Abstract
DCUN1D5 is up-regulated and promotes tumor progression in many cancers such as laryngeal squamous cell carcinoma and breast cancer, but the expression of DCUN1D5 in lung adenocarcinoma and its molecular mechanism are not clear. The differences of DCUN1D5 expression between lung adenocarcinoma and normal tissues were compared by TCGA, GEO and UALCAN databases, and the relationship between DCUN1D5 expression and clinicopathological features of patients was analyzed. The diagnostic and prognostic value of DCUN1D5 in patients with LUAD was analyzed by TCGA, GEPIA and Kaplan–Meier Plotter database. nomogram was constructed to predict the survival probability of patients. The GO, KEGG and GSEA enrichment analysis of DCUN1D5 co-expression genes were completed by R software. R software and GEPIA2 database were used to analyze the relationship between DCUN1D5 expression level and glycolysis-related genes and immune cell infiltration in patients with LUAD. The effects of interfering DCUN1D5 on the biological function and glycolysis level of lung adenocarcinoma cells were evaluated in vitro. The effect of down-regulation of DCUN1D5 on tumor formation in nude mice was studied in animal experiments. The expression of DCUN1D5 was increased in many kinds of tumors, and the expression in lung adenocarcinoma was significantly higher than that in normal tissues. The expression of DCUN1D5 was significantly correlated with TNM and pathological stage. DCUNN1D5 can play a diagnostic role in patients with LUAD and the prognosis of patients with high expression of DCUN1D5 is poor. Functional enrichment of DCUN1D5 co-expression genes involves a variety of biological processes. There is a strong correlation between DCUN1D5 and most glycolysis related genes. In addition, DCUN1D5 also affects tumor immune cell infiltration. In vitro experiments showed that the ability of cell proliferation, migration, invasion and glycolysis were significantly decreased and the ability of apoptosis was enhanced after down-regulation of DCUN1D5. Animal experiments showed that the tumor weight of nude mice decreased significantly after down-regulation of DCUN1D5. DCUN1D5 can be used as a biomarker for diagnosis and prognosis in lung adenocarcinoma. Down-regulation of DCUN1D5 can significantly affect the biological behavior of lung adenocarcinoma cells, which may be related to glycolysis and immune cell infiltration.
Keywords: DCUN1D5, Lung adenocarcinoma, Immune infiltration, Glycolysis
Subject terms: Oncology, Cancer, Lung cancer, Oncogenesis, Diagnostic markers, Prognostic markers
Introduction
Lung cancer is the leading cause of cancer death in men and women aged 50 and older, with far more deaths than breast, prostate and colorectal cancer combined1. Lung cancer consists of different subtypes, including non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC), of which non-small cell lung cancer (NSCLC) accounts for 80–85%2,3. Lung adenocarcinoma is the most common subtype of NSCLC (about 40%), and its occurrence is usually related to gene mutation4. Studies have shown that the clinical prognosis of NSCLC is directly related to the stage of diagnosis5, so early cancer diagnosis is of great significance and urgent need to improve the overall survival rate of patients6. At present, the diagnosis of lung cancer includes different types of imaging examination and pathological evaluation of biopsy7, but the role of these techniques in the early diagnosis of lung cancer is very limited. Although multimodal therapy strategies including targeted therapy, immunotherapy, radiotherapy and non-invasive surgical resection have made great progress in recent decades, the five-year overall survival rate (OS) of lung adenocarcinoma is still about 18%8. Therefore, further understanding of the molecular characteristics of lung adenocarcinoma, looking for new molecular markers and studying new therapeutic targets are of great significance to the diagnosis, treatment and improvement of prognosis of LUAD.
DCUN1D5 (DCN1,defective in cullin neddylation 1, domain containing 5) is located on the chromosome 11q22.3. Can play a role in enabling cullin family protein activity, involvement in cellular responses to DNA damage stimuli, positive regulation of protein neddylation, and regulation of cell growth. Studies have shown that DCUN1D5 is highly expressed in laryngeal squamous cell carcinoma, lung squamous cell carcinoma, breast cancer and other tumors, and as an oncogene to promote the occurrence and development of tumors9–11. However, the function and mechanism of DCUN1D5 in lung adenocarcinoma are not clear.
Tumor microenvironment (TME) includes many types of immune cells, cancer-related fibroblasts, endothelial cells, pericytes and other tissue resident cell types12. Immune cells are an important part of tumor microenvironment. The growth, invasion and metastasis of lung cancer is a complex and dynamic process, which involves not only the genetic abnormalities inherent in the tumor tissue, but also the interaction between the tumor tissue and immune cells in the local microenvironment13. Therefore, the study of immune cell infiltration is of great significance to the diagnosis, treatment and prognosis of cancer.
Glycolysis plays an important role in the occurrence and development of tumors. Studies have shown that glycolysis is the preferred pathway of energy metabolism for cancer cells even when oxygen content is sufficient14. The increased glycolysis of cancer cells and the resulting lactic acidosis can regulate the tumor matrix into a microenvironment that promotes tumorigenesis15. In addition, glycolysis can regulate tumor microenvironment and induce tumor angiogenesis and immune escape16. Therefore, understanding the relationship between immune infiltration and glycolysis can provide a new idea for us to study the molecular mechanism of tumor.
In this study, we explored the expression of DCUN1D5 in LUAD and its relationship with clinicopathological features of patients. The role of DCUN1D5 in diagnosis and prognosis was verified. The possible biological functions and signal pathways of DCUN1D5-related genes were analyzed by bioinformatics. The potential molecular mechanisms of DCUN1D5 involved in immune infiltration and glycolysis were discussed. Finally, DCUN1D5 was down-regulated to observe the changes of biological functions such as proliferation, migration, invasion, apoptosis and glycolysis of LUAD cells. The effect of knockout DCUN1D5 on tumor weight in nude mice was observed in vivo. This study provides a new direction for the diagnosis and treatment of LUAD.
Materials and methods
Data source
The mRNA expression profile data were derived from TCGA database (https://portal.gdc.cancer.gov/) and GSE140797, the dataset of GEO database (https://www.ncbi.nlm.nih.gov/geo/). The clinical data of patients were downloaded from TCGA database to study the relationship between the expression of DCUN1D5 and pathological parameters.
Expression analysis of DCUN1D5
The expression of DCUN1D5mRNA in TCGA and GEO was compared. The expression of DCUN1D5 protein in lung adenocarcinoma and normal tissues was analyzed by bioinformatics database UALCAN17. And the diagnostic value of DCUN1D5 in lung adenocarcinoma was evaluated by drawing ROC curve. Finally, the mRNA and protein expression of DCUN1D5 were detected by qRT-PCR and Western blotting.
Pathological or clinical stage and prognosis
The relationship between DCUN1D5 expression and clinicopathological features of patients with lung adenocarcinoma was analyzed by TCGA database. The correlation between DCUN1D5 and OS in patients with lung adenocarcinoma was analyzed by GEPIA2 (http://gepia2.cancer-pku.cn/#index), Kaplan–Meier Plotter (https://kmplot.com/analysis/) and TCGA database. Furthermore, the effect of DCUN1D5 on OS in patients with different clinical characteristics was studied by TCGA database.
Establishment and evaluation of the nomogram models
All independent clinicopathologic prognostic factors were selected from Cox regression analysis. A nomogram model was constructed using the survival package and rms package of R software. It was used to assess the 1-, 3-, and 5-year OS probabilities for LUAD patients. The accuracy of the nomogram model was validated by comparing the predicted probabilities of the calibrated fold plots with the observed actual probabilities.
Differential expression analysis and gene set enrichment analysis
Differential analysis of DCUN1D5 was performed using the DESeq2 package and edgeR package of the R software. DEGs was screened based on P.adj < 0.05, |log2FC| ≥ 1 and encoded protein gene. Through the data of TCGA, the correlation between DCUN1D5 and other mRNA in LUAD is analyzed. The statistical correlation is verified by Pearson correlation coefficient, and the volcano map and heat map are generated by ggplot2 package of R software for visualization. GO and KEGG enrichment analysis of significantly co-expressed genes was performed using the clusterProfiler package of R software18–20. GSEA enrichment analysis of all DEGs using clusterProfiler package (GSEA, http://www.gsea-msigdb.org/gsea/index.jsp)21. The reference gene set is c2.cp.all.v2022.1.Hs.symbols.gmt.
The relationship between DCUN1D5 expression and glycolysis in LUAD
The relationship between DCUNI1D5 and glycolysis related genes22 was analyzed by R software, and the correlation lollipop map was generated. The correlation between the expression of DCUN1D5 and glycolysis characteristic gene set was analyzed by GEPIA2 database. The scatter plot and survival curve of significant related genes were drawn.
Analysis of immune cell infiltration
The single sample gene set enrichment analysis (ssGSEA) algorithm in R packet GSVA23 was used to calculate the microenvironment level of immune system based on DCUN1D5mRNATPM data, and the relationship between DCUN1D5 expression and these 24 kinds of immune cells was analyzed by Pearson correlation. The relationship between the expression of DCUN1D5 and CD8+ T cells, myeloid dendritic cells (DC), B cells and M2 macrophages was analyzed by TIMER2.0 database (http://timer.cistrome.org/). The LUAD data in TCGA database were divided into DCUN1D5 high expression group and low expression group. R packet GSVA was used to evaluate the difference of immune cell infiltration between high expression group and low expression group.
Cell culture and treatment
HBE and H1299 were acquired from the National Collection of Authenticated Cell Cultures (Shanghai, China). Placed in 37 °C, 5% CO2 incubator, incubated with RPMI-1640 medium with 10% fetal bovine serum. siRNA was designed and constructed by ZHONGSHI TONTRU (Tianjin, China), which was divided into experimental group (si-DCUN1D5) and negative control group (si-NC). The cells in logarithmic growth phase were laid in a six-well plate, and when the degree of cell convergence reached 60–80%, transfection was carried out according to Lipofectamine3000 (Invitrogen, USA) instructions. After 24–48 h of transfection, the silencing efficiency was examined by qRT-PCR and subsequent experiments were performed.
RNA extraction and qRT-PCR analysis
Total RNA of the cells and tissues was extracted by the TRIzol method, reverse transcribed to cDNA using a reverse transcription kit (TIANGEN, China), and then subjected to qRT-PCR reaction according to the instructions of the amplification kit (TIANGEN, China). DCUN1D5 forward primer sequence 5′-GCTCTTCTGCTTGGGAGGACAT-3′, reverse primer sequence 5′-AAGATCAGCATGGACTGTTCTGC-3′. GAPDH forward primer sequence 5′-CGACCACTTTGACAAGCTCA-3′, reverse primer sequence 5′-AGGGGTCTACATGGCAACTG-3′. The relative expression of the gene was expressed by2-ΔΔct.
Western blotting
Proteins were extracted from tissues and protein concentration was determined by BCA method. Total proteins were separated by SDS-PAGE and transferred to a PVDF membrane (Merck, Germany). The membrane was closed in 5% skimmed milk powder milk for 1 h. Rabbit anti-DCUN1D5 antibody (1:1000 dilution, Abmart, China) and rabbit anti-GAPDH antibody(1:4000 dilution, Servicebio, China) were added and incubated overnight, respectively. After thorough washing, HRP-conjugated Goat Anti-Rabbit IgG(H + L) (1:10,000 dilution; Servicebio, China) was incubated for 2 h at room temperature. After washing the membrane, the color was developed in ECL reagent, and the results were scanned and recorded using a chemiluminescent imaging system.
Glucose uptake experiment
The cells of each group after transfection were inoculated in a 6-well plate. 24 h later, the cell culture medium was collected and the cells were counted. According to the instructions of the glucose determination kit (Abbkine, China), the difference between the glucose concentration in the cell culture medium and the initial culture medium is the total glucose consumption. The glucose consumption per 104 cells was calculated.
Lactic acid production experiment
Collect 1 × 106 cells of each group after transfection in a centrifuge tube, wash the cells with cold PBS, discard the supernatant after centrifugation, add 200 μl of LactateAssayBuffer to each tube to sonicate and crush for 5 min in an ice bath (power 200 W, sonicate for 3 s, interval 7 s, repeat 30 times), then centrifuge for 5 min at 12,000g, 4 °C, take the supernatant and put it on The supernatant was taken and placed on ice for measurement. Pipette 50 μl of supernatant and 50 μl of WorkingReagent were mixed well and incubated at 37 °C away from light for 30 min, and the absorbance at 450 nm was measured. The absorbance at 450 nm was measured. A standard curve was made according to the requirements of the kit (Abbkine, China) and the lactic acid content was calculated.
CKK-8 experiment
Cells of each group were collected and inoculated into 96-well plates with 100 μl/2000 cells per well. CCK-8 solution (ReportBitech, China) at 10% of the total volume of culture medium was added to each well at 0 h, 24 h, 48 h and 72 h after plate laying, respectively. After adding the working solution, the 96-well plate was re-placed in the incubator and cultured for 2 h, and the absorbance of each well at 450 nm was detected by enzyme labeling instrument.
Transwell experiment
The ability of cell migration was detected. The cell count after transfection was collected, and 5 × 104 cells were inoculated in the upper chamber with 200 μl serum-free medium, and 750 μl medium containing 20% fetal bovine serum was added to the lower chamber. After 24 h of culture, after being fixed with methanol for 30 min and washed twice, 20 min was stained with 0.1% crystal violet, and the upper unmigrated cells were gently wiped off with cotton swabs. After PBS washing for 3 times, the visual field cells were randomly selected and counted. The ability of cell invasion was detected. Spread the diluted matrigel (Corning, USA) evenly in the upper chamber of transwell and wait for it to solidify. After transfection, 7 × 104 cells were inoculated in the upper chamber in 200 μl serum-free medium, and the rest process was the same as the experiment to detect the migration ability.
Apoptosis experiment
The transfected cells were digested with trypsin without EDTA. The cells were collected and washed with PBS twice in a centrifuge tube, and the supernatant was discarded after centrifugation. The cell precipitation was re-suspended in 100 μl binding buffer (TONBO, USA), 5 μl of AnnexinV-FITC and 5 μl of AAD-7 were added, and 400 μl of binding buffer was added after 15 min of incubation at room temperature. The percentage of apoptosis was detected by flow cytometry.
Establishment of xenograft tumor model of LUAD in nude mice
This animal experiment has been approved by the Ethics Committee of the affiliated Hospital of Chengde Medical College. Mouse xenograft experiments in this study complied with the ARRIVE guidelines and were conducted in accordance with the U.K. Animals (Scientific Procedures) Act, 1986 and associated guidelines. Male Balb/c nude mice weighing 20–30 g at the age of 6–8 weeks were purchased from Sibeifu (Beijing) Biotechnology Limited Company. Balb/c nude mice (Beijing, China) were randomly divided into two groups with 4 mice in each group. The transfected cells in logarithmic phase were digested with trypsin and collected in centrifuge tubes, and the supernatant was discarded after centrifugation, and PBS solution was added to resuscitate the cells to adjust the cell concentration to 1 × 107/ml, and each 0.2 ml was inoculated into the armpit of nude mice. Four weeks after inoculation of cell suspension, the nude mice were killed and the weight of the tumor was measured. Pentobarbital sodium and cervical dislocation euthanasia were performed to decrease animal suffering in the course of the experiment. Mice were deeply anesthetized by intraperitoneal injection of pentobarbital sodium (1%, 50 mg/kg). The standard of deep anesthesia was that nude mice did not respond to limb and head stimuli. Euthanasia for cervical dislocation after being photographed.
Statistical analysis
GraphPadPrism statistical analysis was used. T test for paired or unpaired samples were used for data that met normality and Chi-square for both groups, and Wilcoxon rank sum test were used for information that did not meet normal distribution. Multiple groups were compared by single factor analysis of variance (ANOVA) or two-way ANOVA. Kaplan–Meier curves were compared by Cox test or log-rank test. Univariate and multivariate Cox analysis were used to screen potential prognostic factors. The Benjamini–Hochberg method was used for multiple test correction. Differences were considered statistically significant at p < 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001).
Results
Expression and diagnostic value of DCUN1D5 in LUAD
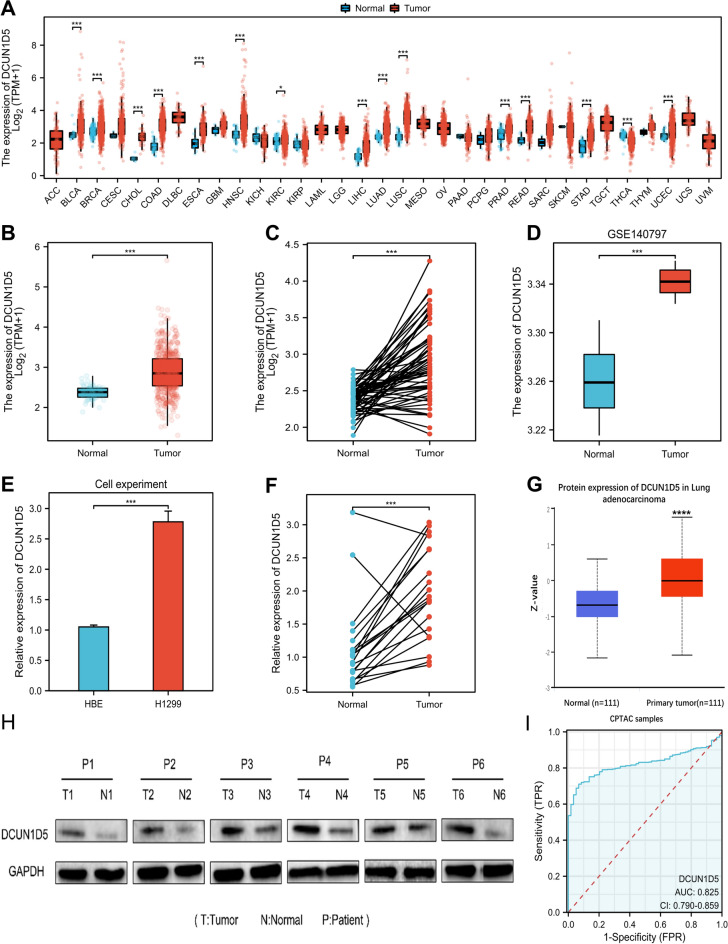
We downloaded data from TCGA to analyze the expression of DCUN1D5 in pan-cancer. The results showed that DCUN1D5 was significantly overexpressed (Fig. 1A) in many kinds of tumors, including LUAD. Further study of the TCGA-LUAD dataset showed that the mRNA expression of DCUN1D5 in lung adenocarcinoma was significantly higher than that in adjacent normal tissues (Fig. 1B,C). To verify our results, the gene expression profile of the GSE140797 data set of lung adenocarcinoma was downloaded from the GEO database. The results showed that the expression of DCUN1D5 in paired cancer tissues was significantly higher than that in paracancerous tissues (Fig. 1D). We used qRT-PCR technique to detect the expression of DCUN1D5 in normal bronchial epithelial cell line HBE and lung adenocarcinoma cell line H1299. The results showed that the high expression of DCUN1D5 in H1299 was higher than that of HBE (Fig. 1E). The qRT-PCR results of lung adenocarcinoma patient tissues showed that the expression of DCUN1D5 protein was significantly elevated in cancer tissues compared to normal tissues (Fig. 1F). In addition, we used the online biological database UALCAN to display the protein level of DCUN1D5, which is expressed higher in cancer tissues, and verified this conclusion using wb experiments (Fig. 1G,H). Finally, we draw a ROC curve to evaluate the diagnostic value of DCUN1D5 in LUAD. The results showed that the value of AUC was 0.825, CI: 0.790–0.895 (Fig. 1I). This shows that DCUN1D5 is of great significance in the diagnosis of LUAD.
Fig. 1.
The expression level of DCUN1D5. (A) Pan-cancer expression of DCUN1D5 mRNA in TCGA database. (B,C) The difference of DCUN1D5 expression in TCGA-LUAD dataset. (D) The difference of DCUN1D5 expression in GSE140797 data set. (E) The expression of DCUN1D5 in HBE and H1299. (F) The expression of DCUN1D5 in cancer tissues and normal tissues. (G) The difference of DCUN1D5 protein expression in cancer and paracancerous tissues in UALCAN database. (H) Differential expression of DCUN1D5 protein in cancer and adjacent tissues of lung adenocarcinoma patients. (I) ROC curve analysis of DCUN1D5 diagnosis in LUAD. *p < 0.05, **p < 0.01, ***p < 0.001.
Correlation between expression of DCUN1D5 and clinicopathological features
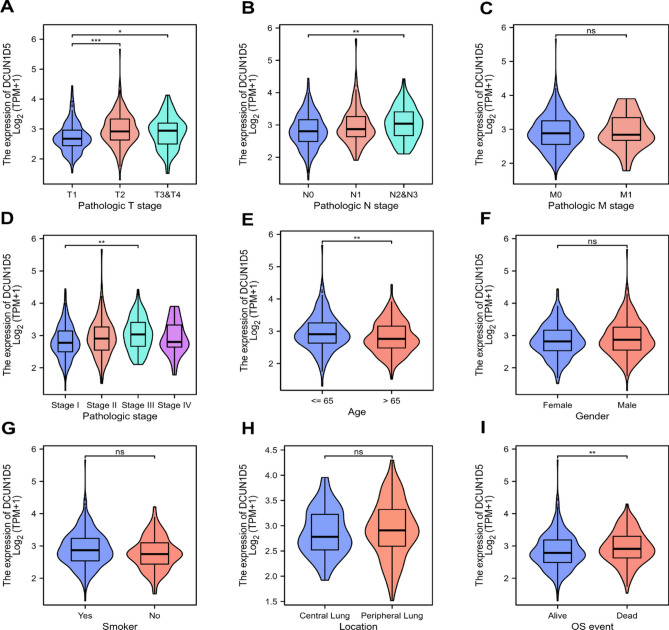
We download the clinical information of LUAD patients through TCGA database to analyze the relationship between different clinicopathological features and DCUN1D5 expression. The results showed that the expression of DCUN1D5 was correlated with T stage, N stage, pathological stage, age and OSevent, but not with M stage, sex, primary site and smoking (Fig. 2). Specifically, compared with T1 stage, the expression of DCUN1D5 in T2 stage and T3&T4 was higher (Fig. 2A). Compared with N0, the expression of N2–N3 increased. Compared with stage I of pathological stage, DCUN1D5 was significantly up-regulated in stage III (Fig. 2D). The expression of DCUN1D5 was increased in patients over 65 years old (Fig. 2E). The expression of DCUN1D5 in survival patients was lower than that in dead patients (Fig. 2I). All these suggest that the high expression of DCUN1D5 plays a certain role in promoting the occurrence and development of tumor.
Fig. 2.
Expression of DCUN1D5 in patients with different clinicopathologic features and the corresponding number of cases. (A) T stage (T1 = 178, T2 = 295, T3&T4 = 68), (B) N stage(N0 = 352, N1 = 99, N2N3 = 77), (C) M stage (M0 = 370,M1 = 25), (D) pathologic stage (I = 298, Stage II = 127, stage III = 85, Stage IV = 26 Stage), (E) age (≤ 65 = 258, > 65 = 267), (F) gender(Female = 291, Male = 253), (G) Smoker (Yes = 453, No = 77), (H) Location (Central lung = 64, peripheral lung = 127), (I) OS event (Alive = 351, Dead = 193). *p < 0.05,**p < 0.01, ***p < 0.001.
High expression of DCUN1D5 is associated with poor prognosis
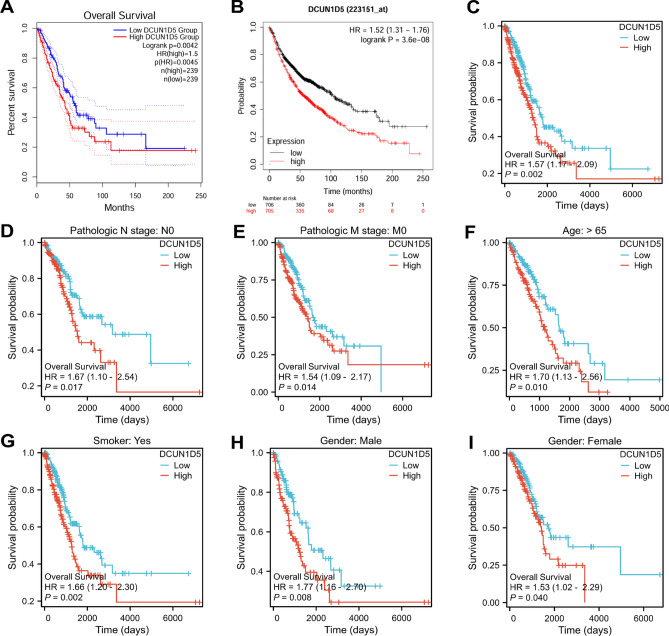
We used GEPIA, Kaplan–Meier Plotter and TCGA databases to analyze the relationship between DCUN1D5 expression and OS. Kaplan–Meier survival analysis based on GEPIA2 database showed that the survival rate (HR = 1.5, p = 0.0042) of patients with high expression of DCUN1D5 was significantly decreased (Fig. 3A). The analysis results of OS (HR = 1.52, p = 3.6e−0.8) of database Kaplan–Meier Plotter and OS (HR = 1.57, p = 0.002) of TCGA are consistent with the results of GEPIA2 (Fig. 3B,C). The results of subgroup survival analysis of different clinicopathological states of LUAD patients were as follows, which showed that the high expression of DCUN1D5 was associated with shorter OS (Fig. 3D–I).
Fig. 3.
The relationship between DCUN1D5 and the prognosis of LUAD. (A) The relationship between the expression of DCUN1D5 in GEPIA2 database and OS in patients with LUAD. (B) The relationship between the expression of DCUN1D5 in Kaplan–Meier Plotter database and OS in LUAD patients. (C) The relationship between the expression of DCUN1D5 in TCGA database and OS in patients with LUAD. (D–I) Prognostic subgroup analysis of LUAD patients with different clinicopathological features. *p < 0.05, **p < 0.01, ***p < 0.001.
Nomogram model of independent prognostic factors for DCUN1D5
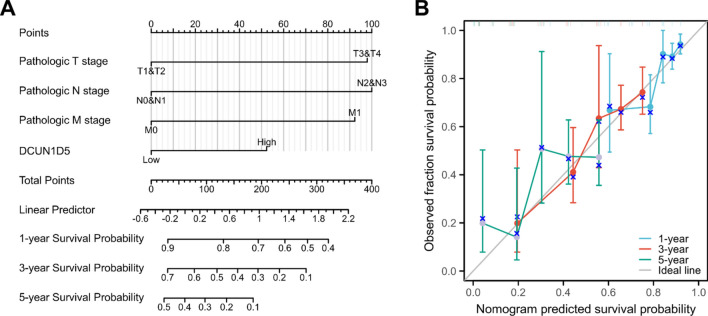
In order to further study the effect of DCUN1D5 expression on the prognosis of LUAD, we carried out univariate and multivariate COX regression analysis of OS. It was determined that T stage, N stage, M stage and the expression of DCUN1D5 were independent prognostic factors (Table 1). Then the independent prognostic factors such as T stage, N stage, M stage and DCUN1D5 were used to construct nomogram, and the correction curve was used to evaluate the prediction accuracy of nomogram model in 1 year, 3 years and 5 years. The results show that DCUN1D5 contributes significantly to the prediction of OS in the nomogram model (Fig. 4A). From the prognostic calibration curve, the predictive model has a more accurate role than a single prognostic factor in predicting OS (Fig. 4B).
Table 1.
Univariate and multivariate Cox regression analyses of clinical characteristics associated with OS of LUAD in TCGA.
| Characteristics | Total (N) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | p value | Hazard ratio (95% CI) | p value | ||
| Age | 520 | ||||
| < = 65 | 257 | Reference | |||
| > 65 | 263 | 1.216 (0.910–1.625) | 0.186 | ||
| Pathologic T stage | 527 | ||||
| T1 and T2 | 461 | Reference | Reference | ||
| T3&T4 | 66 | 2.352 (1.614–3.426) | < 0.001 | 2.067 (1.348–3.168) | < 0.001 |
| Pathologic N stage | 514 | ||||
| N0 and N1 | 441 | Reference | Reference | ||
| N2 and N3 | 73 | 2.360 (1.659–3.358) | < 0.001 | 2.101 (1.398–3.157) | < 0.001 |
| Pathologic M stage | 381 | ||||
| M0 | 356 | Reference | Reference | ||
| M1 | 25 | 2.176 (1.272–3.722) | 0.005 | 1.984 (1.110–3.547) | 0.021 |
| DCUN1D5 | 530 | ||||
| Low | 267 | Reference | Reference | ||
| High | 263 | 1.567 (1.174–2.093) | 0.002 | 1.475 (1.050–2.071) | 0.025 |
Fig. 4.
The nomogram model of LUAD was constructed and evaluated. (A) The nomogram model including TNM staging and DCUN1D5 expression was constructed. (B) The calibration curves of 1 year, 3 years and 5 years were used to evaluate the prediction accuracy of nomogram. *p < 0.05, **p < 0.01, ***p < 0.001.
Enrichment analysis of DCUN1D5 differential genes and related genes
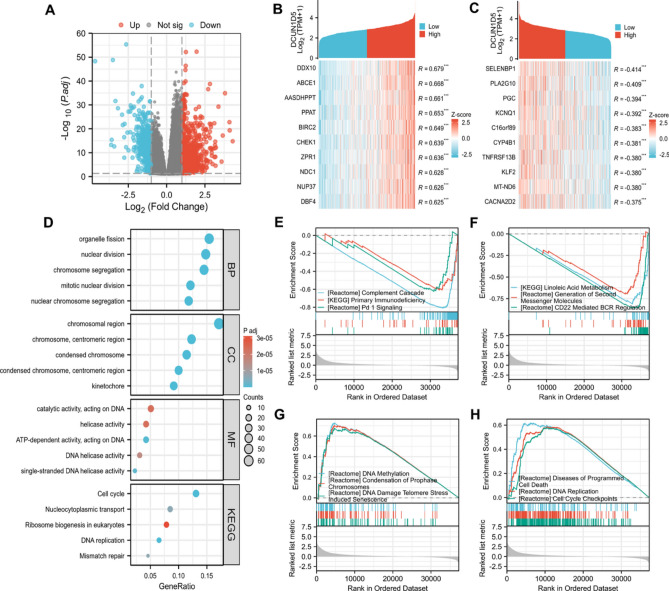
We used R software to analyze the differential genes between DCUN1D5 high expression group and low expression group. Screening was carried out under the conditions of P.adj < 0.05, |log2FC| ≥ 1 and genes encoding proteins. Results 686 genes were up-regulated and 438 genes were down-regulated (Fig. 5A). Then Pearson correlation coefficient analysis was used to test the correlation between DCUN1D5 expression and other molecules in TCGA-LUAD data sets. The top ten positive and negative related genes were selected for visualization (Fig. 5B,C). Using R software, the genes with correlation coefficient R > 0.5 and significant level P.adj < 0.05 were included in GO and KEGG enrichment analysis. DCUN1D5-related genes are involved in 366 biological processes (GO-BP), 102 cellular components (GO-CC), 63 molecular functions (GO-MF) and 11 KEGG. The first five most relevant BP, CC, MF and KEGG were still selected to be displayed in the form of a bubble chart (Fig. 5D). GO functional annotation showed that the related genes were mainly enriched in chromosome segregation, chromosomal region and ATP-dependent activity, acting on DNA. KEGG pathway analysis shows that they are mainly involved in Cell cycle, DNA replication, Nucleocytoplasmic transport, Mismatch repair and Ribosome biogenesis in eukaryotes。In order to further study the function of DCUN1D5, we carried out GSEA enrichment analysis. All the differential genes (DEGs) of DCUN1D5 were included in the analysis. The results showed that there were 220 gene sets under the condition of FDR < 0.25 and P.adj < 0.05. These gene sets mainly include KEGG_REACTOME_PD_1_SIGNALING (NES = − 1.96, P.adj < 0.05) , REACTOME_CD22_MEDIATED_BCR_REGULATION (NES = − 3.39, P.adj < 0.05), REACTOME_DNA_METHYLATION (NES = 1.79, P.adj < 0.05), REACTOME_DNA_REPLICATION (NES = 1.51, P.adj < 0.05) (Fig. 5E–H).
Fig. 5.
Functional enrichment analysis of DCUN1D5. (A) Volcanic map of DCUN1D5 differential genes in TCGA-LUAD data set. (B,C) heat map of 10 genes with the strongest positive and negative correlation with DCUN1D5 expression in TCGA-LUAD data set. (D) GO and KEGG enrichment analysis of DCUN1D5 co-expression genes. (E–H) GSEA enrichment analysis of DCUN1D5D differential genes. *p < 0.05, **p < 0.01, ***p < 0.001.
Effect of DCUN1D5 expression level on glycolysis
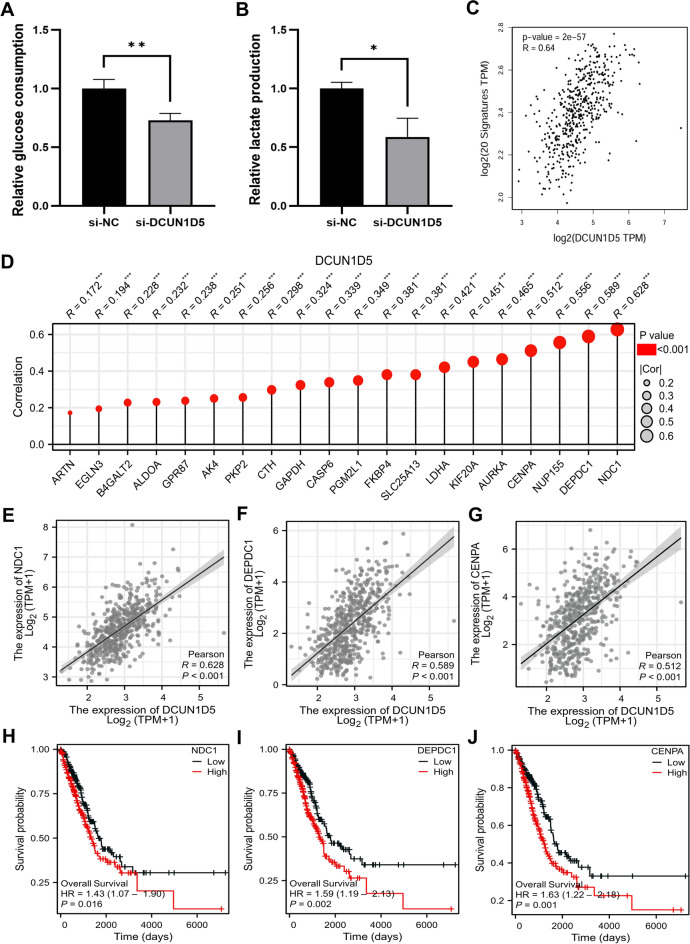
It is well known that glycolysis plays an important role in the occurrence and development of tumors. We studied the effect of DCUN1D5 on glycolysis by knocking down it at the cellular level. The results showed that glucose uptake decreased significantly (Fig. 6A) after down-regulation of DCUN1D5. Compared with the control group, the content of lactic acid in si-DCUN1D5 group decreased (Fig. 6B). GEPIA2 database analysis showed that there was a significant correlation between DCUN1D5 expression in LUAD and glycolysis characteristic gene set (Fig. 6C). We further analyzed the correlation between DCUN1D5 expression and 20 glycolysis genes in TCGA database. The results showed that the expression level of DCUN1D5 was strongly correlated with NDC1, DEPDC1, NUP155 and CENPA (R > 0.5) (Fig. 6D). The scatter plot shows the correlation between DCUN1D5 and NDC1, DEPDC1 and CENPA in TCGA-LUAD (Fig. 6E–G). We further observed that the expression levels of NDC1, DEPDC1 and CENPA had a significant impact on the prognosis (Fig. 6H–J).
Fig. 6.
Effects of DCUN1D5 expression in LUAD on glycolysis. (A) Down-regulation of DCUN1D5 significantly affects glucose uptake. (B) Down-regulation of DCUN1D5 significantly reduces lactate production. (C) Correlation analysis of glycolysis-related gene sets with DCUN1D5 expression in the GEPIA2 database. (D) Correlation lollipop plot of 20 glycolysis-related genes with DCUN1D5 in TCGA database. (E–G) Correlation of DCUN1D5 with NDC1, DEPDC1, and CENPA using scatter plots. (H–J) Relationship between NDC1, DEPDC1, and CENPA expression and patient OS in the TCGA-LUAD dataset. *p < 0.05, **p < 0.01, ***p < 0.001.
Effects of DCUN1D5 expression on immune characterization
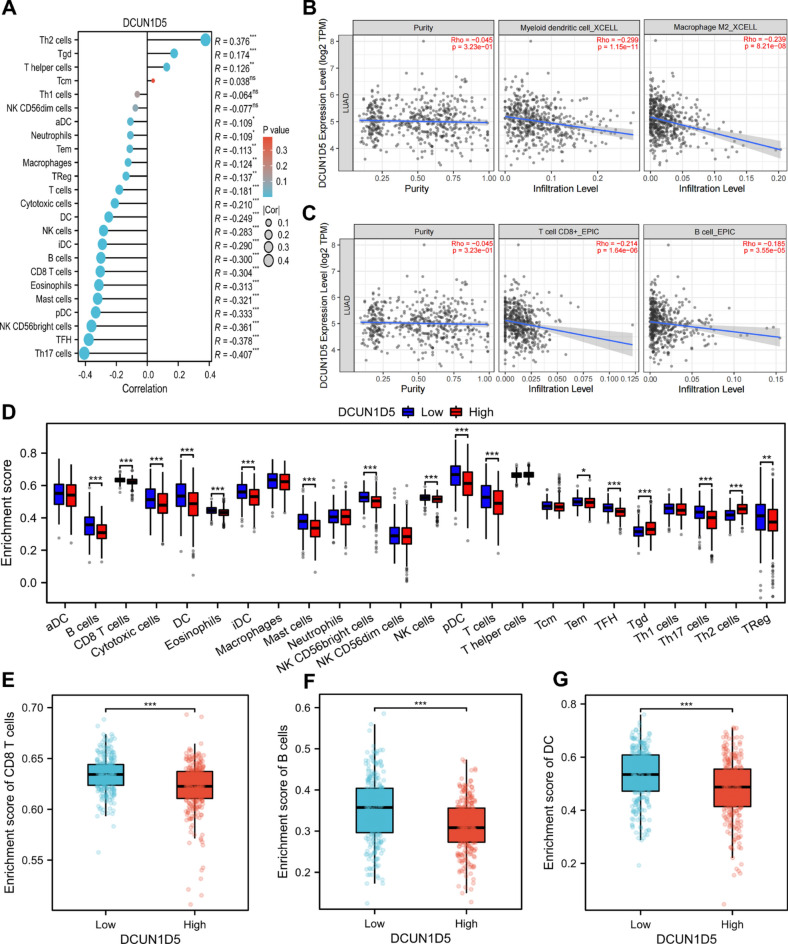
GSEA analysis showed enrichment in immune-related pathways. This indicated that the expression level of DCUN1D5 may be associated with immune characteristics. We analyzed the correlation between DCUN1D5 expression and immune cell infiltration in TCGA-LUAD. The results showed that DCUN1D5 expression was negatively correlated with Th17 cells, pDC, CD8+ T cells, iDC, B cells, macrophages, and NK cells, and positively correlated with Th2 cells (Fig. 7A). We further analyzed the relationship between the expression of DCUN1D5 and the infiltration level of myeloid dendritic cells, M2 macrophages, CD8+ Tcells and Bcells by TIMER2.0. The results showed that there was a significant negative correlation between DCUN1D5 and these cells (Fig. 7B,C). When DCUN1D5 was divided into high expression group and low expression group in TCGA, we found that there were significant differences in the infiltration levels of B cells, CD8+ T cells, DC, Mastcells, pDC, T cells, Th17 cells and Th2 cells (Fig. 7D). The infiltration of CD8+ T cells, B cells and DC decreased in the group with high expression of DCUN1D5 (Fig. 7E–G).
Fig. 7.
Relationship between DCUN1D5 expression and tumor immune cell infiltration. (A) Correlation of DCUN1D5 with different immune infiltrating cells in TCGA-LUAD dataset. (B,C) Correlation of DCUN1D5 expression with four types of immune cell infiltration in the TIMER2.0 database. (D) Different immune cell infiltrations in DCUN1D5 high and low expression groups in the TCGA-LUAD dataset. (E–G) Infiltration levels of CD8+ T cells, B cells, and DC in DCUN1D5 high and low expression groups. *p < 0.05, **p < 0.01, ***p < 0.001.
Effects of DCUN1D5 on the biological behavior of LUAD
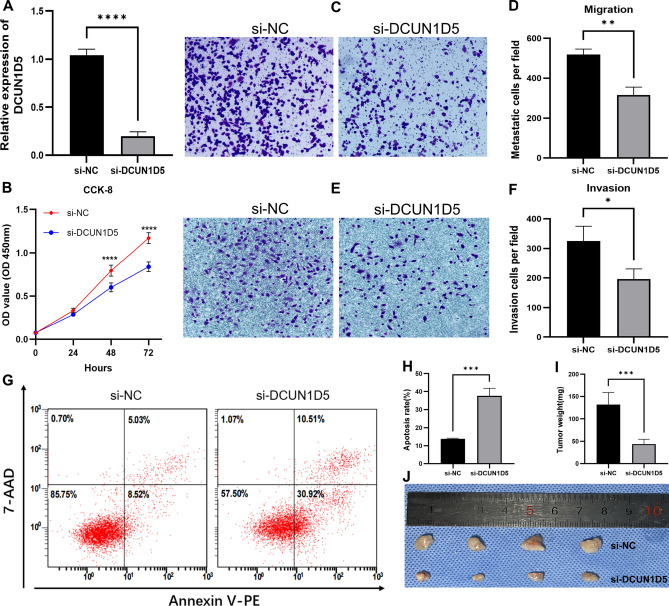
The qRT-PCR results showed that the expression of si-DCUN1D5 group was significantly lower than that of si-NC group after transfection of siRNA, indicating that a model of down-regulation of DCUN1D5 was successfully constructed in H1299 cell line (Fig. 8A). We found that the proliferation ability of lung adenocarcinoma cells H1299 was significantly decreased after down-regulation of DCUN1D5 by in vitro experiments (Fig. 8B). Compared with the control group, the migration and invasion abilities of si-DCUN1D5 group were reduced, and the differences were statistically significant (Fig. 8C–F). We detected the altered apoptotic ability of the cells by flow cytometry. The results showed that the apoptotic ability of cells was significantly increased after knockdown of DCUN1D5 (Fig. 8G,H). In addition, we performed animal experiments to observe the effect of down-regulation of DCUN1D5 on tumor weight in nude mice. The results showed that compared to the control group, the weight of the tumors formed in the si-DCUN1D5 group was significantly lower than the control group (Fig. 8I,J).
Fig. 8.
Effect of silencing DCUN1D5 on the biological behavior of LUAD. (A) qRT-PCR to detect the knockdown efficiency of DCUN1D5 (B) Effect of down-regulation of DCUN1D5 on cell proliferation ability. (C,D) Effect of down-regulation of DCUN1D5 on cell migration ability. (E,F) Effect of down-regulation of DCUN1D5 on cell invasion ability. (G,H) Effect of silencing DCUN1D5 on cell apoptotic ability detected by flow cytometry. (I,J) Silencing DCUN1D5 significantly reduces subcutaneous graft tumor weight in nude mice.*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Discussion
Lung adenocarcinoma remains one of the most aggressive and rapidly fatal tumor types24, and patients often present with diffuse systemic metastases at the time of diagnosis, with an overall survival of less than 5 years. Therefore, the search for potential early diagnostic indicators and specific therapeutic targets is particularly critical.
Human cells express five DCN1-like (defective in cullin neddylation 1) proteins called DCNL1-DCNL5 (also named DCUN1D1–5 for defective in cullin neddylation 1 domain containing protein 1–5), of which DCUN1D5 is a member and plays a unique role in the DNA damage response25. It has been shown that DCUN1D5 is highly expressed in laryngeal squamous cell carcinoma tissues and cells, and overexpression of DCUN1D5 enhances cell proliferation, migration, and invasion and DCUN1D5 may prevent cell cycle progression, thereby promoting repair of damaged DNA9. In oral and lung squamous cell carcinomas, the mRNA level of DCUN1D5 corresponds to the protein level, and its overexpression is associated with reduced disease-specific survival10. Compared to breast cancer without distant metastases DCUN1D5 is more highly expressed in breast cancer with distant metastases, and high DCUN1D5 expression levels are associated with poorer 5-year overall and recurrence-free survival in breast cancer patients11. In addition, DCUN1D5 can be transiently phosphorylated at its N-terminal serine residue by the kinase IKKα during immune signaling, which is involved in the innate immune process in mice26. However, the expression and biological function of DCUN1D5 in lung adenocarcinoma and the molecular mechanism of its action are not yet clear.
In this study, we used TCGA, GEPIA2 and UALCAN database studies to show that DCUN1D5 is highly expressed in LUAD, and further used GSE140797 in GEO database to verify this conclusion. In addition, we also found that DCUN1D5 has a good diagnostic value for LUAD and the overall survival time of patients with high expression of DCUN1D5 is shorter. The co-expression genes of DCUN1D5 were analyzed by GO and KEGG. The results showed that they were mainly enriched in biological processes and pathways such as chromosome segregation, chromosomal region, ATP-dependent activity, acting on DNA, Cell cycle and so on. In order to further explore the biological function of DCUN1D5, we carried out GSEA enrichment analysis. The results show that it mainly involves biological processes such as DNA methylation, DNA replication, CD22 mediated BCR regulation, PD 1 signaling and so on. Previous studies have shown that these biological processes are closely related to the occurrence and development of tumors. For example, studies have found that CLEC14A promoter hypermethylation in LUAD, inhibition of CLEC14A methylation level can hinder LUAD cell replication, promote apoptosis, weaken the ability of cell migration and invasion, and arrest the cell cycle in G0/G1 phase27. Cell cycle checkpoint related gene CDK1 is the central tumor-promoting gene of LUAD. Knocking down CDK1 can inhibit the proliferation of LUAD28. LINC00880 can act as a protein scaffold between CDK1 and PRDX1, forming a ternary complex, which leads to the activation of PI3K/AKT and promotes the occurrence of malignant tumor29. B4GALT1 is highly expressed in LUAD. Inhibition of B4GALT1 can increase the abundance and activity of CD8+ T cells and enhance the anti-tumor immunity of anti-pd-1 therapy in vivo30. Therefore, it is possible for DCUN1D5 to influence the progress of LUAD through these biological processes. The cancer-promoting effect of glycolysis is well known. However, whether DCUN1D5 can regulate the glycolysis level of LUAD has not been reported. We confirmed that knocking down DCUN1D5 can significantly reduce the glucose uptake and lactic acid production of lung adenocarcinoma cells in vitro. This shows that DCUN1D5 can affect the level of glycolysis in some way, but the specific mechanism needs to be further studied. Therefore, we analyzed the correlation between glycolysis related genes and DCUN1D5 expression. The results showed that DCUN1D5 had a strong positive correlation with NDC1, DEPDC1 and CENPA (correlation coefficient R > 0.5). The effects of NDC1, DEPDC1 and CENPA on the prognosis of lung adenocarcinoma were further analyzed in TCGA database. The results showed that OS was shortened in high expression group. Akkafa showed that TMEM48 (NDC1) was highly expressed in the lung adenocarcinoma cell line A549, and miR-421 significantly inhibited the expression of TMEM48 and thus enhanced apoptosis31. Wang showed that the expression of DEPDC1 was upregulated in LUAD tissues and exerted an oncogenic effect in LUAD, and that DEPDC1 inhibited autophagy through the RAS-ERK1/2 signaling pathway32. YU showed that knockdown of CENPA reduced the proliferation of lung adenocarcinoma cells and regulated the characteristics of tumor stem cells33. Whether DCUN1D5 interacts with these factors in a specific way and exerts pro-carcinogenic effects by the mechanisms they have been elucidated will greatly expand our research ideas. Immune cell infiltration is crucial for the tumor immune microenvironment. The results of tumor immune cell infiltration in this experiment showed that the expression of DCUN1D5 was positively correlated with the infiltration of Th2 cells and negatively correlated with the infiltration of Th17 cells, myeloid dendritic cells, M2 macrophages, CD8+ T cells, and B cells. Further studies showed that the level of myeloid dendritic cells, CD8+ T cells and B cells, and NK cells infiltration was decreased while the level of Th2 cells infiltration was increased in the group with high DCUN1D5 expression. Previous studies have shown that in the tumor epithelium and mesenchyme of NSCLC, the majority of CD4+ T cells have a Th2 or Treg phenotype, whereas only a small number of Th1, Th17, and Tfh cells have been observed34. This is consistent with the results from our experiments that the poor prognostic high expression DCUN1D5 group had a higher level of Th2 infiltration and a lower level of Th17 cell infiltration. Study shows that infiltrating CD8+ T cells, interferon-γ (IFNγ)-expressing T helper 1 (TH1) cells, natural killer (NK) cells, and B cells have anti-tumor immune properties35. Dendritic cells (DCs) are crucial for the activation of antigen-specific CD8 T lymphocytes, a pivotal step in the initiation of the innate and adaptive immune responses, which are essential for tumor cell clearance36. Therefore, the low infiltration levels of DC, CD8+ T cells, B cells and NK cells in the high expression group of DCUN1D5 also have implications for our further study. In vitro experiments showed that down-regulation of DCUN1D5 could inhibit the proliferation, migration and invasion of lung adenocarcinoma cells, and promote the apoptosis of lung adenocarcinoma cells. Animal experiments showed that knockout DCUN1D5 could effectively inhibit the growth of subcutaneous transplanted tumor in nude mice. Of course, this study has some limitations. We only use the data in TCGA to analyze the biological function of DCUN1D5, and then we should include more samples from different databases for verification. In addition, only one lung adenocarcinoma cell line was used to explore the effect of DCUN1D5 on the biological function of LUAD, which can increase the number of cell lines for experiments. In this study, we show for the first time that DCUN1D5 may play an important role as an oncogene in lung adenocarcinoma. In addition, this study may provide clues for the diagnosis of LUAD and the search for new therapeutic targets.
Conclusion
Our study shows that DCUN1D5 is highly expressed in lung adenocarcinoma and can be used as a new indicator for the diagnosis and prognosis of lung adenocarcinoma. Silencing DCUN1D5 can affect the biological characteristics of lung adenocarcinoma, and the molecular mechanism of its participation in LUAD may be related to glycolysis and immune infiltration.
Supplementary Information
Abbreviations
- TCGA
The Cancer Genome Atlas database
- GEO
Gene Expression Omnibus
- LUAD
Lung adenocarcinoma
- OS
Overall survival
- qRT‑PCR
Real-time quantitative PCR
- DEGs
Differentially expressed genes
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GSEA
Gene Set Enrichment Analysis
Author contributions
Song Zhao and Zongying Liang designed the experiment. Song Zhao completed the experiment and wrote a manuscript. Xiaoli Han and Jingtao Huang completed the statistical analysis of the data and revised the manuscript. Jingxiong Zheng and Baoshan Zhao revised the manuscript. All the authors contributed to the experiment and approved the submission of the manuscript.
Funding
This study is supported by the Natural Science Foundation of Hebei Province (No. H2021406045) and the Medical Science Research Program of Hebei Province (No. 20220411).
Data availability
All datasets are freely available from public databases. Patients’ clinical information was downloaded from the TCGA database (https://portal.gdc.cancer.gov/). The mRNA expression profile data can be downloaded from the GEO (GSE140797, https://www.ncbi.nlm.nih.gov/geo/) and TCGA (https://portal.gdc.cancer.gov/). The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
All studies involving human tissues were conducted in strict accordance with the Declaration of Helsinki and were approved by the Ethics Committee of the affiliated Hospital of Chengde Medical College. The clinical information of LUAD patients comes from the public database and does not involve ethical issues. The animal experimental research program has been approved by the Ethics Committee of the affiliated Hospital of Chengde Medical College, and the ethical approval number is LL2021028.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-84539-1.
References
- 1.Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin.73(1), 17–48 (2023). [DOI] [PubMed] [Google Scholar]
- 2.Howlader, N. et al. The effect of advances in lung-cancer treatment on population mortality. N. Engl. J. Med.383(7), 640–649 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jha, S. K. et al. Cellular senescence in lung cancer: Molecular mechanisms and therapeutic interventions. Ageing Res. Rev.97, 102315 (2024). [DOI] [PubMed] [Google Scholar]
- 4.Morel, M. & Long, W. FBXL16 promotes cell growth and drug resistance in lung adenocarcinomas with KRAS mutation by stabilizing IRS1 and upregulating IRS1/AKT signaling. Mol. Oncol.18(3), 762–777 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duma, N., Santana-Davila, R. & Molina, J. R. Non-small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin. Proc.94(8), 1623–1640 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Sun, T. et al. Lipidomics reveals new lipid-based lung adenocarcinoma early diagnosis model. EMBO Mol. Med.16(4), 854–869 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nooreldeen, R. & Bach, H. Current and future development in lung cancer diagnosis. Int. J. Mol. Sci.22(16), 8661 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang, H. et al. m6A methylation reader IGF2BP2 activates endothelial cells to promote angiogenesis and metastasis of lung adenocarcinoma. Mol. Cancer22(1), 99 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo, W. et al. In vitro biological characterization of DCUN1D5 in DNA damage response. Asian Pac. J. Cancer Prev.13(8), 4157–4162 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Bommeljé, C. C. et al. Oncogenic function of SCCRO5/DCUN1D5 requires its Neddylation E3 activity and nuclear localization. Clin. Cancer Res.20(2), 372–381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh, J. et al. Widespread alternative splicing changes in metastatic breast cancer cells. Cells10(4), 858 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Visser, K. E. & Joyce, J. A. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell41(3), 374–403 (2023). [DOI] [PubMed] [Google Scholar]
- 13.Wang, S. S. et al. Tumor-infiltrating B cells: their role and application in anti-tumor immunity in lung cancer. Cell. Mol. Immunol.16(1), 6–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, C. et al. Research progress on the mechanism of glycolysis in ovarian cancer. Front. Immunol.14, 1284853 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chelakkot, C. et al. Modulating glycolysis to improve cancer therapy. Int. J. Mol. Sci.24(3), 2606 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu, Y. et al. RNA m1A methylation regulates glycolysis of cancer cells through modulating ATP5D. Proc. Natl. Acad. Sci. USA119(28), e2119038119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandrashekar, D. S. et al. UALCAN: a portal for facilitating Tumor Subgroup Gene expression and survival analyses. Neoplasia19(8), 649–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res.28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci.28, 1947–1951 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res.51, D587–D592 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA102(43), 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huo, C. et al. Glycolysis define two prognostic subgroups of lung adenocarcinoma with different mutation characteristics and immune infiltration signatures. Front. Cell Dev. Biol.9, 645482 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform.14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denisenko, T. V., Budkevich, I. N. & Zhivotovsky, B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis.9(2), 117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keuss, M. J. et al. Characterization of the mammalian family of DCN-type NEDD8 E3 ligases. J. Cell Sci.129(7), 1441–1454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas, Y. et al. The NEDD8 E3 ligase DCNL5 is phosphorylated by IKK alpha during Toll-like receptor activation. PLoS One13(6), e0199197 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su, C. et al. Methylation of CLEC14A is associated with its expression and lung adenocarcinoma progression. J. Cell. Physiol.234(3), 2954–2962 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Ma, X. et al. Transcriptomic analysis of tumor tissues and organoids reveals the crucial genes regulating the proliferation of lung adenocarcinoma. J. Transl. Med.19(1), 368 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng, Y. et al. The super-enhancer-driven lncRNA LINC00880 acts as a scaffold between CDK1 and PRDX1 to sustain the malignance of lung adenocarcinoma. Cell Death Dis.14(8), 551 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui, Y. et al. B4GALT1 promotes immune escape by regulating the expression of PD-L1 at multiple levels in lung adenocarcinoma. J. Exp. Clin. Cancer Res.42(1), 146 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akkafa, F. et al. miRNA-mediated apoptosis activation through TMEM 48 inhibition in A549 cell line. Biochem. Biophys. Res. Commun.503(1), 323–329 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Wang, W. et al. DEPDC1 up-regulates RAS expression to inhibit autophagy in lung adenocarcinoma cells. J. Cell. Mol. Med.24(22), 13303–13313 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu, Q. Y. et al. CENPA regulates tumor stemness in lung adenocarcinoma. Aging (Albany NY).14(13), 5537–5553 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frafjord, A. et al. The immune landscape of human primary lung tumors is Th2 skewed. Front. Immunol.12, 764596 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takacs, G. P., Flores-Toro, J. A. & Harrison, J. K. Modulation of the chemokine/chemokine receptor axis as a novel approach for glioma therapy. Pharmacol. Ther.222, 107790 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, J. B., Huang, X. & Li, F. R. Impaired dendritic cell functions in lung cancer: a review of recent advances and future perspectives. Cancer Commun. (Lond).39(1), 43 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets are freely available from public databases. Patients’ clinical information was downloaded from the TCGA database (https://portal.gdc.cancer.gov/). The mRNA expression profile data can be downloaded from the GEO (GSE140797, https://www.ncbi.nlm.nih.gov/geo/) and TCGA (https://portal.gdc.cancer.gov/). The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.