Abstract
BpeAB-OprB is a multidrug efflux pump of the bacterial pathogen Burkholderia pseudomallei and is responsible for conferring antimicrobial resistance to aminoglycosides and macrolides. Expression of bpeAB-oprB is inducible by its substrate erythromycin and upon entry into stationary phase. BpeR, a member of the TetR family, functions as a repressor of the bpeAB-oprB operon. bpeR expression was similarly induced at stationary phase but lagged behind the induction of bpeAB-oprB expression. The induction of bpeAB-oprB expression could be advanced to the early exponential phase by exogenous addition of the B. pseudomallei autoinducers N-octanoyl-homoserine lactone (C8HSL) and N-decanoyl-homoserine lactone (C10HSL), suggesting that the bpeAB-oprB operon may be quorum regulated. On the other hand, acyl-homoserine lactone (acyl-HSL) production was undetectable in the bpeAB-null mutant and strains which overexpress bpeR. The failure of these strains to produce acyl-HSLs seemed to be at the level of synthesis of acyl-HSLs, as growth-phase-dependent expression of the autoinducer synthase BpsI was abolished in the bpeAB-null mutant. bpsI expression remained growth phase dependent in the bpeR mutant which had functional BpeAB-OprB. BpeAB-OprB function is likewise necessary for optimal production of quorum-sensing-controlled virulence factors such as siderophore and phospholipase C and for biofilm formation. Cell invasion and cytotoxicity towards human lung epithelial (A549) and human macrophage (THP-1) cells were also significantly attenuated in both the bpeAB mutant and bpeR-overexpressing strains, thus suggesting the possibility of attenuating B. pseudomallei virulence using inhibitors of the BpeAB-OprB efflux pump.
Burkholderia pseudomallei is a gram-negative soil saprophyte and the etiological agent of melioidosis in humans and animals. Melioidosis is endemic in southeast Asia, tropical countries, and northern Australia. The bacterium is intrinsically resistant to penicillin, aminopenicillins, narrow-spectrum and expanded-spectrum cephalosporins, most aminoglycosides, macrolides, rifampin, and polymyxins (4). It is, however, susceptible to broad-spectrum cephalosporins such as ceftazidime, cefotaxime, and ceftriaxone; carbapenems; chloramphenicol; tetracyclines; co-trimoxazole; and some fluoroquinolones (4). Ceftazidime is the drug of choice for the treatment of acute severe melioidosis. Prolonged maintenance therapy comprising either co-amoxiclav or co-trimoxazole and doxycycline is recommended, but relapses are common, typically occurring about 21 weeks after therapy (4).
Two multidrug efflux pumps of the resistance-nodulation-division (RND) family, AmrAB-OprA and BpeAB-OprB, have been described for B. pseudomallei, and both are responsible for conferring resistance to the aminoglycosides and macrolides such as gentamicin, kanamycin, streptomycin, spectinomycin, tobramycin, neomycin, erythromycin, and clarithromycin (3, 22). Although it is yet to be verified, AmrAB-OprB and BpeAB-OprB efflux pumps are believed to be regulated by repressor proteins encoded by divergently transcribed amrR and bpeR, respectively. The bpeR gene, located upstream of the bpeAB-oprB operon, encodes a repressor protein of the TetR family.
There are recent reports of a link between homoserine lactone (HSL)-mediated quorum sensing and the mexAB-oprM pump in Pseudomonas aeruginosa (17, 20, 32). P. aeruginosa mexAB-oprM expression was enhanced by the autoinducer N-butyryl-l-homoserine lactone (C4HSL), and this C4HSL-mediated enhancement of mexAB-oprM expression, which was independent of MexR function (32), could be negated by MexT, a positive regulator of the mexEF-oprN operon (20). In this study, we investigated the properties of BpeR as a repressor of the B. pseudomallei BpeAB-OprB efflux pump and demonstrated the interrelationship between the BpeAB-OprB efflux function, quorum sensing, and virulence. We also showed that inhibition of BpeAB-OprB could result in virulence attenuation via the inhibition of quorum sensing in B. pseudomallei.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Unless otherwise stated, the cultures were grown under aerobic conditions at 37°C in Luria-Bertani agar (LA) or Luria-Bertani broth (LB) (Becton Dickinson, Cockeysville, Md.). B. pseudomallei KHW is a virulent clinical isolate which we have used previously (5). Antibiotic concentrations for Escherichia coli, when used, were as follows: ampicillin, 50 μg/ml; gentamicin, 50 μg/ml; trimethoprim, 25 μg/ml; kanamycin, 10 μg/ml; streptomycin, 50 μg/ml; chloramphenicol, 10 μg/ml; tetracycline, 5 μg/ml. For Agrobacterium tumefaciens they were as follows: kanamycin, 50 μg/ml, and tetracycline, 5 μg/ml. For B. pseudomallei they were as follows: kanamycin, 200 μg/ml; trimethoprim, 100 μg/ml; tetracycline, 25 μg/ml. All antibiotics were purchased from Sigma (St Louis, Mo.).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5αλpir | λpir lysogen of DH5α for replication of oriR6K, oriT, and mob region of RP4; Kans | N. Judson, Gibco-BRL |
| HB101(pRK600) | Helper strain for triparental mating; supE44 hsdS20(rB mB) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 Cmr | 8, 31 |
| JB525 | Derivative of E. coli MC1000 harboring plasmid pJBA132 Tetr | 1 |
| A. tumefaciens NTI (traR tra::lacZ749) | pTi58-cured derivative of A. tumefaciens C58; contains pAtC58, pJM649, and pSVB33 | 27 |
| B. pseudomallei | ||
| KHW | Wild-type parental strain, clinical isolate | 5 |
| KHWbpeR::Km | bpeR::Km derivative of KHW; Kanr | This study |
| KHWbpeR(pUCP28TbpeR) | KHWbpeR::Km complemented in trans with pUCP28TpbeR; Kanr Tmpr | This study |
| KHWΔbpeAB | KHWΔbpeAB Kanr | 3 |
| KHWΔbpeAB(pUCP28TbpeAB) | KHWΔbpeAB complemented in trans with pUCP28TbpeAB; Kanr Tmpr | 3 |
| Plasmids | ||
| pGEM-T | Vector for PCR cloning; Ampr | Promega |
| pGEM-TbpeR | pGEM-T carrying the 631-bp bpeR PCR product obtained with AcrRHisF and AcrRHisR primers | This study |
| pGEM-TbpeR::Km | pGEM-TbpeR carrying a 2.3-kbp kanamycin-resistance cassette from pUTKm | This study |
| pJQ200mp18 | Mobilizable allelic-exchange vector; traJ sacB Genr | 29 |
| pJQ200bpeR::Km | pJQ200mp18 carrying a 3-kbp Apal-Spel fragment containing bpeR and kanamycin resistance cassette | This study |
| pUCP28T | Broad-host range vector; incP oriT, pRO1600 ori Tmpr | 39 |
| pUCP28TbpeR | pUCP28T carrying 1.5-kb bpeR PCR product containing bpeR promoter; Tmpr | This study |
| pUCP28TbpeAB | pUCP28T carrying 4.9-kbp bpeAB PCR product containing bpeAB promoter; Tmpr | 3 |
| pUTKm | Source of kanamycin resistance cassette; oriR6K mobRP4 Kanr Ampr | 7 |
| pMC1403 | pBR322-derived plasmid; source of promoterless lacZYA fragment; Ampr | 2 |
| Mini-CTX1 | Broad-host-range plasmid; oriT Tetr | 15 |
| pCYY | Mini-CTX1 carrying promoter less lacZYA cassette from pMC1403 | This study |
| pCYYbpeAB | pCYY ligated to bpeAB promoter fragment | This study |
| pCYYbpeR | pCYY ligated to bpeR promoter fragment | This study |
| pSY1 | pCYY carrying 1.2-kbp bpsIpromoter-lacZ gene fusion; Tetr | 34 |
Kan, kanamycin; Tmp, trimethoprim; Gen, gentamicin; Tet, tetracycline; Cm, chloramphenicol; Amp, ampicillin.
Construction of mutants and strains for complementation.
The bpeR mutant KHWbpeR::Km was derived from B. pseudomallei KHW by insertional mutagenesis and homologous recombination as described previously (3). The 631-bp bpeR product was amplified using the primers AcrRHisF (5′TCAGGATCCGCCAGACGCACGAAGGAGGAA3′) and AcrRHisR (5′CAGAAGCTTCTTGCGCATCGCGGGGCTCGT3′) and ligated with T-tailed pGEM-T (Promega, Madison, Wis.), yielding pGEM-TbpeR. pGEM-TbpeR was linearized with NarI, made blunt ended, and ligated with a 2.3-kbp end-filled EcoRI fragment carrying the kanamycin resistance cassette from pUTKm, yielding pGEM-TbpeR::Km. Underlined sequences refer to restriction endonuclease recognition sites. T4 DNA polymerase (Promega) was used to generate blunt ends in DNA fragments with 3′ or 5′ overhanging ends. pGEM-TbpeR::Km was then digested with ApaI and SpeI, and the 3-kbp ApaI-SpeI fragment carrying bpeR and the kanamycin resistance cassette from pGEM-TbpeR::Km was isolated and made blunt ended before being inserted into SmaI-linearized pJQ200mp18, yielding pJQ200bpeR::Km. pJQ200bpeR::Km was first introduced into E. coli DH5αλpir by electroporation using a MicroPulser (Bio-Rad, Hercules, CA) and then mobilized into B. pseudomallei KHW by triparental mating using E. coli HB101(pRK600) as a helper strain (7). Recombinants which had undergone allelic exchange at bpeR were selected on LA containing kanamycin, streptomycin, and 5% (wt/vol) sucrose and were designated KHWbpeR::Km (Table 1). The bpeR-null mutation was verified by PCR and reverse transcription-PCR (RT-PCR) using primers AcrRHisF and AcrRHisR. PCR yielded a 631-bp fragment from KHW but a 3-kbp fragment from KHWbpeR::Km, which was consistent with the disruption of bpeR by a 2.3-kbp kanamycin resistance cassette (data not shown). RT-PCR using primer pair AcrAHisF (5′TCAGGATCCCGCGTCGAACGGGTTCC3′) and AcrAHisR (5′CAGAAGCTTCCTGTTATTGCGCGCTCG3′) was applied to detect bpeAB expression, and AcrRHisF and AcrRHisR were used to detect bpeR expression (Fig. 1). RT-PCR of 16S RNA using primers 16SF2 (5′GATGACGGTACCGGAAGAATAAGC3′) and 16SR2 (5′CCATGTCAAGGGTAGGTAAGGTTT3′) was included as an internal standard for the amount of template RNA used.
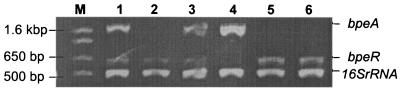
FIG. 1.
Analyses of bpeA and bpeR expression by RT-PCR. RT-PCR results obtained using total RNA isolated from exponential-phase cultures of wild-type parental strain KHW (lane 1), KHWΔbpeAB mutant (lane 2), KHWΔbpeAB(pUCP28TbpeAB) (lane 3), KHWbpeR::Km (lane 4), KHWbpeR::Km(pUCP28TbpeR) (lane 5), and KHW(pUCP28TbpeR) (lane 6). Lane M is 1-kb Plus size markers (Invitrogen, Carlsbad, CA). The bands corresponding to bpeA, bpeR, and 16S rRNA RT-PCR products are indicated on the right. bpeA expression was absent in KHWΔbpeAB (lane 2) but was restored by complementation (lane 3). bpeR expression was absent in KHWbpeR::Km (lane 4), and complementation with pUCP28TbpeR resulted in complete repression of bpeA expression (lane 5). Overexpression of bpeR in KHW also resulted in complete repression of bpeA expression (lane 6).
A 1.5-kb full-length bpeR PCR product inclusive of the putative bpeR promoter was amplified from KHW genomic DNA using the primers AcrRHisF and AcrA5′R (5′GGCCACCGCATCGTCGTA3′). The PCR product was first converted to blunt ends using T4 DNA polymerase before ligation with SmaI-linearized pUCP28T to yield pUCP28TbpeR (Table 1). pUCP28TbpeR was mobilized into KHWbpeR::Km via triparental mating as described previously. PCR using DNA, and RT-PCR using total RNA, isolated from the complemented mutant confirmed the successful transfer of pUCP28TbpeR into KHWbpeR::Km (data not shown).
Construction of promoter-lacZ fusions.
The broad-host-range vector pCYY, carrying a promoterless lacZ cassette, was used to construct bpeRpromoter-lacZ and bpeABpromoter-lacZ fusions. pCYY was derived by ligating a 6.2-kbp EcoRI-SalI promoterless lacZYA cassette from pMC1403 with EcoRI-SalI-linearized mini-CTX1. An 868-bp intergenic fragment, comprising the upstream regulatory sequences of both bpeR and bpeAB, as well as the ATG initiation codons, was amplified from KHW DNA by PCR using the primers AcrABpro (5′TTCCTCCTTCGTGCGTCTGGC3′) and AcrA5′R. Opposite orientations of the blunt-ended PCR product when ligated to SmaI-linearized pCYY yielded either the bpeABpromoter-lacZ fusion (pCYYbpeAB) or the bpeRpromoter-lacZ fusion (pCYYbpeR). The orientations of the inserts were verified by restriction digests (data not shown). Transcriptional expression of bpeAB and bpeR was assayed by measuring β-galactosidase activities in B. pseudomallei strains harboring these reporter plasmids.
DNA and RNA manipulations.
Bacterial genomic DNA was isolated according to the method described by Pitcher et al. (28). Total RNA was extracted from bacteria using the RNeasy Mini Kit (Qiagen, Hilden, Germany) after pretreatment with RNAprotect reagent (Qiagen). PCR was performed in a PTC-100 Peltier thermal cycler (MJ Research, MA) in Mg2+-free buffer containing 100 ng template DNA, 200 μmol (each) deoxynucleoside triphosphates, 50 pmol of each primer, 1.5 mM MgSO4, and 0.5 U Tth polymerase (Biotools, Madrid, Spain) in a total volume of 50 μl. Cycling parameters include 1 cycle (3 min, 94°C) followed by 30 cycles (30 s, 94°C; 30 s of annealing at the respective temperatures; 1 min/kb of product length, 72°C) and a final extension at 72°C for 10 min. RT-PCR was carried out in 10 μl Mg2+-free buffer containing 5 μg template RNA, 200 μM (each) deoxynucleoside triphosphates, 1 μM of each primer, 1 mM MgSO4, 0.1 U Tfl DNA polymerase, and 0.1 U avian myeloblastosis virus reverse transcriptase (Access RT-PCR System; Promega). Cycling parameters include a reverse transcription step at 48°C for 45 min, followed by PCR comprising 1 cycle (2 min, 94°C), 30 cycles (30 s, 94°C; 30 s of annealing at the respective temperatures; 1 min/kb of product length, 68°C), and a final extension at 68°C for 7 min.
Bioassays.
The β-galactosidase assay was performed as described by Miller (21). Briefly, 10 ml AB medium (6) containing either 20 mM glycerol or 20 mM glucose, 0.2% (wt/vol) Casamino Acids, and 25 μg/ml tetracycline was inoculated (1:100) with an overnight culture of B. pseudomallei KHW or its isogenic mutants, harboring either pCYYbpeAB, pCYYbpeR, or the bpsIpromoter-lacZ reporter pSYI. β-Galactosidase activities and cell densities (optical densities at 600 nm [OD600]) were determined at various time intervals during the culture. An 0.1-ml amount of bacterial culture was used for the β-galactosidase assay. Enzyme activity was expressed in Miller units. For assays involving the addition of exogenous compounds, typically a 5-ml bacterial culture was inoculated (1:50) with overnight culture and the compounds to the culture at an OD600 of ∼0.1. N-Octanoyl-l-homoserine lactone (C8HSL) and N-decanoyl-l-homoserine lactone (C10HSL) were purchased from Sigma. β-Galactosidase activities were determined after 4 h of incubation at 37°C, i.e., in early exponential growth phase.
Siderophore production was determined using the chrome azurol assay described by Yang et al. (40). Briefly, 0.1 ml of 24-h-old bacterial culture was added to 0.9 ml chrome azurol solution (pH 5.6) and equilibrated for 2 h before absorbance was read at 630 nm. Siderophore activity was expressed as the change in OD630 readings (ΔOD630) between the test samples and the sample blank and was normalized for cell density by being expressed as a ratio of ΔOD630 to OD600.
Phospholipase C (PLC) activity was determined by the enzymatic hydrolysis of p-nitrophenylphosphorylcholine (NPPC; Sigma) to phosphatidylcholine and p-nitrophenol (18). Briefly, 10 μl of bacterial culture supernatant was mixed with 90 μl of NPPC reagent (250 mM Tris-HCl, pH 7.2; 0.1 mM ZnSO4; 10 mM NPPC; 40% [wt/wt] sorbitol), and the release of p-nitrophenol was detected by reading OD405 after 1 h of incubation at 37°C. The PLC activities shown have been normalized for cell density by being expressed as a ratio of OD410/OD600.
All assays described were performed in triplicate.
Biofilm formation.
Biofilm formation was assayed by the ability of cells to adhere to the wells of 96-well polyvinyl chloride microtiter plates using a modification of the protocol described by O'Toole and Kolter (24). Briefly, 100 μl of a diluted (OD600 of ∼0.05) overnight bacterial culture in AB medium containing 0.2% Casamino Acids and 20 mM glycerol was added into each well of a 96-well microtiter plate. After 20 h of incubation at 30°C and being washed twice with distilled water to remove planktonic cells, 125 μl of 1% (wt/vol) crystal violet (Sigma) was added. After 15 min at room temperature and three rounds of careful washing with distilled water, 300 μl of 95% (vol/vol) ethanol was added to solubilize the stain and the extent of biofilm formation was determined by reading the absorbance of the solution at 595 nm. The assay was performed in triplicate.
MIC and MBC determinations.
MIC determinations were carried out in 96-well microtiter plates by a standard broth microdilution method as described previously (3, 23). The minimal bactericidal concentration (MBC) was defined as the lowest concentration of antibiotic required to kill 99.9% of the inoculum. All antibiotics were purchased from Sigma Co.
Detection of autoinducer production.
The production of acyl-homoserine lactones by the B. pseudomallei strains was detected by cross-streaking against an indicator strain, E. coli JB525 or A. tumefaciens NTI as described previously (34). The bacteria were cross-streaked onto LB agar and observed after incubation at 28°C for 20 h. For detection using A. tumefaciens NTI, the plates were overlaid with 20 μl X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 10 mg/ml) before streaking (30).
Cell invasion and cytotoxicity assays.
Bacterial invasion of A549 and THP-1 cells was performed as described by Elsinghorst except for the following modifications (10). Tetracycline (50 μg/ml) was added to kill extracellular bacteria instead of gentamicin, as KHW is resistant to gentamicin. Mid-log-phase bacteria in LB medium (OD600 of 0.6) were washed and resuspended in an equal volume of 0.85% (wt/vol) NaCl. Twenty-five-microliter aliquots of the bacterial suspension were added to each well of a 24-well tissue culture plate containing 1 × 105 mammalian cells per well (multiplicity of infection = 100). After 2 h of incubation at 37°C in the presence of 5% CO2, the cells in each well were washed three times with phosphate-buffered saline and 1.5 ml of fresh culture medium containing tetracycline (50 μg/ml) was added. After a further 2 h of incubation to kill extracellular bacteria, the wells were again washed three times with phosphate-buffered saline and 1 ml of 0.1% Triton X-100 (Sigma) was added to lyse the mammalian cells. Serial dilutions of the cell lysates were then plated on LA to determine the number of bacteria in the cells after a 2-h exposure. The assays were performed in triplicate. When C8HSL was used, it was added to the mammalian cell culture medium to a final concentration of 100 nM together with the addition of 25 μl of bacterial suspension.
The cytotoxic effects of bacteria on mammalian cells were evaluated by measuring the release of lactate dehydrogenase enzyme using a Cytotoxicity Detection kit (Roche, Mannheim, Germany). Mid-log-phase bacterial cells were added to A549 and THP-1 cells (105 cells/well) cultured in 24-well plates in Dulbecco's modified Eagle's medium (Sigma) and RPMI 1640 (Sigma), respectively, each supplemented with 10% (vol/vol) fetal bovine serum (Sigma). The multiplicity of infection was 100. After 1 h of incubation at 37°C in the presence of 5% CO2, tetracycline (50 μg/ml) was added and the cells were further incubated for 4 h. A 100-μl aliquot of the centrifuged supernatant obtained from each well was used for the lactate dehydrogenase assay. The assays were performed in triplicate. When C8HSL was used, it was added to the mammalian cell culture medium to a final concentration of 100 nM together with 25 μl of bacterial suspension.
RESULTS
BpeR acts as a repressor of the BpeAB-OprB multidrug efflux pump.
BpeR, a member of the TetR repressor family, shared significant similarities in amino acid sequence with MexR of P. aeruginosa (3). We constructed a bpeR-null mutant, KHWbpeR::Km, by insertional mutagenesis. The bpeR-null mutation was verified by RT-PCR and is associated with a corresponding increase in bpeA expression (Fig. 1). Expression of bpeR was restored when KHWbpeR::Km was complemented in trans using pUCP28TbpeR (Fig. 1). Similarly, the expression of bpeA, which was lacking in the mutant KHWΔbpeAB, could also be restored by complementation with pUCP28TbpeAB (Fig. 1). Additionally, bpeA expression was completely repressed when bpeR was overexpressed in both the wild-type strain, KHW(pUCP28TbpeR), and in the bpeR mutant KHWbpeR::Km(pUCP28TbpeR). The effect of bpeR overexpression was the same as that of the bpeAB-null mutation (Fig. 1).
We next determined the MICs and MBCs of antibiotics that are substrates of the BpeAB-OprB efflux pump in wild-type KHW, KHWΔbpeAB, and KHWbpeR::Km, as well as the mutants which have been complemented in trans using pUCP28TbpeAB and pUCP28TbpeR, respectively. Previously, we showed that gentamicin, streptomycin, and erythromycin are substrates of the BpeAB-OprB pump (3). Compared to the wild-type parental strain, the bpeR mutant was 2-, 4-, and 16-fold more resistant to streptomycin, erythromycin, and gentamicin, respectively (Table 2). In contrast, overexpression of bpeR in both KHW and KHWbpeR::Km had the same effect of increasing susceptibility to all three antibiotics. The MICs and MBCs of all three antibiotics on KHWbpeR::Km(pUCP28TbpeR) and KHW(pUCP28TbpeR) were equivalent to that of KHWΔbpeAB (Table 2). These data correlated well with the observed down-regulation of bpeA expression in the complemented bpeR mutant in Fig. 1. We have thus shown that overexpression of bpeR resulted in a significant increase in susceptibility to gentamicin, streptomycin, and erythromycin, while the bpeR-null mutation had the opposite effect of increasing resistance to the antibiotics. The restoration of BpeR repressor function by complementation also verified that the increased antimicrobial resistance of KHWbpeR::Km was indeed a consequence of the bpeR mutation and not due to polar effects on downstream genes or a secondary site mutation.
TABLE 2.
Antimicrobial susceptibilities of B. pseudomallei KHW, KHWΔbpeAB, KHWbpeR::Km, and the complemented mutants KHWbpeR::Km(pUCP28T-bpeR) and KHW(pUCP28T-bpeR)
| Antimicrobial agent | KHW
|
KHWΔbpeAB
|
KHWbpeR::Km
|
KHWbpeR::Km(pUCP28TbpeR)
|
KHW(pUCP28TbpeR)
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| MIC (μg/ml) | MBC (μg/ml) | MIC (μg/ml) | MBC (μg/ml) | MIC (μg/ml) | MBC (μg/ml) | MIC (μg/ml) | MBC (μg/ml) | MIC (μg/ml) | MBC (μg/ml) | |
| Gentamicin | 128 | 256 | 0.125 | 0.5 | 2,048 | 2,048 | 1 | 4 | 1 | 4 |
| Streptomycin | 1,024 | 1,024 | 1 | 1 | 2,048 | 2,048 | 1 | 32 | 1 | 32 |
| Erythromycin | 128 | 1,024 | 0.125 | 0.5 | 512 | 2,048 | 4 | 1,024 | 64 | 1,024 |
BpeAB-OprB is an inducible efflux system.
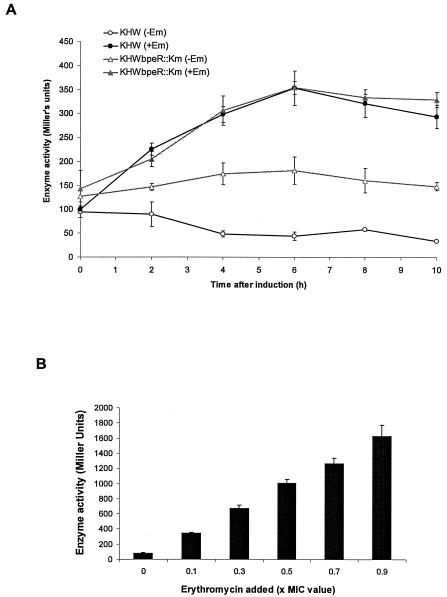
We studied the expression of bpeABpromoter-lacZ in wild type and the bpeR mutant by introducing the plasmid pCYYbpeAB into KHW and KHWbpeR::Km, respectively. Using a time course promoter assay, we demonstrate that basal expression of bpeABpromoter-lacZ in wild type was low but was inducible almost immediately upon exposure to 0.1× MIC (or 10 μg/ml) of erythromycin (Fig. 2A). The induction of bpeAB expression by erythromycin showed a linear dose-dependent relationship with respect to erythromycin (Fig. 2B). As predicted, basal expression of bpeAB was 2.5-fold higher in KHWbpeR::Km than in KHW. However, the higher basal bpeAB expression in KHWbpeR::Km was still inducible in the presence of 0.1× MIC of erythromycin, and it increased to a final level similar to that obtained when erythromycin was added to the culture of KHW (Fig. 2A). We could also induce the expression of bpeRpromoter-lacZ in the presence of 0.1× MIC of erythromycin using KHW(pCYYbpeR), but unlike bpeABpromoter-lacZ expression, the induction of bpeRpromoter-lacZ expression was delayed, occurring only after 6 h of exposure to erythromycin (data not shown). Our data thus provide evidence that the BpeR repressor interacts with the bpeAB-oprB regulatory region to regulate bpeAB expression. Induction of bpeRpromoter-lacZ expression in wild-type cells by erythromycin is also suggestive of a protein-ligand interaction between the BpeR repressor and erythromycin which releases the repression of bpeAB-oprB expression as well as bpeR expression. It appears that an adjacent site, which may be responsive to a different ligand-sensitive regulatory protein, may also be involved in regulating the inducible expression of the bpeAB-oprB operon. This yet unknown BpeR-independent mechanism could explain why, in the bpeR mutant background, bpeAB expression was elevated but not constitutively maximal (Fig. 2A).
FIG. 2.
Effect of erythromycin on expression of bpeABpromoter-lacZ in wild-type B. pseudomallei and the bpeR-null mutant. (A) Effect of bpeR mutation on the basal and inducible expression of bpeAB in KHW and KHWbpeR::Km. β-Galactosidase activities expressed from bpeABpromoter-lacZ fusion were determined in KHW (circles) and KHWbpeR::Km (triangles) harboring the reporter plasmid pCYYbpeAB. Open symbols represent basal expression of bpeAB, while closed symbols represent bpeAB expression after addition of 0.1× MIC of erythromycin (or 10 μg/ml). (B) Dose-dependent induction of bpeAB expression in KHW(pCYYbpeAB) by erythromycin. β-Galactosidase activities were assayed 4 h after the addition of erythromycin to the bacterial cultures in AB medium containing 20 mM glucose, 0.2% Casamino Acids, and 25 μg/ml tetracycline. Error bars represent standard deviations of triplicate β-galactosidase determinations for one typical experiment. Where error bars are not shown, the standard deviation was within the size of the symbol.
Expression of bpeAB-oprB is growth phase dependent while expression of bpeR is inducible upon entry into stationary phase.
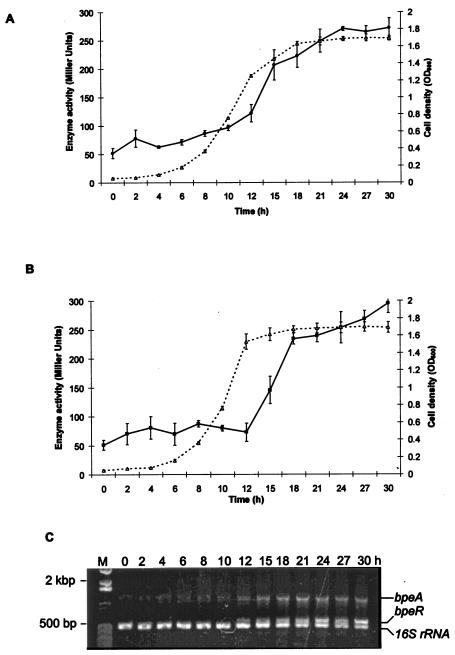
We next studied the expressions of bpeAB and bpeR at different growth phases of the bacterium using KHW(pCYYbpeAB) and KHW(pCYYbpeR), respectively. The expression of bpeABpromoter-lacZ was low from the onset of culture until mid-log phase but then gradually increased to reach maximum at stationary phase (Fig. 3A). In comparison, bpeRpromoter-lacZ expression was low from the onset of culture until late log phase. Induction of bpeRpromoter-lacZ expression began during the early stationary phase and reached maximum during the late stationary phase (Fig. 3B). These data were further confirmed by RT-PCR, which showed undetectable expression of bpeR up to 12 h of culture, followed by maximum expression after 24 h of culture. The detection of the bpeR transcripts at 12 h coincides with entry of the culture into stationary phase. In contrast, bpeAB transcripts were detectable from the onset of culture but started to increase gradually from 12 h onwards and reached maximum in stationary phase (Fig. 3C).
FIG. 3.
Growth-phase-dependent expression of bpeAB and bpeR in wild-type KHW. (A) β-Galactosidase activities were expressed from pCYYbpeAB in KHW. (B) β-Galactosidase activities were expressed from pCYYbpeR in KHW. Closed circles or squares represent β-galactosidase activities in Miller units, while open triangles represent cell densities (OD600). Error bars represent standard deviations of triplicate determinations of cell densities and β-galactosidase activities. Where error bars are not shown, the standard deviation was within the size of the symbol. The bacteria were cultured in AB medium containing 25 μg/ml tetracycline, 20 mM glycerol, and 0.2% (wt/vol) Casamino Acids. (C) Verification of growth-phase-dependent expression of bpeA and bpeR in wild-type cells by RT-PCR. RT-PCR was performed to detect bpeA and bpeR expression from the onset of culture (0 h) to stationary phase (30 h) (upper row). RT-PCR of 16S rRNA was included as an internal control for the reaction and to normalize the amount of total RNA used. Bands corresponding to the bpeA, bpeR, and 16S rRNA transcripts are indicated on the right, while lane M is the 1-kb Plus molecular size markers.
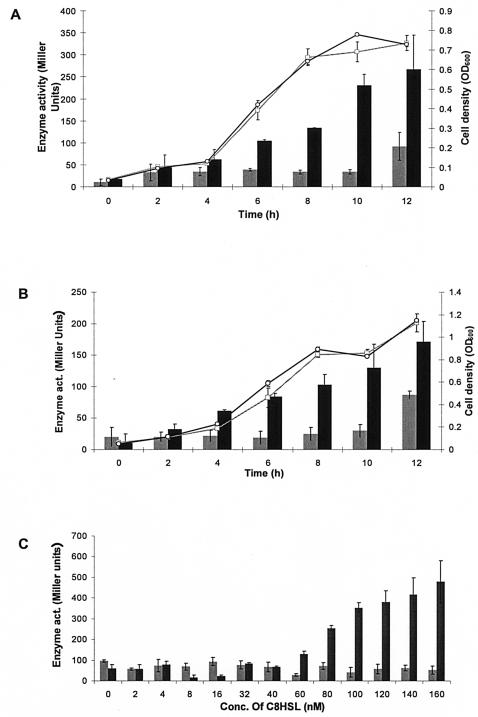
Addition of C8HSL and C10HSL autoinducers advances bpeABpromoter-lacZ expression to early exponential phase.
Since quorum sensing regulates the cell density-dependent expression of many bacterial genes, we then investigated if the expression of bpeAB and bpeR might be regulated by autoinducers. The B. pseudomallei quorum-sensing systems produce several autoinducers during stationary-phase cultures, including C8HSL and C10HSL (34, 37, 38). We wanted to know if the expression of bpeABpromoter-lacZ in wild-type KHW could be advanced to early exponential phase (8 to 12 h) by the addition of cognate autoinducers. C8HSL (100 nM) or C10HSL (100 nM) was added to KHW(pCYYbpeAB) cultured in AB medium containing glucose and Casamino Acids. Maximum expression of bpeABpromoter-lacZ typically attained during stationary-phase culture (24 h) could be prematurely advanced to 12 h by the addition of C8HSL (Fig. 4A). Addition of C10HSL was less effective in advancing the expression of bpeABpromoter-lacZ (Fig. 4B). This suggests that bpeAB-oprB expression might be regulated by quorum sensing and these autoinducers might be effluxed by BpeAB-OprB. The advancement of bpeABpromoter-lacZ expression in KHW to early exponential phase (4 h) was also dependent on the dose of C8HSL added and required at least 80 nM C8HSL to achieve the level of bpeABpromoter-lacZ expression at stationary phase (∼250 Miller units) (Fig. 4C). bpeABpromoter-lacZ expression could not be induced if the concentration of C8HSL was <60 nM (Fig. 4C). This is in contrast to the very low concentrations of C8HSL (0.1 nM) and C10HSL (1 nM), which are required to induce the B. pseudomallei luxI homolog bpsI (34).
FIG. 4.
Advancement of growth-phase-dependent induction of bpeAB-lacZ expression to early exponential phase by exogenous C8HSL and C10HSL. (A) Growth curves (OD600) of KHW in the presence (○) or absence (□) of 100 nM C8HSL. Also shown are the β-galactosidase activities (Miller units) representing bpeAB expression from pCYYbpeAB from onset of culture (0 h) to early exponential phase (12 h) in the absence (grey bars) or presence (black bars) of C8HSL, respectively. (B) Growth curves (OD600) of KHW in the presence (○) or absence (□) of 100 nM C10HSL. Also shown are β-galactosidase activities (Miller units) representing bpeAB expression in the absence (grey bars) or presence (black bars) of C10HSL, respectively. (C) Dose-dependent induction of bpeAB-lacZ expression by exogenous C8HSL. The horizontal axis represents the concentrations of exogenous C8HSL added to the culture medium. Black bars represent cultures to which different amounts of C8HSL were added, while grey bars represent control cultures to which no C8HSL was added. C8HSL was added to the bacterial cultures in AB medium containing 20 mM glucose, 0.2% Casamino Acids, and 25 μg/ml tetracycline about 1 h after inoculation from an overnight culture (OD600 of ∼0.1 to 0.2), and β-galactosidase assays were performed on aliquots of the cultures 4 h after the addition of exogenous C8HSL. Error bars represent standard deviations of triplicate β-galactosidase determinations for one typical experiment.
BpeAB-OprB is required for autoinducer production.
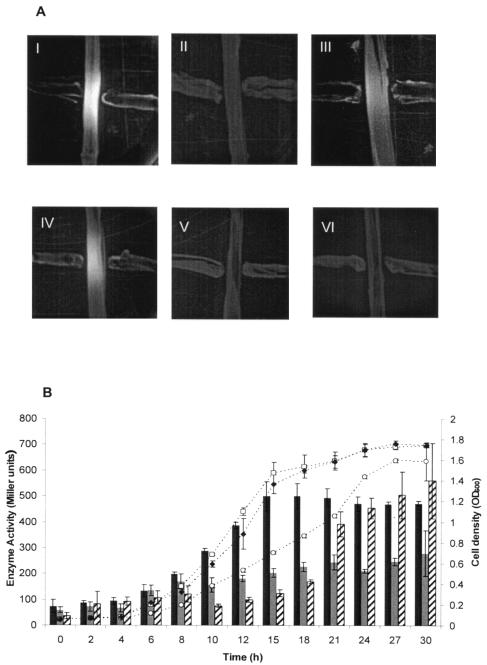
There is also evidence suggesting that multidrug efflux pumps may be involved in the efflux of autoinducers (12, 26). We cross-streaked B. pseudomallei against the reporter strains, E. coli JB525 and A. tumefaciens NTI. Autoinducers produced by KHW were detected by the appearance of green fluorescence on the E. coli JB525 reporter streak (Fig. 5A, panel I). Cross-streaks of KHWΔbpeAB against the E. coli JB525 reporter failed to yield any green fluorescence (Fig. 5A, panel II). Autoinducer production was restored in the complemented bpeAB mutant, KHWΔbpeAB (pUCP28TbpeAB), and in the bpeR mutant, KHWbpeR::Km, which showed that bpeAB-oprB expression was necessary for this process (Fig. 5A, panels III and IV). Additionally, the absence of autoinducer production in the bpeR-overexpressing strains, KHW(pUCPbpeR) and KHWbpeR::Km(pUCP28TbpeR), demonstrates that quorum sensing can be inhibited by overexpressing bpeR (Fig. 5A, panels V and VI, respectively). Similar results were obtained using the A. tumefaciens NTI indicator strain, except that in this case the autoinducers were detected by the appearance of a blue color on the reporter strain streaked on agar overlaid with X-Gal (data not shown).
FIG. 5.
(A) Detection of autoinducer production by B. pseudomallei KHW and its derivatives. Vertical streaks represent the E. coli JB525 reporter strain harboring the luxR-luxIpromoter-GFP plasmid (pJBA132), while the horizontal streaks represent (I) KHW, (II) KHWΔbpeAB mutant, (III) KHWΔbpeAB(pUCP28T-bpeAB)-complemented mutant, (IV) KHWbpeR::Km mutant, (V) KHWbpeR::Km(pUCP28T-bpeR)-complemented mutant, and (VI) KHW(pUCP28TbpeR), respectively. Fluorescence on the vertical streaks indicates the activity of autoinducers produced by the horizontal streaks of tester strains. (B) Effect of BpeAB-OprB on growth-phase-dependent expression of bpsIpromoter-lacZ. The plasmid pSYI carrying bpsIpromoter-lacZ was introduced into KHW, KHWΔbpeAB, and KHWbpeR::Km, respectively, to study the effect of BpeAB-OprB on the expression of the autoinducer synthase BpsI. Aliquots of the bacteria cultured in AB medium containing 20 mM glycerol, 0.2% Casamino Acids, and 25 μg/ml tetracycline were assayed at different time intervals from the onset of culture (0 h) to stationary phase (30 h). Dotted lines represent the cell densities of KHW(pSYI) (□), KHWΔbpeAB(pSYI) (○), and KHWbpeR::Km(pSYI) (⧫), respectively, while β-galactosidase activities are represented as bars [KHW(pSYI), black bars; KHWΔbpeAB(pSYI), gray bars; and KHWbpeR::Km(pSYI), striped bars]. All measurements were done in triplicate; the means and standard errors are shown.
BpeAB-OprB function affects growth-phase-dependent expression of the autoinducer synthase BpsI.
Since a reduction in autoinducer production in KHWΔbpeAB may be the result of either the absence of autoinducer efflux or repression of autoinducer synthase gene expression, we determined the expression of bpsI, which encodes an autoinducer synthase, in KHW and its isogenic mutants (34). It was recently reported that B. pseudomallei has three luxIR homologs which produce a variety of acyl-HSLs, including C8HSL, C10HSL, N-(3-hydroxyoctanoyl)-l-homoserine lactone (3-hydroxy-C8HSL), N-(3-hydroxydecanoyl)-l-homoserine lactone (3-hydroxy-C10HSL), and N-(3-oxotetradecanoyl)-l-homoserine lactone (3-oxo-C14HSL) (37). BpsI synthesizes mainly C8HSL (34).
Wild-type B. pseudomallei KHW entered stationary phase at 15 h after culture in AB medium containing 20 mM glycerol and 0.2% (wt/vol) Casamino Acids (Fig. 5B). In comparison, the doubling time in KHWΔbpeAB was almost twice as long and stationary phase was attained only after 27 h of culture. We introduced the bpsIpromoter-lacZ plasmid, pSYI, into KHW and KHWΔbpeAB to study the effect of BpeAB-OprB on bpsI expression. In the bpeAB mutant, bpsIpromoter-lacZ expression remained low for the duration of culture, finally attaining about half the level of that in KHW. In KHWbpeR::Km, which had functional BpeAB-OprB, the growth properties and levels of bpsIpromoter-lacZ expression after 30 h of culture were similar to those in KHW, which verifies that the absence of growth-phase-dependent expression of bpsI in the bpeAB mutant was indeed a consequence of the defective efflux pump (Fig. 5B). We have also cultured the bpeAB-null mutant under conditions similar to those described in Fig. 5B, but in the presence of 100 nM C8HSL, and observed that the growth of the bpeAB-null mutant was not restored to wild-type level (data not shown). The inclusion of 100 nM C8HSL in the culture medium also did not restore growth-phase-dependent bpsIpromoter-lacZ expression in KHWΔbpeAB, suggesting that intracellular concentrations of C8HSL might be high, possibly due to impaired efflux, and the accumulation of C8HSL in the bpeAB mutant might have resulted in a negative-feedback effect on bpsI expression.
Role of BpeAB-OprB in invasion of epithelial and macrophage cell lines.
Since any effect on quorum sensing could potentially affect virulence, we next determined if BpeAB-OprB would have an effect on the invasion of human lung epithelial (A549) and macrophage (THP-1) cells by B. pseudomallei. In the absence of BpeAB-OprB function, the invasion of both A549 and THP-1 cells by KHWΔbpeAB as well as the bpeR-overexpressing strains KHWbpeR::Km(pUCP28TbpeR) and KHW(pUCP28TbpeR) was significantly attenuated (Table 3). Interestingly, exogenous addition of 100 nM C8HSL to the A549 and THP-1 cell culture media could restore to wild-type levels this defect in cell invasion in the strains KHWΔbpeAB, KHWbpeR::Km(pUCP28TbpeR), and KHW(pUCP28TbpeR). Taken together, these results demonstrate that (i) the BpeAB-OprB efflux function is essential for cell invasion by B. pseduomallei and (ii) the reduced invasiveness of KHWΔbpeAB was probably due to its impaired quorum-sensing mechanism. Additionally, our data also demonstrate that it was possible to attenuate B. pseudomallei virulence by overexpressing the BpeR repressor. It is interesting that KHW, which overexpressed the BpeR repressor, was approximately 10-fold more attenuated in cell invasion of both cell lines than was the bpeAB-null mutant (Table 3). However, upon the addition of exogenous C8HSL, the invasiveness of the bpeR-overexpressing strain and that of the bpeAB-null mutants were equally restored to wild-type levels.
TABLE 3.
Effect of bpeAB and bpeR mutations and exogenous C8HSL (100 nM) on invasion of A549 and THP-1 cells by B. pseudomallei
| Strain | Inoculum (CFU) | Mean no. of cells recovered at 4 ha
|
|||
|---|---|---|---|---|---|
| A549 cells
|
THP-1 cells
|
||||
| −C8HSL | +C8HSL | −C8HSL | +C8HSL | ||
| KHW | 1.5 ± 107 | 7.8 × 104 ± 1.4 × 104 | 8.4 × 104 ± 2.0 × 104 | 8.5 × 104 ± 1.7 × 104 | 6.4 × 104 ± 7.2 × 103 |
| KHWΔbpeAB | 1.4 × 107 | 2.6 × 102 ± 2.3 × 102 | 7.1 × 104 ± 2.1 × 103 | 6.6 × 102 ± 1.0 × 102 | 6.8 × 104 ± 5.3 × 103 |
| KHWΔbpeAB(pUCP28TbpeAB) | 1.5 × 107 | 7.2 × 104 ± 2.2 × 104 | 8.9 × 104 ± 1.5 × 104 | 9.0 × 104 ± 1.2 × 104 | 8.1 × 104 ± 1.5 × 104 |
| KHWbpeR::Km | 1.7 × 107 | 4.3 × 104 ± 9 × 103 | 8.3 × 104 ± 1.5 × 104 | 7.6 × 104 ± 2.0 × 104 | 7.3 × 104 ± 1.6 × 104 |
| KHWbpeR::Km(pUCP28TbpeR) | 1.8 × 107 | 28 ± 17 | 1.1 × 105 ± 1.3 × 104 | 100 ± 15 | 8.3 × 104 ± 1.3 × 104 |
| KHW(pUCP28TbpeR) | 1.6 × 107 | 35 ± 24 | 9.0 × 104 ± 6.6 × 103 | 87 ± 14 | 7.5 × 104 ± 1.4 × 104 |
Values are means ± standard deviations.
Role of BpeAB-OprB in cytotoxicity of B. pseudomallei.
The cytotoxic effects of both KHWΔbpeAB and the bpeR-overexpressing strains, KHWbpeR::Km(pUCP28T-bpeR) and KHW(pUCP28T-bpeR), on both A549 and THP-1 were significantly reduced (Table 4). Forty-four percent of A549 cells and 42% of THP-1 cells were killed after exposure to KHW for 4 h. Compared to KHW, the cytotoxicity of KHWΔbpeAB, KHWbpeR::Km(pUCP28TbpeR), and KHW(pUCP28TbpeR) to A549 was significantly attenuated to 3%, 11%, and 6%, respectively, while their cytotoxicity to THP-1 cells was completely attenuated (Table 4). Addition of exogenous C8HSL could only partially restore the cytotoxicity of KHWΔbpeAB, KHWbpeR::Km(pUCP28TbpeR), and KHW(pUCP28TbpeR) to both types of cells.
TABLE 4.
Effect of bpeAB and bpeR mutations and exogenous C8HSL (100 nM) on cytotoxicity of B. pseudomalleia
| Strain | Inoculum (CFU) | Cytotoxicity (%) in cells:
|
|||
|---|---|---|---|---|---|
| A549
|
THP-1
|
||||
| −C8HSL | +C8HSL | −C8HSL | +C8HSL | ||
| KHW | 1.5 × 107 | 43.9 ± 3.0 | 48.7 ± 1.9 | 41.6 ± 0.6 | 57.1 ± 0.7 |
| KHWΔbpeAB | 1.4 × 107 | 2.8 ± 1.1 | 21.7 ± 4.7 | 0 ± 0.6 | 30.7 ± 3.7 |
| KHWΔbpeAB(pUCP28TbpeAB) | 1.5 × 107 | 50.1 ± 4.9 | 50.6 ± 2.6 | 21.2 ± 1.4 | 55.7 ± 0.7 |
| KHWbpeR::Km | 1.7 × 107 | 52.1 ± 3.8 | 48.8 ± 0.9 | 41.9 ± 0.7 | 55.3 ± 0.8 |
| KHWbpeR::Km(pUCP28TbpeR) | 1.8 × 107 | 11.0 ± 3.2 | 13.7 ± 3.8 | 0.6 ± 1.0 | 27.1 ± 0.5 |
| KHW(pUCP28TbpeR) | 1.6 × 107 | 6.2 ± 6.4 | 9.0 ± 1.6 | 0 ± 0.7 | 27.4 ± 0.5 |
Values are means ± standard deviations.
A difference between the two cell types is that the cytotoxicity towards A549 cells was fully restored to wild-type level when KHWΔbpeAB was complemented in trans with pUCP28TbpeAB, as opposed to its cytotoxicity towards THP-1 cells, which could be only partially restored to 50% of wild-type level in the complemented mutant. The bpeR mutation had no effect on the cytotoxicity of KHW to both types of cells. These data showed that the BpeAB-OprB function is also required for the cytotoxicity of B. pseudomallei towards A549 and THP-1 cells and that overexpression of the BpeR repressor could attenuate cytotoxicity. The attenuated virulence of the strains defective in bpeAB-oprB expression correlates well with their lack of autoinducer production, suggesting the importance of quorum sensing in the cytotoxic effect of B. pseudomallei.
BpeAB-OprB function is required for optimal biofilm formation and for the production of siderophores and phospholipase C.
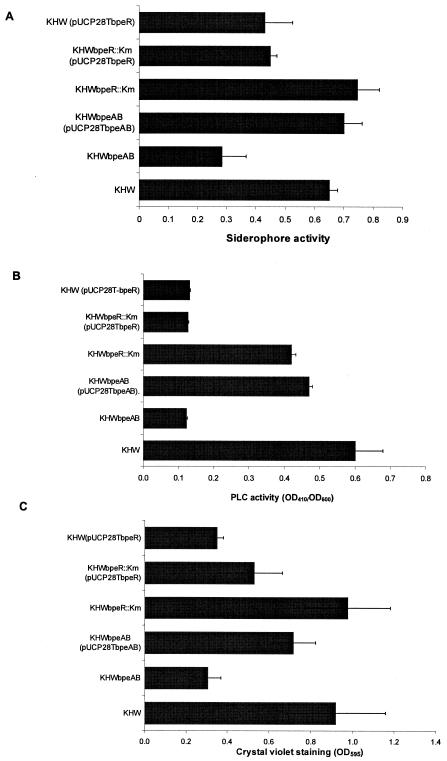
We have previously shown that the B. pseudomallei BpsIR quorum-sensing system regulates siderophore and phospholipase C production (34). We have also observed a reduction in biofilm formation in the bpsI and bpsR mutants (data not shown). Since BpeAB-OprB affects quorum sensing in B. pseudomallei, we were interested to know if these quorum-sensing-controlled cellular processes would also be affected by BpeAB-OprB. Siderophore production was reduced by 50% in the KHWΔbpeAB mutant but was reduced by only 30% in the strains overexpressing bpeR (Fig. 6A). The reduction of PLC production in the bpeAB mutant and the strains overexpressing bpeR was more significant. PLC activity detected in KHWΔbpeAB, KHWbpeR::Km(pUCP28TbpeR), and KHW(pUCP28TbpeR) was only about 20% of the wild-type KHW level (Fig. 6B). Biofilm formation was also significantly reduced in the bpeAB mutant and the bpeR-overexpressing strains. In KHWΔbpeAB, KHWbpeR::Km(pUCP28TbpeR), and KHW(pUCP28TbpeR), biofilm formation was 33%, 58%, and 38% of that in KHW, respectively (Fig. 6C). These data showed that quorum-sensing-controlled processes like optimal siderophore and phospholipase production, as well as biofilm formation, are dependent on BpeAB-OprB function. Overexpression of the BpeR repressor could also partially inhibit these processes.
FIG. 6.
Effect of BpeAB-OprB on siderophore and phospholipase C production and biofilm formation in B. pseudomallei. Siderophore activities, phospholipase C activities, and biofilm formation were measured in KHW, KHWΔbpeAB, KHWΔbpeAB complemented with pUCP28TbpeAB, KHWbpeR::Km, KHWbpeR::Km complemented with pUCP28TbpeR, and KHW harboring pUCP28TbpeR, respectively. (A) Optimal siderophore production in B. pseudomallei is dependent on the BpeAB-OprB function, and overexpression of bpeR in KHWbpeR::Km(pUCP28TbpeR) and KHW(pUCP28TbpeR) reduced siderophore production. Siderophore activities were assayed in the supernatants of 24-h-old cultures and were determined by measuring the differential in OD630 readings between the test and the sample blank. The values shown have been normalized for cell density by being expressed as a ratio of ΔOD630/OD600. (B) Optimal PLC secretion by B. pseudomallei KHW is dependent on BpeAB-OprB, and overexpression of bpeR in KHWbpeR::Km(pUCP28TbpeR) and KHW(pUCP28TbpeR) inhibited PLC secretion. PLC activities were determined in the supernatants of 24-h cultures, and the values shown have been normalized for cell density by being expressed as a ratio of OD410/OD600. (C) Optimal biofilm formation is also dependent on BpeAB-OprB. Biofilm formation was significantly reduced in KHWΔbpeAB and KHW overexpressing bpeR. Each bar is the average reading (± standard deviation) from three independent experiments.
DISCUSSION
The B. pseudomallei BpeAB-OprB efflux system is inducible by its substrate erythromycin. Since subinhibitory concentrations of erythromycin have been reported to induce the expression of luxI and lasI promoters and thus trigger the expression of many quorum-controlled target genes (14), it was necessary to distinguish whether bpeAB expression was induced by erythromycin because of its interaction with the BpeR repressor or if it was due to a more general transcriptional modulation of gene expression by subinhibitory concentrations of wide-ranging antibiotics as described by Goh et al. (14). We addressed this issue by using a subinhibitory concentration (0.1× MIC) of amoxicillin, an antibiotic which is not a substrate of BpeAB-OprB, to study its effect on bpeABpromoter-lacZ expression in KHW. Unlike erythromycin, subinhibitory concentrations of amoxicillin did not induce bpeABpromoter-lacZ and bpeRpromoter-lacZ expression in the wild-type background (data not shown). Thus, the induction of bpeAB expression by low-dose erythromycin appears to be a specific effect of erythromycin on bpeAB expression. Additionally, instead of using subinhibitory concentrations of erythromycin, we also performed transient assays by exposing the KHW(pCYYbpeAB) cultures to high doses of erythromycin at 5× MIC and 10× MIC for 30 min instead of 4 h. At both doses of erythromycin tested, bpeAB expression remained inducible in a dose-dependent manner, also confirming that the induction of bpeAB expression by erythromycin was a substrate-specific event (data not shown).
The low basal bpeAB-oprB expression present in wild-type cells may be attributed to a titration of the BpeR repressor by multiple copies of the bpeABpromoter-lacZ fusion plasmid used in this study (Fig. 2A). KHWbpeR::Km, which lacks the BpeR repressor, showed a basal bpeAB-oprB expression which was about 3.5-fold higher than that of the wild type. This consequently resulted in its relatively higher MICs and MBCs for gentamicin, streptomycin, and erythromycin (Table 2). These data are consistent with the role of BpeR as a repressor of bpeAB-oprB expression. We have also observed a delay in the erythromycin-induced expression of bpeRpromoter-lacZ expression in wild-type cells, compared to that of bpeABpromoter-lacZ expression (data not shown). This supports the view that the BpeR repressor interacts with its ligand to regulate bpeAB-oprB expression as well as its own expression. However, it was unexpected that the bpeAB-oprB expression in KHWbpeR::Km would remain inducible by erythromycin to levels comparable to that of the wild-type parental strain. An explanation would be the participation of another transcriptional regulator which recognizes erythromycin as a ligand and binds to a different site in the bpeAB-oprB regulatory region. Alternatively, a repressor protein of another RND pump (e.g., AmrR) which uses erythromycin as a ligand could share the bpeAB-oprB regulatory region. It has been reported that two promoters transcribe the P. aeruginosa mexAB-oprM operon-one of them regulated by the MexR repressor but the other not (11).
We showed that the expression of bpeAB-oprB is growth phase dependent and that that of bpeR is inducible upon entry into stationary phase. This could be explained if bpeAB-oprB expression is either (i) induced by a metabolite which is a substrate of the pump and accumulates with cell density or (ii) activated by a transcriptional regulator which controls the expression of genes at stationary phase or (iii) regulated by quorum sensing. mexAB-oprM expression in P. aeruginosa is similarly growth phase dependent, and the regulation of its growth-phase-dependent expression does not involve the MexR repressor (13). Our data have shown that bpeAB-oprB expression could be induced by exogenous autoinducers. BpeAB-OprB could be involved in the efflux of autoinducers, and the impairment of this efflux in the bpeAB-null mutant or the bpeR-overexpressing strains would result in an accumulation of autoinducers which would then exert a negative feedback on the expression of the autoinducer synthase. This would explain the attenuated virulence phenotype of the bpeAB-null mutant and the bpeR-overexpressing strains. Three pairs of luxIR homologs and two other luxR homologs have been identified in B. pseudomallei. Together they are responsible for the production of several acyl-homoserine lactone autoinducers: C8HSL, C10HSL, 3-hydroxy-C8HSL, 3-hydroxy-C10HSL, and 3-oxo-C14HSL (34, 37, 38). Using early-exponential-phase cultures when bpeAB-oprB expression was low, we showed that the induction of bpeAB-oprB expression could be advanced to the exponential phase by the exogenous addition of 100 nM C8HSL or C10HSL to the culture medium. It is noted that the response of bpeAB-oprB expression to such high concentrations of exogenous C8HSL or C10HSL might not necessarily imply that bpeAB-oprB expression is directly regulated by quorum sensing, as the acyl-HSLs could have activated a stationary-phase transcriptional regulator. The P. aeruginosa RpoS is an alternative sigma factor responsible for the switching-on of gene expression at stationary phase and is involved in the expression of 40% of quorum-controlled genes (33). rpoS expression is activated by quorum sensing, and among target genes regulated by both quorum sensing and RpoS there are probable RND efflux transporters (19, 33). Since the expression of rpoS in B. pseudomallei is also growth phase dependent (35), it is therefore difficult to distinguish if the induction of bpeAB-oprB expression at stationary phase is due to activation by RpoS or quorum sensing or both.
Although we observed that bpeAB expression could be activated by exogenous autoinducers, it was more responsive to C8HSL than to C10HSL, and only at high concentrations of C8HSL (>80 nM). Likewise, it was recently reported that the expression of P. aeruginosa mexAB-oprM was differentially enhanced by the exogenous addition of C4HSL and 3-oxo-C12HSL (20), although its expression was not regulated by the LasIR quorum-sensing system (13). We have also identified a las/lux box motif in the intergenic region between bpeR and bpeAB-oprB, but the significance of this has not been studied. It could perhaps be addressed by studying the effect of exogenous C8HSL and C10HSL on bpeAB-oprB expression in a quorum-sensing-null genetic background, even though such a mutant could be difficult to construct because of the presence of multiple luxIR homologs in B. pseudomallei (37).
Efflux pumps have also been implicated in the efflux of autoinducers, but direct evidence which demonstrates that autoinducers are indeed substrates of efflux pumps is lacking. It is reported that hyperexpression of P. aeruginosa MexAB-OprM resulted in a decline in N-(3-oxododecanoyl)-homoserine lactone (3-oxo-C12HSL) levels but had no effect on C4HSL levels (12). Consequentially, it is believed that, although C4HSL freely diffuses into and out of P. aeruginosa cells, movement of 3-oxo-C12HSL out of the cells requires active efflux (26). Our data showed that autoinducer production is significantly reduced when BpeAB-OprB function is impaired (Fig. 5A, panels II, V, and VI), and this reduction in autoinducer production seemed to be at the level of autoinducer synthesis, as the expression of the autoinducer synthase was impaired in the bpeAB mutant (Fig. 5B). It is interesting that in KHWbpeR::Km, where bpeAB expression is up-regulated, bpsI expression remained low up to early stationary phase (∼18 h) and then increased to wild-type levels thereafter. This was in contrast to bpsI expression in the KHWΔbpeAB mutant, which was not induced in stationary phase. This again points to other factors besides BpeR being involved in the regulation of bpeAB-oprB expression. We have also measured bpsI expression in the bpeAB-null mutant and found that bpsI expression was not inducible when cells were cultured up to 6 h in the presence of 100 nM exogenous C8HSL (data not shown). This suggests that C8HSL might be effluxed by BpeAB-OprB and that its high intracellular concentration in the bpeAB mutant could have a negative feedback inhibitory effect on the bpsI expression. Any inhibition of autoinducer synthase expression would therefore have a negative effect on quorum-regulated gene expression.
Growth impairment was observed in the KHWΔbpeAB mutant compared to KHW, both when cultured in AB medium supplemented with glycerol and Casamino Acids and when cultured in LB medium. The KHWΔbpeAB mutant had a longer doubling time and attained stationary phase only after >80 h although microscopic examination of the negatively stained cells by transmission electron microscopy showed size and length similar to those of KHW cells (data not shown). The failure to restore the growth impairment of the bpeAB-null mutant to wild-type levels in the presence of 100 nM C8HSL suggests that the cause of the growth defect of the bpeAB-null mutant is probably not regulated by C8HSL. It is also plausible that there is an accumulation in the KHWΔbpeAB mutant of a metabolite which is a physiological substrate of the BpeAB-OprB efflux pump and which has an impact on cell division. Polyamines, whose biosynthesis shares the same pathway as that of acyl-homoserine lactones, are plausible candidates (25, 36). The intracellular level of polyamines, which is tightly regulated, is an important control of cell division in E. coli (16).
Importantly, the impairment of the BpeAB-OprB pump either by the bpeAB-null mutation or by overexpression of bpeR had a dramatic effect on virulence attenuation of B. pseudomallei. These strains showed significant impairment in cell invasion of human lung epithelial (A549) and macrophage (THP-1) cells (Table 3). The cytotoxic effect of these strains on A549 and THP-1 was also significantly attenuated (Table 4). The partial restoration of cytotoxicity of the KHWΔbpeAB mutant and the bpeR-overexpressing strains, KHWbpeR::Km(pUCP28TbpeR) and KHW(pUCP28TbpeR), by the addition of exogenous C8HSL supports the notion that the BpeAB-OprB efflux function is important for optimal autoinducer synthesis. It is possible that the addition of exogenous C8HSL, which failed to induce bpsIpromoter-lacZ expression in the bpeAB mutant, may have a positive effect on the expression of the other luxIR homologs in the efflux-impaired B. pseudomallei. Such an effect could have restored the invasiveness of these efflux-impaired strains. It remains unclear why the addition of exogenous C8HSL autoinducer to the culture medium was able to partially restore the cytotoxic effect of the bpeAB mutant and bpeR-overexpressing strains towards THP-1 cells but had no effect on the cytotoxic effect of the same strains towards A549 cells.
It is also interesting that, in the identification of new regulators that modulate quorum sensing in P. aeruginosa, a mutation in a probable RND-like efflux transporter was found to significantly down-regulate the quorum-sensing-dependent lecA::lux expression (9). Our study has shown that the BpeAB-OprB function is important for optimal production of virulence factors such as siderophore and phospholipase C and for biofilm formation. The BpeAB-OprB function is thus an important virulence determinant of B. pseudomallei.
Future work will include the identification of the physiological substrate of the BpeAB-OprB efflux pump and the BpeR-dependent and BpeR-independent regulatory sites in the bpeA-bpeR intergenic region. Intermediary metabolites in the autoinducer biosynthetic pathway will be examined. Polyamines, such as spermidine and putrescine, which are important for many cellular functions including cell division, are plausible candidates.
Acknowledgments
This work was supported by a grant from the National Medical Research Council of Singapore (NMRC0426/2000). Y. Y. Chan is a recipient of a Research Scholarship from the Faculty of Medicine, National University of Singapore.
REFERENCES
- 1.Andersen, J. B., A. Heydorn, M. Hentzer, L. Eberl, O. Geisenberger, B. B. Christensen, S. Molin, and M. Givskov. 2001. gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl. Environ. Microbiol. 67:575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casadaban, M. J., J. Chou, and S. N. Cohen. 1980. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J. Bacteriol. 143:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan, Y. Y., T. M. Tan, Y. M. Ong, and K. L. Chua. 2004. BpeAB-OprB, a multidrug efflux pump in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 48:1128-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaowagul, W. 2000. Recent advances in the treatment of severe melioidosis. Acta Trop. 74:133-137. [DOI] [PubMed] [Google Scholar]
- 5.Chua, K. L., Y. Y. Chan, and Y. H. Gan. 2003. Flagella are virulence determinants of Burkholderia pseudomallei. Infect. Immun. 71:1622-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark, D., and O. Maaloe. 1967. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23:99-112. [Google Scholar]
- 7.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 9.Diggle, S. P., K. Winzer, A. Lazdunski, P. Williams, and M. Camara. 2002. Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of N-acylhomoserine lactone production and virulence gene expression. J. Bacteriol. 184:2576-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elsinghorst, E. A. 1994. Measurement of invasion by gentamicin resistance. Methods Enzymol. 236:405-420. [DOI] [PubMed] [Google Scholar]
- 11.Evans, K., L. Adewoye, and K. Poole. 2001. MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic region. J. Bacteriol. 183:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans, K., L. Passador, R. Srikumar, E. Tsang, J. Nezezon, and K. Poole. 1998. Influence of the MexAB-OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 180:5443-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans, K., and K. Poole. 1999. The MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa is growth-phase regulated. FEMS Microbiol. Lett. 173:35-39. [DOI] [PubMed] [Google Scholar]
- 14.Goh, E. B., G. Yim, W. Tsui, J. McClure, M. G. Surette, and J. Davies. 2002. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc. Natl. Acad. Sci. USA 99:17025-17030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoang, T. T., A. J. Kutchma, A. Becher, and H. P. Schweizer. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59-72. [DOI] [PubMed] [Google Scholar]
- 16.Inouye, M., and A. B. Pardee. 1970. Requirement of polyamines for bacterial division. J. Bacteriol. 101:770-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohler, T., C. van Delden, L. K. Curty, M. M. Hamzehpour, and J. C. Pechere. 2001. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 183:5213-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurioka, S., and M. Matsuda. 1976. Phospholipase C assay using p-nitrophenylphosphoryl-choline together with sorbitol and its application to studying the metal and detergent requirement of the enzyme. Anal. Biochem. 75:281-289. [DOI] [PubMed] [Google Scholar]
- 19.Latifi, A., M. Foglino, K. Tanaka, P. Williams, and A. Lazdunski. 1996. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21:1137-1146. [DOI] [PubMed] [Google Scholar]
- 20.Maseda, H., I. Sawada, K. Saito, H. Uchiyama, T. Nakae, and N. Nomura. 2004. Enhancement of the mexAB-oprM efflux pump expression by a quorum-sensing autoinducer and its cancellation by a regulator, MexT, of the mexEF-oprN efflux pump operon in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 48:1320-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Moore, R. A., D. DeShazer, S. Reckseidler, A. Weissman, and D. E. Woods. 1999. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 43:465-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard M7-A6 and MIC testing supplemental tables M100-S13, 6th ed., vol. 23, no. 2. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 24.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 25.Parsek, M. R., D. L. Val, B. L. Hanzelka, J. E. Cronan, Jr., and E. P. Greenberg. 1999. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. USA 96:4360-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearson, J. P., C. Van Delden, and B. H. Iglewski. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piper, K. R., S. Beck von Bodman, and S. K. Farrand. 1993. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362:448-450. [DOI] [PubMed] [Google Scholar]
- 28.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 29.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 30.Riedel, K., M. Hentzer, O. Geisenberger, B. Huber, A. Steidle, H. Wu, N. Hoiby, M. Givskov, S. Molin, and L. Eberl. 2001. N-acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147:3249-3262. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Sawada, I., H. Maseda, T. Nakae, H. Uchiyama, and N. Nomura. 2004. A quorum-sensing autoinducer enhances the mexAB-oprM efflux-pump expression without the MexR-mediated regulation in Pseudomonas aeruginosa. Microbiol. Immunol. 48:435-439. [DOI] [PubMed] [Google Scholar]
- 33.Schuster, M., A. C. Hawkins, C. S. Harwood, and E. P. Greenberg. 2004. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 51:973-985. [DOI] [PubMed] [Google Scholar]
- 34.Song, Y., C. Xie, Y. M. Ong, Y. H. Gan, and K. L. Chua. 2005. The BpsIR quorum-sensing system of Burkholderia pseudomallei. J. Bacteriol. 187:785-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subsin, B., M. S. Thomas, G. Katzenmeier, J. G. Shaw, S. Tungpradabkul, and M. Kunakorn. 2003. Role of the stationary growth phase sigma factor RpoS of Burkholderia pseudomallei in response to physiological stress conditions. J. Bacteriol. 185:7008-7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabor, C. W., and H. Tabor. 1985. Polyamines in microorganisms. Microbiol. Rev. 49:81-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ulrich, R. L., D. DeShazer, E. E. Brueggemann, H. B. Hines, P. C. Oyston, and J. A. Jeddeloh. 2004. Role of quorum sensing in the pathogenicity of Burkholderia pseudomallei. J. Med. Microbiol. 53:1053-1064. [DOI] [PubMed] [Google Scholar]
- 38.Valade, E., F. M. Thibault, Y. P. Gauthier, M. Palencia, M. Y. Popoff, and D. R. Vidal. 2004. The PmlI-PmlR quorum-sensing system in Burkholderia pseudomallei plays a key role in virulence and modulates production of the MprA protease. J. Bacteriol. 186:2288-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81-86. [DOI] [PubMed] [Google Scholar]
- 40.Yang, H. M., W. Chaowagul, and P. A. Sokol. 1991. Siderophore production by Pseudomonas pseudomallei. Infect. Immun. 59:776-780. [DOI] [PMC free article] [PubMed] [Google Scholar]