Abstract
Structural studies of the ribosome have benefited greatly from the use of organisms adapted to extreme environments. However, little is known about the mechanisms by which ribosomes or other ribonucleoprotein complexes have adapted to functioning under extreme conditions, and it is unclear to what degree mutant phenotypes of extremophiles will resemble those of their counterparts adapted to more moderate environments. It is conceivable that phenotypes of mutations affecting thermophilic ribosomes, for instance, will be influenced by structural adaptations specific to a thermophilic existence. This consideration is particularly important when using crystal structures of thermophilic ribosomes to interpret genetic results from nonextremophilic species. To address this issue, we have conducted a survey of spontaneously arising antibiotic-resistant mutants of the extremely thermophilic bacterium Thermus thermophilus, a species which has featured prominently in ribosome structural studies. We have accumulated over 20 single-base substitutions in T. thermophilus 16S and 23S rRNA, in the decoding site and in the peptidyltransferase active site of the ribosome. These mutations produce phenotypes that are largely identical to those of corresponding mutants of mesophilic organisms encompassing a broad phylogenetic range, suggesting that T. thermophilus may be an ideal model system for the study of ribosome structure and function.
Members of the bacterial genus Thermus are extreme thermophiles first described by Brock and Freeze in 1969 (2) and have since been found in terrestrial and marine thermal environments throughout the world (60). Together with Deinococcus, Meiothermus, Marinithermus, Oceanithermus, and Vulcanithermus, they form a deeply branching phylum now known to be monophyletic (23, 59). The close affiliation between Thermus and Deinococcus has been confirmed by complete genome sequences of Thermus thermophilus (30) and Deinococcus radiodurans (59). The finding of slightly thermophilic species related to Deinococcus, together with the thermophilic nature of the other genera of this phylum, suggests that thermophily is a primitive character of this clade (59).
Recent advances in structural biology have produced a vast reservoir of high-resolution structural information regarding components of the protein synthetic machinery from members of the Deinococcus-Thermus phylum, including high-resolution crystal structures of the T. thermophilus 30S subunit (47, 61), medium-resolution structures of the entire T. thermophilus 70S ribosome (65), and high-resolution structures of the D. radiodurans 50S subunit (26). The value of these structures is magnified by their interpretation in light of several decades of genetics and biochemistry using ribosomes from mesophiles such as Escherichia coli. However, the ability to make such interpretations is potentially compromised by the absence of genetic and biochemical data obtained for ribosomes from members of the Deinococcus-Thermus phylum. It is assumed that nucleotide sequence conservation within the functional centers reflects conservation of three-dimensional structure. However, it is not known to what degree highly conserved residues in functional sites engage in tertiary interactions specific to adaptation to extreme existence. The resolution of this issue is of some importance, as the general relevance of data obtained from crystal structures of thermophilic ribosomes will be impacted by the extent of structural adaptations unique to thermophily. One approach to assessing the degree of structural divergence or specialization is to characterize the phenotypes of mutations in conserved functional sites and compare them to previously described homologous mutations in a wide range of mesophilic organisms. It is expected that a high degree of structural conservation should be reflected in the appearance of a similar spectra of mutants expressing similar phenotypes. We have previously described T. thermophilus mutations in ribosomal protein S12 (20) and in ribosomal protein L11 and 23S rRNA in the factor-binding region (4), which produced phenotypes similar to those of other species. Here we describe a more extensive analysis targeting the decoding and peptidyltransferase centers of 16S rRNA and 23S rRNA, respectively.
T. thermophilus, in addition to being a valuable source of ribosomes for crystallographic efforts (3, 8, 9, 47, 61, 65), offers several advantages over other species in conducting mutational analysis of rRNA genes. Foremost is the ease of genetic manipulation of this organism (29, 35). The presence of only two copies of rRNA genes (30, 56) facilitates the isolation of rRNA mutants (21), including mutants with homogeneous ribosome populations, most likely resulting from gene conversion between homologous rRNA genes (4). For conducting mutational analysis of ribosomes, antibiotic resistance mutations are advantageous, as T. thermophilus is susceptible to a wide range of antibiotics, providing the opportunity for multiple genetic selections which can target individual functional sites. Further, similar or identical mutations have been identified over a broad phylogenetic range, including bacteria, archaea, eukarya, mitochondria, and chloroplasts (55), consistent with the universal conservation of many rRNA nucleotides located in antibiotic binding sites. Previously identified mutations provide a basis for prediction of phenotypes and the location of mutations arising in a genetic selection.
Here we describe the isolation of a number of antibiotic-resistant mutants of T. thermophilus containing single-base substitutions in 16S and 23S rRNA genes. Characterization of the antibiotic resistance profiles of these mutants indicates that, to a remarkable degree, mutations identified in the thermophile T. thermophilus phenotypically resemble those found in other species, indicating that genetic and biochemical data obtained with ribosomes from mesophilic species can be interpreted using structures of ribosomes derived from T. thermophilus. These results also indicate the potential for this organism to serve as a model system for the genetic analyses of ribosome structure and function.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
All mutants were derived from the Icelandic T. thermophilus strain IB-21 (ATCC 43615) (36). All T. thermophilus cultures were grown aerobically at 72°C in ATCC medium 1598 (Thermus Enhanced Medium [TEM]). Plates contained TEM solidified with 2.8% Difco agar.
Isolation of mutants.
Mutants were isolated by plating 109 cells from a saturated overnight culture onto TEM plates containing antibiotic. Colonies were purified by multiple restreakings. Stocks of isolates were maintained in 25% glycerol frozen at −70°C. Aminoglycosides, all macrolides, clindamycin, lincomycin, chloramphenicol, sparsomycin, and capreomycin sulfate were obtained from Sigma Chemical. The streptogramin B antibiotic pristinamycin (mikamycin B) and the oxazolidinone antibiotic linezolid were generous gifts of Pfizer Pharmaceuticals and Upjohn Pharmaceuticals, respectively.
Antibiotic concentrations used in selections were as follows: kanamycin, 30 μg/ml; paromomycin, 20 and 50 μg/ml; apramycin, 20 μg/ml; neomycin, 5 and 10 μg/ml; gentamicin, 0.5 μg/ml; hygromycin B, 50 and 100 μg/ml; chloramphenicol, 25, 50, 100, and 200 μg/ml; tylosin, 100 μg/ml; and sparsomycin, 0.1 and 0.2 μg/ml.
Identification of mutations.
Segments of relevant genes were amplified by PCR using Taq DNA polymerase (Promega), and PCR products were sequenced manually using Sequenase DNA polymerase (U.S. Biochemical) or by the University of California—Davis sequencing facility. Oligonucleotide primers for PCR and sequencing were obtained from Operon Technologies or Invitrogen (Table 1). Segments of rrsA and rrsB corresponding to 16S rRNA helix 44 were amplified using the oligonucleotides Tth 16S-A and Tth 16S-H and sequenced using oligonucleotide Tth 16S-K. The rpsL gene was amplified using oligonucleotides Tth S12-3 and Tth S12-4. Mutations in and around the peptidyltransferase active site were identified by amplifying rrlA and rrlB with the primer pair Tth 23S-E and Tth 23S-F and sequencing with primers Tth 23S-W, Tth 23S-A, and Tth 23S-J. All mutations are designated using the E. coli rrnB numbering system.
TABLE 1.
Oligonucleotide primers used to amplify and sequence rRNA and ribosomal protein genes
| Name | Sequence | Positiona |
|---|---|---|
| Tth S12-3 | 5′-GTGGTGGCACTGCCGACGATCAATC-3′ | N terminal |
| Tth S12-4 | 5′-GACCTGGTAGTTGGCGCCGCCCAC-3′ | C-terminal |
| Tth 16S-A | 5′-CCCGGGTAGTCCACGCCCTAAACG-3′ | 818 |
| Tth 16S-H | 5′-AGAAAGGAGGTGATCCAGCCGCAC-3′ | 1520 |
| Tth 16S-K | 5′-GTGGGCCCAGTTCGGATTGGGGTCTG-3′ | 1316 |
| Tth 23S-A | 5′-CTCCCACCTATCCTACGCAGGCGC-3′ | 2103 |
| Tth 23S-E | 5′-CGCCAAGGAACTCTGCAAGTTGGC-3′ | 1684 |
| Tth 23S-F | 5′-CCAGAGGTGCGTCCCTTCCGGTCC-3′ | 2663 |
| Tth 23S-J | 5′-CAGGATGCGACGAGCCGACATCGAGG-3′ | 2948 |
| Tth 23S-W | 5′-CCCTCAAGCCTCCTGCGCCC-3′ | 2622 |
Indicates the mature rRNA base located at the end of the oligonucleotide sequence.
Assessment of antibiotic cross-resistance phenotypes.
Resistance to aminoglycosides and capreomycin was assessed by streaking for single colonies using loopfuls of overnight cultures onto TEM plates containing antibiotics at concentration increments of twofold or smaller. Resistance to all drugs was assessed by spreading 100 μl of a saturated overnight culture onto a TEM plate and placing an 0.64-cm-diameter filter disk (Schleicher & Schuell 740-E) containing antibiotic on the plate. Disks contained 100 μg of each antibiotic with the following exceptions: 30 μg neomycin, 20 μg ampicillin, and 1 μg sparsomycin. Plates were incubated at 72°C, and the diameter of the clearing zone produced by the antibiotic was recorded.
RESULTS
Selection of antibiotic-resistant mutants with homogeneous ribosome populations.
Our ability to obtain a large number of rRNA mutants of T. thermophilus is made possible in part by the presence of only two rRNA gene copies. In our earlier search for rRNA mutants, we had obtained the A2058G mutation in 23S rRNA (21). All three isolates of this mutant that we examined proved to contain heterogeneous ribosome populations. In the present study, we identified 21 different base substitutions in 16S and 23S rRNA (Table 2), all but one of which were found to be present in homogeneous ribosome populations, although in some instances, heterogeneous mutants could also be identified. Ribosomal protein mutants produce homogeneous populations of ribosomes due to the single-copy nature of ribosomal protein genes, as recently confirmed by the genome sequences of T. thermophilus HB27 (30) and T. thermophilus HB8 (http://www.ncbi.nlm.nih.gov/genomes). However, pure ribosome populations containing rRNA mutations could arise either from deletions removing the remaining wild-type gene or by gene conversion events between the two homologous rRNA genes (28). We consider the latter explanation more probable, given the highly efficient homologous recombination system of this organism (29, 35). This phenomenon has been previously reported by this laboratory for thiostrepton-resistant 23S rRNA mutants of T. thermophilus HB8 (4), and similar observations have been reported for antibiotic-resistant mutants of Mycobacterium spp. (44). A third possibility, that two independent mutational events produce homogeneous ribosome populations, we consider unlikely on probabilistic grounds and because we have never observed double heterozygotes, e.g., mutants with two different base substitutions, one in each rRNA gene. While most mutants were found to contain pure ribosome populations, those isolated on lower drug concentrations in some instances carried mixed populations of mutant and wild-type ribosomes. This presumably reflects the greater fitness of homogeneous mutants on higher drug concentrations.
TABLE 2.
Mutants of T. thermophilus IB-21 identified in this study, the drug selections used to obtain them, and the number of isolates of each mutant
| rRNA molecule | Mutation | Selection(s) | No. of isolates |
|---|---|---|---|
| 16S | U1406A | Hygromycin B | 1 |
| 16S | U1406C | Hygromycin B | 27 |
| 16S | U1406G | Hygromycin B | 5 |
| 16S | A1408G | Aminoglycosides, capreomycin | 41 |
| 16S | C1409G | Capreomycin | 1 |
| 16S | G1491A | Capreomycin | 18 |
| 16S | U1495C | Hygromycin B | 26 |
| 23S | A2030G | Chloramphenicol | 10 |
| 23S | A2059G | Tylosin | 3 |
| 23S | G2061A | Chloramphenicol | 32 |
| 23S | A2062G | Tylosin, chloramphenicol | 10 |
| 23S | G2447A | Chloramphenicol | 11 |
| 23S | C2452U | Chloramphenicol, sparsomycin | 7 |
| 23S | A2453G | Chloramphenicol | 1 |
| 23S | U2500C | Chloramphenicol | 1 |
| 23S | A2503G | Chloramphenicol | 2 |
| 23S | U2504C | Chloramphenicol | 1 |
| 23S | U2504G | Chloramphenicol | 1 |
| 23S | U2504A | Chloramphenicol | 1 |
| 23S | G2505A | Chloramphenicol | 2 |
Previous reports have not described a large collection of mutations identified in a single study or in a single organism (Table 3). The collection of mutants identified here allows a direct comparison of all these mutations in one unadulterated genetic background, in a bacterial strain obtained from its natural environment without subsequent genetic modification or manipulation. These mutants and their progenitor have not been exposed to any mutagenic agents or repeated subculturing. Furthermore, no plasmids were used in this work, and all mutations were obtained in situ in rRNA genes situated at their native chromosomal loci.
TABLE 3.
Antibiotic resistance mutations, similar or identical to those described in this study, that have been previously identified in other organisms
| rRNA molecule | Mutation | Organisma | Resistance phenotype | Reference |
|---|---|---|---|---|
| 16S | A1408G | Chlamydomonas reinhardtii chloroplast | Kanamycin, neamine | 27 |
| 16S | C1409G | Saccharomyces cerevisiae mitochondria | Paromomycin | 37 |
| 16S | G1491A | Tetrahymena thermophila cytoplasmic | Paromomycin | 50 |
| 16S | U1406C | Mycobacterium smegmatis | Hygromycin B | 42 |
| 16S | U1495C | Tetrahymena thermophila cytoplasmic | Hygromycin B | 50 |
| 23S | A2058G | S. cerevisiae mitochondria | Erythromycin | 49 |
| 23S | A2059G | Nicotiana plumbaginifolia chloroplast | Lincomycin | 11 |
| 23S | A2062C | Halobacterium halobium | Chloramphenicol | 38 |
| 23S | G2447A | S. cerevisiae mitochondria | Chloramphenicol | 15 |
| 23S | C2452A | Mus musculus mitochondria | Chloramphenicol | 1 |
| 23S | C2452U | Halobacterium halobium | Linezolid | 34 |
| 23S | A2453C | Halobacterium sp. | Anisomycin | 31 |
| 23S | A2453G | Halobacterium halobium | Linezolid | 34 |
| 23S | U2500C | Halobacterium halobium | Linezolid | 34 |
| 23S | A2503C | S. cerevisiae mitochondria | Chloramphenicol | 15 |
| 23S | U2504C | M. musculus mitochondria | Chloramphenicol | 33 |
| 23S | U2504C | Halobacterium halobium | Linezolid | 34 |
Species in which mutation was first identified.
Selection of mutations in the decoding center of 16S rRNA.
Spontaneous mutants resistant to the 2-deoxystreptamine aminoglycosides paromomycin, apramycin, neomycin, gentamicin, kanamycin, and hygromycin B were readily obtained. Interestingly, all mutants selected on paromomycin exhibited a drug dependence phenotype (7). Sequencing of rrsA, rrsB, and rpsL indicated the absence of any mutation in 16S rRNA but instead revealed the existence of either the P90L or P90R substitution in ribosomal protein S12. The P90L mutation has previously been found in other organisms and in T. thermophilus IB-21 (20) and T. thermophilus HB8 (9) and is known to confer a streptomycin dependence phenotype.
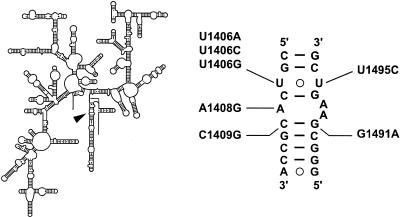
T. thermophilus mutants selected for resistance to the aminoglycoside kanamycin, neomycin, gentamicin, or apramycin were all found to carry the single-base substitution A1408G in 16S rRNA (Fig. 1). This mutation was identified three times on neomycin, once on gentamicin, and four times on apramycin (Table 2). Selection on kanamycin also produced a single isolate containing the U1406C mutation. Selections on hygromycin B provided mutants containing the single-base substitution U1406A, -C, or G or U1495C. These two residues form a noncanonical U-U pair within helix 44 of 16S rRNA, with U1495 directly contacting hygromycin B (3). It seems reasonable that mutations at U1406, which itself does not directly contact hygromycin B, confer resistance by affecting the positioning of U1495.
FIG. 1.
Secondary structure model of the T. thermophilus 16S rRNA molecule (left) (6), with the decoding center indicated by the arrowhead, and expanded (right) to indicate the mutations identified in this study. Mutations at the U1406-U1495 base pair were selected on hygromycin B; the A1408G mutation was isolated on kanamycin; and the A1408G, C1409G, and G1491A mutations were isolated on capreomycin. A1408G, C1409G, and G1491A confer resistance to a number of aminoglycosides as predicted by the phenotypes of homologous mutations in other species.
Capreomycin is a member of the tuberactinomycin family of peptide antibiotics; another member of this class of drugs, viomycin, has been characterized as an inhibitor of the translocation step of protein synthesis (reviewed in reference 18). T. thermophilus IB-21 mutants selected for resistance to capreomycin were found to carry mutations exclusively in 16S rRNA, including A1408G, C1409G, and G1491A. A1408G was again the most frequently identified mutation, with C1409G being isolated only once. Tuberactinomycins, like the aminoglycosides, have been shown to cause mistranslation (39) and compete with aminoglycosides for binding to the ribosome (see reference 18). While these observations suggest the existence of overlapping binding sites for aminoglycosides and tuberactinomycins, there is currently no crystallographic or chemical footprinting evidence for a direct binding of tuberactinomycins to the decoding center.
Cross-resistance phenotypes of decoding site mutants.
The phenotypes of mutants were examined in more detail by assessing their resistance to a number of 30S-targeted antibiotics, including the aminoglycosides, capreomycin, and tetracycline. The most robust resistance that we observed was conferred by the A1408G mutation to kanamycin. While wild-type T. thermophilus IB-21 was completely inhibited by kanamycin at 10 μg/ml, the A1408G mutant was not completely inhibited until 3 mg/ml (Table 4). The C1409G mutant is also quite resistant to kanamycin, being completely inhibited only at 2 mg/ml. Mutations at U1406 also produced substantial kanamycin resistance. As stated above, the U1406C mutation was also independently isolated in a selection for kanamycin-resistant mutants. Resistance to high concentrations of apramycin, neomycin, gentamicin, and kanamycin but not paromomycin conferred by A1408G is consistent with the isolation of this mutation on apramycin, neomycin, gentamicin, and kanamycin but not on paromomycin. Both the A1408G and C1409G mutations confer resistance to a wide range of aminoglycoside antibiotics (Table 5). Although the C1409G and G1491A mutations were isolated by selecting capreomycin resistance, they were found to exhibit cross-resistance to aminoglycosides, as predicted from previous genetic studies. Further, independent isolates of the A1408G mutation exhibited identical cross-resistance patterns, regardless of whether isolated on capreomycin or on aminoglycosides.
TABLE 4.
Antibiotic resistance phenotypes conferred by 16S rRNA mutationsa
| Mutation | Min concn (μg/ml)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Str | Apr | Par | Neo | Kan | Gen | Hyg | Cap | |
| WT | 20 | 2 | 1 | 1 | 10 | 0.5 | 20 | 50 |
| A1408G | 500 | 500 | 20 | 500 | 3,000 | 500 | 20 | >1,000 |
| C1409G | 500 | 200 | 50 | 5 | 2,000 | 100 | 20 | >1,000 |
| G1491A | 20 | 100 | 5 | 2 | 20 | 1 | 20 | 1,000 |
| U1406A | 20 | 2 | 2 | 1 | 500 | >500 | 100 | 50 |
| U1406C | 20 | 10 | 10 | 2 | 500 | 5 | 100 | 50 |
| U1406G | 20 | 5 | 5 | 2 | 100 | 50 | 100 | 100 |
| U1495C | 20 | 20 | 5 | 2 | 20 | 0.5 | 100 | 50 |
Shown are minimal concentrations required to completely inhibit growth as assayed by streaking for single colonies on solid TEM. Abbreviations: WT, wild type; Str, streptomycin; Apr, apramycin; Par, paromomycin; Neo, neomycin; Kan, kanamycin; Gen, gentamicin; Hyg, hygromycin B; Cap, capreomycin.
TABLE 5.
Antibiotic zones of inhibition of 16S rRNA mutants assessed by disc assaya
| Mutation | Diam (mm) of zone of inhibition by:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Str | Apr | Par | Neo | Rib | Nea | Kan | Tob | Gen | Hyg | Cap | Spc | Ksg | Tet | Ery | Rif | Amp | |
| WT | 25 | 38 | 42 | 22 | 37 | 29 | 20 | 32 | 43 | 18 | 13 | − | − | 31 | 28 | 22 | 51 |
| A1408G | − | − | 15 | − | − | − | − | − | − | 22 | − | − | − | 27 | 26 | 14 | 50 |
| C1409G | − | − | − | 14 | − | − | − | − | − | 16 | − | − | − | 30 | 30 | 23 | 50 |
| G1491A | 27 | − | 22 | 18 | 14 | 20 | 17 | 24 | 31 | 16 | − | − | − | 28 | 25 | 18 | 48 |
| U1406A | 26 | 32 | 26 | 17 | 26 | 20 | − | − | − | − | 11 | − | − | 26 | 29 | 22 | 48 |
| U1406C | 28 | 19 | 26 | 17 | 20 | 19 | 13 | 15 | 22 | − | 10 | − | − | 30 | 27 | 19 | 46 |
| U1406G | 28 | 17 | 25 | 19 | 22 | − | − | 19 | 18 | − | 13 | − | − | 32 | 40 | 27 | 49 |
| U1495C | 29 | 21 | 22 | 18 | 24 | 18 | 18 | 24 | 27 | − | 11 | − | − | 26 | 29 | 23 | 41 |
Diameters include diameter of disk, or 6.4 mm. Mutations were isolated on kanamycin (A1408G), capreomycin (A1408G, C1409G, and G1491A), or hygromycin B (U1406C, U1406G, U1406A, and U1495C). Str, streptomycin; Apr, apramycin; Par, paromomycin; Neo, neomycin; Rib, ribostamycin; Nea, neamine; Kan, kanamycin; Tob, tobramycin; Gen, gentamicin; Hyg, hygromycin B; Cap, capreomycin; Spc, spectinomycin; Ksg, kasugamycin; Tet, tetracycline; Ery, erythromycin; Rif, rifampin; Amp, ampicillin; −, no inhibition; WT, wild type.
Mutations at the U1406-U1495 base pair did not exhibit identical antibiotic resistance patterns (Tables 4 and 5). For instance, U1406A confers much stronger resistance to gentamicin than do the other two base changes at this position. This suggests, perhaps not surprisingly, that the geometry of this pair is distorted in qualitatively distinct ways with each substitution. In contrast, the U1495C mutation does not confer detectable gentamicin resistance, despite the presumed loss of the contact between this base and gentamicin as predicted from the A-site rRNA analog-gentamicin nuclear magnetic resonance structure (64).
While mutations in helix 44 have long been known to confer resistance to 2-deoxystreptamine aminoglycosides, they have not been reported to confer resistance to streptomycin. We recently reported the finding of streptomycin resistance conferred by the A1408G mutation and proposed one possible mechanism (22). In the present study, we also observed that the C1409G mutation confers comparable levels of streptomycin resistance (Table 4), presumably by conformational distortion due to the resulting G-G mismatch. In contrast, G1491A does not confer any detectable decrease in streptomycin sensitivity, even though this residue makes direct backbone contact with streptomycin.
None of our mutants exhibited resistance to tetracycline, consistent with the absence of direct contact between tetracycline and helix 44 (3). Further, our attempts to isolate spontaneous tetracycline-resistant mutants were not successful.
Comparison of T. thermophilus 16S rRNA mutations with previously identified decoding site mutations.
The A1408G mutation was first identified in kanamycin- and neamine-resistant mitochondrial mutants of Chlamydomonas reinhardtii (27), while the C1409G and G1491A mutations were originally identified in paromomycin-resistant mitochondrial mutants of Saccharomyces cerevisiae (37) and paromomycin-resistant mutants of Tetrahymena thermophila (50), respectively. The relationship between the decoding center and tuberactinomycin action was previously implied by the isolation of the G1491A mutation in viomycin-resistant mutants of Mycobacterium smegmatis (52). The U1495C mutation was first identified in a hygromycin B-resistant mutant of Tetrahymena (50), and more recently, a U1406C mutation was identified in hygromycin B-resistant mutants of M. smegmatis (42). The U1495C mutation has also been shown to confer hygromycin B resistance in S. cerevisiae cytoplasmic ribosomes (10).
Some of these mutations have been previously introduced by site-directed mutagenesis into a plasmid-encoded E. coli rrnB operon, sometimes producing unexpected phenotypes. Several mutations at the C1409-G1491 base pair produced low-level resistance to a number of aminoglycosides (13); however, the C1409G and G1491A mutations were described as producing dominant lethal phenotypes, whereas the equivalent mutations in T. thermophilus (this study), Tetrahymena (50), and S. cerevisiae (37) are viable. Similarly, the 16S rRNA mutations U1406G and U1495C, -G, or -A were reported as dominant lethal in E. coli (45), even though all three base changes at U1406 are viable in T. thermophilus (this work) and U1406C is viable in M. smegmatis and confers hygromycin B resistance (42). Further, U1495C is viable in T. thermophilus (this work), Tetrahymena (50), and S. cerevisiae (10). On the other hand, A1408G is viable in E. coli (45) as it is in T. thermophilus (this work), Mycobacterium spp. (43), and C. reinhardtii chloroplast (27). U1406A and A1408G both give resistance to a number of aminoglycosides in E. coli (46), as they do in T. thermophilus. The A1408G mutation confers resistance to kanamycin and other 2-deoxystreptamine aminoglycosides in all organisms examined. Thus, in general, the T. thermophilus mutants more closely resemble their mitochondrial, chloroplast, eukaryotic, and bacterial counterparts than do the equivalent E. coli mutants constructed with multicopy plasmids.
Selection of mutations in the peptidyltransferase center of 23S rRNA.
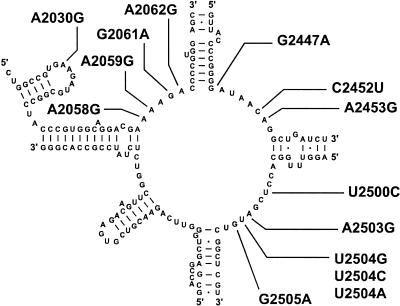
The peptidyltransferase active site is the target of a number of structurally unrelated classes of antibiotics. We previously described the isolation of mutants resistant to the 14-atom macrolide erythromycin by virtue of the A2058G mutation (21). Selections on the 16-atom macrolide tylosin (100 μg/ml) led to the identification of two other mutations in this same region, A2059G and A2062G (Fig. 2; Table 1). While the T. thermophilus A2058G erythromycin resistance mutation was found only in the heterogeneous populations (21), in the present study, the A2059G and A2062G mutations were each found in both pure and mixed populations of ribosomes.
FIG. 2.
Secondary structure model of the peptidyltransferase active site of the T. thermophilus 23S rRNA molecule. Base substitutions characterized in this study are indicated. The A2058G mutation was previously isolated on erythromycin (21), the A2059G mutation was isolated on tylosin, and the A2062G mutation was isolated on tylosin and on chloramphenicol. All others were isolated on chloramphenicol, with C2452U also being isolated on sparsomycin.
Chloramphenicol is a well-documented inhibitor of (peptidyltransferase-catalyzed) peptide bond formation (reviewed in reference 18) and more recently has been shown to cause translational miscoding (54). Footprinting experiments have indicated contact of this drug with several 23S rRNA bases in the peptidyltransferase active site (40), and two 50S subunit-chloramphenicol cocrystal structures have been solved, although these show two distinct chloramphenicol binding states (25, 48). These structural studies are consistent with the fact that mutations conferring chloramphenicol resistance are usually found in and around the active site (55). Selection for T. thermophilus IB-21 mutants resistant to chloramphenicol produced the largest variety of base substitutions (Fig. 2; Table 2), including A2030G, G2061A, A2062G, G2447A, C2452U, A2453G, U2500C, A2503G, U2504A, U2504C, U2504G, and G2505A. Two of the mutations that we detected, A2030G and U2500C, have not previously been reported. As stated above, the A2062G mutation was isolated only on the lowest concentration of chloramphenicol and found to have a low MIC (data not shown). This is consistent with the weak cross-resistance expressed by the same mutant isolated on tylosin (data not shown).
The G2061A mutation was distinct in its extreme slow-growth phenotype, increasing doubling time from 48 min to around 3 h 45 min. It was possible to distinguish this mutant on selection plates as small colonies, and it was among the most abundant mutants. Isolates containing the G2447A mutation were also frequently found among the faster-growing mutants. In the sequencing of over 60 chloramphenicol-resistant mutants, a number of base substitutions were identified only once, indicating that we have not, in all probability, identified all possible mutations in our selections.
Sparsomycin is a peptidyltransferase inhibitor that acts by stabilizing P-site substrate binding (18) and has been shown to stimulate factor-independent translocation (16). Selections for mutants resistant to sparsomycin produced only a single mutation, C2452U, one of the same mutations isolated in chloramphenicol selections.
Cross-resistance phenotypes conferred by peptidyltransferase active site mutations.
We examined all the active site mutants for resistance to the 14-atom macrolides erythromycin, roxithromycin, and oleandomycin; the 16-atom macrolides tylosin, midecamycin, and spiramycin; the lincosamides lincomycin and clindamycin; and the streptogramin B antibiotic pristinamycin (mikamycin B). We also examined cross-resistance to chloramphenicol, sparsomycin, linezolid, and capreomycin. As controls, we also tested responses to streptomycin, ampicillin, and the RNA polymerase inhibitor rifampin.
The A2058G mutation has long been known to confer the so-called MLS (macrolide-lincosamide-streptogramin B) resistance phenotype (55, 57). Unexpectedly, our T. thermophilus A2058G mutant did not exhibit a streptogramin B resistance phenotype. One possible explanation is that streptogramin B resistance may be recessive in this species, and the sensitivity of our A2058G mutant results from the heterozygous state of this allele. The T. thermophilus A2059G mutation also produces a macrolide-lincosamide-resistant, streptogramin B-sensitive phenotype, as it does in mesophiles (57). The T. thermophilus A2059G and A2062G mutations were both found to confer resistance to macrolides, including the 14-atom macrolides erythromycin and roxithromycin and the 16-atom macrolides tylosin, midecamycin, and spiramycin (Table 6). The A2062G mutation produced resistance to macrolides and chloramphenicol but not to lincosamides or streptogramin B. Interestingly, the only mutation to confer resistance to the streptogramin B antibiotic pristinamycin was G2061A.
TABLE 6.
Zones of inhibition of 23S rRNA mutants by 50S subunit inhibitorsa
| Mutation | Diam (mm) of zone of inhibition by:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ery | Rox | Ole | Tyl | Mid | Spi | Lnc | Cln | Prs | Cam | Spr | Lzd | Cap | Str | Rif | Amp | |
| WT | 28 | 28 | − | 35 | 24 | 16 | 38 | 42 | 41 | 32 | 26 | − | 19 | 25 | 22 | 51 |
| A2030G | 20 | 15 | − | 40 | − | 16 | − | − | 28 | − | 36 | − | 24 | 31 | 18 | 53 |
| A2058G | − | − | − | 17 | 9 | − | − | − | 40 | 30 | 25 | − | 22 | 27 | 17 | 52 |
| A2059G | − | − | − | − | − | − | 16 | 42 | 42 | 35 | 25 | − | 17 | 32 | 20 | 55 |
| G2061A | 46 | 44 | − | 42 | 31 | 36 | 24 | 43 | − | − | 15 | − | 17 | 27 | 24 | 51 |
| A2062G | − | − | − | − | − | − | 43 | 45 | 30 | − | 25 | − | 21 | 31 | 16 | 53 |
| G2447A | 28 | 31 | − | 39 | 22 | 20 | 38 | 45 | 32 | − | 24 | − | 16 | 32 | 20 | 50 |
| C2452U | 23 | 20 | − | 38 | 23 | 26 | 17 | 29 | 38 | − | − | − | 17 | 32 | 17 | 47 |
| A2453G | 29 | 31 | − | 41 | 19 | 22 | 36 | 52 | 40 | − | 14 | − | 24 | 33 | 17 | 49 |
| U2500C | 20 | 24 | − | 34 | 21 | 21 | 39 | 45 | 37 | − | 27 | − | 19 | 35 | 20 | 51 |
| A2503G | 25 | 25 | − | 29 | − | − | 35 | 41 | 35 | − | 22 | − | 15 | 34 | 20 | 56 |
| U2504A | 18 | 25 | − | 43 | 20 | 21 | 48 | 45 | 33 | − | 30 | − | 25 | 34 | 28 | 52 |
| U2504C | 15 | 17 | − | 36 | 19 | 17 | 35 | 38 | 37 | − | 21 | − | 14 | 31 | 17 | 51 |
| U2504G | 22 | 19 | − | 33 | 22 | 18 | − | 36 | 30 | − | 20 | − | 13 | 34 | 19 | 53 |
| G2505A | − | − | − | 37 | 31 | − | − | 39 | 27 | − | 17 | − | 18 | 33 | 18 | 55 |
A2058G was isolated on erythromycin, and A2059G was isolated on tylosin; all others were isolated on chloramphenicol. Abbreviations: WT, wildtype; −, no inhibition; Ery, erythromycin; Rox, roxithromycin; Ole, oleandomycin; Tyl, tylosin; Mid, midecamycin; Spi, spiramycin; Lnc, lincomycin; Cln, clindamycin; Prs, pristinamycin; Cam, chloramphenicol; Spr, sparsomycin; Lzd, linezolid; Cap, capreomycin; Str, streptomycin; Rif, rifampin; Amp, ampicillin.
The G2505A mutation was found to confer resistance to 14-atom macrolides and the 16-atom macrolide spiramycin but not to the 16-atom macrolides tylosin and midecamycin. G2505 contacts the desosamine sugar moiety of erythromycin and roxithromycin via its phosphate (48), and resistance presumably occurs via local backbone distortion. G2505 also contacts the mycarose sugar moiety of tylosin and spiramycin via its O2′ (24), but G2505A confers resistance to spiramycin and not tylosin. Mutations at U2504 all confer resistance to chloramphenicol. However, U2504G was distinct from the other two substitutions in conferring resistance to lincomycin.
Of all the active site mutations, only C2452U showed detectable resistance to sparsomycin, consistent with this mutation being the only one isolated on this drug. The Haloarcula marismortui 50S subunit-sparsomycin cocrystal structure shows C2452 to be in proximity to this drug (25).
Comparison of T. thermophilus 23S rRNA mutants with those of other species.
As stated earlier, many of the base substitutions identified in this study were originally found in various other organisms, particularly in mitochondria and chloroplasts (55). Perhaps the most frequently reported base substitutions in and around the active site are at A2058 (57), first identified in S. cerevisiae mitochondria (49). The T. thermophilus A2058G mutation confers resistance to 14-atom and 16-atom macrolides, although resistance to tylosin was relatively weak (Table 6). This mutation has been reported to not confer resistance to tylosin in Arcanobacterium pyogenes (32). The adjacent A2059G mutation, originally identified in lincomycin-resistant Nicotiana plumbaginifolia chloroplasts (11), confers weak lincomycin resistance in T. thermophilus (Table 6). This mutation also confers macrolide resistance, as has been reported for several other species (57).
We did not detect mutations at the universally conserved residue A2451, despite the identification of a chloramphenicol resistance mutation at this position in mouse mitochondria (33) and the direct contact between this base and chloramphenicol as detected by chemical footprinting (40). While it may be that mutations at this position produce dominant lethal phenotypes in T. thermophilus as they do in E. coli (41, 53), it should be remembered that several mutations that are lethal in E. coli are viable in other species, including T. thermophilus. Further, a number of chloramphenicol resistance mutations found in this study were isolated only once, indicating that we have almost certainly not identified all possible chloramphenicol resistance mutations in this organism. Mutations at A2451 may simply be too infrequent for us to detect at our current depth of screening.
Of the mutations identified in chloramphenicol selections, the vast majority have been observed in other species. An A2062C mutation has been reported to confer resistance to chloramphenicol in Halobacterium halobium (38) and to 16-atom macrolides and streptogramins in Streptococcus pneumoniae (12). Our similar mutation, A2062G, does not confer lincosamide resistance in T. thermophilus, as is the case with Mycoplasma hominis (17). Interestingly, A2062 becomes covalently linked to 16-atom macrolides as observed in the crystal structure of the 50S subunit of H. marismortui (24). The U2504C mutation has been identified as a chloramphenicol resistance mutation in Mus musculus (33) and S. cerevisiae mitochondria (58). We have found that all three base substitutions at this position confer chloramphenicol resistance in T. thermophilus.
A2030G was isolated 10 times in our selections (Table 2), but as this mutation has not been reported previously, no comparisons can be made. G2032A confers lincomycin resistance in Nicotiana chloroplasts (11) and chloramphenicol (14) or linezolid (54, 63) resistance in E. coli. A2030G confers resistance to lincomycin, clindamycin, and chloramphenicol in T. thermophilus, suggesting a similar effect of these mutations on local rRNA structure. Neither A2030 nor G2032 makes direct contact with chloramphenicol or lincomycin (48), and mutations in this loop presumably disrupt tertiary structure to cause resistance.
G2447A was first found as a chloramphenicol resistance mutation in S. cerevisiae mitochondria (15) and has since been constructed by site-directed mutagenesis in E. coli, where it produces a deleterious phenotype but confers chloramphenicol resistance to ribosomes translating in vitro (53).
G2061A has recently been identified in clindamycin-resistant plastid mutants of the protist Toxoplasma gondii (5). However, the T. thermophilus mutant showed no resistance to either lincomycin or clindamycin. Lincosamide resistance has not otherwise been previously reported for this mutation, and no interaction between clindamycin and G2061 was noted in the D. radiodurans crystal structure (48). We therefore cannot explain this phenotypic difference at this time.
Mutations similar to those identified in this study that have also been found in other organisms include C2452A in Mus mitochondria (1), A2453C in H. halobium (31), and A2503C in S. cerevisiae mitochondria (15). In the case of A2453C, H. halobium mutants were selected for resistance to anisomycin, a drug which binds to the active site (25). Selection of T. thermophilus mutants on sparsomycin gave rise to C2452U, which confers sparsomycin resistance in H. halobium (51). However, while U2500C confers weak sparsomycin resistance in H. halobium, we did not detect resistance conferred by this mutation in T. thermophilus.
A number of mutations, similar or identical to those appearing in our selections, including A2062C, C2452U, A2453G, U2500C, and U2504C, have been shown to confer resistance to linezolid in H. halobium (34). As linezolid does not inhibit T. thermophilus, we were unable to assess cross-resistance to this drug. With regard to the large subunit, again T. thermophilus mutants appear to be consistent with mitochondria, chloroplasts and most bacteria.
DISCUSSION
Genetic selections for spontaneously arising rRNA mutants of T. thermophilus recapitulate many of the mutations found in a phylogenetically diverse range of species, exhibiting remarkable phenotypic congruity over great evolutionary distances. This is particularly striking given the extreme environments to which Thermus spp. are specifically adapted and the structural constraints presumed to be placed on ribosomes functioning at high temperatures. The deeply branching nature of the Deinococcus-Thermus phylum and its position closer to the root of the universal phylogenetic tree may, in part, be responsible for the similarity of T. thermophilus mutants to those of such a broad range of taxa. At the same time, our findings provide a powerful demonstration of the extreme conservation of ribosome active site structure throughout evolutionary history.
The results of this survey also suggest the absence of a large number of uniquely thermophilic structural adaptations in and around T. thermophilus ribosomal active sites. Phylogenetic analysis based on rRNA sequences led to the hypothesis which states that the last common ancestor of all extant life was thermophilic (62), although this model has not been without challenge (19). One corollary of the thermophilic last common ancestor hypothesis is that the conserved structural core of all ribosomes, including the decoding and peptidyltransferase active sites, should be intrinsically thermostable as they have not significantly changed since their divergence from the last common ancestor. One implication of this notion is that the conserved core elements of mesophilic and psychrophilic ribosomes should also be thermostable, while the less conserved supporting structures have adapted to function at lower temperatures. Kinetic analyses comparing the temperature dependence of partial reactions catalyzed by ribosomes of thermophilic, mesophilic, and psychrophilic species might help to resolve this issue.
Antibiotic resistance mutations arising in rRNA genes of a number of organisms have been reconstructed by site-directed mutagenesis of plasmid-encoded E. coli rRNA operons in order to take advantage of the sophisticated genetics of this organism and to examine these mutations in a bacterial context (13, 14, 41, 45, 46, 53, 63). As stated earlier, some of these mutations, notably from mitochondria and the protist Tetrahymena, did not produce the expected resistance phenotypes in E. coli but were instead found to be dominantly lethal (13, 45). These observations might be interpreted as defining structural divergence among bacterial, mitochondrial, and eukaryotic ribosomes. However, more recently isolated bacterial mutants (42, 52), including those described in the present study, have been shown to behave similarly to the equivalent mitochondrial and eukaryotic counterparts, arguing against structural divergence among ribosomes as a cause for the incongruous observations made in the E. coli system. While the inconsistent results attained with E. coli might have their basis in some innate peculiarity of this species, it seems more plausible that they are indicative of physiological artifacts arising from the use of multicopy, plasmid-encoded rRNA operons necessitated by the existence of seven chromosomal rRNA operons. Thus, interpretation of the phenotypes of rRNA mutants of E. coli should take into account the potential impact of hyperexpression of rRNA on regulation of ribosome synthesis and overall growth physiology. It is perhaps pertinent that we have not observed any such phenotypic distinctions between ribosomal protein mutants of T. thermophilus and E. coli (7, 20).
The results of this study provide support for the use of T. thermophilus as a model organism for the study of ribosome structure and function using mutational approaches. Further, our results indicating the phenotypic similarity of mutants of T. thermophilus to those of mesophilic species argue for the validity of using the current crystal structures of T. thermophilus ribosomes in interpreting genetic and biochemical data previously obtained with other organisms. Our data suggest that genetic manipulation of T. thermophilus ribosomes will have significant relevance to our understanding of ribosome structure and function and that this species represents a useful alternative to other organisms as a model system for such experiments. These properties provide an incentive for the development of more sophisticated genetic approaches to T. thermophilus ribosome structure and function.
Acknowledgments
This work was supported by a grant, GM19756, from the NIH to A.E.D.
We are especially grateful to Jill Thompson for numerous discussions and comments on the manuscript.
REFERENCES
- 1.Blanc, H., C. T. Wright, M. J. Bibb, D. C. Wallace, and D. A. Clayton. 1981. Mitochondrial DNA of chloramphenicol-resistant mouse cells contains a single nucleotide change in the region encoding the 3′ end of the large ribosomal RNA. Proc. Natl. Acad. Sci. USA 78:3789-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brock, T. D., and H. Freeze. 1969. Thermus aquaticus gen. n. and sp. n., a non-sporulating extreme thermophile. J. Bacteriol. 98:289-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodersen, D. E., W. M. Clemons, Jr., A. P. Carter, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103:1143-1154. [DOI] [PubMed] [Google Scholar]
- 4.Cameron, D., J. Thompson, S. T. Gregory, P. E. March, and A. E. Dahlberg. 2004. Thiostrepton-resistant mutants of Thermus thermophilus. Nucleic Acids Res. 32:3220-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camps, M., G. Arrizabalaga, and J. Boothroyd. 2002. An rRNA mutation identifies the apicoplast as the target for clindamycin in Toxoplasma gondii. Mol. Microbiol. 43:1309-1318. [DOI] [PubMed] [Google Scholar]
- 6.Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. Shang, N. Yu, and R. R. Gutell. 2002. The Comparative RNA Web (CRW) Site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr, J. F., S. T. Gregory, and A. E. Dahlberg. 2005. Severity of the streptomycin resistance and streptomycin dependence phenotypes of ribosomal protein S12 of Thermus thermophilus depends on the identity of highly conserved amino acid residues. J. Bacteriol. 187:3548-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter, A. P., W. M. Clemons, D. E. Brodersen, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340-348. [DOI] [PubMed] [Google Scholar]
- 9.Carter, A. P. 2002. Ph.D. thesis. Cambridge University, Cambridge, United Kingdom.
- 10.Chernoff, Y. O., A. Vincent, and S. W. Liebman. 1994. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J. 13:906-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cseplö, A., T. Etzold, J. Schell, and P. H. Schreier. 1988. Point mutations in the 23S rRNA genes of four lincomycin resistant Nicotiana plumbaginifolia mutants could provide new selectable markers for chloroplast transformation. Mol. Gen. Genet. 214:295-299. [DOI] [PubMed] [Google Scholar]
- 12.Depardieu, F., and P. Courvalin. 2001. Mutation in 23S rRNA responsible for resistance to 16-membered macrolides and streptogramins in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:319-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeStasio, E. A., and A. E. Dahlberg. 1990. Effects of mutagenesis of a conserved base-paired site near the decoding region of Escherichia coli 16S ribosomal RNA. J. Mol. Biol. 212:127-133. [DOI] [PubMed] [Google Scholar]
- 14.Douthwaite, S. 1992. Functional interactions within 23S rRNA involving the peptidyltransferase center. J. Bacteriol. 174:1333-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dujon, B. 1980. Sequence of the intron and flanking exons of the mitochondrial 21S rRNA gene of yeast strains having different alleles at the omega and rib-1 loci. Cell 20:185-197. [DOI] [PubMed] [Google Scholar]
- 16.Fredrick, K., and H. F. Noller. 2003. Catalysis of ribosomal translocation by sparsomycin. Science 300:1159-1162. [DOI] [PubMed] [Google Scholar]
- 17.Furneri, P. M., G. Rappazzo, M. P. Musumarra, P. Di Piettro, L. S. Catania, and L. S. Roccasalva. 2001. Two new point mutations at A2062 associated with resistance to 16-membered macrolide antibiotics in mutant strains of Mycoplasma hominis. Antimicrob. Agents Chemother. 45:2958-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gale, E. F., E. Cundliffe, P. E. Reynolds, M. H. Richmond, and M. J. Waring. 1981. The molecular basis of antibiotic action, p. 402-547. John Wiley & Sons, London, United Kingdom.
- 19.Galtier, N., N. Tourasse, and M. Gouy. 1999. A nonhyperthermophilic common ancestor to extant life forms. Science 283:220-221. [DOI] [PubMed] [Google Scholar]
- 20.Gregory, S. T., J. H. D. Cate, and A. E. Dahlberg. 2001. Streptomycin-resistant and streptomycin-dependent mutants of the extreme thermophile Thermus thermophilus. J. Mol. Biol. 309:333-338. [DOI] [PubMed] [Google Scholar]
- 21.Gregory, S. T., J. H. D. Cate, and A. E. Dahlberg. 2001. Spontaneous erythromycin resistance mutations in a 23S rRNA gene, rrlA, of the extreme thermophile Thermus thermophilus IB-21. J. Bacteriol. 183:4382-4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gregory, S. T., J. F. Carr, and A. E. Dahlberg. 2005. A mutation in the decoding center of Thermus thermophilus 16S rRNA suggests a novel mechanism of streptomycin resistance J. Bacteriol. 187:2200-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffiths, E., and R. S. Gupta. 2004. Distinctive protein signatures provide molecular markers and evidence for the monophyletic nature of the Deinococcus-Thermus phylum. J. Bacteriol. 186:3097-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen, J. L., J. A. Ippolito, N. Ban, P. Nissen, P. B. Moore, and T. A. Steitz. 2002. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell 10:117-128. [DOI] [PubMed] [Google Scholar]
- 25.Hansen, J. L., P. B. Moore, and T. A. Steitz. 2003. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 330:1061-1075. [DOI] [PubMed] [Google Scholar]
- 26.Harms, J., F. Schluenzen, R. Zarivach, A. Bashan, S. Gat, I. Agmon, H. Bartels, F. Franceschi, and A. Yonath. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107:679-688. [DOI] [PubMed] [Google Scholar]
- 27.Harris, E. H., B. D. Burkhart, N. W. Gillham, and J. E. Boynton. 1989. Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics 123:281-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashimoto, J. G., B. S. Stevenson, and T. M. Schmidt. 2003. Rates and consequences of recombination between rRNA operons. J. Bacteriol. 185:966-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hashimoto, Y., T. Yano, S. Kuramitsu, and H. Kagamiyama. 2001. Disruption of Thermus thermophilus genes by homologous recombination using a thermostable kanamycin-resistant marker. FEBS Lett. 506:231-234. [DOI] [PubMed] [Google Scholar]
- 30.Henne, A., H. Bruggemann, C. Raasch, A. Wiezer, T. Hartsch, H. Liesegang, A. Johann, T. Lienard, O. Gohl, R. Martinez-Arias, C. Jacobi, V. Starkuviene, S. Schlenczeck, S. Dencker, R. Huber, H. P. Klenk, W. Kramer, R. Merkl, G. Gottschalk, and H. J. Fritz. 2004. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 22:547-553. [DOI] [PubMed] [Google Scholar]
- 31.Hummel, H., and A. Böck. 1987. 23S ribosomal RNA mutations in halobacteria conferring resistance to the anti-80S ribosome targeted antibiotic anisomycin. Nucleic Acids Res. 15:2431-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jost, B. H., H. T. Trinh, J. G. Songer, and S. J. Billington. 2004. Ribosomal mutations in Arcanobacterium pyogenes confer a unique spectrum of macrolide resistance. Antimicrob. Agents Chemother. 48:1021-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kearsey, S. E., and I. W. Craig. 1981. Altered ribosomal RNA genes in mitochondria from mammalian cells with chloramphenicol resistance. Nature 290:607-608. [DOI] [PubMed] [Google Scholar]
- 34.Kloss, P., L. Xiong, D. L. Shinabarger, and A. S. Mankin. 1999. Resistance mutations in 23S rRNA identify the site of action of the protein synthesis inhibitor linezolid in the ribosomal peptidyl transferase center. J. Mol. Biol. 294:93-101. [DOI] [PubMed] [Google Scholar]
- 35.Koyama, Y., T. Hoshino, N. Tomizuka, and K. Furukawa. 1986. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J. Bacteriol. 166:338-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kristjansson, J. K., G. O. Hreggvidsson, and G. A. Alfredsson. 1986. Isolation of halotolerant Thermus spp. from submarine hot springs in Iceland. Appl. Environ. Microbiol. 52:1313-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, M., A. Tzagoloff, K. Underbrink-Lyon, and N. C. Martin. 1982. Identification of the paromomycin-resistance mutation in the 15S rRNA gene of yeast mitochondria. J. Biol. Chem. 257:5921-5928. [PubMed] [Google Scholar]
- 38.Mankin, A. S., and R. A. Garrett. 1991. Chloramphenicol resistance mutations in the single 23S rRNA gene of the archaeon Halobacterium halobium. J. Bacteriol. 173:3559-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marrero, P., M. J. Cabanas, and J. Modolell. 1980. Induction of translational errors (misreading) by tuberactinomycins and capreomycins. Biochem. Biophys. Res. Commun. 97:1047-1042. [DOI] [PubMed] [Google Scholar]
- 40.Moazed, D., and H. F. Noller. 1987. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie 69:879-884. [DOI] [PubMed] [Google Scholar]
- 41.Muth, G. W., L. Ortoleva-Donnelly, and S. A. Strobel. 2000. A single adenosine with a neutral pKa in the ribosomal peptidyl transferase center. Science 289:947-950. [DOI] [PubMed] [Google Scholar]
- 42.Pfister, P., M. Risch, D. E. Brodersen, and E. C. Bottger. 2003. Role of 16S rRNA helix 44 in ribosomal resistance to hygromycin B. Antimicrob. Agents Chemother. 47:1496-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prammananan, T., P. Sander, B. A. Brown, K. Frischkorn, G. O. Onyi, Y. Zhang, E. C. Bottger, and R. J. Wallace, Jr. 1998. A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J. Infect. Dis. 177:1573-1581. [DOI] [PubMed] [Google Scholar]
- 44.Prammananan, T., P. Sander, B. Springer, and E. C. Bottger. 1999. RecA-mediated gene conversion and aminoglycoside resistance in strains heterozygous for rRNA. Antimicrob. Agents Chemother. 43:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Recht, M. I., S. Douthwaite, K. D. Dahlquist, and J. E. Puglisi. 1999. Effect of mutations in the A site of 16S rRNA on aminoglycoside antibiotic-ribosome interaction. J. Mol. Biol. 286:33-43. [DOI] [PubMed] [Google Scholar]
- 46.Recht, M. I., and J. D. Puglisi. 2001. Aminoglycoside resistance with homogeneous and heterogeneous populations of antibiotic-resistant ribosomes. Antimicrob. Agents Chemother. 45:2414-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schluenzen, F., A. Tocilj, R. Zaravich, J. Harms, M. Gluehmann, D. Janell, A. Bashan, H. Bartels, I. Agmon, F. Franceschi, and A. Yonath. 2000. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell 102:615-623. [DOI] [PubMed] [Google Scholar]
- 48.Schlunzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 49.Sor, F., and H. Fukuhara. 1982. Identification of two erythromycin resistance mutations in the mitochondrial gene coding for the large subunit ribosomal RNA in yeast. Nucleic Acids Res. 10:6571-6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spangler, E. A., and E. H. Blackburn. 1985. The nucleotide sequence of the 17S ribosomal RNA gene of Tetrahymena thermophila and the identification of point mutations resulting in resistance to the antibiotics paromomycin and hygromycin. J. Biol. Chem. 260:6334-6340. [PubMed] [Google Scholar]
- 51.Tan, G. T., A. DeBlasio, and A. S. Mankin. 1996. Mutations in the peptidyl transferase center of 23S rRNA reveal the site of action of sparsomycin, a universal inhibitor of translation. J. Mol. Biol. 261:222-230. [DOI] [PubMed] [Google Scholar]
- 52.Taniguchi, H., B. Chang, C. Abe, Y. Nikaido, Y. Mizuguchi, and S. Yoshida. 1997. Molecular analysis of kanamycin and viomycin resistance in Mycobacterium smegmatis by use of the conjugation system. J. Bacteriol. 179:4795-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson, J., D. F. Kim, M. O'Connor, K. R. Lieberman, M. A. Bayfield, S. T. Gregory, R. Green, H. F. Noller, and A. E. Dahlberg. 2001. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc. Natl. Acad. Sci. USA 98:9002-9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson, J., M. O'Connor, J. A. Mills, and A. E. Dahlberg. 2002. The protein synthesis inhibitors, oxazolidinones and chloramphenicol, cause extensive translational inaccuracy in vivo. J. Mol. Biol. 322:273-279. [DOI] [PubMed] [Google Scholar]
- 55.Triman, K. L., A. Peister, and R. A. Goel. 1998. Expanded versions of the 16S and 23S ribosomal RNA mutation databases (16SMDBexp and 23SMDBexp). Nucleic Acids Res. 26:280-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ulbrich, N., I. Kumagai, and V. A. Erdmann. 1984. The number of ribosomal RNA genes in Thermus thermophilus HB8. Nucleic Acids Res. 12:2055-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vester, B., and S. Douthwaite. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiss-Brummer, B., A. Zollner, A. Haid, and S. Thompson. 1995. Mutation of a highly conserved base in the yeast mitochondrial 21S rRNA restricts ribosomal frameshifting. Mol. Gen. Genet. 248:207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, K. W. Minton, R. D. Fleischmann, K. A. Ketchum, K. E. Nelson, S. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams, R., and R. Sharp. 1995. The taxonomy and identification of Thermus, p. 1-42. In R. Sharp and R. Williams (ed.), Thermus species. Plenum Press, New York, N.Y.
- 61.Wimberly, B. T., D. E. Brodersen, W. M. Clemons, Jr., R. J. Morgan-Warren, A. P. Carter, C. Vonrhein, T. Hartsch, and V. Ramakrishnan. 2000. Structure of the 30S ribosomal subunit. Nature 407:327-339. [DOI] [PubMed] [Google Scholar]
- 62.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiong, L., P. Kloss, S. Douthwaite, N. M. Andersen, S. Swaney, D. L. Shinabarger, and A. S. Mankin. 2000. Oxazolidinone resistance mutations in 23S rRNA of Escherichia coli reveal the central region of domain V as the primary site of drug action. J. Bacteriol. 182:5325-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoshizawa, S., D. Fourmy, and J. D. Puglisi. 1998. Structural origins of gentamicin antibiotic action. EMBO J. 17:6437-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 A resolution. Science 292:883-896. [DOI] [PubMed] [Google Scholar]