Abstract
Conjugation is a major mechanism for disseminating genetic information in bacterial populations, but the signal that triggers it is poorly understood in gram-negative bacteria. F-plasmid-mediated conjugation requires TraM, a homotetramer, which binds cooperatively to three binding sites within the origin of transfer. Using in vitro assays, TraM has previously been shown to interact with the coupling protein TraD. Here we present evidence that F conjugation also requires TraM-TraD interactions in vivo. A three-plasmid system was used to select mutations in TraM that are defective for F conjugation but competent for tetramerization and cooperative DNA binding to the traM promoter region. One mutation, K99E, was particularly defective in conjugation and was further characterized by affinity chromatography and coimmunoprecipitation assays that suggested it was defective in interacting with TraD. A C-terminal deletion (S79*, where the asterisk represents a stop codon) and a missense mutation (F121S), which affects tetramerization, also reduced the affinity of TraM for TraD. We propose that the C-terminal region of TraM interacts with TraD, whereas its N-terminal domain is involved in DNA binding. This arrangement of functional domains could in part allow TraM to receive the mating signal generated by donor-recipient contact and transfer it to the relaxosome, thereby triggering DNA transfer.
Bacterial conjugation is a major mechanism for diversifying bacterial genomes and transmitting traits of medical and industrial importance. It is defined as the unidirectional transfer of single-stranded DNA from a donor to a recipient cell in response to an uncharacterized mating signal generated by mating pair formation during intimate cell-to-cell contact (43). This process requires the formation of a cytoplasmic protein-plasmid DNA complex called the relaxosome and a transmembrane multiprotein complex called the transferosome, which belongs to the type IV secretion system family (13, 24).
The F-plasmid is the paradigm for a large group of conjugative plasmids and integrative, conjugative elements (2) that carry genes important for human and veterinary medicine, such as those for antibiotic resistance and toxin production (20). F-plasmid traM encodes a 127-amino-acid protein essential for conjugative DNA transfer (13, 22). TraM forms tetramers and cooperatively binds to three sites (sbmA, -B, and -C) at the origin of transfer (oriT) in the F-plasmid (8, 12). Both tetramerization and DNA binding are essential for TraM function during F conjugation (28). TraM autoregulates its expression by binding to sbmA and -B, which overlap the two tandem traM promoters collectively called PtraM. Using in vitro assays, TraM has been shown to interact with TraD, an inner membrane component of the transferosome encoded by the F-plasmid (1, 9). TraM is essential neither for the nicking reaction that involves other components of the relaxosome nor for F-pilus assembly or mating-pair formation that involve most of the transferosome components (10, 11, 19, 22, 32). Therefore, TraM was proposed to transmit a signal between the cytoplasmic relaxosome and the transferosome during F conjugation (13, 43).
Similar to TraM, F-plasmid TraD is essential for conjugation but is not required for pilus synthesis or for the nicking reaction (10, 22, 33). TraD belongs to the “TraG family” of coupling proteins, including RP4 TraG, R388 TrwB, and VirD4 in the Agrobacterium tumefaciens T-DNA transfer system, which are proposed to couple the relaxosome to the DNA transport site during conjugation (4). TraG family proteins interact with both relaxosome and transferosome components, supporting this model (9, 14, 25, 39, 41). Coupling proteins might also serve as a pump to propel the single-stranded DNA through the mating pore (15, 24). TraD has two N-terminal membrane-spanning regions such that the large carboxyl-terminal region is in the cytoplasm (23). In the F-like R1 plasmid transfer system, the C-terminal 38 residues of TraD can bind to TraM (1), agreeing with genetic evidence that the C-terminal region of TraD defines its specificity for the F-like rather than the RP4-like conjugation systems (38).
Although TraM-TraD interaction is an intriguing finding (1, 9), no direct evidence has been presented that it is important for F-plasmid transfer. Similarly, the region of TraM involved in this interaction has not been defined. By screening for TraM mutants defective for F conjugation but competent for autoregulation, we isolated a C-terminal TraM mutant, K99E, which appears to have a decreased affinity for TraD but was competent for DNA binding and tetramerization, supporting a model whereby TraM links the relaxosome to the coupling protein, TraD.
MATERIALS AND METHODS
Growth media and bacterial strains.
Cells were grown in LB (Luria-Bertani) broth or on LB solid medium containing appropriate antibiotics or other supplements (37). Antibiotics were used at the following final concentrations: ampicillin (Amp), 50 μg/ml; kanamycin (Km), 25 μg/ml; spectinomycin (Spc), 100 μg/ml; nalidixic acid (Nal), 40 μg/ml; and chloramphenicol, 50 μg/ml. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactoside), glucose, and IPTG (isopropylthio-β-d-galactoside) were used at final concentrations of 100 μg/ml, 0.4% (wt/vol), and 1 mM, respectively. The following Escherichia coli strains were used: XK1200 [F− ΔlacU124 Δ(nadA gal attλ bio) gyrA (Nalr)] (31), ED24 (F− Lac− Spcr) (44), DH5α [ΔlacU169 (Φ80dlacZΔM15) supE44 hsdR17 recA1 endA1 gyrA96 (Nalr) thi-1 relA1] (17), and BL21(DE3) [F− dcm ompT hsdS (rB− mB−) gal λ(DE3)] (Stratagene).
DNA manipulation, PCR, DNA sequencing, and sequence analysis.
DNA purification, manipulation, and PCR(s) followed standard procedures (37) or protocols from the manufacturers. Vent DNA polymerase (New England BioLabs) was used for all PCRs except for random PCR mutagenesis that used Taq DNA polymerase. DNA sequencing was performed using the Amersham DYEnamic ET terminator cycle sequencing kit and an Applied Biosystems 373-S DNA sequencer with an XL upgrade. DNA and protein sequences were compiled and analyzed using Genetool and Peptool software.
Plasmids and plasmid construction.
All plasmids and oligonucleotides used in this work are listed in Table 1. The 0.7-kb EcoRI-HindIII fragments of the PCR products generated by random PCR mutagenesis of traM (see below) were ligated to a 2.5-kb EcoRI-HindIII fragment of pRFM200, resulting in pRFM200 derivatives named after the corresponding traM mutations. A 0.7-kb BstBI-KpnI fragment from the pRFM200 derivative containing mutation K99E was used to replace the 0.7-kb BstBI-KpnI fragment in pJLM102, resulting in pJLM105. The 2.5-kb EcoRI-BamHI fragment from pT7-7 was ligated to the 0.7-kb EcoRI-BamHI fragment of DNA amplified from pRFM200 or its derivatives containing mutations S79* (where the asterisk represents a stop codon), K99E, or F121S using primers JLU205 and JLU4, resulting in pJLM200, pJLM201, pJLM202, or pJLM203, respectively, which have a FLAG epitope tag fused to the N terminus of TraM. The 2.5-kb EcoRI-BamHI vector fragment from pT7-7 was ligated to the 2.2-kb EcoRI-BamHI fragment of DNA containing traD that was amplified from pNLK5 using JLU208 and JLU207 as primers, resulting in pJLHD001. A 2.2-kb EcoRI-HindIII fragment from pNLK5 was cloned into the EcoRI-HindIII sites of pBAD33, resulting in pJLD331.
TABLE 1.
Plasmids and oligonucleotides
| Plasmid or oligonucleotide | Description (source or reference) |
|---|---|
| pACPM24fs::lacZ | pACYC184 with a -1 frameshifted PtraM-traM24-lacZ fusion (26) |
| pBluescript KS+ | Cloning vector; high-copy-number, pMB1-derived replicon; Ampr (40) |
| pJLD331 | pBAD33 with F traD (this work) |
| pJLHD001 | pT7-7 with a His6-tagged traD (this work) |
| pJLM102 | pT7-4 with a DraI-BglII fragment containing F-plasmid PtraM and traM (28) |
| pJLM104 | pJLM102 derivative with a traM missense mutation, I109T (28) |
| pJLM105 | pJLM102 derivative with a traM missense mutation, K99E (this work) |
| pJLM200 | pT7-7 with FLAG-tagged traM (this work) |
| pJLM201 | pJLM200 derivative with a nonsense mutation, S79*, in traM (this work) |
| pJLM202 | pJLM200 derivative with a missense mutation, K99E, in traM (this work) |
| pJLM203 | pJLM200 derivative with a missense mutation, F121S, in traM (this work) |
| pNLK5 | pBAD18 with F traD (23) |
| pOX38-Km | Tra+ FinO−, transfer-derepressed F derivative, Kmr (6) |
| pOX38-MK3 | Tra+ FinO− TraM−, transfer-deficient F derivative, Kmr (35) |
| pRF940 | pBR322-derived pBend2 with sbmABC from the F-plasmid (12) |
| pRFM200 | pT7-5 with an F BstBI-BglII fragment from traM to PfinP (26) |
| pRFM200-Mdel | pRFM200 derivative with a deletion containing most of traM (26) |
| pT7-4, -5, and -7 | Cloning vectors; pMB1 replicons; Ampr (42) |
| JLU3 | 5′CTATAGGGAGACCGGAATTCG3′; including the EcoRI site (underlined) in pT7-5 |
| JLU4 | 5′CGATAAGCTTGGGCTGCAGG3′, including the HindIII site (underlined) in pT7-5 |
| JLU80 | 5′TAGGCGTATCACGAGGCCC3′, 5′ beginning at nucleotide 4328 in pBR322 |
| JLU81 | 5′GGTGCCTGACTGCGTTAGC3′, 5′ complementing nucleotide 64 in pBR322 |
| JLU205 | 5′TAGAATTCGCgactacaaagacgacgatgacaagGCTAAGGTGAACCTGTATATC3′; 5′ end starting 2 nucleotides upstream of the EcoRI site (underlined) in pT7-7 plus a FLAG tag (lowercase) and the 21 nucleotides after the start codon in F traM |
| JLU207 | 5′TGGGGATCCTGAGAATTGAAGACTGGAG3′, complementary to nucleotide 40 downstream of the 5′ end of F traD with 3 bases changed to produce a BamHI site (underlined) |
| JLU208 | 5′TAGAATTcaccatcaccatcaccatATGAGTTTTAACGCAAAGGATATG3′, 5′ end starting 2 nucleotides upstream of the EcoRI site (underlined) in pT7-7 plus a His6 tag (lowercase) and the first 24 nucleotides of F traD |
traM mutagenesis.
Random PCR mutagenesis was performed by using oligonucleotides JLU3 and JLU4 as primers to amplify traM under error-prone PCR conditions as previously described (26). The traM fragments generated by random PCR mutagenesis were cloned to form pRFM200 derivatives and transformed into DH5α cells carrying pOX38-MK3 and pACPM24fs::lacZ. The transformed cells were grown on LB plates with X-Gal, ampicillin, chloramphenicol, and kanamycin at 37°C for 24 h. Each light-blue colony was patched on a Km-Amp plate and a Km-Spc plate covered with fresh ED24 cells. The light-blue transformants that did not produce transconjugants on Km-Spc plates were selected for further characterization. Plasmid DNA was extracted and sequenced using primers JLU3 and JLU4 to locate mutations in traM.
β-Galactosidase assays.
A fresh, single colony was inoculated into LB broth containing appropriate antibiotics and grown at 37°C with shaking for 16 h. A 200-μl sample was used for determining β-galactosidase activity as described by Miller (30) and reported as Miller units. The values were calculated using the equation 1,000(A420/tvOD600), where t is the time of reaction (minutes), v is the volume of culture added (ml), and OD600 is the optical density at 600 nm.
Donor ability assays.
E. coli XK1200 and ED24 were used as donor and recipient strains, respectively. The mating experiments were performed as previously described (27). Donor ability was calculated as the number of transconjugants divided by the number of donors.
SDS-polyacrylamide gel electrophoresis and immunoblot analysis.
Exponentially growing cells (0.1 OD600 unit) or specified amounts of pure proteins were separated by a 15% sodium dodecyl sulfate (SDS)-polyacrylamide gel with a 7% stacking gel. TraM and TraD were assayed by immunoblot as described by Penfold et al. (35) using rabbit anti-TraM antiserum (8) and rabbit anti-TraD (34), respectively, at 1:10,000 dilutions.
Purification of TraM and its mutants.
The purification of TraM and its mutants used procedures including salting out (ammonium sulfate), cation exchange chromatography (MonoS HR 5/5; Amersham), and size exclusion chromatography (SEC) (Hiload 16/60 Superdex 75 prep grade; Amersham) as previously described (28).
Analytical SEC.
Purified TraM and K99E (5 μg) were brought to volumes of 1 ml with SEC buffer (50 mM sodium phosphate, 150 mM NaCl, pH 7.2) and loaded onto a Hiload 16/60 Superdex 75 prep grade column at 4°C with fast protein liquid chromatography. The column was eluted at 0.5 ml/min with 120 ml (1 column volume) of SEC buffer, and the eluate was collected in 2-ml fractions. Samples (10 μl) from different fractions were separated on a 15% SDS-polyacrylamide gel and analyzed by immunoblotting with anti-TraM antiserum. The column was calibrated with molecular weight markers (Sigma) under the same chromatographic conditions.
Electrophoretic mobility shift assays (EMSA).
DNA fragments containing sbmABC were amplified by PCR from pRF940 by use of primers JLU80 and JLU81. Purification and quantification of sbmABC and EMSA procedures were performed as previously described (28).
Overexpression and solubilization of His6-TraD.
BL21(DE3) cells containing pJLHD were grown in 500 ml of LB broth containing ampicillin at 37°C with vigorous shaking. After 3 h, IPTG was added to the culture to a final concentration of 1 mM, and the culture was grown for another 2 h before harvesting. His6-TraD was dissolved following previously described procedures (34). Approximately 250 OD600 units of cells was pelleted and suspended in 10 ml buffer A (50 mM Tris-HCl [pH 7.8], 0.4 mg/ml lysozyme [Sigma]) plus one tablet of Complete Mini protease inhibitor cocktail (Roche). The suspension was incubated at 37°C for 30 min until the suspension became viscous. DNase (50 units; Roche) and 150 μl of MgCl2 (1 M) were added to the suspension and incubated at 37°C for 15 min until the suspension was no longer viscous. The suspension was then lysed by sonication on ice for 3 min (30 seconds with a 30-second break, repeated six times) at maximum output. The unlysed cells were removed by a low-speed centrifugation (2,500 × g, 4°C, 15 min), and the membrane fraction was collected by a high-speed centrifugation (17,500 × g, 4°C, 30 min). The collected membrane fraction pellet was homogenized in 7.5 ml of buffer B (50 mM Tris-HCl [pH 7.8], 10 mM imidazole, 10% glycerol, 1.5% Triton X-100, 300 mM NaCl) plus one tablet of Complete Mini protease inhibitor cocktail. The suspension was incubated at 4°C for 5 h with gentle shaking and was centrifuged at medium speed (5,000 × g, 4°C, 15 min) to clear the undissolved membrane fraction. The supernatant, which contained solubilized TraD, was used immediately for affinity chromatography or stored at −80°C for later use.
Analysis of TraM-TraD interactions using affinity chromatography.
Solubilized TraD extract (1.5 ml) was mixed with 20 μl of 50% Ni-nitrilotriacetic acid (NTA) agarose resin (QIAGEN) gently at 4°C overnight. The resin was pelleted by centrifugation at 15,000 × g for 10 seconds and was washed two times with 100 μl of cold buffer C (50 mM Tris-HCl [pH 7.8], 20 mM imidazole, 1% Triton X-100, 300 mM NaCl). The washed resin was suspended in 1 ml of buffer D (50 mM Tris-HCl [pH 7.8], 10 mM imidazole, 1% Triton X-100, 150 mM NaCl) plus one-fifth of a tablet of Complete Mini protease inhibitor cocktail (Roche). Bovine serum albumin (BSA) (0.6 nmol) and 0.02 nmol of purified TraM (or one of its mutant proteins) were added to the suspension and mixed gently at 4°C for 5 h. The resin was pelleted by centrifugation at 15,000 × g for 10 seconds and washed with 100 μl of cold buffer C three times. The washed resin was eluted three times with 20 μl of elution buffer (50 mM Tris-HCl [pH 7.8], 250 mM imidazole, 1% Triton X-100, 300 mM NaCl). A 2-μl volume out of the pooled 60-μl eluate was run on a 15% SDS-polyacrylamide gel and assayed by immunoblotting with anti-TraM or anti-TraD antiserum. His6-tagged TraK (L. S. Frost and J. Manchak, unpublished data) was used as a negative control.
Analysis of TraM-TraD interactions using coimmunoprecipitation assays.
BL21(DE3) cells containing pJLD331 and a plasmid expressing FLAG-tagged TraM (or one of its mutant proteins) were grown in 10 ml of LB containing chloramphenicol and ampicillin at 37°C with vigorous shaking. After 2 h, arabinose was added to the culture to a final concentration of 0.1%, and the culture was grown for another 3 h before harvesting. Approximately 5 OD600 units of cells was pelleted and suspended in 1 ml of immunoprecipitation buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% Triton X-100, 5 mM EDTA, 10% glycerol) plus one-fifth of a tablet of Complete Mini protease inhibitor cocktail (QIAGEN). Lysozyme (10 μg) was added and mixed at 4°C for 30 min. The suspension was lysed by sonication on ice for 1 min (10 seconds with a 15-second break, repeated six times) at a medium output. The lysate was centrifuged at 15,000 × g and 4°C for 15 min, and the supernatant was transferred to a tube containing 20 μl of 50% anti-FLAG M2 agarose (Sigma). The mixture was incubated at 4°C for 3 h with gentle shaking. The resin was pelleted by centrifugation at 10,600 × g for 5 seconds and washed three times with 0.5 ml of TBS buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl). The washed resin was mixed with 100 μl of glycine-HCl (0.1 M; pH 3.5) and incubated for 5 min at room temperature. The mixture was centrifuged at 10,600 × g for 5 seconds, and the supernatant was transferred into a fresh tube containing 10 μl of 10× TBS buffer. Either 2 μl or 10 μl of supernatant was separated by 15% SDS-polyacrylamide gel and assayed by immunoblotting with anti-TraM or anti-TraD antiserum, respectively.
RESULTS
Selection of autoregulation-competent TraM mutants that are defective for F conjugation.
TraM-TraD interactions have been demonstrated using in vitro techniques with the C-terminal region of TraD implicated in this process (9, 38). Mutations in traM were sought that would demonstrate whether TraM-TraD interactions are important for conjugation as well as identify amino acids in TraM that are involved in these interactions. Since tetramerization and the ability to bind DNA are required for repression of PtraM by TraM (28), we used a three-plasmid system to select randomly generated traM mutations that affect F conjugation but not autoregulation. In this system, pRFM200 constitutively expresses TraM at a low level, whereas pACPM24fs::lacZ, carrying PtraM fused to lacZ, allows for the detection of autoregulation-defective TraM mutants as previously described (26). A third plasmid, pOX38-MK3 (a traM-deficient F derivative), was used in complementation assays to determine the function of TraM or its mutants during F conjugation.
DNA fragments generated by random PCR mutagenesis of traM were cloned into pT7-5 to give mutant derivatives of pRFM200 in cells containing pOX38-MK3 and pJLPM24fs::lacZ (see Materials and Methods). Approximately 1% of clones were deficient for autoregulation and have been characterized elsewhere (28). The remaining colonies formed light-blue colonies that synthesized TraM (or its mutants) capable of repressing PtraM in pJLPM24::lacZ. Approximately 6,000 of these colonies were patched onto solid medium coated with a layer of fresh ED24 recipient cells. Three of these colonies did not produce transconjugants, indicating that they were defective in conjugation but competent for autoregulation. Plasmid DNA from these three colonies was isolated, and sequence analysis found three missense mutations in traM, i.e., Q78H, K99E, and V106A (Table 2).
TABLE 2.
Properties of TraM and its mutantsa
| TraM or its mutant | Codon change | Expression of TraM or its mutantb | lacZ activity (Miller units)c | pOX38-MK3 complementation (T/D)d |
|---|---|---|---|---|
| TraM | None | + | 8.3 ± 0.3 | 5 × 10−1 |
| Q78H | CAA to CAU | + | 10.3 ± 1.6 | 1 × 10−3 |
| K99E | AAA to GAG | + | 8.1 ± 0.2 | <1 × 10−7 |
| V106A | GUU to GCU | + | 8.9 ± 0.7 | 5 × 10−5 |
| Mdel | traM deletion | − | 43.5 ± 1.7 | <1 × 10−7 |
TraM or its mutants were expressed by pRFM200 or its derivatives, respectively. Mutants are named after their codon changes.
Determined by immunoblot analysis of cells (0.1 OD600unit) containing pRFM200 or one of its mutant derivatives using anti-TraM antiserum. +; immunoblot bands with intensities comparable to that of cells (0.1 OD600 unit) containing pRFM200 were considered; −, no detectable bands were considered.
Determined by assaying the β-galactosidase activity of equivalent numbers of cells containing pOX38-MK3, pACPM24fs::lacZ, and pRFM200 or one of its derivatives.
Determined by assaying donor ability of cells containing pOX38-MK3, pACPM24fs::lacZ, and pRFM200 or one of its mutant derivatives. <1 × 10−7 means that there was no detectable donor ability.
Functional analysis of TraM mutants.
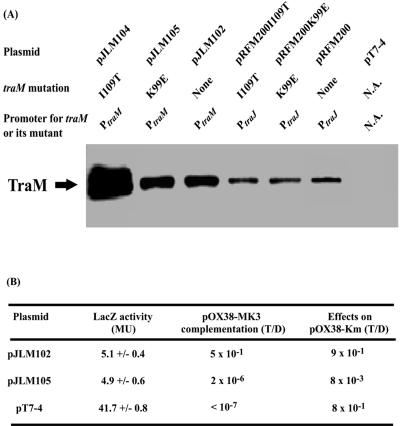
When coresident with pACPM24fs::lacZ, all three mutants repressed PtraM to levels similar to that of wild-type TraM, as determined by β-galactosidase assays (Table 2). Donor ability assays indicated that these mutants, especially K99E, did not complement pOX38-MK3 in transfer ability assays (Table 2). Immunoblot analysis with anti-TraM antiserum determined that TraM and the three mutants were expressed at similar intracellular levels, indicating that the decreased ability to complement pOX38-MK3 did not result from lowered protein stability (Fig. 1A and Table 2).
FIG. 1.
Characterization of the wild type and selected mutants of TraM. (A) Comparison of levels of TraM and its mutants by immunoblot analysis with anti-TraM antiserum when expressed from its native promoters (PtraM) or from an attenuated foreign promoter (PtraJ). N.A., not applicable. (B) Autorepression of PtraM was determined by assaying the β-galactosidase activity of cells containing pACPM24fs::lacZ and pJLM102 and its derivative pJLM105 (K99E). Complementation of pOX38-MK3 was determined by measuring the donor ability of cells containing pOX38-MK3 and pJLM102 or pJLM105. <10−7 means that there was no detectable donor ability. Negative dominance was assayed by measuring the donor ability of pOX38-Km and pJLM102 or pJLM105. pT7-4 was the vector control.
TraM K99E was then cloned downstream of PtraM to give pJLM105 to further determine its ability to autoregulate PtraM and support F conjugation. Under PtraM, pJLM105 expressed K99E at levels similar to that of wild-type TraM (pJLM102) but gave levels lower than that of I109T (pJLM104), an autoregulation-defective TraM mutant (Fig. 1A) (28). This indicated that K99E was able to autoregulate PtraM. pJLM105 complemented pOX38-MK3 at a very low level (2 × 10−6 transconjugants per donor [T/D]), which was slightly higher than those of pRFM200K99E (<10−7 T/D), probably due to greater K99E expression (Table 2 and Fig. 1A and B). When coresident with pOX38-Km (a wild-type F-plasmid derivative), pJLM105 decreased the transfer frequency of pOX38-Km by approximately 100-fold (Fig. 1B), suggesting negative dominance of K99E over wild-type TraM.
Characterization of the tetramerization and DNA-binding abilities of K99E.
To further confirm whether or not K99E was defective in tetramerization or DNA binding, analytical SEC and EMSA were used to compare K99E with the wild-type TraM.
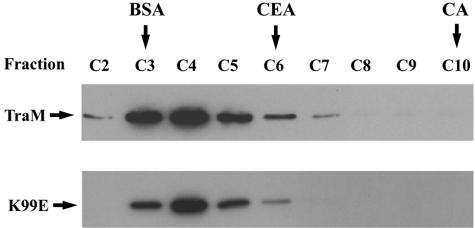
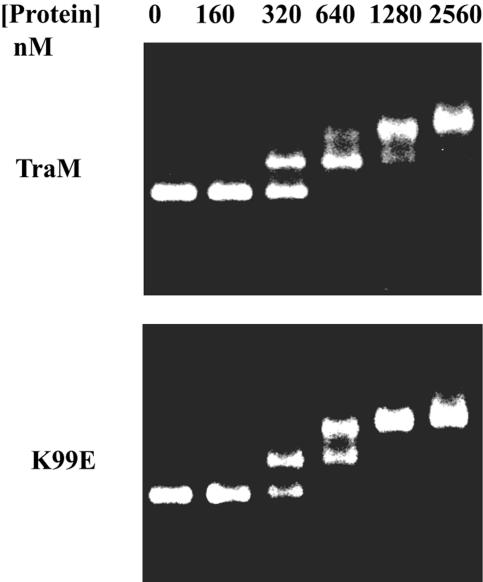
In analytical SEC, TraM and K99E gave a single peak at fraction C4 (Fig. 2), which corresponds to the size of TraM tetramers (28), indicating that K99E was a tetramer. EMSA showed that K99E bound to a DNA fragment containing sbmABC at concentrations similar to that of wild-type TraM, indicating that K99E had wild-type DNA-binding affinity (Fig. 3). Increasing amounts of both TraM and K99E shifted sbmABC sequentially to distinct positions which represent different affinities to sbmA, -B, and -C, a characteristic of wild-type TraM binding. Therefore, since the K99E mutation did not appear to affect tetramerization or DNA binding, it could have affected a different property of TraM, which is also required for F conjugation, with the most likely candidate being the ability to interact with TraD.
FIG. 2.
Analytical SEC fractions of purified TraM and K99E. Ten microliters from each fraction was separated by 15% SDS-polyacrylamide gels and visualized by immunoblot analysis with anti-TraM antiserum. Fraction numbers are indicated above each lane. The positions of different marker proteins are shown above the figure. BSA is bovine serum albumin monomer (66 kDa); CEA represents chicken egg albumin (45 kDa); CA represents carbonic anhydrase (29 kDa).
FIG. 3.
Binding of TraM or K99E to sbmABC as determined by EMSA. Increasing concentrations of TraM or K99E (in nM) are shown above the figures. Each reaction contained 40 nM of sbmABC, which contains all three TraM binding sites.
Affinity chromatography analysis of TraM-TraD interactions.
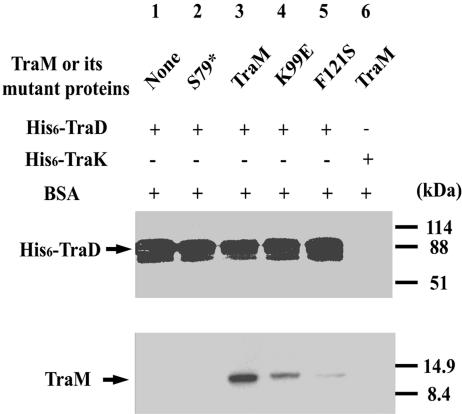
Affinity chromatography was used to determine whether K99E is defective in TraM-TraD interactions. All three mutations (Q78H, K99E, and V106A), which affected TraM function in conjugation but not in autoregulation, are located within the C-terminal half of TraM (Table 2). We also tested S79*, a truncated TraM mutant lacking the C-terminal residues 79 to 127, and F121S, a C-terminal TraM mutant that is defective in tetramerization (28).
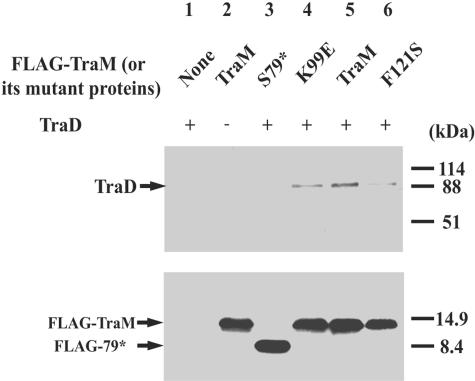
His6-tagged TraD was overproduced and bound to Ni-NTA agarose at saturating levels. Equivalent molar quantities of purified wild-type or mutant TraM were used in each assay (see Materials and Methods). After extensive washing, the bound His6-TraD as well as the proteins associated with TraD was eluted from the Ni-NTA agarose, using a buffer containing a high concentration of imidazole. More wild-type TraM was coeluted with TraD than with any of the mutants (Fig. 4, bottom), indicating that the mutations had affected TraM-TraD interaction. In a control experiment, no detectable TraM was coeluted with His6-tagged TraK (also a transferosome component), indicating the specificity of TraM for TraD. Whereas detectable amounts of K99E or F121S were coeluted with TraD, no S79* was detected, indicating that the deletion of the C-terminal region had a greater effect than did single-point mutations. Amino acid K99 appeared to affect the interaction with TraD directly, whereas F121S, which affects tetramerization, suggested that this property is also required for interacting with TraD.
FIG. 4.
His6-TraD and TraM (or its mutant proteins) interaction as determined by affinity chromatography. Equivalent amounts of purified TraM or selected mutants were incubated with BSA and His6-TraD (or His6-TraK)-saturated Ni-NTA agarose resin as indicated. Eluted protein was detected by immunoblotting using anti-TraD antiserum (top panel) or anti-TraM antiserum (bottom panel). Protein molecular weight markers are indicated to the right of the figure. Lane 1, His6-TraD and BSA; lane 2, mixture in lane 1 plus S79*; lane 3, mixture in lane 1 plus TraM; lane 4, mixture in lane 1 plus K99E; lane 5, mixture in lane 1 plus F121S; lane 6, His6-TraK, BSA, and TraM. +, present; −, not present.
Coimmunoprecipitation analysis of TraM and TraD.
To verify the in vitro results from affinity chromatography, coimmunoprecipitation assays were used to compare the affinity of TraD for TraM or its mutants in vivo. FLAG epitope tags were fused to the N terminus of TraM and its derivatives to ensure equal immunoreactivities among all the TraM variants (Table 1). TraD and FLAG-tagged TraM or its mutants were coexpressed in cells, and M2 anti-FLAG monoclonal antibodies were used to coprecipitate FLAG-TraM and TraD. TraD was coprecipitated from cells expressing both TraD and FLAG-TraM but not from cells expressing TraD alone, indicating that TraD was coprecipitated with FLAG-TraM (Fig. 5, top). Less TraD was coprecipitated with FLAG-K99E or FLAG-F121S, and no detectable TraD was coprecipitated with FLAG-S79*. These results confirmed that K99E, F121S, and S79* had reduced affinities for TraD compared to that of wild-type TraM.
FIG. 5.
TraD and TraM interactions assayed by coimmunoprecipitation. TraD and FLAG-tagged TraM (or one of its mutants) were coexpressed in E. coli cells. The complex of TraD and FLAG-tagged TraM was coprecipitated by anti-FLAG M2 agarose from the cell extract. The amounts of TraD and FLAG-tagged TraM (or one of its mutants) were determined by immunoblotting with anti-TraD antiserum (top panel) or anti-TraM antiserum (bottom panel). Protein molecular weight markers are indicated to the right of the figure. Lane 1, TraD; lane 2, FLAG-TraM; lane 3, TraD and FLAG-S79*; lane 4, TraD and FLAG-K99E; lane 5, TraD and FLAG-TraM; lane 6, TraD and FLAG-F121S. +, interaction; −, no interaction.
DISCUSSION
F-plasmid traM was randomly mutated using PCR mutagenesis to select for mutations that were competent for DNA binding and tetramerization but defective for conjugation. Three mutants were identified, with K99E being the most defective for F conjugation. Although other explanations for the inability of K99E to support conjugation are possible, the reduced affinity of K99E for TraD suggests that TraM-TraD interaction is required for F conjugation. K99E did not lose the ability to interact with TraD completely, and increasing K99E expression partially restored its function during F conjugation. This suggests that amino acid K99 might be only one of several key ligands for TraM interaction with TraD. Deletion of the C-terminal region (S79*) or disruption of oligomerization (F121S) was also deleterious for TraM-TraD interaction. Thus, TraM oligomerization, as well as direct contact with TraD, appears to be important for conjugation.
The C-terminal region of TraD corresponds to a large cytoplasmic domain projecting from the inner membrane (23). All known F-like TraD proteins have a C-terminal domain that determines plasmid specificity during mobilization (27). A deletion of this domain in F TraD increases the range of plasmids mobilized at the expense of a lowering of DNA transfer frequency (38). R388 TrwA, which has been suggested to perform a function similar to that of F TraM, interacts with TrwB, a homolog of F TraD that lacks the C-terminal domain and has a broader specificity for plasmid mobilization (25). Our unsuccessful attempts to use the bacterial two-hybrid system (21) to detect TraM-TraD interactions might be explained by the requirement for unencumbered C-terminal domains in both proteins (data not shown). Because this in vivo protein interaction system requires tagging one target protein at the N-terminal end and the other at the C-terminal end or vice versa, one of the tags could have prevented interactions between the two proteins. Recently, the region containing the C-terminal 38 residues of TraD has been shown to interact with TraM (1). TraM might also interact with other domains of TraD, since the deletion of this domain reduced but did not abolish TraD function during F conjugation (38). Although the C-terminal 38 amino acids of TraD can bind to TraM, the complete cytoplasmic fragment of TraD did have approximately 10-fold higher affinity for TraM, suggesting that other domains of TraD might be involved (1).
The F relaxosome is a double-stranded DNA (dsDNA)-protein complex, in which dsDNA appears to exist in an equilibrium between nicked and ligated states, with the relaxase (TraI) bound at the nic site (3, 45). Transfer of the nicked strand does not occur until after donor-recipient cell contact and the initiation of conjugative DNA synthesis, which is thought to require unwinding of the DNA (22). Although binding of the relaxase at oriT requires limited denaturation of the DNA around nic, the extent of the melted region does not appear to be enough to support helicase activity (16, 46). Unwinding the DNA is an activity of the helicase domain of TraI in the F-plasmid (29), which requires over 30 nucleotides of single-stranded DNA upstream of the 5′ end of the nic site in vivo (7). Therefore, a “mating signal” must be produced by donor-recipient cell contact, resulting in localized melting at oriT that initiates unwinding by TraI.
TraM-TraD interactions could be a key step in triggering DNA unwinding during F conjugation. TraM binds to plasmid DNA near the nic site through its N-terminal domain (28, 36), leaving the C-terminal domain to mediate specific interactions with the C-terminal cytoplasmic region of TraD. TraD is bound to the inner membrane through two transmembrane domains near the N terminus (23). Gilmour et al. (14) have shown that TraG (the coupling protein of plasmid R27; F TraD homolog) interacts with R27 TrhB (an inner membrane transferosome component; F TraB homolog), with F TraB interacting with the secretin-like TraK (18). Another homolog of F TraB, VirB10, has been shown to have TonB-like activity that could transmit a signal from the outer membrane to TraD in the inner membrane (5). Thus, a cascade of interactions between the transferosome and TraD, TraD and TraM, and TraM and the relaxosome could constitute at least part of the pathway for the unknown mating signal generated by donor-recipient contact. This implicates TraD as a protein essential for DNA unwinding, agreeing with previous studies suggesting that TraD is important for efficient conjugative DNA synthesis (22).
Single residue substitutions at different sites within the C-terminal region of TraM can cause major conformational changes in TraM, thereby changing its ability to bind DNA (28). Since TraD appears to interact with multiple amino acids at the C-terminal region of TraM, this interaction might cause a conformational change in TraM that affects its function. Because TraM binds to three sites upstream of nic and probably forms a nucleosome-like structure similar to that proposed for RP4 TraK (8, 12, 47), conformational changes in TraM might lead to localized DNA denaturation upstream of the nic site, resulting in the activation of the TraI helicase (7). Relaxosome-transferosome interactions might also involve interactions between TraD and TraI or plasmid DNA (25, 34), which could also trigger denaturation of DNA at oriT.
Although this model is still hypothetical, the nature of the bacterial mating signal is becoming clearer. As suggested by Willetts and Wilkins (43), TraM appears to be an essential component in this signaling pathway whereby successful mating pair formation triggers DNA synthesis and transfer during F conjugation.
Acknowledgments
This work is supported by a grant to L.S.F. from the Canadian Institutes for Health Research. J.L. was supported by a studentship from the Alberta Ingenuity Fund.
We thank Jan Manchak for technical assistance.
REFERENCES
- 1.Beranek, A., M. Zettl, K. Lorenzoni, A. Schauer, M. Manhart, and G. Koraimann. 2004. Thirty-eight C-terminal amino acids of the coupling protein TraD of the F-like conjugative resistance plasmid R1 are required and sufficient to confer binding to the substrate selector protein TraM. J. Bacteriol. 186:6999-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boltner, D., and A. M. Osborn. 2004. Structural comparison of the integrative and conjugative elements R391, pMERPH, R997, and SXT. Plasmid 51:12-23. [DOI] [PubMed] [Google Scholar]
- 3.Byrd, D. R., and S. W. Matson. 1997. Nicking by transesterification: the reaction catalysed by a relaxase. Mol. Microbiol. 25:1011-1022. [DOI] [PubMed] [Google Scholar]
- 4.Cabezon, E., J. I. Sastre, and F. de la Cruz. 1997. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol. Gen. Genet. 254:400-406. [DOI] [PubMed] [Google Scholar]
- 5.Cascales, E., and P. J. Christie. 2004. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc. Natl. Acad. Sci. USA 101:17228-17233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandler, M., and D. J. Galas. 1983. Cointegrate formation mediated by Tn9. II. Activity of IS1 is modulated by external DNA sequences. J. Mol. Biol. 170:61-91. [DOI] [PubMed] [Google Scholar]
- 7.Csitkovits, V. C., D. Dermic, and E. L. Zechner. 2004. Concomitant reconstitution of TraI-catalyzed DNA transesterase and DNA helicase activity in vitro. J. Biol. Chem. 279:45477-45484. [DOI] [PubMed] [Google Scholar]
- 8.Di Laurenzio, L., L. S. Frost, and W. Paranchych. 1992. The TraM protein of the conjugative plasmid F binds to the origin of transfer of the F and ColE1 plasmids. Mol. Microbiol. 6:2951-2959. [DOI] [PubMed] [Google Scholar]
- 9.Disque-Kochem, C., and B. Dreiseikelmann. 1997. The cytoplasmic DNA-binding protein TraM binds to the inner membrane protein TraD in vitro. J. Bacteriol. 179:6133-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett, R., and N. Willetts. 1980. Characterization of an in vivo system for nicking at the origin of conjugal DNA transfer of the sex factor F. J. Mol. Biol. 136:129-150. [DOI] [PubMed] [Google Scholar]
- 11.Fekete, R. A., and L. S. Frost. 2000. Mobilization of chimeric oriT plasmids by F and R100-1: role of relaxosome formation in defining plasmid specificity. J. Bacteriol. 182:4022-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fekete, R. A., and L. S. Frost. 2002. Characterizing the DNA contacts and cooperative binding of F plasmid TraM to its cognate sites at oriT. J. Biol. Chem. 277:16705-16711. [DOI] [PubMed] [Google Scholar]
- 13.Frost, L. S., K. Ippen-Ihler, and R. A. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilmour, M. W., J. E. Gunton, T. D. Lawley, and D. E. Taylor. 2003. Interaction between the IncHI1 plasmid R27 coupling protein and type IV secretion system: TraG associates with the coiled-coil mating pair formation protein TrhB. Mol. Microbiol. 49:105-116. [DOI] [PubMed] [Google Scholar]
- 15.Gomis-Ruth, F. X., G. Moncalian, R. Perez-Luque, A. Gonzalez, E. Cabezon, F. de la Cruz, and M. Coll. 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409:637-641. [DOI] [PubMed] [Google Scholar]
- 16.Guasch, A., M. Lucas, G. Moncalian, M. Cabezas, R. Perez-Luque, F. X. Gomis-Ruth, F. de la Cruz, and M. Coll. 2003. Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nat. Struct. Biol. 10:1002-1010. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 18.Harris, R. L., V. Hombs, and P. M. Silverman. 2001. Evidence that F-plasmid proteins TraV, TraK and TraB assemble into an envelope-spanning structure in Escherichia coli. Mol. Microbiol. 42:757-766. [DOI] [PubMed] [Google Scholar]
- 19.Howard, M. T., W. C. Nelson, and S. W. Matson. 1995. Stepwise assembly of a relaxosome at the F plasmid origin of transfer. J. Biol. Chem. 270:28381-28386. [PubMed] [Google Scholar]
- 20.Ippen-Ihler, K. A., and R. A. Skurray. 1993. Genetic organization of transfer-related determinants on the sex factor F and related plasmids, p. 23-52. In D. B. Clewell (ed.), Bacterial conjugation. Plenum Publishing Corp., New York, N.Y.
- 21.Karimova, G., J. Pidoux, A. Ullmann, and D. Ladant. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA 95:5752-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kingsman, A., and N. Willetts. 1978. The requirements for conjugal DNA synthesis in the donor strain during Flac transfer. J. Mol. Biol. 122:287-300. [DOI] [PubMed] [Google Scholar]
- 23.Lee, M. H., N. Kosuk, J. Bailey, B. Traxler, and C. Manoil. 1999. Analysis of F factor TraD membrane topology by use of gene fusions and trypsin-sensitive insertions. J. Bacteriol. 181:6108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Llosa, M., F. X. Gomis-Ruth, M. Coll, and F. de la Cruz. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 25.Llosa, M., S. Zunzunegui, and F. de la Cruz. 2003. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc. Natl. Acad. Sci. USA 100:10465-10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu, J., R. A. Fekete, and L. S. Frost. 2003. A rapid screen for functional mutants of TraM, an autoregulatory protein required for F conjugation. Mol. Genet. Genomics 269:227-233. [DOI] [PubMed] [Google Scholar]
- 27.Lu, J., J. Manchak, W. Klimke, C. Davidson, N. Firth, R. A. Skurray, and L. S. Frost. 2002. Analysis and characterization of the IncFV plasmid pED208 transfer region. Plasmid 48:24-37. [DOI] [PubMed] [Google Scholar]
- 28.Lu, J., W. Zhao, and L. S. Frost. 2004. Mutational analysis of TraM correlates oligomerization and DNA binding with autoregulation and conjugative DNA transfer. J. Biol. Chem. 279:55324-55333. [DOI] [PubMed] [Google Scholar]
- 29.Matson, S. W., J. K. Sampson, and D. R. Byrd. 2001. F plasmid conjugative DNA transfer: the TraI helicase activity is essential for DNA strand transfer. J. Biol. Chem. 276:2372-2379. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Moore, D., J. H. Wu, P. Kathir, C. M. Hamilton, and K. Ippen-Ihler. 1987. Analysis of transfer genes and gene products within the traB-traC region of the Escherichia coli fertility factor, F. J. Bacteriol. 169:3994-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson, W. C., M. T. Howard, J. A. Sherman, and S. W. Matson. 1995. The traY gene product and integration host factor stimulate Escherichia coli DNA helicase I-catalyzed nicking at the F plasmid oriT. J. Biol. Chem. 270:28374-28380. [PubMed] [Google Scholar]
- 33.Panicker, M. M., and E. G. Minkley, Jr. 1985. DNA transfer occurs during a cell surface contact stage of F sex factor-mediated bacterial conjugation. J. Bacteriol. 162:584-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panicker, M. M., and E. G. Minkley, Jr. 1992. Purification and properties of the F sex factor TraD protein, an inner membrane conjugal transfer protein. J. Biol. Chem. 267:12761-12766. [PubMed] [Google Scholar]
- 35.Penfold, S. S., J. Simon, and L. S. Frost. 1996. Regulation of the expression of the traM gene of the F sex factor of Escherichia coli. Mol. Microbiol. 20:549-558. [DOI] [PubMed] [Google Scholar]
- 36.Polzleitner, E., E. L. Zechner, W. Renner, R. Fratte, B. Jauk, G. Hogenauer, and G. Koraimann. 1997. TraM of plasmid R1 controls transfer gene expression as an integrated control element in a complex regulatory network. Mol. Microbiol. 25:495-507. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Sastre, J. I., E. Cabezon, and F. de la Cruz. 1998. The carboxyl terminus of protein TraD adds specificity and efficiency to F-plasmid conjugative transfer. J. Bacteriol. 180:6039-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schroder, G., S. Krause, E. L. Zechner, B. Traxler, H. J. Yeo, R. Lurz, G. Waksman, and E. Lanka. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184:2767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Short, J. M., J. M. Fernandez, J. A. Sorge, and W. D. Huse. 1988. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 16:7583-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szpirer, C. Y., M. Faelen, and M. Couturier. 2000. Interaction between the RP4 coupling protein TraG and the pBHR1 mobilization protein Mob. Mol. Microbiol. 37:1283-1292. [DOI] [PubMed] [Google Scholar]
- 42.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willetts, N., and B. Wilkins. 1984. Processing of plasmid DNA during bacterial conjugation. Microbiol. Rev. 48:24-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willetts, N. S., and D. J. Finnegan. 1970. Characteristics of E. coli K12 strains carrying both an F prime and an R factor. Genet. Res. 16:113-122. [DOI] [PubMed] [Google Scholar]
- 45.Zechner, E. L., H. Pruger, E. Grohmann, M. Espinosa, and G. Hogenauer. 1997. Specific cleavage of chromosomal and plasmid DNA strands in gram-positive and gram-negative bacteria can be detected with nucleotide resolution. Proc. Natl. Acad. Sci. USA 94:7435-7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, S., and R. J. Meyer. 1995. Localized denaturation of oriT DNA within relaxosomes of the broad-host-range plasmid R1162. Mol. Microbiol. 17:727-735. [DOI] [PubMed] [Google Scholar]
- 47.Ziegelin, G., W. Pansegrau, R. Lurz, and E. Lanka. 1992. TraK protein of conjugative plasmid RP4 forms a specialized nucleoprotein complex with the transfer origin. J. Biol. Chem. 267:17279-17286. [PubMed] [Google Scholar]