Abstract
We previously identified and characterized a two-component regulatory system in the meningococcus with homology to the phoP-phoQ system in salmonella and showed that allele replacement of the NMB0595 regulator gene led to loss of virulence, sensitivity to antimicrobial peptides, perturbed protein expression, and magnesium-sensitive growth. On the basis of these findings we proposed that the system should be designated the meningococcal PhoPQ system. Here we further characterized the NMB0595 mutant and demonstrated that it had increased membrane permeability and was unable to form colonies on solid media with low magnesium concentrations, features that are consistent with disruption of PhoPQ-mediated modifications to the lipooligosaccharide structure. We examined the transcriptional profiles of wild-type and NMB0595 mutant strains and found that magnesium-regulated changes in gene expression are completely abrogated in the mutant, indicating that, similar to the salmonella PhoPQ system, the meningococcal PhoPQ system is regulated by magnesium. Transcriptional profiling of the mutant indicated that, also similar to the salmonella PhoPQ system, the meningococcal system is involved in control of virulence and remodeling of the bacterial cell surface in response to the host environment. The results are consistent with the hypothesis that the PhoP homologue plays a role in the meningococcus similar to the role played by PhoP in salmonella. Elucidating the role that the PhoPQ system and PhoPQ-regulated genes play in the response of the meningococcus to the host environment may provide new insights into the pathogenic process.
Neisseria meningitidis is the major cause of epidemic meningitis worldwide. Vaccines based on the capsular polysaccharide of the meningococcus are effective for protecting against serogroup A and C disease but provide only short-lived immunity (36). A conjugated serogroup C polysaccharide vaccine has recently been introduced, and its introduction has been associated with a dramatic reduction in serogroup C meningococcal disease in the United Kingdom (3). However, the serogroup B polysaccharide is poorly immunogenic in humans, and no vaccine is currently available for this serogroup. The development of a vaccine for serogroup B disease is thus a priority for many areas of the world.
Protective antigens recognized during natural infection have escaped detection so far, possibly because they are expressed in vivo only during infection. Successful pathogens, such as Salmonella, Shigella, and Listeria, are known to express virulence genes in response to the host environment (33). Environmentally determined gene regulation is also seen in the meningococcus, which leads, for example, to the expression in vivo of iron-regulated proteins and to modulation of capsule and pilus expression that alters epithelial adherence and invasion (22, 42, 42, 46, 46), but the regulators are unknown.
Two-component regulatory systems control gene expression in many bacteria in response to environmental signals. These systems comprise a membrane-associated sensor kinase protein and a cytoplasmic transcriptional regulator. The membrane-located sensor autophosphorylates at a conserved histidine residue in response to external stimuli. The high-energy phosphate group is then transferred to a conserved aspartate residue in the response regulator, which causes it to undergo a conformational change that alters its ability to bind target DNA at specific promoter sites and thus acts as a transcriptional regulator. Two-component regulators respond to a wide variety of environmental signals, including magnesium, calcium, iron, osmolarity, and pH, and they orchestrate changes in cell structure, metabolism, and motility in response to these environmental cues. Many two-component systems are also involved in controlling virulence gene expression in response to changes in magnesium levels and pH. The PhoPQ system found in Salmonella and other enteric bacteria, such as Escherichia coli and Shigella, regulates more than 40 different genes in Salmonella, and these genes are designated PhoP-activated (pag) and PhoP-repressed (prg) genes (14, 14-16, 16, 34, 34) In Salmonella enterica serovar Typhimurium these genes affect survival within macrophages, resistance to host antimicrobial peptides and acid pH, invasion of epithelial cells, and antigen presentation. Inactivation of the PhoP/PhoQ system attenuates virulence more than 10,000-fold (34, 35, 35). PhoP/PhoQ homologues have been shown to be involved in control of virulence in a range of bacteria (25, 37, 37, 50), and detection of genes regulated by PhoP has been shown to be a powerful method for identification of genes involved in virulence (1, 1, 7, 7, 19).
Regulation involving two-component regulators often involves complex regulatory networks or cascades of regulation (9, 30). Most bacteria possess many two-component regulator systems (the E. coli genome encodes about 30) that interact to control the activity of many genes in response to a variety of environmental signals. For example, expression of the ugd gene in S. enterica is controlled by the PhoP/PhoQ system in response to low magnesium levels, by the PmrA/PmrB system in response to iron and pH, and by the RcsC/YojN/RcsB system in response to envelope stress (38). Lipopolysaccharide (LPS) modification in salmonella is also subject to complex regulation in which modifications to the lipid A portion of the molecule are controlled by the PhoP/PhoQ system but modifications to the core polysaccharide portion of the molecule are controlled the PmrAB system, which is itself under the control of the PhoPQ system (20, 21, 43).
We previously described a two-component regulatory system in the meningococcus (27) encoded by the adjacent NMB0595 and NMB0594 genes and characterized a knockout mutant of the regulator gene (NMB0595) in a group C strain, M96255789. The mutant had many characteristics in common with salmonella PhoPQ mutants. For example, magnesium-regulated changes in protein expression were abrogated in the mutant; the mutant grew poorly at low concentrations of magnesium; and the mutant was sensitive to defensins and was attenuated in the ability to travel through a layer of human epithelial cells (27). The M96255789 wild-type strain was found to be avirulent in a mouse model of infection, so the same mutation was introduced into another group C N. meningitidis strain that was virulent in mice, strain L91543. The L91543/NMB0595 knockout mutant strain was found to be sensitive to the cationic peptide polymyxin and was avirulent in a mouse model of infection (39). The features shared with Salmonella phoP mutants suggested that the NMB0595/NMB0594 system is a functional homologue of the Salmonella PhoPQ system, which led us to propose the designation PhoPQ for the meningococcal system. Recently, other researchers (52) inactivated the NMB0595 gene in their group B strain (designated NMB) of the meningococcus and confirmed that the mutant was sensitive to polymyxin. The NMB/NMB0595 mutant strain did not have a magnesium-dependent growth phenotype. Analysis of lipooligosaccharide (LOS) from the knockout mutant demonstrated that it had lost phosphoethanolamine (PEtn) decoration at the heptose (HepII) residue of the LOS inner core.
LOS (LPS in most other bacteria) is a key component of pathogenesis in meningococci. As an endotoxin, it is a major inflammatory mediator and contributes to much of the pathology; however, it also influences colonization and resistance to bactericidal antibody. Its structure is subject to strain variation, which allows meningococci to be divided into 12 immunotypes on the basis of the inner core structure (26). The structure is also phase variable such that a single strain may express more than one immunotype (28). As a target of host immunity, LOS is regarded as a candidate vaccine component (17, 44), so the finding that NMB0595 may be involved in promoting phase and strain variation of its structure is of considerable interest for vaccine development. lgtG, encoding a glucosyl transferase specific to the inner core, was significantly upregulated in the NMB/NMB0595 mutant (52). These findings, together with the lack of a magnesium-sensitive phenotype, led Tzeng et al. to argue against functional homology with the salmonella PhoP/PhoQ system, and they proposed an alternative nomenclature, misR (NMB0595)/misS (NMB0594), for the regulator and sensor of the meningococcal inner core structure (52).
In the current study our aim was to clarify these issues by further exploring the meningococcal NMB0595/NMB0594 two-component regulatory system at the physiological and transcriptome levels. Identifying the functions of genes regulated by this two-component system is also likely to be a route for determining the virulence mechanisms of this important pathogen.
MATERIALS AND METHODS
Bacterial strains.
N. meningitidis strains L91543 (C2a P1.2) (39) and M96255789 (C2a nontypeable) (27) used in this study were supplied by the Health Protection Agency, Manchester, United Kingdom. Organisms were routinely grown on Columbia agar base (Oxoid) plates supplemented with 6% (vol/vol) defibrinated horse blood (TC Supplies) (CB agar) and Mueller-Hinton broth (Oxoid). The peptone broth used for growth in some experiments contained 5 g neutralized bacteriological peptone (Oxoid) per liter, 1 g Na2HPO4 per liter, and 1% (vol/vol) glucose, and the pH was adjusted to 7.35.
Microarray design and construction.
The pan-Neisseria multistrain-multispecies microarray was designed using the approach described previously (24) to cover the genome sequences of three strains of N. meningitidis and one strain of Neisseria gonorrhoeae. Generation of a gene pool that was representative of all the genes in each of the four genomes covered by the microarray was the first step in this process. N. meningitidis MC58 (51) was used as the base strain for the microarray, and therefore all genes (with designations beginning with NMB) from this strain were included in the gene pool. The MC58 genes were then used as a reference to identify unique or divergent genes present in the other strains using a cumulative BLAST filtering process. An iterative approach was used to sequentially add to the gene pool any strain- or species-specific genes identified in N. meningitidis Z2491 (41), N. meningitidis FAM18 (NMC; The Wellcome Trust Sanger Institute, unpublished data) and N. gonorrhoeae FA1090 (NG; University of Oklahoma, unpublished data). Several potential PCR products were designed for each gene in the gene pool, and the optimal product was selected, based on the results of the BLAST analysis, to ensure minimal cross-hybridization to other genes in the gene pool. PCR products were amplified using a RoboAmp 4200 (MWG Biotech, Germany), and each product was verified by agarose gel electrophoresis to ensure that there was amplification of a single product of the correct size. PCR failures were subjected to further rounds of PCR amplification with modified conditions until a single product of the correct size was obtained. Additional validation of the array elements was achieved by sequencing 5% of the PCR products. Microarrays were constructed by robotic spotting of the PCR products in duplicate on GAPSII aminosilane glass slides (Corning, Corning, N.Y.) using a MicroGrid II (BioRobotics, United Kingdom) (23). The microarrays were postprint processed according to the slide manufacturer's instructions before hybridization.
Transcriptomics methodology.
Bacteria from the blood agar plates (CB agar) were harvested into RNA-Later (Ambion) and incubated at room temperature for 15 min. For liquid cultures the bacteria were harvested into 0.2 volume of 95% ethanol-5% RNA phenol (Sigma Aldrich) and incubated on ice for 30 min (49). For both methods after centrifugation the pellets were resuspended in Trizol (Invitrogen). Chloroform was added, and after centrifugation at 8,000 rpm and 4°C the aqueous phase was precipitated with isopropanol. The RNA was extracted using RNeasy kits (Qiagen) with on-column DNase treatment. The integrity of purified RNA was checked using an Agilent 2100 Bioanalyser. cDNA was synthesized from total RNA from wild-type and phoP mutant strains using SuperScript III reverse transcriptase and random hexamer primers (Invitrogen) and was labeled with either Cy3- or Cy5-dCTP (GE Healthcare). At least three biological replicates were used for each experimental condition, each with dye swaps, which gave a minimum of six arrays per condition. Hybridization was carried out in a hybridization chamber. After washing, the arrays were scanned with an Axon Genepix 4000B microarray scanner using the Genepix software.
Data analysis.
Microarray data were analyzed using the GeneSpring (SiliconGenetics) software package. Only data points that were flagged as present or marginal were included in the analysis. The data were normalized using the intensity-dependent Lowess normalization, with 50% of the data used for smoothing. Average normalized expression ratios (either mutant/wild type or 2 mM Mg2+/20 mM Mg2+) were calculated for each gene. To avoid excessive variability of normalized ratios for genes with very low (and proportionally highly variable) control channel values, control channel values that were less than 100 were set to 100. The data were then filtered by using Student t test P values to identify the genes whose expression differed significantly from unity and applying the Benjamini-Hochberg false discovery rate multiple-test correction to obtain a false discovery rate of 0.001, which corresponded to an uncorrected t test P value of approximately 0.0001.
Gene lists were compared with SG3b- in Bioscript Library 2.0 of GeneSpring by comparing user-generated lists with gene lists grouped into functional categories as part of the annotation of the genome of N. meningitidis serogroup A strain Z2491 (NMA) (41). A number of additional functional groups were generated, including membrane (any gene with “membrane” in its annotation), periplasmic (any gene with “periplasmic” in its annotation), signal sequence (any gene with “signal sequence” in its annotation), and cell envelope (any gene with “membrane,” “capsule,” “periplasmic,” or “transport” in its annotation).
Electron microscopy.
Bacteria were harvested from CB agar plates and fixed in 2.5% paraformaldehyde. After 30 min of incubation, the bacteria were washed twice in phosphate-buffered saline to remove the paraformaldehyde and then postfixed in 2.5% glutaraldehyde in cacodylate buffer (0.1 M) and in 1% buffered osmium tetroxide (OsO4), stained with 2% uranyl acetate, dehydrated in an ethanol series, and embedded in Spurr resin. Ultrathin sections were obtained with an Ultracut E (Reichert) ultramicrotome and counterstained with lead citrate. The sections were observed using an Hitachi H-7000 transmission electron microscope.
RESULTS
Phenotypic analysis: LOS-related phenotype. (i) Permeability.
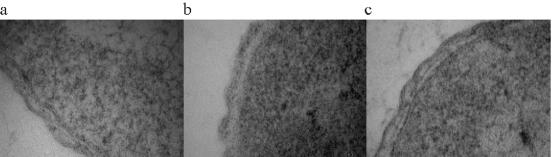
LPS/LOS forms the outer envelope of gram-negative bacteria, and the structure of the molecules is critically involved in cell permeability. PhoPQ-mediated changes in LPS structure are known to modulate cell permeability, which correlates with polymyxin sensitivity, in salmonella. To investigate membrane permeability, we examined cells of the wild type and the L91543/NMB0595 mutant by electron microscopy after OsO4 fixation followed by uranyl acetate staining. First, the cells were incubated with the dye before the sections were cut so that the stain had to penetrate through the cell's outer layer to stain the peptidoglycan layer and inner membrane. Second, the cells were stained after sectioning to allow the stain to access all membrane layers. As Fig. 1a shows, the stain was able to penetrate the cell wall of L91543/NMB0595 mutant cells so that a clear black line (which we believe to be the peptidoglycan layer) adjacent to the edge of the cytoplasm was visible. In contrast, the stain was less able to penetrate wild-type cells (Fig. 1b), and a clear zone was adjacent to the cytoplasm. This difference was attributed to the difference in permeability of intact cells (rather than intrinsic differences in stain binding) since no difference in morphology was seen in cells that were sectioned prior to staining (Fig. 1c).
FIG. 1.
Transmission electron microscopy of the meningococcal cell wall from the wild type and the L91543/NMB0595 mutant. (a and b) OsO4-uranyl acetate staining of L91543/NMB0595 cells (a) and wild-type L91543 cells (b) before the cells were sectioned. (c) Wild-type L91543 cells sectioned prior to staining.
(ii) Growth on low levels of magnesium.
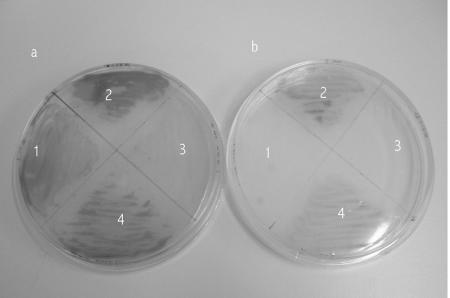
A related characteristic of salmonella phoPQ and pmrAB mutants is that they are unable to form colonies on solid media containing low concentrations of magnesium (45). Magnesium stabilizes the bacterial outer membrane by interacting with and neutralizing the phosphate groups in LPS/LOS (6). If insufficient magnesium is available to neutralize the negative charges, then the membrane may become unstable. The mutually negative charges in the outer membrane of adjacent cells is thought to inhibit colony formation. To counteract this, the salmonella PhoPQ system responds to a low magnesium concentration by activating genes encoding enzymes that covalently modify LPS to reduce the negative repulsive forces and thereby stabilize colony formation. phoPQ (and pmrAB) mutants are unable to mediate these modifications and thus cannot form colonies on solid media containing low magnesium concentrations (45). To test if the meningococcal NMB0595 mutants expressed the same phenotype, we plated the wild type and knockout mutants (both the initial M96255789/NMB0595 mutant strain that we constructed [27] and the L91543/NMB0595 mutant strain constructed later[39]) on solid minimal media containing 1 mM and 50 mM magnesium. As Fig. 2 shows, both parent strains were able to grow with both high and low levels of magnesium. In contrast, the M96255789/NMB0595 phoP mutant strain grew poorly on solid medium containing the high level of magnesium (despite the fact that it grew well in a broth culture containing magnesium at this concentration) and not at all with low magnesium concentrations. The L91543/NMB0595 mutant strain had a phenotype similar to that reported for salmonella since it was unable to form colonies on solid media containing low levels of magnesium.
FIG. 2.
Growth of the wild-type and NMB0595 mutant strains on solid media with two concentrations of magnesium. The meningococcal strains were grown overnight on peptone agar plates supplemented with either 50 mM (a) or 1 mM (b) magnesium chloride. In order to better visualize colony growth, the colonies were exposed to an oxidase reagent to increase the contrast between the growth and the agar before the plates were photographed. Quadrant 1, L91543/NMB0595 mutant; quadrant 2, wild-type strain L91543; quadrant 3, M96255789/NMB0595 mutant; quadrant 4, wild-type strain M96255789.
Transcriptome analysis.
A key feature of any regulator is the nature of the signal(s) to which it responds, which was investigated next by transcriptome analysis. Microarrays were constructed using the genome sequence of the group B MC58 strain as the base sequence. Additional neisserial strain- and species-specific genes, including genes from the group C strain N. meningitidis FAM18 (NMC; The Wellcome Trust Sanger Institute, unpublished data), were added to the array. It is important to note that for biosafety reasons, all our experiments were performed with group C strains. It is therefore likely that the gene complement of our experimental strain was different and/or divergent from that of the base strain used to construct the microarray. However, since group B and group C strains are very closely related (more than 95% DNA homology), this would not be expected to significantly affect the analysis.
(i) Blood agar-grown cells.
To explore as close to the full range of signals to which the NMB0595/NMB0594 system might respond as possible, we initially performed a transcriptome analysis of the wild-type and mutant L91543/NMB0595 strains grown on blood agar plates. Blood agar is a standard (rich) growth medium for N. meningitidis and is likely to contain many of the possible compounds to which the system responds. Both strains grew well on this medium, although the colonies were slightly smaller for the mutant strain. A total of 281 genes were identified as genes that were significantly up- or downregulated in the mutant compared to the wild type when the organisms were grown on blood agar (see Table S1 in the supplemental material). Of these, 129 were upregulated in the mutant and 152 were downregulated in the mutant.
(ii) Magnesium-dependent gene regulation.
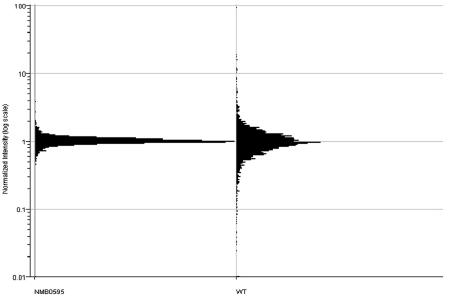
Regulation of the salmonella PhoPQ system is usually demonstrated with between micromolar and millimolar concentrations of magnesium. However, we found that neither the wild-type nor the mutant meningococcal strains grew well at micromolar concentrations of magnesium, so we compared magnesium regulation with 2 mM magnesium to magnesium regulation with 20 mM magnesium. Both strains grew reasonably well under these conditions. Meningococcal strains (wild type and mutant) were grown in magnesium-supplemented broth and harvested, and the RNA was extracted for transcriptome analysis. Two-color experiments were performed for the magnesium parameter, so that the ratio of gene expression with 20 mM magnesium to gene expression with 2 mM magnesium was determined for both the wild type and the L91543/NMB0595 mutant. As Fig. 3 shows, there was a wide range of ratios of gene expression levels in the wild-type strain in response to magnesium, but nearly all genes showed expression ratios close to unity in the L91543/NMB0595 mutant strain. Altogether, 106 genes were identified as genes that were significantly regulated by magnesium in the wild-type strain; 79 genes were upregulated at a magnesium concentration of 20 mM, whereas 27 genes were downregulated at a magnesium concentration of 20 mM (see Table S2 in the supplemental material). The levels of relative gene expression ranged from 0.2 to 19. No genes were identified as genes that were significantly regulated by magnesium in the L91543/NMB0595 mutant strain. One of the genes that was upregulated in the wild type at a magnesium concentration of 2 mM was NMB0595, the phoP-like regulatory gene, which gave an average normalized expression ratio (20 mM magnesium/2 mM magnesium) of 0.489 (P = 1.1 × 10−5), indicating that this gene is approximately twofold upregulated with 2 mM magnesium. The normalized expression for the gene encoding the cognate sensor protein, NMB0594, was also upregulated (expression ratio, 0.65), but the level of significance, P = 0.003, was below the significance threshold.
FIG. 3.
Magnesium-dependent gene expression: frequency plot of normalized expression ratios for all meningococcal genes for wild-type strain L91543 (WT) and L91543/NMB0595 mutant (NMB0595) cells grown with 20 mM and 2 mM magnesium.
Nature of genes whose expression was perturbed in the NMB0595 mutant strain. (i) Gene regulators.
Gene regulators are often connected in interacting networks and cascades of regulation so it was of interest to investigate whether the meningococcal NMB0595/NMB0594 system is involved in the regulation of any other regulator. The meningococcal genome encodes only three complete putative two-component regulatory systems in addition to the NMB0595/NMB0594 system, the NMB1607/NMB1606, NMB0115/NMB0114, and NMB1250/NMB1249 systems. One other gene, NMB1792, encodes a putative sensor kinase, but there is no obvious adjacent regulator gene. As determined by the significance criteria that we adopted, the expression of only one of the two-component regulator genes, the phoQ-like sensor gene NMB0594, was significantly changed (normalized expression, 0.342; P = 4 × 10−6) in the phoP mutant grown on blood agar, indicating that there was either a polar effect of the NMB0595 mutation or autoregulation of the system. The expression of NMB1250 (a regulator component) was slightly reduced (normalized expression, 0.71), but the significance level (P = 0.004) was below the cutoff. The same gene was significantly regulated by magnesium (normalized expression, 2.3), and its cognate sensor, NMB1249, had an expression level of 2.8 (although at a significance value of P = 0.0002, which was just below the cutoff). This magnesium-related difference in gene expression was not observed in the L91543/NMB0595 mutant. Note that the direction of expression of the NMB1250/NMB1249 system in response to magnesium was opposite that of the NMB0595/NMB0594 system (as described above, both NMB0595 and NMB0594 were downregulated in medium containing 20 mM magnesium in the wild-type strain). The expression levels of the following six other regulatory genes were significantly perturbed in the L91543/NMB0595 strain grown on blood agar: NMB0398, a putative ArsR family regulator (normalized expression, 0.49); NMB1405, a putative RTX family regulator (normalized expression, 2.0); NMB1711, a GntR family regulator; NMB1711 (normalized expression, 0.58); cspA, a putative transcriptional regulator containing a cold shock domain (normalized expression, 0.43); and NMB0556, a putative repressor (normalized expression, 2.3).
(ii) Genes involved in LOS synthesis and modification.
There were 10 genes involved in synthesis of the lipid A part (PEtn-Kdo2-lipid A) of the LOS molecule, and the expression of 3 of these genes was determined to be significantly downregulated in the L91543/NMB0595 mutant grown on blood agar; these genes were kdsA, lpxC, and lptA. kdsA and lpxC (but not lptA) were also magnesium regulated, although in opposite directions (lpxC was upregulated with 2 mM magnesium). kdsA and lpxC are both involved in the early steps in lipid A biosynthesis, whereas LptA is responsible for adding PEtn to the lipid A region of LOS. The expression of only 1 gene (NMB0828, a putative rfaD gene) of the 13 genes involved in synthesis and modification of the core polysaccharide structure of LOS was modified in the L91543/NMB0595 mutant.
(iii) Other functional gene categories.
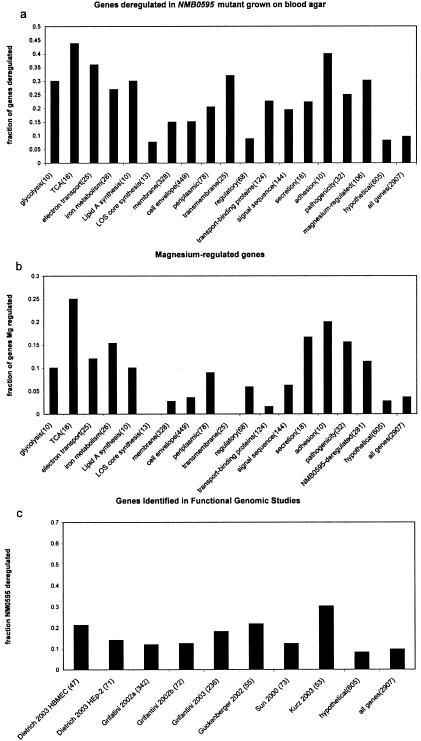
The list of genes that were identified as genes that were either up- or downregulated in the L91543/NMB0595 mutant was compared with lists of genes categorized into functional groups, based largely on the genome annotation (41). The functional categories whose members significantly overlapped the list of genes perturbed in the L91543/NMB0595 mutant and genes regulated by magnesium in the wild-type strain are shown in Fig. 4a and b, respectively. A clear preponderance of genes involved in carbon source utilization and energy metabolism was apparent among genes affected by the NMB0595 mutation, including genes involved in oxidative phosphorylation (P = 6 × 10−9), respiration (P = 1 × 10−8), electron transport (P = 4 × 10−5), and the tricarboxylic acid cycle (P = 0.0006). Many of these genes were clustered into operons in which most or all of the genes were significantly deregulated, including the nuo operon (nuoABCDEFGHIJKLMN) encoding subunits of NADH dehydrogenase, the pet operon (petABC) encoding cytochrome c subunits, and the sdh operon (sdhABCD) encoding subunits of a putative succinate dehydrogenase. There was also a very significant overlap with genes involved in the structure and dynamics of the meningococcal cell surface (transport-binding proteins, potentially secreted genes, membrane proteins, periplasmic proteins, lipid A synthesis). A significant overlap was also seen with genes involved in pathogenicity, adhesion, and iron metabolism. The pathogenicity genes whose expression was changed in the mutant included several genes encoding products that have been investigated as potential vaccine components, including the adhesin NspA gene (32), the NadA gene (5), opa (2), and NMA2175, which encodes a homologue of VapA, a Rhodococcus equi antigen that has been investigated as a candidate vaccine protein for infection of horses (48). The iron uptake (a key component of meningococcal virulence) genes that were perturbed included the bacterioferritin-encoding genes bfrA and bfrB and hemO encoding a heme utilization protein. Several genes involved in lactate metabolism, which has recently been associated with virulence in the meningococcus (47), were also highly upregulated, including genes encoding two putative lactate permease proteins, NMB1712 and NMB0543. NMB1712 was particularly striking in that it was expressed at negligible levels in the wild-type strain but was quite highly expressed in the L91543/NMB0595 mutant.
FIG. 4.
Functional classification of NMB0595-deregulated and magnesium-regulated genes. The fractions of genes identified as deregulated in the L91543/NMB0595 mutant grown on blood agar (a) and differentially expressed in wild-type L91543 cells grown in 20 mM magnesium compared to cells grown in 2 mM magnesium (b) were plotted for each functional category. The numbers in parentheses indicate the total number of genes in each functional category. TCA, tricarboxylic acid. (c) Comparison of genes deregulated in L91543/NMB0595 cells grown on blood agar to lists of genes that were found to be regulated upon exposure of meningococci to either HEp-2 epithelial cells or HBMEC (8), human bronchial epithelial cell line 16HBE14 (12), human serum (31), iron (13), or heat shock (18) or identified as genes that are involved in virulence by transposon mutagenesis (47). For each graph, the fraction of expected genes can be estimated by examining the “all genes” and “hypotheticals” categories.
(iv) Comparison of NMB0595-perturbed and magnesium-regulated genes.
There was a highly significant association between magnesium-regulated genes and genes which were identified as genes that were deregulated in the mutant grown on blood agar (P = 7 × 10−7), but only about 12% of the genes whose expression was deregulated in the L91543/NMB0595 mutant grown on blood agar were regulated by magnesium in the wild type (Fig. 4b), at least under the conditions that we tested using the significance criteria which we adopted. Several discordant sets of genes were identified. For instance, both the succinate dehydrogenase operon (sdh) and the NADH dehydrogenase operon (nuo) were significantly upregulated in the L91543/NMB0595 mutant grown on blood agar, but only the sdh operon was regulated by magnesium. There was a significant association between magnesium regulation and genes involved in pathogenicity, adhesion, and secretion (Fig. 4b). The magnesium-regulated pathogenicity-associated genes included nspA, the mafA adhesin gene, and the putative hemolysin gene, NMB1646.
(v) Comparison with genes identified in other functional genomic studies.
Sun et al. (47) used transposon mutagenesis to identify 73 genes that were required for virulence in a rat model of infection. Expression of nine of these genes was significantly perturbed in the L91543/NMB0595 mutant, including NMB0543 encoding lactate permease and genes encoding several other transport or putative transport proteins. Several authors have performed transcriptome analyses with the meningococcus exposed to different environments, including human serum (31), contact with either HEp-2 epithelial cells or human brain microvascular endothelial cells (HBMEC) (8), contact with the human bronchial epithelial cell line 16HBE14 (12), contact the cell line 16HBE14 for N. meningitidis and Neisseria lactamica (11), in vitro exposure to different levels of iron (13), and heat shock (18). The degree of overlap (number of genes identified in both lists) between genes identified in these studies and genes identified as genes that were deregulated in the L91543/NMB0595 mutant grown on blood agar is shown in Fig. 4c. The highest degree of overlap (16 of 53 genes) was found with genes identified as genes that were upregulated upon exposure of the meningococcus to human serum (31), but other associations were found. For instance, six of seven genes identified as genes that were downregulated upon exposure of the meningococcus to HBMEC were upregulated in the NMB0595 mutant (the seventh gene was not detected).
In silico analysis of meningococcal PhoP protein.
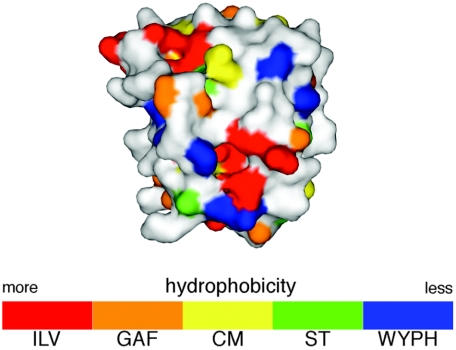
The regulator components of bacterial two-component response regulators from Bacillus subtilis and E. coli have been classified into a number of groups on the basis of the surface composition of the α1 helix-α5 helix interface, a surface that is important in response regulator-HisKA/Hpt domain contacts (29). A PSI-BLAST search used to determine the structural templates from which to model NMB0595 revealed that the NMB0595 sequence was most similar to the sequences of response regulators in the OmpR subclass that includes E. coli PhoP. Furthermore, an in silico structural analysis of the protein encoded by NMB0595 based on surface characteristics of the receiver domain confirmed that its predicted structure is consistent with the structure of E. coli OmpR subclass E (29), which includes PhoP (Fig. 5).
FIG. 5.
In silico model of NMB0595. The model was generated as described previously (29) and was color coded according to a numerically weighted hydrophobicity scale for each amino acid, as shown in the color bar at the bottom (single-letter code).
DISCUSSION
We previously demonstrated that the NMB0595 mutation confers susceptibility to cationic antimicrobial peptides (CAMPs), including polymyxin (27, 39). We demonstrate here that, as in Salmonella, this resistance is associated with increased membrane permeability, at least to the osmium tetroxide-uranyl acetate electron microscope stain used in this study. This difference in membrane permeability is likely to be caused by a loss of PhoP-mediated modification to the LOS structure in the mutant, leading to changes in outer membrane structure and hydrophobicity. These results suggest that a similar mechanism of PhoPQ-mediated modification to the LOS structure underpins polymyxin resistance in the meningococcus. We found several genes involved in LOS synthesis whose expression was significantly modified in the L91543/NMB0595 mutant, but, in contrast to the findings of Tzeng et al. (52), these genes did not include lgtG. Tzeng et al. (52) proposed that the LgtG-mediated loss of PEtn modification to the inner core was responsible for conferring polymyxin sensitivity on their NMB0595 mutant. This was, however, unlikely to account for polymyxin sensitivity in our L91543/NMB0595 mutant since lgtG gene expression was unchanged. It is possible that strain variation might account for these differences since the core polysaccharide structure of meningococcal LOS is highly variable and this variation might influence NMB0595/NMB0594-mediated modification to the LOS structure.
A key aspect of any regulatory system is the identity of the environmental signal to which the system responds. Gene expression was first examined for wild-type and L91543/NMB0595 mutant cells grown on blood agar, and a large number of genes (281 genes) were identified whose expression was significantly perturbed in the mutant. There are at least three possible reasons that can account for this kind of perturbation. First, expression of a perturbed gene could be under the direct control of the inactivated gene in wild-type cells. Second, a perturbed gene could be under the control of genes whose expression is under the control of the inactivated gene in wild-type cells. And third, the expression of a perturbed gene may be a response by the cell to stress induced by loss of the inactivated gene. With the caveats described above in mind, there are a number of interesting features of the set of genes identified as genes that were deregulated in the mutant. In common with the salmonella PhoPQ system, the meningococcal NMB0595/NMB0594 system appears to regulate the expression of several virulence genes, including nspA, which is a homologue of the pagM gene regulated by the salmonella phoPQ system (4). A clear preponderance of genes are involved in carbon source utilization and respiration. This may indicate that respiratory genes are targets of NMB0595 gene regulation, but it may also indicate that there are indirect effects. Respiratory enzymes are located on the inner membrane and are responsible for generating the proton motive force that is used to drive ATP synthesis. We demonstrated that the L91543/NMB0595 mutant is characterized by increased outer membrane permeability, and it is possible this may affect the efficiency of the respiratory pathways located on the inner membrane and thereby generate compensatory gene-regulatory events. However, genes encoding the F1 and F0 ATP synthase subunits were also all downregulated upon contact of either N. meningitidis or N. lactamica with 16HBE14 epithelial cells (11). Also, transposon mutagenesis of the meningococcus identified genes involved in lactate metabolism as genes that are also involved in virulence (47). Together, these results suggest that control of central metabolism may be a feature of the response of this pathogen to the host environment.
Among the genes deregulated in the L91543/NMB0595 mutant, there is a very significant association with genes involved in the dynamics of the meningococcal cell surface, including genes encoding adhesins, membrane proteins, transport protein, and potentially secreted protein. Many of these genes (e.g., the mafA adhesin gene) have been identified in several other studies (11) as genes that are regulated in response to the host environment. This suggests that, like the Salmonella PhoPQ system (10), the NMB0595/NMB0594 system is involved in remodeling the bacterial cell surface in response to the host environment and the immune response.
Oshima et al. (40) performed a transcriptome analysis of all two-component regulatory systems in E. coli. The functional groups of genes perturbed in the meningococcal L91543/NMB0595 mutant (Fig. 5) overlap the genes identified as genes that are perturbed in several two-component regulatory mutants of E. coli, including the genes encoding the PhoPQ system, the ArcAB system, the CpxRA system, and the BasRS system (homologous to the Salmonella PmrAB system). These systems differ in both the signals to which they respond and their target genes. PhoPQ responds to magnesium and calcium; ArcAB responds to anaerobic conditions; the CpxRA system responds to a variety of envelope stresses, including membrane protein damage, starvation, and high osmolarity; and the BasRS system (PmrAB in Salmonella) responds to iron and pH. The transcriptome analysis described here established that the NMB0595/NMB0594 system responds to magnesium, demonstrating that it sees the same signal as the Salmonella PhoPQ system. NMB0595 is itself magnesium regulated in the wild type, indicating that, like other two-component regulators, the NMB0595/NMB0594 system is likely to be autoregulated.
We examined another magnesium-dependent phenotype in Salmonella, the inability to form colonies on solid media containing low magnesium concentrations (45). Our results indicate that both the NMB0595 mutants have this phenotype (Fig. 2). The mechanism underlying this phenotype in Salmonella is known to be disruption of PhoPQ-mediated modifications to the LPS structure in mutant strains, providing further evidence that modifications to LOS in the meningococcus are controlled by the NMB0595/NMB0594 system (52). However, in contrast to both NMB0595 mutants generated in our laboratory, the NMB/NMB0595 mutant strain (52) did not appear to have a magnesium-sensitive growth phenotype. Once again, the reason for the discrepancy between the two studies may be strain variation. The Salmonella PhoPQ system controls expression of the mgtA gene, encoding a magnesium transporter, in response to low levels of magnesium. On the basis of finding no change in “mgtA” gene expression in their NMB/NMB0595 mutant, Tzeng et al. (52) argued that the NMB0595/NMB0594 system is not functionally homologous to the Salmonella PhoPQ system. However, no gene is currently designated mgtA in the meningococcus. A BLAST search of the meningococcal genome with the salmonella mgtA gene identified two genes with high similarity scores, NMB1325 and NMB1042, but the level of homology (about 35% identity at the amino acid level) is not high enough to be sure of functional homology. We found that expression of neither gene was significantly altered in our L91543/NMB0595 mutant grown on blood agar or in the wild type in response to magnesium. However, expression of salmonella mgtA is stimulated at micromolar magnesium concentrations, but we were unable to grow the meningococcus at magnesium concentrations less than 1 mM, so it is possible that the magnesium transport system may not be active in either the wild-type or mutant strain under the conditions tested.
Tzeng et al. (52) proposed that the NMB0595/NMB0594 system should be renamed MisRS, for the reasons described above. We do not agree with this change in nomenclature for the following reasons. First, there is no evidence in our data that the LOS inner core structure is a target for NMB0595/NMB0594-mediated modification. In any case, the Salmonella PhoPQ system is also involved in modification of the inner core structure of LPS through addition of phosphate and PEtn to the 2-keto-3-deoxyoctulosonic acid and heptose residues of the inner core (10). Although these modifications are only indirectly controlled by the PhoPQ system, through its downstream regulation of the PmrAB system, it is not clear from either set of data whether the NMB0595/NMB0594 system, or a system controlled by NMB0595/NMB0594, is responsible for the observed differences in modifications to the meningococcal LOS inner core. It should also be noted that the PhoPQ system of Yersinia pestis is also involved in modifying the LOS inner core structure of that pathogen (25). Second, the additional findings described here (that the system is regulated by magnesium, that NMB0595 mutants exhibit a magnesium-dependent inability to form colonies on solid media, and that in silico analysis indicates that the NMB0595 regulator exhibits structural homology with E. coli PhoP) indicate that the NMB0595/NMB0594 system is indeed a functional homologue of the salmonella PhoPQ system, and its nomenclature should reflect that fact.
In conclusion, we demonstrated that the meningococcal PhoPQ system is a magnesium-sensing two-component regulatory system. Inactivation of the PhoPQ system leads to increased membrane permeability and an inability to form colonies at low magnesium concentrations. These observations are consistent with a role in regulating modifications to the meningococcal LOS structure in response to the host environment. phoP gene inactivation perturbed expression of a large number of genes, many of which are involved in the synthesis of components of the meningococcal cell surface, such as LOS, and several previously identified virulence genes and/or vaccine candidates; however, many of the genes have unknown functions. Elucidating the role that the PhoPQ system and PhoPQ-regulated genes play in the response of the meningococcus to the host environment may provide new insights into the pathogenic process.
Supplementary Material
Acknowledgments
We acknowledge BμG@S (the Bacterial Microarray Group at St. George's Hospital Medical School) for supplying the microarray and for advice. We thank Doug Kojetin and John Cavanagh (North Carolina State University) for assistance with the subclassification process, supported by a grant to John Cavanagh from NIH (grant GM55769). We thank Ray Borrow and Helen Findlow (Vaccine Evaluation Department, Manchester Medical Microbiology Partnership) for immunotyping meningococcal strains. We thank Francisca Cardoso (Servei de Microscopia Universitat Autonoma de Barcelona) for assistance with the electron microscopy.
This work was supported by the Meningitis Trust. We acknowledge The Wellcome Trust for funding the multicollaborative microbial pathogen microarray facility under its Functional Genomics Resources Initiative. E. Mendoza acknowledges the Generalitat de Catalunya for support from the Nanotec postdoctoral fellowship program.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adams, P., R. Fowler, N. Kinsella, G. Howell, M. Farris, P. Coote, and C. D. Connor. 2001. Proteomic detection of PhoPQ- and acid-mediated repression of Salmonella motility. Proteomics 1:597-607. [DOI] [PubMed] [Google Scholar]
- 2.Aho, E. L., J. A. Dempsey, M. M. Hobbs, D. G. Klapper, and J. G. Cannon. 1991. Characterization of the opa (class 5) gene family of Neisseria meningitidis. Mol. Microbiol. 5:1429-1437. [DOI] [PubMed] [Google Scholar]
- 3.Balmer, P., R. Borrow, and E. Miller. 2002. Impact of meningococcal C conjugate vaccine in the UK. J. Med. Microbiol. 51:717-722. [DOI] [PubMed] [Google Scholar]
- 4.Belden, W. J., and S. I. Miller. 1994. Further characterization of the PhoP regulon: identification of new PhoP-activated virulence loci. Infect. Immun. 62:5095-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comanducci, M., S. Bambini, B. Brunelli, J. du-Bobie, B. Arico, B. Capecchi, M. M. Giuliani, V. Masignani, L. Santini, S. Savino, D. M. Granoff, D. A. Caugant, M. Pizza, R. Rappuoli, and M. Mora. 2002. NadA, a novel vaccine candidate of Neisseria meningitidis. J. Exp. Med. 195:1445-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coughlin, R. T., S. Tonsager, and E. J. McGroarty. 1983. Quantitation of metal cations bound to membranes and extracted lipopolysaccharide of Escherichia coli. Biochemistry 22:2002-2007. [DOI] [PubMed] [Google Scholar]
- 7.Detweiler, C. S., D. B. Cunanan, and S. Falkow. 2001. Host microarray analysis reveals a role for the Salmonella response regulator phoP in human macrophage cell death. Proc. Natl. Acad. Sci. USA 98:5850-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dietrich, G., S. Kurz, C. Hubner, C. Aepinus, S. Theiss, M. Guckenberger, U. Panzner, J. Weber, and M. Frosch. 2003. Transcriptome analysis of Neisseria meningitidis during infection. J. Bacteriol. 185:155-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eguchi, Y., T. Okada, S. Minagawa, T. Oshima, H. Mori, K. Yamamoto, A. Ishihama, and R. Utsumi. 2004. Signal transduction cascade between EvgA/EvgS and PhoP/PhoQ two-component systems of Escherichia coli. J. Bacteriol. 186:3006-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ernst, R. K., T. Guina, and S. I. Miller. 2001. Salmonella typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes Infect. 3:1327-1334. [DOI] [PubMed] [Google Scholar]
- 11.Grifantini, R., E. Bartolini, A. Muzzi, M. Draghi, E. Frigimelica, J. Berger, F. Randazzo, and G. Grandi. 2002. Gene expression profile in Neisseria meningitidis and Neisseria lactamica upon host-cell contact: from basic research to vaccine development. Ann. N. Y. Acad. Sci. 975:202-216. [DOI] [PubMed] [Google Scholar]
- 12.Grifantini, R., E. Bartolini, A. Muzzi, M. Draghi, E. Frigimelica, J. Berger, G. Ratti, R. Petracca, G. Galli, M. Agnusdei, M. M. Giuliani, L. Santini, B. Brunelli, H. Tettelin, R. Rappuoli, F. Randazzo, and G. Grandi. 2002. Previously unrecognized vaccine candidates against group B meningococcus identified by DNA microarrays. Nat. Biotechnol. 20:914-921. [DOI] [PubMed] [Google Scholar]
- 13.Grifantini, R., S. Sebastian, E. Frigimelica, M. Draghi, E. Bartolini, A. Muzzi, R. Rappuoli, G. Grandi, and C. A. Genco. 2003. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. USA 100:9542-9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groisman, E. A. 1998. The ins and outs of virulence gene expression: Mg2+ as a regulatory signal. Bioessays 20:96-101. [DOI] [PubMed] [Google Scholar]
- 15.Groisman, E. A., E. Chiao, C. J. Lipps, and F. Heffron. 1989. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc. Natl. Acad. Sci. USA 86:7077-7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groisman, E. A., and M. H. Saier. 1990. Salmonella virulence: new clues to intramacrophage survival. Trends Biochem. Sci. 15:30-33. [DOI] [PubMed] [Google Scholar]
- 17.Gu, X. X., and C. M. Tsai. 1993. Preparation, characterization, and immunogenicity of meningococcal lipooligosaccharide-derived oligosaccharide-protein conjugates. Infect. Immun. 61:1873-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guckenberger, M., S. Kurz, C. Aepinus, S. Theiss, S. Haller, T. Leimbach, U. Panzner, J. Weber, H. Paul, A. Unkmeir, M. Frosch, and G. Dietrich. 2002. Analysis of the heat shock response of Neisseria meningitidis with cDNA- and oligonucleotide-based DNA microarrays. J. Bacteriol. 184:2546-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guina, T., E. C. Yi, H. Wang, M. Hackett, and S. I. Miller. 2000. A PhoP-regulated outer membrane protease of Salmonella enterica serovar Typhimurium promotes resistance to alpha-helical antimicrobial peptides. J. Bacteriol. 182:4077-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunn, J. S. 2001. Bacterial modification of LPS and resistance to antimicrobial peptides. J. Endotoxin Res. 7:57-62. [PubMed] [Google Scholar]
- 21.Guo, L., K. B. Lim, J. S. Gunn, B. Bainbridge, R. P. Darveau, M. Hackett, and S. I. Miller. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250-253. [DOI] [PubMed] [Google Scholar]
- 22.Hammerschmidt, S., R. Hilse, J. P. van Putten, R. Gerardy Schahn, A. Unkmeir, and M. Frosch. 1996. Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J. 15:192-198. [PMC free article] [PubMed] [Google Scholar]
- 23.Hinds, J., K. G. Laing, J. A. Mangan, and P. Butcher. 2002. Glass slide microarrays for bacterial genomes, p. 83-99. In B. W. Wren and N. Dorrell (ed.), Methods in microbiology: functional microbial genomics. Academic Press, London, United Kingdom.
- 24.Hinds, J., A. A. Witney, and J. K. Vass. 2002. Microarray design for bacterial genomes, p. 67-82. In B. W. Wren and N. Dorrell (ed.), Methods in microbiology: functional microbial genomics. Academic Press, London, United Kingdom.
- 25.Hitchen, P. G., J. L. Prior, P. C. Oyston, M. Panico, B. W. Wren, R. W. Titball, H. R. Morris, and A. Dell. 2002. Structural characterization of lipo-oligosaccharide (LOS) from Yersinia pestis: regulation of LOS structure by the PhoPQ system. Mol. Microbiol. 44:1637-1650. [DOI] [PubMed] [Google Scholar]
- 26.Jennings, M. P., Y. N. Srikhanta, E. R. Moxon, M. Kramer, J. T. Poolman, B. Kuipers, and L. P. van der. 1999. The genetic basis of the phase variation repertoire of lipopolysaccharide immunotypes in Neisseria meningitidis. Microbiology 145:3013-3021. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, C. R., J. Newcombe, H. A. Borde, A. R. Gorringe, S. G. P. Funnell, and J. McFadden. 2001. Generation and characterisation of a phop homologue mutant of Neisseria meningitidis. Mol. Microbiol. 39:1345-1355. [DOI] [PubMed] [Google Scholar]
- 28.Kahler, C. M., and D. S. Stephens. 1998. Genetic basis for biosynthesis, structure, and function of meningococcal lipooligosaccharide (endotoxin). Crit. Rev. Microbiol. 24:281-334. [DOI] [PubMed] [Google Scholar]
- 29.Kojetin, D. J., R. J. Thompson, and J. Cavanagh. 2003. Sub-classification of response regulators using the surface characteristics of their receiver domains. FEBS Lett. 554:231-236. [DOI] [PubMed] [Google Scholar]
- 30.Kox, L. F., M. M. Wosten, and E. A. Groisman. 2000. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 19:1861-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurz, S., C. Hubner, C. Aepinus, S. Theiss, M. Guckenberger, U. Panzner, J. Weber, M. Frosch, and G. Dietrich. 2003. Transcriptome-based antigen identification for Neisseria meningitidis. Vaccine 21:768-775. [DOI] [PubMed] [Google Scholar]
- 32.Martin, D., B. R. Brodeur, J. Hamel, F. Couture, A. De, Z. Lian, S. Martin, D. Andrews, and R. W. Ellis. 2000. Candidate Neisseria meningitidis NspA vaccine. J. Biotechnol. 83:27-31. [DOI] [PubMed] [Google Scholar]
- 33.Mekalanos, J. J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 174:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A 2-component regulatory system (Phop-Phoq) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, S. I., W. P. Loomis, C. Alpuche Aranda, I. Behlau, and E. Hohmann. 1993. The PhoP virulence regulon and live oral Salmonella vaccines. Vaccine 11:122-125. [DOI] [PubMed] [Google Scholar]
- 36.Morley, S. L., and A. J. Pollard. 2001. Vaccine prevention of meningococcal disease, coming soon? Vaccine 20:666-687. [DOI] [PubMed] [Google Scholar]
- 37.Moss, J. E., P. E. Fisher, B. Vick, E. A. Groisman, and A. Zychlinsky. 2000. The regulatory protein PhoP controls susceptibility to the host inflammatory response in Shigella flexneri. Cell. Microbiol. 2:443-452. [DOI] [PubMed] [Google Scholar]
- 38.Mouslim, C., and E. A. Groisman. 2003. Control of the Salmonella ugd gene by three two-component regulatory systems. Mol. Microbiol. 47:335-344. [DOI] [PubMed] [Google Scholar]
- 39.Newcombe, J., L. Eales-Reynolds, L. Wootton, A. R. Gorringe, S. P. G. Funnell, S. G. Taylor, and J. J. McFadden. 2004. Infection with an avirulent phoP mutant of Neisseria meningitidis confers broad cross-reactive immunity. Infect. Immun. 72:338-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oshima, T., H. Aiba, Y. Masuda, S. Kanaya, M. Sugiura, B. L. Wanner, H. Mori, and T. Mizuno. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 46:281-291. [DOI] [PubMed] [Google Scholar]
- 41.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 42.Pettersson, A., A. Maas, D. van Wassenaar, P. van der Ley, and J. Tommassen. 1995. Molecular characterization of FrpB, the 70-kilodalton iron-regulated outer membrane protein of Neisseria meningitidis. Infect. Immun. 63:4181-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raetz, C. R. 2001. Regulated covalent modifications of lipid A. J. Endotoxin Res. 7:73-78. [PubMed] [Google Scholar]
- 44.Rahman, M. M., C. M. Kahler, D. S. Stephens, and R. W. Carlson. 2001. The structure of the lipooligosaccharide (LOS) from the alpha-1,2-N-acetyl glucosamine transferase (rfaK(NMB)) mutant strain CMK1 of Neisseria meningitidis: implications for LOS inner core assembly and LOS-based vaccines. Glycobiology 11:703-709. [DOI] [PubMed] [Google Scholar]
- 45.Soncini, F. C., V. E. Garcia, F. Solomon, and E. A. Groisman. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178:5092-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stojiljkovic, I., V. Hwa, L. de Saint Martin, P. O'Gaora, X. Nassif, F. Heffron, and M. So. 1995. The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol. Microbiol. 15:531-541. [DOI] [PubMed] [Google Scholar]
- 47.Sun, Y. H., S. Bakshi, R. Chalmers, and C. M. Tang. 2000. Functional genomics of Neisseria meningitidis pathogenesis. Nat. Med. 6:1269-1273. [DOI] [PubMed] [Google Scholar]
- 48.Taouji, S., I. Nomura, S. Giguere, S. Tomomitsu, T. Kakuda, V. Ganne, and S. Takai. 2004. Immunogenicity of synthetic peptides representing linear B-cell epitopes of VapA of Rhodococcus equi. Vaccine 22:1114-1123. [DOI] [PubMed] [Google Scholar]
- 49.Tedin, K., and U. Blasi. 1996. The RNA chain elongation rate of the lambda late mRNA is unaffected by high levels of ppGpp in the absence of amino acid starvation. J. Biol. Chem. 271:17675-17686. [DOI] [PubMed] [Google Scholar]
- 50.Teng, F., L. Wang, K. V. Singh, B. E. Murray, and G. M. Weinstock. 2002. Involvement of PhoP-PhoS homologs in Enterococcus faecalis virulence. Infect. Immun. 70:1991-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. JeffriesK. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1815. [DOI] [PubMed] [Google Scholar]
- 52.Tzeng, Y. L., A. Datta, K. Ambrose, M. Lo, J. K. Davies, R. W. Carlson, D. S. Stephens, and C. M. Kahler. 2004. The MisR/MisS two-component regulatory system influences inner core structure and immunotype of lipooligosaccharide in Neisseria meningitidis. J. Biol. Chem. 279:35053-35062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.