Abstract
Streptococcus mutans UA159, the genome sequence reference strain, exhibits nonlantibiotic bacteriocin (mutacin) activity. In this study, we have combined bioinformatic and mutational analyses to identify the ABC transporter designated NlmTE, which is required for mutacin biogenesis in strain UA159 as well as in another mutacin producer, S. mutans N.
The oral bacterium Streptococcus mutans produces small (<10-kDa), ribosomally synthesized antimicrobial peptides (bacteriocins) termed mutacins, which are generally divided into two categories: (i) the posttranslationally modified, lanthionine-containing (lantibiotic) mutacins (6, 11, 18, 20, 21) and (ii) the unmodified mutacins (3, 4, 9, 22). A general feature of the secretion leader sequences of many peptide bacteriocins produced by gram-positive bacteria (8, 10, 15), as well as some produced by gram-negative species (17), is a highly conserved double-glycine (GG) motif. This GG motif immediately precedes the site where specific proteolytic cleavage of the signal peptide occurs during export of the bacteriocin by ATP-binding cassette (ABC) transport systems (8, 10). The genes encoding the regulatory, biosynthetic, export, and immunity elements of the lantibiotic mutacins are entirely contained within multigene modules (6, 11, 18, 20, 21), similar to those of other lantibiotic systems (15, 16, 25). In contrast, the currently defined genetic loci for nonlantibiotic mutacins do not appear to contain any dedicated export- or immunity-associated genes juxtaposed with the mutacin structural genes (9, 22).
S. mutans UA159, the genome sequence reference strain, does not possess any genetic loci that encode lantibiotic mutacins (1). However, we have previously reported that strain UA159 produces the nonlantibiotic mutacin IV (22), as well as an additional, as yet unidentified, inhibitory agent(s) (J. D. F. Hale et al., submitted for publication). We report here the use of bioinformatic and mutational analyses to identify the locus encoding the ABC transport system responsible for mutacin processing and export in S. mutans strains UA159 and N (producer of the nonlantibiotic mutacin N) (3, 9).
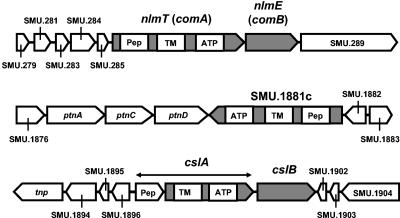
Typical nonlantibiotic bacteriocin export systems consist of a ca. 700-amino-acid ABC transporter and a ca. 400-amino-acid accessory protein (8, 10). The ABC transporter component is characteristically composed of three domains: (i) an N-terminal peptidase domain, (ii) a membrane-spanning permease, and (iii) a C-terminal ATPase domain containing the characteristic Walker motifs (8, 10). When a BLAST (2) search of the S. mutans UA159 genome sequence was conducted by using the amino acid sequences of several known nonlantibiotic bacteriocin ABC exporters (e.g., PlnG from Lactobacillus plantarum [GenBank accession no. CAA64189] and CbnT from Carnobacterium piscicola [GenBank accession no. AAB81307]) as query sequences, the translated products of only two open reading frames were found to possess the characteristics described above (Fig. 1): SMU.286 (comA), encoding the 760-amino-acid ComA ABC transporter that plays a role in biofilm formation (27), and SMU.1881c (designated orf763), which specifies a 763-amino-acid protein with 73% identity (86% similarity) to ComA. While the gene immediately downstream of comA was comB (encoding the ABC transporter accessory protein), no comB counterpart adjacent to orf763 was found (Fig. 1). Although not detected by the BLAST algorithm, a third ABC transporter/accessory protein-encoding locus, cslAB (SMU.1897 to SMU.1900 [Fig. 1]), has previously been reported to be essential for natural transformation in S. mutans (19).
FIG. 1.
Genomic organization of the open reading frames (symbolized by the filled pentagons) encoding the three putative ABC transport systems investigated in this study. The N-terminal peptidase (Pep), transmembrane permease (TM), and C-terminal ATP-binding (ATP) domains of the ABC transporter component are also shown. Note that the CslA ABC transporter appears to be the product of two open reading frames (SMU.1897 and SMU.1898). Genes encoding hypothetical proteins are identified by their GenBank locus tags, e.g., SMU.279. The translational orientation of each open reading frame is also indicated. nlmTE, ABC transporter required for export of nonlantibiotic mutacins (see the text); ptnACD, components of the mannose-specific phosphotransferase system; tnp, putative transposase of ISSmu1 (1); cslAB, ABC transport system required for natural transformation (19).
In the present study, the comA, comB, orf763, cslA, and cslB genes of S. mutans strain UA159 were individually inactivated by allelic replacement with the erythromycin resistance determinant ermAM (5) by using the PCR ligation mutagenesis strategy described by Lau et al. (13), ultimately generating UAΔComA, UAΔComB, UAΔORF763, UAΔCslA, and UAΔCslB, respectively. The source of ermAM was pSLER1 (pSL1190 [Pharmacia] containing ermAM cloned into the NdeI site). ermAM was inserted in the same transcriptional orientation as the gene of interest, and the absence of a transcription terminator downstream of ermAM was expected to preclude any polar effects. All PCR primers (Invitrogen) used in this study are listed in Table 1. Transformants were selected on brain heart infusion agar (Becton Dickinson) supplemented with 0.5% (wt/vol) yeast extract containing 2.5 μg/ml erythromycin. The presence of the desired specific mutations was confirmed by Southern hybridization (23) and by sequencing of the PCR products generated by using the appropriate primer combinations (Table 1).
TABLE 1.
PCR primers used in this study
| Primer | Nucleotide sequence (5′-3′)a | Location of primer (nucleotides) |
|---|---|---|
| ComAUpF | CAAAAATCATAGCAATAT | 2410-2427 |
| ComAUpR | CCCCCCGAATTCATAAATAACTTGTTTCAT | 3378-3395 |
| ComADwF | AAAACTGCAGATTATAACCTGTTTAATT | 5641-5658 |
| ComADwR | TTTGCTATTTTTCTTAGA | 6457-6476 |
| ComBUpF | TTTAGATGGTGATATTTCGTTTGA | 4946-4969 |
| ComBUpR | CCCCCCGAATTCTTTTTCTTTTCCTTCTCTTCTCG | 6007-6029 |
| ComBDwF | AAAACTGCAGAAAGGGGCAAACCGCTCGTCT | 6315-6335 |
| ComBDwR | TTTTGATTTGGTTGCCTGAAGC | 7361-7382 |
| ORF763UpF | GGTGAATTCACTCGTGTCTTGGGGTATTGGTGG | 7551-7572 |
| ORF763UpR | ACCTCTAGACATCTATGTAGTCAGGCTAGTGCTC | 6965-6991 |
| ORF763DwF | CCGTCGCTGCCATAATTTTCTGCAGTGTTAGTT | 4883-4516 |
| ORF763DwR | GGCTTCTCGAGCAATATCAGGAAACATCCTAGG | 3681-3702 |
| CslAUpF | CATAATCTAGAGACACCCTTATTTGTGAACGACC | 4222-4255 |
| CslAUpR | GGATGAATTCACTACCGTTTACGATATTGCTG | 5277-5307 |
| CslADwF | CCAAACTGCAGTCTGCCAGAGTGGCTAATAA | 6816-6846 |
| CslADwR | CAGTCTTGTCTGCCATGTGCAGCAGGTTAT | 7461-7490 |
| CslBUpRb | ATGATAGCGTCTTCGGTAGAATTCAGCG | 7652-7679 |
| CslBDwF | GCATCAGTGTCGTCTTACCCTGCCTCAG | 8738-8765 |
| CslBDwR | ACATCTGCTCTTAAAGATACTCCTATTGCC | 9561-9590 |
| ErmInv1 | CCAGTTCGCGTTAAATGCCCTTTACCTG | 374-403 |
| ErmInv2 | CTTACCCGCCATACCACAGATGTTCCAGAT | 784-813 |
The ComAx primers were adapted from those described by Yoshida and Kuramitsu (27). The ComBx, ORF763x, CslAx/Bx, and ErmInv primers were designed based on sequences with GenBank accession numbers AE014877, AE015014, AE015015, and Y00116, respectively. Restriction sites for EcoRI (GAATTC), PstI (CTGCAG), and XbaI (TCTAGA) incorporated into the primer are underlined.
The partner for this primer during PCR was CslADwF.
The various mutants chosen for further study were then tested for mutacin production by use of a standard deferred antagonism protocol (3) against a panel of 85 indicator bacteria (Table 2), 64 of which have previously been shown to be sensitive to mutacin IV, while the remaining 21 (comprising mainly nonstreptococcal strains, e.g., Lactococcus lactis and Micrococcus luteus) are inhibited by an additional, as yet unidentified inhibitory agent(s) produced by strain UA159 (Hale et al., submitted). Inactivation of either comA or comB resulted in complete abrogation of bacteriocin elaboration by S. mutans UA159, whereas deletion of orf763, cslA, or cslB had no discernible effect (Table 2), indicating that the ComAB ABC transporter alone was essential for the export of both mutacin IV and the additional inhibitory agent(s). Such a result was not unexpected, as a survey of mutacin-like prepeptides potentially encoded by the S. mutans UA159 genome reveals significant similarity between their secretion signal peptides and that of the NlmA peptide (22) of mutacin IV (Fig. 2).
TABLE 2.
Inhibitory spectra of wild-type S. mutans strains UA159 and N and their mutants
| Indicator bacteriuma | No. of indicator strains inhibited by:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| UA159 | UAΔNlmT (UAΔComA) | UAΔNlmE (UAΔComB) | UAΔORF763 | UAΔCslA | UAΔCslB | N | NΔNlmT | |
| Lactococcus lactis (7) | 7 | 0 | 0 | 7 | 7 | 7 | 7 | 0 |
| Micrococcus luteus (1) | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 |
| Streptococcus constellatus (10) | 10 | 0 | 0 | 10 | 10 | 10 | 10 | 0 |
| Streptococcus gordonii (7) | 7 | 0 | 0 | 7 | 7 | 7 | 7 | 0 |
| “Streptococcus milleri” (3) | 3 | 0 | 0 | 3 | 3 | 3 | 3 | 0 |
| Streptococcus mitis (12) | 12 | 0 | 0 | 12 | 12 | 12 | 12 | 0 |
| Streptococcus oralis (10) | 10 | 0 | 0 | 10 | 10 | 10 | 10 | 0 |
| Streptococcus pyogenes (10) | 10 | 0 | 0 | 10 | 10 | 10 | 10 | 0 |
| Streptococcus salivarius (10) | 10 | 0 | 0 | 10 | 10 | 10 | 10 | 0 |
| Streptococcus sanguinis (5) | 5 | 0 | 0 | 5 | 5 | 5 | 5 | 0 |
| Streptococcus uberis (10) | 10 | 0 | 0 | 10 | 10 | 10 | 10 | 0 |
| Streptococcus mutans (5) | 0 | NTb | NT | NT | NT | NT | 5 | 0 |
The numbers of strains tested are shown in parentheses.
NT, not tested.
FIG. 2.
Alignment of NlmA and NlmB (the prepeptides of the two-component nonlantibiotic mutacin IV [22]) with other putative prepeptides (in strain UA159) containing the double-glycine motif. BLAST similarities were obtained only when NlmA was used as the query sequence. The site where peptidase cleavage occurs during export is indicated by the inverted arrow. For simplicity, only the first 10 amino acids of each putative mature peptide are shown. ComC, the prepeptide which is processed to yield the competence-stimulating peptide, is included for additional comparison. The consensus sequence of the signal peptide displays only those amino acids present in at least five of the peptides listed.
The development of natural competence for genetic transformation in S. mutans is believed to be analogous to that in Streptococcus pneumoniae (7, 12). In S. pneumoniae, competence development occurs in two stages. The first (early) stage consists of a quorum-sensing signal transduction circuit, initiated by the binding of competence-stimulating peptide (CSP) to its cognate cell surface receptor ComD (a histidine kinase), which is then perpetuated by ComE, the global transcriptional regulator of competence (12). The second (late) stage involves the synthesis and assembly of the DNA uptake and processing machinery (12). The precursor of CSP, ComC, contains the GG motif in its signal peptide, and its export involves an ABC transport system which is designated ComAB in both S. pneumoniae and Streptococcus gordonii (12, 14). In the case of S. mutans, however, there appears to be some confusion in the nomenclature in that SMU.286/SMU.287 has been designated comAB (27) although cslAB has been previously shown to be essential for natural transformation (19). It is noteworthy that the role of comAB in natural competence in S. mutans has not been confirmed (27). Furthermore, the signal peptide of ComC differs from those of mutacin IV and other mutacin-like peptides (Fig. 2). In order to resolve this apparent discrepancy, we assessed the transformabilities of strains UA159, UAΔComA, UAΔComB, UAΔCslA, and UAΔCslB essentially as described by Petersen and Scheie (19) with minor modifications. Briefly, donor DNA (1 μg/ml) was added to cultures at an optical density at 600 nm of 0.15 to 0.20, and incubation was continued until the culture attained an optical density at 600 nm of 0.85 to 0.90, at which point 100-μl aliquots of appropriate dilutions were plated onto Todd-Hewitt agar containing 600 μg/ml kanamycin. The donor DNA was plasmid pFX-ErmKan, which consists of the replicative backbone of pFX3 (26) and the genetic determinants for erythromycin (ermAM [5]) and kanamycin (aphA3 [24]) resistance.
As shown in Table 3, the transformation frequencies attained by the comA and comB mutants were comparable to that of the wild type. In contrast, dramatic reductions (>99%) in the transformabilities of both UAΔCslA and UAΔCslB were observed (Table 3), which corroborates previous findings obtained with csl mutants (19), suggesting that the csl locus is essential for competence in S. mutans. The addition of 500 ng/ml synthetic CSP (NH2-SGSLSTFFRLFNRSFTQALGK-COOH; microcollections GmbH, Germany) boosted the transformation frequency of strain UAΔCslA by >30-fold (data not shown), further supporting a role for CslAB as the transport system responsible for CSP secretion (19). Due to the role of SMU.286 (comA) and SMU.287 (comB) in mutacin production but not in genetic transformation, we propose that SMU.286 and SMU.287 should be redesignated nlmT (nonlantibiotic mutacin transporter) and nlmE (nlmT accessory protein), respectively.
TABLE 3.
Transformation properties of S. mutans UA159 and its ABC transporter mutants
| Strain | Transformation frequency (104 kanamycin-resistant CFU/ml)a |
|---|---|
| Wild-type UA159 | 1.5 (1.4-1.7) |
| UAΔNlmT (UAΔComA) | 1.0 (0.95-1.1) |
| UAΔNlmE (UAΔComB) | 1.4 (1.3-1.5) |
| UAΔCslA | 0.015 (0.011-0.2) |
| UAΔCslB | 0.014 (0.01-0.02) |
The average values (ranges) of three independent experiments are shown.
In light of the findings described above, we decided to extend our study to inactivate (by allelic replacement with ermAM) the nlmT homologue in S. mutans N, a strain that produces the nonlantibiotic mutacin N (3, 4) and does not appear to contain any lantibiotic-associated genes (J. D. F. Hale, unpublished data). The inhibitory spectrum of mutacin N is distinctive compared to that of other mutacins in its ability to inhibit certain S. mutans strains (3, 4). The resulting nlmT-deficient mutant, NΔNlmT, failed to express any inhibitory activity (Table 2), indicating that the NlmTE ABC transporter is also required for nonlantibiotic mutacin export in S. mutans strain N.
In conclusion, we have determined that the nlmTE locus (SMU.286/SMU.287, previously designated comAB) encodes the ABC transporter required for nonlantibiotic mutacin biogenesis in S. mutans strains UA159 and N. Furthermore, we have resolved an apparent discrepancy in the nomenclature and function of two ABC-transporter-encoding loci in natural competence. While cslAB (19) is clearly involved in genetic transformation (possibly as the export mechanism for CSP), our results do not support a similar role for comAB.
Acknowledgments
We are grateful to Ann Holmes (Department of Oral Sciences, University of Otago School of Dentistry) and Mogens Kilian (University of Aarhus, Denmark) for the provision of bacterial strains, Dennis Cvitkovitch (University of Toronto, Canada) for the gift of synthetic CSP, and Annalee O'Rourke (Department of Microbiology and Immunology) for supplying plasmid pFX-ErmKan.
This study was financially supported by the Health Research Council of New Zealand and the Otago Medical Research Foundation. J.D.F.H. was a recipient of a University of Otago Postgraduate Scholarship.
REFERENCES
- 1.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. K. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Balakrishnan, M., R. S. Simmonds, A. Carne, and J. R. Tagg. 2000. Streptococcus mutans strain N produces a novel low molecular mass non-lantibiotic bacteriocin. FEMS Microbiol. Lett. 183:165-169. [DOI] [PubMed] [Google Scholar]
- 4.Balakrishnan, M., R. S. Simmonds, M. Kilian, and J. R. Tagg. 2002. Different bacteriocin activities of Streptococcus mutans reflect distinct phylogenetic lineages. J. Med. Microbiol. 51:941-948. [DOI] [PubMed] [Google Scholar]
- 5.Brehm, J., G. Salmond, and N. Minton. 1987. Sequence of the adenine methylase gene of the Streptococcus faecalis plasmid pAMβ1. Nucleic Acids Res. 15:3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, P., F. Qi, J. Novak, and P. W. Caufield. 1999. The specific genes for lantibiotic mutacin II biosynthesis in Streptococcus mutans T8 are clustered and can be transferred en bloc. Appl. Environ. Microbiol. 65:1356-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cvitkovitch, D. G., Y. H. Li, and R. P. Ellen. 2003. Quorum sensing and biofilm formation in streptococcal infections. J. Clin. Investig. 112:1626-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eijsink, V. G. H., L. Axelsson, D. B. Diep, L. S. Håvarstein, H. Holo, and I. F. Nes. 2002. Production of class II bacteriocins by lactic acid bacteria; an example of biological warfare and communication. Antonie Leeuwenhoek 81:639-654. [DOI] [PubMed] [Google Scholar]
- 9.Hale, J. D. F., M. Balakrishnan, and J. R. Tagg. 2004. Genetic basis for mutacin N and of its relationship to mutacin I. Indian J. Med. Res. 119:247-251. [PubMed] [Google Scholar]
- 10.Håvarstein, L. S., D. B. Diep, and I. F. Nes. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16:229-240. [DOI] [PubMed] [Google Scholar]
- 11.Hillman, J. D., J. Novak, E. Sagura, J. A. Gutierrez, T. A. Brooks, P. J. Crowley, M. Hess, A. Azizi, K. Leung, D. Cvitkovitch, and A. S. Bleiweis. 1998. Genetic and biochemical analysis of mutacin 1140, a lantibiotic from Streptococcus mutans. Infect. Immun. 44:141-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lacks, S. A. 2004. Transformation, p. 89-115. In E. I. Tuomanen, T. J. Mitchell, D. A. Morrison, and B. G. Spratt (ed.), The pneumococcus. ASM Press, Washington, D.C.
- 13.Lau, P. C., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193-205. [DOI] [PubMed] [Google Scholar]
- 14.Lunsford, R. D., and J. London. 1996. Natural genetic transformation in Streptococcus gordonii: comX imparts spontaneous competence on strain Wicky. J. Bacteriol. 178:5831-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McAuliffe, O., R. P. Ross, and C. Hill. 2001. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 25:285-308. [DOI] [PubMed] [Google Scholar]
- 16.McLaughlin, R. E., J. J. Ferretti, and W. L. Hynes. 1999. Nucleotide sequence of the streptococcin A-FF22 lantibiotic regulon: model for production of the lantibiotic SA-FF22 by strains of Streptococcus pyogenes. FEMS Microbiol. Lett. 175:171-177. [DOI] [PubMed] [Google Scholar]
- 17.Michiels, J., G. Dirix, J. Vanderleyden, and C. Xi. 2001. Processing and export of peptide pheromones and bacteriocins in gram-negative bacteria. Trends Microbiol. 9:164-168. [DOI] [PubMed] [Google Scholar]
- 18.Mota-Meira, M., C. Lacroix, G. LaPointe, and M. C. Lavoie. 1997. Purification and structure of mutacin B-Ny266: a new lantibiotic produced by Streptococcus mutans. FEBS Lett. 410:275-279. [DOI] [PubMed] [Google Scholar]
- 19.Petersen, F. C., and A. A. Scheie. 2000. Genetic transformation in Streptococcus mutans requires a peptide secretion-like apparatus. Oral Microbiol. Immunol. 15:329-334. [DOI] [PubMed] [Google Scholar]
- 20.Qi, F., P. Chen, and P. W. Caufield. 1999. Purification of mutacin III from group III Streptococcus mutans UA787 and genetic analyses of mutacin III biosynthesis genes. Appl. Environ. Microbiol. 65:3880-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi, F., P. Chen, and P. W. Caufield. 2000. Purification and biochemical characterization of mutacin I from the group I strain of Streptococcus mutans, CH43, and genetic analysis of mutacin I biosynthesis genes. Appl. Environ. Microbiol. 66:3221-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi, F., P. Chen, and P. W. Caufield. 2001. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl. Environ. Microbiol. 67:15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Trieu-Cuot, P., and P. Courvalin. 1983. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5"-aminoglycoside phosphotransferase type III. Gene 23:331-341. [DOI] [PubMed] [Google Scholar]
- 25.Upton, M., J. R. Tagg, P. Wescombe, and H. F. Jenkinson. 2001. Intra- and interspecies signaling between Streptococcus salivarius and Streptococcus pyogenes mediated by SalA and SalA1 lantibiotic peptides. J. Bacteriol. 183:3931-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu, F., L. E. Pearce, and P.-L. Yu. 1991. Construction of a family of lactococcal vectors for gene cloning and translational fusions. FEMS Microbiol. Lett. 77:55-60. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida, A., and H. K. Kuramitsu. 2002. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 68:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]