Abstract
N-acyl homoserine lactone (AHL) is required by Erwinia carotovora subspecies for the expression of various traits, including extracellular enzyme and protein production and pathogenicity. Previous studies with E. carotovora subsp. carotovora have shown that AHL deficiency causes the production of high levels of RsmA, an RNA binding protein that functions as a global negative regulator of extracellular enzymes and proteins and secondary metabolites (Rsm, regulator of secondary metabolites). We document here that ExpR, a putative AHL receptor belonging to the LuxR family of regulators, activates RsmA production. In the absence of AHL, an ExpR+ E. carotovora subsp. carotovora strain compared to its ExpR− mutant, produces higher levels of rsmA RNA and better expresses an rsmA-lacZ transcriptional fusion. Moreover, the expression of the rsmA-lacZ fusion in Escherichia coli is much higher in the presence of expR71 (the expR gene of E. carotovora subsp. carotovora strain Ecc71) than in its absence. We also show that purified preparation of MBP-ExpR71 binds (MBP, maltose binding protein) rsmA DNA. By contrast, MBP-ExpR71 does not bind ahlI (gene for AHL synthase), pel-1 (gene for pectate lyase), or rsmB (gene for regulatory RNA that binds RsmA), nor does ExpR71 activate expression of these genes. These observations strongly suggest transcriptional activation of rsmA resulting from a direct and specific interaction between ExpR71 and the rsmA promoter. Several lines of evidence establish that N-3-oxohexanoyl-l-homoserine lactone (3-oxo-C6-HL), the major AHL analog produced by E. carotovora subsp. carotovora strain Ecc71, inhibits ExpR71-mediated activation of rsmA expression. These findings for the first time establish that the expR effect in E. carotovora subsp. carotovora is channeled via RsmA, a posttranscriptional regulator of E. carotovora subspecies, and AHL neutralizes this ExpR effect.
Erwinia carotovora subspecies cause soft-rotting or tissue macerating disease in many plants and plant organs. Their capacity to cause rotting depends heavily upon their ability to produce secreted proteins and enzymes (2, 6). These bacteria utilize Type I, Type II, and Type III secretion systems to translocate proteins into the milieu. Proteases (Prt) are secreted by the Type I system, pectate lyase (Pel), polygalacturonase (Peh), and cellulase (Cel) are exported via the Type II system, and Harpin and other putative effectors are secreted by the Type III system (18, 48). Of the effectors and enzymes secreted by these bacteria, especially significant in the context of plant tissue maceration are the pectinases such as Pel, Peh, and pectin lyase (Pnl). These enzymes acting in concert with Prt and Cel cause degradation of plant cell wall components triggering cell separation and cell death. Harpins are required for the elicitation of the hypersensitive response and also for symptom production in Arabidopsis thaliana (38, 46). Aside from extracellular proteins, many of these bacteria produce antibiotics and other secondary metabolites (5).
Extracellular proteins are coregulated in E. carotovora subsp. carotovora and most likely in other E. carotovora subspecies by plant signals, quorum sensing signals (QSS), and an assortment of transcriptional factors and posttranscriptional regulators (4, 12, 13, 17, 21, 28, 45, 55). While many of the regulatory steps await identification and characterization, several generalizations are possible. As an example, posttranscriptional regulation by the RsmA-RsmB pair is of paramount importance in the expression of exoprotein genes and their secretion pathways. RsmA is a small RNA-binding protein that promotes decay of RNA (4, 9). rsmB specifies an untranslated regulatory RNA that binds RsmA and neutralizes its negative regulatory effect (28). Many of the transcriptional factors, known to regulate extracellular protein production, actually act via these posttranscriptional regulators (8, 10, 24, 29, 39, 40).
N-acyl homoserine lactones (AHLs) function as cell density (quorum) sensing signals (reviewed in references 14, 30, 52, 54, 59, and 61). Since its discovery in the marine bacterium Vibrio fischeri, where the signal is responsible for the expression of lux genes controlling light production, such signals have now been detected in many bacteria. These molecules control diverse phenotypes, including bacteria-microbe and bacteria-plant/animal interactions, conjugation, production of secreted proteins, extracellular polysaccharides, antibiotics, pigments, and other secondary metabolites (see 11, 43, 44, 54, 59, 61, 62, 63 and references cited therein). Extensive studies of the lux genes and other AHL-regulated genes have demonstrated that signaling systems minimally comprise biosynthetic precursors (acyl-ACP [acyl-carrier protein] and SAM [S-adenosyl-l-methionine]); LuxI or similar proteins that function as AHL synthases; and LuxR and LuxR-like protein receptors of AHL that bind target DNAs and activate transcription (30, 54, 59). Many homologs of LuxR are now known that function as transcriptional activators upon AHL-binding (see, for example, 42, 57, and 58). While most QSS systems conform to this pattern, there are instances in which LuxR-like proteins are known to bind DNA in their native state (i.e., in the absence of AHL) and modulate transcription. One noteworthy example is the AHL-regulated production of capsular polysaccharide (CPS) in Pantoea (Erwinia) stewartii subsp. stewartii (35, 36). In this instance, EsaR, a LuxR homolog, governs the expression of the cps locus. EsaR directly represses the transcription of the rcsA gene which encodes an essential coactivator for RcsA/RcsB-mediated transcription of cps genes. The repressor activity of EsaR is compromised by AHL binding, thereby relieving repression of the cps genes.
In E. carotovora, AHL controls extracellular enzymes, antibiotic production, and pathogenicity (20, 21, 44, 45, 53). Two different transcriptional regulators that also function as AHL receptors are believed to operate in E. carotovora subsp. carotovora. One such regulator, CarR, a LuxR-like transcriptional factor, controls the production of the antibiotic carbapenem. Binding of AHL to CarR imparts specificity onto this transcriptional factor. It also is known that CarR occurs as dimers and that AHL-binding converts the dimers to higher order multimers. The CarR-AHL complex binds DNA and activates the transcription of the carA-H operon required for the biosynthesis of the antibiotic (20, 32, 60). A similar regulatory mechanism most likely controls antibiotic production in E. carotovora subsp. betavasculorum (7). It should be noted that neither the CarR nor the CarR-AHL complex control exoenzyme or AHL production (61).
The LuxR homologs, ExpR and RexR, are the other AHL receptors of E. carotovora subsp. carotovora (1, 15). The initial studies with ExpR of E. carotovora subsp. carotovora strain SCC3193 (ExpR3193) did not establish a clear role of the AHL receptor in exoprotein production. For example, inactivation of expR had very little effect on extracellular pectate lyase production or AHL synthesis. However, multiple copies of expR had some inhibitory effect on extracellular enzyme production. von Bodman et al. (58) have documented that ExpR3193 is a DNA-binding protein and that its DNA-binding property is inhibited by AHL. Thus, ExpR, like other members of the LuxR family of regulators, is believed to function as a transcriptional factor, although the identity of the target(s) of ExpR in E. carotovora subsp. carotovora has until now remained obscure. In this report we document that (i) ExpR activates expression of rsmA; (ii) ExpR specifically binds rsmA DNA; and (iii) AHL prevents ExpR-rsmA binding and ExpR-mediated activation of rsmA transcription. Our findings also explain how AHL deficiency causes RsmA overproduction responsible for the inhibition of exoprotein and secondary metabolite production in E. carotovora subsp. carotovora.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids are described in Table 1. All the wild-type strains were maintained on Luria-Bertani (LB) agar. The strains carrying antibiotic markers were maintained on LB agar containing appropriate antibiotics.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| Erwinia carotovora subsp. atroseptica | ||
| Eca12 | Wild type | 64 |
| Erwinia carotovora subsp. carotovora | ||
| Ecc71 | Wild type | 64 |
| AC5006 | Lac− derivative of Ecc71 | 41 |
| AC5091 | AhlI− derivative of AC5006 | 4 |
| AC5094 | AhlI− derivative of Ecc71 | 4 |
| AC5098 | ExpR− derivative of AC5006 | This study |
| AC5099 | AhlI− ExpR− derivative of AC5006 | This study |
| SCC3193 | Wild type | 45 |
| EC153 | Wild type | Laboratory collection |
| Escherichia coli | ||
| DH5α | supE44 ΔlacU169 φ80lacZΔM15 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Gibco BRL |
| MC4100 | araD139 Δ(lacIPOZYA)U169 recA+thi Strr | 27 |
| VJS533 | ara Δ(lac-proAB) rpsL φ80lacZΔM15 recA56 | 16 |
| Plasmids | ||
| pDK6 | Kmr | 23 |
| pLARF5 | Tcr | 22 |
| pMAL-c2g | Apr, protein expression vector | New England Biolabs |
| pMP220 | Tcr, promoter-probe vector | 51 |
| pCL1920 | Spr, Smr | 26 |
| pAKC781 | Apr, peh-1+ DNA in pBluescriptSK(+) | 27 |
| pAKC783 | Apr, pel-1+ DNA in pBluescriptSK(+) | 27 |
| pAKC851 | Tcr, ahlI+ and expR+ DNA in pLARF5 | 4 |
| pAKC856 | Apr, ahlI+ DNA in pBluescriptSK(+) | 4 |
| pAKC875 | Tcr, rsmA+ DNA in pLARF5 | 4 |
| pAKC882 | Apr, rsmA coding region in pT7-7 | 38 |
| pAKC935 | Spr, expR3193+ DNA of SCC3193 in pCL1920 | This study |
| pAKC936 | Spr, expR71+ DNA in pCL1920 | This study |
| pAKC1002 | Tcr, rsmB-lacZ in pMP220 | 28 |
| pAKC1034 | Apr, 200-bp celV fragment in pGEM-T Easy | 29 |
| pAKC1100 | Tcr, rsmA-lacZ in pMP220 | 3 |
| pAKC1201 | Kmr, ptac-ahlI in pDK6 | This study |
| pAKC1202 | Tcr, ahlI-lacZ in pMP220 | This study |
| pAKC1203 | Tcr, pel-lacZ in pMP220 | This study |
| pAKC1204 | Tcr, rsmA+ DNA of SCC3193 in pLARF5 | This study |
| pAKC1205 | Tcr, rsmA+ DNA of Eca12 in pLARF5 | This study |
| pAKC1206 | Tcr, rsmA+ DNA of EC153 in pLARF5 | This study |
| pAKC1212 | Kmr, Tcr, ExpR− derivative of pAKC851 | This study |
| pAKC1220 | Apr, expR71 coding region in pMAL-c2g | This study |
| pHV200 | Apr, 8.8-kb SalI fragment containing the lux operon | 16 |
| pHV2001 | Apr, frameshift mutant of luxI in pHV200 | E. P. Greenberg |
The composition of LB, minimal salts, and minimal salts plus celery extract media have been described in previous publications (4, 41). When required, antibiotics were supplemented as follows: ampicillin (Ap), 100 μg/ml; kanamycin (Km), 50 μg/ml; spectinomycin (Sp), 50 μg/ml; and tetracycline (Tc), 10 μg/ml. Media were solidified using 1.5% (wt/vol) agar.
The media compositions for agarose plate enzymatic activity assays have been described by Chatterjee et al. (4).
Extracellular enzyme assays.
The extracellular pectate lyase (Pel), polygalacturonase (Peh), protease (Prt), and cellulase (Cel) activities in the culture supernatants were tested according to procedures published previously (4). Bacterial cultures were grown at 28°C in minimal salts medium with sucrose or sucrose plus celery extract. Supernatants were collected by centrifugation (10,000 rpm, 10 min; Beckman J2-21 centrifuge) and used for assays. Halos around the wells on the assay plates indicate the enzymatic activities.
Nucleotide sequence determination of expR71 and alignment of ExpRs.
The nucleotide sequence of expR71 was determined from pAKC851 using primers based on the nucleotide sequence of the ahlI gene of Ecc71, which overlapped with expR71. Sequence alignment was performed using ClustalW at www.expasy.ch, and default parameters were used. Domain search was performed using rpsblast at www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi.
DNA techniques.
Standard procedures were used in the isolation of plasmids and chromosomal DNA, gel electrophoresis, and DNA ligation (49). Restriction and modification enzymes were obtained from Promega Biotec (Madison, WI). Prime-a-Gene DNA labeling system (Promega Biotec) was used for labeling DNA probes. Southern blot analysis was carried out at high-stringency conditions (hybridization at 65°C in 6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5× Denhardt's, 0.5% [wt/vol] sodium dodecyl sulfate [SDS], and 100 μg/ml denatured salmon sperm DNA; washing at 65°C with 2× SSC for 30 min, 1× SSC plus 0.1% [wt/vol] SDS for 30 min followed with 0.1× SSC plus 0.1% [wt/vol] SDS for 30 min) as well as at low-stringency conditions (hybridization and washing conditions are the same as high-stringency conditions except the temperature was at 55°C). For expR3193 probe, a 500-bp BamHI-ClaI fragment from pAKC935 was used.
Construction of plasmids and mutant strains used.
A fragment containing the entire open reading frame (ORF) of expR3193 was PCR amplified from SCC3193 using the primers 5′-TGTAAGCTTAGGTTACTATGGATATACGTAT-3′ and 5′-TGTGGATCCAGGTTGCGAGAGTGGCCACTG-3′ and cloned in the BamHI-HindIII site of pCL1920 to yield pAKC935. The expR71 fragment was amplified from Ecc71 using the primers designed based on Ecc71 expR nucleotide sequences 5′-TGCAAGCTTGCCCTTAGCATTGTCGTTCGATTG-3′ and 5′-TGCGGATCCCGCTATTGCACAGGCTTGATGAGC-3′. The PCR product was digested with BamHI-HindIII and cloned in a BamHI-HindIII site of pCL1920 to yield pAKC936. The plasmids pAKC1204, pAKC1205, and pAKC1206 were obtained from genomic libraries of E. carotovora subsp. carotovora strain SCC3193, E. carotovora subsp. atroseptica strain Eca12 and E. carotovora subsp. carotovora strain EC153 and by in situ colony hybridization using rsmA probe of Ecc71. AC5098 and AC5099 were constructed by marker exchange of AC5006 and AC5091 with pAKC1212, respectively. The procedures for marker exchange have been described in Chatterjee et al. (4). Inactivation of expR in mutants was confirmed by Northern blot analysis.
Bioluminescence assay for AHL estimation.
Culture supernatants and high-performance liquid chromatography (HPLC) fractions were assayed for bioluminescence using an E. coli based bioassay system as described by Chatterjee et al. (4). E. coli strain VJS533 harboring pHV200I was used as a biosensor indicator. There is a linear relationship between the quantity of AHL present in the samples and the emission of bioluminescence. Relative light unit (RLU) is expressed as counts per minute per milliliter of culture.
Production and fractionation of AHLs.
Ecc71 was grown in 2.5 liters of minimal salts medium supplemented with sucrose (0.5%, wt/vol) at 28°C to a Klett value of ca. 200. The culture supernatant was collected by centrifugation at 10,000 rpm for 10 min (Beckman J2-21 centrifuge) and extracted with an equal volume of ethyl acetate. The ethyl acetate extract was evaporated to dryness, and the residue was dissolved in 5 ml of distilled water and subjected to HPLC according to the method of Morin et al. (37) modified as follows. The residue in distilled water was loaded on a C-18 reverse-phase column (Jupiter 5U C-18 300A; 250 by 4.6 mm; Phenomenex). The column was eluted with a linear gradient from 0 to 50% methanol in water over 60 min at a flow rate of 1 ml/min. Detection was by UV light at 210 nm. The eluted fractions were assayed for bioluminescence activity according to Chatterjee et al. (4). Active fractions (71AHL) were pooled, concentrated, and rechromatographed under similar conditions for further purification.
Detection of AHLs by analytical thin-layer chromatography (TLC).
A modification of the procedure described by Holden et al. (19) was used for the detection of AHLs by analytical TLC. Crude ethyl acetate extract or the HPLC fraction was applied in volumes of 1 to 5 μl to a C-18 reverse-phase TLC plate (150 μm adsorbent layer thickness; Sigma-Aldrich, St. Louis, MO), and the chromatogram was developed with methanol:water (60:40, vol/vol). N-3-oxohexanoyl-l-homoserine lactone (3-oxo-C6-HL) [N-(β-ketocaproyl)-dl-homoserine lactone; purchased from Sigma] and N-3-oxooctanoyl-l-homoserine lactone (3-oxo-C8-HL, kindly provided by Paul Williams, University of Nottingham, United Kingdom) were used as standards. The plate was dried and overlaid with the indicator bacterium E. coli VJS533 carrying pHV200I. The biosensor overlay was prepared as follows: the indicator bacterium was grown in LB medium containing ampicillin at 28°C to a Klett value of ca. 200 and was then mixed with an equal volume of warm LB medium containing 1.5% (wt/vol) agar. The overlaid plate was incubated for 3 h at 28°C and exposed to X-ray film to record bioluminescent spots.
Mass spectrometry.
All mass spectrometry experiments were performed on a Thermo-Finnigan TSQ7000 triple-quadrupole mass spectrometer with the API2 source and Performance Pack (ThermoFinnigan, San Jose, CA) using electrospray ionization (ESI). The heated inlet capillary was maintained at 250°C; the voltage on the stainless steel electrospray needle was 4.5 kV. All other voltages were optimized to maximize ion transmission and minimize unwanted fragmentation and were determined during the regular tuning and calibration of the instrument. For mass spectrometry/mass spectrometry (MS/MS) experiments, argon was used as the collision gas, and collision energies ranged from 20 to 40eV.
For MS and MS/MS experiments, samples were infused at a rate of 10 μl/min using a syringe pump (Harvard Apparatus, Holliston, MA). Nitrogen sheath gas was provided to the ESI source at 80 lb/in2. The spectra acquired for each sample are an average of 150 individual scans or spectra.
The mass spectrometer is connected to an integrated Thermo-Finnigan liquid chromatography (LC) system consisting of a P4000 quaternary LC pump and SCM1000 vacuum degasser, an AS3000 autosampler, and a UV6000LP diode-array detector. This system was used for all LCMS and LCMS/MS experiments.
Northern and Western blot analyses.
Bacterial cultures were grown at 28°C in minimal salts medium supplemented with sucrose (0.5%, wt/vol), sucrose, and celery extract or LB medium with appropriate antibiotics as described in figure legends. Cells were collected while cultures reached a Klett value of ca. 150 or 200. RNA isolation and Northern blot analysis were performed as described in Liu et al. (28). The probes used were the 183-bp NdeI-SalI fragment of rsmA from pAKC882, 314-bp EcoRV-KpnI fragment of pel-1 from pAKC783, 743-bp HindIII fragment of peh-1 from pAKC781, 386-bp DraI-EcoRI fragment of ahlI from pAKC856, and 200-bp EcoRI fragment of celV from pAKC1034. For Western blot analysis, bacterial cells were collected, suspended in 1× SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer (49), and boiled. To determine total bacterial protein concentrations, samples were precipitated with trichloroacetic acid. The protein concentrations were determined with a bicinchoninic acid protein assay kit (Pierce, Rockford, IL) according to the manufacturer's specifications. Western blot analysis of the total bacterial protein was performed as described in Mukherjee et al. (38). The antiserum raised against RsmA of Ecc71 (10) was used as probe.
Expression and purification of MBP-ExpR71 protein.
A fragment containing the entire coding region of expR71 was PCR amplified from pAKC851 by using primers 5′-TGTGGATCCATGTCGCCATTATTCTCCAGTAGC-3′ and 5′-TGTAAGCTTCTATTGCACAGGCTTGATGAGCTG-3′. The fragment was digested with BamHI and HindIII and cloned into pMAL-c2g vector (New England Biolabs, Beverly, MA) to yield pAKC1220.
E. coli strain DH5α carrying pAKC1220 was grown in LB medium supplemented with glucose (0.2%, wt/vol) and ampicillin at 37°C. When the culture reached an A600 value of 0.6, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to yield a final concentration of 1 mM. Three hours after IPTG addition, bacterial cells were collected by centrifugation. MBP-ExpR71 fusion protein was purified by amylose resin (New England Biolabs) affinity chromatography according to the protocol provided by the company. Protein concentration was determined by using a CB-X Protein Assay kit (Geno Technology, Inc., St. Louis, MO). Crude extracts and purified MBP-ExpR71 were analyzed on SDS-PAGE in a 10% (wt/vol) polyacrylamide gel.
Gel mobility shift assays.
The DNA fragments were amplified from Ecc71 by PCR using primers listed as follows: rsmA, 5′-GCGGATCCGGCAAGCAGGATAGAA-3′ and 5′-GATTATAAAGAGTCGGGTCTCTCTGT-3′ (corresponding to −245 to −16 from the putative translational start site); pel-1, 5′-TGAGGATCCATGTTTCATCCGCAATACATTTAAC-3′ and 5′-TGATCTAGATATTTCATTATCACTGTCTCCTTG-3′ (corresponding to −131 to +8 from the putative translational start site); ahlI, 5′-TGCTCTAGATTCTGAGGGTAAATAGCTCTTCTG-3′ and 5′-TGCAGATCTGAGTAATATAAAAATCCGGAATCG-3′ (corresponding to −220 to +63 from the putative translational start site); and rsmB (nontranslatable RNA regulator), 5′-CGAAGCTTAAGTTAGTAACCGGTTACAGTG-3′ and 5′-TGTGAGAGATCTCTTACATTCTC-3′ (corresponding to −180 to +41 from the transcriptional start site). The DNA fragments were purified using Wizard SV Gel and PCR Clean-Up System (Promega Biotec, Madison, WI) and end labeled with [α-32P]dATP and Klenow fragment. Protein-DNA interaction was assayed in 20 μl of binding buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 50 mM KCl, 1 mM dithiothreitol, 0.1 mM EDTA, and 5% [wt/vol] glycerol) containing 1 μg of salmon sperm DNA, 2 μg of bovine serum albumin (BSA), and purified MBP-ExpR71 protein with or without competitors. The reaction mixtures were incubated at room temperature for 20 min and subjected to electrophoresis in 5% (wt/vol) polyacrylamide (acryl:bis-acyl, 29:1) gels. The gels were dried and exposed to X-ray film.
β-Galactosidase assays.
Bacterial constructs were grown at 28°C in minimal salts plus sucrose or LB media supplemented with appropriate antibiotics and purified AHL from Ecc71 (71AHL) as described in figure legends or footnotes to tables. The β-galactosidase assays were performed according to Miller (33).
Cell densities of bacterial cultures used in Northern blot and Western blot analyses as well as for exoenzyme and β-galactosidase assays are represented as Klett values. Klett values of 150, 200, and 250 are approximately equal to 1.6, 2.3, and 2.8 absorbance at A600.
The experiments were performed at least two to three times, and the results were reproducible.
Nucleotide sequence accession number. The sequence of the expR gene of Ecc71 has been deposited in the GenBank databases under accession number L40174.
RESULTS
Occurrence of expR in E. carotovora subsp. carotovora strain Ecc71.
To examine the occurrence of an expR homolog in E. carotovora subsp. carotovora strain Ecc71, Southern blot analysis of EcoRI-digested Ecc71 chromosomal DNA and a previously isolated AhlI+ plasmid, pAKC851, using expR3193 DNA as the probe, was performed under high-stringency conditions as well as low-stringency conditions. Hybridization bands with weak signals were detected with a chromosomal DNA fragment of Ecc71 and pAKC851 under low-stringency conditions, but no signal was detected under high-stringency conditions (data not shown). To estimate the extent of sequence divergence, we sequenced Ecc71 expR (expR71; accession no. AY894425) and compared sequences of expR71 with sequences of expR genes available in databases. Alignment of ExpR amino acid sequences (unpublished data) revealed that Ecc71 and E. carotovora subsp. betavasculorum strain Ecb168 (accession no. AF001050) share 93% identity, but both strains show ca. 60% identity with ExpR proteins of E. carotovora subsp. carotovora strains EC153 (accession no. AY894424), SCC3193 (accession no. X80475), and E. carotovora subsp. atroseptica strain CFBP6272 (accession no. AJ580600). Interestingly, significant differences in the ExpR sequences are present only in the N-terminal domain, the putative AHL binding region (residues 11 to 160). By contrast, all these ExpR homologs share high homology in the putative HTH domain located in the C terminal (residues 175 to 232).
Characterization of AHLs produced by Ecc71.
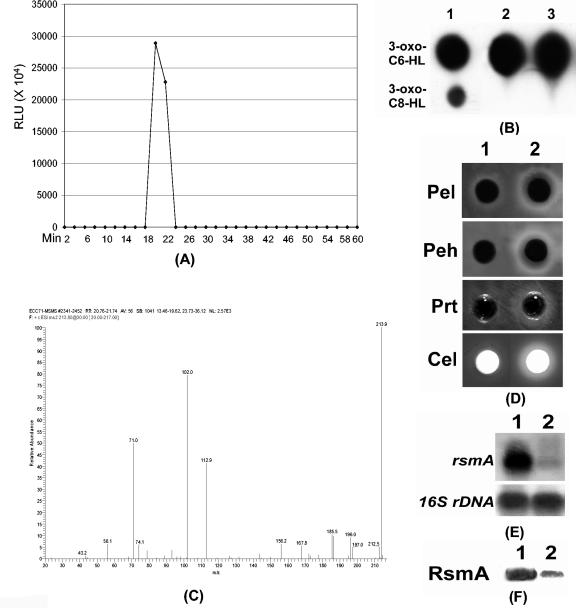
We prepared ethyl acetate extracts of spent cultures of Ecc71 fractionated by HPLC on a C-18 reverse phase column. The fractions were assayed for bioluminescence as described in the Materials and Methods section. A single peak (designated as 71AHL) showing bioluminescence activity was observed in ethyl acetate extracts of Ecc71 with a retention time of about 20 min (Fig. 1A). TLC analysis of this active fraction as well as ethyl acetate extract showed one major spot which had a retardation factor identical to that of 3-oxo-C6-HL (Fig. 1B). To confirm the identity of this AHL, active fractions containing 71AHL were analyzed by LCMS and LCMS/MS (Fig. 1C), and the results were compared to standard 3-oxo-C6-HL. The retention time, parent ions observed, and product ion spectrum for 71AHL matched those for the standard.
FIG. 1.
(A) RLUs produced by HPLC fractions of crude AHL extract from Ecc71 in E. coli strain VJS533 harboring pHV200I. (B) TLC analysis of AHLs. Lane 1, mixture of synthetic 3-oxo-C6-HL and 3-oxo-C8-HL; lane 2, crude AHL extract of Ecc71; and lane 3, HPLC purified 71AHL (ca. 7 nmol). (C) LCMS/MS scan for HPLC-fractionated 71AHL. (D) Agarose plate assays for Pel, Peh, Prt, and Cel activities of AC5094 (ExpR+ AhlI−). Thirty microliters of culture supernatant was applied in each well. (E and F) Northern blot analysis and Western blot analysis of rsmA or RsmA of AC5094. Each lane contained 10 μg of total RNA for Northern blot analysis, and 10 μg of total protein for Western blot analysis. (D, E, and F) Lane 1, in the absence of AHL (i.e., same volume of water was added); lane 2, in the presence of 71AHL (300 μl of 1 mM 71AHL added to a 6-ml culture to yield a final concentration of 50 μM). Bacteria were inoculated in minimal salts plus sucrose (0.5%, wt/vol) medium supplemented with or without 71AHL. Total RNAs and proteins were extracted after 5 h incubation at 28°C, and after 8 h incubation culture supernatants were collected by centrifugation (10,000 rpm, 10 min) for exoenzyme assays.
Having established the chemical nature of the AHL fraction from Ecc71, we tested the purified preparation for biological activity, i.e., for the restoration of exoenzyme production in an AhlI− mutant of Ecc71. The data in Fig. 1D show that 71AHL restored Pel, Peh, Cel, and Prt production in an AhlI− strain of Ecc71, AC5094.
Previous studies (3, 24) in E. carotovora subsp. carotovora strains Ecc71 and SCC3193 documented that the expression of rsmA is higher in the AHL-deficient mutants than in the parents. To test if the effects of AHL deficiency on rsmA expression could be neutralized by adding the fractionated 71AHL, we compared the levels of rsmA transcript and RsmA protein in AC5094 (AhlI− mutant of Ecc71) in the absence or in the presence of 71AHL. The Northern blot analysis (Fig. 1E) and Western blot analysis (Fig. 1F) results reveal that the levels of rsmA transcript and RsmA produced by AC5094 were much lower in the presence of 71AHL than in the absence of 71AHL.
Purification of MBP-ExpR71.
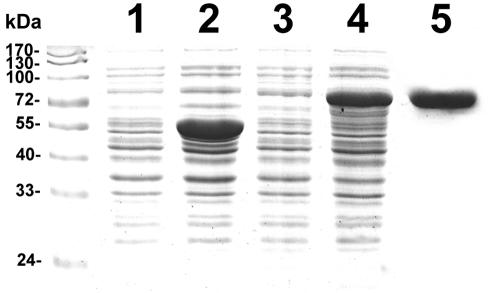
To overexpress and purify ExpR71 for DNA binding studies, we initially tried to express ExpR71 as ExpR71-6His fusion protein, but our attempts were hampered by protein insolubility. We therefore attempted to express ExpR71 as MBP-ExpR71 fusion protein. For this, the coding region of expR71 was PCR amplified and cloned into the pMAL-c2g vector to yield pAKC1220. After IPTG induction, a protein of ca. 78 kDa was overproduced by E. coli strain DH5α carrying pAKC1220 (Fig. 2, lane 4). The apparent molecular mass of ca. 78 kDa matched well with the mass of 27.95 kDa of the polypeptide deduced from the expR71 sequence plus the mass of 50.84 kDa of MBP2-β-galactosidase α fragment made from the pMAL-c2g vector (Fig. 2, lane 2), indicating that the overproduced protein is MBP-ExpR71 fusion protein. The MBP-ExpR71 protein was purified by amylose resin affinity chromatography, which yielded a preparation of above 90% purity judged by the SDS-PAGE analysis (Fig. 2, lane 5).
FIG. 2.
Overexpression and purification of MBP-ExpR71 protein. Lanes 1 and 2, lysates of E. coli DH5α carrying the vector (pMAL-c2g) without or with 1 mM IPTG; lanes 3 and 4, lysates of DH5α carrying pAKC1220 (expR71 in pMAL-c2g) without or with 1 mM IPTG; and lane 5, purified MBP-ExpR71. Lanes 1 to 4 contained 5 μg of total bacterial proteins, and lane 5 contained 2 μg of purified MBP-ExpR71. Samples were analyzed by SDS-PAGE in a 10% (wt/vol) polyacrylamide gel.
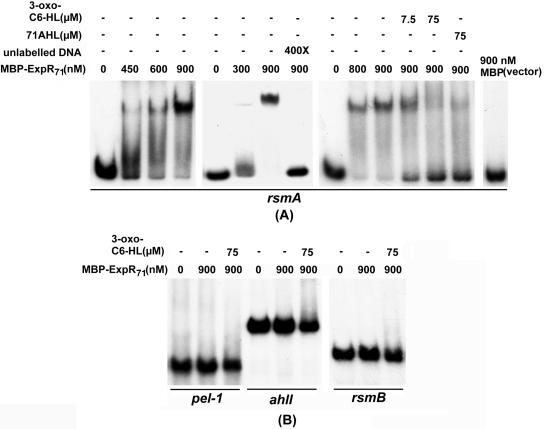
Gel mobility shift assays with MBP-ExpR71.
Gel mobility shift assays were carried out to determine interaction of purified MBP-ExpR71 protein with DNA segments containing promoter regions of ahlI, pel-1, rsmA, and rsmB. α-32P-labeled DNA fragments of ahlI, pel-1, rsmA, and rsmB and MBP-ExpR71 protein were incubated in the DNA binding buffer and resolved on a nondenaturing 5% polyacrylamide gel. Results in Fig. 3A reveal that MBP-ExpR71 binds the rsmA DNA segment. The extent of MBP-ExpR71-rsmA band shift is proportional to the concentration of protein added, and the excess of cold rsmA DNA abolishes the retarded band, indicating that the binding is specific. Amylose resin affinity chromatography purified MBP2-β-galactosidase α fragment protein from IPTG induced cells of DH5α carrying pMAL-c2g vector did not bind the rsmA probe, indicating that the binding is between ExpR71 protein and rsmA DNA (Fig. 3A). MBP-ExpR71 does not bind the DNA fragments of ahlI, pel-1, and rsmB in the presence or in the absence of 3-oxo-C6-HL (Fig. 3B).
FIG. 3.
Gel mobility shift assays for binding of purified MBP-ExpR71 protein to (A) rsmA and (B) pel-1, ahlI, and rsmB DNAs. DNA fragments were end-labeled with [α-32P]dATP. Two nanograms of probe DNAs was added in each reaction. The amounts of proteins, unlabeled DNA, and 71AHL or synthetic 3-oxo-C6-HL used in each reaction are indicated on the top of each figure.
Effects of 3-oxo-C6-HL on MBP-ExpR71-rsmA binding.
We tested the effects of 71AHL (3-oxo-C6-HL) on the binding of rsmA and ExpR71-MBP by adding the fractionated 71AHL or commercially available 3-oxo-C6-HL to the binding reactions. Figure 3A shows that 71AHL or synthetic 3-oxo-C6-HL prevented the MBP-ExpR71-rsmA binding.
ExpR activates transcription of rsmA.
Previous studies (3, 24) in Ecc71 and SCC3193 have demonstrated that under conditions of AHL deficiency, rsmA transcript levels are high, but the basis for this effect was not known. Gel mobility shift assay results demonstrated that MBP-ExpR71 binds the promoter region of rsmA and that the binding is prevented by 71AHL (3-oxo-C6-HL). We therefore considered the possibility that ExpR in the absence of AHL could activate rsmA expression. We present below several lines of evidence that support this hypothesis.
(i) Effects of expR71 on expression of rsmA-lacZ fusion in E. coli.
Results of rsmA-ExpR71 binding assays strongly suggested a direct effect of ExpR on rsmA transcription. To further examine this possibility, we used E. coli strains as surrogate hosts. We argued that the activation of rsmA expression in the presence of expR71 but no activation in its absence would provide strong evidence that ExpR acts in the absence of any other Erwinia-specific factor. We transferred the rsmA-lacZ transcriptional fusion plasmid, pAKC1100, into E. coli strain MC4100 carrying vector (pCL1920) or expR71 (pAKC936). β-Galactosidase assay data (Table 2) revealed that the expression of rsmA-lacZ in MC4100 was higher in the presence of expR71 (ca. threefold) than in MC4100 carrying rsmA-lacZ fusion (pAKC1100) and vector, i.e., in the absence of expR71. β-Galactosidase assay results of MC4100 carrying ahlI-lacZ, pel-lacZ, and rsmB-lacZ fusions with vector or expR71 revealed that expR71 had no effect on the expression of these fusions (Table 2).
TABLE 2.
Expression of rsmA-lacZ, pel-lacZ, ahlI-lacZ, and rsmB-lacZ fusions in E. coli in the presence of expR71
| Bacterial constructa | Relevant characteristicb | β-Galactosidase activityc |
|---|---|---|
| MC4100(pCL1920+pMP220) | vectors | 49 ± 3 |
| MC4100(pAKC936+pMP220) | expR71 + vector | 52 ± 3 |
| MC4100(pCL1920+pAKC1100) | vector + rsmA-lacZ | 620 ± 13 |
| MC4100(pAKC936+pAKC1100) | expR71 + rsmA-lacZ | 1,905 ± 17 |
| MC4100(pCL1920+pAKC1202) | vector + ahlI-lacZ | 262 ± 18 |
| MC4100(pAKC936+pAKC1202) | expR71 + ahlI-lacZ | 269 ± 14 |
| MC4100(pCL1920+pAKC1203) | vector + pel-lacZ | 173 ± 7 |
| MC4100(pAKC936+pAKC1203) | expR71 + pel-lacZ | 187 ± 6 |
| MC4100(pCL1920+pAKC1002) | vector + rsmB-lacZ | 943 ± 27 |
| MC4100(pAKC936+pAKC1002) | expR71 + rsmB-lacZ | 961 ± 34 |
Bacteria were grown at 28°C in LB supplemented with spectinomycin and tetracycline to a Klett value of ca 250, and the cultures were used for assay.
The relevant characteristics of the genes carried by bacteria are given.
Expressed as Miller units.
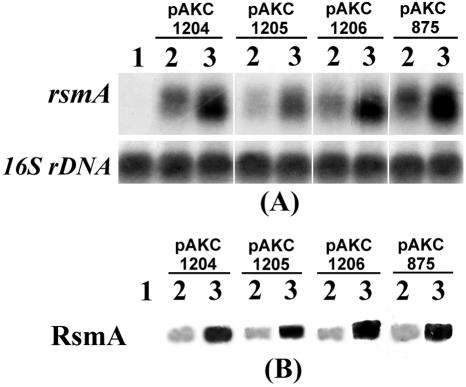
Those data revealed that the transcription of Ecc71 rsmA was activated by the expR71 plasmid in E. coli. To determine the effects of expR71 DNA on the expression of rsmA genes from other Erwinia carotovora strains, we transferred rsmA plasmids of E. carotovora subsp. carotovora strains Ecc71, SCC3193, and EC153 as well as E. carotovora subsp. atroseptica strain Eca12 into E. coli strain DH5α carrying vector or expR71 plasmid. The results of Northern blot analysis (Fig. 4A) and Western blot analysis (Fig. 4B) demonstrate that the production of rsmA RNAs and RsmA proteins was activated in the presence of the expR71 gene in E. coli DH5α.
FIG. 4.
(A) Northern blot analysis and (B) Western blot analysis of E. coli strain DH5α carrying pCL1920 or pAKC936 (expR71) with rsmA plasmids from SCC3193 (pAKC1204), Eca12 (pAKC1205), EC153 (pAKC1206), and Ecc71 (pAKC875). Total RNAs and proteins were extracted from bacteria grown at 28°C in LB medium supplemented with spectinomycin and tetracycline to a Klett value of ca. 150. For Northern blot analysis, each lane contained 10 μg of total RNA, and for Western blot analysis, each lane contained 5 μg of total bacterial protein. Equal loading of RNA was checked by hybridization of the blot with a probe corresponding to 16S rRNA (rDNA). Lane 1, DH5α carrying pLARF5 plus pCL1920 (cloning vectors); 2, DH5α carrying pCL1920 and rsmA+ plasmids; and 3, DH5α carrying expR71+ (pAKC936) and rsmA+ plasmids.
(ii) AHL neutralizes expR71 effects on expression of rsmA-lacZ in E. coli.
The data presented above demonstrated that expression of the rsmA-lacZ fusion is stimulated in the presence of expR71 in E. coli, and the MBP-ExpR71-rsmA binding is prevented by adding 71AHL (3-oxo-C6-HL). Therefore, it was of interest to test the effects of 71AHL on ExpR-mediated activation of rsmA expression in E. coli strain MC4100. The results of the β-galactosidase assay in Table 3 revealed that in the presence of 71AHL (3-oxo-C6-HL), expression of rsmA-lacZ in MC4100 carrying pAKC1100 and pAKC936 (expR71+ plasmid) was reduced to the basal level (i.e., the level in MC4100 carrying pAKC1100 and pCL1920). In addition, we tested expression of the rsmA-lacZ fusion in MC4100 carrying pAKC936 in the presence or absence of ahlI+ plasmid pAKC1201. MC4100 carrying pAKC1201 produced AHL, and the levels of this metabolite were comparable to that in Ecc71 (data not shown). The results of β-galactosidase activity (Table 3) indicated that rsmA expression was higher in MC4100 carrying pAKC936 in the absence of pAKC1201 than in the presence of pAKC1201. These results demonstrate that in E. coli, expR71 stimulates the expression of rsmA-lacZ in the absence of AHL and the effects are neutralized by AHL.
TABLE 3.
Effects of expR71 plasmid on the expression of rsmA-lacZ fusion in E. coli in the presence of 3-oxo-C6-HL or ahlI plasmid
| Bacterial construct | Relevant characteristicc | AHL | β-galactosidase activityd |
|---|---|---|---|
| MC4100(pCL1920+pAKC1100)a | vector + rsmA-lacZ | 548 ± 18 | |
| MC4100(pCL1920+pAKC1100)a | vector + rsmA-lacZ | +71AHL | 535 ± 19 |
| MC4100(pAKC936+pAKC1100)a | expR71 + rsmA-lacZ | 1,801 ± 27 | |
| MC4100(pAKC936+pAKC1100)a | expR71 + rsmA-lacZ | +71AHL | 584 ± 20 |
| MC4100(pCL1920+pAKC1100+pDK6)b | vector + rsmA-lacZ + vector | 624 ± 14 | |
| MC4100(pCL1920+pAKC1100+pAKC1201)b | vector + rsmA-lacZ + ahlI | 601 ± 27 | |
| MC4100(pAKC936+pAKC1100+pDK6)b | expR71 + rsmA-lacZ + vector | 1,942 ± 28 | |
| MC4100(pAKC936+pAKC1100+pAKC1201)b | expR71 + rsmA-lacZ + ahlI | 669 ± 19 |
Bacteria were grown at 28°C in LB supplemented with spectinomycin and tetracycline to a Klett value of ca 100 and divided into two flasks. 71AHL (to a final concentration of 50 μM) was added to one, and the other was used as control (added water). After an additional 3 h of incubation at 28°C, cultures were used for assay.
Bacteria were grown at 28°C in LB supplemented with spectinomycin, tetracycline, and kanamycin to a Klett value of ca 250. Cultures were used for assay.
The relevant characteristics of the genes carried by bacteria are given.
Expressed as Miller units.
(iii) Effects of expR71 on rsmA expression in E. carotovora subsp. carotovora.
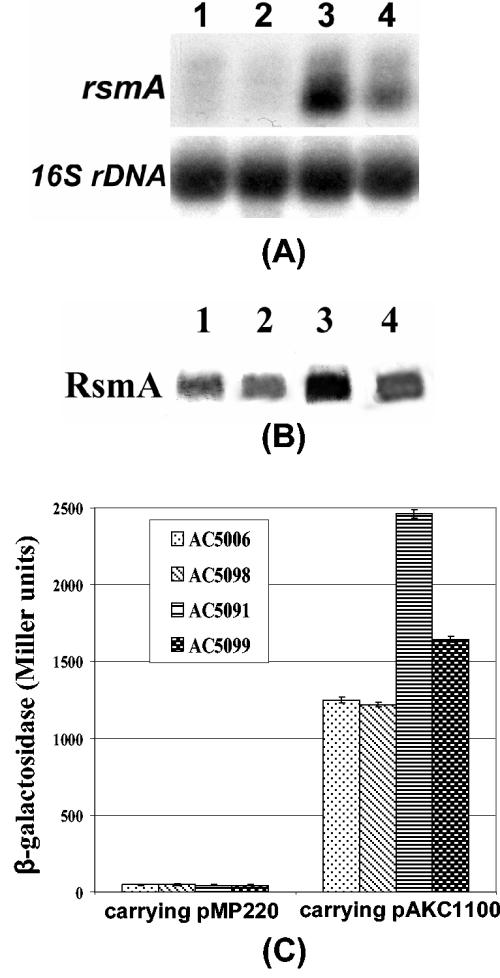
We compared the expression of rsmA in AC5006, a LacZ− derivative of Ecc71 and its mutants, AhlI− ExpR+ (AC5091), AhlI+ ExpR− (AC5098), and AhlI− ExpR− (AC5099). The data in Fig. 5A and B reveal that rsmA RNA and RsmA protein were overproduced by the AhlI− ExpR+ mutant (lane 3) compared to the AhlI+ ExpR+ parent (lane 1). The levels of rsmA transcripts and RsmA protein were lower in the AhlI− ExpR− mutant than that in the AhlI− ExpR+ strain (lanes 3 and 4). Furthermore, the β-galactosidase assay data (Fig. 5C) revealed that (i) expression of an rsmA-lacZ fusion also was higher in the AhlI− ExpR+ mutant than in the parent, and (ii) expression of the rsmA-lacZ in the AhlI− ExpR− strain was lower than that in the AhlI− ExpR+ strain.
FIG. 5.
(A) Northern blot analysis of rsmA and (B) Western blot analysis of RsmA of E. carotovora subsp. carotovora strains. Lane 1, AC5006 (AhlI+ ExpR+); 2, AC5098 (AhlI+ ExpR−); 3, AC5091 (AhlI− ExpR+); and 4, AC5099 (AhlI− ExpR−). Total RNAs and proteins were extracted from bacteria grown at 28°C in minimal salts plus sucrose (0.5%, wt/vol) and celery extract medium to a Klett value of ca. 200. Each lane contained 10 μg of total RNA for Northern blot analysis and 10 μg of total protein for Western blot analysis. Equal loading of RNA was checked by hybridization of the blot with a probe corresponding to 16S rRNA (rDNA). (C) β-Galactosidase assays of AC5006, AC5098, AC5091, and AC5099 carrying pMP220 (vector) or pAKC1100 (rsmA-lacZ fusion). Bacterial constructs were grown at 28°C in minimal salts medium plus sucrose and tetracycline to a Klett value of ca. 200 and harvested for the assays. Bars represent standard errors.
Effects of ExpR71 deficiency on exoenzyme production in the absence of AHL.
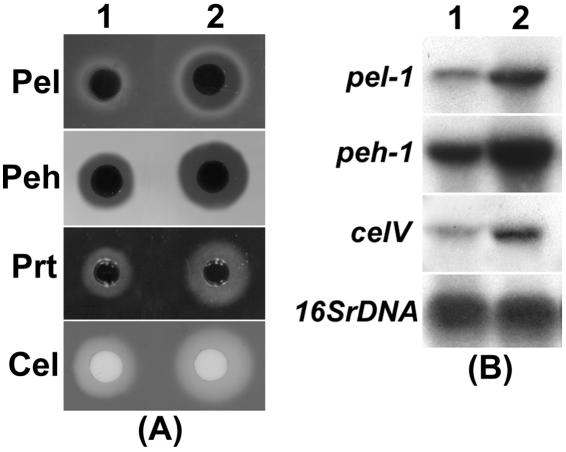
Based upon the observations (i) that AHL is required for exoenzyme production in Ecc71; (ii) that RsmA negatively affects exoenzyme production; and (iii) that ExpR71 activates expression of rsmA in the absence of AHL, it was predicted that AhlI− ExpR71− strain would produce higher levels of exoenzymes than the AhlI− ExpR+ strain. To test this prediction, we grew E. carotovora subsp. carotovora strains in minimal salts medium supplemented with sucrose and celery extract and assayed for exoenzyme production and transcripts levels of several exoenzyme genes. Figure 6 shows that the levels of Pel, Peh, Prt, and Cel as well as pel-1, peh-1, and celV transcripts in AhlI− ExpR− strains (column 2) were higher than that in AhlI− ExpR+ strains (column 1).
FIG. 6.
(A) Agarose plate assays for Pel, Peh, Prt, and Cel activities of AC5091 (AhlI− ExpR+, lane 1) and AC5099 (AhlI− ExpR−, lane 2). Fifty microliters of culture supernatant was applied in each well. (B) Northern blot analysis of pel-1, peh-1, and celV of AC5091 (1) and AC5099 (2). Each lane contained 10 μg of total RNA. Equal loading of RNA was checked by hybridization of the blot with a probe corresponding to 16S rRNA (rDNA). Bacteria were grown in minimal salts plus sucrose (0.5%, wt/vol) and celery extract medium at 28°C to a Klett value of ca. 200 for total RNAs extraction. Culture supernatants were used for exoenzyme assays.
DISCUSSION
In this report we present two significant findings: (i) ExpR activates transcription of rsmA, which specifies an RNA-binding protein known to inhibit exoprotein and secondary metabolite production; and (ii) AHL inhibits this ExpR function, including ExpR binding its target DNA. Findings with various LuxR homologs have established that in a vast majority of cases protein-AHL complexes activate transcription of target genes or operons. In the absence of AHL, the N-terminal domains of LuxR family proteins inhibit DNA binding by the C-terminal DNA binding domains (34). However, this inhibition is relieved upon binding of AHLs to LuxR homologs. Thus, activator functions of many LuxR proteins depend upon their interaction with the cognate signals. There are several instances, including ExpR (this report), where this model of LuxR action does not apply. A well-studied example is that of EsaR. Studies by von Bodman and her associates (35, 36, 58) have demonstrated that EsaR, a LuxR homolog produced by Pantoea stewartii, acts as a repressor by binding the targeted promoter (lux box) in an AHL-independent manner. The EsaR action is neutralized by its interaction with AHL. Another example comprises SmaR, a LuxR homolog of Serratia sp. strain ATCC39006. In this case genetic evidence suggests that SmaR is a repressor of the carA-H operon (50). Furthermore, QscR in Pseudomonas aeruginosa functions as a repressor of various virulence genes (25). ExpR of E. carotovora subspecies now adds to the growing list of QSS systems where the AHL receptors function as transcriptional regulators in the absence of the signal.
The effect of ExpR on rsmA is established by the levels of rsmA transcripts, expression of a rsmA-lacZ fusion, the levels of RsmA protein, and DNA binding studies. Comparative analysis of AhlI− ExpR+ and AhlI− ExpR− strains of Ecc71 demonstrates an activator function of ExpR71 (Fig. 5). It should be noted that the levels of rsmA RNA and RsmA protein as well as the expression of the rsmA-lacZ fusion were very similar in the AhlI+ ExpR− and AhlI+ ExpR+ parent strain (Fig. 5). We attribute these responses to neutralization of ExpR action by AHL in the parent strain to the extent that it behaves phenotypically as an ExpR-deficient strain. Likewise, studies with E. coli strains, apparently lacking ahlI genes as well as an AHL producing system, strongly suggest activation of rsmA expression by ExpR. We have documented a higher level of expression of an rsmA-lacZ fusion in E. coli MC4100 in the presence of expR71+ DNA than in its absence. Moreover, in E. coli DH5α, ExpR71 activates expression of rsmA genes from several other E. carotovora subsp. carotovora strains, including EC153, SCC3193, and E. carotovora subsp. atroseptica strain Eca12 (Fig. 4). Gel mobility shift assays demonstrate that ExpR specifically binds rsmA DNA (Fig. 3). This observation, taken along with transcript assay data and expression of an rsmA-lacZ fusion, establishes that rsmA overexpression in the absence of AHL is due to activation of rsmA promoter by ExpR.
The DNA-binding properties of the LuxR family of proteins are known to be modified by interaction with small molecules such as AHL (56). The results of gel mobility shift data (Fig. 3) clearly demonstrate inhibition of ExpR binding to rsmA DNA in the presence of 3-oxo-C6-HL produced by E. carotovora subsp. carotovora strain Ecc71. A previous study of von Bodman et al. (58) has revealed that EsaR, a LuxR homolog of Pantoea stewartii subsp. stewartii, binds lux box sequences. This binding is neutralized by 3-oxo-C6-HL as indicated by in vivo β-galactosidase assays using an artificial 35LB10-lacZ promoter fusion containing lux box sequences. Interestingly, the addition of the AHL did not promote EsaR-DNA complex dissociation in their DNA mobility shift assay. However, they also reported that AHL interacts specifically with EsaR protein and induces structural changes that may neutralize its DNA binding affinity (36). In Serratia sp. strain ATCC 39006, BHL [N-(butanoyl)-l-homoserine lactone] inhibits DNA binding ability of SmaR, thereby relieving SmaR-mediated repression of the carA-H operon (50). We also document here a marked reduction in the expression of an rsmA-lacZ fusion in E. coli MC4100 in the presence of the expR71 gene and AHL. We attribute this reduction to the loss of activator function of ExpR due to the formation of ExpR-AHL complex. Another line of evidence for a negative effect of AHL comes from studies done with the Ahl+ and Ahl− strains of Ecc71. rsmA RNA and RsmA protein levels were higher in AhlI− strains than in AhlI+ bacteria (Fig. 5A and B). The expression of an rsmA-lacZ fusion was higher in an AhlI− strain than in the AhlI+ parent (Fig. 5C). Moreover, an AhlI− ExpR− double mutant produced reduced levels of rsmA RNA and RsmA protein compared to the levels in an AhlI− ExpR+ strain (Fig. 5A and B). These data collectively point to a conditional effect of ExpR in that it manifests under conditions of AHL deficiency or AHL limitation. We should recall that AHL deficiency has been reported to result in high levels of rsmA transcripts in several E. carotovora subsp. carotovora strains (3, 24). Our data on the activation of rsmA transcription in the presence of ExpR71 but in the absence of AHL now explain the basis for this finding.
Studies with E. chrysanthemi ExpR (ExpREch) have demonstrated that it binds to the promoter regions of the five major pel genes and expI. The ExpREch-DNA band shift profiles changed in the presence of AHL (42, 47). There also is some evidence suggesting activation of the promoters of some of these genes by ExpREch. Our data with E. carotovora subsp. carotovora present a very different picture regarding the action of ExpREcc. First, it activates expression of rsmA but has no direct effect on pel-1 or ahlI. Second, ExpREcc specifically binds rsmA DNA but not the DNA fragments of pel-1 or ahlI. Third, ExpREcc activates rsmA transcription and binds rsmA DNA in the absence of AHL. In fact, AHL quite effectively inhibits these ExpREcc actions. Based upon these observations we conclude that the modus operandi of the quorum-sensing systems are fundamentally different in two groups of soft-rotting bacteria, namely E. carotovora subspecies and E. chrysanthemi. We have initiated studies of cross-species effects of ExpR proteins and their chimeric derivatives to better understand the molecular bases for the observed differences.
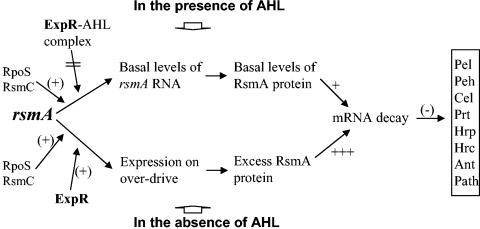
After taking into consideration the data presented here and the literature on ExpR and bacteria deficient in AHL or ExpR, we conclude that the primary quorum-sensing signaling system in E. carotovora subsp. carotovora comprises three major components: AHL, ExpR, and RsmA. As depicted in Fig. 7, ExpR and ExpR-AHL complex modulate the levels of RsmA which, in turn, is responsible for the expression of an array of genes known to be expressed in a growth phase or cell density dependent manner. In several E. carotovora subspecies, as in P. stewartii (36), AHL production is constitutive and not autoregulated (unpublished data). At low cell density, AHL concentration in the niche remains low (4, 31, 39), which cannot support high levels of target (exoprotein) gene expression. As the cell population increases, AHL concentration also rises, promoting activation of exoprotein gene expression. In the absence of AHL, free ExpR, the activator of rsmA transcription, is the dominant species. Under these conditions rsmA transcription is in overdrive (lower pathway, Fig. 7), causing a marked increase in the RsmA pool, ensuring complete or near-complete inhibition of exoprotein and secondary metabolite production. On the other hand, in the presence of high levels of AHL (upper pathway, Fig. 7), most, if not all, ExpR exists as ExpR-AHL complex, lacking this activator function. Thus, the main physiological function of ExpR appears to activate rsmA expression in the absence of AHL and AHL modulates this activation. This model explains the basis for the pleiotropic effects of AHL deficiency and the requirement of AHL for the expression of genes for exoproteins and secondary metabolites.
FIG. 7.
A speculative model depicting the regulatory events in the expression of rsmA in E. carotovora subsp. carotovora. The basal level of rsmA expression (upper pathway) is controlled by several factors, including RpoS and RsmC. Increased pool of free ExpR but not ExpR-AHL complex (lower pathway) causes overproduction of RsmA, which in turn suppresses exoprotein and antibiotic production and pathogenicity by controlling mRNA stability.
We now also understand the basis for the apparent paradox presented by the ExpR-deficient bacteria in that this deficiency did not yield a recognizable change in bacterial phenotype. There is a substantial basal level of RsmA in AHL+ and ExpR-deficient bacteria, and this basal level of RsmA could be responsible for the lack of a recognizable phenotype in ExpR-deficient E. carotovora subsp. carotovora mutants. In this context we should recognize that RsmA pool size is determined by several other regulators, including RsmC (10), RpoS (40), and rsmB RNA (28), the latter controlled by regulators such as GacA (8) and KdgR (29).
Acknowledgments
Our work was supported by the National Science Foundation (grants MCB-9728505) and the Food for the 21st Century program of the University of Missouri.
We thank Judy Wall for critically reviewing the manuscript, P. Williams for providing 3-oxo-C8-HL, T. Palva for providing strain SCC3193, and S. B. von Bodman for helping us with ExpR overexpression and purification.
REFERENCES
- 1.Andersson, R. A., A. R. B. Eriksson, R. Heikinheimo, A. Mäe, M. Pirhonen, V. Kõiv, H. Hyytiäinen, A. Tuikkala, and E. T. Palva. 2000. Quorum sensing in the plant pathogen Erwinia carotovora subsp. carotovora: the role of expR (Ecc). Mol. Plant-Microbe Interact. 13:384-393. [DOI] [PubMed] [Google Scholar]
- 2.Barras, F., F. Van Gijsegem, and A. K. Chatterjee. 1994. Extracellular enzymes and pathogenesis of soft-rot Erwinia. Annu. Rev. Phytopathol. 32:201-234. [Google Scholar]
- 3.Chatterjee, A., Y. Cui, and A. K. Chatterjee. 2002. RsmA and the quorum-sensing signal, N-[3-oxohexanoyl]-L-homoserine lactone, control the levels of rsmB RNA in Erwinia carotovora subsp. carotovora by affecting its stability. J. Bacteriol. 184:4089-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterjee, A., Y. Cui, Y. Liu, C. K. Dumenyo, and A. K. Chatterjee. 1995. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density sensing signal, N-(3-oxohexanoyl)-L-homoserine lactone. Appl. Environ. Microbiol. 61:1959-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatterjee, A. K., C. K. Dumenyo, Y. Liu, and A. Chatterjee. 2000. Erwinia: genetics of pathogenicity factors, p. 236-260. In J. Lederberg (ed.), Encyclopedia of microbiology, 2nd ed., vol. 2. Academic Press, New York, N.Y.
- 6.Collmer, A., and N. T. Keen. 1986. The role of pectic enzymes in plant pathogenesis. Annu. Rev. Phytopathol. 24:383-409. [Google Scholar]
- 7.Costa, J. M., and J. E. Loper. 1997. EcbI and EcbR: homologs of LuxI and LuxR affecting antibiotic and exoenzyme production by Erwinia carotovora subsp. betavasculorum. Can. J. Microbiol. 43:1164-1171. [DOI] [PubMed] [Google Scholar]
- 8.Cui, Y., A. Chatterjee, and A. K. Chatterjee. 2001. Effects of the two-component system comprising GacA and GacS of Erwinia carotovora subsp. carotovora on the production of global regulatory rsmB RNA, extracellular enzymes, and harpinEcc. Mol. Plant-Microbe Interact. 14:516-526. [DOI] [PubMed] [Google Scholar]
- 9.Cui, Y., A. Chatterjee, Y. Liu, C. K. Dumenyo, and A. K. Chatterjee. 1995. Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp. carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-L-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. J. Bacteriol. 177:5108-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui, Y., A. Mukherjee, C. K. Dumenyo, Y. Liu, and A. K. Chatterjee. 1999. rsmC of the soft-rotting bacterium Erwinia carotovora subsp. carotovora negatively controls extracellular enzyme and HarpinEcc production and virulence by modulating the levels of regulatory RNA (rsmB) and RNA binding protein (RsmA). J. Bacteriol. 181:6042-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eriksson, A. R., R. A. Andersson, M. Pirhonen, and E. T. Palva. 1998. Two-component regulators involved in the global control of virulence in Erwinia carotovora subsp. carotovora. Mol. Plant-Microbe Interact. 11:743-752. [DOI] [PubMed] [Google Scholar]
- 13.Frederick, R. D., J. Chiu, J. L. Bennetzen, and A. K. HandA. 1997. Identification of a pathogenicity locus, rpfA, in Erwinia carotovora subsp. carotovora that encodes a two-component sensor-regulator protein. Mol. Plant-Microbe Interact. 10:407-415. [DOI] [PubMed] [Google Scholar]
- 14.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoerine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 15.Gray, K. M., and J. R. Garey. 2001. The evolution of bacterial LuxI and LuxR quorum sensing regulators. Microbiology 147:2379-2387. [DOI] [PubMed] [Google Scholar]
- 16.Gray, K. M., and E. P. Greenberg. 1992. Physical and functional maps of the luminescence gene cluster in an autoinducer-deficient Vibrio fischeri strain isolated from a squid light organ. J. Bacteriol. 174:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris, S. J., Y. L. Shih, S. D. Bentley, and G. P. C. Salmond. 1998. The hexA gene of Erwinia carotovora encodes a LysR homologue and regulates motility and the expression of multiple virulence determinants. Mol. Microbiol. 28:705-717. [DOI] [PubMed] [Google Scholar]
- 18.He, S. Y. 1998. Type III protein secretion systems in plant and animal pathogenic bacteria. Annu. Rev. Phytopathol. 36:363-392. [DOI] [PubMed] [Google Scholar]
- 19.Holden, M. T. G., S. R. Chabra, R. de Nys, P. Stead, N. J. Bainton, P. J. Hill, M. Manefield, N. Kumar, M. Labatte, D. England, S. Rice, M. Givskov, G. P. C. Salmond, G. S. A. B. Stewart, B. W. Bycroft, S. Kjelleberg, and P. Williams. 1999. Quorum-sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other gram-negative bacteria. Mol. Microbiol. 33:1254-1266. [DOI] [PubMed] [Google Scholar]
- 20.Holden, M. T. G., S. J. McGowan, B. W. Bycroft, G. S. A. B. Stewart, P. Williams, and G. P. C. Salmond. 1998. Cryptic carbapenem antibiotic production genes are widespread in Erwinia carotovora-facile trans activation by the carR transcriptional regulator. Microbiology 144:1495-1508. [DOI] [PubMed] [Google Scholar]
- 21.Jones, S., B. Yu, N. J. Bainton, M. Birdsall, B. W. Bycroft, S. R. Chhabra, A. J. R. Cox, P. Golby, P. J. Reeves, S. Stephens, M. K. Winson, G. P. C. Salmond, G. S. A. B. Stewart, and P. Williams. 1993. The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J. 12:2477-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 23.Kleiner, D., W. Paul, and M. J. Merrick. 1988. Construction of multicopy expression vectors for regulated overproduction of proteins in Klebsiella pneumoniae and other enteric bacteria. J. Gen. Microbiol. 134:1779-1784. [DOI] [PubMed] [Google Scholar]
- 24.Kõiv, V., and A. Mäe. 2001. Quorum sensing controls the synthesis of virulence factors by modulating rsmA gene expression in Erwinia carotovora subsp. carotovora. Mol. Genet. Genomics 265:287-292. [DOI] [PubMed] [Google Scholar]
- 25.Ledgham, F., I. Ventre, C. Soscia, M. Foglino, J. N. Sturgis, and A. Lazdunski. 2003. Interactions of the quorum sensing regulator QscR: interaction with itself and the other regulators of Pseudomonas aeruginosa LasR and RhlR. Mol. Microbiol. 48:199-210. [DOI] [PubMed] [Google Scholar]
- 26.Lerner, C. G., and M. Inouye. 1990. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 18:4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, Y., A. Chatterjee, and A. K. Chatterjee. 1994. Nucleotide sequence and expression of a novel pectate lyase gene (pel-3) and a closely endopolygalacturonase gene (peh-1) of Erwinia carotovora subsp. carotovora 71. Appl. Environ. Microbiol. 60:2545-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, Y., Y. Cui, A. Mukherjee, and A. K. Chatterjee. 1998. Characterization of a novel RNA regulator of Erwinia carotovora ssp. carotovora that controls production of extracellular enzymes and secondary metabolites. Mol. Microbiol. 29:219-234. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Y., G.-Q. Jiang, Y. Cui, A. Mukherjee, W.-L. Ma, and A. K. Chatterjee. 1999. kdgREcc negatively regulates genes for pectinases, cellulase, protease, harpinEcc, and a global RNA regulator in Erwinia carotovora subsp. carotovora. J. Bacteriol. 181:2411-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loh, J., E. A. Pierson, L. S. Pierson III, G. Stacey, and A. K. Chatterjee. 2002. Quorum sensing in plant associated bacteria. Curr. Opin. Plant Biol. 5:285-290. [DOI] [PubMed] [Google Scholar]
- 31.Mäe, A., M. Montesano, V. Koiv, and E. T. Palva. 2001. Transgenic plants producing the bacterial pheromone N-acyl-homoserine lactone exhibit enhanced resistance to the bacterial phytopathogen Erwinia carotovora. Mol. Plant-Microbe Interact. 14:1035-1042. [DOI] [PubMed] [Google Scholar]
- 32.McGowan, S., M. Sebaihia, S. Jones, B. Yu, N. Bainton, P. Chan, B. Bycroft, G. S. A. B. Stewart, P. Williams, and G. P. C. Salmond. 1995. Carbapenem antibiotic production in Erwinia carotovora regulated by CarR, a homologue to the LuxR transcriptional activator. Microbiology 141:541-550. [DOI] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 35.Minogue, T. D., A. L. Carlier, M. D. Koutsoudis, and S. B. von Bodman. 2005. The cell density-dependent expression of stewartan exopolysaccharide in Pantoea stewartii ssp. stewartii is a function of EsaR-mediated repression of the rcsA gene. Mol. Microbiol. 56:189-203. [DOI] [PubMed] [Google Scholar]
- 36.Minogue, T. D., M. Wehland-von Trebra, F. Bernhard, and S. B. von Bodman. 2002. The autoregulatory role of EsaR, a quorum sensing regulator in Pantoea stewartii subsp. stewartii: evidence for a repressor function. Mol. Microbiol. 44:1625-1635. [DOI] [PubMed] [Google Scholar]
- 37.Morin, D., B. Grasland, K. Vallée-Réhel, C. Dufau, and D. Haras. 2003. On-line high-performace liquid chromatography-mass spectrometric detection and quantification of N-acylhomoserine lactones, quorum sensing signal molecules, in the presence of biological matrices. J. Chromatogr. A 1002:79-92. [DOI] [PubMed] [Google Scholar]
- 38.Mukherjee, A., Y. Cui, Y. Liu, and A. K. Chatterjee. 1997. Molecular characterization and expression of the Erwinia carotovora hrpNEcc gene, which encodes an elicitor of the hypersensitive reaction. Mol. Plant-Microbe Interact. 10:462-471. [DOI] [PubMed] [Google Scholar]
- 39.Mukherjee, A., Y. Cui, W. Ma, Y. Liu, and A. K. Chatterjee. 2000. hexA of Erwinia carotovora ssp. carotovora strain Ecc71 negatively regulates production of RpoS and rsmB RNA, a global regulator of extracellular proteins, plant virulence and the quorum-sensing signal, N-(3-oxohexanoyl)-L-homoserine lactone. Environ Microbiol. 2:203-215. [DOI] [PubMed] [Google Scholar]
- 40.Mukherjee, A., Y. Cui, W.-L. Ma, Y. Liu, A. Ishihama, A. Eisenstark, and A. K. Chatterjee. 1998. RpoS (Sigma-S) controls expression of rsmA, a global regulator of secondary metabolites, harpin, and extracellular proteins in Erwinia carotovora. J. Bacteriol. 180:3629-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murata, H., J. L. McEvoy, A. Chatterjee, A. Collmer, and A. K. Chatterjee. 1991. Molecular cloning of an aepA gene that activates production of extracellular pectolytic, cellulolytic, and proteolytic enzymes in Erwinia carotovora subsp. carotovora. Mol. Plant-Mircobe Interact. 4:239-246. [Google Scholar]
- 42.Nasser, W., M. L. Bouillant, G. P. C. Salmond, and S. Reverchon. 1998. Characterization of the Erwinia chrysanthemi expI-expR locus directing the synthesis of two N-acyl-homoserine lactone signal molecules. Mol. Microbiol. 29:1391-1405. [DOI] [PubMed] [Google Scholar]
- 43.Pemberton, C. L., N. A. Whitehead, M. Sebaihia, K. S. Bell, L. J. Hyman, S. J. Harris, A. J. Matlin, N. D. Robson, P. R. J. Birch, J. P. Carr, I. K. Toth, and G. P. C. Salmond. 2005. Novel quorum-sensing-controlled genes in Erwinia carotovora subsp. carotovora: identification of a fungal elicitor homologue in a soft-rotting bacterium. Mol. Plant-Microbe Interact. 18:343-353. [DOI] [PubMed] [Google Scholar]
- 44.Pierson, L. S., III, D. W. Wood, and S. B. von Bodman. 1999. Quorum sensing in plant-associated bacteria, p. 101-116. In G. M. Dunney and S. C. Winans (ed.), Cell-cell signaling. American Society for Microbiology Press, Washington, D.C.
- 45.Pirhonen, M., D. Flego, R. Heikinheimo, and E. T. Palva. 1993. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 12:2467-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rantakari, A., O. Virtaharju, S. Vähämiko, S. Taira, E. T. Palva, H. Saarilahti, and M. Romantschuk. 2001. Type III secretion contributes to the pathogenesis of the soft-rot pathogen Erwinia carotovora: partial characterization of the hrp gene cluster. Mol. Plant-Microbe Interact. 14:962-968. [DOI] [PubMed] [Google Scholar]
- 47.Reverchon, S., M. L. Bouillant, G. Salmond, and W. Nasser. 1998. Integration of the quorum-sensing system in the regulatory networks controlling virulence factor synthesis in Erwinia chrysanthemi. Mol. Microbiol. 29:1407-1418. [DOI] [PubMed] [Google Scholar]
- 48.Salmond, G. P. C. 1994. Secretion of extracellular virulence factors by plant pathogenic bacteria. Annu. Rev. Phytopathol. 32:181-200. [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Slater, H., M. Crow, L. Everson, and G. P. C. Salmond. 2003. Phosphate availability regulates biosynthesis of two antibiotics, prodigiosin and carbapenem, in Serratia via both quorum-sensing-dependent and -independent pathways. Mol. Microbiol. 47:303-320. [DOI] [PubMed] [Google Scholar]
- 51.Spaink, H. P., R. J. H. Okker, C. A. Wijffelman, E. Pees, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 52.Swift, S., A. V. Karlyshev, L. Fish, E. L. Durant, M. K. Winson, S. R. Chhabra, P. Williams, S. Macintyre, and G. S. A. B. Stewart. 1997. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida-identification of the luxRI homologs ahyRI and asaRI and their cognate N-acylhomoserine lactone signal molecules. J. Bacteriol. 179:5271-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swift, S., J. P. Throup, P. Williams, G. P. C. Salmond, and G. S. A. B. Stewart. 1996. Quorum sensing: a population-density component in the determination of bacterial phenotype. Trends Biochem. Sci. 21:214-219. [PubMed] [Google Scholar]
- 54.Taga, M. E., and B. L. Bassler. 2003. Chemical communication among bacteria. Proc. Natl. Acad. Sci. USA 100:14549-14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomson, N. R., A. Cox, B. W. Bycroft, G. S. Stewart, P. Williams, and G. P. C. Salmond. 1997. The rap and hor proteins of Erwinia, Serratia and Yersinia: a novel subgroup in a growing superfamily of proteins regulating diverse physiological processes in bacterial pathogens. Mol. Microbiol. 26:531-544. [DOI] [PubMed] [Google Scholar]
- 56.Throup, J. P., M. Camara, G. S. Briggs, M. K. Winson, S. R. Chhabra, B. W. Bycroft, P. Williams, and G. S. A. B. Stewart. 1995. Characterization of the yenl/yenr locus from Yersinia enterocolitica mediating the synthesis of two N-acylhomoserine lactone signal molecules. Mol. Microbiol. 17:345-356. [DOI] [PubMed] [Google Scholar]
- 57.Urbanowski, M. L., C. P. Lostroh, and E. P. Greenberg. 2004. Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein. J. Bacteriol. 186:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.von Bodman, S. B., J. K. Ball, M. A. Faini, C. M. Herrera, T. D. Minogue, M. L. Urbanowski, and A. M. Stevens. 2003. The quorum sensing negative regulators EsaR and ExpR(Ecc), homologues within the LuxR family, retain the ability to function as activators of transcription. J. Bacteriol. 185:7001-7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.von Bodman, S. B., W. D. Bauer, and D. L. Coplin. 2003. Quorum sensing in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 41:455-482. [DOI] [PubMed] [Google Scholar]
- 60.Welch, M., D. E. Todd, N. A. Whitehead, S. J. Mcgowan, B. W. Bycroft, and G. P. C. Salmond. 2000. N-acyl homoserine lactone binding to the CarR receptor determines quorum-sensing specificity in Erwinia. EMBO J. 19:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whitehead, N. A., J. T. Byers, P. Commander, M. J. Corbett, S. J. Coulthurst, L. Everson, A. K. P. Harris, C. L. Pemberton, N. J. L. Simpson, H. Slater, D. S. Smith, M. Welch, N. Williamson, and G. P. C. Salmond. 2002. The regulation of virulence in phytopathogenic Erwinia species: quorum sensing, antibiotics and ecological considerations. Antonie van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 81:223-231. [DOI] [PubMed] [Google Scholar]
- 62.Withers, H., S. Swift, and P. Williams. 2001. Quorum sensing as an integral component of gene regulatory networks in gram-negative bacteria. Curr. Opin. Microbiol. 4:186-193. [DOI] [PubMed] [Google Scholar]
- 63.Wood, D. W., F. Gong, M. M. Daykin, P. Williams, and L. S. Pierson III. 1997. N-acyl-homoserine lactone-mediated regulation of phenazine gene expression by Pseudomonas aureofaciens 30-84 in the wheat rhizosphere. J. Bacteriol. 179:7663-7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zink, R. T., R. J. Kemble, and A. K. Chatterjee. 1984. Transposon Tn5 mutagenesis in Erwinia carotovora subsp. carotovora and Erwinia carotovora subsp. atroseptica. J. Bacteriol. 157:809-814. [DOI] [PMC free article] [PubMed] [Google Scholar]