Abstract
Hierarchical control ensures that facultative bacteria preferentially use the available respiratory electron acceptor with the most positive standard redox potential. Thus, nitrate is used before other electron acceptors such as fumarate for anaerobic respiration. Nitrate regulation is mediated by the NarX-NarL two-component system, which activates the transcription of operons encoding nitrate respiration enzymes and represses the transcription of operons for other anaerobic respiratory enzymes, including enzymes involved in fumarate respiration. These are fumarate reductase (encoded by the frdABCD operon), fumarase B, which generates fumarate from malate, and the DcuB permease for fumarate, malate, and aspartate. The transcription of the corresponding structural genes is activated by the DcuS-DcuR two-component system in response to fumarate or its dicarboxylate precursors. We report results from preliminary transcription microarray experiments that revealed two previously unknown members of the NarL regulon: the aspA gene encoding aspartate-ammonia lyase, which generates fumarate; and the dcuSR operon encoding the dicarboxylate-responsive regulatory system. We measured beta-galactosidase expression from monocopy aspA-lacZ, frdA-lacZ, and dcuS-lacZ operon fusions in response to added nitrate and fumarate and with respect to the dcuR and narL genotypes. Nitrate, acting through the NarX-NarL regulatory system, repressed the transcription of all three operons. Only frdA-lacZ expression, however, was responsive to added fumarate or a dcuR+ genotype. Phospho-NarL protein protected operator sites in the aspA and dcuS promoter regions from DNase I cleavage in vitro. The overall results are consistent with the hypothesis that nitrate represses frdA operon transcription not only directly, by repressing frdA promoter activity, but also indirectly, by repressing dcuS promoter activity.
As facultative aerobes, enterobacteria such as Escherichia coli use a variety of electron acceptors for anaerobic respiration. The synthesis of terminal respiratory enzymes is subject to hierarchical control (22) to ensure that respiratory oxidants are consumed preferentially, with oxygen used first, nitrate (NO3−) second, and finally, the other acceptors—nitrite (NO2−), a group of S and N oxides exemplified by dimethyl sulfoxide, and fumarate (51, 61). Respiratory chain composition thereby is adjusted in response to the environment to maintain optimal proton motive force (60).
The first level of hierarchical control involves response to oxygen. Aerobic cultures synthesize only basal levels of anaerobic respiratory enzymes, but in anaerobic cultures the oxygen-sensitive Fnr protein activates anaerobic respiratory gene transcription (19, 30). The second level of hierarchical control, superimposed on Fnr control, involves response to nitrate. This efficient respiratory oxidant induces synthesis of nitrate respiratory enzymes and simultaneously represses synthesis of enzymes for respiring the lower-potential acceptors. Additionally, nitrate reduction results in an accumulation of nitrite, which induces synthesis of respiratory nitrite reductase after nitrate is depleted (13, 55, 62). Nitrate and nitrite regulation is effected by the paralogous NarX-NarL and NarQ-NarP sensor-regulator two-component systems (57).
Aerobic and anaerobic respiration are intertwined with the citrate cycle, and enterobacteria regulate citrate cycle enzyme synthesis in response to both carbon source and respiratory oxidant. Enzyme synthesis is activated by the cyclic AMP receptor protein (Crp; also known as catabolite activator protein, Cap). Thus, glucose-grown E. coli synthesizes certain citrate cycle enzymes at low levels and thereby excretes substantial amounts of carbon from glucose as acetate rather than as CO2 (12). Citrate cycle enzyme synthesis is also decreased during anaerobic growth due to transcriptional repression by the ArcB-ArcA two component system (23). In particular, the pathway from 2-oxoglutarate to fumarate is sharply decreased in anaerobic cultures, in part to prevent the accumulation of ubiquinol (QH2) resulting from succinate oxidation. Instead, the cycle is reconfigured to form the reductive branch operating from oxaloacetate to succinyl-coenzyme A (Fig. 1A). The reductive branch generates metabolic precursors (aspartate and succinyl-coenzyme A) and also helps to maintain overall redox balance by regenerating NAD+ (12, 51).
FIG. 1.
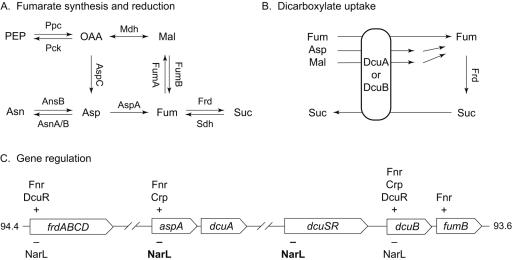
Reductive branch of the citrate cycle. (A) Pathways to fumarate. Reactions for both the reductive and oxidative modes of the citrate cycle are shown. For simplicity, additional reaction components such as NADH are not shown. PEP, phosphoenol pyruvate; OAA, oxaloacetate; Mal, malate; Fum, fumarate; Suc, succinate; Ppc, PEP carboxylase; Pck, PEP carboxykinase; Mdh, malate dehydrogenase; AnsB, asparaginase II; AsnA/B, asparagine synthetases I and II; Frd, fumarate reductase; Sdh, succinate dehydrogenase; AspC, aspartate-ammonia lyase; FumA and FumB, fumarases A and B, respectively. (B) The DcuA and DcuB antiporters catalyze the exchange of fumarate, malate, and aspartate in exchange for succinate. (C) Regulators of gene expression. The region between 93.6 and 94.4 centisomes on the E. coli genome is shown; broken lines indicate intervening genes not depicted. Positive and negative transcriptional regulation is indicated by + and −, respectively. Repression of aspA and dcuS transcription by NarL protein, reported in this study, is indicated in boldface.
Fumarate reduction to succinate functions in anaerobic respiration (59, 60). Therefore, an additional task of the citrate cycle reductive branch is to produce fumarate as a respiratory acceptor. In carbon-rich environments, such as LB broth, fumarate is manufactured from C4-dicarboxylates and related compounds, including oxaloacetate, malate, and aspartate (Fig. 1A). Feeding these pathways are the Dcu carriers (Fig. 1B), which import aspartate, malate, and fumarate in exchange for succinate (25).
Synthesis of enzymes for C4-dicarboxylate utilization is controlled by the DcuS-DcuR two-component system (17, 67). The DcuS sensor responds to fumarate and other C4-dicarboxylates to control the phosphorylation of the DcuR response regulator (2, 24). The phospho-DcuR protein is known to activate the transcription of three operons: the dcuB-fumB operon encoding the DcuB carrier and fumarase B, the frdABCD operon encoding fumarate reductase (Fig. 1C), and the dctA gene encoding the aerobic C4-dicarboxylate transporter (2, 17, 18, 67).
Like oxygen, nitrate can serve as an acceptor for QH2 oxidation, and therefore nitrate-respiring cultures use the conventional citrate cycle during growth conditions where catabolite repression does not inhibit enzyme synthesis (44). For example, anaerobic repression of succinate dehydrogenase synthesis is relieved during nitrate respiration (21). Additionally, nitrate, acting via the NarL response regulator, represses the transcription of the frdA and dcuB-fumB operons (Fig. 1C) (18, 22, 27, 53). Therefore, this aspect of hierarchical control serves also to minimize respiratory fumarate reduction, a competitor to the more favorable respiratory succinate oxidation.
Here we report results that extend our understanding of hierarchical control in the context of fumarate respiration. First, we found that nitrate represses dcuSR operon transcription in narL+ strains. This suggests that phospho-NarL may repress frdA operon expression indirectly by repressing synthesis of the DcuS-DcuR activator of frdA operon transcription initiation in addition to directly repressing transcription from the frdA promoter. Second, we found that nitrate represses aspA gene transcription, providing yet another means to limit the availability of fumarate for respiration (Fig. 1). Finally, our results indicate that the DcuS-DcuR system does not control aspA or dcuSR operon transcription. The absence of dcuSR autoregulation was previously suggested by Abo-Amer et al. (2).
MATERIALS AND METHODS
Strains and plasmids.
Strains and plasmids are listed in Table 1. Control region sequences for the aspA and dcuSR operons are depicted in Fig. 2. Genetic crosses were performed by use of P1kc-mediated generalized transduction (43). Null alleles of nar regulatory genes (Table 1) have been described previously (47). Standard methods were used for restriction endonuclease digestion, ligation, transformation, and PCR amplification of DNA (37).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype | Reference or source |
|---|---|---|
| E. coli | ||
| IMW205 | dcuR::Km | 67 |
| VJS632 | F− λ− prototroph | 56 |
| VJS676 | As VJS632 but Δ(argF-lacIZYA)U169 | 56 |
| Derivatives of strain VJS676 | ||
| VJS2197 | λΦ(narG-lacZ) | 46 |
| VJS4033 | λΦ(narG-lacZ) ΔnarX242 narL505 ychO2084::Ω-Cm narQ251::Tn10d(Tc) | 46 |
| VJS4339 | λΦ(fdnG-lacZ) Δ(narXL-ychO)240::Km narQ251::Tn10d(Tc) narP253::Tn10d(Cm) | 47 |
| VJS4341 | λΦ(fdnG-lacZ) narQ251::Tn10d(Tc) narP253::Tn10d(Cm) | 47 |
| VJS3046 | λΦ(frdA-lacZ) | 47 |
| VJS3052 | λΦ(frdA-lacZ) narL215::Tn10 | 47 |
| VJS4330 | λΦ(frdA-lacZ) narP253::Tn10d(Cm) | 47 |
| VJS4331 | λΦ(frdA-lacZ) narL215::Tn10 narP253::Tn10d(Cm) | 47 |
| VJS8515 | λ− Δ(att-lom)::bla {Φ(aspA-lacZ)} | This study |
| VJS8535 | λΦ(frdA-lacZ) dcuR::Km | This study |
| VJS8537 | λ− Δ(att-lom)::bla {Φ(aspA-lacZ)}/dcuR::Km | This study |
| VJS8559 | λ− Δ(att-lom)::bla {Φ(aspA-lacZ)}/narL215::Tn10 | This study |
| VJS8561 | λ− Δ(att-lom)::bla {Φ(aspA-lacZ)}/narL215::Tn10 narP253::Tn10d(Cm) | This study |
| VJS8566 | λ− Δ(att-lom)::bla {Φ(aspA-lacZ)}/narP253::Tn10d(Cm) | This study |
| VJS9009 | λ− Δ(att-lom)::bla {Φ(dcuS-lacZ)} | This study |
| VJS9010 | λ− Δ(att-lom)::bla {Φ(dcuS-lacZ)}/narL215::Tn10 | This study |
| VJS9011 | λ− Δ(att-lom)::bla {Φ(dcuS-lacZ)}/narP253::Tn10d(Cm) | This study |
| VJS9012 | λ− Δ(att-lom)::bla {Φ(dcuS-lacZ)}/dcuR::Km | This study |
| VJS9016 | λ− Δ(att-lom)::bla {Φ(dcuS-lacZ)}/narL215::Tn10 narP253::Tn10d(Cm) | This study |
| Plasmids | ||
| pRS415 | Apr; lacZ operon fusion vector | 49 |
| pVJS3266 | Apr; Δ(lacY lacA cynX tet) derivative of pRS415 | This study |
| pVJS4042 | As pVJS3266 but Φ(aspA-lacZ) | This study |
| pVJS4043 | As pVJS3266 but Φ(dcuS-lacZ) | This study |
FIG. 2.
Control region sequences. (A) aspA-fxsA intergenic region. (B) dcuS-yjdI intergenic region. Regions protected by phospho-NarL protein from DNase I cleavage (Fig. 3) are indicated with thick lines. Flanking dashed lines for the aspA sequence indicate uncertainty about the exact boundaries of protection. Numbering is with respect to transcription initiation points, indicated by asterisks, for aspA (18) and dcuS (Fig. 4). Promoter elements (−10 and −35) are indicated with thick underlines or overlines, depending on orientation, and translation initiation sequences (Shine-Dalgarno element and initiation codon) are indicated with thin underlines or overlines. Sequences for binding the Fnr, Crp, and NarL proteins are boxed; consensus sequences are shown below. Restriction sites (EcoRI and BamHI) introduced for constructing Φ(aspA-lacZ) and Φ(dcuS-lacZ) operon fusions are indicated in lowercase.
Culture media and conditions.
Defined, complex, and indicator media for genetic manipulations were used as described previously (37). Defined medium to grow cultures for enzyme assays and for RNA extraction was buffered with 3-{N-morpholino} propanesulfonic acid (MOPS) as previously described (56). The initial pH of this medium is set at 8.0 in order to ameliorate nitrite toxicity (58). Because the pKa′ of MOPS is 7.2, the buffering capacity of this medium continually increases as acidic fermentation products accumulate; at harvest, cultures typically had pH values of about 7.5.
Medium for batch cultures grown to the mid-exponential phase contained glucose (80 mM) as the carbon source, and the respiratory oxidants NaNO3 and sodium fumarate were added to 40 mM and 50 mM, respectively, as indicated below.
Cultures were grown at 37°C. Culture densities were monitored with a Klett-Summerson photoelectric colorimeter (Klett Manufacturing Co., New York, N.Y.) equipped with a number 66 (red) filter. Anaerobic cultures for enzyme assays and for RNA extraction were grown in screw-cap tubes as described previously (56).
Enzyme assay.
β-Galactosidase activities were determined at room temperature (approximately 21°C) by following the hydrolysis of O-nitrophenyl-β-d-galactoside in CHCl3-sodium dodecyl sulfate-permeabilized cells. Specific activities are expressed in arbitrary (Miller) units (43). All cultures were assayed in duplicate, and reported values are averaged from at least two independent experiments.
Gene and operon fusions.
The Φ(frdA-lacZ) gene fusion was previously described (46). Briefly, the frdA control region and amino-terminal coding sequence were cloned as a 1.4-kb HindIII-BglII fragment into the gene fusion plasmid pRS414, the resulting construct was crossed into bacteriophage λRS45, and a monocopy lysogen was isolated (49).
The Φ(aspA-lacZ) and Φ(dcuS-lacZ) operon fusions were constructed by cloning the corresponding control regions into plasmid pVJS3266, a Δ(lacYA) derivative of operon fusion plasmid pRS415. This plasmid is analogous to the previously described plasmid pVJS3253, a Δ(lacYA) derivative of plasmid pRS414 (54). Constructs were crossed into bacteriophage λInCh and placed in monocopy in the host chromosome as described previously (8, 54). Control region sequences were isolated as PCR products with introduced EcoRI and BamHI sites as shown in Fig. 2. The veracity of each cloned insert was confirmed by DNA sequencing.
Transcript microarray analysis.
Relative mRNA levels were determined by parallel two-color hybridization to DNA microarrays following previously described methods (29, 69). Glass-slide microarrays spotted with PCR products representing virtually all E. coli genes (6) were generous gifts from the laboratory of Carol Gross, University of California, San Francisco.
Total RNA was extracted from early-exponential-phase cultures and analyzed as described previously (28). Reverse transcriptase (Stratascript; Stratagene, La Jolla, Calif.) synthesis of cDNA employed 500 ng (experiment 1) or 10 μg (experiment 2) of random hexamers (Amersham Biosciences, Piscataway, N.J.) hybridized to 15 μg of total RNA (28). Direct labeling (experiment 1) was done by incorporating Cy3- and Cy5-labeled dUTP (Amersham Biosciences) during cDNA synthesis as described previously (29). Indirect labeling (experiment 2) was done by incorporating 5-(3-aminoallyl)-2′-deoxyuridine 5′-triphosphate (Sigma-Aldrich, St. Louis, Mo.) during cDNA synthesis and subsequently labeling with Cy3 and Cy5 dyes (Amersham Biosciences) as described previously (28).
Labeled cDNA probes were hybridized to the microarrays as described previously (28). Fluorescence intensities of microarray spots were measured using a Genepix 4000 scanner and Genepix 3.0 software (Axon Instruments, Foster City, Calif.). The quality of hybridization spots was assessed, and data were normalized as red fluorescence/green fluorescence (R/G) median ratios, as described previously (28, 29). Results were also evaluated qualitatively by visualizing hybridization spots in genome order (68).
Phospho-NarL footprint analysis.
DNase I protection experiments (15) were performed essentially as described previously (20). Briefly, DNA templates were cleaved with one restriction endonuclease, end labeled by incubation with T4 polynucleotide kinase in the presence of [γ-32P]ATP, and cleaved with a second restriction endonuclease to remove the label from one end of the double-stranded template. Template reaction mixtures were extracted with phenol-chloroform and then passaged over a nucleotide removal spin column (QIAquick; QIAGEN, Inc., Valencia, CA). DNA binding reactions and DNase I digestions were performed essentially as described previously (20). Maltose binding protein (MBP)-NarL protein was phosphorylated by including 100 mM acetyl phosphate in each binding reaction mixture (34). Digestion products were separated by electrophoresis on thin sequencing gels (8 M urea-8% polyacrylamide) and visualized by PhosphorImager analysis (ImageQuant 5.0; Molecular Dynamics, Sunnyvale, CA). A G+A sequencing ladder (39) served to locate the protected regions.
Transcription initiation analysis.
Primer extension experiments were performed essentially as described previously (66). The primer (5′-AAGCTTAGTGAATCCGTAATCATGGTCATAG) corresponds to lacZ codons 1 to 8 in the template strand of the Φ(trpB′-lacZ)W209 operon fusion vector pVJS3266, and therefore its proximal (3′) end is 132 nucleotides (nt) downstream of the transcription initiation site shown in Fig. 2B. A strain carrying the Φ(dcuS-lacZ) operon fusion plasmid pVJS4043 was cultured to the mid-exponential phase anaerobically in the absence of nitrate. Total RNA was extracted and mixed with 5′-end-radiolabeled primer for extension reactions. Products were resolved on a thin polyacrylamide-urea slab gel and visualized by PhosphorImager analysis. The same primer was used for parallel DNA sequencing reactions to provide size markers for the extension products.
RESULTS
Transcript microarray rationale.
As described in Materials and Methods, analyses employed glass slides made through a consortium of university laboratories in northern California (29, 69). Our initial experiments probed the NarX-NarL regulon to validate procedures in our laboratory. This served as a prelude to our ongoing microarray analyses of the E. coli EnvZ-OmpR regulon (16). We chose to begin with the NarX-NarL regulon for four reasons. First, several Nar-regulated operons have been identified previously and therefore provide a benchmark for evaluating our results. Second, multisubunit respiratory enzymes are encoded by operons of three or more genes, and we reasoned that our conclusions from microarray data would be bolstered if all genes of an operon exhibited coordinate expression (68). Third, many respiratory operons exhibit 10-fold-or-greater regulation in response to nitrate, so we anticipated robust transcriptional responses. Finally, we were interested to learn new features of the NarX-NarL regulon.
Our approaches followed logic used by G. Zimmer et al. in their study of the Ntr regulon (36, 69). cDNA copies of transcripts from one strain representing the stimulated state (labeled with the fluorescent dye Cy-5) were competed against those from a second strain representing the unstimulated state (labeled with the fluorescent dye Cy-3). Both strains were cultured in defined media of identical composition to minimize differences arising from indirect physiological effects. All strains carried null alleles of the narQ and narP genes, so results reflect only the action of the NarX-NarL system. Strains also carried monocopy Φ(narG-lacZYA) or Φ(fdnG-lacZYA) fusions to monitor LacZ enzyme activities in cultures used for mRNA extraction, so lacZYA ratios provided additional signals for nitrate induction. We also observed hybridization signals for the cynX gene, which results from transcriptional readthrough from the lacZYA operon (68). (Part of the cynX gene, located immediately downstream of the lacA gene, is included in the operon fusion constructs) (49).
The first experiment compared the transcript abundance from strain VJS4033, whose genotype results in nitrate-independent narG operon expression (ΔnarX narL505), to that from the wild-type strain VJS2097 (narX+ narL+). During growth in the absence of nitrate, constitutive target operon expression mimics the stimulated state, whereas the wild-type strain represents the unstimulated state. A single experiment using this approach identified the expected nitrate-induced operons (see below). However, the narL505 allele exhibits a weak phenotype with respect to frdA operon repression (46), and therefore this operon and others were not identified among those subject to nitrate repression. Therefore, the second experiment compared transcripts from the narX+ narL+ strain VJS4341 to those from the Δ(narXL) null strain VJS4339. During growth in the presence of nitrate, the wild-type strain represents the stimulated state, whereas the null strain mimics the unstimulated state. A single experiment using this approach identified the expected nitrate-repressed operons in addition to the nitrate-induced operons (see below).
Our criteria for identifying regulated operons followed those of Zimmer et al. (69). Operons whose expression was activated by the NarX-NarL system each contained at least one gene whose mRNA level was ≥2.5-fold higher in the stimulated culture than in the unstimulated culture (normalized R/G median ratio ≥2.5; data not shown). Likewise, operons whose expression was repressed by the NarX-NarL system each contained at least one gene whose mRNA level was ≥2.5-fold lower in the stimulated culture than in the unstimulated culture (normalized G/R median ratio ≥2.5; data not shown). Independently, we also evaluated gene expression qualitatively by viewing the hybridization spots in genome order (not shown) (68).
Transcript microarray results.
Because of the small number of experiments, we have not attempted to conduct a detailed statistical analysis or to generate an exhaustive list of NarX-NarL-regulated genes. Nevertheless, the following genes in operons known to be activated at least fivefold by the NarX-NarL system during anaerobic batch growth in defined glucose medium were identified: narGHJI (10, 50), fdnGHI (3), narK (7, 31), and nirBDC (62) (data not shown). Likewise, genes in operons whose expression is repressed at least fivefold were identified as follows: dmsABC (11), frdABCD (22, 27, 53), adhE (33, 42), focA-pflBA (26), and dcuB-fumB (18) (data not shown). Finally, the genes aspA and dcuS were newly identified as subject to nitrate repression (see below). The microarrays did not identify Nar regulon members whose expression is only weakly regulated under the culture conditions employed. For example, the napFDAGHBC and nrfABCDEFG operons encoding periplasmic nitrate and nitrite reductases, respectively, are not induced strongly by nitrate during growth in batch culture (62).
In E. coli, several anaerobic respiratory enzymes are encoded by duplicate operons. In these cases, the primary operon, regulated by the Fnr and Nar systems, is expressed at high levels, whereas the secondary operon is expressed at low levels during exponential growth (1, 9, 35). Despite this low level of unregulated expression, we observed ≥2.5-fold differences in transcript abundances (data not shown) for genes in three of these secondary operons: narZYWV, homologous to narGHJI; fdoGHI, homologous to fdnGHI; and ynfEFGH, homologous to dmsABC. We attribute these results to cross-hybridization between PCR products of the secondary operons and cDNA derived from the expression of the homologous primary operons.
We also observed ≥2.5-fold differences in transcript abundances, in at least one of the two experiments (data not shown), for genes whose functions are not defined. These apparently activated genes were yeaR-yoaG (b1797 and b1796), and the apparently repressed genes were yegE (b2067) and yeiTA (b2146 and b2147). We did not study these further.
Nitrate repression of Φ(aspA-lacZ) and Φ(frdA-lacZ) expression.
The aspA gene, not previously known to be subject to Nar regulation, was among those whose transcription apparently was repressed (data not shown). We therefore constructed a monocopy Φ(aspA-lacZ) operon fusion as described in Materials and Methods in order to evaluate these microarray results. We also monitored expression from a previously constructed Φ(frdA-lacZ) gene fusion for which both Dcu-mediated activation and Nar-mediated repression are well documented. We examined the effects of the inducer fumarate and the corepressor nitrate as well as a null allele of the dcuR gene encoding the dicarboxylate-responsive transcription activator. Results are presented in Table 2.
TABLE 2.
Effects of nitrate, fumarate, and dcuR null allele on Φ(aspA-lacZ), Φ(frdA-lacZ), and Φ(dcuS-lacZ) expression
| Fusion | dcuR | LacZ sp acta
|
Fold induction by fumarate
|
Fold repression by NO3−
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| −NO3−
|
+NO3−
|
||||||||
| −Fum | +Fum | −Fum | +Fum | −NO3− | +NO3− | −Fum | +Fum | ||
| Φ(aspA-lacZ) | + | 220 | 150 | 19 | 18 | 0.7 | 0.9 | 12 | 8.3 |
| − | 180 | 190 | 25 | 19 | 1.1 | 0.8 | 7.2 | 10 | |
| Φ(frdA-lacZ) | + | 130 | 280 | 10 | 8 | 2.2 | 0.8 | 13 | 35 |
| − | 100 | 80 | 10 | 8 | 0.8 | 0.8 | 10 | 10 | |
| Φ(dcuS-lacZ) | + | 86 | 110 | 25 | 33 | 1.3 | 1.3 | 3.4 | 3.3 |
| − | 95 | 110 | 26 | 25 | 1.2 | 1.0 | 3.7 | 3.4 | |
Strains were cultured anaerobically to the mid-exponential phase in MOPS medium containing glucose as the carbon source.
In cultures with no added fumarate, nitrate was an equally effective corepressor for both Φ(aspA-lacZ) and Φ(frdA-lacZ) expression (12- and 13-fold, respectively, with dcuR+; 7.2- and 10-fold, respectively, with dcuR). These results substantiate the microarray results for (dcuR+) cultures grown in the same medium (data not shown).
In cultures with no added nitrate, fumarate weakly induced Φ(frdA-lacZ) expression through the DcuS-DcuR system (2.2-fold in dcuR+ versus 0.8-fold in dcuR), as expected for glucose-grown cultures (67). Nitrate repressed Φ(frdA-lacZ)-directed β-galactosidase activity by 35-fold in the fumarate-induced dcuR+ strain but only by 10- to 13-fold in the other three cultures where fumarate was either absent or ineffective due to the dcuR null allele (Table 2). This threefold-enhanced repression in fumarate-grown dcuR+ cultures suggests that nitrate repression of Φ(frdA-lacZ) expression might result from antagonism of fumarate induction, in addition to the previously documented direct repression of frdA operon transcription.
In cultures with no added nitrate, fumarate had little effect on Φ(aspA-lacZ) expression (0.7-fold in dcuR+ versus 1.1-fold in dcuR), and the magnitudes by which nitrate repressed Φ(aspA-lacZ) expression were similar irrespective of fumarate or dcuR genotype (7.2- to 12-fold). Therefore, nitrate repression of Φ(aspA-lacZ) expression is solely attributable to direct repression of aspA gene transcription.
Nitrate repression of Φ(dcuS-lacZ) expression.
The results summarized above suggested that nitrate repression of frdA operon transcription might be due in part to antagonism of fumarate induction. Furthermore, the second transcript microarray experiment revealed 2.6-fold repression by nitrate of dcuS gene transcription (data not shown). Together, these results suggested that nitrate represses expression of the dcuSR operon encoding the fumarate-responsive DcuS-DcuR system. We therefore constructed a monocopy Φ(dcuS-lacZ) operon fusion and evaluated its expression as described above for analyzing Φ(frdA-lacZ) and Φ(aspA-lacZ) expression. Indeed, nitrate repressed Φ(dcuS-lacZ) expression by about 3.5-fold (Table 2). As with Φ(aspA-lacZ) expression, Φ(dcuS-lacZ) expression was indifferent to added fumarate or dcuR genotype.
Nitrate repression in narP and narL null mutants.
Previous studies determined that the narL+ but not the narP+ gene is required for nitrate repression of Φ(frdA-lacZ) (47) and Φ(dcuB-lacZ) (18) expression. For the present study, we conducted similar assays with Φ(aspA-lacZ) and Φ(dcuS-lacZ) operon fusion strains. The results were analogous to those previously obtained for Φ(frdA-lacZ) expression: nitrate repression was essentially abolished in narL null strains and unaffected in narP null strains (data not shown). This indicates that the NarX-NarL system is solely responsible for nitrate repression of these four operons (dcuB-fumB, aspA, frdABCD, and dcuSR) involved in fumarate respiration.
Binding of phospho-NarL protein to the aspA and dcuS control regions.
We employed DNase I protection assays with purified NarL protein to identify NarL binding sites in the upstream control regions. Sequences are shown in Fig. 2. Previously, the aspA transcription initiation site was determined by primer extension, and the locations of binding sites for Fnr and Crp proteins were deduced from sequence inspection (18). The promoter for the divergently transcribed fxsA gene has been identified previously (63).
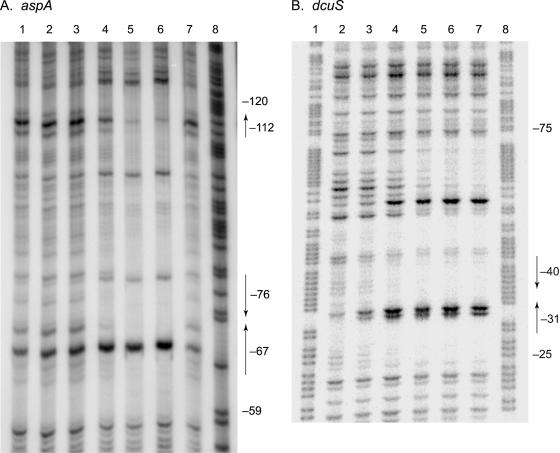
Phospho-NarL protein protected at least 53 nt of the aspA control region nontranscribed (template) strand (Fig. 3A). The protected region extended approximately from positions −60 to −115 with respect to the aspA transcription initiation site (Fig. 2A). DNase I made relatively few cuts at the borders of the protected region, which may be as large as 70 nt.
FIG. 3.
Interaction of phospho-NarL protein with control regions. Templates were labeled on the nontranscribed (coding) strand and subjected to DNase I protection as described in Materials and Methods. Numbers indicate positions relative to the transcription initiation sites, and arrows indicate locations of NarL heptamer sequences (Fig. 2). (A) aspA. Lanes 1 to 6, phospho-MBP-NarL dimers at 0.2, 0.4, 0.8, 1.6, 3.2, and 6.4 μM, respectively; lane 7, no protein; lane 8, G+A sequence. (B) dcuS. Lanes 1 and 8, G+A sequence; lane 2, no protein; lanes 3 to 7, phospho-MBP-NarL dimers at 0.2, 0.4, 0.8, 1.6, and 3.2 μM, respectively.
The transcription initiation site for the dcuSR operon has not been reported. The initiation codons for the divergent dcuS and yjdI genes are separated by only 180 nt, and a potential σ70-dependent promoter for dcuS transcription consists of potential −35 (TTGAAT; consensus, TTGACA) and −10 (GATAAT; consensus, TATAAT) elements separated by 16 nt (Fig. 2B). These elements and the spacing between them are exactly conserved in the Salmonella enterica LT-2 dcuS upstream region (6, 40), whereas sequence further upstream is divergent.
We performed primer extension analysis on mRNA isolated from a strain carrying the Φ(dcuS-lacZ) operon fusion plasmid as described in Materials and Methods. The extension product (Fig. 4) comigrated with the sequencing product representing the T residue in the nontranscribed (template) strand. This T residue corresponds to the transcribed strand A residue denoted with an asterisk in Fig. 2B. This result indicates that dcuSR operon transcription initiates from the predicted promoter.
FIG. 4.

Primer extension analysis of dcuS transcription initiation. The 5′-end-labeled primer was complementary to the lacZ sequence in a plasmid-borne Φ(dcuS-lacZ) operon fusion construct. Details are presented in Materials and Methods. Lanes are for the DNA sequence ladder (A, C, G, and T) and the primer extension reaction (PE), which was loaded in a double-width lane. The corresponding sequence of the template (noncoding) strand is indicated. The asterisk denotes the position of the T residue complementary to the A residue assigned as +1 (Fig. 2B).
Phospho-NarL protein protected approximately 51 nt of the dcuS control region nontranscribed (template) strand (Fig. 3B), extending from position −25 through position −75 with respect to the assigned transcription initiation site (Fig. 2B).
DISCUSSION
The repression of fumarate reductase synthesis by nitrate has long been established as an element in the hierarchical control of respiratory enzyme synthesis (64). Nitrate repression is mediated by the NarX-NarL regulatory system (22, 27, 53) and results from binding of phospho-NarL protein to a site overlapping the frdA operon transcription initiation point (34). More recently, the DcuS-DcuR regulatory system has been identified as mediating dicarboxylate-responsive activation of frdA operon transcription (17, 67). Results reported here indicate that the NarX-NarL system also represses synthesis of the DcuS-DcuR system (Fig. 1C). Furthermore, nitrate repression of frdA operon expression was more pronounced in the presence of added fumarate (35-fold) than in its absence (13-fold) (Table 2). Thus, in addition to direct repression, the hierarchical control of fumarate respiration may also involve a second level of regulation whereby decreased DcuS-DcuR synthesis renders fumarate induction less effective.
Nitrate repression of dcuSR operon transcription.
Expression from a monocopy Φ(dcuS-lacZ) operon fusion revealed three- to fourfold repression by nitrate (Table 2), virtually all of which can be attributed to the NarX-NarL regulatory system rather than the paralogous NarQ-NarP system (data not shown). This level of repression is lower than that of other operons, such as frdABCD, whose expression is repressed by nitrate about 12-fold during growth in the absence of added fumarate (Table 2). Nevertheless, relatively small changes in regulatory gene expression can have significant effects on target gene regulation (4).
DNA sites for phospho-NarL binding consist of 7-nt sequences (NarL heptamers) usually arranged as inverted repeats with 2-nt spacing (7-2-7 heptamer pairs). The heptamer consensus is TACYYMT, where Y stands for C or T and M stands for A or C (13). These sites have been identified in various configurations in a number of Nar-regulated control regions (57).
DNase I protection identified a region for phospho-NarL binding upstream of the probable dcuS transcription initiation site (Fig. 2B, 3B, and 4). At the downstream end of this region, a consensus NarL heptamer is centered at position −31 with respect to the proposed transcription initiation site, and a second near-consensus heptamer (six matches to consensus) is centered at position −40. Thus, the dcuSR operon control region contains an authenticated 7-2-7 heptamer pair for phospho-NarL binding overlapping the likely −35 region of the promoter (Fig. 2B) and therefore positioned appropriately to effect the observed repression (Table 2).
The dcuSR control region footprint spanned more than 50 nt, suggesting that at least two dimers of phospho-NarL protein were bound. Additional NarL heptamer sequences within this upstream protected region are not evident from sequence inspection. However, in several other control regions studied, binding of a phospho-NarL dimer to one strong site promotes cooperative binding to adjacent weak sites (13, 34). Strong DNase I-hypersensitive sites were induced by phospho-NarL binding (Fig. 3B). This previously has been observed for other phospho-NarL binding regions and likely results from DNA bending around the bound phospho-NarL dimer (38).
The expression of the Φ(dcuS-lacZ) operon fusion was unaffected by added fumarate and independent of the dcuR+ allele (Table 2), suggesting that dcuSR operon transcription is not autoregulated. This conclusion was previously suggested upon finding that phospho-DcuR protein failed to interact specifically with dcuS upstream DNA (2).
Thus, synthesis of one two-component regulatory system (DcuS-DcuR) is regulated by a second (NarX-NarL). Other documented examples of such two-component cascades include PhoR-PhoP regulation of ResE-ResD synthesis in Bacillus subtilis (5) and EnvZ-OmpR regulation of SsrB-SsrA synthesis in Salmonella enterica (14, 32).
Nitrate repression of aspA gene transcription.
Previous analysis of expression from an Φ(aspA-lacZ) monocopy construct, determined from cultures in LB broth, showed that transcription from the aspA promoter is activated by both the Crp and Fnr proteins (65). Our analysis, determined from cultures in defined medium with glucose as carbon source, revealed 10- to 15-fold repression by nitrate (Table 2), essentially all of which can be attributed to the NarX-NarL regulatory system (data not shown). This level of repression is similar to that of the frdA operon during growth in the absence of added fumarate (see Table 2).
Expression of the Φ(aspA-lacZ) operon fusion was indifferent to added fumarate and independent of the dcuR+ allele (Table 2), suggesting that aspA gene transcription is not induced by C4-dicarboxylates. This was surprising, since aspartate is equivalent to fumarate, malate, and other C4-dicarboxylates as an inducer of DcuR-dependent gene expression (67). Perhaps the apparent DcuS-DcuR response to added aspartate reflects conversion to succinate, which is excreted (Fig. 1B) (25).
AspA enzyme plays a central role in amino acid catabolism (41), and aspartate is preferentially consumed during growth on complex media, such as tryptone (45) The regulation of AspA enzyme synthesis therefore must account for this catabolic function as well as its role in anaerobic fumarate respiration.
DNase I protection identified a region of at least 53 nt for phospho-NarL binding, upstream of the aspA transcription initiation site (Fig. 2A and 4A). At the downstream end of this region, a consensus NarL heptamer is centered at position −67 with respect to the transcription initiation site, and a second near-consensus heptamer (five matches to consensus) is centered at position −76. Thus, the aspA operon control region contains an authenticated 7-2-7 heptamer pair for phospho-NarL binding. At the other end of this region, a near-consensus heptamer (five matches) is centered at position −112, although a partner heptamer to form a 7-2-7 pair is not evident from sequence inspection (Fig. 2A and 4A). Again, cooperative interactions might contribute to phospho-NarL binding to the upstream end of this region.
This location for phospho-NarL binding, from about 60 to 120 nt upstream of the transcription initiation site, does not match expectations for a direct negative control (operator) site. Thus, nitrate repression of aspA transcription may be indirect. For example, bound phospho-NarL might interfere with transcription activation mediated by the Crp and Fnr proteins (48). A series of experiments will be necessary to determine the exact role of phospho-NarL in controlling aspA gene expression.
In E. coli, the fxsA gene is transcribed divergently from the aspA gene (Fig. 2B). We did not examine the regulation of fxsA gene expression, although conceivably it could be regulated by the NarL, Fnr, and Crp binding sites that control aspA transcription. The FxsA protein was identified as an overproduction suppressor of F-mediated exclusion of bacteriophage T7, and therefore the fxsA gene promoter was identified by a promoter-up substitution (63).
Nitrate repression of fumarate respiration.
Nitrate represses the transcription of several operons encoding components of the fumarate respiration pathway (Fig. 1): frdABCD, encoding fumarate reductase (22, 27, 53); dcuB-fumB, encoding a dicarboxylate carrier and fumarase B (18); aspA, encoding aspartate-ammonia lyase (as described in this work); and dcuSR, encoding the dicarboxylate-responsive regulatory system (as described in this work). Thus, all specific aspects of fumarate respiration—transport, synthesis, reduction, and regulation—are inhibited by the preferred acceptor nitrate. The NarX-NarL system is necessary and sufficient for full nitrate repression of this fumarate respiration pathway, whereas the paralogous NarQ-NarP system plays no significant role (18, 47; also described in this study). This is consistent with the notion that the NarX-NarL system broadly controls cellular physiology during growth conditions where nitrate respiration is the predominant avenue for energy production. By contrast, the NarQ-NarP system may play a more restricted role in controlling gene expression when respiratory electron acceptors are limiting (52).
Acknowledgments
We thank Sydney Kustu, in whose laboratory this study was initiated; Virgil Rhodius and Carol Gross for providing helpful technical advice and for sharing materials; Fritz Unden for providing the dcuR::Km allele; and Alex Appleman and Radomir Schmidt for helpful comments on a draft version of this work.
This study was supported by Public Health Service grants GM36877 and GM48591 from the National Institute of General Medical Sciences (awarded to V.S. and M.M.I., respectively).
REFERENCES
- 1.Abaibou, H., J. Pommier, S. Benoit, G. Giordano, and M. A. Mandrand-Berthelot. 1995. Expression and characterization of the Escherichia coli fdo locus and a possible physiological role for aerobic formate dehydrogenase. J. Bacteriol. 177:7141-7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abo-Amer, A. E., J. Munn, K. Jackson, M. Aktas, P. Golby, D. J. Kelly, and S. C. Andrews. 2004. DNA interaction and phosphotransfer of the C4-dicarboxylate-responsive DcuS-DcuR two-component regulatory system from Escherichia coli. J. Bacteriol. 186:1879-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, B. L., and V. Stewart. 1990. Structural genes for nitrate-inducible formate dehydrogenase in Escherichia coli K-12. Genetics 125:691-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bijlsma, J. J., and E. A. Groisman. 2003. Making informed decisions: regulatory interactions between two-component systems. Trends Microbiol. 11:359-366. [DOI] [PubMed] [Google Scholar]
- 5.Birkey, S. M., W. Liu, X. Zhang, M. F. Duggan, and F. M. Hulett. 1998. Pho signal transduction network reveals direct transcriptional regulation of one two-component system by another two-component regulator: Bacillus subtilis PhoP directly regulates production of ResD. Mol. Microbiol. 30:943-953. [DOI] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Bonnefoy, V., and J. A. DeMoss. 1992. Identification of functional cis-acting sequences involved in regulation of narK gene expression in Escherichia coli. Mol. Microbiol. 6:3595-3602. [DOI] [PubMed] [Google Scholar]
- 8.Boyd, D., D. S. Weiss, J. C. Chen, and J. Beckwith. 2000. Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J. Bacteriol. 182:842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, L., L. I. Wei, J. P. Audia, R. A. Morton, and H. E. Schellhorn. 1999. Expression of the Escherichia coli NRZ nitrate reductase is highly growth phase dependent and is controlled by RpoS, the alternative vegetative sigma factor. Mol. Microbiol. 34:756-766. [DOI] [PubMed] [Google Scholar]
- 10.Chippaux, M., V. Bonnefoy-Orth, J. Ratouchniak, and M.-C. Pascal. 1981. Operon fusions in the nitrate reductase operon and study of the control gene nirR in Escherichia coli. Mol. Gen. Genet. 182:477-479. [DOI] [PubMed] [Google Scholar]
- 11.Cotter, P. A., and R. P. Gunsalus. 1989. Oxygen, nitrate, and molybdenum regulation of dmsABC gene expression in Escherichia coli. J. Bacteriol. 171:3817-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cronan, J. E., Jr., and D. LaPorte. 1996. Tricarboxylic acid cycle and glyoxylate bypass, p. 206-216. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 13.Darwin, A. J., K. L. Tyson, S. J. Busby, and V. Stewart. 1997. Differential regulation by the homologous response regulators NarL and NarP of Escherichia coli K-12 depends on DNA binding site arrangement. Mol. Microbiol. 25:583-595. [DOI] [PubMed] [Google Scholar]
- 14.Feng, X., R. Oropeza, and L. J. Kenney. 2003. Dual regulation by phospho-OmpR of ssrA/B gene expression in Salmonella pathogenicity island 2. Mol. Microbiol. 48:1131-1143. [DOI] [PubMed] [Google Scholar]
- 15.Galas, D. J., and A. Schmitz. 1978. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 5:3157-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goh, E. B., D. F. Siino, and M. M. Igo. 2004. The Escherichia coli tppB (ydgR) gene represents a new class of OmpR-regulated genes. J. Bacteriol. 186:4019-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golby, P., S. Davies, D. J. Kelly, J. R. Guest, and S. C. Andrews. 1999. Identification and characterization of a two-component sensor-kinase and response-regulator system (DcuS-DcuR) controlling gene expression in response to C4-dicarboxylates in Escherichia coli. J. Bacteriol. 181:1238-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golby, P., D. J. Kelly, J. R. Guest, and S. C. Andrews. 1998. Transcriptional regulation and organization of the dcuA and dcuB genes, encoding homologous anaerobic C4-dicarboxylate transporters in Escherichia coli. J. Bacteriol. 180:6586-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guest, J. R. 1992. Oxygen-regulated gene expression in Escherichia coli. The 1992 Marjory Stephenson Prize Lecture. J. Gen. Microbiol. 138:2253-2263. [DOI] [PubMed] [Google Scholar]
- 20.Huang, K. J., and M. M. Igo. 1996. Identification of the bases in the ompF regulatory region, which interact with the transcription factor OmpR. J. Mol. Biol. 262:615-628. [DOI] [PubMed] [Google Scholar]
- 21.Iuchi, S., A. Aristarkhov, J. M. Dong, J. S. Taylor, and E. C. C. Lin. 1994. Effects of nitrate respiration on expression of the Arc-controlled operons encoding succinate dehydrogenase and flavin-linked l-lactate dehydrogenase. J. Bacteriol. 176:1695-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iuchi, S., and E. C. C. Lin. 1987. The narL gene product activates the nitrate reductase operon and represses the fumarate reductase and trimethylamine N-oxide reductase operons in Escherichia coli. Proc. Natl. Acad. Sci. USA 84:3901-3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iuchi, S., and E. C. C. Lin. 1995. Signal transduction in the Arc system for control of operons encoding aerobic respiratory enzymes, p. 223-231. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 24.Janausch, I. G., I. Garcia-Moreno, and G. Unden. 2002. Function of DcuS from Escherichia coli as a fumarate-stimulated histidine protein kinase in vitro. J. Biol. Chem. 277:39809-39814. [DOI] [PubMed] [Google Scholar]
- 25.Janausch, I. G., E. Zientz, Q. H. Tran, A. Kroger, and G. Unden. 2002. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta 1553:39-56. [DOI] [PubMed] [Google Scholar]
- 26.Kaiser, M., and G. Sawers. 1995. Nitrate repression of the Escherichia coli pfl operon is mediated by the dual sensors NarQ and NarX and the dual regulators NarL and NarP. J. Bacteriol. 177:3647-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalman, L. V., and R. P. Gunsalus. 1988. The frdR gene of Escherichia coli globally regulates several operons involved in anaerobic growth in response to nitrate. J. Bacteriol. 170:623-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khodursky, A. B., J. A. Bernstein, B. J. Peter, V. Rhodius, V. F. Wendisch, and D. P. Zimmer. 2003. Escherichia coli spotted double-strand DNA microarrays: RNA extraction, labeling, hybridization, quality control, and data management. Methods Mol. Biol. 224:61-78. [DOI] [PubMed] [Google Scholar]
- 29.Khodursky, A. B., B. J. Peter, N. R. Cozzarelli, D. Botstein, P. O. Brown, and C. Yanofsky. 2000. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:12170-12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiley, P. J., and H. Beinert. 1998. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22:341-352. [DOI] [PubMed] [Google Scholar]
- 31.Kolesnikow, T., I. Schröder, and R. P. Gunsalus. 1992. Regulation of narK gene expression in Escherichia coli in response to anaerobiosis, nitrate, iron, and molybdenum. J. Bacteriol. 174:7104-7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, A. K., C. S. Detweiler, and S. Falkow. 2000. OmpR regulates the two-component system SsrA-SsrB in Salmonella pathogenicity island 2. J. Bacteriol. 182:771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leonardo, M. R., P. R. Cunningham, and D. P. Clark. 1993. Anaerobic regulation of the adhE gene, encoding the fermentative alcohol dehydrogenase of Escherichia coli. J. Bacteriol. 175:870-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, J., S. Kustu, and V. Stewart. 1994. In vitro interaction of nitrate-responsive regulatory protein NarL with DNA target sequences in the fdnG, narG, narK and frdA operon control regions of Escherichia coli K-12. J. Mol. Biol. 241:150-165. [DOI] [PubMed] [Google Scholar]
- 35.Lubitz, S. P., and J. H. Weiner. 2003. The Escherichia coli ynfEFGHI operon encodes polypeptides which are paralogues of dimethyl sulfoxide reductase (DmsABC). Arch. Biochem. Biophys. 418:205-216. [DOI] [PubMed] [Google Scholar]
- 36.Magasanik, B. 2000. Global regulation of gene expression. Proc. Natl. Acad. Sci. USA 97:14044-14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maloy, S. R., V. J. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria. A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Maris, A. E., M. R. Sawaya, M. Kaczor-Grzeskowiak, M. R. Jarvis, S. M. Bearson, M. L. Kopka, I. Schröder, R. P. Gunsalus, and R. E. Dickerson. 2002. Dimerization allows DNA target site recognition by the NarL response regulator. Nat. Struct. Biol. 9:771-778. [DOI] [PubMed] [Google Scholar]
- 39.Maxam, A. M., and W. Gilbert. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65:499-560. [DOI] [PubMed] [Google Scholar]
- 40.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 41.McFall, E., and E. B. Newman. 1996. Amino acids as carbon sources, p. 358-379. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 42.Membrillo-Hernández, J., and E. C. C. Lin. 1999. Regulation of expression of the adhE gene, encoding ethanol oxidoreductase in Escherichia coli: transcription from a downstream promoter and regulation by Fnr and RpoS. J. Bacteriol. 181:7571-7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 44.Prohl, C., B. Wackwitz, D. Vlad, and G. Unden. 1998. Functional citric acid cycle in an arcA mutant of Escherichia coli during growth with nitrate under anoxic conditions. Arch. Microbiol. 170:1-7. [DOI] [PubMed] [Google Scholar]
- 45.Pruss, B. M., J. M. Nelms, C. Park, and A. J. Wolfe. 1994. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J. Bacteriol. 176:2143-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rabin, R. S., and V. Stewart. 1992. Either of two functionally redundant sensor proteins, NarX and NarQ, is sufficient for nitrate regulation in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 89:8419-8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rabin, R. S., and V. Stewart. 1993. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J. Bacteriol. 175:3259-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott, S., S. Busby, and I. Beacham. 1995. Transcriptional co-activation at the ansB promoters: involvement of the activating regions of CRP and FNR when bound in tandem. Mol. Microbiol. 18:521-531. [DOI] [PubMed] [Google Scholar]
- 49.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 50.Stewart, V. 1982. Requirement of Fnr and NarL functions for nitrate reductase expression in Escherichia coli K-12. J. Bacteriol. 151:1320-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stewart, V. 1988. Nitrate respiration in relation to facultative metabolism in enterobacteria. Microbiol. Rev. 52:190-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stewart, V. 2003. Nitrate- and nitrite-responsive sensors NarX and NarQ of proteobacteria. Biochem. Soc. Trans. 31:1-10. [DOI] [PubMed] [Google Scholar]
- 53.Stewart, V., and B. L. Berg. 1988. Influence of nar (nitrate reductase) genes on nitrate inhibition of formate-hydrogen lyase and fumarate reductase synthesis in Escherichia coli K-12. J. Bacteriol. 170:4437-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stewart, V., and P. J. Bledsoe. 2003. Synthetic lac operator substitutions to study the nitrate- and nitrite-responsive NarX-NarL and NarQ-NarP two-component regulatory systems of Escherichia coli K-12. J. Bacteriol. 185:2104-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stewart, V., Y. Lu, and A. J. Darwin. 2002. Periplasmic nitrate reductase (NapABC enzyme) supports anaerobic respiration by Escherichia coli K-12. J. Bacteriol. 184:1314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stewart, V., and J. Parales, Jr. 1988. Identification and expression of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J. Bacteriol. 170:1589-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stewart, V., and R. S. Rabin. 1995. Dual sensors and dual response regulators interact to control nitrate- and nitrite-responsive gene expression in Escherichia coli, p. 233-252. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington D.C.
- 58.Tomsett, A. B., and R. H. Garrett. 1980. The isolation and characterization of mutants defective in nitrate assimilation in Neurospora crassa. Genetics 95:649-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tran, Q. H., J. Bongaerts, D. Vlad, and G. Unden. 1997. Requirement for the proton-pumping NADH dehydrogenase I of Escherichia coli in respiration of NADH to fumarate and its bioenergetic implications. Eur. J. Biochem. 244:155-160. [DOI] [PubMed] [Google Scholar]
- 60.Tran, Q. H., and G. Unden. 1998. Changes in the proton potential and the cellular energetics of Escherichia coli during growth by aerobic and anaerobic respiration or by fermentation. Eur. J. Biochem. 251:538-543. [DOI] [PubMed] [Google Scholar]
- 61.Unden, G., and J. Bongaerts. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320:217-234. [DOI] [PubMed] [Google Scholar]
- 62.Wang, H., and R. P. Gunsalus. 2000. The nrfA and nirB nitrite reductase operons in Escherichia coli are expressed differently in response to nitrate than to nitrite. J. Bacteriol. 182:5813-5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, W. F., X. Cheng, and I. J. Molineux. 1999. Isolation and identification of fxsA, an Escherichia coli gene that can suppress F exclusion of bacteriophage T7. J. Mol. Biol. 292:485-499. [DOI] [PubMed] [Google Scholar]
- 64.Wimpenny, J. W. T., and J. A. Cole. 1967. The regulation of metabolism in facultative bacteria. 3. The effect of nitrate. Biochim. Biophys. Acta 148:233-242. [DOI] [PubMed] [Google Scholar]
- 65.Woods, S. A., and J. R. Guest. 1987. Differential roles of the Escherichia coli fumarases and fnr-dependent expression of fumarase B and aspartase. FEMS Microbiol. Lett. 48:219-224. [Google Scholar]
- 66.Wu, S. Q., W. Chai, J. T. Lin, and V. Stewart. 1999. General nitrogen regulation of nitrate assimilation regulatory gene nasR expression in Klebsiella oxytoca M5al. J. Bacteriol. 181:7274-7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zientz, E., J. Bongaerts, and G. Unden. 1998. Fumarate regulation of gene expression in Escherichia coli by the DcuSR (dcuSR genes) two-component regulatory system. J. Bacteriol. 180:5421-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zimmer, D. P., O. Paliy, B. Thomas, P. Gyaneshwar, and S. Kustu. 2004. Genome image programs: visualization and interpretation of Escherichia coli microarray experiments. Genetics 167:2111-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khodursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 97:14674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]