Abstract
The Liverpool epidemic strain (LES) of Pseudomonas aeruginosa is a transmissible aggressive pathogen of cystic fibrosis (CF) patients. We compared transcriptome profiles of two LES isolates with each other and with a laboratory and genetic reference strain (PAO1) after growth to late exponential phase and following exposure to oxidative stress. Both LES isolates exhibited enhanced antimicrobial resistances linked to specific mutations in efflux pump genes. Although transcription of AmpC β-lactamase was up-regulated in both, one LES isolate contained a specific mutation rendering the ampC gene untranslatable. The virulence-related quorum-sensing (QS) regulon of LES431, an isolate that caused pneumonia in the non-CF parent of a CF patient, was considerably up-regulated in comparison to either isolate LES400, associated with a chronic CF infection, or strain PAO1. Premature activation of QS genes was detected in isolates from both non-CF parents and the CF patient in a previously reported infection episode. LES isolates lacking the up-regulated QS phenotype contained different frameshift mutations in lasR. When fed to Drosophila melanogaster, isolate LES431 killed the fruit flies more readily than either isolate LES400 or strain PAO1, indicating that virulence varies intraclonally. The LES may represent a clone with enhanced virulence and antimicrobial resistance characteristics that can vary or are lost due to mutations during long-term colonization but have contributed to the successful spread of the lineage throughout the CF population of the United Kingdom.
Pseudomonas aeruginosa, the most common pathogen associated with morbidity and mortality in cystic fibrosis (CF), causes chronic lung infections that once established are impossible to eradicate. For many years, the prevailing view was that individual CF patients acquired P. aeruginosa infections separately and thus carried their own unrelated strains. In 1996, Cheng et al. (6) reported the spread of a drug-resistant strain of P. aeruginosa among patients in a children's CF center in Liverpool. Since then, there have been other reports of CF “epidemic” strains (14, 45). An analysis of post-2000 patient samples has indicated that of 80 CF patients (from a total of 92 sampled) in the Liverpool adult center colonized with P. aeruginosa, 79% carry the Liverpool epidemic strain (LES) (35). In a recent study (45) involving over 1,200 isolates from 31 CF centers in England and Wales, the LES was identified as the most common clone, present in 48% of CF centers and accounting for 11% of the isolates. The LES appears to be more aggressive than other strains of P. aeruginosa. It has been able to replace previously established strains of P. aeruginosa (superinfection) (28) and has infected the non-CF parents of a CF patient, causing significant morbidity (29). Furthermore, there is greater morbidity among CF patients colonized with the LES than among those carrying nonepidemic strains of P. aeruginosa (1). Clearly, this strain represents a transmissible and aggressive clone well adapted to the CF environment.
The genome of P. aeruginosa displays a mosaic structure, with all strains possessing a highly conserved backbone comprising the vast majority of their genome and including the recognized virulence factors (9, 56). Variations between strains include the presence or absence of genomic islands, and there is some evidence to suggest that the LES possesses such islands (36). However, variations in gene expression have been shown to influence the virulence of an autoaggressive and highly adherent small-colony variant of a CF isolate, with significant up-regulation of virulence-related genes occurring (51). The success of the LES may be due either to the prior acquisition of genes or islands, to transcriptional variations in gene expression, or to a combination of both. We hypothesize that such changes contribute to greater colonization and/or transmissibility of the strain, enhancing its ability to cause CF infections, and may lead to enhanced virulence manifesting itself in episodes such as the infection of non-CF parents.
To test the hypothesis that the additional properties of the LES may be due to variations in gene expression, we compared the transcriptional signature of two LES isolates (associated with chronic and acute infections, respectively) to each other and to the laboratory and genetic reference strain PAO1 under two different growth conditions designed to maximize the number of expressed genes: late exponential phase and stationary phase with exposure to hydrogen peroxide. Here, we report variations in the expression of virulence-related genes not only between LES isolates and strain PAO1 but between the two LES isolates.
MATERIALS AND METHODS
Bacterial strains.
Isolate LES400 is our laboratory reference isolate for the LES, used in previous genetic analysis (36), and was isolated from a CF patient who had been colonized for at least 6 months (chronic infection). It has an identical pulsed-field gel electrophoresis (PFGE) pattern to the earliest-known LES isolate from 1988. The other LES isolates were sputum culture isolates taken from a case involving respiratory tract infection of the non-CF parents of a CF patient (29). Isolates LES431 and LES430 were both isolated from the CF patient's father, who presented with pneumonia. Isolate LES417 was isolated from the patient's mother, who presented with pyrexia and bilateral chest wheezes. Isolates LES416 and LESB44 were isolated from the CF patient. Isolates LES416, LES417, LES430, and LES431 share an identical PFGE pattern. Strain PAO1 is a widely studied laboratory strain of P. aeruginosa for which the entire genome sequence is known (47). PFGE analysis of SpeI-digested genomic DNA was carried out using 1% (wt/vol) agarose gels and interpreted according to the protocol of Tenover et al. (49).
Growth conditions and RNA isolation for microarray analysis.
In the absence of stress, P. aeruginosa strains were grown up to late exponential phase (optical density at 600 nm [OD600] of 2.7 to 3.0) in Luria broth (LB). For exposure of bacteria to hydrogen peroxide, stationary phase-grown cultures (3 × 1010 cells) were resuspended in fresh LB and kept in a dialysis tube (14-kDa cutoff; 25 mm) with an effective length of 6 cm for the exchange of fluids. Then, the dialysis tube was resuspended in a 1-liter Erlenmeyer flask containing 600 ml of LB with or without 10 mM hydrogen peroxide (Sigma-Aldrich) to generate oxidative stress. The flasks were incubated at 37°C and 200 rpm on a rotary shaker for 2 h.
Total RNA was extracted from approximately 3 × 1010 cells by a modified hot-phenol method (48). The procedure for RNA isolation, purification, and quantification has been described previously (51). For each GeneChip experiment, the culturing of bacteria, exposure to hydrogen peroxide or LB control, and subsequent RNA isolation were performed in triplicate on the same day. Equal amounts of each of three preparations were then pooled to a total of 10 μg for cDNA synthesis and hybridization onto a single GeneChip. This procedure was duplicated. Thus, ultimately, two GeneChips per strain and growth condition were scanned at 570 nm with 3-μm resolution by the Affymetrix scanner.
GeneChip microarray analysis.
The generation of cDNA and subsequent biotin-ddUTP terminal-labeling steps were performed as described in the manufacturer's instructions for the P. aeruginosa GeneChip (Affymetrix), using the 10 μg of total RNA mixed with random primers (Invitrogen) and control in vitro transcripts of 10 non-Pseudomonas gene sequences (kindly provided by S. Lory and coworkers, University of Washington), as described previously (51). GeneChip hybridization and washing were carried out following the manufacturer's instructions (Affymetrix) and as described previously (51).
The P. aeruginosa PAO1 GeneChip contains oligonucleotide probes for 5,549 protein-coding genes, 18 tRNA genes, a representative rRNA cluster, and 199 intergenic regions selected from the annotated genome of P. aeruginosa strain PAO1 (47). In addition, there are probes for 117 genes from P. aeruginosa strains other than PAO1 and 14 genes from other species, which can serve as controls (31). Data analysis was performed using the Affymetrix Microarray Suite software (version 5.0) with Affymetrix default parameters. The average microarray hybridization signal intensity was scaled to 150. Two GeneChips for each strain per condition were compared by the four-comparison survival method (3, 5) as follows. The data were imported into a Microsoft Access database capable of searching for genes that significantly changed their signal intensities by the Wilcoxon rank test, with a minimum of a twofold change in all four comparisons. The arithmetic average and the standard deviation of the four comparisons were calculated. As an independent criterion for significantly changed signal intensities, a Bonferroni correction of the signal ratios obtained from the MicroArray Suite software was applied to account for the number of tests (40), which in this case was the total number of open reading frames (ORFs) on the chip. First, the ratio of calibrated and corrected hybridization signals per gene (Si) obtained from cultures grown under identical conditions was verified to follow a Gaussian distribution, and the variance (σ) was calculated. mRNA transcript levels of a gene (i) were considered to be significantly differentially expressed, if the ratio S(i)A/S(i)B or S(i)B/S(i)A exceeds the threshold (1 + uσ), whereby the factor u defines that upper boundary of the normalized Gaussian integral Φ(u) where Φ(u) = xn matches the Bonferroni-corrected 95% confidence interval in the expression (1 − α) = xn (here, n = 5,900, α = 0.025, and 0.975 ≪ x <1.0).
In summary, changes were only classified as significant if they fulfilled the criteria of the four-comparison survival method and exceeded the threshold of the Bonferroni correction for multiple testing. Data were combined with the latest annotation (15 December 2004) from the website of the P. aeruginosa PAO1 sequence and the community annotation project provided at http://www.pseudomonas.com.
Generation of targets from genomic DNA hybridization.
For hybridization of genomic DNA from isolates LES400 and LES431 on the PAO1 GeneChip, 25 μg of genomic DNA from stationary-phase-grown cells was fragmented with 7.5 U of DNase I (Amersham) at 37°C for 10 min. This enzyme produced a majority of fragments in the range of 50 to 200 bp, which is suitable for GeneChip hybridization. The fragmented DNA from two independent genomic DNA preparations for each LES isolate was denatured at 95°C, labeled, and then hybridized on a GeneChip as described for the cDNA expression analysis. The absence or presence of genes was classified as described previously (56).
Exoproduct secretion assays.
All cultures were inoculated in either King's A medium or in LB to an OD600 of approximately 0.05 and then incubated at 37°C with shaking (300 rpm). The amount of pyocyanin in culture supernatants was quantified by measuring the OD695 value (16). LasA protease and elastase activities were measured by determination of the ability of P. aeruginosa culture supernatants to lyse boiled Staphylococcus aureus cells, leading to a decrease in OD600 (15), and by the elastin Congo red assay (38), respectively. In the latter assay, insoluble elastin Congo red was removed by centrifugation, and the increase in the OD495 value in supernatants was used as a measure of elastase activity.
Antimicrobial sensitivity tests and β-lactamase activities.
MICs for antimicrobial agents were determined using E-test strips according to the manufacturer's instructions (AB Biodisk).
PCR amplification and nucleotide sequencing.
Details of the oligonucleotide primers (Sigma-Genosys) used in PCR assays and for nucleotide sequencing will be made available on request. PCR amplicons were purified using S-400 microspin columns (Amersham-Pharmacia Biotech) and sequenced by Lark Technologies, Inc., using the same oligonucleotide primers employed in the PCR amplification and internal primers.
Virulence against fruit flies.
Virulence against Drosophila melanogaster was assessed essentially as described by Chugani et al. (8). The fruit flies were obtained from Blades Biological, Ltd. (Cowden, Kent, United Kingdom), and three independent assays were run consecutively.
RESULTS AND DISCUSSION
Whole-genome comparisons.
PFGE of the three isolates used in the microarray analysis indicated that the LES isolates shared few bands in common with strain PAO1 and differed from each other by two bands (Fig. 1). Size estimations using PFGE gels indicated that the LES did not have a larger genome than strain PAO1.
FIG. 1.
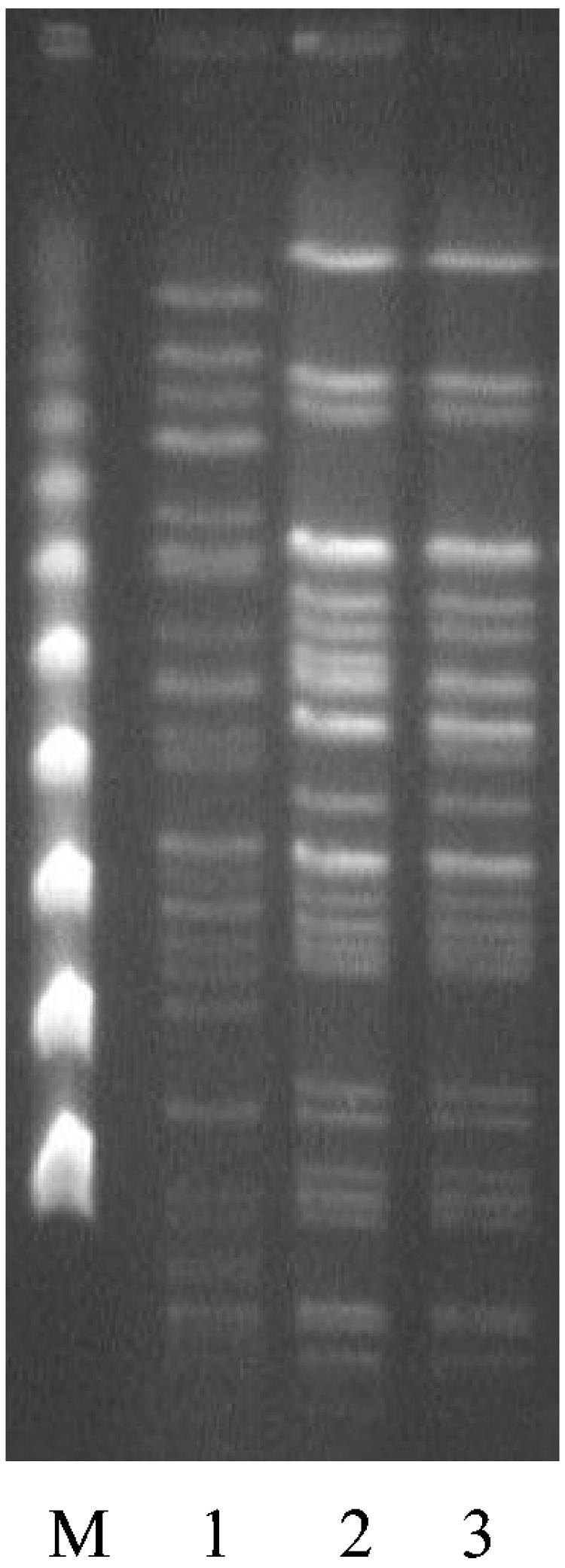
PFGE comparisons of LES isolates and strain PAO1. M, pulse marker, 50 to 1,000 kb (Sigma-Aldrich); lane 1, PAO1; lane 2, LES400; lane 3, LES431.
GeneChip analysis using DNA from isolates LES400 and LES431 indicated the presence in LES strains of at least 95% of PAO1 genes and confirmed the additional presence of O6 antigen genes and PAGI-1 genes, as reported previously (36). In accordance with the known pyoverdine type of the LES (type III) (36), PAO1 pyoverdine sythesis and receptor genes were identified as absent from LES isolates. Among other absent genes/gene clusters were a transport-related cluster (PA0202 to PA0206), two bacteriophage-related clusters (PAO632 to PA0648 and PA0715 to PA0759) and a lipopolysaccharide-related cluster (PA3142 to PA3160). Also absent from LES isolates was the putative polysaccharide-related cluster PA1378 to PA1393, a cluster of type IV pilus genes (PA4525 to PA4527; pilABC), and several other clusters of unknown function (PA0980 to PA0985, PA2100 to PA2106, PA2218 to PA2228, PA2730 to PA2736, and PA3500 to PA3513). PCR assays targeting PA0203, PA0641, and PA1384 (galE) supported the notion that these genes were absent from the LES isolates. Variations in the pilABC cluster between strains of P. aeruginosa have been reported previously (7, 18). Initially, PCR amplification assays using primers designed to the PA4526 (pilB) sequence of strain PAO1 were negative for both LES isolates. However, when primers designed on the basis of conservation between strains PAO1 and PA14 were used (PILBF2 and PILBR2), isolates LES400 and LES431 were PCR positive for pilB, suggesting that the pil genes are present in LES isolates but vary in sequence from their equivalents in strain PAO1.
Although PAGI-1 coding sequences 1 to 31 were detected in the LES isolates, coding sequences 34 to 51 were lacking. This observation was supported by PCR assays for one of the missing genes, suggesting that the LES carries a truncated version of the PAGI-1 island, lacking the region with a G+C content significantly below that of the genome average (23). This island is also only partially present in the other P. aeruginosa strain that has been genome sequenced (PA14). Only minor differences in gene content were detected between the two LES isolates.
Global GeneChip expression analysis.
Gene expression microarrays were used to compare the transcriptomes of the two LES isolates against strain PAO1 and against each other, following growth to late exponential phase in LB and after exposure to hydrogen peroxide. Tables listing all those genes identified as up- or down-regulated are available at C. Winstanley's website (http://www.liv.ac.uk/mmgum/). A summary of the numbers of differentially expressed genes in the various comparisons is shown in Table 1. The following text serves to highlight some of the main features of the expression analysis.
TABLE 1.
Numbers of differentially expressed genes
| Growth condition and changeb | No. of expressed genesa
|
||
|---|---|---|---|
| LES400 vs PAO1 | LES431 vs PAO1 | LES431 vs LES400 | |
| LB up | 222 | 225 | 211 |
| QS activated | 8 | 127 | 142 |
| QS repressed | 5 | 6 | 4 |
| Antimicrobial | 12 | 8 | 5 |
| LB down | 119 (137) | 103 (112) | 137 |
| QS activated | 10 | 6 | 1 |
| QS repressed | 11 | 9 | 3 |
| Antimicrobial | 1 | 1 | 3 |
| H2O2 up | 146 | 349 | 219 |
| QS activated | 18 | 70 | 6 |
| QS repressed | 1 | 3 | 0 |
| Antimicrobial | 5 | 12 | 3 |
| H2O2 down | 350 (367) | 251 (299) | 90 |
| QS activated | 8 | 9 | 11 |
| QS repressed | 10 | 21 | 5 |
| Antimicrobial | 0 | 1 | 3 |
Values for down-regulated genes when LES isolates were compared to strain PAO1 were adjusted by subtracting genes that were recorded as absent from LES isolates. The figure prior to this adjustment is shown in parentheses.
Up- or down-regulation under either growth condition (LB or H2O2 exposed) was at least twofold. QS, number of quorum-sensing regulated genes previously reported as either QS activated or QS repressed (43, 51); antimicrobial, genes associated with antimicrobial susceptibility.
The most striking finding was that in isolate LES431 grown under LB growth conditions and compared to either strain PAO1 or isolate LES400, the majority (56% and 67% for strain PAO1 and isolate LES400, respectively) of up-regulated genes were among those reported previously as activated by quorum sensing (QS) (Tables 1 to 3). Thus, isolate LES431 expressed substantially elevated levels of mRNA transcripts for many of the known P. aeruginosa virulence genes (44, 52), including those encoding alkaline protease, elastase, LasA protease, Clp proteases, CbpD, phenazine biosynthesis, hydrogen cyanide synthesis, aminopeptidase, and lectin. Notably, mRNA levels were elevated to similar ratios whether compared to another member of the same clone (isolate LES400) or to a member of another clone (strain PAO1).
TABLE 3.
Summary of quorum-sensing-related genes that were down regulated in (i) isolate LES400 compared to strain PAO1, (ii) isolate LES431 compared to strain PAO1, and (iii) isolate LES431 compared to isolate LES400
| ORFa | Gene name | LB LES400 ↓ vs PAO1 | H2O2 LES400 ↓ vs PAO1 | LB LES431 ↓ vs PAO1 | H2O2 LES431 ↓ vs PAO1 | LB LES431 ↓ vs 400 | H2O2 LES431 ↓ vs 400 | Product description |
|---|---|---|---|---|---|---|---|---|
| QS activated | ||||||||
| PA0026 | 4.3 | Hypothetical protein | ||||||
| PA0105 | coxB | 6.6 | Cytochrome c oxidase subunit II | |||||
| PA0107 | 14 | Conserved hypothetical protein | ||||||
| PA1317 | cyoA | 13 | Cytochrome o ubiquinol oxidase subunit II | |||||
| PA1319 | cyoC | 4.56 | Cytochrome o ubiquinol oxidase subunit III | |||||
| PA1404 | 4.8 | Hypothetical protein | ||||||
| PA1431 | rsaL | 35 | Regulatory protein RsaL | |||||
| PA1432 | lasI | 12 | Autoinducer synthesis protein LasI | |||||
| PA2365 | 3.2 | Conserved hypothetical protein | ||||||
| PA2445 | gcvP2 | 3.8 | 2.9 | Glycine cleavage system protein P2 | ||||
| PA2446 | gcvH2 | 3.9 | 2.2 | Glycine cleavage system protein H2 | ||||
| PA3181 | 9.7 | 2-Keto-3-deoxy-6-phosphogluconate aldolase | ||||||
| PA3182 | pgl | 2.6 | 6-Phosphogluconolactonase | |||||
| PA3183 | zwf | 5.9 | 2.1 | Glucose-6-phosphate 1-dehydrogenase | ||||
| PA3188 | 74 | 26 | Probable permease of ABC sugar transporter | |||||
| PA3190 | 46 | 9.6 | 38 | 34 | Probable binding protein component of ABC sugar transporter | |||
| PA3195 | gapA | 6.0 | Glyceraldehyde 3-phosphate dehydrogenase | |||||
| PA3369 | 4.0 | Hypothetical protein | ||||||
| PA3418 | ldh | 6.4 | Leucine dehydrogenase | |||||
| PA3691 | 3.6 | Hypothetical protein | ||||||
| PA3692 | 4.5 | Probable outer membrane protein precursor | ||||||
| PA3923 | 5.9 | Hypothetical protein | ||||||
| PA4131 | 7.2 | Probable iron-sulfur protein | ||||||
| PA4133 | 4.1 | Cytochrome c oxidase subunit (cbb3 type) | ||||||
| PA4311 | 5.7 | Conserved hypothetical protein | ||||||
| PA4496 | 8.1 | 4.9 | 2.5 | 11 | Probable binding protein component of ABC transporter | |||
| PA4498 | 14 | 34 | 3.1 | 31 | Probable metallopeptidase | |||
| PA4876 | osmE | 8.0 | Osmotically inducible lipoprotein OsmE | |||||
| PA4880 | 3.3 | Probable bacterioferritin | ||||||
| PA4916 | 12 | Hypothetical protein | ||||||
| PA4917 | 5.0 | Hypothetical protein | ||||||
| PA5027 | 9.5 | Hypothetical protein | ||||||
| QS repressed | ||||||||
| PA0509 | nirN | 4.4 | 12 | Probable c-type cytochrome | ||||
| PA0510 | 40 | Probable uroporphyrin-III c-methyltransferase | ||||||
| PA0512 | 8.4 | 15 | 17 | Conserved hypothetical protein | ||||
| PA1559 | 3.9 | |||||||
| PA2007 | maiA | 18 | Maleylacetoacetate isomerase | |||||
| PA2008 | fahA | 19 | Fumarylacetoacetase | |||||
| PA2009 | hmgA | 7.8 | Homogentisate 1,2-dioxygenase | |||||
| PA2259 | ptxS | 2.8 | 5.1 | Transcriptional regulator PtxS | ||||
| PA2260 | 4.3 | Hypothetical protein | ||||||
| PA2261 | 11 | Probable 2-ketogluconate kinase | ||||||
| PA2540 | 4.8 | Conserved hypothetical protein | ||||||
| PA3174 | 3.2 | Probable transcriptional regulator | ||||||
| PA3205 | 2.6 | Hypothetical protein | ||||||
| PA3364 | amiC | 24 | 4.6 | 12 | Aliphatic amidase expression-regulating protein | |||
| PA3365 | 6.6 | 5.7 | 5.1 | 10 | Probable chaperone | |||
| PA3391 | nosR | 13 | 38 | Regulatory protein NosR | ||||
| PA3392 | nosZ | 62 | 23 | 42 | 56 | Nitrous oxide reductase precursor | ||
| PA3393 | nosD | 5.9 | 4.6 | 7.3 | NosD protein | |||
| PA3394 | nosF | 8.1 | NosF protein | |||||
| PA3575 | 2.7 | Hypothetical protein | ||||||
| PA3662 | 4.2 | 7.7 | 12 | 20 | Hypothetical protein | |||
| PA3790 | oprC | 9.7 | 37 | 7.4 | Putative copper transport outer membrane porin OprC precursor | |||
| PA3872 | narI | 2.8 | Respiratory nitrate reductase gamma chain | |||||
| PA3877 | narK1 | 9.1 | Nitrite extrusion protein 1 | |||||
| PA3913 | 10 | 4.7 | 9.7 | Probable protease | ||||
| PA4442 | cysN | 2.3 | ATP sulfurylase GTP-binding subunit/APS kinase | |||||
| PA4587 | ccpR | 10 | 3.6 | 5.6 | 44 | 12 | Cytochrome c551 peroxidase precursor | |
| PA4770 | lldP | 2.9 | l-lactate permease | |||||
| PA4918 | 3.2 | 4.5 | 35 | 43 | 8.3 | Hypothetical protein |
These genes have been reported previously as being regulated by quorum sensing (either QS activated or QS repressed) (43, 51).
Among the genes up-regulated in isolates LES400 and LES431 compared to strain PAO1 under both growth conditions were several associated with antimicrobial susceptibility, including the ampC β-lactamase gene, the MexAB-OprM and MexXY efflux pumps (Table 4), and the pyochelin biosynthesis genes (PA4221 to PA4231). Regulatory genes associated with alginate production (algU and mucA) were up-regulated following growth in LB only in isolate LES400. Notable genes up-regulated in isolate LES431 following oxidative stress compared to either strain PAO1 or isolate LES400 included a bacteriophage-related gene cluster (PA0611to PA0628).
TABLE 4.
Summary of expression variations for genes associated with antimicrobial susceptibilitya
| ORF | Gene name | LB LES400 ↑ vs PAO1 | H2O2 LES400 ↑ vs PAO1 | LB LES400 ↓ vs PAO1 | LB LES431 ↑ vs PAO1 | H2O2 LES431 ↑ vs PAO1 | LB LES431 ↓ vs PAO1 | H2O2 LES431 ↓vs PAO1 | LB LES431 ↑ vs 400 | H2O2 LES431 ↑ vs 400 | LB LES431 ↓ vs 400 | H2O2 LES431 ↓ vs 400 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PA0424 | mexR | 18 | 9.0 | 7.2 | 10 | |||||||
| PA0425 | mexA | 3.4 | 2.6 | |||||||||
| PA0426 | mexB | 2.4 | ||||||||||
| PA0427 | oprM | 3.9 | 2.9 | |||||||||
| PA0958 | oprD | 16 | 5.0 | 12 | ||||||||
| PA2018 | mexY | 7.9 | 11 | |||||||||
| PA2019 | mexX | 8.0 | 17 | 4.1 | 33 | |||||||
| PA2020 | mexZ | 3.2 | ||||||||||
| PA2493 | mexE | 16 | 66 | 18 | 295 | 5.1 | ||||||
| PA2494 | mexF | 57 | ||||||||||
| PA2495 | oprN | 6.0 | ||||||||||
| PA4110 | ampC | 12 | 46 | 137 | 178 | 11 | 4.0 | |||||
| PA4205 | mexG | 45 | 8.7 | 5.9 | ||||||||
| PA4206 | mexH | 11 | 6.7 | 9.2 | ||||||||
| PA4207 | mexI | 15 | 8.4 | 6.2 | 4.9 | |||||||
| PA4208 | opmD | 4.3 | 8.0 | 5.5 | ||||||||
| PA4599 | mexC | 28 | 10 | 32 | 43 | |||||||
| PA4776 | pmrA | 6.7 | 6.6 | |||||||||
| PA4777 | pmrB | 9.2 |
Arrows indicate isolates in which the gene is up-or down-regulated. LB or H2O2 indicate the two growth conditions used in this study. Values represent the average change in gene expression from replicate experiments.
Genes down-regulated in both LES isolates compared to strain PAO1 under both growth conditions included genes from clusters involved in anaerobic metabolism (nirJLFCMSQ [PA0511 to PA0520], norBC [PA0523 to PA0524], nosRZDF [PA3391 to PA3394], and arcDABC [PA5170 to PA5173]), motility/chemotaxis (flgBCDEGJK [PA1077 to PA1086], fliCD [PA1092 to PA1096], and pctCAB [4307 to 4310]), and twitching motility (pilGIJK [PA0408 to PA0412]). Type IV pilus-associated genes of the cluster pilMNOPQ were down-regulated in isolate LES400 under both growth conditions but in isolate LES431 only under stress. The presence of this cluster has been associated with CF and may confer an early colonization or persistence advantage (18). We confirmed that following growth on Luria agar both LES isolates, unlike strain PAO1, lacked any visible flagella or pili, were nonmotile, and lacked twitching activity (data not shown).
Under oxidative stress, a large cluster of ribosomal proteins (PA4237 to PA4274) was down-regulated in isolate LES400 when compared to either strain PAO1 or isolate LES431. Genes associated with ribosomal biogenesis are known to be down-regulated following exposure to hydrogen peroxide (34). Similarly, several general secretory pathway genes (secA, secB, secE, and secY) were down-regulated in isolate LES400 in comparison to strain PAO1 or isolate LES431. Of the 13 nuo genes mostly clustered at PA2638 to PA2649 and encoding NADH dehydrogenase complex I, 10 were down-regulated in isolate LES400 and 5 were down-regulated in isolate LES431 when compared to strain PAO1. nuoAL genes were down-regulated in isolate LES400 compared to isolate LES431. F1F0 ATP synthase genes (PA5554 to PA5560) were also down-regulated in isolate LES400 compared to either strain PAO1 or isolate LES431. These data indicate a somewhat enhanced oxidative stress response, especially in isolate LES400, compared to strain PAO1.
Under stress, there was modest up-regulation (2.2 to 4.9 fold) of genes associated with DNA repair (recA, lexA, recN; PA0670 to PA0671) and of sodB (superoxide dismutase; 13 fold) in isolate LES431 compared to strain PAO1. recA was also up-regulated in isolate LES400.
Up-regulation of genes involved in antimicrobial susceptibility in LES isolates.
We reported previously that antibiotic susceptibility profiles of LES isolates sharing PFGE genotypes can vary widely (35). The sensitivities of both LES isolates used in the microarray analysis and strain PAO1 to a number of antimicrobial agents are detailed in Table 5. Both LES isolates were less sensitive than strain PAO1 to β-lactams, aminoglycosides, and quinolones but with some notable variations between isolates LES400 and LES431. In particular, isolate LES431 was more resistant to the β-lactams piperacillin (in combination with the β-lactamase inhibitor tazocin) and imipenem (Table 5). Changes in gene expression that are likely to contribute to these variations in antimicrobial susceptibility are shown in Table 4 (12, 13, 19, 33, 50, 58).
TABLE 5.
Antimicrobial agent susceptibility profiles of the isolates
| Antimicrobial agent | MIC value (μg/ml)
|
||
|---|---|---|---|
| LES400 | LES431 | PAO1 | |
| Piperacillin/tazocin | 8 | 64 | 1 |
| Aztreonam | 48 | 32 | 0.75 |
| Ceftazidime | 16 | 16 | 0.5 |
| Imipenem | 0.2 | 4 | 0.38 |
| Meropenem | 6 | 6 | 0.25 |
| Imipenem (+ EDTA) | 0.2 | 1 | <1 |
| Gentamicin | 4 | 2 | 0.5 |
| Amikacin | 16 | 12 | 1 |
| Tobramycin | 1 | 0.75 | 0.19 |
| Colistin | 0.19 | 0.19 | 0.19 |
| Ofloxacin | 4 | 3 | 0.5 |
| Ciprofloxacin | 0.5 | 1 | 0.032 |
| Cotrimoxazole | 2 | 0.25 | 0.15 |
We found a number of mutations in loci associated with antimicrobial susceptibility (summarized in Table 6). Expression of ampC was up-regulated in both LES isolates compared to strain PAO1, but the level of expression in isolate LES431 was >10-fold higher (Table 4). To resolve the variation in ampC expression between the two LES isolates, we sequenced the ampR genes and ampR-ampC intergenic regions, including the start of ampC. The predicted AmpR protein sequence for the LES isolates differed in one internal position compared to PAO1 (D135→G). A similar mutation (D135→N) has been reported previously in a strain exhibiting high levels of β-lactamase activity (2). However, the major difference was the replacement of the ampR ATG start codon with the sequence TTG, a mutation likely to render the sequence untranslatable. Since AmpR is reported to be a positive regulator for ampC (24), this observation was counter to what might be expected. It has been reported that mutations in AmpR can lead to enhanced expression of chromosomal β-lactamase in P. aeruginosa (2), but isolate LES431 in particular is a strain with high constitutive expression of ampC that lacks a functional AmpR, which suggests that activation by AmpR is not a prerequisite for high levels of AmpC.
TABLE 6.
Summary of mutations in regions associated with antimicrobial susceptibility and QS
| Locus | LES isolate(s)a | Mutation(s) | Comment(s) | Relevant reference(s) |
|---|---|---|---|---|
| Antimicrobial | Susceptibility | |||
| mexR | LES400 and LES431 | R83→C | Mutation not reported previously | 13, 24, 32, 57 |
| mexA-R intergenic | LES400 and LES431 | Ribosome-binding site for mexA AGGA → AGGG | Previous report of mutation in the ribosome-binding site leading to down-regulation (AGGA → CGGA) | 13 |
| ampR | LES400 and LES431 | D135→G; | Previous report of D135→N mutation in strain with high levels of β-lactamase activity; disruption of AmpR translation | 2 |
| ampR-C intergenic | LES400 only | ATG→TTG (start codon) −71 with respect to the start codon of ampG5 C→T | Effect of mutation is unknown | |
| ampC | LES400 only | ATG→ATT (start codon) | Disruption of AmpC translation | |
| ampD | LES400 and LES431 | G148→A and S175→L | Same substitutions observed previously in strains with high AmpC activity but discounted as cause of up-regulation | 2, 19 |
| PA4523-ampD intergenic | LES400 and LES431 | 76 bp upstream of the ampD start codon; A→C | Effect of mutation is unknown | |
| mexZ | LES400 and LES431 | Q164→termination (CAA→TAA) | Similar though not identical truncations associated with aminoglycoside resistance among CF isolates | 49 |
| QS related | ||||
| rhlR-rhlI intergenic | LES400 and LES431 | 30 bp upstream of the rhlI start codon, TTTTTTTTCTC → TTTTTTT-CTC | Effect of mutation is unknown | |
| rhlB | LES400 and LES431 | Downstream of P3 (one rhlR transcription start site), AGGGAGGGGGATGCTC → AGGGAGGGGGATGCGC | Effect of mutation is unknown | |
| lasR | LES400 only | Repetition of GGTGCTC leading to divergence from G123 onward | Loss of the HTH DNA-binding LuxR motif | |
| lasR | LES430 only | Deletion of GTGGATGCTC leading to divergence from position W152 onward | Loss of the HTH DNA-binding LuxR motif | |
| lasR | LESB44 only | Insertion of GAAG leading to divergence from position I35 onward | Loss of most of LasR | |
| rsaL-lasI intergenic | LES400 and LES431 | In between the lux-box-like sequence NNCT-(N)12-AGNN and the lasI start codon, located 6 bp away from the lux box-like sequence | Effect of mutation is unknown | |
| PA2587 and gacA intergenic | LES400 and LES431 | 6 bp different from PAO1; nearest mutation was 160 bp upstream from the gacA start codon | Effect of mutation is unknown |
LES isolates refers to LES400 and LES431 only, with the exception of the mutations found in the lasR gene of isolates LES430 and LESB44.
Intriguingly, there were two further mutations in isolate LES400 when compared to both isolate LES431 and strain PAO1, one in the ampR-ampC intergenic region and another leading to mutation of the ampC start codon (Table 6). Thus, although expression of ampC transcription may be up-regulated in isolate LES400 compared to that in strain PAO1, it is likely that the AmpC protein cannot be translated.
Inactivation of the ampD gene has been associated with increased levels of AmpC β-lactamase in P. aeruginosa (2, 19). We amplified and sequenced the ampD genes and the PA4523-ampD intergenic regions of isolates LES400 and LES431 and found them to be identical. The AmpD predicted protein sequence differed in two positions when compared to strain PAO1 (Table 6). Identical substitutions have been observed in more than one other strain associated with high basal levels of AmpC activity (19). However, the same mutations were also present in strains with low basal levels and inducible AmpC, suggesting that such mutations cannot explain the high basal levels observed in strain LES431 (19). In addition, there was a single nucleotide variation between LES strains and strain PAO1 in the PA4523-ampD intergenic region (Table 6).
In support of the gene expression data, using the chromogenic cephalosporin nitrocefin we were able to detect strong β-lactamase activity in isolate LES431. In contrast, β-lactamase activity in isolate LES400 was barely detectable (unpublished data). It has been reported previously that imipenem is not a substrate for the MexAB-OprM or MexXY efflux pumps (27). Since imipenem resistance changes in the presence of a metallo-β-lactamase inhibitor (EDTA) (Table 5), it seems more likely that imipenem resistance and the variation in MICs between the LES isolates are largely due to the production of β-lactamase from the ampC gene. MICs for ceftazidime were the same for LES400 and LES431, suggesting that although ceftazidime resistance has been used to measure of ampC activity (2), other factors can contribute. It has been reported previously that some P. aeruginosa isolates can be more susceptible to β-lactam antibiotics such as ceftazidime despite overproduction of both AmpC and MexAB-OprM, although the mechanisms behind this are unclear (12).
Overproduction of MexAB-OprM can significantly enhance resistance by P. aeruginosa to a range of drugs (17, 22, 32, 57, 58). A number of different mutations associated with overexpression of the MexAB-OprM efflux pump have been reported. nalB mutants arise from nucelotide sequence variations in the adjacent mexR repressor gene (37, 39, 46, 58). Cao et al. (4) have reported that nalC mutants carry mutations in the gene PA3721 (renamed nalC), whose product appears to repress the genes PA3720 and PA3719. Up-regulation of these genes may contribute to the nalC mutant phenotype. However, we did not observe any alterations in expression between LES strains and PAO1 for the PA3719 to PA3721 genes.
We sequenced the mexR gene and the mexA-mexR intergenic region of the two LES isolates. Both LES400 and LES431 carry a single amino acid change in the predicted MexR protein when compared to PAO1. Isolates LES400 and LES431 also carry a single nucleotide change in the intergenic region between mexR and mexA in the putative ribosome-binding site for mexA. In a previous study, Hocquet et al. (12) complemented various mexR mutations but concluded that this efflux pump contributed only marginally to β-lactam and fluoroquinilone resistance. However, it has been suggested that the MexAB-OprM pump plays a role in P. aeruginosa invasiveness and may be involved in the delivery of virulence factors to host cells (11). Hocquet et al. (12) have speculated that overexpression of this efflux system may contribute to the success of an epidemic clone by playing an important role in virulence rather than antibiotic resistance. It has been reported that a QS autoinducer can enhance mexAB-oprM without MexR-mediated regulation (41). This activity is repressed by MexT (PA2492). However, we observed no variation in expression of mexT.
In a study of P. aeruginosa clinical isolates, carbapenem resistance was linked with the loss of or decreased levels of OprD (33). In isolates LES400 and LES431, expression of oprD was down-regulated (Table 4) compared to PAO1. MICs of meropenem were reported to be two to four times higher for isolates expressing MexAB-OprM in a background of low OprD levels (33). It seems likely that these variations in gene expression also contributed to the 24-fold increase in MICs for meropenem in the LES strains compared to PAO1.
The MexXY system enables P. aeruginosa to become resistant to aminoglycosides, tetracyclines, and macrolides (13, 26) and has been specifically implicated in the emergence of resistance to aminoglycosides in CF isolates (50). There was evidence of up-regulation of the MexXY system in LES strains compared to PAO1 under the conditions used for growth in this study (Table 4). Expression of this system is normally inducible by aminoglycosides under the control of the MexZ repressor. It has been demonstrated that mutations in mexZ are associated with overproduction of the MexXY efflux system and increased resistance to aminoglycosides (50), and CF isolates have been shown to overproduce this system constitutively (53). Sequencing of the mexZ genes and mexZ-mexX intergenic regions of isolates LES400 and LES431 revealed that the gene was identical in the two LES isolates, differing by five nucleotides from the PAO1 sequence. Although four of these five mutations were synonymous nucleotide substitutions, the fifth introduced a stop codon leading to premature termination of the MexZ protein after 163 amino acids, compared to the length in strain PAO1 of 210 amino acid residues (Table 6). Similar although not identical truncations have been reported previously and implicated strongly in the development of stable aminoglycoside resistance among CF isolates of P. aeruginosa (50). The mexZ-mexX intergenic regions were identical in the three strains. These data suggest that the mutation in mexZ contributes to the up-regulation of the MexXY system and the greater resistance to aminoglycosides in the LES isolates.
Clinical strains that simultaneously overproduce the MexAB-OprM and MexXY efflux pumps have been reported previously (25). The LES isolates overexpress both efflux pumps and the AmpC β-lactamase. It has been suggested that simultaneous expression of two or three Mex pumps (MexAB-OprM, MexCD-OprJ, and MexEF-OprN) has an additive effect on the MICs of relevant antimicrobial agents (21). Clearly, the LES produces a considerable armory with which to defend itself from antimicrobial agents. Yet as well as some isolates displaying this prowess to the full, LES populations include isolates, such as LES400, with mutations removing some of these weapons.
Premature expression of QS-regulated genes in some LES isolates.
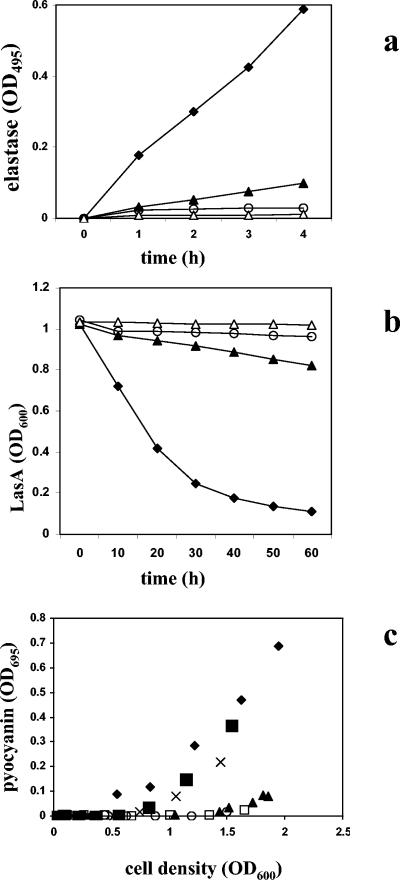
In P. aeruginosa, numerous genes, including many known virulence genes, are regulated by the two lux-like QS systems rhl and las (44, 52), and these genes were up-regulated in isolate LES431 (Tables 1 to 3). Assays for elastase, LasA, and pyocyanin confirmed the high-level expression of these QS-regulated activities in isolate LES431 and indicated premature induction of the QS system (Fig. 2). Interestingly, we detected premature pyocyanin production in an isolate from the infected non-CF father of a CF patient (LES431) and in isolates from the non-CF mother (LES417) and CF patient (LES416) from the same infection episode (Fig. 2c) (29).
FIG. 2.
Expression of QS-regulated phenotypes. Production of elastase (a), LasA (b), and pyocyanin (c) in P. aeruginosa PAO1 (▴), LES400 (○), LES431 (⧫), LES416 (▪), LES417 (x), and LES430 (□). ▵, LB control.
To identify mutations in important QS regulatory genes that could account for the observed variations in transcription of QS-related genes, we sequenced the lasR-rsaL-lasI, rhlIR, vfr, gacA, and qscR regions of the LES400 and LES431 isolates. The mutations found are summarized in Table 6. Significantly, in isolate LES400 alone, there was a repetition of a heptanucleotide sequence (GGTGCTC) within the coding region of LasR, leading to divergence from the normal LasR sequence from position G123 onwards, resulting in the complete loss of the HTH DNA-binding LuxR motif. Interestingly, a second isolate from the non-CF father in the infection episode (LES430) did not share the QS phenotype of isolate LES431 (Fig. 2c). In isolate LES430, there was a 10-bp deletion within the LasR-coding sequence, again leading to loss of the DNA-binding domain (Table 6). We also identified an LES isolate from the CF patient (LESB44) that lacked the unusual QS phenotype and carried a different mutation in lasR (Table 6). Isolates LES430 and LESB44 both had pyocyanin and elastase activities that were similar to those of isolate LES400. The only other difference between LES isolates and PAO1 in the lasR-rsaL-lasI region was a mutation close to the lux-box-like element thought to control both rsaL and lasI expression (Table 6) (54). This mutation was also carried by isolates LES416, LES417, and LES430.
A number of previous studies have identified genes with a role in the regulation of the QS system, including qscR (8), gacA (20), vfr (30), rpoS (43, 55), and rpoN (10). Neither nucleotide sequencing nor analysis of gene expression data yielded any compelling evidence for the involvement of these genes in the QS phenotype of isolate LES431. We did identify in both LES isolates a 6-bp difference in the intergenic region between PA2587 and gacA. gacA is up-regulated in isolate LES400 compared to strain PAO1 (2.4 fold) following growth in LB and in isolate LES431 compared to strain PAO1 following oxidative stress (3.2 fold), but these variations cannot account for the QS phenotype of isolate LES431.
Although the predicted RhlI and RhlR protein sequences for the LES isolates were identical to those of strain PAO1, there was a 1-bp deletion in both LES400 and LES431 30 bp upstream of the rhlI start codon (Table 6). The intergenic region between rhlB and rhlR was identical in all three strains. However, it has been reported that the P. aeruginosa PAO1 rhlR gene has four transcription start sites (P1 to P4), two of which are within the rhlB coding region (30). We observed a 1-bp difference between the LES strains and strain PAO1 near the end of the rhlB coding region (Table 6). This mutation is also present in the sequence reported by Medina et al. (30) as PAO1, even though it differs from the strain PAO1 genome sequence. Since during growth in LB medium, rhlR is expressed from promoter P2, and promoter P3 is thought to be σ54 dependent (30), this mutation is unlikely to play a role in the observed differences in transcription during growth in LB.
A recent report highlighted the existence of QS-deficient clinical isolates of P. aeruginosa (42), concluding that QS-deficient strains are capable of causing infections. In the study by Schaber et al. (42), PCR assays suggested that two of five QS-deficient strains lacked lasR and rhlR genes. In our study, we show that the same strain (LES) can exhibit phenotypes from premature and excessive production of the QS system to QS deficiency, the latter being due to naturally occurring frameshift mutations in the lasR gene. Although we do not unequivocally identify mutations that account for the up-regulation in LES431 and other isolates, the mutation upstream of lasI may play a role in this phenotype.
Evidence from an infection model of the greater virulence of isolate LES431.
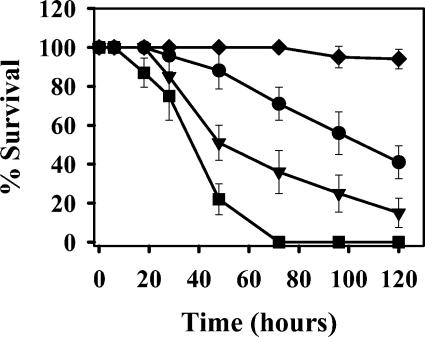
Chugani et al. (8) demonstrated the increased virulence of a qscR mutant of strain PAO1 compared to the wild type in a D. melanogaster infection assay. We compared the virulence of isolate LES431, which shares a similar QS phenotype to qscR mutants, with isolate LES400, which carries a lasR mutation, and strain PAO1. Isolate LES431 clearly gave the phenotype described previously as hypervirulence (Fig. 3). It is interesting that isolate LES400 was more virulent than strain PAO1 in this infection model, which suggests that even LES isolates lacking the QS-regulated virulence genes cannot be considered avirulent.
FIG. 3.
Virulence of P. aeruginosa in fruit flies. Deaths of flies over time when fed P. aeruginosa PAO1 (•), LES400 (▾), LES431 (▪), or buffer (⧫) are shown.
The LES may represent a transmissible, hypervirulent clone.
In common with chronic CF infections by P. aeruginosa, the LES exhibits considerable phenotypic diversity in properties such as antimicrobial sensitivity and colony morphology (34). What distinguishes the LES from most other P. aeruginosa strains is its inherent ability to transmit between CF patients and, in the case of isolate LES431, to cause infections in non-CF parents of CF patients. Although our study does not definitively identify the factors responsible for these additional abilities, we demonstrate the flexibility and adaptability of this strain. In particular, isolate LES431, which because of its history might be considered a highly virulent variant of the clone, has high levels of β-lactamase activity coupled with up-regulation of QS-regulated virulence genes. In contrast, isolate LES400, associated with a chronic CF infection, carries a specific LasR mutation leading to loss of QS activity and has also lost AmpC β-lactamase activity, due to a point mutation in the ampC start codon. It may be that either during the course of chronic CF infections or during laboratory culture, isolate LES400 lost the requirement for these and other properties. However, both LES isolates retained resistance to several other antimicrobials, some of which are due to shared mutations.
von Götz et al. (51) have demonstrated previously that CF strains can evolve into variants expressing higher levels of virulence genes, contradicting the general assumption that selection during CF will have a tendency to reduce such expression. It is possible that the QS phenotype displayed by isolate LES431 may have contributed to its aggressive abilities. This raises the possibility that isolate LES431 represents a novel variant of LES that has evolved from a clone already known to be a successful colonizer of CF patients. However, we have observed that the QS phenotype of isolate LES431 is shared by not only isolates from other CF patients, but also the oldest known LES isolate (from 1988). This suggests that rather than evolving from a successful CF clone, the novel QS phenotype was an intrinsic characteristic of the LES that has been lost subsequently by some isolates, due to mutations such as those we have observed with the lasR gene. It may be that the hypervirulence phenotype is a rare, short-lived phenomenon creating an opportunity for transmission and infection beyond what is normal but placing a burden on the bacterium, leading to the selection for subsequent mutations and then loss of the QS system. In the context of CF infections, the emergence of a hypervirulent, transmissible strain constitutes a potential concern for infection control, which at present is aimed solely at CF patients and not at non-CF parents and health workers.
TABLE 2.
Summary of quorum sensing-related genes that were up-regulated in (i) isolate LES400 compared to strain PAO1, (ii) isolate LES431 compared to strain PAO1, and (iii) isolate LES431 compared to isolate LES400
| ORFa | Gene name | LB LES400 ↑ vs PAO1 | H2O2 LES400 ↑ vs PAO1 | LB LES431 ↑ vs PAO1 | H2O2 LES431 ↑ vs PAO1 | LB LES431 ↑ vs LES400 | H2O2 LES431 ↑ vs LES400 | Product description |
|---|---|---|---|---|---|---|---|---|
| QS activated | ||||||||
| PA0026 | 2.6 | 11 | Hypothetical protein | |||||
| PA0050 | 3.3 | Hypothetical protein | ||||||
| PA0059 | osmC | 4.6 | 6.3 | Osmotically inducible protein OsmC | ||||
| PA0106 | coxA | 64 | 26 | Cytochrome c oxidase, subunit 1 | ||||
| PA0107 | 24 | 21 | 5.0 | Conserved hypothetical protein | ||||
| PA0122 | 15 | 14 | Conserved hypothetical protein | |||||
| PA0143 | nuh | 11 | 24 | Nonspecific ribonucleoside hydrolase | ||||
| PA0176 | 3.1 | Probable chemotaxis transducer | ||||||
| PA0355 | pfpI | 2.8 | Protease PfpI | |||||
| PA0364 | 5.4 | 4.6 | Probable oxidoreductase | |||||
| PA0366 | 4.0 | 3.3 | Probable aldehyde dehydrogenase | |||||
| PA0447 | gcdH | 2.4 | Glutaryl-CoA dehydrogenase | |||||
| PA0567 | 16 | 9.2 | Conserved hypothetical protein | |||||
| PA0586 | 4.3 | 8.2 | Conserved hypothetical protein | |||||
| PA0588 | 5.2 | 6.9 | Conserved hypothetical protein | |||||
| PA0852 | cbpD | 44 | 55 | Chitin-binding protein CbpD precursor | ||||
| PA0996 | pqsA | 3.5 | 9.3 | 16 | Probable coenzyme A ligase | |||
| PA0997 | pqsB | 6.5 | 5.8 | 7.3 | Homologous to β-ketoacyl-acyl carrier protein synthase | |||
| PA0998 | pqsC | 4.6 | 6.5 | 5.0 | Homologous to β-ketoacyl-acyl carrier protein synthase | |||
| PA0999 | pqsD | 5.3 | 5.3 | 6.5 | 3-Oxoacyl-acyl carrier protein synthase III | |||
| PA1000 | pqsE | 6.2 | Quinolone signal response protein | |||||
| PA1001 | phnA | 12.1 | 4.6 | Anthranilate synthase component I | ||||
| PA1173 | napB | 5.6 | Cytochrome c-type protein NapB precursor | |||||
| PA1174 | napA | 3.5 | 4.4 | Periplasmic nitrate reductase protein NapA | ||||
| PA1175 | napD | 3.0 | 5.0 | NapD protein of periplasmic nitrate reductase | ||||
| PA1176 | napF | 6.4 | Ferredoxin protein NapF | |||||
| PA1177 | napE | 5.8 | 4.4 | Periplasmic nitrate reductase protein NapE | ||||
| PA1247 | aprE | 7.5 | Alkaline protease secretion protein AprE | |||||
| PA1248 | aprF | 12 | Alkaline protease secretion outer membrane protein AprF precursor | |||||
| PA1250 | aprI | 16 | 7.5 | Alkaline proteinase inhibitor AprI | ||||
| PA1323 | 11 | 6.7 | Hypothetical protein | |||||
| PA1324 | 7.5 | 5.3 | 3.3 | 5.8 | Hypothetical protein | |||
| PA1404 | 16.9 | 4.1 | 3.8 | 3.6 | Hypothetical protein | |||
| PA1431 | rsaL | 20 | 14 | 655 | 16 | Regulatory protein RsaL | ||
| PA1432 | lasI | 21 | Autoinducer synthesis protein LasI | |||||
| PA1656 | 3.3 | Hypothetical protein | ||||||
| PA1657 | 14 | 11 | Conserved hypothetical protein | |||||
| PA1658 | 4.4 | 24 | Conserved hypothetical protein | |||||
| PA1660 | 11 | Hypothetical protein | ||||||
| PA1662 | 15 | Probable ClpA/B-type protease | ||||||
| PA1667 | 12 | Hypothetical protein | ||||||
| PA1784 | 3.6 | 3.6 | 5.9 | Hypothetical protein | ||||
| PA1869 | 78 | 28 | 68 | Probable acyl carrier protein | ||||
| PA1871 | lasA | 4.9 | 5.2 | 12 | LasA protease precursor | |||
| PA1874 | 5.3 | Hypothetical protein | ||||||
| PA1881 | 31 | 3.8 | Probable oxidoreductase | |||||
| PA1891 | 5.9 | Hypothetical protein | ||||||
| PA1893 | 6.2 | Hypothetical protein | ||||||
| PA1894 | 3.4 | 11 | 4.8 | 6.1 | Hypothetical protein | |||
| PA1895 | 8.9 | 3.8 | Hypothetical protein | |||||
| PA1896 | 11 | Hypothetical protein | ||||||
| PA1897 | 4.2 | 13 | 24 | Hypothetical protein | ||||
| PA1901 | phzC2 | 38 | 126 | Phenazine biosynthesis protein PhzC | ||||
| PA1902 | phzD2 | 49 | 77 | Phenazine biosynthesis protein PhzD | ||||
| PA1903 | phzE2 | 31 | 35 | Phenazine biosynthesis protein PhzE | ||||
| PA1904 | phzF2 | 86 | 91 | Probable phenazine biosynthesis protein | ||||
| PA1905 | phzG2 | 22 | 70 | Probable pyridoxamine 5′-phosphate oxidase | ||||
| PA1999 | 2.8 | 2.6 | Probable CoA transferase, subunit A | |||||
| PA2000 | 2.9 | 2.8 | Probable CoA transferase, subunit B | |||||
| PA2001 | atoB | 3.5 | 2.5 | Acetyl-CoA acetyltransferase | ||||
| PA2030 | 15 | 10 | Hypothetical protein | |||||
| PA2031 | 6.2 | 10 | Hypothetical protein | |||||
| PA2066 | 6.0 | 4.4 | Hypothetical protein | |||||
| PA2067 | 8.5 | 11 | Probable hydrolase | |||||
| PA2068 | 20 | 18 | Probable MFS transporter | |||||
| PA2069 | 25 | 45 | Probable carbamoyl transferase | |||||
| PA2080 | 5.4 | Hypothetical protein | ||||||
| PA2143 | 33 | Hypothetical protein | ||||||
| PA2146 | 29 | 13 | 18 | 14 | Conserved hypothetical protein | |||
| PA2157 | 4.1 | Hypothetical protein | ||||||
| PA2159 | 13 | 3.8 | 12 | Conserved hypothetical protein | ||||
| PA2165 | 5.4 | 5.0 | Probable glycogen synthase | |||||
| PA2166 | 6.5 | 117 | 7.0 | 18 | Hypothetical protein | |||
| PA2171 | 14 | 5.4 | 12 | Hypothetical protein | ||||
| PA2172 | 6.5 | 3.2 | Hypothetical protein | |||||
| PA2180 | 2.3 | Hypothetical protein | ||||||
| PA2190 | 16 | 25 | Conserved hypothetical protein | |||||
| PA2193 | hcnA | 58 | 35 | Hydrogen cyanide synthase HcnA | ||||
| PA2194 | hcnB | 15 | 111 | Hydrogen cyanide synthase HcnB | ||||
| PA2195 | hcnC | 12 | 40 | Hydrogen cyanide synthase HcnC | ||||
| PA2274 | 4.1 | 2.6 | 6.7 | Hypothetical protein | ||||
| PA2300 | chiC | 7.7 | 7.0 | Chitinase | ||||
| PA2305 | 9.4 | 23 | Probable nonribosomal peptide synthetase | |||||
| PA2328 | 4.3 | Hypothetical protein | ||||||
| PA2329 | 4.9 | Probable ATP-binding component of ABC transporter | ||||||
| PA2330 | 2.5 | Hypothetical protein | ||||||
| PA2331 | 3.6 | Hypothetical protein | ||||||
| PA2345 | 2.8 | Conserved hypothetical protein | ||||||
| PA2365 | 32 | 12 | 7.3 | Conserved hypothetical protein | ||||
| PA2366 | 13 | 7.7 | 6.3 | 13 | Conserved hypothetical protein | |||
| PA2367 | 15 | 6.2 | Hypothetical protein | |||||
| PA2423 | 5.6 | Hypothetical protein | ||||||
| PA2433 | 15 | 6.9 | 2.9 | 2.8 | Hypothetical protein | |||
| PA2512 | antA | 7.6 | Anthranilate dioxygenase large subunit | |||||
| PA2513 | antB | 18 | 50 | Anthranilate dioxygenase small subunit | ||||
| PA2552 | 2.2 | Probable acyl-CoA dehydrogenase | ||||||
| PA2553 | 2.7 | Probable acyl-CoA thiolase | ||||||
| PA2587 | pqsH | 5.7 | 28 | Probable FAD-dependent mono-oxygenase | ||||
| PA2588 | 26 | 22 | Probable transcriptional regulator | |||||
| PA2591 | vqsR | 6.0 | 12 | Probable transcriptional regulator | ||||
| PA2592 | 4.3 | 5.4 | 4.9 | Probable periplasmic spermidine/putrescine-binding protein | ||||
| PA2747 | 6.6 | 5.4 | Hypothetical protein | |||||
| PA2939 | 147 | 134 | Probable aminopeptidase | |||||
| PA3032 | snr1 | 10 | 5.9 | Cytochrome c Snr1 | ||||
| PA3104 | xcpP | 3.7 | Secretion protein XcpP | |||||
| PA3181 | 5.4 | 3.5 | 10 | 2-Keto-3-deoxy-6-phosphogluconate aldolase | ||||
| PA3182 | pgl | 5.7 | 3.8 | 4.8 | 6-Phosphogluconolactonase | |||
| PA3183 | zwf | 6.2 | 2.9 | 11 | Glucose-6-phosphate 1-dehydrogenase | |||
| PA3326 | 14 | 11 | 14 | Probable Clp family ATP-dependent protease | ||||
| PA3327 | 5.6 | Probable nonribosomal peptide synthetase | ||||||
| PA3328 | 6.1 | 9.1 | 8.1 | Probable FAD-dependent mono-oxygenase | ||||
| PA3329 | 37 | 34 | 33 | Hypothetical protein | ||||
| PA3330 | 34 | 46 | 55 | Probable short-chain dehydrogenase | ||||
| PA3331 | 6.1 | 16 | 8.6 | Cytochrome P450 | ||||
| PA3332 | 26 | 30 | 13 | Conserved hypothetical protein | ||||
| PA3333 | fabH2 | 6.6 | 52 | 6.0 | 3-Oxoacyl-acyl carrier protein synthase III | |||
| PA3334 | 20 | Probable acyl carrier protein | ||||||
| PA3335 | 19 | Hypothetical protein | ||||||
| PA3347 | 3.1 | Hypothetical protein | ||||||
| PA3361 | lecB | 26 | 6.0 | Fucose-binding lectin PA-IIL | ||||
| PA3369 | 9.4 | 11 | 6.3 | Hypothetical protein | ||||
| PA3370 | 5.2 | 4.8 | Hypothetical protein | |||||
| PA3418 | ldh | 10 | 4.2 | 3.3 | Leucine dehydrogenase | |||
| PA3476 | rhlI | 11 | Autoinducer synthesis protein RhlI | |||||
| PA3477 | rhlR | 6.5 | 2.7 | 12 | Transcriptional regulator RhlR | |||
| PA3478 | rhlB | 12 | 9.3 | 57 | Rhamnosyltransferase chain B | |||
| PA3479 | rhlA | 28 | 16 | 46 | Rhamnosyltransferase chain A | |||
| PA3520 | 38 | 9.0 | Hypothetical protein | |||||
| PA3535 | 10 | 12 | Probable serine protease | |||||
| PA3688 | 4.0 | Hypothetical protein | ||||||
| PA3691 | 11 | 8.7 | 2.9 | 8.5 | Hypothetical protein | |||
| PA3692 | 8.2 | 6.5 | 7.8 | Probable outer membrane protein precursor | ||||
| PA3724 | lasB | 413 | 4.0 | 597 | Elastase LasB | |||
| PA3888 | 3.4 | 2.8 | Probable permease of ABC transporter | |||||
| PA3904 | 9.9 | 21 | Hypothetical protein | |||||
| PA3906 | 9.5 | 21 | Hypothetical protein | |||||
| PA3907 | 7.4 | 24 | Hypothetical protein | |||||
| PA3923 | 5.9 | 27 | Hypothetical protein | |||||
| PA4117 | 3.2 | Probable bacteriophytochrome | ||||||
| PA4129 | 6.7 | 4.1 | Hypothetical protein | |||||
| PA4130 | 7.2 | 3.8 | Probable sulfite or nitrite reductase | |||||
| PA4131 | 6.6 | 7.0 | Probable iron-sulfur protein | |||||
| PA4133 | 3.9 | 28 | 16 | Cytochrome c oxidase subunit (cbb3 type) | ||||
| PA4134 | 4.9 | 42 | Hypothetical protein | |||||
| PA4139 | 6.9 | 3.8 | Hypothetical protein | |||||
| PA4141 | 66 | 33 | 102 | Hypothetical protein | ||||
| PA4142 | 9.6 | 5.5 | Probable secretion protein | |||||
| PA4171 | 10 | 13 | Probable protease | |||||
| PA4175 | prpL | 19 | 52 | Pvds-regulated endoprotease, lysyl class | ||||
| PA4205 | mexG | 45 | 8.7 | 5.9 | Hypothetical protein | |||
| PA4206 | mexH | 11 | 6.7 | 9.2 | Probable RND efflux membrane fusion protein precursor | |||
| PA4207 | mexI | 15 | 8.4 | 6.2 | 4.9 | Probable RND efflux transporter | ||
| PA4208 | opmD | 4.3 | 8.0 | 5.5 | Probable outer membrane protein precursor | |||
| PA4209 | phzM | 31 | 37 | Probable phenazine-specific methyltransferase | ||||
| PA4210 | phzA1 | 19 | 27 | Probable phenazine biosynthesis protein | ||||
| PA4211 | phzB1 | 124 | 525 | Probable phenazine biosynthesis protein | ||||
| PA4217 | phzS | 55 | 69 | Flavin-containing mono-oxygenase | ||||
| PA4296 | 4.1 | 8.1 | Probable two-component response regulator | |||||
| PA4306 | 9.0 | Hypothetical protein | ||||||
| PA4311 | 13 | 4.0 | 2.9 | Conserved hypothetical protein | ||||
| PA4496 | 3.4 | Probable binding protein component of ABC transporter | ||||||
| PA4498 | 4.6 | Probable metallopeptidase | ||||||
| PA4590 | pra | 27 | 10 | Protein activator | ||||
| PA4648 | 10 | 18 | Hypothetical protein | |||||
| PA4649 | 19 | Hypothetical protein | ||||||
| PA4738 | 7.8 | 19 | 5.0 | 18 | Conserved hypothetical protein | |||
| PA4739 | 13 | 26 | 8.1 | 18 | Conserved hypothetical protein | |||
| PA4778 | 6.4 | 6.6 | Probable transcriptional regulator | |||||
| PA4869 | 5.4 | Hypothetical protein | ||||||
| PA4876 | osmE | 8.3 | 4.6 | Osmotically inducible lipoprotein OsmE | ||||
| PA4880 | 6.1 | 8.2 | Probable bacterioferritin | |||||
| PA5058 | phaC2 | 3.1 | 4.1 | Poly(3-hydroxyalkanoic acid) synthase 2 | ||||
| PA5061 | 3.1 | 2.9 | Conserved hypothetical protein | |||||
| PA5220 | 26 | 21 | Hypothetical protein | |||||
| PA5481 | 38 | 8.7 | 29 | Hypothetical protein | ||||
| PA5482 | 19 | 12 | 29 | Hypothetical protein | ||||
| QS repressed | ||||||||
| PA0887 | acsA | 3.5 | Acetyl-coenzymeA synthetase | |||||
| PA1559 | 15 | 4.1 | Hypothetical protein | |||||
| PA2007 | maiA | 15 | 16 | Maleylacetoacetate isomerase | ||||
| PA2008 | fahA | 8.1 | 6.9 | Fumarylacetoacetase | ||||
| PA2009 | hmgA | 7.8 | 9.5 | Homogentisate 1,2-dioxygenase | ||||
| PA2540 | 28 | Conserved hypothetical protein | ||||||
| PA3038 | 2.3 | 2.8 | Probable porin | |||||
| PA3234 | 3.6 | 4.0 | Probable sodium:solute symporter | |||||
| PA3235 | 2.9 | 2.9 | 5.4 | Conserved hypothetical protein | ||||
| PA3205 | 6.7 | 2.7 | Hypothetical protein |
These genes have been reported previously as being regulated by quorum sensing (either QS regulated or QS repressed) (43, 51). CoA, coenzyme A; MFS, major facilitator superfamily; FAD, flavin adenine dinucleotide.
Acknowledgments
We acknowledge the Pseudomonas Genome Project and the Pseudomonas aeruginosa Community Annotation Project. We are grateful for the excellent technical assistance of Tanja Toepfer during microarray hybridization and to John Corkill for assistance with PFGE.
C.A.H. and C.W. acknowledge funding from the United Kingdom Cystic Fibrosis Trust.
REFERENCES
- 1.Al Aloul, M., J. Crawley, C. Winstanley, C. A. Hart, M. J. Ledson, and M. J. Walshaw. 2004. Increased morbidity associated with chronic infection by an epidemic Pseudomonas aeruginosa strain in CF patients. Thorax 59:334-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagge, N., O. Ciofu, M. Hentzer, J. I. Campbell, M. Givskov, and N. Hoiby. 2002. Constitutive high expression of chromosomal β-lactamase in Pseudomonas aeruginosa caused by a new insertion sequence (IS1669) located in ampD. Antimicrob. Agents Chemother. 46:3406-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakay, M. Y., Y. W. Chen, R. Borup, P. Zhao, K. Nagaraju, and E. P. Hoffman. 2002. Sources of variability and effect of experimental approach on expression profiling data interpretation. BMC Bioinformatics 3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao, L., R. Srikumar, and K. Poole. 2004. MexAB-OprM hyperexpression in NalC-type multidrug-resistant Pseudomonas aeruginosa: identification and characterization of the nalC gene encoding a repressor of PA3720-PA3719. Mol. Microbiol. 53:1423-1436. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Y. W., P. Zhao, R. Borup, and E. P. Hoffman. 2000. Expression profiling in the muscular dystrophies: identification of novel aspects of molecular pathophysiology. J. Cell Biol. 151:1321-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, K., R. L. Smyth, J. R. Govan, C. Doherty, C. Winstanley, N. Denning, D. P. Heaf, H. van Saene, and C. A. Hart. 1996. Spread of beta-lactam-resistant Pseudomonas aeruginosa in a cystic fibrosis clinic. Lancet 348:639-642. [DOI] [PubMed] [Google Scholar]
- 7.Choi, J. Y., C. D. Sifri, B. C. Goumnerov, L. G. Rahme, F. M. Ausubel, and S. B. Calderwood. 2002. Identification of virulence genes in a pathogenic strain of Pseudomonas aeruginosa by representational difference analysis. J. Bacteriol. 184:952-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chugani, S. A., M. Whiteley, K. M. Lee, D. D'Argenio, C. Manoil, and E. P. Greenberg. 2001. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 98:2752-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst, R. K., D. A. D'Argenio, J. K. Ichikawa, M. G. Bangera, S. Selgrade, J. L. Burns, P. Hiatt, K. McCoy, M. Brittnacher, A. Kas, D. H. Spencer, M. V. Olson, B. W. Ramsey, S. Lory, and S. I. Miller. 2003. Genome mosaicism is conserved but not unique in Pseudomonas aeruginosa isolates from the airways of young children with cystic fibrosis. Environ. Microbiol. 5:1341-1349. [DOI] [PubMed] [Google Scholar]
- 10.Heurlier, K., V. Denervaud, G. Pessi, C. Reimmann, and D. Haas. 2003. Negative control of quorum sensing by RpoN (σ54) in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:2227-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirakata, Y., R. Srikumar, K. Poole, N. Gotoh, T. Suematsu, S. Kohno, S. Kamihira, R. E. Hancock, and D. P. Speert. 2002. Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J. Exp. Med. 196:109-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hocquet, D., X. Bertrand, T. Kohler, D. Talon, and P. Plesiat. 2003. Genetic and phenotypic variations of a resistant Pseudomonas aeruginosa epidemic clone. Antimicrob. Agents Chemother. 47:1887-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hocquet, D., C. Vogne, F. El Garch, A. Vejux, N. Gotoh, A. Lee, O. Lomovskaya, and P. Plesiat. 2003. MexXY-OprM efflux pump is necessary for adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 47:1371-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, A. M., J. R. Govan, C. J. Doherty, M. E. Dodd, B. J. Isalska, T. N. Stanbridge, and A. K. Webb. 2001. Spread of a multiresistant strain of Pseudomonas aeruginosa in an adult cystic fibrosis clinic. Lancet 358:557-558. [DOI] [PubMed] [Google Scholar]
- 15.Kessler, E., M. Safrin, J. C. Olson, and D. E. Ohman. 1993. Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J. Biol. Chem. 268:7503-7508. [PubMed] [Google Scholar]
- 16.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 17.Kohler, T., M. Michea-Hamzehpour, U. Henze, N. Gotoh, L. K. Curty, and J. C. Pechere. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23:345-354. [DOI] [PubMed] [Google Scholar]
- 18.Kus, J. V., E. Tullis, D. G. Cvitkovitch, and L. L. Burrows. 2004. Significant differences in type IV pilin allele distribution among Pseudomonas aeruginosa isolates from cystic fibrosis (CF) versus non-CF patients. Microbiology 150:1315-1326. [DOI] [PubMed] [Google Scholar]
- 19.Langaee, T. Y., L. Gagnon, and A. Huletsky. 2000. Inactivation of the ampD gene in Pseudomonas aeruginosa leads to moderate-basal-level and hyperinducible AmpC β-lactamase expression. Antimicrob. Agents Chemother. 44:583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ledgham, F., I. Ventre, C. Soscia, M. Foglino, J. N. Sturgis, and A. Lazdunski. 2003. Interactions of the quorum sensing regulator QscR: interaction with itself and the other regulators of Pseudomonas aeruginosa LasR and RhlR. Mol. Microbiol. 48:199-210. [DOI] [PubMed] [Google Scholar]
- 21.Lee, A., W. Mao, M. S. Warren, A. Mistry, K. Hoshino, R. Okumura, H. Ishida, and O. Lomovskaya. 2000. Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J. Bacteriol. 182:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, X. Z., H. Nikaido, and K. Poole. 1995. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang, X., X. Q. Pham, M. V. Olson, and S. Lory. 2001. Identification of a genomic island present in the majority of pathogenic isolates of Pseudomonas aeruginosa. J. Bacteriol. 183:843-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindquist, S., F. Lindberg, and S. Normark. 1989. Binding of the Citrobacter freundii AmpR regulator to a single DNA site provides both autoregulation and activation of the inducible ampC β-lactamase gene. J. Bacteriol. 171:3746-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Llanes, C., D. Hocquet, C. Vogne, D. Benali-Baitich, C. Neuwirth, and P. Plesiat. 2004. Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob. Agents Chemother. 48:1797-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCallum, S. J., J. Corkill, M. Gallagher, M. J. Ledson, C. A. Hart, and M. J. Walshaw. 2001. Superinfection with a transmissible strain of Pseudomonas aeruginosa in adults with cystic fibrosis chronically colonised by P aeruginosa. Lancet 358:558-560. [DOI] [PubMed] [Google Scholar]
- 29.McCallum, S. J., M. J. Gallagher, J. E. Corkill, C. A. Hart, M. J. Ledson, and M. J. Walshaw. 2002. Spread of an epidemic Pseudomonas aeruginosa strain from a patient with cystic fibrosis (CF) to non-CF relatives. Thorax 57:559-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medina, G., K. Juarez, R. Diaz, and G. Soberon-Chavez. 2003. Transcriptional regulation of Pseudomonas aeruginosa rhlR, encoding a quorum-sensing regulatory protein. Microbiology 149:3073-3081. [DOI] [PubMed] [Google Scholar]
- 31.Ochsner, U. A., P. J. Wilderman, A. I. Vasil, and M. L. Vasil. 2002. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45:1277-1287. [DOI] [PubMed] [Google Scholar]
- 32.Okamoto, K., N. Gotoh, and T. Nishino. 2001. Pseudomonas aeruginosa reveals high intrinsic resistance to penem antibiotics: penem resistance mechanisms and their interplay. Antimicrob. Agents Chemother. 45:1964-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pai, H., J. Kim, J. Kim, J. H. Lee, K. W. Choe, and N. Gotoh. 2001. Carbapenem resistance mechanisms in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 45:480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palma, M., D. DeLuca, S. Worgall, and L. E. Quadri. 2004. Transcriptome analysis of the response of Pseudomonas aeruginosa to hydrogen peroxide. J. Bacteriol. 186:248-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panagea, S., C. Winstanley, Y. N. Parsons, M. J. Walshaw, M. J. Ledson, and C. A. Hart. 2003. PCR-based detection of a cystic fibrosis epidemic strain of Pseudomonas aeruginosa. Mol. Diagn. 7:195-200. [DOI] [PubMed] [Google Scholar]
- 36.Parsons, Y. N., S. Panagea, C. H. Smart, M. J. Walshaw, C. A. Hart, and C. Winstanley. 2002. Use of subtractive hybridization to identify a diagnostic probe for a cystic fibrosis epidemic strain of Pseudomonas aeruginosa. J. Clin. Microbiol. 40:4607-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poole, K., K. Tetro, Q. Zhao, S. Neshat, D. E. Heinrichs, and N. Bianco. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rust, L., C. R. Messing, and B. H. Iglewski. 1994. Elastase assays. Methods Enzymol. 235:554-562. [DOI] [PubMed] [Google Scholar]
- 39.Saito, K., H. Yoneyama, and T. Nakae. 1999. nalB-type mutations causing the overexpression of the MexAB-OprM efflux pump are located in the mexR gene of the Pseudomonas aeruginosa chromosome. FEMS Microbiol. Lett. 179:67-72. [DOI] [PubMed] [Google Scholar]
- 40.Salunkhe, P., F. von Götz, L. Wiehlmann, J. Lauber, J. Buer, and B. Tümmler. 2002. GeneChip expression analysis of the response of Pseudomonas aeruginosa to paraquat-induced superoxide stress. Genome Lett. 4:165-174. [Google Scholar]
- 41.Sawada, I., H. Maseda, T. Nakae, H. Uchiyama, and N. Nomura. 2004. A quorum-sensing autoinducer enhances the mexAB-oprM efflux-pump expression without the MexR-mediated regulation in Pseudomonas aeruginosa. Microbiol. Immunol. 48:435-439. [DOI] [PubMed] [Google Scholar]
- 42.Schaber, J. A., N. L. Carty, N. A. McDonald, E. D. Graham, R. Cheluvappa, J. A. Griswold, and A. N. Hamood. 2004. Analysis of quorum sensing-deficient clinical isolates of Pseudomonas aeruginosa. J. Med. Microbiol. 53:841-853. [DOI] [PubMed] [Google Scholar]
- 43.Schuster, M., A. C. Hawkins, C. S. Harwood, and E. P. Greenberg. 2004. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 51:973-985. [DOI] [PubMed] [Google Scholar]
- 44.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott, F. W., and T. L. Pitt. 2004. Identification and characterization of transmissible Pseudomonas aeruginosa strains in cystic fibrosis patients in England and Wales. J. Med. Microbiol. 53:609-615. [DOI] [PubMed] [Google Scholar]
- 46.Srikumar, R., C. J. Paul, and K. Poole. 2000. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 182:1410-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 48.Tao, H., C. Bausch, C. Richmond, F. R. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogne, C., J. R. Aires, C. Bailly, D. Hocquet, and P. Plesiat. 2004. Role of the multidrug efflux system MexXY in the emergence of moderate resistance to aminoglycosides among Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob. Agents Chemother. 48:1676-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Götz, F., S. Haussler, D. Jordan, S. S. Saravanamuthu, D. Wehmhoner, A. Strussmann, J. Lauber, I. Attree, J. Buer, B. Tummler, and I. Steinmetz. 2004. Expression analysis of a highly adherent and cytotoxic small colony variant of Pseudomonas aeruginosa isolated from a lung of a patient with cystic fibrosis. J. Bacteriol. 186:3837-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whiteley, M., and E. P. Greenberg. 2001. Promoter specificity elements in Pseudomonas aeruginosa quorum-sensing-controlled genes. J. Bacteriol. 183:5529-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whiteley, M., M. R. Parsek, and E. P. Greenberg. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 182:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolfgang, M. C., B. R. Kulasekara, X. Liang, D. Boyd, K. Wu, Q. Yang, C. G. Miyada, and S. Lory. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:8484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao, Q., X. Z. Li, A. Mistry, R. Srikumar, L. Zhang, O. Lomovskaya, and K. Poole. 1998. Influence of the TonB energy-coupling protein on efflux-mediated multidrug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:2225-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ziha-Zarifi, I., C. Llanes, T. Kohler, J. C. Pechere, and P. Plesiat. 1999. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob. Agents Chemother. 43:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]