis of the data collected was performed on log2-transformed signal ratios (log2 [test strain/MSB8]) by using GACK analysis software (6) (http://falkow.stanford.edu/whatwedo/software/software.html), which provides an estimate of the probability (%EPP) that any given gene is present in the test strain compared to the control. The %EPP, ranging from −0.5 (high likelihood of divergence) to 0.5 (high likelihood of presence), was then transformed to an estimated probability of divergence (%EPD), ranging from 0 to 100 (highest likelihood of divergence). Based on the CGH data and the GACK analysis, genes were considered to be shared between T. maritima MSB8 and the test strain when the signal ratios were less than 3 and divergent for signal ratios greater than 10. Hierarchical clustering of the CGH data, as presented in Fig. 1B, was performed using the TIGR-MEV package (http://www.tigr.org/software/).
FIG. 1.
Relatedness of the Thermotoga strains used in this study. (A) Small-subunit rRNA gene phylogeny. *, strains that were compared in the CGH study. (B) CGH results and hierarchical clustering based on the CGH data.
Amplification and sequencing of divergent regions.
One of the main disadvantages of CGH studies is that although single genes or regions that are present in the reference genome can be identified as absent from the test strains, it is impossible to tell whether the same region has been replaced by a foreign piece of DNA in the test strain or if the same gene/region has become highly divergent and is therefore no longer detectable under the conditions used for hybridization. In the present study, the main question concerned genetic exchange in this lineage and, as such, it was necessary to conduct a detailed analysis of the larger regions that we identified as being absent from one of the test strains, RQ2 in this case. Strain RQ2 was chosen for the detailed analysis, as this strain appeared to be highly metabolically divergent and was also the main focus of the SSH- and lambda-based studies of Nesbo et al. (14, 16). The regions that were chosen for detailed analysis, size of product, and possibility of an insertion or deletion event are presented in Tables S1 and S3. Whenever the same region was found to be divergent throughout the majority of the strains, this region was amplified in all of the relatives. These regions were then sequenced either by walking directly on the PCR product or through the sequencing of microlibraries (12). The χ2 analysis was performed as previously described (9).
Nucleotide sequence accession numbers.
The nucleotide sequences of the strain RQ2 regions presented in Fig. 3 and Tables S1 and S3 have been deposited in GenBank under the following accession numbers: DQ073429, DQ073430, DQ073431, DQ073432, DQ073433, DQ073434, DQ073435, and DQ073436.
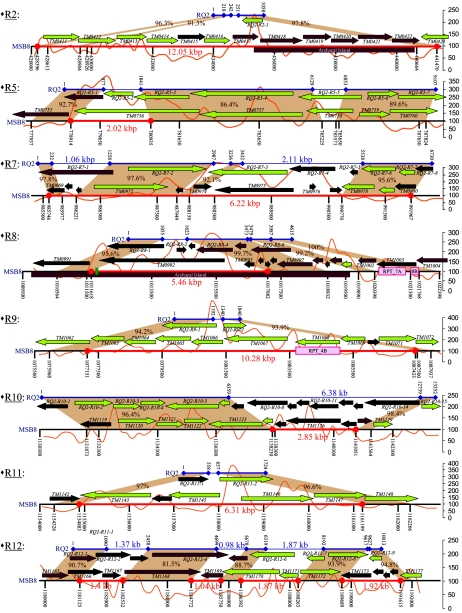
FIG. 3.
Analysis of the sequenced RQ2 regions compared to their MSB8 counterparts. For each RQ2 region that was sequenced to closure and analyzed, the blue line (top) represents the PCR product obtained for RQ2, whereas the black line (bottom) represents the same region in MSB8. Red lines delimited by two red filled circles were drawn to give length indications of remarkable sequences of the MSB8 genome. The coordinates refer to the start of the PCR product for RQ2 and to the whole genome for MSB8. MSB8 genes (above the black line for MSB8) and RQ2 genes (underneath the blue line for RQ2) are color coded according to their best match. Green, best match to bacterial species; brown, best match to bacterial species; black, hypothetical proteins (no match). The orange line represents the χ2 analysis of the G+C content of the MSB8 region (a scale is shown on the right). The brown areas represent the nucleotide identities (values in dark brown) between the MSB8 and RQ2 sequences. Finally, remarkable features are represented, such as predicted “archaeal islands” in the MSB8 genome (brown boxes), repeats (pink boxes).
RESULTS
Relatedness of the Thermotoga strains by 16S rDNA and CGH analysis.
A phylogenetic representation of the 16S rDNA sequences for the strains that were used in this study is presented in Fig. 1A. All 10 Thermotoga strains clustered very closely but were found to separate into three distinct groups, one of which is more closely related to the sequenced MSB8, and another to Thermotoga neapolitana strain NS-E. The strains that clustered with the sequenced strain MSB8 include NE2x/L8B, NE7/L9B, and S1/L12B, which shared 100% identity to the 16S rDNA sequence, and strain RQ2, which had one nucleotide difference over 1,221 bp compared to strain MSB8 (see Table S2 in the supplemental material). Strains LA4, LA10, VMA1/L12B, and RQ7 formed a separate cluster closer to the 16S rDNA sequence derived from T. neapolitana NS-E. Finally, strain PB1platt clustered separately from the other strains used in the CGH study, sharing 99.51% homology (six mismatches in 1,221 bp) with strain MSB8. Strain PB1platt was isolated from an oil field in Alaska, a location that is the most distant from the places of isolation of the other Thermotoga strains and clustered with Thermotoga strain RKU-10, which was isolated from a deep subterranean oil reservoir in Japan. These two species shared 100% identity over 1,221 bp of the 16S rDNA gene.
The results from the CGH analysis were in agreement with the results of the phylogenetic reconstruction of the 16S rDNA sequences presented above. Hierarchical clustering of the data from the CGH analysis of the different Thermotoga strains (Fig. 1B), compared to the reference genome, MSB8, revealed groupings into two main clusters. Strains NE2x/L8B (99.84% of the genes shared with MSB8), RQ2 (93.08% shared), S1/L12B (92.02% shared), NE7/L9B (93.42% shared), and PB1platt (90.03% shared) all clustered together. The other isolates (LA10, 23.43%; LA4, 16.57%; RQ7, 25.73%; and VMA1/L12B, 30.62%) share lower levels of similarity with the reference genome, MSB8, and appear to be more closely related to T. neapolitana NS-E (data not presented). Strains NE7/L9B, S1/L12B, and NE2x/L8B were isolated from the same location as strain NS-E, i.e., Naples, Italy (Table 1), and clustered together in both the CGH and the 16S rDNA analyses (Fig. 1). Strains LA4 and LA10 were isolated from Lake Abbe, Djibouti (Table 1) and clustered together in the CGH analysis (Fig. 1B). On the other hand, the two strains isolated from the Azores (Table 1), i.e., RQ2 and RQ7, clustered separately (Fig. 1B).
These two patterns of hybridization become more evident when the CGH data are aligned to a circular representation of the T. maritima MSB8 genome (Fig. 2). More remarkably, it becomes evident that most of the genes that are absent or divergent from genes in strain MSB8 are not distributed randomly over the bacterial chromosomes but, rather, group to form large islands. For example, in the strain RQ2 genome, 106 of the estimated 129 divergent ORFs occur as islands ranging in size from 2 to 38 kb. Similarly, for strain S1/L12B, 15 islands larger than 2 kb could be found within the set of 149 divergent ORFs.
FIG. 2.
Circular representation of the T. maritima MSB8 genome and CGHs using microarrays. Outer circle and second circle, predicted coding regions on the plus and the minus strand, respectively, color coded by role categories (10). Third circle, atypical nucleotide composition. Fourth circle, “archaeal islands” as predicted by Nelson et al. (9). Fifth circle, repeats associated with CRISPR elements (red; CRISPR locus numbers from I to VIII are listed), CRISPR-associated sequence loci (half-sized ticks, orange), transposons (blue), and repeat elements (three-quarter-sized ticks, cyan). Sixth circle, CGH regions R1 to R12 (see Tables S1 and S3). The circles 7 to 24 show the CGH results (comparison to MSB8) for strains RQ2 (circles 7 and 8), S1/L12B (circles 9 and 10), PB1platt (circles 11 and 12), NE7/L9B (circles 13 and 14), NE2x/L8B (circles 15 and 16), LA10 (circles 17 and 18), LA4 (circles 19 and 20), RQ7 (circles 21 and 22), and VMA1/L12B (circles 23 and 24). For each strain, the outermost circle represents the hybridization ratio, R (see Materials and Methods); gray, R < 3 (genes shared); yellow, 3 < R < 5; orange, 5 < R < 7; red, 7 < R < 10; brown, R > 10 (genes divergent). The innermost blue circle displays the %EPD for each gene compared to MSB8. The %EPD could not be calculated for strains LA10 (circle 18), LA4 (circle 20), RQ7 (circle 22), and VMA1/L12B (circle 24).
Of all of the strains included in this study that appear to be closely related to strain MSB8, strain PB1platt has the highest level of divergence and is missing a number of regions that encode pathways for the uptake and metabolism of various substrates in strain MSB8. In total, 186 (almost 10%) of the MSB8 ORFs do not have homologs in strain PB1platt (Fig. 1B). Included in these 186 genes, found almost exclusively in 19 islands scattered over the chromosome, are a divergon that encodes a xylose repressor and iron sulfur cluster binding protein (TM0032 to TM0034); a region containing an endo-1,4-beta-xylanase (TM0070) and three adjacent transporters (TM0071 to TM0073); the transcriptional regulator, endoglucanase, and sugar transport region (TM0299 to TM0305); a cation-transporting ATPase system (TM0313 to TM0318); as well as 22 genes in a contiguous region (TM1194 to TM1217) that encode an endoglucanase (TM1201), oligopeptide ABC transporter (TM1194 and TM1196 to TM1199), a maltose transporter (TM1202 to TM1204), and 6 putative NADH dehydrogenases (TM1211 to TM1216). The transcripts for the oligopeptide ABC transporter and for the maltose ABC transporter have been shown to be upregulated in the presence of lactose and unchanged in the presence of maltose, compared to glucose (17). Therefore, it seems more likely that they are involved in the transport of lactose or a related carbohydrate. Interestingly, the latter region (TM1194 to TM1217) was one of those proposed to be an “archaeal island” from the original analysis of the MSB8 genome, is absent from strain RQ2, and appears to have been acquired by strain MSB8 as two independent events (see below). One large region that is also absent from strain PB1platt (TM0411 to TM0437; also variable in strain RQ2) includes genes for the metabolism of tagatose, two sugar transporters, a pectate lyase, glycerol dehydrogenase, as well as other genes that could be involved in the metabolism of xylan. Also missing in strain PB1platt are genes associated with the pentose phosphate pathway and the uptake and metabolism of glycerol and ribose (TM0945 to TM0967); a glycerol-3-phosphate transport region (TM1120 to TM1126, which is also absent from strains NE7/L9B and S1/L12B); a beta-galactosidase and associated transporter region (TM1194 to TM1199, also absent from NE7/L9B and S1/L12B); and a region that encodes an arabinogalactan-galactosidase, maltose transporters, and six NADH dehydrogenases (TM1201 to TM1218). Finally, of all the strains included in this study, PB1platt is the only one that does not have the CRISPR region TM1788 to TM1802 found in the reference MSB8 genome but, rather, has a cassette of genes that have features of the CAS proteins from T. neapolitana (data not presented).
Strains S1/L12B and NE7/L9B have essentially identical patterns of hybridization to the Thermotoga MSB8 array. In total, 149 (8%) of the ORFs in strain MSB8 do not have homologs in the strain S1/L12B genome. Of these, 37 occur as single ORFs, and the remainder occur in 15 islands larger than 2 kb. In addition, 6.9% are devoted to transport. When comparing strains S1/L12B and NE7/L9B to each other, the only apparent differences are one chemotaxis and flagellar biosynthesis operon (TM0698 to TM0705) that is absent from strain NE7/L9B, as well as a large section of contiguous genes (TM0966 to TM1005) that encodes only hypothetical and conserved hypothetical proteins and that is also absent from strain NE7/L9B.
In addition to the regions described above, the lipopolysaccharide biosynthesis operon and surrounding regions that includes TM0611 through TM0653 in the reference strain MSB8 genome has variable levels of hybridization in all the strains tested, including strain NE2x/L8B, which otherwise is identical to strain MSB8. Also, a number of single genes that are randomly distributed on the reference genome appear to be absent from the genome of NE2x/L8B. These include an indole-3-glycerol phosphate synthase, an orotate phosphoribosyltransferase, a threonine dehydratase, a fructokinase, xylose repressor, a cold shock protein, and a number of conserved hypothetical proteins. As these genes represent individual changes, it is also possible that they represent genes that are evolving at a higher rate and cannot be detected using the array.
Finally, Thermotoga strains LA10, LA4, RQ7, and VMA1/L12B appear to be divergent from the reference strain MSB8, sharing only 23.43%, 16.57%, 25.73%, and 30.62% of their genes, respectively, with the reference T. maritima MSB8 (Fig. 1B). The low number of genes in common between the reference MSB8 and the strains LA10, LA4, RQ7, and VMA1/L12B, when hybridizing against the MSB8 microarray, is most likely the result of gene sequences that are too divergent in these strains compared to the MSB8 sequences, rather than a total absence. Nevertheless, most of the genes that are conserved between these four strains and MSB8 are grouped in three large islands, with a size ranging approximately from 13 kb to 81 kb (Fig. 2). Surprisingly, these conserved genes are mostly hypothetical proteins. These three regions also contain a putative polysaccharide export protein (TM0638), a putative NH3-dependent NAD+ synthetase (TM0645), a glutamine synthetase (TM0943), and four ORFs coding for subunits of a ribose ABC transporter (TM0955, TM0956, TM0958, and TM0959). Thermotoga strains LA10, LA4, RQ7, and VMA1/L12B are closely related to T. neapolitana NS-E. A whole-genome alignment between T. neapolitana NS-E and T. maritima MSB8 revealed that these two genomes are, on average, 80.4% identical and surprisingly syntenic, with only a few insertions/deletions and inversions present (data not shown). Considering the percent identity cutoff for hybridization against the MSB8 array, i.e., approximately 85% (nucleotide level), it is logical to assume that strain NS-E, as well as LA10, LA4, RQ7, and VMA1/L12B, are likely to hybridize poorly to the MSB8 array. However, the three regions described above were found to be highly conserved between NS-E and MSB8, with an average percent identity of well above 90%. Because most of the genes present in these regions seem to be hypothetical, it is likely that they carry functions that are essential for these species but that are yet to be characterized.
Analysis of the genomic islands divergent in Thermotoga sp. strain RQ2.
A more detailed analysis was performed for nine regions that were predicted as being absent or highly divergent in strain RQ2. These regions were PCR amplified using primers designed for the flanking genes in strain MSB8. The PCR products were subsequently sequenced and assembled to closure (results presented in Table S1 in the supplemental material). In all cases, the predictions from the CGH analysis of gene absence or variability were correct. In addition, this analysis revealed at least three major types of gene transfer events. The first represents events whereby large regions (up to 12 kb in size) that are present in the reference strain MSB8 genome are missing in their entirety from the genome of strain RQ2 (regions RQ2-R2, RQ2-R9, and RQ2-R11 in Fig. 3). These appear to be large gene insertion events in strain MSB8 that have all occurred in intergenic regions, and the genes flanking these regions have remained with a high degree of conservation between strains RQ2 and MSB8. In region RQ2-R2 (corresponding to TM0411 to TM0423), for example, strain RQ2 lacks genes coding for the transport and metabolism of tagatose, a putative alpha-glucosidase, and three sugar ABC transporters. In strain MSB8, one subset of genes, TM0417 to TM0422, was predicted to be an “archaeal island,” and another subset of genes (TM0411 through TM0416) are best aligned to genes in bacterial species (data not shown). This entire region is also absent from strain PB1platt but is present in all other Thermotoga strains that are closely related to MSB8, i.e., S1/L12B, NE2x/L8B, and NE7/L9B. It is now apparent that this entire region was acquired by strain MSB8 in two independent events, from both bacterial and archaeal donors. Similarly, region RQ2-R9 is a 10-kb stretch comprising nine genes (TM1063 through TM1071) that is missing from strain RQ2 compared to MSB8. These nine genes code for five oligopeptide ABC transporters (TM1063 to TM1067), two proteins involved in sugar metabolism (TM1068 and TM1071), one transcriptional regulator belonging to the DeoR family (sugar catabolism), and one hypothetical protein. These genes are entirely conserved in the genomes of the strains S1/L12B, NE2x/L8B, NE7/L9B, with one of the oligopeptide subunits (TM1067) being absent from strain PB1platt. The variable region (TM1063 to TM1071) that corresponds to RQ2-R9 encodes an “oligopeptide transporter,” which may possibly be a sugar transporter, as this transporter seems to be part of an operon also comprising an alpha-glucosidase and a transcriptional regulator from the DeoR family (TM1069). In strain RQ2, this region is completely absent and corresponds to a 100-bp piece of “unique” DNA. Finally, strain RQ2 does not have a region (TM1261 to TM1271) that encodes a phosphate transport system, nor does it have one of the two DNA mismatch repair proteins in the reference strain MSB8 (these regions were not amplified).
The second type of gene transfer event relates to major rearrangements with individual genes rather than deletion events associated with entire operons or large contiguous cassettes of genes. This is evident in regions RQ2-R5, RQ2-R7, RQ2-R8, and RQ2-R10 (Fig. 3), which all appear to have undergone complex rearrangements/gene insertion/deletion events. In RQ2-R5, for example, there are two small regions (2.02 kb and 346 bp) (Fig. 3 and Tables S1 and S3 in the supplemental material) of the MSB8 genome that have been replaced in the strain RQ2 genome by an 864-bp and a 724-bp unique sequence, respectively. What makes these two rearrangements remarkable is that they have occurred in each case within the predicted ORFs, not in intergenic regions. The N terminus ends of TM0756 and RQ2-R5-3 are conserved, but the C terminus ends of these two genes are different, leading to two different predicted proteins, as follows: TM0756 codes for a galactosyltransferase, whereas RQ2-R5-3 is a glycosyltransferase-fusion protein (Table S3 in the supplemental material). Similarly, TM0758 and RQ2-R5-5 have conserved N- and C-terminal ends, but the middle portions of the two strains are completely different. While the resulting two genes appear to encode the same flagellin, their differences in activity, if any, remain to be seen. Another example is region RQ2-R7; a 244-bp region of the MSB8 genome has been replaced in the RQ2 genome by a 1.06-kb unique sequence. Although the C terminus of RQ2-R7-1 and TM0969 are conserved, RQ2-R7-1 codes for a much larger protein, TM0969, with a unique N terminus giving the protein a different function from its ortholog in MSB8. TM0969 is a small hypothetical protein, whereas RQ2-R7-1 is a putative archaeal ATPase. Downstream of TM0969, strain MSB8 lacks a small 249-bp region compared to RQ2, and a 2.62-kb sequence in MSB8 has been replaced by a unique 2.11-kb region in RQ2, leading to three different predicted genes in RQ2, namely, a putative methyl-accepting chemotaxis protein (RQ2-R7-3), an HD domain protein (RQ2-R7-4) and a hypothetical protein (RQ2-R7-5). In regions RQ2-R8 and RQ2-R10, a 5.46-kb and 2.85-kb region, respectively, containing hypothetical proteins in MSB8 (TM0992 and TM1125 to TM1127), was replaced in strain RQ2 by a 568-bp and 6.38-kb region, where the RQ2 ORFs are also all hypothetical. Elsewhere in region RQ2-R8, RQ2 lacks two small regions of 958 bp and 1.1 kb compared to MSB8, along with four hypothetical proteins (TM0999 through TM1002) and one transposase-related protein (TM1003). Again, it is noticeable that almost the entire RQ2-R8 region was originally predicted to be an “archaeal island.”
Finally, the third type of gene transfer variant relates to RQ2-R12. On the array, this region in strain RQ2 appears to be divergent from MSB8. However, after sequencing and annotation, most of the genes in this region in strain MSB8 (TM1165 to TM1172) appear to be conserved in RQ2, have diverged to a certain extent, and are therefore not similar enough to give a positive result by microarray hybridization. For example, TM1166 only has 85.6% identity with its RQ2 counterpart (RQ2-R12-2). Three of the RQ2-R12 ORFs not only are divergent from their MSB8 homologs but also are smaller in size. RQ2-R12-5, RQ2-R12-6, and RQ2-R12-7 cover only 85.69%, 78.02%, and 90.58% of the length of TM1169, TM1170, and TM1171, respectively (Fig. 3; Table S3 in the supplemental material). This does not seem to have any effect concerning the predicted function of RQ2-R12-5 and RQ2-R12-7, compared to TM1169 and TM1171. However, RQ2-R12-6 is predicted to be a putative response regulator/HD domain protein, and its homolog TM1170 in strain MSB8 was annotated as an ABC transporter. RQ2 lacks a 1.92-kb region compared to MSB8, along with four genes (TM1173 to TM1176).
CRISPR sequences across the Thermotoga strains.
Region R1 was selected for analysis across all of the strains, because the results of the CGH comparisons suggested that it was divergent across all of the strains. In MSB8, region R1 is comprised of two long DNA repeats, LR1 and RPT5A (Fig. S1 in the supplemental material), separated by a reiterated 30-bp repeat and unique intervening sequences. These repeat features are hallmarks of CRISPR (10, 18), and region R1 is one of the eight CRISPR loci found in the genome of MSB8 (Table 2). The role of CRISPR in microbial genomes is still not known (5, 20), but we have hypothesized that the variable presence and absence of CRISPR elements in the microbial lineages is the result of gene transfer events as well as intrachromosomal recombination.
TABLE 2.
Number of shared spacer sequences between the eight CRISPR loci within the MSB8 genome
| CRISPR locus | No. of shared spacer sequences for CRISPR locus indicateda
|
|||||||
|---|---|---|---|---|---|---|---|---|
| I (40) | II (8) | III (8) | IV (24)b | V (2) | VI (8) | VII (4) | VIII (12) | |
| I | ||||||||
| II | 0 | |||||||
| III | 0 | 0 | ||||||
| IV | 0 | 0 | 0 | |||||
| V | 0 | 0 | 0 | 0 | ||||
| VI | 0 | 0 | 0 | 0 | 0 | |||
| VII | 0 | 0 | 0 | 0 | 0 | 0 | ||
| VIIIb | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
The number of spacer sequences in each CRISPR locus of the MSB8 strain is shown in parentheses.
Locus IV (n = 24) and locus VIII (n = 12) share one spacer sequence.
Sequence analysis of R1 revealed that the Thermotoga strains studied have different numbers of the CRISPR repeats and spacer sequences, so that the R1 region is variable in size across the different strains, from 101 bp for NE7/L9B up to 2.69 kb for NE2x/L8B (Fig. S1 in the supplemental material). A reasonable explanation for this variability in size is that there has been recombination between homologous DNA repeats. However, diversity in the R1 region of Thermotoga strains is not a case of simple expansion or contraction of the DNA repeats. Rather, there is a remarkable degree of sequence variation between strains, so that unique spacer sequences are found in strains with R1 regions of dissimilar size (Table 3). This observation contrasts with the strong conservation of spacer sequences previously observed with M. tuberculosis complex strains, for which successive deletions appear to generate CRISPR elements of variable lengths (20). Instead, the situation for Thermotoga strains is more like that for diverse Campylobacter strains (18). The observation of unique spacer sequences, which were found in all of the Thermotoga strains, suggests that the R1 region might provide a relatively compact genetic marker to predict similarities based on global distribution of the strains.
TABLE 3.
Number of shared spacer sequences in the CRISPR locus I of various Thermotoga strains
| Strain | No. of shared spacer sequences for strain indicateda
|
|||||
|---|---|---|---|---|---|---|
| MSB8 (40) | NE2x/L8B (41)b | S1/L12B (2)c | NE7/L9B (2) | RQ2 (28) | PB1platt (10) | |
| MSB8 | ||||||
| NE2x/L8Bb | 40 | |||||
| S1/L12B | 0 | 0 | ||||
| NE7/L9Bc | 0 | 0 | 2 | |||
| RQ2 | 0 | 0 | 0 | 0 | ||
| PB1plattd | 1 | 1 | 0 | 0 | 0 | |
The number of spacer sequences in the CRISPR locus I (MSB8 genomic region R1) for each strain is shown in parentheses.
NE2x/L8B is nearly identical to MSB8, except that the 18th spacer sequence was likely duplicated and inserted between the 10th and 11th spacers.
S1/L12B and NE7/L9B have identical spacer sequences in CRISPR locus I.
PB1platt shares one spacer sequence with both MSB8 and NE2x/L8B.
A possible relatedness can be deciphered from the R1 regions from the different strains that are of similar sizes, as strains that have similar-sized CRISPR elements have identical spacer sequences (Table 3 and Fig. S1 in the supplemental material). For example, strains NE7/L9B and S1/L12B, which were isolated from the same geographic locale (Table 1) and share spacer sequences, display a comparable degree of global dissimilarity from the reference strain in the CGH analysis. Similarly, strains MSB8 and NE2x/L8B, which share spacer sequences and were isolated from locations that are very close to each other (Table 1), also correlate closely in the CGH analysis.
DISCUSSION
The results presented in this study highlight the dynamic nature of the genome of members of this genus and support the idea that there has been extensive gene transfer in the Thermotoga lineage. This genome variability is independent of the closeness of strains to each other based on 16S rRNA phylogenetic analysis, and it highlights the limitations of using 16S rDNA sequencing and analysis as a tool to describe microbial species diversity. The mechanisms for gene transfer in this lineage remain to be elucidated, as none of the usual obvious mechanisms for gene transfer, such as phage, plasmid sequences, or transposable elements could be identified within the flanking regions that appear to have been acquired by strain MSB8 as a result of gene transfer. The repetitive nature of the CRISPR elements, their locations on the chromosome, and their variability among different strains suggest that they may somehow be involved in the mobility of DNA. Preliminary studies show that some of the regions that are inverted in the recently sequenced genome of T. neapolitana NS-E, compared to the chromosome of strain MSB8, are flanked by copies of the 30-bp repeat that is described above as being present in R1 (Nelson et al., unpublished results). Although T. maritima has so far not been shown to be competent, most certainly due to the lack of efficient molecular biology tools, various type II secretion pathway proteins and type IV pilin-related proteins that function in natural competence in other bacterial species could be identified in the T. maritima MSB8 genome (9). Homologs of various competence genes could also be identified, suggesting that there may be an inherent system for the uptake of exogenous DNA, thereby facilitating the exchange of DNA with other organisms.
From the whole-genome CGH study presented here, similarities in the total number of genes that are variable when compared to the reference strain MSB8 become evident. For example, of the more closely related strains, it appears that strain PB1platt, although similar to strain MSB8 on the array, is most divergent in terms of metabolic capabilities with respect to the metabolism of carbohydrates. In contrast to T. maritima strain MSB8, strain PB1platt either does not use the plant polymers pectin or xylan, or glycerol, maltose, tagatose, or cellobiose for energy, or it uses systems that are divergent from those employed by strain MSB8 and were therefore not detectable using microarrays. Compared to the other Thermotoga strains used in this study, strain PB1platt was isolated from a unique environment, the upcoming produced fluids (oil-water-gas mixtures) from the Prudhoe Bay oil fields. These geothermally heated reservoirs may represent isolated pockets of microbial communities situated deep down below the permafrost soil hostile to hyperthermophilic life. Therefore, microorganisms such as PB1platt could represent survivors from the times where this crude oil had been formed. It is also possible that they have invaded their hot biotope very recently during the procedure used for secondary oil recovery, i.e., when seawater (which may possibly harbor some dormant hyperthermophiles which had originated from submarine vents) is pumped down into the oil reservoirs. In both hypotheses, strain PB1platt had to adapt to an environment in which sugars and plant polymers are not (or are no longer) available. Therefore, gene transfer and genome plasticity are important features for genetic evolution of the Thermotogales, in order to ensure adaptability to changing environments.
Of additional significance are the changes that have occurred within the gene sequences of the isolates that have most likely resulted in some selective advantage in terms of efficiency of the resultant proteins. This was seen in at least two situations in the comparisons between strains MSB8 and RQ2 (the galactosyltransferase/glycosyltransferase and the flagellins) and has most likely occurred in many other genes (this will become evident if we obtain the complete genome sequence of strain RQ2, for example). There have been studies that have demonstrated that the nature of the carboxyl-terminal domain of bacterial topoisomerases strongly determine their DNA binding efficiency and cleavage (21). Domain exchange between the Thermus aquaticus DNA polymerase and the 3′-to-5′ exonuclease domain of the homologous mesophile Escherichia coli DNA polymerase I and the homologous T. neapolitana DNA polymerase resulted in variable chimeras that had characteristics from the parental polymerases (variable in temperature and high polymerase activity, processivity, 3′-to-5′ exonuclease activity and proofreading function) (22). A study by Tsoka and Ouzounis also revealed that metabolic enzymes seem to exhibit a much higher tendency to participate in multiple gene fusion events than any other proteins (19). This suggests that the changes that have happened within genes in the order Thermotogales have somehow conveyed a selective advantage to the species that have acquired the change, allowing them to increase their metabolic fitness.
It is obvious that lateral gene transfer is a powerful evolutionary force that has played a significant role in microbial species evolution. Codon analysis and conservation of gene order based on the complete genome sequence gave the initial clues to the promiscuity demonstrated by the Thermotogales. Although it has been argued that Thermotoga is a deep-branching bacterium and that some of these archaeal-like genes are ancestral genes that have since been lost in other bacterial lineages (15), many of the gene sequences that were thought to be lost or gained in this study appear to be associated with particular biological processes and may therefore be a reflection of the environmental niche where these individual species are residing. Based on the Thermotoga analysis, it is evident that substrate availability may be one of the main reasons for loss and gain of genetic material. The most striking example is strain PB1platt. This Thermotoga strain was isolated from the most distant location, i.e., from an oil field at the Prudhoe Bay, Alaska. Although it shares a high level of genome similarity based on the CGH analysis, it does not group with the other T. maritima or T. neapolitana strains included in this study based on 16S rDNA phylogeny but, rather, appears to be more closely related to strain RKU, which was isolated from the same type of environment, i.e., a deep oil field.
It is also possible that shared regulatory elements/promoters among hyperthermophilic species have enabled the efficient activity of acquired genes. Alternatively, regulatory elements from other locations in the chromosome can be tapped to regulate these acquired genes and/or pathways, allowing for the success of these transfer events. The regions that have been subject to gene transfer events are scattered over the chromosome, and there does not appear to be any bias for gene loss or gain of particular regions of the chromosome.
The value of CGH in analyzing potential gene transfer events has been highlighted in this study. It is now evident that the SSH study of Nesbo and colleagues (15) may have overestimated the number of genes that are unique to strain RQ2, as CGH analysis does not suggest such a high level of diversity between the two species. Ultimately, however, whole-genome comparisons remain the most reliable way to detect the subtle genetic differences between closely related species.
Supplementary Material
Acknowledgments
This project was funded by a grant (DE-FG02-01ER63133) from the U.S. Department of Energy.
We thank Bruce Weaver and Joanne Emerson for support with various aspects of this work.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Agron, P. G., M. Macht, L. Radnedge, E. W. Skowronski, W. Miller, and G. L. Andersen. 2002. Use of subtractive hybridization for comprehensive surveys of prokaryotic genome differences. FEMS Microbiol. Lett. 211:175-182. [DOI] [PubMed] [Google Scholar]
- 2.Boucher, Y., C. J. Douady, R. T. Papke, D. A. Walsh, M. E. Boudreau, C. L. Nesbo, R. J. Case, and W. F. Doolittle. 2003. Lateral gene transfer and the origins of prokaryotic groups. Annu. Rev. Genet. 37:283-328. [DOI] [PubMed] [Google Scholar]
- 3.Futterer, O., A. Angelov, H. Liesegang, G. Gottschalk, C. Schleper, B. Schepers, C. Dock, G. Antranikian, and W. Liebl. 2004. Genome sequence of Picrophilus torridus and its implications for life around pH 0. Proc. Natl. Acad. Sci. USA 101:9091-9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huber, R., T. A. Langworthy, H. Konig, M. Thomm, C. R. Woese, U. B. Sleytr, and K. O. Stetter. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch. Microbiol. 144:324-333. [Google Scholar]
- 5.Jansen, R., J. D. Embden, W. Gaastra, and L. M. Schouls. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43:1565-1575. [DOI] [PubMed] [Google Scholar]
- 6.Kim, C. C., E. A. Joyce, K. Chan, and S. Falkow. 2002. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 3:research0065.1-0065.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koonin, E. V. 2003. Horizontal gene transfer: the path to maturity. Mol. Microbiol. 50:725-727. [DOI] [PubMed] [Google Scholar]
- 8.Lawrence, J. G., and H. Hendrickson. 2003. Lateral gene transfer: when will adolescence end? Mol. Microbiol. 50:739-749. [DOI] [PubMed] [Google Scholar]
- 8a.Mongodin, E., J. Finan, M. W. Climo, A. Rosato, S. Gill, and G. L. Archer. 2003. Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 185:4638:4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, J. A. Eisen, C. M. Fraser, et al. 1999. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 10.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson, K. E., A. N. Pell, P. Schofield, and S. Zinder. 1995. Isolation and characterization of an anaerobic ruminal bacterium capable of degrading hydrolyzable tannins. Appl. Environ. Microbiol. 61:3293-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. Chris Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. A. Eisen, K. N. Timmis, A. Dusterhoft, B. Tummler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 13.Nelson, K. E., S. H. Zinder, I. Hance, P. Burr, D. Odongo, D. Wasawo, A. Odenyo, and R. Bishop. 2003. Phylogenetic analysis of the microbial populations in the wild herbivore gastrointestinal tract: insights into an unexplored niche. Environ. Microbiol. 5:1212-1220. [DOI] [PubMed] [Google Scholar]
- 14.Nesbo, C. L., and W. F. Doolittle. 2003. Targeting clusters of transferred genes in Thermotoga maritima. Environ. Microbiol. 5:1144-1154. [DOI] [PubMed] [Google Scholar]
- 15.Nesbo, C. L., S. L'Haridon, K. O. Stetter, and W. F. Doolittle. 2001. Phylogenetic analyses of two “archaeal” genes in Thermotoga maritima reveal multiple transfers between archaea and bacteria. Mol. Biol. Evol. 18:362-375. [DOI] [PubMed] [Google Scholar]
- 16.Nesbo, C. L., K. E. Nelson, and W. F. Doolittle. 2002. Suppressive subtractive hybridization detects extensive genomic diversity in Thermotoga maritima. J. Bacteriol. 184:4475-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen, T. N., A. D. Ejaz, M. A. Brancieri, A. M. Mikula, K. E. Nelson, S. R. Gill, and K. M. Noll. 2004. Whole-genome expression profiling of Thermotoga maritima in response to growth on sugars in a chemostat. J. Bacteriol. 186:4824-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schouls, L. M., S. Reulen, B. Duim, J. A. Wagenaar, R. J. Willems, K. E. Dingle, F. M. Colles, and J. D. Van Embden. 2003. Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. J. Clin. Microbiol. 41:15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsoka, S., and C. A. Ouzounis. 2000. Prediction of protein interactions: metabolic enzymes are frequently involved in gene fusion. Nat. Genet. 26:141-142. [DOI] [PubMed] [Google Scholar]
- 20.van Embden, J. D., T. van Gorkom, K. Kremer, R. Jansen, B. A. van Der Zeijst, and L. M. Schouls. 2000. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 182:2393-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viard, T., R. Cossard, M. Duguet, and C. B. de La Tour. 2004. Thermotoga maritima-Escherichia coli chimeric topoisomerases. Answers about involvement of the carboxyl-terminal domain in DNA topoisomerase I-mediated catalysis. J. Biol. Chem. 279:30073-30080. [DOI] [PubMed] [Google Scholar]
- 22.Villbrandt, B., H. Sobek, B. Frey, and D. Schomburg. 2000. Domain exchange: chimeras of Thermus aquaticus DNA polymerase, Escherichia coli DNA polymerase I and Thermotoga neapolitana DNA polymerase. Protein Eng. 13:645-654.11054459 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.