ABSTRACT
Introduction
Antimicrobial resistance (AMR) is one of the leading public health threats globally. AMR genes can be transferred between bacteria through lateral gene transfer, and AMR organisms can spread through environments by contaminated water, agriculture and animals. Thus, widespread environmental dissemination of bacteria and lateral gene transfer facilitate AMR transmission pathways. Farm environments in dairy and calf production are known to harbour AMR bacteria that pose a risk for food contamination and to workers in direct or indirect contact with animals. Escherichia coli is present in farm environments and is known to participate in lateral gene transfer, providing a good marker of resistance genes in each environment.
Methods
In this study, E. coli from nine cohorts of calves was isolated at different time points from nine barns, nine trailers and one slaughterhouse environment in a single special‐fed veal calf production facility. The antimicrobial susceptibility to 15 antimicrobials, classified as highly or critically important by the World Health Organization, was characterised for E. coli isolates using Kirby–Bauer disk diffusion.
Results
The highest proportion of isolates showing multidrug resistance was present in barn environments (51.7%), where calves were housed from their arrival at < 2 weeks of age until they were transported to slaughter. Additionally, 15 E. coli isolates were resistant to 11 of the 15 antimicrobials tested. Trailer and slaughterhouse environments had greater prevalence of resistance after accommodating calves, including resistance to third‐generation cephalosporins.
Conclusion
These data highlight the importance of calf environments in the dissemination of resistant bacteria and gives insight into where interventions could be most effective in combatting antimicrobial‐resistant bacteria that could infect humans and livestock.
Keywords: antimicrobial resistance, calves, environment, Escherichia coli, livestock, veal production
Summary.
Barn environments had the greatest proportion of multidrug‐resistant E. coli (51.7%) compared to slaughterhouse (35.7%) and trailer (33.7%) environments. This could suggest an environmental source of AMR for calves.
Prevalence of AMR in trailers and slaughterhouse increase as calves move through these environments.
Implementing stricter cleaning and disinfection protocols in barn environments could be highly effective in reducing the transmission of AMR elements through environments and into the food supply.
1. Introduction
Antimicrobial resistance (AMR), or the ability of microorganisms to survive treatment with drugs, is one of the most important public health threats of the century (Lammie and Hughes 2016; McEwen and Collignon 2018; Murray et al. 2022; WHO 2021). Approximately 35,000 people in the United States die annually due to antimicrobial‐resistant infections (CDC 2019). Bacteria acquire resistance through de novo mutations or lateral gene transfer (Komp Lindgren, Åsa, and Hughes 2003). Through various interactions between humans, animals and the environment, such as the use of manure for fertilisation or direct handling of animals on a farm, resistant organisms can spread among environments and animal hosts (Checcucci et al. 2020; McEwen and Collignon 2018). Livestock are known reservoirs of AMR genes and antimicrobial‐resistant bacteria, so understanding transmission through veal calf environments is critical to identifying prevention measures (Smith et al. 2002).
Calf production systems are particularly important reservoirs of AMR. Indeed, the frequency of resistance in commensal Escherichia coli (E. coli) has been shown to be greater in environmental and faecal samples from calves relative to adult cattle partially due to higher antimicrobial usage in calves as they are most susceptible to infection. (Agga et al. 2022; Gaire et al. 2021; Hanon et al. 2015; Um et al. 2015; Vinayamohan et al. 2022). Special‐fed veal calves (calves that are fed a balanced milk or soy‐based diet) are generally housed individually for up to 10 weeks before transitioning to group housing (Veal Quality Assurance Program 2018). At around 20 weeks, veal calves are sent for slaughter (USDA 2013). Prior to slaughter, these calves progress through multiple environments including barns, transport trailers and slaughterhouse holding pens. As with other animal production systems, antimicrobials are sometimes used to prevent and treat disease in veal calves, which can facilitate selection of AMR bacteria (Cheng et al. 2022; USDA 2013). However, AMR can still be found to antimicrobials not licensed for use in veal calves (like fluoroquinolones and chloramphenicol), suggesting other sources of AMR gene acquisition (Afema, Davis, and Sischo 2019; Food and Drug Administration 2022).
Some antimicrobial classes deemed critically important for human health (e.g., third‐generation cephalosporins, fluoroquinolones and macrolides) are commonly used in livestock to treat infection or prevent disease (Collignon and McEwen 2019; McEwen and Collignon 2018; WHO 1970). High infectious disease pressure sometimes necessitates high levels of antimicrobial use in preweaned calves (Cheng et al. 2022). The level of AMR in commensal E. coli from veal calves has previously been documented (Hutchinson et al. 2017). Preharvest environments (where calves are present up until slaughter) represent a key conduit for the dissemination of AMR from farms into the food supply. Therefore, this cohort study aimed to describe AMR in E. coli recovered from the environments of barns, trailers and the slaughterhouse in a veal calf production system. Additionally, it aimed to evaluate changes in prevalence of AMR before and after arrival of calf cohorts.
2. Materials and Methods
2.1. Study Design and Setting
Samples were collected from the environment of nine cohorts of veal calves as part of a larger prospective cohort study of veal calves conducted from November 2018 to October 2019 (Locke et al. 2022). Nine cohorts of special‐fed veal calves raised in a single vertically integrated production system were enrolled, and environmental samples were collected from barns, trailers and the slaughterhouse holding pens. Cohorts were selected by convenience sampling based on the processor schedule and the proximity of the farm to the slaughterhouse. Dairy‐breed calves were sourced by the production company from dairy farms and auctions in the Midwest and Eastern United States and transported to grower barns in Ohio. Within this production setting, barns remained empty for approximately 3 weeks before shipments of calves, generally < 1 week of age, arrived over 1 or 2 days to populate the barns. Cohorts of approximately 200 calves spent their first weeks in individual stalls with slatted flooring before moving to group housing between 8 and 10 weeks of age. Proprietary milk replacer and a texturized starter grain were fed to calves until slaughter at about 20–24 weeks of age and weighing approximately 227 kg.
Calves were transported from the barn to the slaughterhouse using double decker trailers hauled by semitrucks that were owned by a third‐party contractor. Trailer density ranged from 80 to 82 calves, where calves spent 1–4 h between loading and unloading. Calves spent 30 min–3 h in the slaughterhouse holding pens after unloading and before slaughter.
2.2. Sample Collection
A total of 198 boot swabs (Solar Biologicals Inc., Ogdensburg, NY) were collected from three environments that calves inhabited: nine grower barns, nine livestock trailers and the holding pens of one slaughterhouse. These will be referred to as barn, trailer and slaughterhouse hereafter. A scheme was generated using BioRender (n.d.) and is shown below (Figure 1). For all samples, plastic boot covers were worn in combination with latex gloves that were changed between samples. Boot swabs were premoistened in buffered peptone water (BPW) and worn on the feet or hands of the individual collecting the sample. Barn samples were collected in environments where the calves were housed prior to transportation, including walls (n = 1), pen floors (n = 2), gate (n = 1) and alleyways (n = 2). The livestock trailers had two levels from which floor (n = 1) and wall (n = 1) samples were individually collected on each floor, totalling four samples. The entire length of walls and floor of the trailer were swabbed. From the slaughterhouse, samples were collected from gates (n = 1), walls (n = 1) and floors by the entrance (n = 2). In all cohorts, 22 samples were collected from the three environments described above. In the trailer and slaughterhouse, samples were collected at two different time points: before (n = 4) and after (n = 4) calves passed through the environment (total = 8). Samples were collected from the barns at one time point as described above (n = 6). Boot swabs were placed in a bag before being mailed on ice overnight to the Wisconsin Veterinary Diagnostic Laboratory (WVDL). Ethics approvals were not required, since the research used only environmental samples from animal environments. Likewise, human subjects were not involved with the research.
FIGURE 1.
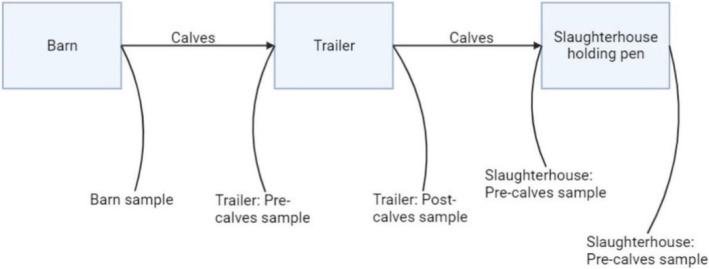
Diagram showing the veal calf environments sampled for antimicrobial‐resistant bacteria and time points for sampling in each environment. Samples were collected from barns where calves were first housed (n = 9), then from trailers that transported them (n = 9) and holding pens of the slaughterhouse (n = 1) before and after they passed through.
2.3. Isolation and Identification of E. coli
At the WVDL, boot swabs were preenriched in BPW (1:10) and incubated at 36 ± 2°C for 18–24 h in accordance with the bovine environmental sampling protocol (Markey et al. 2013; Miller et al. 1991; USDA 2017; Williams 2016; University of Wisconsin, 2018). 10 mL aliquots from BPW were then dispensed into culture vials shipped back to The Ohio State University on ice, where they were aliquoted into 1 mL cryovials and stored at −80°C until analysis. To revive samples, frozen stock was inoculated in tryptic soy broth (TSB) (Oxoid, Hants, UK) at 37°C for 24 h. The samples were then streaked for colony isolation on CHROMagar (CHROMagar, Paris, France) and incubated at 37°C for 24 h. Four blue colonies on the media (presumptive E. coli) were selected and inoculated in tryptic soy broth at 37°C for 24 h (i.e., four isolates per sample). The TSB cultures were then streaked on MacConkey agar (Oxoid, Hants, UK) and incubated at 37°C for 24 h. The isolates were identified as E. coli based on their colony morphology on CHROMagar and MacConkey agar.
2.4. Antimicrobial Susceptibility Testing by Disk Diffusion
Isolates from the MacConkey agar were added to phosphate‐buffered saline to achieve an optical density of 0.650–0.750. Each standardised solution was swabbed onto Mueller Hinton agar (Difco, Le Pont de Claix, France) and subjected to disk diffusion susceptibility testing, using the Clinical & Laboratory Standards Institute (CLSI) guidelines from the M100 and VET01S to determine breakpoints for Enterobacterales (CLSI 2023a, 2023b). For the purposes of statistical analysis, results that tested in the ‘intermediate’ range were considered resistant. Escherichia coli isolates were tested against the following antimicrobials, classified as critically or highly important by the World Health Organization: amoxicillin/clavulanic acid (AmC), 20 μg/10 μg; ampicillin (AM), 10 μg; azithromycin (AZM), 15 μg; cefoxitin (FOX), 30 μg; ceftiofur (TIO), 30 μg; ceftriaxone (CRO), 30 μg; chloramphenicol (CHL), 30 μg; ciprofloxacin (CIP), 5 μg; gentamicin (GM), 10 μg; meropenem (MEM), 10 μg; nalidixic acid (NA), 30 μg; streptomycin (S), 10 μg; sulfisoxazole (SOX); tetracycline (Te), 30 μg; trimethoprim/sulfamethoxazole (SXT) and 1.25 μg/23.75 μg (WHO 2018). All antimicrobials were sourced from Becton Dickinson (BD, Sparks, MD).
2.5. Whole Genome Sequencing
Escherichia coli isolates resistant to third‐generation cephalosporins and with reduced susceptibility to fluoroquinolones were submitted for whole genome sequencing. Not all samples were available for sequencing, so four of seven were submitted. DNA isolation was completed using DNeasy PowerSoil Pro Kit (Qiagen, Valencia, CA, USA) per the manufacturer's instruction. The isolated DNA samples were then delivered to The Ohio State University Applied Microbiology Services Laboratory for whole genome sequencing using the Nextseq 2000 Illumina platform (San Diego, CA), 2 × 150 bp paired‐end reads, 50 × coverage. The Nextera library kit and IDT for Illumina DNA/RNA UD Indexes(R) (Illumina, San Diego, CA) were used for DNA library construction. Raw reads were run through the Global Health Research Unit (GHRU) pipeline for quality control and assembly (Underwood 2020). Resulting assemblies were then analysed using ResFinder and PlasmidFinder (Zankari et al. 2012; Carattoli et al. 2014; Bortolaia et al. 2020) hosted online by the Center for Genomic Epidemiology (n.d.) (Center for Genomic Epidemiology, Kongens Lyngby, Denmark).
2.6. Statistical Analysis
Data analysis was completed using R Studio (v4.2.1; R Core Team 2022). Descriptive analysis of the zone diameters for AMR was made using the AMR R package (v1.8.1; Berends et al. 2022). Data tables and figures were generated using Microsoft Excel (Microsoft Corporation). Multidrug resistance (MDR) was defined as resistance to at least one drug in three or more antimicrobial classes (Magiorakos et al. 2012). SAS (v9.4; SAS Institute, Cary, NC) was used for the construction of logistic regression models.
PROC GLIMMIX, a procedure provided by SAS, was used for constructing generalised linear mixed models. Because the calf cohorts were only sampled once at the barns, the effect of environment (barns, trailers, slaughterhouse holding pens) and time point (before or after calves loaded into each environment) on the MDR of isolates was investigated using two models. For the effect of environment, the univariable ‘environment model’ included MDR (yes/no) as the outcome and the environment (barns/trailers/slaughterhouse) as the independent variable. To further investigate the interaction of environment and sampling time, samples collected at trailers and slaughterhouse pens (i.e., barn samples excluded) were fit into the multivariable ‘interaction model’ with MDR as the outcome and the environment, time point and their interaction as independent variables. A two‐factor hierarchical random effect of the variable ‘sample identification’ nested within the ‘calf cohort’ was included in both models to account for the dependence between isolates cultured from the same samples and between samples collected for the same calf cohorts. The intraclass correlation coefficients (ICC) for the proportion of the variance explained by each hierarchy of the random effect were calculated based on the residual variance of the standard logistic distribution (π 2/3 = 3.29), that is, the latent variable approach (Goldstein, Browne, and Rasbash 2002). Degrees of freedom and standard error corrections were estimated using the Kenward–Roger approximation (Kenward and Roger 1997).
Results were reported as estimated odds ratios of an isolate having MDR for combinations of environment and time point of sample collection. Multiple comparisons were adjusted using a Bonferroni procedure to prevent inflation of the Type I error (Bland & Altman, 1995). Additionally, model‐based estimates for the probability of MDR with corresponding 95% confidence intervals were also reported. The significance level (α) of 0.05 was used and effects with p < 0.05 were considered statistically significant.
3. Results
3.1. Descriptive Statistics
Twelve of the samples in cohort one showed no growth on CHROMagar after two attempts, so they were omitted. Some presumptive E. coli colonies were determined later to not be E. coli based on their morphology on MacConkey agar and were thus not tested against antimicrobials. From the 198 environmental samples, a total of 679 isolates were confirmed as E. coli and subjected to AMR characterisation (Barn = 172, Slaughterhouse = 249, Trailer = 258).
Of all isolates tested, 432 (63.6%) were resistant to at least one antimicrobial. The greatest proportion of resistant isolates was observed for tetracycline (56.6%) and ampicillin (43.4%) (Table 1). In trailers and slaughterhouse, resistant isolates (n = 149 and n = 152, respectively) were resistant to a median of four antimicrobials. In barns (n = 131), the median was six.
TABLE 1.
Frequency of AMR to critically or highly important antimicrobials among E. coli isolates recovered from barn, trailer and slaughterhouse environments for nine cohorts of special‐fed veal calves.
| Antimicrobial | Barn% (No.) | Trailer% (No.) | Slaughterhouse% (No.) | All samples% (No.) |
|---|---|---|---|---|
| Tetracycline | 72.1 (124) | 49.2 (127) | 53.4 (133) | 56.6 (384) |
| Ampicillin | 56.4 (97) | 39.1 (101) | 38.7 (96) | 43.4 (294) |
| Sulfisoxazole | 55.2 (95) | 34.9 (90) | 40.2 (100) | 42.0 (285) |
| Streptomycin | 54.1 (93) | 32.9 (85) | 32.9 (82) | 38.3 (260) |
| Chloramphenicol | 50.0 (86) | 27.5 (71) | 29.7 (74) | 34.0 (231) |
| Sulfamethoxazole/Trimethoprim | 41.3 (71) | 16.7 (43) | 23.8 (59) | 25.5 (173) |
| Amoxicillin‐Clavulanic Acid | 21.6 (37) | 18.7 (47) | 17.3 (43) | 18.9 (127) |
| Azithromycin | 11.0 (19) | 4.7 (12) | 3.2 (8) | 5.8 (39) |
| Cefoxitin | 10.5 (18) | 4.7 (12) | 3.6 (9) | 5.7 (39) |
| Ceftriaxone | 12.8 (22) | 3.5 (9) | 3.2 (8) | 5.7 (39) |
| Ceftiofur | 12.5 (20) | 3.5 (9) | 3.3 (8) | 5.6 (37) |
| Nalidixic acid | 7.0 (12) | 4.7 (12) | 1.6 (4) | 4.1 (28) |
| Ciprofloxacin | 7.6 (13) | 2.7 (7) | 1.2 (3) | 3.4 (23) |
| Gentamicin | 6.4 (11) | 0.8 (2) | 2.0 (5) | 2.7 (18) |
| Meropenem | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) |
| Number of samples | 160–172 | 251–258 | 246–249 | 664–679 |
Note: Not every isolate was tested against every antimicrobial.
The most common resistance phenotype was Te (13.4%), followed by Am‐Chl‐S‐Sox‐Sxt‐Te (8.1%) and Am‐Amc‐Chl‐S‐Sox‐Te (7.9%) (Table 2). All phenotypes present in more than 2% of the isolates are also shown in Table 2. Phenotypes present in more than 2% of isolates within each environment are shown in Table S1.
TABLE 2.
Frequency of antimicrobial resistance phenotypes (present in 2% or more of isolates) among E. coli recovered from environmental samples collected from barn, trailer and slaughterhouse in a special‐fed veal calf production system.
| Phenotype | % (No.) (n = 679) |
|---|---|
| Te | 13.4 (58) |
| Am‐Chl‐S‐Sox‐Sxt‐Te | 8.1 (35) |
| Am‐Amc‐Chl‐S‐Sox‐Te | 7.9 (34) |
| Am‐Amc‐Chl‐Sox‐Te | 4.9 (21) |
| Am‐Chl‐Sox‐Te | 4.9 (21) |
| S‐Te | 4.9 (21) |
| Am‐Amc‐Chl‐S‐Sox‐Sxt‐Te | 4.4 (19) |
| Am‐S‐Te | 2.5 (11) |
| Am | 2.3 (10) |
| Am‐S‐Sox‐Sxt‐Te | 2.1 (9) |
| Am‐Te | 2.1 (9) |
Abbreviations: Am, ampicillin; Amc, amoxicillin‐clavulanic acid; Azm, azithromycin; Chl, chloramphenicol; Cip, ciprofloxacin; Cro, ceftriaxone; Fox, cefoxitin; Gm, entamicin; Na, nalidixic acid; No., number of resistant isolates; S, streptomycin; Sox, sulfisoxazole; Sxt, sulfamethoxazole/trimethoprim; Te, tetracycline; Tio, ceftiofur.
3.2. Multidrug‐Resistant E. coli
The proportion of isolates resistant to three or more classes of antimicrobials (i.e., MDR) was 51.7% (89/172) in barns, 33.7% (87/258) in trailers and 35.7% (89/249) in slaughterhouse pens. Differences were observed between location (Figure 2), time points (Figure 3) and cohorts (Table 5).
FIGURE 2.
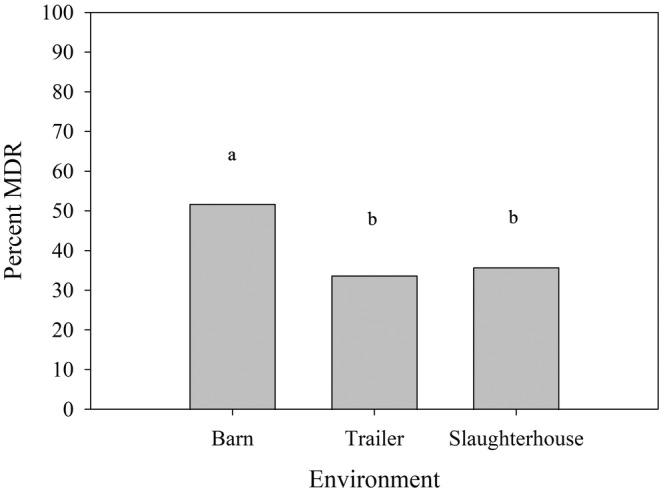
Empirical proportion of environmental E. coli isolates displaying MDR in each of the three environments veal calves were housed and transported through in the longitudinal cohort study. Overall, 51.7% (89/172), 33.7% (87/258) and 35.7% (89/249) isolates in barns, trailers and slaughterhouse pens, respectively, displayed MDR. Statistical differences among environments were determined by an univariable logistic regression model (Table 3), where those with a common letter on top of the bars are not significantly different (Bonferroni‐adjusted p > 0.05).
FIGURE 3.
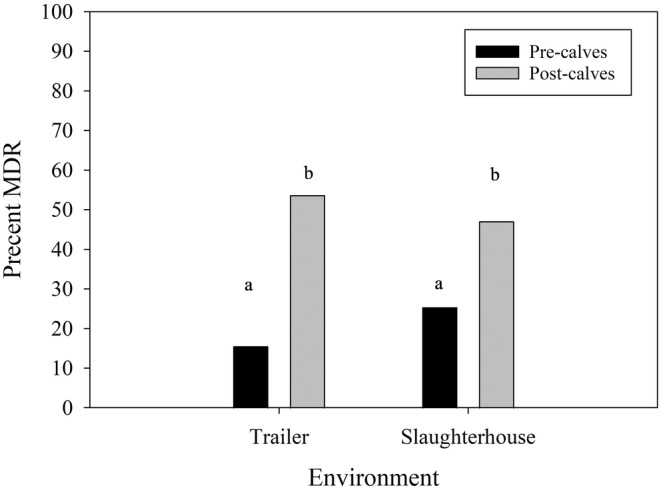
Empirical proportion of environmental E. coli isolates displaying MDR in trailers and slaughterhouse pens, sampled before and after the cohorts had entered the environments. Overall, 15.6% (21/135) and 53.7% (66/123) isolates sampled before and after entering trailers displayed MDR, while 25.4% (33/130) and 47.1% (56/119) for slaughterhouse pens. Statistical differences among environments and time points were determined by a multivariable logistic regression model (Table 4), where those with a common letter on top of the bars are not significantly difference (Bonferroni‐adjusted p > 0.05).
TABLE 5.
Frequency of MDR in E. coli isolates recovered from swabs taken from environments of nine different cohorts of veal calves before and after calves entered each environment in a longitudinal study. Barn samples were all taken at a single time point. Isolates that did not grow or were not confirmed as E. coli were not tested, resulting in differences in the total number of tested isolates.
| Cohort | Barn | Trailer | Slaughterhouse | ||
|---|---|---|---|---|---|
| Overall% (No./total) | Pre% (No./total) | Post% (No./total) | Pre% (No./total) | Post% (No./total) | |
| 1 | 100.0 (1/1) | 100.0 (11/11) | 66.7 (8/12) | 100.0 (12/12) | — (0/0) |
| 2 | 40.7 (11/27) | 25.0 (3/12) | 46.7 (7/15) | 0.0 (0/14) | 21.4 (3/14) |
| 3 | 0.0 (0/4) | 6.3 (1/16) | 50.0 (8/16) | 54.6 (6/11) | 50.0 (7/14) |
| 4 | 70.8 (17/24) | 6.3 (1/16) | 68.8 (11/16) | 18.8 (3/16) | 12.5 (2/16) |
| 5 | 29.2 (7/24) | 18.8 (3/16) | 7.1 (1/14) | 0.0 (0/14) | 31.3 (5/16) |
| 6 | 61.9 (13/21) | 6.3 (1/16) | 40.0 (2/5) | 26.7 (4/15) | 36.4 (4/11) |
| 7 | 69.6 (16/23) | 0.0 (0/16) | 85.7 (12/14) | 43.8 (7/16) | 81.3 (13/16) |
| 8 | 37.5 (9/24) | 0.0 (0/16) | 37.5 (6/16) | 0.0 (0/16) | 50.0 (8/16) |
| 9 | 62.5 (15/24) | 6.3 (1/16) | 73.3 (11/15) | 6.3 (1/16) | 87.5 (14/16) |
| Mean | 52.5 | 18.8 | 52.9 | 27.8 | 41.1 |
The univariable environment model reported a significant effect of the environment (barn/trailer/slaughterhouse) on the probability of MDR within E. coli isolates (Table 3). Bonferroni‐adjusted pairwise comparisons showed the probability for barn samples was significantly greater than those from trailers (odds ratio [OR] = 13.9, 95% confidence interval [CI] = [4.5, 43.2], p ≤ 0.0001) and slaughterhouse pens (OR = 6.9, 95% CI = [2.4, 19.6], p ≤ 0.0001). The ICC of the random effect was estimated as 39.4% and 17.4% for MDR probability among calf cohorts and among samples in the same cohort, respectively.
TABLE 3.
Final univariable logistic regression analysis of the environment of sample collection (at barns of original dairy farms, at trailers and at holding pens of the slaughterhouse) associated with the multidrug resistance (MDR) within E. coli isolates.
| Outcome | Variable | Odds ratio a (95% CI) | p |
|---|---|---|---|
| MDR (Yes/No) | Environment | ||
| Barn | Reference | ||
| Trailer | 0.07 (0.03–0.18) | < 0.0001 | |
| Slaughterhouse | 0.15 (0.06–0.34) | < 0.0001 |
Odds ratio represents the fold change of odds of the MDR within E. coli isolates.
The multivariable interaction model reported a significant effect of the time of sampling (OR = 6.78, 95% CI = [3.40, 13.51], p < 0.0001) on the probability of MDR within E. coli isolates (Table 4). Pairwise comparisons reported isolates collected after calves had been in the trailers (OR = 9.8, 95% CI = [3.7, 26.3], p < 0.0001) and slaughter facility (OR = 4.7, 95% CI = [1.8, 12.1], p = 0.002) had a greater odds of being MDR than isolates taken prior to calf entry from the same environments. The interaction between environment and sampling time had a nonsignificant effect on the probability of MDR (p = 0.28). The ICC of the random effect was estimated as 23.5% and 26.3% for MDR probability among calf cohorts and among samples in the same cohort, respectively.
TABLE 4.
Final multivariable logistic regression analysis of interaction between the environment (at trailers and at holding pens of the slaughterhouse) and time point (before and after calves entered the environment) of sample collection associated with the multidrug resistance (MDR) within E. coli isolates. Samples were only collected once at barns and were excluded from the analysis.
| Outcome | Variable | Odds ratio a (95% CI) | p |
|---|---|---|---|
| MDR (Yes/No) | Environment | ||
| Trailer | Reference | ||
| Slaughterhouse | 1.32 (0.67–2.60) | 0.21 | |
| Sampling time | |||
| Before calves entered | Reference | ||
| After calves entered | 6.78 (3.40–13.51) | < 0.001 | |
| Environment × Time | 0.48 (0.12–1.85) | 0.28 |
Odds ratio represents the fold change of odds of the MDR within E. coli isolates.
3.3. Resistance to Critically Important Antimicrobials
Of all E. coli isolates (n = 679), 52.3% (n = 355) were resistant to at least one critically important antimicrobial as defined by the WHO (ceftiofur, ceftriaxone, azithromycin, ciprofloxacin, nalidixic acid, ampicillin, amoxicillin‐clavulanic acid, gentamycin, streptomycin, meropenem). In barns, 65.7% of isolates (113/172) were resistant to at least one critically important antimicrobial, followed by 48.4% in trailers (125/258), and 47.0% in the slaughterhouse (117/249).
There were 88 isolates showing resistance to highest priority critically important antimicrobials. Notably, there were four isolates resistant to more than three of these antimicrobials, one of which was resistant to all five. Also notable was that seven isolates were resistant to both third‐generation cephalosporins and the fluoroquinolone ciprofloxacin (Table S2). Whole genome sequencing was performed for four of these isolates. Three isolates carried qnrS1, a plasmid‐mediated gene known to confer resistance to fluoroquinolones, while all isolates carried bla TEM‐1B, an extended‐spectrum beta‐lactamase (ESBL) gene that confers resistance to oxyimino‐cephalosporins, such as cefotaxime and ceftriaxone. Carriage of other bla genes was also detected for three isolates (Table S3). All isolates carried acrAB, marR, rpoB, soxS and tolC which are involved with reduced susceptibility to ciprofloxacin (van der Putten et al. 2018). No isolates carried known point mutations that mediate fluoroquinolone resistance; however, unknown point mutations were identified in parC for all isolates, and in gyrA for two isolates (196‐1, 216‐1). Known plasmids, including IncFIB, IncFIC, IncFII, IncHI2, IncHI2A, IncN and IncY, were identified in three out of four isolates. The isolate that did not carry any known plasmids also lacked qnrS1, while all other isolates carried multiple plasmids.
4. Discussion
This study aimed to characterise levels of AMR in environments of veal calf production. Our study found large variance in resistance between calf cohorts and time points. Additionally, coresistance to fluoroquinolones and third‐generation cephalosporins (Table S2) was found, suggesting an important public health concern as these antimicrobials are commonly prescribed antimicrobials for human medicine. Also, resistance was found to ciprofloxacin (n = 23, 3.4%) and chloramphenicol (n = 231, 34.0%) (Table 1) despite these antimicrobials not being available for use in veal calves in the United States (Food and Drug Administration 2022; Lambrecht et al. 2018).
Resistance in this study was lower than one previous study that found 74.6% (n = 406/544) of environmental dairy calf faecal isolates resistant to chloramphenicol and 25.4% (n = 138/544) to ciprofloxacin, plus greater prevalence of resistance to AMP, GM, NA, S and TIO than our study (Afema, Davis, and Sischo 2019). Another study found carbapenemase‐producing Enterobacteriaceae in cattle raised ‘antimicrobial‐free’ (Vikram and Schmidt 2018). Such variation implies levels of resistance to these antimicrobials is not consistent from farm to farm, which could be due to different practices in each farm/region, the different ages of calves sampled in each study, or methodological differences in studies. These data also show persistence of AMR is not solely dependent on antimicrobial use. For instance, environments within some farms such as water troughs and calving areas have been shown by Watson et al. (2011) to harbour extended‐spectrum beta‐lactamase (ESBL) producing E. coli, indicating transmission of ESBL‐producing bacteria across sites or between farms is likely common. Additionally, faecal samples from wild birds tested positive for the same CTX‐M ESBL strain detected on calf farms, indicating that by contaminating the environment, wild birds are a vector for AMR bacteria (Watson et al. 2011).
Resistant organisms from environmental sources, including wildlife such as wild birds, rodents and wildlife faeces, can be transmitted to calves, contaminate food and water and transfer resistance genes to pathogenic organisms (Davies and Wales 2019). We previously showed that environmental exposures preharvest were likely the cause of some Salmonella enterica subsp. enterica lymph node infections found in calves based on genetic comparison (Locke et al. 2022). Importantly, prevalence of resistant S. enterica infections has been shown to correlate with the resistance of commensal E. coli in the environment (DeFrancesco et al. 2004). Additionally, Kim, Van Kessel, and Haley (2021) showed that highly resistant E. coli encoding transferrable resistance genes were present in veal calf faeces, including strains associated with disease in humans (Kim, Van Kessel, and Haley 2021). Combined with our data that suggest transmission of AMR genes through environments by veal calves, calves spreading resistance acquired from environmental sources is a plausible reason for the differences in MDR isolates between environments sampled (Tables 3, 4, 5). It appears likely that the barn is the most important source of resistance, and that introduction of calves into an environment can be expected to result in increased levels of resistance (Tables 3 and 4).
This study only looked at one transport company and one harvest facility's holding pens. Results are likely influenced by geographic location, facility practices and protocols and types of livestock on farms and hauled by the transport company. Additionally, it is important to note that data presented in Figures 2 and 3 are merely descriptive and inferences should be made from results of the appropriately constructed model. Control measures focused on environments that previously housed calves, like cleaning and disinfection or biosecurity practices related to controlling contact with wildlife and their faeces, may help lower the risk of these environments as a source of AMR (Davies and Wales 2019; Watson et al. 2011). Stricter protocols, like consistent use of footbaths or use of low‐pressure foamers over pressure washing may reduce spread of antimicrobial‐resistant bacteria by decreasing the probability of calves or their hides becoming contaminated with AMR bacteria (Van Os 2021). In our results, a large variation in MDR levels between cohorts (the proportion of MDR E. coli ranged from 0% to 100% between cohorts in 3/5 time points) may suggest specific conditions or practices are associated with greater prevalence of resistance. Antimicrobial use data were not available on the cohorts in this study; however, antimicrobial use data from similar cohorts has been previously described (Cheng et al. 2022). Thus, additional work is necessary to find additional interventions that could reduce drug resistance in these environments. Given our data, external biosecurity measures to reduce the introduction of AMR into barns as well as internal control measures in barns (specifically cleaning and disinfection) may be most promising for future studies.
5. Conclusions
The findings in this study show that AMR, including MDR, in preharvest environments, including the trailer and slaughterhouse, increases as calves pass through these environments and gives a better understanding of specific types of resistance present. Substantial inter‐cohort variance suggests cohort‐level practices may affect AMR, giving incentive to investigate on‐farm interventions that will reduce spread of drug‐resistant organisms. The presence of coresistance to fluoroquinolones and third‐generation cephalosporins is a point of concern, and the epidemiology of these AMR bacteria must be understood to limit their dissemination.
Conflicts of Interest
The authors declare no conflicts of interest.
Supporting information
Table S1. Phenotypes of environmental E. coli from the environments where calves were housed and transported in the longitudinal study. AMR phenotypes with a frequency of over 2% in each environment are shown. For trailer and slaughterhouse isolates, phenotypes are shown for samples taken before and after calves passed through the environment.
Table S2. Phenotypes of isolates showing resistance to the fluoroquinolone ciprofloxacin and 3rd generation cephalosporins. Isolates in bold were sequenced by whole genome sequencing.
Table S3. Presence of genes (green) identified from sequencing for environmental E. coli isolates showing co‐resistance to third‐generation cephalosporins and fluoroquinolones.
Acknowledgements
We would like to thank the veal producers, slaughterhouse personnel and livestock trailer drivers for their collaboration.
Funding: This work was supported by the USDA National Institute of Food and Agriculture, grant number: 2018‐68003‐27466.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Afema, J. A. , Davis M. A., and Sischo W. M.. 2019. “Antimicrobial Use Policy Change in Pre‐Weaned Dairy Calves and Its Impact on Antimicrobial Resistance in Commensal Escherichia coli: A Cross Sectional and Ecological Study.” BMC Microbiology 19, no. 1: 217. 10.1186/s12866-019-1576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agga, G. E. , Galloway H. O., Netthisinghe A. M. P., Schmidt J. W., and Arthur T. M.. 2022. “Tetracycline‐Resistant, Third‐Generation Cephalosporin–Resistant, and Extended‐Spectrum β‐Lactamase–Producing Escherichia coli in a Beef Cow‐Calf Production System.” Journal of Food Protection 85, no. 11: 1522–1530. 10.4315/jfp-22-178. [DOI] [PubMed] [Google Scholar]
- Berends, M. S. , Luz C. F., Friedrich A. W., Sinha B. N., Albers C. J., and Glasner C.. 2022. “Amr: An R Package for Working With Antimicrobial Resistance Data.” Journal of Statistical Software 104, no. 3: 1–31. 10.18637/jss.v104.i03. [DOI] [Google Scholar]
- BioRender . n.d. “Scientific Image and Illustration Software.” https://www.biorender.com/.
- Bland, J. M. , and Altman D. G.. 1995. “Statistics Notes: Multiple Significance Tests: The Bonferroni Method.” BMJ 310, no. 6973: 170. 10.1136/bmj.310.6973.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolaia, V. , Kaas R. S., Ruppe E., et al. 2020. “ResFinder 4.0 for Predictions of Phenotypes From Genotypes.” Journal of Antimicrobial Chemotherapy 75, no. 12: 3491–3500. 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli, A. , Zankari E., García‐Fernández A., et al. 2014. “ In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing.” Antimicrobial Agents and Chemotherapy 58, no. 7: 3895–3903. 10.1128/aac.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center for Genomic Epidemiology . n.d. “Center for Genomic Epidemiology.” https://www.genomicepidemiology.org/.
- CDC . 2019. Antibiotic Resistance Threats in the United States. Atlanta, GA: U.S. Department of Health and Human Services. https://www.cdc.gov/drugresistance/biggest‐threats.html. [Google Scholar]
- Checcucci, A. , Trevisi P., Luise D., et al. 2020. “Exploring the Animal Waste Resistome: The Spread of Antimicrobial Resistance Genes Through the Use of Livestock Manure.” Frontiers in Microbiology 11: 1416. 10.3389/fmicb.2020.01416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, T. , Almeida B. G., Pempek J. A., Masterson M. A., and Habing G. G.. 2022. “The Use of Common Antimicrobial Agents in US Veal Calves.” Zoonoses and Public Health 69, no. 4: 359–369. 10.1111/zph.12928. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute . 2023a. CLSI M100. 33rd ed. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Clinical and Laboratory Standards Institute . 2023b. CLSI VET01S. 6th ed. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Collignon, P. , and McEwen S.. 2019. “One Health—Its Importance in Helping to Better Control Antimicrobial Resistance.” Tropical Medicine and Infectious Disease 4, no. 1: 22. 10.3390/tropicalmed4010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, R. , and Wales A.. 2019. “Antimicrobial Resistance on Farms: A Review Including Biosecurity and the Potential Role of Disinfectants in Resistance Selection.” Comprehensive Reviews in Food Science and Food Safety 18, no. 3: 753–774. 10.1111/1541-4337.12438. [DOI] [PubMed] [Google Scholar]
- DeFrancesco, K. A. , Cobbold R. N., Rice D. H., Besser T. E., and Hancock D. D.. 2004. “Antimicrobial Resistance of Commensal Escherichia coli From Dairy Cattle Associated With Recent Multi‐Resistant Salmonellosis Outbreaks.” Veterinary Microbiology 98, no. 1: 55–61. 10.1016/j.vetmic.2003.10.017. [DOI] [PubMed] [Google Scholar]
- FDA . 2022. Extralabel use and antimicrobials. Silver Spring, MD: U.S. Food and Drug Administration. https://www.fda.gov/animal‐veterinary/antimicrobial‐resistance/extralabel‐use‐and‐antimicrobials. [Google Scholar]
- Gaire, T. N. , Scott H. M., Sellers L., Nagaraja T. G., and Volkova V. V.. 2021. “Age Dependence of Antimicrobial Resistance Among Fecal Bacteria in Animals: A Scoping Review.” Frontiers in Veterinary Science 7: 622495. 10.3389/fvets.2020.622495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, H. , Browne W., and Rasbash J.. 2002. “Partitioning Variation in Multilevel Models.” Understanding Statistics 1: 223–231. 10.1207/S15328031US0104_02. [DOI] [Google Scholar]
- Hanon, J.‐B. , Jaspers S., Butaye P., et al. 2015. “A Trend Analysis of Antimicrobial Resistance in Commensal Escherichia coli From Several Livestock Species in Belgium (2011–2014).” Preventive Veterinary Medicine 122, no. 4: 443–452. 10.1016/j.prevetmed.2015.09.001. [DOI] [PubMed] [Google Scholar]
- Hutchinson, H. , Finney S., Muñoz‐Vargas L., Feicht S., Masterson M., and Habing G.. 2017. “Prevalence and Transmission of Antimicrobial Resistance in a Vertically Integrated Veal Calf Production System.” Foodborne Pathogens and Disease 14, no. 12: 711–718. 10.1089/fpd.2017.2310. [DOI] [PubMed] [Google Scholar]
- Kenward, M. G. , and Roger J. H.. 1997. “Small Sample Inference for Fixed Effects From Restricted Maximum Likelihood.” Biometrics 53, no. 3: 983–997. 10.2307/2533558. [DOI] [PubMed] [Google Scholar]
- Kim, S. W. , Van Kessel J. A., and Haley B. J.. 2021. “Genome Sequences of Antimicrobial‐Resistant Escherichia coli Isolated From Veal Calves in the USA.” Journal of Global Antimicrobial Resistance 26: 69–73. 10.1016/j.jgar.2021.04.024. [DOI] [PubMed] [Google Scholar]
- Komp Lindgren, P. , Åsa K., and Hughes D.. 2003. “Mutation Rate and Evolution of Fluoroquinolone Resistance in Escherichia coli Isolates From Patients With Urinary Tract Infections.” Antimicrobial Agents and Chemotherapy 47, no. 10: 3222–3232. 10.1128/aac.47.10.3222-3232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrecht, E. , Van Meervenne E., Boon N., et al. 2018. “Characterization of Cefotaxime‐ and Ciprofloxacin‐Resistant Commensal Escherichia coli Originating From Belgian Farm Animals Indicates High Antimicrobial Resistance Transfer Rates.” Microbial Drug Resistance 24, no. 6: 707–717. 10.1089/mdr.2017.0226. [DOI] [PubMed] [Google Scholar]
- Lammie, S. L. , and Hughes J. M.. 2016. “Antimicrobial Resistance, Food Safety, and One Health: The Need for Convergence.” Annual Review of Food Science and Technology 7, no. 1: 287–312. 10.1146/annurev-food-041715-033251. [DOI] [PubMed] [Google Scholar]
- Locke, S. R. , Pempek J. A., Meyer R., et al. 2022. “Prevalence and Sources of Salmonella Lymph Node Infection in Special‐Fed Veal Calves.” Journal of Food Protection 85, no. 6: 906–917. 10.4315/jfp-21-410. [DOI] [PubMed] [Google Scholar]
- Magiorakos, A.‐P. , Srinivasan A., Carey R. B., et al. 2012. “Multidrug‐Resistant, Extensively Drug‐Resistant and Pandrug‐Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance.” Clinical Microbiology and Infection 18, no. 3: 268–281. 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- Markey, B. K. , Leonard F., Archambault M., Culinane A., and Maguire D.. 2013. Clinical Veterinary Microbiology. Second Edition Edinburgh, UK: Mosby. [Google Scholar]
- McEwen, S. A. , and Collignon P. J.. 2018. “Antimicrobial Resistance: A One Health Perspective.” Microbiology Spectrum 6, no. 2: 2017. 10.1128/microbiolspec.arba-0009-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, R. G. , Tate C. R., Mallinson E. T., and Scherrer J. A.. 1991. “Xylose‐Lysine‐Tergitol 4: An Improved Selective Agar Medium for the Isolation of Salmonella .” Poultry Science 70, no. 12: 2429–2432. 10.3382/ps.0702429. [DOI] [PubMed] [Google Scholar]
- Murray, C. J. , Ikuta K. S., Sharara F., et al. 2022. “Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis.” Lancet 399, no. 10325: 629–655. 10.1016/s0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. L. , Harris A. D., Johnson J. A., Silbergeld E. K., and Morris J. G.. 2002. “Animal Antibiotic Use Has an Early but Important Impact on the Emergence of Antibiotic Resistance in Human Commensal Bacteria.” Proceedings of the National Academy of Sciences 99, no. 9: 6434–6439. 10.1073/pnas.082188899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA . 2017. National Poultry Improvement Plan Program Standards. 2017. Washington, DC: U.S. Department of Agriculture, Animal and Plant Health Inspection Service. https://www.poultryimprovement.org/documents/ProgramStandardsJanuary2017.pdf. [Google Scholar]
- Um, M. M. , Barraud O., Kérourédan M., et al. 2015. “Comparison of the Incidence of Pathogenic and Antimicrobial‐Resistant Escherichia coli Strains in Adult Cattle and Veal Calf Slaughterhouse Effluents Highlighted Different Risks for Public Health.” Water Research 88: 30–38. 10.1016/j.watres.2015.09.029. [DOI] [PubMed] [Google Scholar]
- Underwood, A. 2020. “GHRU (Genomic Surveillance of Antimicrobial Resistance) Retrospective 1 Bioinformatics Methods v.4.” Center for Genomic Pathogen Surveillance. https://www.protocols.io/view/ghru‐genomic‐surveillance‐of‐antimicrobial‐resista‐bp2l6b11kgqe/v4.
- University of Wisconsin . 2018. Bovine Environmental Sampling Instructions for Salmonella spp. Madison: University of Wisconsin. [Google Scholar]
- USDA Food Safety and Inspection Service . 2013. “Safe Food Handling and Preparation | Food Safety and Inspection Service.” https://www.fsis.usda.gov/food‐safety/safe‐food‐handling‐and‐preparation.
- van der Putten, B. C. , Remondini D., Pasquini G., Janes V. A., Matamoros S., and Schultsz C.. 2018. “Quantifying the Contribution of Four Resistance Mechanisms to Ciprofloxacin Mic in Escherichia coli: A Systematic Review.” Journal of Antimicrobial Chemotherapy 74, no. 2: 298–310. 10.1093/jac/dky417. [DOI] [PubMed] [Google Scholar]
- Van Os, J. 2021. Two Heads Are Better Than One: A Starter Guide to Pairing Dairy Calves. Madison: University of Wisconsin. [Google Scholar]
- Veal Quality Assurance Program . 2018. Veal Quality Assurance Certification Resource Manual. Gladstone: Veal Quality Assurance Program. [Google Scholar]
- Vikram, A. , and Schmidt J. W.. 2018. “Functional bla kpc‐2 Sequences Are Present in U.S. Beef Cattle Feces Regardless of Antimicrobial Use.” Foodborne Pathogens and Disease 15, no. 7: 444–448. 10.1089/fpd.2017.2406. [DOI] [PubMed] [Google Scholar]
- Vinayamohan, P. G. , Locke S. R., Portillo‐Gonzalez R., Renaud D. L., and Habing G. G.. 2022. “Antimicrobial Use and Resistance in Surplus Dairy Calf Production Systems.” Microorganisms 10, no. 8: 1652. 10.3390/microorganisms10081652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, E. , Jeckel S., Snow L., et al. 2011. “Epidemiology of Extended Spectrum Beta‐Lactamase E. coli (CTX‐M‐15) on a Commercial Dairy Farm.” Veterinary Microbiology 154, no. 3–4: 339–346. 10.1016/j.vetmic.2011.07.020. [DOI] [PubMed] [Google Scholar]
- Williams, S. M. 2016. A Laboratory Manual for the Isolation, Identification and Characterization of Avian Pathogens. Jacksonville, FL: American Association of Avian Pathologists. [Google Scholar]
- WHO . 1970. Critically Important Antimicrobials for Human Medicine. 1970. Geneva, Switzerland: World Health Organization. https://apps.who.int/iris/handle/10665/312266. [Google Scholar]
- WHO . 2018. Critically Important Antimicrobials for Human Medicine 24–25. Geneva, Switzerland: World Health Organization. [Google Scholar]
- WHO . 2021. Antimicrobial Resistance. Geneva, Switzerland: World Health Organization. https://www.who.int/news‐room/fact‐sheets/detail/antimicrobial‐resistance. [Google Scholar]
- Zankari, E. , Hasman H., Kaas R. S., et al. 2012. “Genotyping Using Whole‐Genome Sequencing Is a Realistic Alternative to Surveillance Based on Phenotypic Antimicrobial Susceptibility Testing.” Journal of Antimicrobial Chemotherapy 68, no. 4: 771–777. 10.1093/jac/dks496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Phenotypes of environmental E. coli from the environments where calves were housed and transported in the longitudinal study. AMR phenotypes with a frequency of over 2% in each environment are shown. For trailer and slaughterhouse isolates, phenotypes are shown for samples taken before and after calves passed through the environment.
Table S2. Phenotypes of isolates showing resistance to the fluoroquinolone ciprofloxacin and 3rd generation cephalosporins. Isolates in bold were sequenced by whole genome sequencing.
Table S3. Presence of genes (green) identified from sequencing for environmental E. coli isolates showing co‐resistance to third‐generation cephalosporins and fluoroquinolones.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


