Abstract
Conjugative transfer of Enterococcus faecalis plasmid pCF10 is induced by the heptapeptide pheromone cCF10. cCF10 produced by plasmid-free recipient cells is detected by pCF10-containing donor cells, which respond by induction of plasmid-encoded transfer functions. The pCF10-encoded membrane protein PrgY is essential to prevent donor cells from responding to endogenously produced pheromone while maintaining the ability to respond to pheromone from an exogenous source; this function has not been identified in any nonenterococcal prokaryotic signaling system. PrgY specifically inhibited endogenous cCF10 and cPD1 (a pheromone that induces transfer of closely related plasmid pPD1) but not cAD1 (which is specific for less-related plasmid pAD1). Ectopic expression of PrgY in plasmid-free recipient cells reduced pheromone activity in culture supernatants and reduced the ability of these cells to acquire pCF10 by conjugation but did not have any effect on the interaction of these cells with exogenously supplied cCF10. The cloned prgY gene could complement a pCF10 prgY null mutation, and complementation was used to identify point mutations impairing PrgY function. Such mutations also abolished the inhibitory effect of PrgY expression in recipients on pheromone production and on acquisition of pCF10. Most randomly generated point mutations identified in the genetic screen mapped to a predicted extracellular domain in the N terminus of PrgY that is conserved in a newly identified family of related proteins from disparate species including Borrelia burgdorferi, Archaeoglobus fulgidus, Arabidopsis thaliana, and Homo sapiens. The combined genetic and physiological data suggest that PrgY may sequester or inactivate cCF10 as it is released from the membrane.
Expression of genes required for conjugative horizontal transfer of certain Enterococcus faecalis plasmids is activated through cell-cell signaling by peptide sex pheromones (10). This form of intercellular communication differs significantly from many quorum-sensing systems in both gram-positive and gram-negative species. In quorum sensing, all members of a microbial population participate in both production of the extracellular signal and response to the signal, such that expression of the biological behavior controlled by the quorum-sensing system is dependent on population density. In such systems, the biosynthetic machinery for producing the signal and the sensing machinery for signal detection are typically encoded by the same genetic unit (normally the chromosome), and often both sets of genes are linked. In contrast, the enterococcal pheromone systems utilize plasmid-encoded signal-sensing machinery that is genetically distinct from the chromosomally encoded signal molecule. Therefore, communication occurs not within a homogeneous population but between two distinct cell types, the plasmid-free recipient cell (signal producer) and the plasmid-containing donor cell (responder). The pheromone plasmids have evolved unique mechanisms, not found in other cell-cell signaling systems, to avoid induction of plasmid transfer by endogenous pheromone while retaining the ability to detect and respond to the same molecule from an exogenous source. This allows these plasmids to spread efficiently without unnecessarily extracting a metabolic burden from their host cells unless potential recipients are in close proximity.
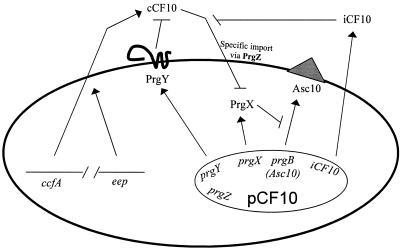
This paper describes the results of experiments designed to examine the role of the pheromone plasmid-encoded membrane protein PrgY in blocking self-induction of the conjugal transfer genes of tetracycline resistance plasmid pCF10 (15) by endogenous pheromone in donor cells. Plasmid pCF10 confers a response to the heptapeptide (LVFLVTV) pheromone cCF10. When a cell carrying pCF10 encounters cCF10 in the growth medium, the concerted activity of the pCF10-encoded pheromone binding protein PrgZ and the chromosomal oligopeptide permease (Opp) system results in import of cCF10 (26). Recent data indicate that interaction of internalized cCF10 with the cytoplasmic repressor protein PrgX disrupts a negative control circuit that ultimately leads to expression of conjugation functions (7, 24). The most obvious phenotypic change resulting from pheromone induction is visible aggregation of induced cells in liquid culture. Aggregation is mediated by a pCF10-encoded surface protein called aggregation substance or Asc10 (23); Asc10 binds to the cell wall of all E. faecalis cells, allowing formation of effective mating pairs between donors and recipients. The structural gene for Asc10, prgB, is subject to very strong regulation by cCF10 such that it is completely off at both the mRNA and protein levels in uninduced donor cells and strongly activated after pheromone induction. Thus, either Asc10-mediated clumping responses or reporter gene fusions to prgB are often used to monitor induction status. The key regulatory circuits controlling Asc10 expression are illustrated in Fig. 1.
FIG. 1.
Model of pCF10 pheromone production, control, and response. ccfA, encoding the pheromone cCF10, and Eep are both expressed from the chromosome. Mature cCF10 is processed from the signal sequence of the lipoprotein CcfA (5) by the membrane protease Eep (4) as it exits the cell. PrgY, PrgX, PrgZ, Asc10 (aggregation substance, expressed from prgB), and iCF10 are all encoded within pCF10. In the absence of PrgY or the inhibitor peptide iCF10, endogenous mature cCF10 can reinternalize via PrgZ into donor cells to continually induce pCF10 transfer proteins. PrgY and the inhibitor peptide iCF10 prevent this self-induction by endogenous cCF10. iCF10 neutralizes endogenous cCF10 in the medium, and PrgY sequesters or blocks the activity of endogenous cCF10. PrgX functions as the on-off switch for induction and upon interaction with pheromone releases repression of Asc10 expression (7, 23). Asc10 mediates aggregation of the cells, which is necessary for efficient transfer of pCF10 to recipient cells.
We recently determined that cCF10 is produced by posttranslational proteolytic processing of the signal peptide of a putative secreted lipoprotein of unknown function encoded by the ccfA gene (5). All enterococcal sex pheromones studied thus far are seven- or eight-amino-acid hydrophobic peptides that are produced by processing of lipoprotein signal sequences (3, 5, 20). An et al. have identified a putative membrane protease called Eep, which seems to play an important role in the processing of several pheromones (4). A number of different families of pheromone plasmids, each encoding a response to specific pheromone, have been identified, with pCF10, pAD1, and pPD1 being the most heavily studied. Most strains of E. faecalis produce at least six active pheromones from their chromosome that each activate a specific cognate plasmid (12). The systems are all highly specific, such that in cells carrying more than one of these plasmids, there is no cross-induction of transfer of a noncognate plasmid by any pheromone (16).
When pCF10 transfers into a new host strain, this recipient cell becomes a potential donor and expresses conjugation functions only when exposed to exogenous pheromone. Genetic studies have shown that two pCF10 gene products are involved in preventing self-induction by endogenous pheromone. The heptapeptide iCF10 (AITLIFI) encoded by the prgQ locus is secreted into the extracellular medium at concentrations that are just sufficient to neutralize the cCF10 secreted into the medium by the same cell (30, 31). This balance between the two peptides can be overcome by addition of exogenous cCF10 at less than five molecules per responder cell (29). In addition to iCF10, expression of a membrane protein, PrgY, is also required to block spontaneous self-induction. Our previous results indicated that plasmid-free recipient cells contain substantial quantities of cCF10 in the cell wall fraction, in addition to the pheromone released into the growth medium (9). Comparative analysis of pCF10 and a pCF10 derivative, pCF389 (containing a Tn917 insertion in prgY), suggested that PrgY might block an autocrine induction loop in donor cells by somehow sequestering or blocking the activity of cell-associated endogenous cCF10 (9). These studies did not yield any insights into the molecular mechanism of PrgY action, and initial attempts to carry out complementation analysis and biochemical studies were unsuccessful because of problems in cloning and expression of prgY. The pAD1 and pPD1 plasmids encode prgY homologs that have been designated traB (2, 32). The cumulative results from several groups indicated that this family of related proteins probably has a common function, but there was some evidence to suggest that the TraB proteins negatively regulated production of pheromone excreted into the growth medium while PrgY had a larger effect on cell wall-associated pheromone.
In this report, we describe the use of the cloned prgY gene in functional complementation and mutational analyses of several E. faecalis strains that produce different levels of cCF10. We also report the results of attempts to carry out complementation of a prgY null mutation with the traB genes and vice versa. These data suggest that PrgY and the TraB proteins act similarly. There is functional conservation between prgY and traB of pPD1, but not between either of these genes and traB of pAD1. In addition, we describe a previously unreported family of PrgY-like proteins in species representing all biological kingdoms. PrgY expression in recipient cells reduces conjugative transfer into these cells, and point mutations that abolish control of self-induction of donor cells also abolish the inhibitory effect of ectopic PrgY expression in recipients on conjugative transfer. However, PrgY expression in recipients does not affect binding or uptake of exogenous cCF10. An N-terminal conserved segment predicted to lie outside of the cell membrane plays a key role in PrgY function. The data suggest that this protein functions by sequestering or degrading pheromone or propheromone as it is exported from the membrane.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. faecalis was grown at 37°C in Todd-Hewitt broth (THB; Difco) or M9-YE glucose medium (Difco) supplemented with antibiotics at the following concentrations: tetracycline, 10 μg/ml; erythromycin, 10 μg/ml; spectinomycin, 1,000 μg/ml; kanamycin, 1,000 μg/ml; rifampin, 50 μg/ml. Nisin (Sigma) was used at 25 ng/ml in broth and 50 ng/ml in agar plates unless otherwise stated. Physiological experiments in this study were conducted with E. faecalis strain OG1RF or isogenic, protease-deficient strain TX5128 (32). Escherichia coli was grown at 37°C with shaking in Luria-Bertani medium at the following antibiotic concentrations: erythromycin, 200 μg/ml, spectinomycin, 50 μg/ml.
DNA manipulation.
Plasmids were isolated with the QIAGEN mini kit as recommended by the manufacturer. Digested DNA products were purified using a QIAquick gel extraction kit (QIAGEN), and PCR products were purified using a QIAquick PCR purification kit (QIAGEN). Plasmid constructs were verified using restriction enzyme digestion. Restriction enzymes were purchased from Promega and New England BioLabs. PCR was performed with a Perkin-Elmer Gene Amp PCR system or an Eppendorf Mastercycler using either BioXact DNA polymerase (Bioline) or Platinum Pfx polymerase (Invitrogen). All sequencing was done by the Advanced Genetic Analysis Center at the University of Minnesota. Primers were purchased from Integrated DNA Technologies, Inc. E. faecalis was transformed with DNA as previously described (6). For all plasmids constructed using a PCR-generated fragment, the DNA was sequenced to confirm that no mutation was incorporated by PCR.
Plasmids.
The plasmids used in this study are listed in Table 1. pMSP3545S was constructed by cloning the PCR-amplified spectinomycin resistance gene from pFW11 (33) with flanking XhoI and XbaI restriction sites into pMSP3545 (8). The prgY gene from pCF10 was PCR amplified with flanking XbaI and NcoI restriction sites and cloned upstream of the spectinomycin resistance gene in pMSP3545S, resulting in a translational fusion with the nisin promoter. The pAD1 and pPD1 traB genes were similarly PCR amplified and cloned into pMSP3545S to construct the pMSP3545S, pAD1, and pPD1 traB plasmids. Plasmid pMSP6043-1 was made by ligating the EcoRI fragment from PrgN through the middle of PrgY from pINY6023 (34) into the EcoRI site of pDL276 (19). This was checked by restriction digestion and Western blotting and was found to express PrgZ but no detectable PrgY (data not shown).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Reference | Description |
|---|---|---|
| E. faecalis strains | ||
| OG1RF | 17 | GelE+ SprE+; rifampin and fusidic acid resistant |
| OG1SSp | 17 | Streptomycin and spectinomycin resistant |
| FA2-2 | 13 | GelE− SprE− rifampin and fusidic acid resistant |
| OG1X | 22 | GelE− SprE− streptomycin resistant |
| TX5128 | 35 | GelE− SprE−; mγδ gelE insertion in OG1RF |
| Plasmids | ||
| pCF10 | 15 | Pheromone-inducible plasmid |
| pCF389 | 11 | pCF10 with Tn917 insertion in prgY after amino acid 310 of 384 |
| pMSP3545 | 8 | Nisin-inducible cloning shuttle vector |
| pMSP3545S | This work | Spectinomycin resistance gene from pFW11 cloned into XbaI XhoI sites of pMSP3545 |
| pDL276 | 19 | Gram-positive cloning shuttle vector |
| pMSP6043 | 21 | prgN,-O,-P,-W,-Z, and-Y in pDL276 |
| pMSP6049 | 21 | prgN,-O,-P,-W, and-Y in pDL276 |
| pMSP6043-1 | This work | prgN,-O,-P,-W, and-Z in pDL276 |
| pMSP3545Sa | ||
| pMSP3545S-1 | This work | Cloned E. faecalis/pCF10 prgY |
| pMSP3545S-2 | This work | Cloned E. faecalis/pPD1 traB |
| pMSP3545S-3 | This work | Cloned E. faecalis/pAD1 traB |
| pMSP3545S-4 | This work | Cloned Borrelia burgdorferi traB |
| Random mutations in pMSP3545S-1 | ||
| pMSP3545S-1 H21R | This work | A → G change at base 61 |
| pMSP3545S-1 E45L | This work | G → A change at base 133 |
| pMSP3545S-1 W46R | This work | T → C change at base 135 and silent T → C change at base 789 |
| pMSP3545S-1 R50Q | This work | G → A change at base 149 |
| pMSP3545S-1 E101K | This work | G → A change at base 301 |
| pMSP3545S-1 L142P | This work | T → C change at base 425 |
| pMSP3545S-1 G367R | This work | G → A change at base 1102 |
| pMSP3545S-1 G367E | This work | G → A change at base 1103 |
| Engineered mutations in pMSP3545S-1 | ||
| pMSP3545S-1 G18A | This work | G → C change at base 53 |
| pMSP3545S-1 S23A | This work | T → G change at base 67 |
| pMSP3545S-1 D120A | This work | A → C change at base 359 |
| pMSP3545S-1 R121A | This work | AG → GC change at bases 361 and 362, respectively |
| pMSP3545S-1 T126A | This work | A → G change at base 376 |
| pMSP3545S-1 R129A | This work | CG → GC change at bases 385 and 386, respectively |
| pMSP3545S-1 R190A | This work | CG → GC change at bases 568 and 569, respectively |
Genes were cloned into pMSP3545S using the NcoI and XbaI sites.
Mutagenesis and screening.
Random mutations in prgY were generated by PCR amplification of prgY in pMSP3545S-1, including the flanking XbaI and NcoI sites, using Bio-X-Act polymerase. This PCR product was purified, digested with XbaI and NcoI, and ligated into pMSP3545S cut with XbaI and NcoI. This ligation was transformed into E. coli, and the resulting colonies were scraped, pooled, and harvested for plasmid. This heterogeneous plasmid pool was transformed into E. faecalis(pCF389) and plated on THB agar containing tetracycline, spectinomycin, and 50 ng nisin/ml. Single colonies were then screened visually on the plates for a crumbly dry texture that corresponded to a clumpy phenotype in broth. Dry colonies were transferred onto another plate and inoculated into individual wells of a 96-well plate containing THB with tetracycline, spectinomycin, and nisin and grown with shaking overnight to confirm the phenotype. Wells with clumpy cultures were then screened for production of PrgY by Western blotting, and those that were positive for PrgY expression were streaked for isolation and rechecked for both loss of function and protein expression. Mutant derivatives of plasmid pMSP3545S-1 were subsequently sequenced directly from E. faecalis plasmid DNA amplified by PCR using Pfx polymerase. These plasmids were then transformed back into E. faecalis OG1RF(pCF389) to reconfirm the loss of function and positive protein expression. Site-directed prgY mutants were made in pMSP3545S-1 using the GeneTailor site-directed mutagenesis system (Invitrogen) as recommended by the manufacturer. Potential clones were sequenced directly to screen for the desired single change in the prgY gene, and G18A, S23A, R121A, R129A, and R190A were subcloned into cut pMSP3545S using XbaI and NcoI.
Determination of pheromone activity in cell culture supernatants.
Strains were grown at 37°C without shaking in THB overnight and then diluted 1:10 into fresh medium plus antibiotics and 25 ng nisin/ml and grown an additional 4 to 6 h. The optical density at 600 nm was determined for each sample, and equivalent cell volumes were harvested by centrifugation at 8,000 rpm (Beckman J2-21 centrifuge, JA20 rotor) for 10 min. The supernatant from each sample was collected and autoclaved for 15 min at 121°C and 15 lb/in2 and then precipitated with 5% trichloroacetic acid, neutralized with NaOH, and resuspended in the same final volume of THB (approximately 0.4 ml). These were subjected to a clumping assay (9) to detect pheromone activity. Briefly, twofold serial dilutions of precipitated supernatant samples were made in M9-YE medium in a round-bottom well microtiter plate. Ten microliters of a 15-h OG1RF(pCF10) indicator culture was added to each well. Samples were incubated with shaking at 37°C for 2 h, and positive clumping reactions were scored. The titer is reported as the reciprocal of the highest dilution which showed a positive clumping reaction. Results reported are representative of at least two independent experiments done on separate days.
Plasmid transfer.
Overnight cultures (15 h) grown at 37°C in THB plus the proper antibiotics were diluted 1:10 in fresh medium plus 25 ng nisin/ml and/or 10 ng/ml cCF10 where stated. These were incubated for 1 h at 37°C and then combined in a 1:9 ratio of donors to recipients and incubated for an additional 30 min or the time indicated before plating. Transconjugants were enumerated by serial dilution of THB agar with rifampin, tetracycline, and kanamycin (for pDL276-derived constructs) or rifampin, tetracycline, and erythromycin (for pMSP3545-derived constructs). Measurements reported represent means from duplicate assays, and error bars represent 1 standard deviation from the mean. Assays were repeated at least two times in duplicate; representative results from one experiment, performed in duplicate, are presented.
Assay of pheromone binding.
To assay for pheromone binding, a synthetic pheromone preparation was diluted in M9-YE medium to give a titer in a clumping-inducing microtiter plate assay (18) of 64, as determined experimentally. A volume of 100 μl of this diluted material was mixed with an equal volume of medium containing 6 × 108 exponentially growing E. faecalis cells per ml to be tested for pheromone binding. After a 15-min incubation at 37°C, the bacteria were removed by centrifugation in an Eppendorf Microfuge at 13,000 rpm. The pheromone activity remaining in the supernatant was assayed with a standard microtiter plate clumping assay (18) and compared with that in control preparations diluted and incubated in parallel in the absence of bacteria.
Protein preparation, SDS-PAGE, and immunoblotting.
To prepare PrgY protein from whole cells for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and immunoblotting, cells were grown overnight in THB (for initial screening of random PCR mutants) or M9-YE (for all others) stationary at 37°C, diluted 1:5 into fresh THB with 25 ng nisin/ml, and then grown for 2.5 h. An equivalent amount of cells was collected by centrifugation and treated with lysozyme solution (10 mg/ml lysozyme, 10 mM Tris-HCl [pH 8.0], 50 mM NaCl, 10 mM EDTA) at 37°C for 15 min. The resulting protoplasts were resuspended in sample buffer for SDS-PAGE (31.3 mM Tris [pH 6.8], 5% glycerol, 0.125% bromphenol blue, 5% β-mercaptoethanol, 4% SDS), boiled for 5 min, and separated on a denaturing 10% polyacrylamide gel and transferred onto a nitrocellulose membrane. PrgY was detected with a polyclonal antibody (8) at a dilution of 1:1,000. Detection was performed with the enhanced-chemiluminescence protocol (Pierce). To prepare Asc10 protein for Western analysis of whole-cell extracts spotted onto a membrane, cells were grown stationary at 37°C overnight, diluted 1:10 into fresh THB plus the indicated concentration of nisin and cCF10 (where applicable), and grown for 6 h at 37°C. An equivalent amount of cells was collected by centrifugation, washed twice with phosphate-buffered saline, and resuspended in 200 μl phosphate-buffered saline plus 1% SDS. These samples were boiled for 5 min and spotted onto a nitrocellulose membrane at a 1:100 dilution. Asc10 was detected with a polyclonal antibody constructed against the N-terminal domain of Asc10 (28) at a dilution of 1:2,500, and detection was performed with the enhanced-chemiluminescence protocol (Pierce).
Computer sequence analysis.
blastp version 2.2.9 (1) (1 May 2004) was used for searching the nr database through the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST/, accessed 30 July 2004). Default settings were used, except that mask for lookup table only was specified. The dendrogram of the PrgY family was done using ClustalW version 1.81 (36), accessed through the Supercomputer Laboratory at the Institute for Chemical Research, Kyoto University (http://clustalw.genome.ad.jp/), using default settings, except that the gap open penalty was changed from 10 to 5. The multiple-sequence alignment (MSA) of representatives of the PrgY family was done with the ClustalW version 1.81 program hosted by the European Bioinformatics Institute (http://www.ebi.ac.uk/clustalw) using default settings, except that the gap open penalty was changed from 10 to 5 and the gap extension penalty was changed from 0.05 to 0.5. The PrgY family members were further characterized through the Washington University Pfam database of protein families (http://pfam.wustl.edu/). Transmembrane topology was predicted using HMMTOP version 2.0 (38) (http://www.enzim.hu/hmmtop/) and TMHMM version 2.0 (25).
RESULTS
Functional complementation of PrgY.
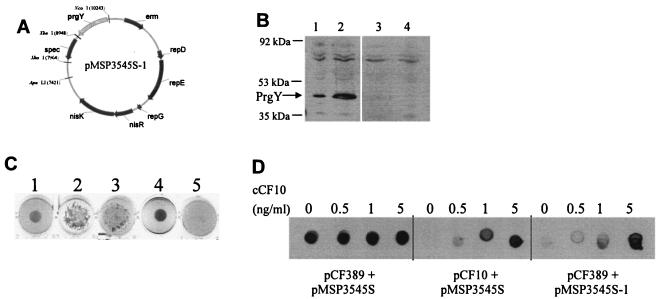
In order to study the mechanism of PrgY action, a PrgY expression construct was made by fusing the prgY gene in frame with the nisin-inducible promoter and ribosomal binding site of the pMSP3545S vector (8) (see Materials and Methods; Fig. 2A). Inducible expression of PrgY from pMSP3545S-1 in E. faecalis was confirmed by Western blot analysis (Fig. 2B). Expression of PrgY from this construct was almost completely shut off in the absence of nisin (Fig. 2B, lane 3) and was induced 5- to 10-fold over wild-type PrgY expression from pCF10 with nisin induction (Fig. 2B, lane 2).
FIG. 2.
Inducible expression of functional PrgY in E. faecalis. (A) PrgY was expressed from the nisin-inducible vector pMSP3545S-1, whose map is depicted. (B) PrgY expression in whole-cell extracts was analyzed by Western blotting with a polyclonal anti-PrgY antibody. PrgY expression from E. faecalis strain OG1RF harboring pCF10 (lane 1) or pMSP3545S-1 induced with nisin (lane 2) or not induced with nisin (lane 3) and OG1RF harboring the pCF10-derived prgY mutant pCF389 (lane 4). The arrow at 43 kDa corresponds to the expected size of PrgY. (C) Clumping assay to assess PrgY function in trans. Strains induced for plasmid transfer express Asc10 and clump in liquid culture (see text), which causes uneven settling at the bottom of the well. This is depicted in wells 2 and 3 (compare with nonclumpy well 1). Strains shown in wells 1 to 5 are OG1RF(pCF10) (well 1), OG1RF(pCF10) induced with cCF10 (well 2), the constitutively clumpy prgY mutant OG1RF(pCF389) (see text) (well 3), and OG1RF(pCF389/pMSP3545S-1) plus nisin for expression of PrgY (well 4) or without nisin (well 5). Cultures were grown with shaking overnight in THB or in THB with synthetic cCF10 (10 ng/ml) or nisin (25 ng/ml). (D) PrgY-mediated trans-acting regulation of Asc10 expression. Asc10 expression was measured by immunoblot analysis of whole-cell extracts spotted onto a membrane and detected with a polyclonal anti-Asc10 antibody. The cCF10 concentration used to treat each sample is indicated above each sample spot, and the strain is indicated below. pMSP3545S is the vector control, and PrgY was expressed from pMSP3545S-1. All strains were treated with the indicated cCF10 concentration and 5 ng nisin/ml for 6 h following 1:10 dilution of an overnight culture into fresh medium. Cell extracts were washed twice before harvesting. An equivalent amount of protein was spotted for each sample.
A strain harboring pCF389, a pCF10 derivative with a Tn917 insertion in prgY, is constitutively clumpy in broth culture due to constitutive induction by self-pheromone (11, 21). Due to the presence of PrgY and the inhibitor peptide iCF10, a strain harboring the wild-type pCF10 plasmid is not clumpy unless exogenous cCF10 is added. Plasmid pMSP3545S-1 was transformed into E. faecalis OG1RF(pCF389) to determine whether PrgY expressed from pMSP3545S-1 could reverse the constitutively clumpy phenotype of this strain. The resulting strain carrying both of these plasmids was tested for clumping using a biological clumping assay, where the clumpy phenotype of OG1RF(pCF389) and OG1RF(pCF10) induced with cCF10 was apparent by the heavy aggregation and irregular settling of cells after overnight growth with shaking (Fig. 2C, lanes 2 and 3) and the nonclumpy phenotype of uninduced OG1RF(pCF10) is demonstrated by regular concentrated settling of cells (Fig. 2C, lane 1). Expression of PrgY in trans from the pMSP3545S-1 plasmid abolished the constitutively clumpy phenotype of pCF389 (compare Fig. 2C, lanes 3 and 4), and the cells were nonclumpy, similar to uninduced OG1RF(pCF10), which settled in an even, small, circular mass at the bottom of the well. In the absence of nisin induction and PrgY expression, strain OG1RF(pCF389/pMSP3545S-1) was slightly clumpy (Fig. 2C, lane 5), indicating that PrgY was expressed at a low level in the absence of nisin induction, in agreement with Western blot analysis (Fig. 2B, lanes 2 and 3).
To confirm the results of the clumping assays, expression of aggregation substance (Asc10; the protein that mediates clumping) was measured by spot blotting using a polyclonal anti-Asc10 antibody. Expression was found to be constitutively high in OG1RF(pCF389) cells and inducible by cCF10 in OG1RF(pCF10) cells (Fig. 2D). When PrgY expression was induced by nisin in OG1RF(pCF389/pMSP3545S-1), Asc10 expression levels in response to increasing cCF10 levels were similar to those of OG1RF(pCF10) (Fig. 2D), indicating that PrgY-mediated trans-acting regulation was sensitive to exogenous cCF10.
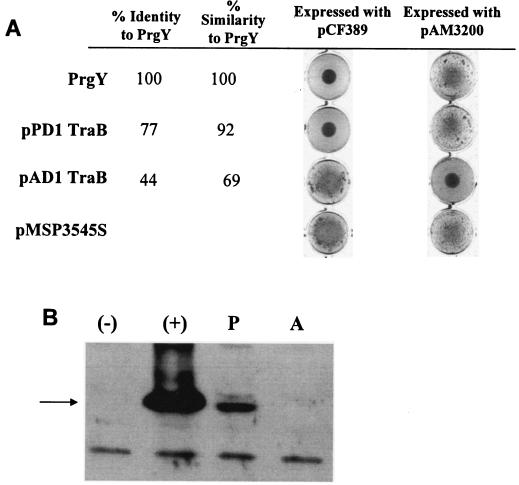
pPD1 TraB controls endogenous cCF10.
The TraB proteins encoded by the pAD1 and pPD1 plasmids share high identity with PrgY. To determine their ability to function in the pCF10 system, the traB genes were each cloned into pMSP3545S and tested for the ability to complement the pCF10 derivative (pCF389) containing a prgY mutation by using the clumping assay. As shown in Fig. 3A, pPD1 traB complemented pCF389 and controlled endogenous cCF10 whereas pAD1 traB did not. Not surprisingly, the polyclonal anti-PrgY antibody detected the pPD1 TraB protein, although at a much lower level than the wild-type PrgY protein, but did not detect the pAD1 TraB protein (Fig. 3B), supporting the notion that pPD1 TraB and PrgY share antigenic motifs.
FIG. 3.
PrgY/TraB functional assays in pCF10 and pAD1 backgrounds. (A) prgY or traB was cloned into pMSP3545S and coexpressed with the indicated plasmid in E. faecalis strain OG1RF. The clumpy phenotype is represented by strains harboring the pMSP3545S vector control (bottom) and indicates constitutive induction by endogenous pheromone. Cultures were grown overnight with shaking in THB with 25 ng nisin/ml for induction of protein expression. (B) Western blot assay showing cross-reactivity of the TraB protein from pPD1 (P) and pAD1 (A) with the PrgY polyclonal antibody. The TraB proteins were expressed from pMSP3545S in strain OG1RF(pCF389). Extracts from OG1RF(pCF389/pMSP3545S-1) expressing wild-type prgY (+) or from strain OG1RF(pCF389/pMSP3545S) carrying the vector (−) were run for comparison. The arrow at 43 kDa corresponds to the expected size of PrgY, and the band below is a nonspecific cross-reacting band. Overnight cultures were diluted 1:5 in fresh medium plus 25 ng nisin/ml and grown 2.5 h before harvesting.
A strain harboring a derivative of the pAD1 plasmid with an insertion in traB, pAM3200, is clumpy in broth culture, similar to the pCF10 derivative with a mutation in prgY, pCF389 (2). This pAM3200 construct was used to test whether prgY or pPD1 traB expression from pMSP3545S can complement a pAD1 traB mutation. pAD1 traB complemented pAM3200 (Fig. 3A), as expected (this result confirmed that failure of the same chimeric plasmid to complement pCF389 was not due to lack of expression of the cloned gene). However, neither pPD1 traB nor prgY could complement pAM3200, indicating that some specific function of pAD1 TraB protein is distinct from those of the other two proteins.
Expression of PrgY in protease-deficient E. faecalis strains reduces the amount of cCF10 released into the growth medium.
We sought to further elucidate the functional overlap between PrgY and the TraB proteins by direct comparative examination of their effects on levels of pheromone activity expressed by various E. faecalis strains. Low pheromone activity in the culture supernatants of proteolytic E. faecalis strain OG1RF made it difficult to examine prgY/traB effects without laborious extraction and concentration of pheromones from various cell fractions. We previously showed that most of the pheromone activity produced by OG1RF is cell associated and that the effect of PrgY expression on endogenous pheromone activity could only be demonstrated by assaying concentrated cell wall extracts (9). To avoid this problem, plasmids expressing prgY or one of the traB genes were moved into the isogenic serine protease (sprE) and gelatinase (gelE) mutant strain TX5128 (35). TX5128 was previously shown to have increased pheromone activity in the supernatant due to absence of the pheromone-degradative activities of the two secreted proteases (39); this higher level of pheromone in TX5128 culture supernatants made it possible to document effects of PrgY and the TraB proteins on pheromone production by simply assaying these supernatants. TX5128 derivatives expressing PrgY from pMSP3545S-1 (Table 2) or pCF10 (data not shown) had reduced supernatant pheromone activity compared with the vector or plasmid-free strains. This effect was also seen in two other GelE-deficient strains, FA2-2 and OG1X (data not shown). Expression of each of the pPD1 and pAD1 TraB proteins in TX5128 demonstrated a similar reduction in its cognate supernatant pheromone activity (Table 2). Consistent with the high level of sequence identity between PrgY and pPD1 TraB, we observed that PrgY reduced the supernatant activity of both cCF10 and cPD1, while pPD1 TraB reduced cPD1 and partially reduced cCF10 (starred titers, Table 2); neither protein reduced cAD1, and less-related pAD1 TraB had no effect on either cCF10 or cPD1. These results demonstrate that PrgY reduces pheromone activity in the supernatant of some E. faecalis strains and that there is specificity of the interaction of PrgY/TraB proteins with their cognate pheromone, with some functional overlap between PrgY and pPD1 TraB.
TABLE 2.
Reduction of endogenous pheromone activity in recipient cells by PrgY or the TraB proteins is specific
| Plasmida | Cloned gene | Pheromone activityb
|
||
|---|---|---|---|---|
| cCF10 | cPD1 | cAD1 | ||
| pMSP3545S | 64 | 8 | 8 | |
| pMSP3545S-1 | pCF10 prgY | 4 | <1c | 8 |
| pMSP3545S-2 | pPD1 traB | 16c | <1 | 8 |
| pMSP3545S-3 | pAD1 traB | 64 | 8 | <1 |
Plasmids were expressed in strain TX5128.
Supernatants (grown 6 h from a 10% overnight inoculum induced with 25 ng nisin/ml) were diluted twofold, and cCF10, cAD1, or cPD1 activity is reported as the inverse of the largest dilution that was able to aggregate an OG1RF(pCF10), OG1RF(pAD1) or OG1RF(pPD1) indicator strain. The relative amounts of pheromone are representative of at least two independent experiments. A titer of <1 indicates no active pheromone was observed at the dilutions examined.
Cross-specificity of the pPD1 and pCF10 systems; see text.
Expression of PrgY in recipient cells reduces conjugative transfer of pCF10.
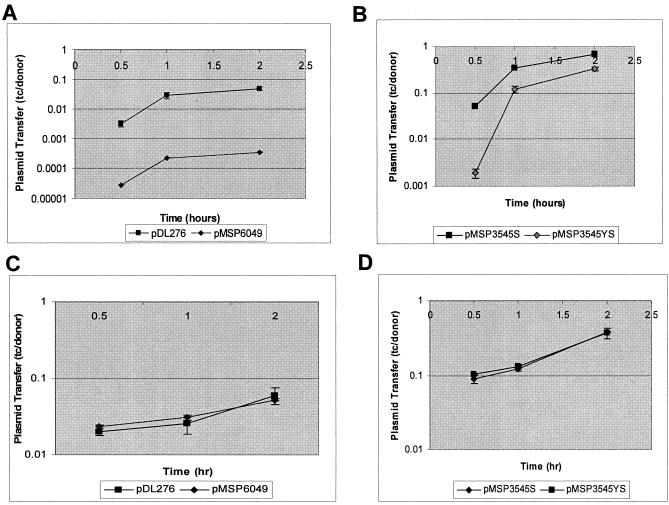
Previously, PrgY was found to reduce endogenous pheromone activity in the cell wall of strain OG1RF while not affecting supernatant pheromone activity (9), so we sought further evidence that PrgY expression has detectable biological consequences in OG1RF. To do this, transfer of pCF10 from an E. faecalis donor strain into a PrgY-expressing strain was compared with transfer into strains carrying the vector alone. For these experiments, donor strain OG1SSp(pCF10) was used because it carries antibiotic resistance markers (Table 1) that can be selected against when enumerating pCF10 transconjugants with OG1RF as a recipient strain. The donor strain was not induced with exogenous cCF10, so that any pheromone induction that might occur would be from cCF10 produced by the recipient cells in the mating mixtures. pCF10 transfer into an OG1RF strain expressing PrgY from its native promoter (pMSP6049) was reduced at least 100-fold compared with OG1RF harboring the vector alone (Fig. 4A). Transfer of pCF10 into nisin-induced OG1RF cells carrying pMSP3545S-1 was also reduced relative to an isogenic strain carrying the pMSP3545S vector alone (Fig. 4B). Addition of cCF10 to the donor culture prior to mating abolished the PrgY-dependent reduction in mating frequency (Fig. 4C and D), indicating that PrgY expression in recipient cells does not inhibit their ability to form a mating pair with an induced donor cell, and the reductions in mating frequency by PrgY observed in these experiments is related to a reduction in pheromone produced by the recipient cells during the course of the mating.
FIG. 4.
Conjugative transfer of pCF10 into PrgY-expressing E. faecalis recipients. pCF10 transfer into OG1RF recipient strains expressing PrgY from its native promoter (pMSP6049, A and C) or from a nisin-inducible construct (pMSP3545S-1, B and D). The construct expressing PrgY (pMSP6049 or pMSP3545S-1) is depicted with filled diamonds, and the vector (pDL276 or pMSP3545S) is depicted with filled squares. OG1SSp(pCF10) was the donor strain for these experiments. Results are expressed as transconjugants (tc) per donor. Conjugative transfer of pCF10 was done in broth culture in the absence of exogenous cCF10 for the time indicated (y axis) after donor and recipient cells were combined (A and B) or in the presence of 10 ng/ml exogenous cCF10, added immediately after the donor and recipient cells were combined (C and D). The measurements reported are representative of at least two independent experiments done in duplicate. Error bars represent 1 standard deviation of the mean.
Effects of PrgZ and PrgY on the ability of E. faecalis cells to bind exogenous cCF10.
E. faecalis cells expressing PrgZ bind a significant amount of exogenous pheromone from the culture medium (34), and this binding activity is required for the specific import of cCF10 into pheromone-responding donor cells. To determine whether PrgY could also bind exogenous pheromone or whether PrgY can alter PrgZ binding of exogenous pheromone, PrgY and PrgZ were expressed together or separately in OG1RF and the exogenous pheromone-binding capacity was quantified. For these experiments, PrgZ and PrgY were expressed from their native promoter in the pDL276 vector. The results (Table 3) indicate that cells expressing PrgZ bind a significant amount of exogenous cCF10, as previously shown (34), and that PrgY expression does not alter binding by PrgZ or increase exogenous pheromone binding when expressed by itself.
TABLE 3.
Exogenous pheromone binding by PrgY and PrgZa
| Plasmid content | Relevant prg gene content | Exogenous pheromone-binding capacity |
|---|---|---|
| None | None | 0 |
| pDL276 | None | 0 |
| pMSP6049 | prgY | 0 |
| pMSP6023ΔY | prgZ | 64 |
| pMSP6043 | prgY prgZ | 64 |
E. faecalis OG1RF harboring the plasmids indicated was assayed for binding by assessing the ability of the bacteria to reduce the pheromone titer of a preparation of cCF10 (previously diluted to a titer of 64) by the assay described in Materials and Methods. The values listed for binding capacity indicate the reduction in pheromone titer observed after incubation of the pheromone with the bacterial strain indicated. This experiment was repeated twice in duplicate with the same results both times.
Construction and characterization of random PrgY point mutations.
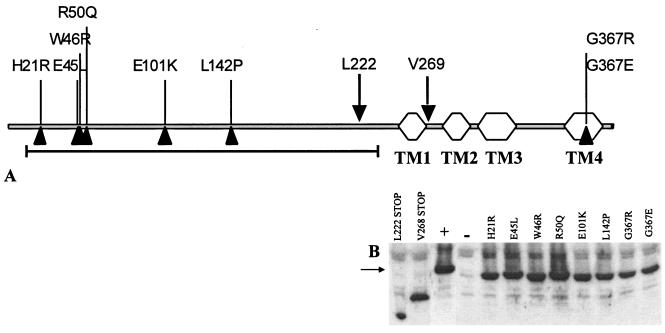
In order to further characterize the control of endogenous pheromone by PrgY, pCF389 complementation was used as a screen to identify random prgY point mutations that failed to prevent self-induction. Briefly, the prgY gene was PCR amplified to incorporate random mutations and cloned into pMSP3545S. The cloned product was then transformed into E. faecalis OG1RF(pCF389) and plated on THB agar containing 50 ng nisin/ml to induce PrgY expression. Nonfunctional PrgY mutants were identified on the plate by their dry-colony phenotype that corresponds to clumping in broth culture, and these were further screened for PrgY protein expression by Western blotting. Those plasmids that expressed a nonfunctional PrgY protein were sequenced to identify the location of the incorporated sequence error. The positions of eight point mutations and two truncation mutations generated by this method are indicated in Fig. 5A. Six of the eight point mutations were located in the N-terminal half of PrgY, which is predicted to lie on the outside of the membrane. All of the PrgY mutant proteins identified by this screen were expressed at the same level as wild-type PrgY (Fig. 5B) but were completely abolished for functional complementation of pCF389 in OG1RF.
FIG. 5.
(A) Positions of random mutations in prgY shown on a linear map of the gene. The Pfam TraB domain (Pfam 01963) is indicated with a black line; predicted transmembrane domains (TM) are indicated by hexagons. The mutations at L222 and V269 result in truncations (down-pointing triangles). (B) Protein expression from whole-cell extracts of prgY derivatives. The arrow at 43 kDa corresponds to the expected size of PrgY. OG1RF(pCF389) was the strain background in these experiments. Plasmid pMSP3545S-1 expressing wild-type prgY (+) and the pMSP3545S vector used for expression of wild-type and prgY derivatives (−) were used for comparison. All strains were treated with 25 ng nisin/ml for 2.5 h. An equivalent amount of protein was loaded for each sample.
PrgY mutations abolishing function map to conserved residues in a newly identified family of proteins.
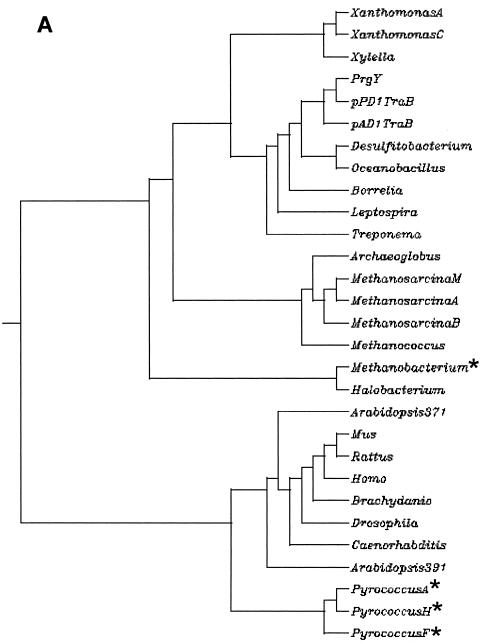
A family of proteins that share identity with PrgY and the pAD1 and pPD1 TraB proteins has been revealed by recent sequencing efforts. These proteins are encoded by species including gram-negative and gram-positive bacteria, archaea, plants, and mammals (Fig. 6A). We compiled a list of 29 nonredundant family members (see Materials and Methods) based on sequence identity and found that each of these proteins has the TraB domain identified by a Pfam hidden Markov model (HMMs; Pfam 01963) (27). We found only one species, Arabidopsis thaliana, to encode more than one prgY-like gene. Surprisingly, prgY-like genes were not found in any sequenced gram-positive organisms that are closely related to E. faecalis or that have characterized peptide pheromone systems similar to the pheromone-inducible plasmid systems of E. faecalis, such as Staphylococcus and Streptococcus species. Analysis of the family of protein sequences for transmembrane domains with TMHMM (25) used by the Pfam database and the HMMTOP (38) program indicated that the prokaryotic family members share three or four transmembrane domains that are fairly conserved in location. These transmembrane domains are also shared by some of the archaea, but there is lower conservation among the eukaryotes. Several of the PrgY family members had no identifiable transmembrane domains using both programs and may be cytoplasmic (starred, Fig. 6A).
FIG. 6.
The PrgY family. (A) Protein sequences of PrgY family members were acquired from the nonredundant database using the National Center for Biotechnology Information blastp search engine (1) with default parameters and mask for the lookup table only specified. The dendrogram was created from an MSA of all of the sequences using ClustalW version 1.81 (36) hosted at http://clustalw.genome.ad.jp/. Stars indicate those sequences that are predicted to be cytoplasmic (see text) by both the TMHMM (25) and HMMTOP (38) programs. Abbreviations are for (in order from the top) Xanthomonas axonopodis, Xanthomonas campestris, Xylella fastidiosa, Enterococcus faecalis/pCF10 PrgY, E. faecalis/pPD1 TraB, E. faecalis/pAD1 TraB, Desulfitobacterium hafniense, Oceanobacillus iheyensis, Borrelia burgdorferi, Leptospira interrogans, Treponema pallidum, Archaeoglobus fulgidus, Methanosarcina mazei, Methanosarcina acetivorans, Methanosarcina barkeri, Methanococcus jannaschii, Methanobacterium thermautotrophicum, Halobacterium sp., Arabidopsis thaliana (protein At1g05270, 371 amino acids long), Mus musculus, Rattus norvegicus, Homo sapiens, Brachydanio rerio, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana (protein AT5g52030, 391 amino acids long), Pyrococcus abyssi, Pyrococcus horikoshii, and Pyrococcus furiosus. (B) MSA of PrgY with the TraB proteins from the E. faecalis pAD1 and pPD1 plasmids, Borrelia burgdorferi, Archaeoglobus fulgidus, Arabidopsis thaliana (protein At1g05270, 371 amino acids long), and Mus musculus. Locations of conserved residues and site-directed mutants are shown. Identical residues in this alignment are indicated with a star below the residue, a colon indicates a conserved substitution, and a period indicates a semiconserved substitution. Regions shaded in gray indicated predicted transmembrane domains (see Materials and Methods). Conserved residues throughout the entire PrgY protein family are in bold. Residues changed through random or site-directed mutagenesis are indicated above the residues that are changed. The A's that are in bold indicate conserved residues that were changed to alanine by site-directed mutagenesis. All other substitutions were generated by random PCR mutagenesis. Truncation mutants generated by random PCR mutagenesis are indicated at their truncation sites (STOP).
From the 29 currently available PrgY family member sequences, a multiple sequence alignment (MSA) was made using ClustalW (see Materials and Methods). An MSA of a few representatives is shown in Fig. 6B. From the full alignment, eight conserved residues were identified (in bold, Fig. 6B). When the cytoplasmic proteins were removed from the MSA, two more conserved residues were identified, R121 and R129 (also in bold, Fig. 6B). As shown in Fig. 6B, two of the random mutations (E45L and R50Q) mapped to residues that are conserved throughout the entire PrgY family. In addition, the G367 residue, where two different random mutations were isolated, is conserved throughout all of the membrane-spanning prokaryotic family members, and the other mutations identified through random mutagenesis also mapped to fairly well-conserved residues. These results imply conservation of function within the PrgY-like family of proteins, which is interesting, considering the specific role of PrgY with the regulation of cCF10-inducible plasmid transfer.
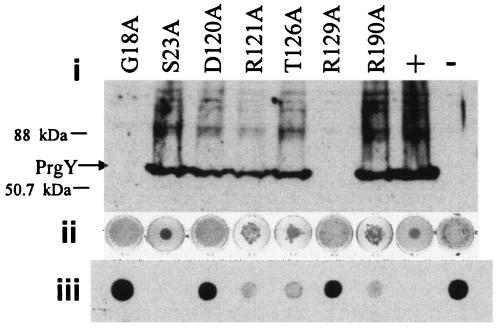
To further determine the contribution of these conserved residues to the function of PrgY, site-directed mutagenesis was used to change seven of the remaining conserved residues to alanine to determine whether these changes abolished function in E. faecalis. Two of these site-directed mutants no longer expressed stable protein (Fig. 7, G18A and R129A), demonstrating the importance of these specific amino acids in the stability of the protein. One mutant, S23A, retained wild-type control of endogenous pheromone. The other four mutations reduced or completely abolished pCF389 complementation as determined by an overnight clumping assay and Asc10 spot blotting (Fig. 7) while retaining production of stable protein. The random and engineered mutant proteins were expressed in cells carrying wild-type pCF10 to determine whether any exhibited a dominant negative phenotype, and none of them were found to be dominant negative.
FIG. 7.
Engineered mutants with reduced or abolished function. Engineered mutants with reduced or abolished function were analyzed by (i) Western blotting using a polyclonal anti-PrgY antibody with an arrow at 43 kDa corresponding to the expected size of PrgY, (ii) clumping assay depicting the level of Asc10 shutdown as measured by aggregation of cells in an overnight shaking broth culture, and (iii) Western analysis of Asc10 expression using a polyclonal anti-Asc10 antibody. All strains were treated with 25 ng nisin/ml for 2.5 h (i), overnight (ii), or for 6 h (iii). OG1RF(pCF389) was the strain background in these experiments. Plasmid pMSP3545S-1 expressing wild-type prgY (+) and the pMSP3545S vector used for expression of wild-type and prgY derivatives (−) were used for comparison. Cell extracts were washed twice before harvesting. An equivalent amount of protein was loaded or spotted for each sample.
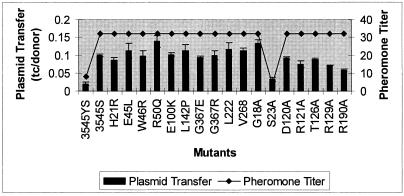
Random and specific mutations fail to reduce supernatant pheromone activity or pCF10 transfer.
The random and directed PrgY mutants were tested for the ability to reduce pheromone activity and pCF10 transfer. Because pheromone activity is easily assayed in TX5128 cell supernatants, the plasmids expressing prgY mutant derivatives were moved to this strain for both experiments so the results could be compared. pCF10 transfer into strain TX5128 was higher than that into OG1RF, as previously described (39); therefore, expression of PrgY in TX5128 resulted in only a fivefold reduction in pCF10 transfer rates. All of the random mutants and all of the directed mutants that were abolished for complementation of pCF389 also failed to reduce supernatant pheromone activity and pCF10 transfer (Fig. 8). Directed mutant S23A, which exhibited wild-type control of endogenous pheromone in donors, also demonstrated wild-type reduction of excreted pheromone and reduced acquisition of pCF10 in recipients. Certain mutations that partially reduced complementation, including R190A, exhibited an intermediate recipient phenotype in the mating assay. In the pheromone assays, which are less sensitive and precise, all protein-expressing mutants showed elevated pheromone production, with the exception of S23A (which was identical to the wild type in all assays). Since all prgY point mutations affecting the phenotype in one assay showed parallel changes in other assays, we think that a single functional activity of the protein is responsible for all of the phenotypic effects associated with PrgY expression.
FIG. 8.
Supernatant pheromone activity and pCF10 transfer into strains expressing PrgY mutants. Plasmid transfer levels are expressed as transconjugants (tc) per donor. prgY and its derivatives, or the vector pMSP3545S, were expressed in TX5128 (GelE− SprE−) for both experiments. OG1Ssp(pCF10) was used as the donor strain for pCF10 transfer. Pheromone titers represent cCF10 activity that was detected by microtiter clumping assays; the titers reported represent the reciprocal of the highest dilution that induced clumping (see Materials and Methods) and are representative of at least two independent experiments done on separate days. Plasmid transfer rates are representative of two independent experiments done on separate days in duplicate. The amino acid substitution S23A generated a wild-type phenotype in the pCF389 complementation assay, whereas all the other substitutions shown resulted in partial or complete loss of pCF389 complementation ability (Fig. 5 and 7).
DISCUSSION
This study used a combination of genetic and physiological experiments to show that PrgY can control endogenous cCF10 pheromone activity in pCF10-containing donor cells without abolishing the ability of these cells to sense the presence of the same peptide pheromone from an exogenous source, e.g., a potential recipient cell. We also used computer analysis of sequence databases to look for related proteins in other organisms. The most significant results were as follows. (i) PrgY expression in the absence of any other pCF10 proteins reduced the cCF10 activity in the supernatant of E. faecalis cells and impaired the ability of these cells to acquire pCF10 by conjugation. The activity of PrgY is specific. (ii) Analysis of prgY point mutants obtained in a complementation-based genetic screen indicated that a single functional activity (reduction of endogenous pheromone production) mediated by PrgY is responsible for all effects of the protein observed in both donor and recipient cells. (iii) PrgY is representative of a large family of related proteins found in all biological kingdoms (but not in all organisms), and random mutations abolishing its function map to highly conserved residues, suggesting the possibility of a common biological function for many members of this family. (iv) The collective data were used to formulate a hypothetical mechanism for PrgY function (discussed below and depicted in Fig. 8).
While a previous study showed that PrgY reduced endogenous cell-associated cCF10 activity (9), results reported here demonstrate that PrgY can also mediate a specific, significant reduction of pheromone activity in the culture supernatants of protease-deficient strains of E. faecalis. The biological relevance of PrgY-mediated pheromone reduction was demonstrated by the observation of reduced pCF10 transfer into recipient strains expressing PrgY (Fig. 5 and 8). We also found that PrgY expression does not increase exogenous pheromone binding or affect exogenous binding by PrgZ (Table 3), indicating that the PrgY-mediated reduction in pheromone activity occurs at the level of pheromone excretion by the host cell and not through rebinding of previously released extracellular pheromone. This PrgY-mediated reduction in excreted pheromone activity was not previously seen in strain OG1RF, most likely because secreted serine and gelatinase proteases in OG1RF reduce pheromone activity in cell supernatant significantly (39) and mask any further reduction by expressed PrgY protein. Since GelE is released into the growth medium, it seems likely that excreted pheromone is more sensitive to GelE degradation than cell-associated pheromone, which would explain the GelE-related differences we observed in the various strains.
Analysis of mutations in prgY that abolish control of endogenous pheromone in donor strains also suggests that PrgY functions by reducing excreted pheromone (Fig. 8). The prgY mutations affecting endogenous pheromone control were located primarily in the N-terminal region of PrgY and map to conserved residues in a family of PrgY-like proteins (Fig. 6B). These mutations are part of a domain that is predicted to be on the outside of the cell membrane by the HMMTOP method of transmembrane domain prediction (37) and overlap the TraB domain defined by Pfam's hidden Markov model (Pfam 01963). Our preliminary unpublished experimental data also support this model for PrgY topology. The results reported here strongly indicate that this domain is directly involved in pheromone reduction activity.
The PrgY and TraB proteins demonstrate specificity for their cognate pheromone in the absence of other plasmid-encoded proteins (Table 2), implying that there may be a direct interaction between these proteins and the pheromone, a notion that has not been previously explored. Considering that PrgY is a membrane protein, an interaction of PrgY with pheromone would most likely occur at the cytoplasmic membrane surface, where the pheromone is secreted following processing from the signal peptide of the lipoprotein CcfA (Fig. 9). Since the mutations in PrgY mapped to a region of PrgY that is predicted to be on the outside of the membrane, it is possible that PrgY interacts with nascent cCF10 after it is processed by signal peptidase II (SPII) and by Eep and immediately following its release from the membrane. The work presented here indicates that PrgY specificity may extend to the cPD1 peptide (Table 2), and in support of this we found 45% identity between the signal sequences of the lipoproteins encoding cPD1 and cCF10 and no significant homology with the signal sequence encoding cAD1 (data not shown). These findings support a model where the PrgY/TraB proteins directly interact with mature pheromone or with some form of the signal peptide precursor to prevent the release of pheromone into the supernatant.
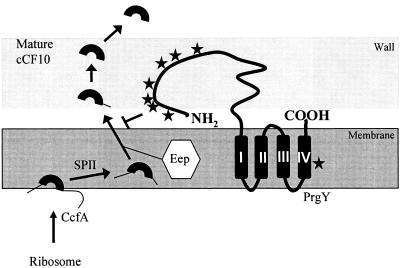
FIG. 9.
Model of PrgY-mediated cCF10 inhibition. Mature cCF10 (top left) in the cell wall or supernatant is encoded within the signal sequence of host-encoded lipoprotein CcfA (5). Signal peptidase II (SPII) removes the entire signal peptide from CcfA, and Eep further processes the signal peptide to expose the cCF10 peptide. Active export or further proteolytic processing by host-encoded proteins may occur at this stage. PrgY (center) is a predicted transmembrane protein with four membrane-spanning segments in the C-terminal portion of the protein, designated I to IV, predicted by the HMMTOP program (http://www.enzim.hu/hmmtop/). PrgY specifically blocks production of active cCF10, probably by degrading or modifying newly processed or preprocessed cCF10 in the cell wall or by blocking processing or release into the cell medium (see Discussion). The mutations identified in this study (stars) that still produce full-length protein but can no longer block production of active cCF10 and prevent self-induction are indicated with stars (for locations, see Fig. 6B). The majority of these mutations map to the N-terminal half of the predicted extracellular portion of PrgY.
The mechanism by which release of active pheromone is inhibited by PrgY and the TraB proteins is not clear, but we can begin to speculate based on recent data on pheromone processing (4, 5) and results reported here. At the cell surface, PrgY/TraB could degrade or modify processed or preprocessed pheromone through a direct interaction or redirect the prepheromone to be alternately processed before or after release from SPII and Eep. We do not favor an alternative model where PrgY/TraB would block processing of pheromone by inhibiting SPII or Eep, given our finding in this work that PrgY activity is specific, while Eep and SPII are known to process a variety of lipoprotein signal sequences, including that for cAD1 (4, 14). It is still possible, however, that the PrgY/TraB proteins function through an interaction with Eep. An interaction between the membrane-spanning domains of PrgY and Eep could serve to position the extracellular domain of PrgY ideally for the immediate capture of pheromone as it is released from the membrane, which probably occurs concomitantly with proteolytic processing of cCF10 (4). This type of model is also in agreement with the extracellular domain of PrgY participating directly in endogenous cCF10 control without interacting with exogenously supplied cCF10. The model makes testable predictions, including that of a direct physical interaction between cCF10 (or a precursor) and PrgY and implies that there are specificity determinants for this interaction that lie within the ccfA signal peptide sequence and possibly within the cCF10 sequence itself.
Since PrgY is a specific regulator of a unique peptide pheromone signaling system, it is interesting to find this protein in such a disparate range of species (Fig. 6A). Two lines of evidence from this study support the notion that this family may share function: (i) random and engineered PrgY functional mutants demonstrated the functional and structural importance of residues that are conserved throughout the entire PrgY family, and (ii) the PrgY-like protein encoded by the pPD1 plasmid was found to complement a prgY mutant in pCF10.
In pCF10, prgY is flanked by genes encoding the surface pheromone binding protein PrgZ and the pheromone response regulator PrgX. There do not appear to be any prgX or prgZ homologs or other pCF10-specific genes directly surrounding the PrgY-like genes in the chromosomes of the organisms analyzed, with the exception of Borrelia burgdorferi, which encodes a gene with very low homology to PrgX and in the same orientation as PrgX with respect to PrgY. The low homology between pCF10-encoded PrgX and its closely related pAD1 and pPD1 counterpart TraA make it difficult to determine the significance of this finding. There has only been one documented example of oligopeptide signaling in gram-negative bacteria (40), and to our knowledge this type of signaling has not been observed in the spirochetes encoding PrgY-like proteins. In addition, the gene encoding the PrgY-like protein in B. burgdorferi is found on the chromosome instead of one of its many plasmids, a surprising finding since the only known function of PrgY is in regulation of plasmid transfer. The role PrgY homologs play in B. burgdorferi and the other spirochetes is unknown. The existence of this PrgY-like family of proteins and their potential functional overlap indicate two equally intriguing possibilities; either the PrgY-like proteins have a function in this species separate from regulation of peptide signaling, or these proteins are involved in regulation of uncharacterized peptide signaling systems in these organisms.
Acknowledgments
This research is supported by NIH grant GM49530 to G.M.D. J.R.C. is a trainee funded by NIH MinnCResT training grant T32DE07288.
We gratefully acknowledge Lynda Ellis for providing training in protein sequence analysis.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, F. Y., and D. B. Clewell. 1994. Characterization of the determinant (traB) encoding sex pheromone shutdown by the hemolysin/bacteriocin plasmid pAD1 in Enterococcus faecalis. Plasmid 31:215-221. [DOI] [PubMed] [Google Scholar]
- 3.An, F. Y., and D. B. Clewell. 2002. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. J. Bacteriol. 184:1880-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An, F. Y., M. C. Sulavik, and D. B. Clewell. 1999. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J. Bacteriol. 181:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antiporta, M. H., and G. M. Dunny. 2002. ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF. J. Bacteriol. 184:1155-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bae, T., B. Kozlowicz, and G. M. Dunny. 2002. Two targets in pCF10 DNA for PrgX binding: their role in production of Qa and prgX mRNA and in regulation of pheromone-inducible conjugation. J. Mol. Biol. 315:995-1007. [DOI] [PubMed] [Google Scholar]
- 7.Bae, T., B. K. Kozlowicz, and G. M. Dunny. 2004. Characterization of cis-acting prgQ mutants: evidence for two distinct repression mechanisms by Qa RNA and PrgX protein in pheromone-inducible enterococcal plasmid pCF10. Mol. Microbiol. 51:271-281. [DOI] [PubMed] [Google Scholar]
- 8.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183-190. [DOI] [PubMed] [Google Scholar]
- 9.Buttaro, B. A., M. H. Antiporta, and G. M. Dunny. 2000. Cell-associated pheromone peptide (cCF10) production and pheromone inhibition in Enterococcus faecalis. J. Bacteriol. 182:4926-4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandler, J. R., and G. M. Dunny. 2004. Enterococcal peptide sex pheromones: synthesis and control of biological activity. Peptides 25:1377-1388. [DOI] [PubMed] [Google Scholar]
- 11.Christie, P. J., and G. M. Dunny. 1986. Identification of regions of the Streptococcus faecalis plasmid pCF-10 that encode antibiotic resistance and pheromone response functions. Plasmid 15:230-241. [DOI] [PubMed] [Google Scholar]
- 12.Clewell, D. B., F. Y. An, S. E. Flannagan, M. Antiporta, and G. M. Dunny. 2000. Enterococcal sex pheromone precursors are part of signal sequences for surface lipoproteins. Mol. Microbiol. 35:246-247. [DOI] [PubMed] [Google Scholar]
- 13.Clewell, D. B., P. K. Tomich, M. C. Gawron-Burke, A. E. Franke, Y. Yagi, and F. Y. An. 1982. Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J. Bacteriol. 152:1220-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dev, I. K., and P. H. Ray. 1990. Signal peptidases and signal peptide hydrolases. J. Bioenerg. Biomembr. 22:271-290. [DOI] [PubMed] [Google Scholar]
- 15.Dunny, G., C. Funk, and J. Adsit. 1981. Direct stimulation of the transfer of antibiotic resistance by sex pheromones in Streptococcus faecalis. Plasmid 6:270-278. [DOI] [PubMed] [Google Scholar]
- 16.Dunny, G. M., M. H. Antiporta, and H. Hirt. 2001. Peptide pheromone-induced transfer of plasmid pCF10 in Enterococcus faecalis: probing the genetic and molecular basis for specificity of the pheromone response. Peptides 22:1529-1539. [DOI] [PubMed] [Google Scholar]
- 17.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. USA 75:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunny, G. M., R. A. Craig, R. L. Carron, and D. B. Clewell. 1979. Plasmid transfer in Streptococcus faecalis: production of multiple sex pheromones by recipients. Plasmid 2:454-465. [DOI] [PubMed] [Google Scholar]
- 19.Dunny, G. M., L. N. Lee, and D. J. LeBlanc. 1991. Improved electroporation and cloning vector system for gram-positive bacteria. Appl. Environ. Microbiol. 57:1194-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flannagan, S. E., and D. B. Clewell. 2002. Identification and characterization of genes encoding sex pheromone cAM373 activity in Enterococcus faecalis and Staphylococcus aureus. Mol. Microbiol. 44:803-817. [DOI] [PubMed] [Google Scholar]
- 21.Hedberg, P. J., B. A. Leonard, R. E. Ruhfel, and G. M. Dunny. 1996. Identification and characterization of the genes of Enterococcus faecalis plasmid pCF10 involved in replication and in negative control of pheromone-inducible conjugation. Plasmid 35:46-57. [DOI] [PubMed] [Google Scholar]
- 22.Ike, Y., R. A. Craig, B. A. White, Y. Yagi, and D. B. Clewell. 1983. Modification of Streptococcus faecalis sex pheromones after acquisition of plasmid DNA. Proc. Natl. Acad. Sci. USA 80:5369-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kao, S. M., S. B. Olmsted, A. S. Viksnins, J. C. Gallo, and G. M. Dunny. 1991. Molecular and genetic analysis of a region of plasmid pCF10 containing positive control genes and structural genes encoding surface proteins involved in pheromone-inducible conjugation in Enterococcus faecalis. J. Bacteriol. 173:7650-7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozlowicz, B. K., T. Bae, and G. M. Dunny. 2004. Enterococcus faecalis pheromone-responsive protein PrgX: genetic separation of positive autoregulatory functions from those involved in negative regulation of conjugative plasmid transfer. Mol. Microbiol. 54:520-532. [DOI] [PubMed] [Google Scholar]
- 25.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 26.Leonard, B. A., A. Podbielski, P. J. Hedberg, and G. M. Dunny. 1996. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc. Natl. Acad. Sci. USA 93:260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchler-Bauer, A., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, C. J. Lanczycki, C. A. Liebert, C. Liu, T. Madej, G. H. Marchler, R. Mazumder, A. N. Nikolskaya, A. R. Panchenko, B. S. Rao, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, S. Vasudevan, Y. Wang, R. A. Yamashita, J. J. Yin, and S. H. Bryant. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCormick, J. K., H. Hirt, C. M. Waters, T. J. Tripp, G. M. Dunny, and P. M. Schlievert. 2001. Antibodies to a surface-exposed, N-terminal domain of aggregation substance are not protective in the rabbit model of Enterococcus faecalis infective endocarditis. Infect. Immun. 69:3305-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mori, M., Y. Sakagami, Y. Ishii, A. Isogai, C. Kitada, M. Fujino, J. C. Adsit, G. M. Dunny, and A. Suzuki. 1988. Structure of cCF10, a peptide sex pheromone which induces conjugative transfer of the Streptococcus faecalis tetracycline resistance plasmid, pCF10. J. Biol. Chem. 263:14574-14578. [PubMed] [Google Scholar]
- 30.Nakayama, J., G. M. Dunny, D. B. Clewell, and A. Suzuki. 1995. Quantitative analysis for pheromone inhibitor and pheromone shutdown in Enterococcus faecalis. Dev. Biol. Stand. 85:35-38. [PubMed] [Google Scholar]
- 31.Nakayama, J., R. E. Ruhfel, G. M. Dunny, A. Isogai, and A. Suzuki. 1994. The prgQ gene of the Enterococcus faecalis tetracycline resistance plasmid pCF10 encodes a peptide inhibitor, iCF10. J. Bacteriol. 176:7405-7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakayama, J., K. Yoshida, H. Kobayashi, A. Isogai, D. B. Clewell, and A. Suzuki. 1995. Cloning and characterization of a region of Enterococcus faecalis plasmid pPD1 encoding pheromone inhibitor (ipd), pheromone sensitivity (traC), and pheromone shutdown (traB) genes. J. Bacteriol. 177:5567-5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Podbielski, A., B. Spellerberg, M. Woischnik, B. Pohl, and R. Lutticken. 1996. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177:137-147. [DOI] [PubMed] [Google Scholar]
- 34.Ruhfel, R. E., D. A. Manias, and G. M. Dunny. 1993. Cloning and characterization of a region of the Enterococcus faecalis conjugative plasmid, pCF10, encoding a sex pheromone-binding function. J. Bacteriol. 175:5253-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh, K. V., X. Qin, G. M. Weinstock, and B. E. Murray. 1998. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J. Infect. Dis. 178:1416-1420. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tusnady, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]
- 38.Tusnady, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 39.Waters, C. M., M. H. Antiporta, B. E. Murray, and G. M. Dunny. 2003. Role of the Enterococcus faecalis GelE protease in determination of cellular chain length, supernatant pheromone levels, and degradation of fibrin and misfolded surface proteins. J. Bacteriol. 185:3613-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoon, H. S., and J. W. Golden. 1998. Heterocyst pattern formation controlled by a diffusible peptide. Science 282:935-938. [DOI] [PubMed] [Google Scholar]