Abstract
MtrR represses expression of the Neisseria gonorrhoeae mtrCDE multidrug efflux transporter genes. MtrR displays salt-dependent DNA binding, a stoichiometry of two dimers per DNA site, and, for a protein that was expected to be essentially all helical, a high percentage of random coil and possibly β-sheet structure.
In order to colonize human mucosal membranes, Neisseria gonorrhoeae must overcome host defense mechanisms that include exposure to potentially lethal levels of antimicrobial hydrophobic agents. Early studies implicated the multiple transferable resistance (mtr) locus as a key determinant in resistance that initially was thought to play a role in the modification of the gonococcal cell envelope (11). Later studies demonstrated that this locus encoded a three-gene operon, designated mtrCDE, which forms an energy-dependent efflux system that expels multiple hydrophobic agents (3, 13). The MtrD protein is a multidrug efflux transporter that belongs to the resistance/nodulation/division transporter family (12). The MtrC protein belongs to the membrane fusion protein family that links MtrD with MtrE, an outer membrane protein, which serves as a channel for export of antimicrobials to the extracellular environment (3).
Due to the broad substrate specificity of multidrug efflux transporters, which could result in the accidental efflux of needed metabolic intermediates, the expression of their genes is regulated tightly (10). Transcription of the mtrCDE operon is controlled by both cis- and trans-acting factors under the influence of the mtrR gene (14). The presence of missense or deletion mutations of the mtrR gene in clinical isolates leads to increased transcription of mtrCDE and a consequential increase in antimicrobial resistance, thus confirming the repression of MtrCDE efflux pump transcription by MtrR (24). The mtrR gene is located ∼250 bp upstream of the mtrCDE genes and is transcribed divergently from that operon. The mtrR gene encodes the 210-amino-acid-residue, ∼23-kDa protein, MtrR. MtrR contains a putative N-terminal helix-turn-helix motif and amino acid sequence similarity to several members of the TetR/CamR family, e.g., 53% identity and 78% homology to AcrR (2, 17, 20, 23). Footprinting and DNase I protection experiments established that MtrR protects a 22- to 27-base-pair region upstream of mtrC (17). Although a DNA binding site was identified and the DNA binding domain bears strong homology to those of other TetR family members, the stoichiometry of MtrR binding to DNA is unknown. TetR family members have shown variability in their DNA binding oligomerization states (5, 9, 15, 21).
In order to understand better the DNA binding properties of MtrR and its role in Neisseria gonorrhoeae resistance against hydrophobic agents and other antibiotics, we carried out a biophysical and biochemical characterization of this multidrug efflux pump gene repressor. These studies included the determination of the length of cognate DNA required for optimal MtrR binding, the effect of NaCl concentration on DNA binding affinity, the stoichiometry of binding, and the secondary structure of MtrR in the presence or absence of cognate DNA. Unanticipated differences between MtrR and the TetR family member QacR, the Staphylococcus aureus multidrug binding transcription repressor (8), were observed.
Cloning, expression, and purification of MtrR.
The 633-base-pair mtrR gene from N. gonorrhoeae strain FA19 was PCR amplified from chromosomal DNA by using primers that contained engineered restriction sites NdeI and BamHI. After digestion with NdeI and BamHI, the fragment was cloned into a pET-15b ampicillin-resistant vector containing an N-terminal hexahistidine affinity tag followed by a thrombin cleavage site. The vector was sequenced to ensure fidelity and transformed into Rosetta-gami B(DE3)pLysS cells resistant to chloramphenicol. One-liter cultures were grown in Luria-Bertani broth containing 100 μg/ml ampicillin and 50 μg/ml chloramphenicol at 37°C to an optical density at 600 nm of 0.6 AU (absorbance unit), at which time cells were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h. Cells were then centrifuged and resuspended in 20 mM Tris, pH 7.6, 500 mM NaCl, 10% glycerol, and 1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) as a reducing agent. The cells were lysed by French press, after which the lysate was centrifuged and the supernatant loaded onto a Ni2+-nitrilotriacetic acid column. Pure hexahistidine-tagged MtrR was eluted with buffer A (100 mM Na+/K+ phosphate buffer, pH 8.5, 300 mM NaCl, 5% glycerol, 1 mM TCEP) containing 500 mM imidazole (data not shown). Fractions were analyzed by quantitative time of flight mass spectrometry and sodium dodecyl sulfate-polyacrylamide gel electrophoresis before dialyzing overnight into 200 mM Na+/K+ phosphate, pH 7.5, containing 10% glycerol, and 1 mM TCEP (phosphate storage buffer [PSB]). Specific and complete cleavage of the hexahistidine tag was attained. However, the stability of MtrR was compromised, and therefore, only purified His-tagged MtrR was used in our studies.
DNA binding affinity and binding stoichiometry.
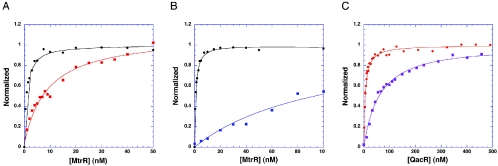
A fluorescence polarization-based assay was used to determine the DNA binding affinity of MtrR for a pair of oligodeoxynucleotides from the mtrCDE promoter. These oligodeoxynucleotides were purchased from Oligos Etc. (Wilsonville, OR) and were 27 and 22 base pairs long with fluorescein covalently attached to their 5′ end by a hexamethylene linker. Each oligodeoxynucleotide encompassed the direct repeat that in previous footprinting studies was protected to different extents by MtrR. The sequences of one strand of the 27-mer and one strand of the 22-mer are 5′-TTTTTATCCGTGCAATCGTGTATGTAT and 5′-ATCCGTGCAATCGTGTATGTAT, respectively, with the pseudo direct repeats underlined (17). The standard DNA binding solution used in these studies was 20 mM Tris-HCl, pH 7.4, 100 mM NaCl, 0.1 nM fluoresceinated DNA (or a higher concentration, provided the Kd was ≥10 times the DNA concentration), and 1 μg of poly(dI-dC), as nonspecific DNA. MtrR in PSB was titrated into the binding mixture until the millipolarization (mP) no longer rose. All experiments were carried out at 25°C. The excitation wavelength was 490 nm, and units of fluorescence polarization (millipolarization) were read at 530 nm. The data were plotted and analyzed by using the following equation: P = {(Pbound − Pfree)[protein])/Kd + [protein]} + Pfree, where P is the polarization measured at a given total protein concentration, Pfree is the initial polarization of free fluorescein-labeled DNA, Pbound is the maximum polarization of specifically bound DNA, and [protein] is the protein concentration. The free and total protein concentrations are assumed to be equal because the concentration of fluorescein-labeled DNA is 10-fold lower than the Kd. The generated hyperbolic curves are fit by nonlinear least-squares regression analysis, assuming a bimolecular model such that the Kd values represent the protein concentrations at half-maximal ligand binding and plotted by using the graphing program Kaleidograph (17). The longer oligodeoxynucleotide (Kd = 0.9 nM) bound ∼9-fold better than the shorter oligodeoxynucleotide (Kd = 7.8 nM) (Fig. 1A). Increasing the oligodeoxynucleotide length to 31 base pairs did not result in higher affinity (data not shown). Consequently, the remaining DNA binding experiments used the 27-mer.
FIG. 1.
Binding isotherms of MtrR and QacR. (A) MtrR binding to its 22-mer (red curve) and 27-mer (black curve) cognate oligodeoxynucleotides. (B) MtrR binding to the 27-mer cognate oligodeoxynucleotide in the presence of 100 mM NaCl (black curve) and 200 mM NaCl (blue curve). (C) QacR binding to its 28-base-pair high-affinity DNA binding site (IR1) in the presence of 100 NaCl (orange curve) and 200 mM NaCl (violet curve). The sequence of the one of the IR1 strands is 5′-CTTATAGACCGATCGCACGGTCTATAAG-3′. The binding data displayed in each panel have been normalized to the calculated binding maximum millipolarization of each curve.
The length of the higher-affinity DNA binding site of MtrR is nearly identical to the high-affinity DNA binding site (IR1) of QacR. Two dimers of QacR bind the 28-base-pair IR1, which is located in the promoter region of the qacA multidrug efflux pump gene, and although pseudopalindromic, IR1 contains four pseudo direct repeats that interact with QacR (9, 21). To determine whether MtrR employs the same stoichiometry or a different stoichiometry of binding to the mtrCDE promoter, a fluorescence polarization assay was utilized. The binding buffer and conditions were identical to those used in the binding affinity determination experiments except that the concentration of the 27-mer was increased to 20 nM, i.e., >20-fold higher than the Kd, thereby ensuring stoichiometric binding. MtrR was titrated into the binding solution until the total protein concentration (in monomers) reached 200 nM. The graph of the resulting data shows a linear increase in the observed millipolarization until saturation of the high affinity DNA sites, after which low-affinity DNA binding takes place (Fig. 2). The inflection point occurs at an MtrR monomer concentration of 80 nM, which, when divided by the concentration of cognate DNA (20 nM), indicates a stoichiometry of four protomers, presumably two dimers per DNA site.
FIG. 2.
Determination of the stoichiometry of MtrR-DNA binding. Note the inflection point at an MtrR monomer concentration of 80 nM (black arrow), indicating the shift from high- to low-affinity binding (indicated with black lines).
In a parallel approach to determine the oligomerization state of DNA-bound and unbound MtrR, a series of dynamic light scattering (DLS) experiments were done. DLS measures the inherent light scattering of a macromolecule, which fluctuates due to the Brownian motion of the macromolecule, as a function of time. From these measurements, the translational diffusion coefficient (DT) of the macromolecule can be calculated, which in turn allows the determination of the hydrodynamic radius of the average scattering particle (RH) via the following equation: DT = kT/6πηRH, where k is the Boltzmann constant, T is the temperature in kelvin units, and η is the solvent viscosity (7). RH can be used to estimate the molecular mass. DLS experiments were carried out at 24°C on MtrR (50 μl of a 0.4 mM dimer solution in PSB) and revealed a molecular mass (mean ± standard deviation) of 40 kDa ± 15 kDa, which is consistent with an MtrR dimer. DLS studies on the DNA-bound form of MtrR (50 μl of a solution containing 0.1 mM dimer MtrR and duplex 27-mer) revealed a species with a molecular mass of 110 kDa ± 10 kDa, which can be explained by the binding of four MtrR protomers (4 protomers · 24 kDa/protomer = 96 kDa) to one 27-bp oligodeoxynucleotide (27 bp · 660 Da/bp = 18 kDa), i.e., 96 kDa plus 18 kDa equals 114 kDa. The DNA binding data, combined with the results from the DLS experiments, indicate that two MtrR dimers bind the mtrCDE promoter. This DNA binding stoichiometry is the same as that utilized by TetR family member QacR but contrasts with those of family members TetR and EthR, which bind one and four dimers, respectively, to their operators (5, 15).
To characterize the DNA binding mechanism of MtrR further, the effect of salt concentration on affinity was examined. DNA binding was affected significantly by increasing the NaCl concentration with the Kd increasing over 100-fold (from 0.9 nM to 99.0 nM) by simply doubling the NaCl concentration from 100 mM, the more physiologically relevant concentration, to 200 mM (Fig. 1B). By contrast, when the same experiment was carried out with QacR binding to IR1, only a fourfold effect was observed, whereby the Kd in 100 mM NaCl was 5.7 nM and that in 200 mM NaCl was 22.5 nM (Fig. 1C). In an attempt to provide a molecular understanding for the different salt effects of MtrR and QacR, both of which bind two dimers to their cognate DNA, the sequences of the MtrR and QacR DNA binding domains were aligned and an analysis of potential protein-DNA ionic interactions was done by homology modeling (data not shown). QacR engages in only three side chain-phosphate backbone ionic interactions per subunit, thereby providing a reasonable chemical rationale for the modest effect of higher salt concentrations on binding affinity. If MtrR were to bind its cognate DNA site in a manner similar to QacR, only those interactions made by QacR would again be made by MtrR, as the sequence alignment and homology modeling do not reveal any potential additional ionic interactions within the established QacR DNA binding domain. However, while the DNA binding domain of QacR begins with its most N-terminal residue, MtrR has eight additional residues (1-MRKTKTEA-8) that are N-terminal to the beginning of the consensus TetR family DNA binding domain. Moreover, MtrR residue 10 is a lysine, and the corresponding residue in QacR is an asparagine. Thus, 5 of the first 10 residues of MtrR are basic and not present in QacR, and their presence suggests that 1 or more of these basic residues engages in electrostatic interaction with the mtrCDE DNA.
Secondary structure determination.
As a member of the TetR family, MtrR is expected to be predominantly α helical. To quantify the secondary-structure content of MtrR, circular dichroism (CD) studies were done on MtrR and, for further comparison, on QacR. Because the crystal structures of apo QacR (apo is the drug- and DNA-free protein) and a QacR-DNA complex are known (21, 22), quantification of its secondary-structure content by CD in these states provides a good idea of the accuracy and precision of this approach in determining the helicity of MtrR. Spectra of the apo and DNA-bound proteins were taken in order to determine whether or not DNA binding significantly alters the secondary-structure content of MtrR. Spectra of the apo proteins and their DNA-bound complexes, in PSB, were taken from 190 nm to 300 nm in a 0.4-ml cell at 25°C and analyzed for secondary-structure content with the deconvolution program K2D (1). The dimer concentrations of MtrR and QacR in all spectral measurements were 4.0 μM and 5.6 μM, respectively. These assured stoichiometric DNA binding under the buffer conditions employed in the experiment.
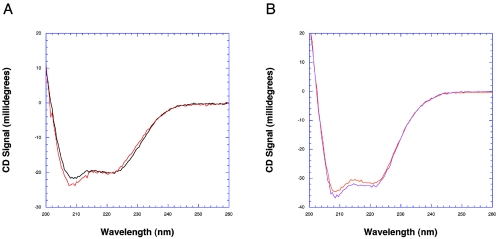
The analysis of the CD spectra of MtrR reveals a helical content of 38% that does not increase or decrease significantly upon DNA binding (Fig. 3A; Table 1). This helical content of MtrR is significantly lower than that observed for QacR in solution (∼60%) (Fig. 3B), which is underestimated by ∼20% compared to the crystal structure (∼75%). In addition, QacR is known to lose helicity upon DNA binding according to the crystal structure, a result that is not evident in the CD spectra of the QacR-DNA complex (Fig. 3B; Table 1). MtrR has unanticipated high random coil content (40 to 45%) and β-sheet structure (∼18%) that do not appear to change significantly upon DNA binding. Perhaps unstructured regions of MtrR play a role in the binding of small-molecule inducers and undergo coil-to-helix transitions after binding these coeffectors. Indeed, induction of α helicity in the drug-binding domain of thiostrepton-binding transcription regulator TipAS upon binding drug, as well as in the multidrug binding domain of QacR upon multidrug binding, has been observed (16). Contributing in part to the lower-than-expected helicity and higher random coil content and apparent β structure of MtrR might be its inherent instability, as this protein loses activity over a period of days when stored at 4°C (data not shown). Therefore, all CD spectra of MtrR were collected within one hour of its purification. Regardless, the finding of β-sheet and significant random coil structures in MtrR makes this TetR regulator unusual, as the three-dimensional structures of all other TetR family members, including TetR (19), QacR (21, 22), CprB (18), and EthR (4, 6), are essentially all helical.
FIG. 3.
Circular dichroism spectra of MtrR and QacR. (A) MtrR in its apo (black spectra) and DNA-bound (red spectra) forms. (B) QacR in its apo (orange spectra) and IR1-bound (violet spectra) forms.
TABLE 1.
Secondary-structure contents (percent) of MtrR and QacR
| Protein or complex | % Secondary structure content
|
||
|---|---|---|---|
| α helix | β structure | Random coil | |
| MtrR | 37.7 | 18.3 | 45.1 |
| MtrR-DNA | 38.8 | 19.4 | 40.6 |
| QacR | 58.3 | 7.9 | 33.5 |
| QacR-DNA | 60.0 | 8.0 | 32.1 |
The data presented here provide a biochemical characterization of MtrR binding to the mtrCDE promoter and an assessment of its solution state secondary structure in the presence and absence of cognate DNA. Unexpectedly, MtrR contains a large amount of random coil and β sheet, seemingly beyond the error associated with our CD experiments. The latter secondary structure has never been observed in a TetR family member, and confirmation of its existence will require additional structural studies. The differences between MtrR and QacR, TetR, and EthR underscore the wide variety of DNA binding mechanisms of the TetR family. The crystallizations and X-ray structure determinations of MtrR and its DNA complex will be necessary to understand fully the DNA binding mechanism of this TetR family member and are under way.
Acknowledgments
This work was supported by grants from the American Heart Association (0310050Z to K.M.H.) and the National Institutes of Health (AI21150 to W.M.S. and AI48953 to R.G.B.). W.M.S. is the recipient of a Senior Research Career Scientist Award from the VA Medical Research Service.
We thank Maria A. Schumacher for pointing out the basic nature of the N terminus of MtrR.
REFERENCES
- 1.Andrade, M. A., P. Chacon, J. J. Merelo, and F. Moran. 1993. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 6:383-390. [DOI] [PubMed] [Google Scholar]
- 2.Aramaki, H., N. Yagi, and M. Suzuki. 1995. Residues important for the function of a multihelical DNA binding domain in the new transcription factor family of Cam and Tet repressors. Protein Eng. 8:1259-1266. [DOI] [PubMed] [Google Scholar]
- 3.Delahay, R. M., B. D. Robertson, J. T. Balthazar, W. M. Shafer, and C. A. Ison. 1997. Involvement of the gonococcal MtrE protein in the resistance of Neisseria gonorrhoeae to toxic hydrophobic agents. Microbiology 143:2127-2133. [DOI] [PubMed] [Google Scholar]
- 4.Dover, L. G., P. E. Corsino, I. R. Daniels, S. L. Cocklin, V. Tatituri, G. S. Besra, and K. Futterer. 2004. Crystal structure of the TetR/CamR family repressor Mycobacterium tuberculosis EthR implicated in ethionamide resistance. J. Mol. Biol. 340:1095-1105. [DOI] [PubMed] [Google Scholar]
- 5.Engohang-Ndong, J., D. Baillat, M. Aumercier, F. Bellefontaine, G. S. Besra, C. Locht, and A. R. Baulard. 2004. EthR, a repressor of the TetR/CamR family implicated in ethionamide resistance in mycobacteria, octamerizes cooperatively on its operator. Mol. Microbiol. 51:175-188. [DOI] [PubMed] [Google Scholar]
- 6.Frenois, F., J. Engohang-Ndong, C. Locht, A. R. Baulard, and V. Villeret. 2004. Structure of EthR in a ligand bound conformation reveals therapeutic perspectives against tuberculosis. Mol. Cell 16:301-307. [DOI] [PubMed] [Google Scholar]
- 7.Garcia de la Torre, J. G., and V. A. Bloomfield. 1981. Hydrodynamic properties of complex, rigid, biological macromolecules: theory and applications. Q. Rev. Biophys. 14:81-139. [DOI] [PubMed] [Google Scholar]
- 8.Grkovic, S., M. H. Brown, N. J. Roberts, I. T. Paulsen, and R. A. Skurray. 1998. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J. Biol. Chem. 273:18665-18673. [DOI] [PubMed] [Google Scholar]
- 9.Grkovic, S., M. H. Brown, M. A. Schumacher, R. G. Brennan, and R. A. Skurray. 2001. The staphylococcal QacR multidrug regulator binds a correctly spaced operator as a pair of dimers. J. Bacteriol. 183:7102-7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grkovic, S., M. H. Brown, and R. A. Skurray. 2002. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66:671-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guymon, L. F., D. L. Walstad, and P. F. Sparling. 1978. Cell envelope alterations in antibiotic-sensitive and-resistant strains of Neisseria gonorrhoeae. J. Bacteriol. 136:391-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagman, K. E., C. E. Lucas, J. T. Balthazar, L. Snyder, M. Nilles, R. C. Judd, and W. M. Shafer. 1997. The MtrD protein of Neisseria gonorrhoeae is a member of the resistance/nodulation/division protein family constituting part of an efflux system. Microbiology 143:2117-2125. [DOI] [PubMed] [Google Scholar]
- 13.Hagman, K. E., W. Pan, B. G. Spratt, J. T. Balthazar, R. C. Judd, and W. M. Shafer. 1995. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 141:611-622. [DOI] [PubMed] [Google Scholar]
- 14.Hagman, K. E., and W. M. Shafer. 1995. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J. Bacteriol. 177:4162-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hillen, W., and C. Berens. 1994. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 48:345-369. [DOI] [PubMed] [Google Scholar]
- 16.Kahmann, J. D., H. J. Sass, M. G. Allan, H. Seto, C. J. Thompson, and S. Grzesiek. 2003. Structural basis for antibiotic recognition by the TipA class of multidrug-resistance transcriptional regulators. EMBO J. 22:1824-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucas, C. E., J. T. Balthazar, K. E. Hagman, and W. M. Shafer. 1997. The MtrR repressor binds the DNA sequence between the mtrR and mtrC genes of Neisseria gonorrhoeae. J. Bacteriol. 179:4123-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Natsume, R., Y. Ohnishi, T. Senda, and S. Horinouchi. 2004. Crystal structure of a gamma-butyrolactone autoregulator receptor protein in Streptomyces coelicolor A3(2). J. Mol. Biol. 336:409-419. [DOI] [PubMed] [Google Scholar]
- 19.Orth, P., D. Schnappinger, W. Hillen, W. Saenger, and W. Hinrichs. 2000. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat. Struct. Biol. 7:215-219. [DOI] [PubMed] [Google Scholar]
- 20.Pan, W., and B. G. Spratt. 1994. Regulation of the permeability of the gonococcal cell envelope by the mtr system. Mol. Microbiol. 11:769-775. [DOI] [PubMed] [Google Scholar]
- 21.Schumacher, M. A., M. C. Miller, S. Grkovic, M. H. Brown, R. A. Skurray, and R. G. Brennan. 2002. Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J. 21:1210-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schumacher, M. A., M. C. Miller, S. Grkovic, M. H. Brown, R. A. Skurray, and R. G. Brennan. 2001. Structural mechanisms of QacR induction and multidrug recognition. Science 294:2158-2163. [DOI] [PubMed] [Google Scholar]
- 23.Stapleton, P., V. Adams, R. Pike, V. Lucas, G. Roberts, P. Mullany, R. Rowbury, M. Wilson, and H. Richards. 2004. Characterisation of viridans group streptococci with different levels of Tet(M)-mediated tetracycline resistance. Int. J. Antimicrob. Agents 24:439-443. [DOI] [PubMed] [Google Scholar]
- 24.Veal, W. L., A. Yellen, J. T. Balthazar, W. Pan, B. G. Spratt, and W. M. Shafer. 1998. Loss-of-function mutations in the mtr efflux system of Neisseria gonorrhoeae. Microbiology 144:621-627. [DOI] [PubMed] [Google Scholar]