Abstract
The EAL domain (also known as domain of unknown function 2 or DUF2) is a ubiquitous signal transduction protein domain in the Bacteria. Its involvement in hydrolysis of the novel second messenger cyclic dimeric GMP (c-di-GMP) was demonstrated in vivo but not in vitro. The EAL domain-containing protein Dos from Escherichia coli was reported to hydrolyze cyclic AMP (cAMP), implying that EAL domains have different substrate specificities. To investigate the biochemical activity of EAL, the E. coli EAL domain-containing protein YahA and its individual EAL domain were overexpressed, purified, and characterized in vitro. Both full-length YahA and the EAL domain hydrolyzed c-di-GMP into linear dimeric GMP, providing the first biochemical evidence that the EAL domain is sufficient for phosphodiesterase activity. This activity was c-di-GMP specific, optimal at alkaline pH, dependent on Mg2+ or Mn2+, strongly inhibited by Ca2+, and independent of protein oligomerization. Linear dimeric GMP was shown to be 5′pGpG. The EAL domain from Dos was overexpressed, purified, and found to function as a c-di-GMP-specific phosphodiesterase, not as a cAMP-specific phosphodiesterase, in contrast to previous reports. The EAL domains can hydrolyze 5′pGpG into GMP, however, very slowly, thus implying that this activity is irrelevant in vivo. Therefore, c-di-GMP is the exclusive substrate of EAL. Multiple-sequence alignment revealed two groups of EAL domains hypothesized to correspond to enzymatically active and inactive domains. The domains in the latter group have mutations in residues conserved in the active domains. The enzymatic inactivity of EAL domains may explain their coexistence with GGDEF domains in proteins possessing c-di-GMP synthase (diguanulate cyclase) activity.
The approximately 250-amino-acid protein domain EAL (www.sanger.ac.uk/Software/Pfam), also referred to as domain of unknown function 2 or DUF2 (http://smart.embl-heidelberg.de), is conserved in the Bacteria. The domain name originates from one of the most conserved amino acid signature motifs, EAL (Glu-Ala-Leu). EAL is encoded by most sequenced genomes in all branches of the bacterial phylogenetic tree, which implies that EAL-containing proteins play important roles in Bacteria (8, 9). This domain is not encoded in the genomes of Archaea or Eukarya, except for two putative proteins of Anopheles gambiae, which probably originated from bacterial contamination (19). EAL is often linked to sensory and/or output (signal transduction) domains and is, in fact, one of the most ubiquitous bacterial signal transduction domains whose biochemical activity has not been characterized yet (8, 9). In many proteins, EAL is located C terminal of the approximately 170-amino-acid GGDEF domain (also known as DUF1). For example, of the 17 EAL domain-containing proteins identified in the Escherichia coli genome, 7 also contain GGDEF.
Benziman and colleagues were the first researchers to reveal the link between the GGDEF and EAL domains and the unusual cyclic dinucleotide cyclic bis(3′→5′) dimeric GMP (c-di-GMP). They characterized c-di-GMP as an activator of cellulose synthase in Gluconacetobacter xylinus (formerly Acetobacter xylinum) (13, 16, 18). Subsequent characterization by this group of researchers of the enzymes involved in c-di-GMP synthesis (diguanylate cyclase [DGC]) and hydrolysis (phosphodiesterase [PDE]), along with massive genome sequencing, paved the way for determination of the global role of c-di-GMP in Bacteria (8, 9). c-di-GMP is emerging as a novel, widely distributed bacterial signaling molecule (19). It controls cellular processes related to bacterial life on surfaces that often involve multicellular behavior, including biofilm formation (4, 10), surface motility (3), development (1), regulation of gene expression (23), etc. (reviewed in references 7, 12, and 15a).
Paradoxically, three orthologous DGCs and three orthologous PDEs from G. xylinus described by Benziman and colleagues had similar domain organizations in that they all contained GGDEF and EAL domains arranged in tandem (22). The DGC activity of the GGDEF domain was predicted based on the sequence similarity to mammalian adenylate cyclases (15). This activity was subsequently demonstrated both in vivo (2, 21, 24) and in vitro (14, 19). A DGC catalyzes synthesis of c-di-GMP from two molecules of GTP. The GGDEF domain is sufficient for DGC activity when it is present as a dimer in the proper conformation, which depends on the conformation of the sensory domains (5, 19).
The EAL domain has been predicted to possess PDE activity (8, 9). The best indirect evidence linking an EAL domain with c-di-GMP-specific PDE activity came recently from studies that involved proteins containing EAL but not GGDEF domains, including Salmonella enterica YhjH (21) and Vibrio cholerae VieA (24). These proteins, when overexpressed, decreased intracellular levels of c-di-GMP in their host bacteria. Whether they hydrolyzed c-di-GMP directly or indirectly, alone or by interacting with other proteins, is not known, and the products of their activity are also not known. The biochemical activity of an individual EAL domain was not assayed prior to this study. So far, only PDEs containing both GGDEF and EAL domains have been studied in vitro; these PDEs include three PDEs from G. xylinus, PdeA1 to PdeA3 (22), and the E. coli direct oxygen sensor Dos (20). PdeA1 to PdeA3 were shown to cleave a single phosphodiester bond in c-di-GMP, yielding linear dimeric GMP (l-di-GMP). This activity was designated PDE-A. The subsequent hydrolysis of l-di-GMP into two GMP molecules was attributed to a different, uncharacterized enzyme(s) designated PDE-B (16, 18). Dos, on the other hand, was reported to function as a cyclic AMP (cAMP)-specific PDE (20).
There are several important questions concerning EAL domains that hamper progress in the field of c-di-GMP-dependent signal transduction. These questions, summarized below, prompted us to undertake the present study. Is an EAL domain sufficient for PDE activity, or does this activity require GGDEF? What is the substrate specificity of EAL domains (c-di-GMP, cAMP, or other nucleotides)? Can substrate specificity be predicted from the primary sequence of the EAL domain? Do EAL domain-containing proteins express only PDE-A activity or both PDE-A and PDE-B activities? Why do some proteins containing GGDEF and EAL domains function as DGCs, while others function as PDEs?
MATERIALS AND METHODS
Sequence analysis.
Sequences of the EAL (DUF2) domains were obtained from the Pfam (www.sanger.ac.uk/Software/Pfam) and SMART (www.smart.embl-heidelberg.de) databases. For multiple-sequence alignment, Clustal W 1.8 (11), available at http://searchlauncher.bcm.tmc.edu/, was used. The alignment was subsequently manually adjusted.
Construction of plasmids for protein overexpression.
Genomic DNA of E. coli MG1655 was purified from bacterial cells using a Bactozol kit (Molecular Research Center, Cincinnati, Ohio). The E. coli yahA gene and DNA fragments encoding individual EAL domains of YahA (residues 77 to 362) and Dos (residues 540 to 807) were PCR amplified using E. coli genomic DNA, Pfu Hotstart DNA polymerase (Stratagene, La Jolla, Calif.), and gene-specific primers (whose sequences are available upon request). The PCR fragments were gel purified, digested with the appropriate restriction enzymes (whose sites were created), and cloned into vector pET-23a (EMD Biosciences, San Diego, Calif.). The plasmids that were constructed were used for overexpression of proteins and EAL protein domains as C-terminal His6 fusions.
Protein overexpression and purification.
For purification of YahA::His6, EALYahA::His6, and EALDos::His6, the following protocol was used. Briefly, isopropyl-β-d-thiogalactopyranoside (IPTG) (final concentration, 0.2 mM) was added to exponentially growing E. coli DH5α cells containing appropriate plasmids (optical density at 600 nm, 0.6 to 0.8; 30°C; LB broth). After 2 to 6 h of induction, the cells were chilled to 4°C and collected by centrifugation. The cell pellets were resuspended in buffer consisting of 200 mM NaCl, 0.5 mM EDTA, 5 mM MgCl2, 20 mM Tris-HCl (pH 7.6), and 5% glycerol that also contained protease inhibitors (phenylmethylsulfonyl fluoride and P8849) at the concentrations specified by the manufacturer (Sigma, St. Louis, MO). The cell suspensions were passed through a French pressure mini-cell (Spectronic Instruments, Rochester, NY), and this was followed by brief sonication using a Sonifier 250 (Branson Ultrasonics, Danbury, Conn.). The crude cell extracts were centrifuged at 15,000 × g for 45 min. Soluble protein fractions were collected and mixed with preequilibrated Ni2+ resin (QIAGEN, Valencia, Calif.) for 1 h at 4°C, which was placed into a column and extensively washed with buffer containing 50 mM Tris-HCl (pH 8.0), 350 mM NaCl, 0.5 mM EDTA, 10% glycerol, and 5 mM MgCl2. The proteins were subsequently eluted using 200 mM imidazole. The concentration of imidazole was decreased from 200 mM to 60 mM stepwise (step 1, 100 mM; step 2, 60 mM) using Slide-A-Lyzer dialysis cassettes (Pierce Biotechnology, Rockford, Ill.). Lowering the imidazole concentration also resulted in protein precipitation; therefore, concentrated protein solutions (2 to 5 mg ml−1) were maintained as frozen (−80°C) aliquots in 60 mM imidazole. Protein purity was assessed using capillary electrophoresis (Bioanalyzer; Agilent Technologies, Wilmington, DE). The protein concentration was determined using a BCA protein assay kit (Pierce Biotechnology). When necessary, proteins were subsequently purified using fast protein liquid chromatography. All proteins were purified to >95% purity.
Enzymatic assays.
Chemically synthesized c-di-GMP was a gift from M. Benziman (Hebrew University, Israel); other nucleotides were purchased from Sigma (St. Louis, MO). The optimized composition of the PDE assay buffer was 50 mM Tris-HCl (pH 9.35 at room temperature), 5 mM MgCl2, 0.5 mM EDTA, 50 mM NaCl, and the appropriate nucleotide substrate at a concentration of 100 μM. The reaction (final volume, 100 μl) was started by addition of the enzyme (final concentration, 0.08 μM unless otherwise indicated) and was allowed to proceed at 37°C for various times (usually 20 s to 2 min for c-di-GMP and up to 16 h for ATP, GTP, CTP, TTP, cyclic AMP [cAMP], or cGMP). The reaction was stopped by addition of CaCl2 (final concentration, 10 mM). The sample was immediately boiled for 3 min and subsequently centrifuged. The supernatant was then filtered through a 0.22-μm filter, and the reaction products were analyzed by reversed-phase high-performance liquid chromatography (HPLC).
The pH dependence and divalent metal cation dependence assays were performed in PDE assay buffer that had the composition described above except that the pH was different or the buffer contained divalent metal chlorides in place of MgCl2. The maximum velocity (Vmax) and equilibrium constant (Km) were calculated for each protein at pH 9.35 by measuring the rate of the reaction using concentrations of c-di-GMP ranging from 20 to 250 μM.
A biochemical analysis of l-di-GMP was performed with mung bean nuclease (New England Biolabs, Beverly, Mass.) and shrimp alkaline phosphatase (U.S. Biochemical Corp., Cleveland, Ohio) using buffers and conditions specified by the manufacturers.
HPLC.
Reaction samples (100 μl) were injected into a Supelcosil LC-18-T column (15 by 4.6 cm; Sigma-Aldridge, St. Louis, MO) and separated by reversed-phase HPLC (Summit HPLC system; Dionex, Sunnyvale, Calif.) using the buffer system and gradient program described previously (19). Products were detected at 254 nm.
Fast protein liquid chromatography.
Purification of proteins and determination of their oligomeric states were performed essentially as described previously (19) using a Superdex 200 10/300 GL gel filtration column (Amersham Biosciences, Piscataway, NJ) equilibrated with PDE assay buffer.
Mass spectrometry.
HPLC-purified l-di-GMP was concentrated by drying and diluted 1:10 in a solution containing 10 mg ml−1 α-cyano-4-hydroxycinnamic acid, 0.1% trifluoroacetic acid, and 50% acetonitrile, and 1 μl was spotted onto a matrix-assisted laser desorption ionization—time of flight plate. The sample was analyzed in the negative ion mode with a Voyager DE PRO mass spectrometer (Applied Biosystems, Foster City, CA). The resulting spectra were calibrated against bradykinin fragment 1-7 (molecular weight, 757.3997) and des-Arg-bradykinin (molecular weight, 904.4681) by close external calibration.
RESULTS AND DISCUSSION
E. coli YahA is a c-di-GMP-specific phosphodiesterase, PDE-A.
To directly test whether the EAL domain possesses PDE activity, we overexpressed, purified, and characterized in vitro the E. coli EAL domain-containing protein with an unknown function, YahA. This 362-amino-acid protein contains an EAL domain but lacks GGDEF. The EAL domain of YahA is linked to the N-terminal putative DNA-binding domain.
The full-length YahA protein was overexpressed as a YahA::His6 fusion and was purified to apparent homogeneity (Fig. 1). Its enzymatic activity was assayed using the cyclic nucleotides c-di-GMP, cAMP, and cGMP, as well as the four nucleotide triphosphates. YahA was found to possess c-di-GMP-specific PDE-A activity (Fig. 2). The rate of hydrolysis of nucleotide triphosphates was negligible; it was approximately 3 to 5 orders of magnitude lower than the rate of hydrolysis of c-di-GMP, whereas the rate of hydrolysis of cAMP or cGMP was below the detection limit (not shown). Upon prolonged incubation, slow hydrolysis of l-di-GMP into GMP (i.e., PDE-B activity) was observed (Fig. 2). The rate of this reaction was almost 3 orders of magnitude lower than that of the PDE-A reaction (not shown). This implies that the PDE-B activity of YahA is probably physiologically irrelevant.
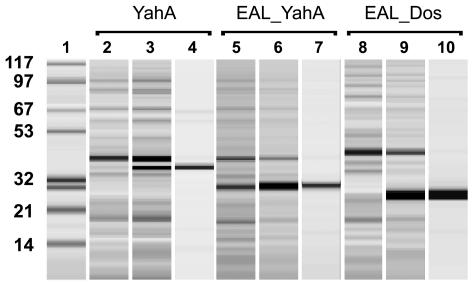
FIG. 1.
Overexpression and purification of the E. coli YahA protein, the EAL domain of YahA, and the EAL domain of E. coli Dos (protein chip, Bioanalyzer). Lane 1, molecular mass markers (molecular masses [in kDa] are indicated on the left); lanes 2, 5, and 8, crude extracts of E. coli DH5α cells prior to induction of expression of the fusion proteins; lanes 3, 6, and 9, crude extracts of E. coli cells after 2 h of induction with IPTG; lanes 4, 7, and 10, purified recombinant proteins.
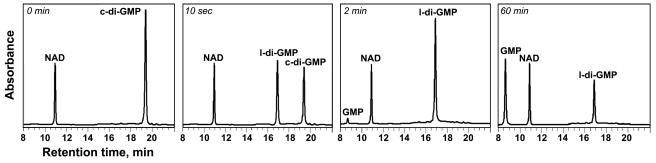
FIG. 2.
HPLC chromatograms showing the progression (0, 10 s, 2 min, 60 min) of c-di-GMP hydrolysis by YahA (final concentration of protein, 5 μM). For quantification purposes, the same amount of NAD was added to each sample prior to injection into the column.
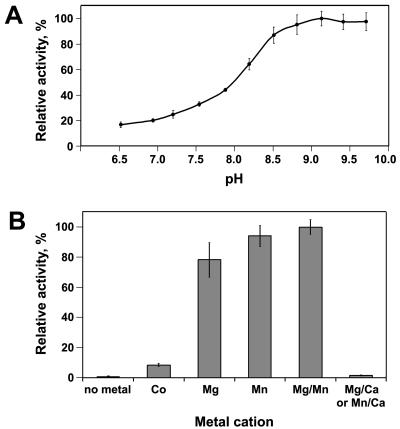
We investigated the pH dependence of the PDE-A activity of YahA and found a strong preference for alkaline conditions. The pH optimum was found to be 9.35. The enzymatic activity declined only slightly when the pH was increased to 10 (Fig. 3A). We also tested the dependence of the PDE-A activity on divalent metal cations (Fig. 3B). We found that this activity was absolutely dependent on either Mg2+ or Mn2+. The only other cation that supported some PDE-A activity was Co2+ (Fig. 3B), while Fe2+, Ni2+, and Ca2+ did not support activity (<1% relative activity [not shown]). Benziman and colleagues noticed that Ca2+ strongly and specifically inhibited PDE-A activity of the “washed membrane fractions” from G. xylinus that served as a source of PDE-A (17, 18). Consistent with this observation, we found that the PDE-A activity of YahA was strongly inhibited by Ca2+ (Fig. 3B). This property was subsequently used as a method of stopping PDE-A reactions.
FIG. 3.
(A) pH dependence of the PDE-A activity of YahA. (B) Dependence of the PDE-A activity of YahA on divalent metal cations. The PDE-A reaction buffer (pH 9.35) contained different divalent metal cations (final concentration, 5 mM) instead of Mg2+. The bar on the right shows inhibition of the PDE-A activity in the standard reaction buffer containing 5 mM (final concentration) MgCl2 or 5 mM (final concentration) MnCl2 by Ca2+ (5 mM [final concentration] CaCl2). The reaction with Co2+ was carried out in PDE buffer at pH 8.4 to avoid precipitation of Co2+ complexes at higher pH values. The error bars indicate the standard deviations calculated from at least three replicates.
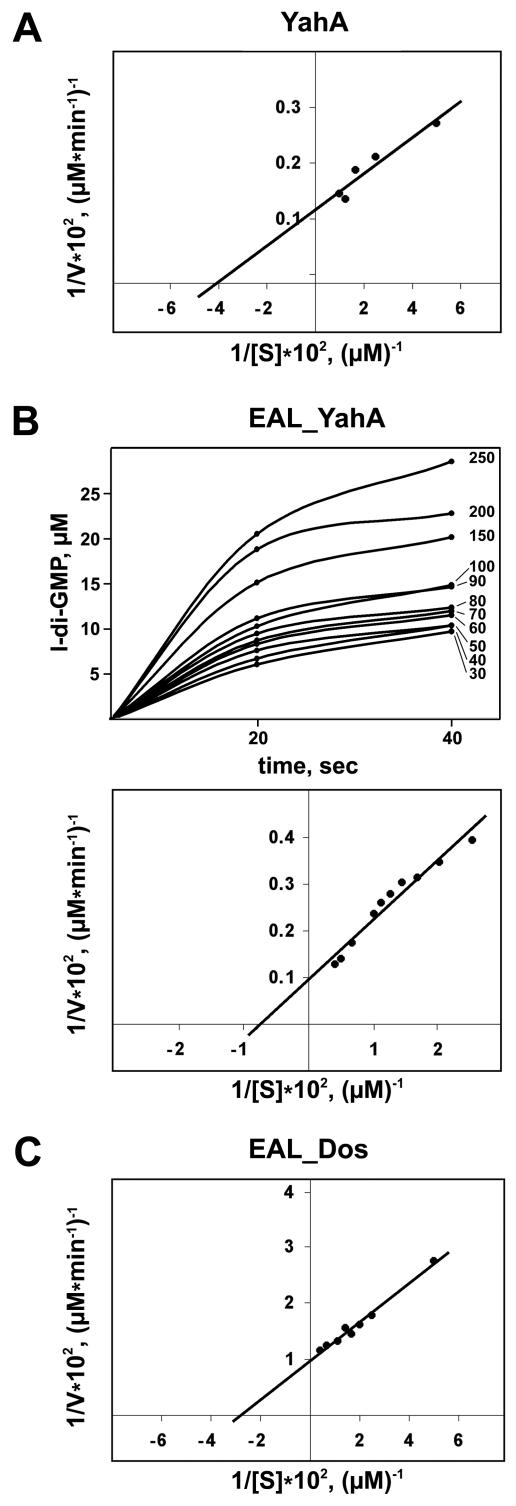
The Vmax and Km of the PDE-A reaction by YahA were calculated at the optimal pH using various c-di-GMP concentrations. The Vmax was found to be 750 mol l-di-GMP min−1 mol YahA−1, and the Km was 25 μM, suggesting that YahA is a very efficient c-di-GMP-specific PDE-A (Fig. 4A).
FIG. 4.
Lineweaver-Burk double-reciprocal plots for YahA (A), the EAL domain of YahA (B), and the EAL domain of Dos (C), showing the dependence of the initial reaction rate (V) on the substrate concentration (S). The initial rates of the reactions were calculated using the dependence of l-di-GMP formation on time. The plot showing this dependence for EALYahA is present in panel B for illustrative purposes; the initial c-di-GMP concentration (μM) for each curve is indicated on the right.
These results indicate that (i) YahA possesses c-di-GMP-specific PDE-A activity, (ii) PDE-A activity is dominant over PDE-B activity and the latter is probably catalyzed in vivo by a different enzyme(s), and (iii) substrate specificity is encoded within the EAL domain and GGDEF is not required.
Analysis of the product of PDE-A activity.
The product of c-di-GMP hydrolysis, l-di-GMP, observed by Benziman and colleagues using G. xylinus “washed membrane fractions” was identified as 5′pGpG (16, 17). To investigate whether the same product is generated by YahA::His6, we purified l-di-GMP by HPLC and subjected it to mass spectroscopy. l-di-GMP appeared as a peak at a molecular mass of 706.79 Da (data not shown). The calculated molecular mass of pGpG is 706.45 Da, while c-di-GMP, whose calculated molecular mass is 688.45 Da, appeared as a peak at 689.32 Da (19). The greater molecular mass of l-di-GMP than of c-di-GMP corresponds well to the addition of one H2O molecule upon c-di-GMP hydrolysis.
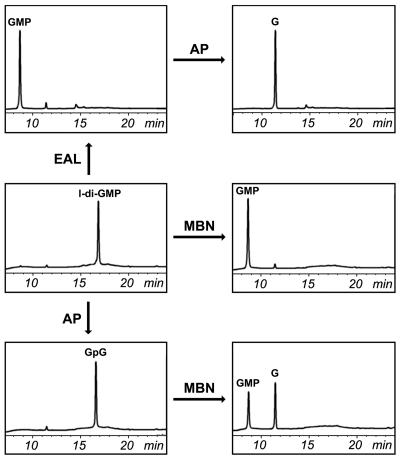
To investigate the structure of l-di-GMP, we subjected it to a series of enzymatic conversions, as summarized in Fig. 5. Treatment of l-di-GMP with mung bean nuclease resulted in GMP, as judged by its comigration with chemically synthesized GMP. To distinguish between 5′pGpG-OH3′ and 5′OH-GpGp3′, hydrolysis of c-di-GMP by YahA::His6 was allowed to proceed until full conversion into GMP. The latter compound was purified and treated with shrimp alkaline phosphatase, which specifically removes 5′ phosphates. A phosphate was removed, thus demonstrating that GMP, and hence l-di-GMP, had a 5′ phosphate. Treatment of l-di-GMP with shrimp alkaline phosphatase resulted in a product with a different mobility on HPLC, presumably GpG. Subsequent treatment of GpG with mung bean nuclease yielded a 1:1 mixture of GMP and guanosine. This mixture was converted to guanosine by treatment with shrimp alkaline phosphatase. Taken together, these data unambiguously show that the reaction product of PDE-A activity is 5′pGpG.
FIG. 5.
Biochemical analysis of l-di-GMP. G, guanosine; AP, shrimp alkaline phosphatase; MBN, mung bean nuclease.
EAL domain of YahA is sufficient for c-di-GMP-specific PDE-A activity.
Since we could not exclude the possibility that the N-terminal domain of YahA is somehow involved in its activity or in determination of substrate specificity, we tested the activity of the EAL domain from YahA. We cloned and overexpressed this domain as a C-terminal His6 fusion. The EALYahA::His6 fusion was purified to apparent homogeneity (Fig. 1), and its activity was tested by using the procedure used for the full-length protein. EALYahA::His6 retained PDE-A activity, as well as the substrate specificity of full-length YahA. Furthermore, the Vmax characteristic of EALYahA (1,000 mol l-di-GMP min−1 mol EALYahA−1) was similar to that of full-length YahA, while the Km (130 μM) was severalfold higher (Fig. 4B). The parameters for pH and metal dependence and inhibition were virtually identical to those determined for YahA (not shown). This suggests that the N-terminal domain is not required for the PDE-A activity per se. The role of this domain is currently being investigated.
Knowing that the GGDEF domain encoding DGC requires dimerization for activity (19), we investigated the oligomeric state of EALYahA::His6, as well as that of YahA::His6. As judged by the results of size exclusion chromatography, both proteins exist primarily as monomers (not shown). This suggests that di- or oligomerization is not required for PDE-A activity.
Therefore, we present here the first biochemical evidence that an individual EAL domain contains substrate specificity determinants and is sufficient to encode c-di-GMP-specific PDE-A activity.
Enzymatically active and inactive EAL proteins.
The sufficiency of the EAL domain for c-di-GMP-specific PDE-A activity along with sufficiency of the GGDEF domain for DGC activity shown by us previously (19) brought up the question of how the proteins containing GGDEF and EAL domains operate. The incompatibility of the GGDEF and EAL domain activities prompted us to explore the possibility that EAL domains exist in two forms, an enzymatically active form and an inactive form. This hypothesis was supported by the observation that three DGCs from G. xylinus, DgcA1 to DgcA3, which contain GGDEF and EAL domains, possess DGC activity but not PDE-A activity (22). Furthermore, the coexistence of DGC and PDE-A activities in a single protein containing both GGDEFand EAL domains has never been reported, nor is it supported by our unpublished data on the Rhodobacter sphaeroides proteins containing GGDEF and EAL domains.
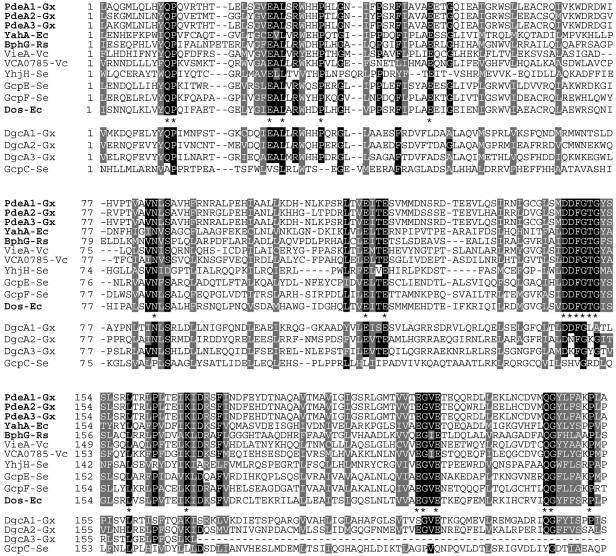
If the hypothesis that in any protein containing GGDEF and EAL domains one domain is enzymatically inactive were correct, could one distinguish an active EAL from an inactive EAL based on the primary sequence? To answer this question, we constructed and analyzed a multiple-sequence alignment of enzymatically active EAL domains, both verified in vitro and predicted (Fig. 6). Based on the PDE-A activity of the EAL domain of YahA, we assumed that EAL domains of PdeA1 to PdeA3 are also enzymatically active. This is in agreement with our observation (unpublished) that the EAL domain of the R. sphaeroides GGDEF- and EAL domain-containing protein BphG possesses PDE-A activity. We added the EAL domain of BphG to the alignment of the enzymatically active EAL domains. We also assumed that the EAL domains of S. enterica YhjH (21) and V. cholerae VieA (24), which were shown to decrease c-di-GMP levels when they were overexpressed, are active. The VCA0785 protein could replace VieA in a functional assay; therefore, it is likely to contain an enzymatically active EAL domain (24). The EAL domains of these proteins from four bacterial species were aligned (Fig. 6, upper panels). The alignment revealed several highly conserved regions that contained invariable residues. The EAL motif, which gave the name to this protein domain family, must be redefined as EXL, where X is a hydrophobic aliphatic residue. There appear to be more extended conserved motifs than EAL that are apparently essential for PDE-A activity (e.g., DDFGTG).
FIG. 6.
Two proposed classes of EAL domains, enzymatically active and inactive. The multiple-sequence alignment of the enzymatically active EAL domains (above the consensus line) was generated by using Clustal W (11) and was adjusted manually. Residues identical in >80% of the active EAL domains are indicated by a black background. Residues conserved in 100% of the active EAL domains are indicated in the consensus line by asterisks. Similar residues present in 100% of the active EAL domains are indicated by a grey background. The proposed enzymatically inactive EAL domains are below the consensus line. Proteins whose activities have been tested in vitro are indicated by boldface type. Ec, E. coli; Gx, G. xylinus; Rs, R. sphaeroides; Se, S. enterica; Vc, V. cholerae.
Following our hypothesis, we assumed that the EAL domains of G. xylinus DgcA1 to DgcA3 are inactive. When the EAL domains of DgcA1 to DgcA3 were aligned with the enzymatically active EAL domains, we found that they lacked several conserved residues present in the active EAL domains; e.g., each DgcA1 to DgcA3 protein contained at least one substitution in the most conserved DDFGTG motif, in addition to not containing other conserved residues present in the active EAL domains (Fig. 6). These deviations from the consensus may explain the enzymatic inactivity of the EAL domains of DgcA1 to DgcA3.
We extended our analysis to EAL domain-containing proteins whose activities can be predicted with less reliability than the activities discussed above. The activities of three GGDEF and EAL domain-containing proteins from S. enterica were predicted based on their effects on c-di-GMP-dependent biofilm formation (10). GcpC is likely to possess DGC activity, suggesting that its EAL domain is enzymatically inactive. GcpC was found to lack several invariable residues present in the enzymatically active EAL domains, which is consistent with its identification as an inactive protein (Fig. 6). GcpE is likely to possess PDE-A activity (10). In agreement with this, the EAL domain sequence of GcpE contains all invariable residues of active EAL domains (Fig. 6). The GcpF protein was predicted to possess DGC activity. However, the sequence of its EAL domain contains all of the invariable residues present in active EAL domains and almost all semiconserved residues (Fig. 6). At present, we do not know whether a mutation(s) in the nonconserved residue(s) inactivated the PDE-A activity of the EAL domain of GcpF or whether the prediction derived from phenotypic observations was inaccurate. These questions can be resolved only by additional experimentation. Importantly, however, GcpF is the only protein whose activity could not be assigned unambiguously. For the majority of EAL domain-containing proteins whose activities can be deduced from in vivo studies, our sequence-based predictions proved to be accurate.
We believe that this validates our hypothesis that there are enzymatically inactive EAL domains in bacterial proteins. The conserved residues and motifs identified in Fig. 6 may be useful for predicting the PDE-A activity of an EAL domain-containing protein. However, additional experimentation is needed to identify residues essential for PDE-A activity. Using the same approach, it must be possible to place GGDEF domains into enzymatically active (DGC) and inactive groups. Such an analysis would allow workers to predict activities of the GGDEF and EAL domain-containing proteins from genome sequences.
E. coli Dos is a c-di-GMP-specific PDE-A.
When the sequence of the EAL domain of the E. coli direct oxygen sensor Dos, which contains both GGDEF and EAL domains, was added to the EAL domain alignment, it clearly fell into the group of active EAL domains (Fig. 6). Dos has been reported to possess cAMP-specific PDE activity, which is regulated by oxygen through the heme-containing PAS domain (20). We failed to identify any obvious differences in the EAL domain sequence from Dos that separated it from the rest of the active EAL domains. The inability to gain insight into the different substrate specificity of Dos prompted us to test its substrate specificity.
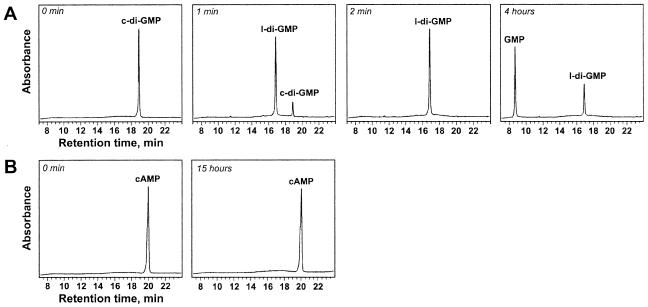
We cloned, overexpressed, and purified the EALDos::His6 fusion (Fig. 1) and assayed its substrate specificity. EALDos::His6 was found to be exclusively c-di-GMP specific (Fig. 7). The calculated constants Vmax and Km were found to be 100 mol l-di-GMP min−1 mol EALDos−1 and 36 μM, respectively (Fig. 4C). Similar to what was observed for YahA and its EAL domain, the EAL domain of Dos hydrolyzed 5′pGpG into GMP significantly more slowly; the rate was approximately 3% of the rate of c-di-GMP hydrolysis. This showed that E. coli Dos is a c-di-GMP-specific PDE-A. Thus, Dos, whose overall sequence similarity to G. xylinus PdeA1 to PdeA3 has long been noted (6), performs the same function as the G. xylinus proteins.
FIG. 7.
Substrate specificity of the EAL domain of Dos (final concentration, 0.2 μM). (A) c-di-GMP; (B) cAMP.
No cAMP or cGMP hydrolysis by EALDos::His6 was detected in our studies, even at high enzyme and/or substrate concentrations (Fig. 7B and data not shown). The rate of cAMP hydrolysis by full-length Dos reported by Sasakura et al. (20) is approximately 3 orders of magnitude lower than the rate of c-di-GMP hydrolysis observed here. Our inability to detect even low-level cAMP-dependent activity probably resulted from our use of cAMP instead of the fluorescent cAMP analog, 2′-O-anthraniloyl-cAMP, used by Sasakura et al., which may be a somewhat better substrate for Dos than cAMP.
The fact that the EAL domain of Dos possesses c-di-GMP-specific PDE-A activity validates our sequence-based prediction and suggests that c-di-GMP is the sole substrate of EAL domains.
Conclusions.
The following conclusions regarding the EAL domain were drawn from our study. (i) EAL domains are likely to be either enzymatically active or inactive. Inactive EAL domains are hypothesized to be present in DGCs containing both GGDEF and EAL domains. Several key residues identified in this work can help distinguish one group from another. (ii) Enzymatically active EAL domains possess c-di-GMP-specific PDE-A activity. c-di-GMP is the only substrate for this activity. (iii) c-di-GMP is hydrolyzed by the enzymatically active EAL domains into 5′pGpG. While subsequent hydrolysis to GMP does take place, because of its much lower rate it is likely to be irrelevant in vivo.
Acknowledgments
This work was supported by NSF grant MCB-0316270.
We are indebted to the late Moshe Benziman and to Haim Weinhoiz, Hebrew University of Israel, for chemically synthesized c-di-GMP, as well as for sharing assay protocols. We thank D. Gene Corson (supported by the University of Wyoming NSF EPSCoR Summer Research Scholarship for undergraduates) for participation in cloning of the yahA gene.
Footnotes
This paper is dedicated to the memory of Professor Moshe Benziman, Hebrew University, Israel, in whose laboratory cyclic diguanylate and enzymes involved in its synthesis and hydrolysis were discovered.
REFERENCES
- 1.Aldridge, P., R. Paul, P. Goymer, P. Rainey, and U. Jenal. 2003. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 47:1695-1708. [DOI] [PubMed] [Google Scholar]
- 2.Ausmees, N., R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. Lindberg. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol. Lett. 204:163-167. [DOI] [PubMed] [Google Scholar]
- 3.Boles, B. R., and L. McCarter. 2002. Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J. Bacteriol. 184:5946-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bomchil, N., P. Watnick, and R. Kolter. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J. Bacteriol. 185:1384-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, C., R. Paul, D. Samoray, N. C. Amiot, B. Giese, U. Jenal, and T. Schirmer. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. USA 101:17084-17089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, A. L., J. R. Tuckerman, G. Gonzalez, R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. A. Gilles-Gonzalez. 2001. Phosphodiesterase A1, a regulator of cellulose synthesis in Acetobacter xylinum, is a heme-based sensor. Biochemistry 40:3420-3426. [DOI] [PubMed] [Google Scholar]
- 7.D'Argenio, D. A., and S. I. Miller. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology 150:2497-2502. [DOI] [PubMed] [Google Scholar]
- 8.Galperin, M. Y. 2004. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6:552-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 10.Garcia, B., C. Latasa, C. Solano, F. Garcia-del Portillo, C. Gamazo, and I. Lasa. 2004. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol. Microbiol. 54:264-277. [DOI] [PubMed] [Google Scholar]
- 11.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 12.Jenal, U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7:185-191. [DOI] [PubMed] [Google Scholar]
- 13.Mayer, R., P. Ross, H. Weinhouse, D. Amikam, G. Volman, P. Ohana, R. D. Calhoon, H. C. Wong, A. W. Emerick, and M. Benziman. 1991. Polypeptide composition of bacterial cyclic diguanylic acid-dependent cellulose synthase and the occurrence of immunologically crossreacting proteins in higher plants. Proc. Natl. Acad. Sci. USA 88:5472-5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pei, J., and N. Grishin. 2001. GGDEF domain is homologous to adenylyl cyclase. Proteins 42:210-216. [DOI] [PubMed] [Google Scholar]
- 15a.Römling, U., M. Gomelsky, and M. Y. Galperin. C-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol., in press. [DOI] [PubMed]
- 16.Ross, P., H. Weinhouse, Y. Aloni, D. Michaeli, P. Weinberger-Ohana, R. Mayer, S. Braun, E. de Vroom, G. A. van der Marel, J. H. van Boom, and M. Benziman. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanilic acid. Nature 325:279-281. [DOI] [PubMed] [Google Scholar]
- 17.Ross, P., R. Mayer, H. Weinhouse, D. Amikam, Y. Huggirat, M. Benziman, E. de Vroom, A. Fidder, P. de Paus, L. A. Sliedregt, G. A. van der Marel, and J. H. van Boom. 1990. The cyclic diguanylic acid regulatory system of cellulose synthesis in Acetobacter xylinum. Chemical synthesis and biological activity of cyclic nucleotide dimer, trimer, and phosphothioate derivatives. J. Biol. Chem. 265:18933-18943. [PubMed] [Google Scholar]
- 18.Ross, P., Y. Aloni, H. Weinhouse, D. Michaeli, P. Weinberger-Ohana, R. Mayer, and M. Benziman. 1986. Control of cellulose synthesis in Acetobacter xylinum. A unique guanyl oligonucleotide is the immediate activator of the cellulose synthase. Carbohydr. Res. 149:101-117. [Google Scholar]
- 19.Ryjenkov, D. A., M. Tarutina, O. V. Moskvin, and M. Gomelsky. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasakura, Y., S. Hirata, S. Sugiyama, S. Suzuki, S. Taguchi, M. Watanabe, T. Matsui, I. Sagami, and T. Shimizu. 2002. Characterization of a direct oxygen sensor heme protein from Escherichia coli. Effects of the heme redox states and mutations at the heme-binding site on catalysis and structure. J. Biol. Chem. 277:23821-23827. [DOI] [PubMed] [Google Scholar]
- 21.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Römling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123-1134. [DOI] [PubMed] [Google Scholar]
- 22.Tal, R., H. C. Wong, R. Calhoon, D. Gelfand, A. L. Fear, G. Volman, R. Mayer, P. Ross, D. Amikam, H. Weinhouse, A. Cohen, S. Sapir, P. Ohana, and M. Benziman. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 180:4416-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas, C., C. R. Andersson, S. R. Canales, and S. S. Golden. 2004. PsfR, a factor that stimulates psbAI expression in the cyanobacterium Synechococcus elongatus PCC 7942. Microbiology 150:1031-1040. [DOI] [PubMed] [Google Scholar]
- 24.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]