Abstract
The mismatch repair (MMR) system plays a crucial role in the maintenance of DNA replication fidelity and genomic stability. The clinical value of the MMR molecular marker as an immunotherapy for advanced solid tumors has been documented. However, this therapy is not effective in some patients. This study aimed to develop an MMR-related molecular prognostic model for identifying appropriate populations of stomach adenocarcinoma (STAD) for better treatment outcome. The MMR genes expression data were downloaded from TCGA and CCLE databases. The expression of four MMR genes, construction of a prognostic risk model, and assessment of immune infiltration in STAD were performed using Xiantao online tool. GEPIA2 was used to explore the association of MMR genes expression with clinical stage and overall survival. The frequency and prognostic value of MMR genes in STAD were conducted on the cBioPortal. The MLH1 co-expression network was established based on the LinkedOmics database. This study found that the expression of MSH2, MSH6 and PMS2 was up-regulated in STAD tissues. Moreover, differential MMR genetic expression levels were not significantly correlated with the clinical stages of STAD. Besides, no significant difference in PFS or OS was observed in STAD patients with or without MMR genetic alteration. Moreover, MLH1 and MSH2 were used to establish a prognostic risk model. The immune infiltration levels of most immune cells were upregulated in the high-risk group with elevated expression of PDCD1 and low TMB score. Finally, we found that MLH1 was an independent predictor of STAD prognosis among the four MMR genes. An MMR-related prognostic model for STAD was constructed based on genes. This model provides a new therapeutic concept for the diagnosis and treatment of STAD.
Keywords: Stomach adenocarcinoma, Mismatch repair, MLH1, MSH2, Prognosis
Subject terms: Cancer, Computational biology and bioinformatics, Genetics, Immunology, Systems biology, Oncology
Introduction
Gastric cancer is one of the most commonly diagnosed malignant tumors of the digestive system worldwide. According to the latest GLOBOCAN data, gastric cancer ranks fifth in cancer incidence with approximately 1.03 million new cases and fourth in cancer-related mortality with an estimated 728,685 deaths reported1. The development of gastric cancer is a multi-factor, multi-step complex biological process, but its exact pathogenesis is not fully understood. Evidence suggests that genetic and environmental factors may independently or jointly contribute to the development of gastric cancer. Due to the lack of early specific symptoms, most patients are diagnosed at advanced stage and their 5-year survival rate is below 30%2. Despite remarkable progress in the formulation of diagnostic and treatment technologies, the prognosis of gastric cancer is poor, which places a heavy burden on families and society3. Thus, further studies are needed to explore the molecular mechanisms underlying gastric cancer development to accelerate the formulation of strategies for the diagnosis and treatment of gastric cancer.
Microsatellites, also defined as short tandem repeats (1–6 nucleotides), are randomly distributed throughout the human genome and prone to high mutation rates4. The mismatch repair (MMR) system consists of a series of mismatch repair proteins that include the MutL homolog 1 (MLH1), MutS homolog 2 (MSH2), MutS homolog 6 (MSH6), post meiotic segregation increased 2 (PMS2), among others. Previous studies found that the MMR system may be involved in the maintenance of the DNA replication fidelity and genomic stability. Therefore, impaired MMR system may result in abnormal replication of microsatellite sequences and will further lead to microsatellite instability (MSI). Notably, the MSI is classified as microsatellite Instability High (MSI-H), microsatellite Instability Low (MSI-L) and microsatellite Stability (MSS), depending on the stable states. The available techniques for detecting microsatellite status include immunohistochemistry (IHC), polymerase chain reaction (PCR) and next Generation Sequencing (NGS). However, the expensive cost limits the clinical promotion of NGS. Clinical evidence indicates a high concordance (90–95%) between IHC and PCR. Because of its simplicity, convenience, low price, high specificity and sensitivity, IHC is more commonly applied to detect the microsatellite status. Under the normal expression of the four MMR proteins (MLH1, NSH2, MSH6 and PMS2), the states of MMR (pMMR) and microsatellite stability (MSS) or microsatellite instability Low (MSI-L) exist in a stable homeostasis. Loss of one or more of these four proteins was found to result in deficient MMR (pMMR) and microsatellite instability High (MSI-H)5,6.
MSI was first identified in hereditary nonpolyposis colorectal cancer (HNPCC)7and has since been detected in other cancers such as endometrial carcinoma, gastric cancer and colorectal cancer8. Considering its high heterogeneity, traditional histological classification may not be adequate in guiding the clinical treatment of gastric cancer. According to the Cancer Genome Atlas in 2014, gastric cancer is classified into four subtypes: Genomically Stable (GS), Microsatellite Instability (MSI), Epstein-Barr Virus-infected (EBV), and Chromosomal Instability (CIN)9. As one of the classic biomarkers of gastrointestinal tumors, the MMR system has been used to screen for Lynch syndrome and guide the treatment of patients with stage II colorectal cancer. For instance, the KEYNOTE-158 study confirmed the clinical role of MSI-H molecular marker in the application of immunotherapy in advanced solid tumors10. However, some patients fail to derive satisfactory clinical benefit from immunotherapy. Thus, more effective prognostic molecular biomarkers are needed to facilitate the screening of susceptible patient populations. In this study, we established a prognostic model based on MMR proteins to identify appropriate populations of gastric cancer to improve better treatment outcomes.
Methods
Data resource collection
The complete clinical data of 407 stomach adenocarcinoma (STAD) tissues and 32 paracancerous tissues were searched and downloaded from the TCGA database (https://portal.gdc.cancer.gov/). Data for each STAD cell line was derived from the CCLE database (https://portals.broadinstitute.org/ccle/). The Xiantao online tool (https://www.xiantaozi.com/) was employed to transform and visualize the clinical characteristic data and RNA transcriptome data, construct a risk model, and analyze the immune status of STAD. The association of MMR genes expression with clinical stage and overall survival was explored using the GEPIA2 (http://gepia2.cancer-pku.cn/#index). The frequency and prognosis of MMR genes in STAD were determined using the cBioPortal (http://cbioportal.org).
Construction of clinical predictive model
LASSO Cox regression was adopted to construct the prognostic risk model. The risk score was calculated through Xiantao online tool using the following formula: Risk score = 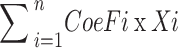 (
(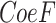 represents gene coefficient,
represents gene coefficient, 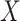 represents gene expression level). Next, we categorized the STAD samples into high and low risk groups based on median of risk score. Kaplan-Meier survival curve and receiver operating characteristic (ROC) curve were employed to evaluate the prediction potential of the model. After integrating the risk scores and clinical features, a nomogram was designed and calibration curves were employed to evaluate the accuracy of the risk model prediction using the Xiantao online tool.
represents gene expression level). Next, we categorized the STAD samples into high and low risk groups based on median of risk score. Kaplan-Meier survival curve and receiver operating characteristic (ROC) curve were employed to evaluate the prediction potential of the model. After integrating the risk scores and clinical features, a nomogram was designed and calibration curves were employed to evaluate the accuracy of the risk model prediction using the Xiantao online tool.
Immune infiltration in tumor microenvironment of STAD
The stromal score, immune score, and ESTIMATE score for each sample in different risk groups were calculated using the Xiantao online tool. The immune infiltration of different risk groups was further integrated with the ssGSEA algorithm in Xiantao online tool based on the immune cell surface markers. Somatic mutation data of STAD were acquired to calculate TMB scores in high and low risk groups.
Gene function enrichment analysis
Functional enrichment analysis for the MMR gene was performed via the Xiantao online tool. Besides, an MLH1 co-expression network was constructed on the LinkedOmics database (http://www.linkedomics.org). Meanwhile, Gene Ontology (Biological Process) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of MLH1 co-expression genes were performed on the same database11–13.
Statistical analysis
Differential distribution of clinical characteristic data in subgroups was analyzed using the Wilcoxon test. Kaplan-Meier survival analysis and log-rank test were employed to determine the survival for the various risk groups. Univariate and multivariate analysis were performed using Cox proportional hazard regression models. Correlation analysis between genes and the prognosis was determined with the Spearman test. Statistical significance was defined as p-value < 0.05.
Results
Differential mRNA expression of four MMR genes in STAD
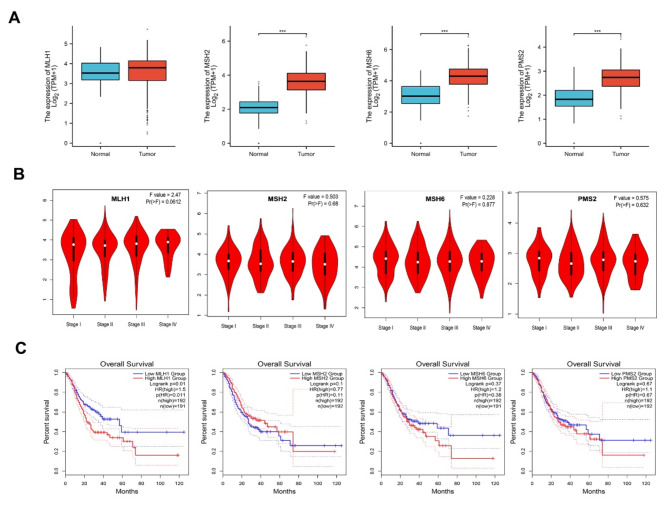
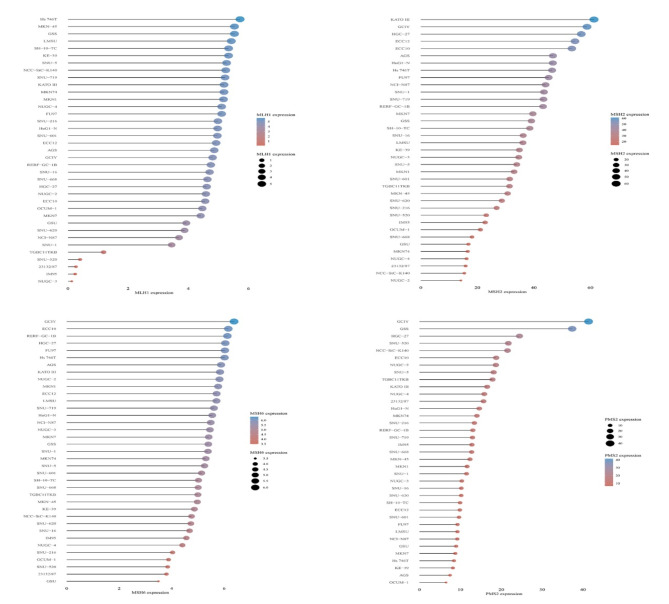
The mRNA expression levels of four MMR genes in STAD and normal tissues were determined on the TCGA database. The results showed that MSH2, MSH6 and PMS2 expression were significantly up-regulated in STAD tissues. There was no significant difference in MLH1 expression between tumor and normal tissues (Fig. 1A). Moreover, there was no significant association between differential MMR genetic expression levels and the clinical stages of STAD (Fig. 1B). Kaplan-Meier survival analysis revealed that only lower MLH1 expression was associated with favorable overall survival, whereas there was no significant relationship between other three MMR mRNA expression and the overall survival outcome (Fig. 1C). The mRNA expression of four MMR proteins in 37 cell lines of STAD was determined using the Cell dataset (Fig. 2).
Fig. 1.
The mRNA expression profile of the four MMR genes in STAD. A : The mRNA expression of MLH1, MSH2, MSH6 and PMS2 in STAD and normal tissues in TCGA database (*** p < 0.001). B : MLH1, MSH2, MSH6 and PMS2 expression at various clinical stages of STAD. C : The effect of MLH1, MSH2, MSH6 and PMS2 expression on overall survival in STAD. Lower MLH1 expression was associated with better overall survival. MSH2, MSH6 and PMS2 were not significantly associated with the overall survival ( p < 0.05).
Fig. 2.
The expression profile of the four MMR genes in STAD cell lines.
The MMR genetic variations in STAD
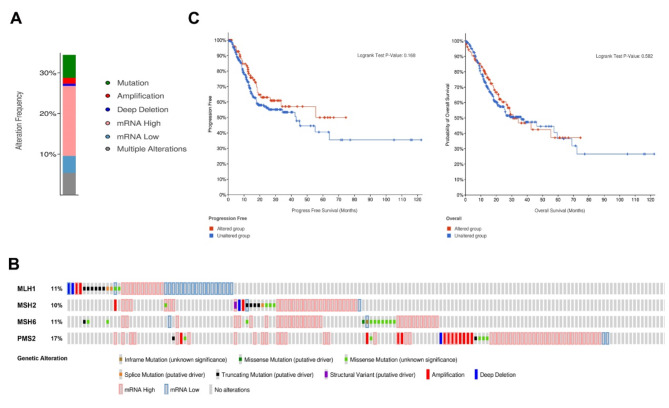
Next, the role of these four MMR genetic alterations in STAD progression was investigated on the cBioportal database (TCGA, PanCancer Atlas). The results showed that the MMR genetic changes occurred in 116 (29%) of STAD patients, including mRNA high (15.23%), gene mutation (6.14%), gene multiple alterations (4.17%), gene amplification (1.97%), gene deep deletion (0.74%) and mRNA low (0.25%) (Fig. 3A). Gene variation rates were detected in 6% (MLH1), 9% (MSH2), 9% (MSH6) and 14% (PMS2), respectively (Fig. 3B). However, no significant difference in PFS or OS was observed in STAD patients with or without genetic alterations (Fig. 3C).
Fig. 3.
The four MMR genetic variations in STAD. A , B : The summary of genetic alteration frequency in MMR proteins in STAD. C : Kaplan-Meier curve showing no significant differences in PFS and OS of STAD with or without genetic alteration.
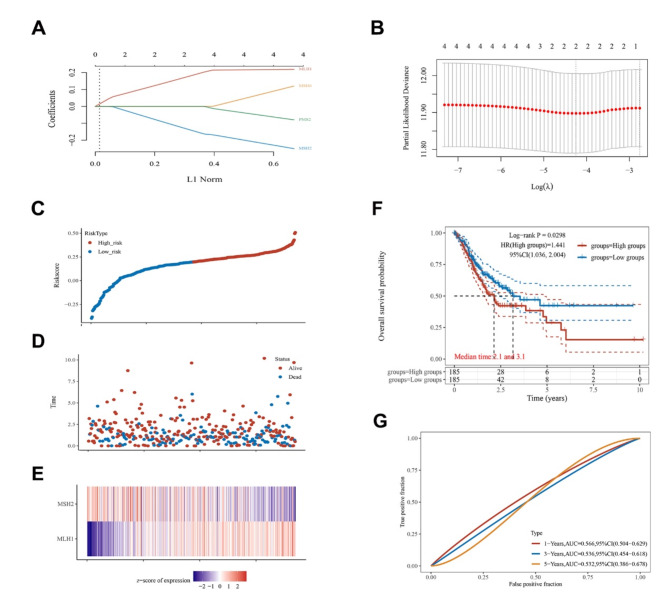
Prognosis value of MMR genetic risk model in STAD
To test the prognosis value of the four MMR genes in STAD, we established a prognostic risk model using the LASSO regression. The following formula was used: Risk Score = (0.1746)*MLH1+(−0.1332)*MSH2 (Fig. 4A-B). The STAD patients were classified into high risk and low risk group based on the median value of risk score. The death frequencies increased with risk score, and the survival rate of high-risk group was worse compared with that of the low-risk group (Fig. 4C-D). The mRNA expression of MLH1 and MSH2 was presented using heat maps (Fig. 4E). Kaplan-Meier survival curve analysis demonstrated that patients in the high-risk group had significantly poorer prognosis compared to those in the low- risk group (Fig. 4F). Analysis of the ROC curve for the efficacy of risk model prediction, yielded the AUC values of 0.566 (1-years OS), 0.536 (3-years OS), and 0.532 (5-years OS) (Fig. 4G).
Fig. 4.
The prognostic performance of MMR genes in STAD. A : LASSO regression coefficient distribution. B : The optimal parameters determined by cross-validated LASSO regression. C : Risk scores in patients with STAD in high and low risk group. D : Heat maps showing the survival status of STAD patients. E : MLH1 and MSH2. F : Kaplan-Meier survival curve illustrating that patients in high-risk group had worse prognosis than those in low- risk group. G : The AUC values were 0.566 (1-years OS), 0.536 (3-years OS), and 0.532 (5-years OS).
Establishment of nomogram for risk model in STAD
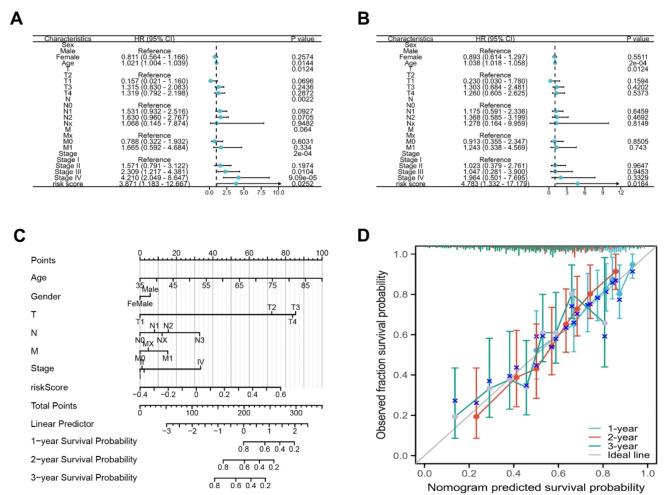
Further results showed that the risk score was as an independent prognostic factor for STAD progression as evidenced by results of the univariate and multivariate Cox regression analyses (Fig. 5A-B). Subsequently, a nomogram was constructed to predict the survival outcomes of patient in low and high-risk groups (Fig. 5C). Calibration curves revealed that the prediction results of the nomogram were in agreement with the observed fraction survival probability (Fig. 5D).
Fig. 5.
Establishment of nomogram for risk model in STAD. A : Forest plot of univariate Cox regression analysis between risk score and clinical data. B : Forest plot of multivariate Cox regression analysis between risk score and clinical data. C : A nomogram model for predicting STAD based on risk score and clinical data. D : The calibration curves of nomogram predicted survival probability agreed well with observed fraction survival probability at 1-year, 2-year and 3-year survival.
Analysis of immune status in high and low risk groups of STAD
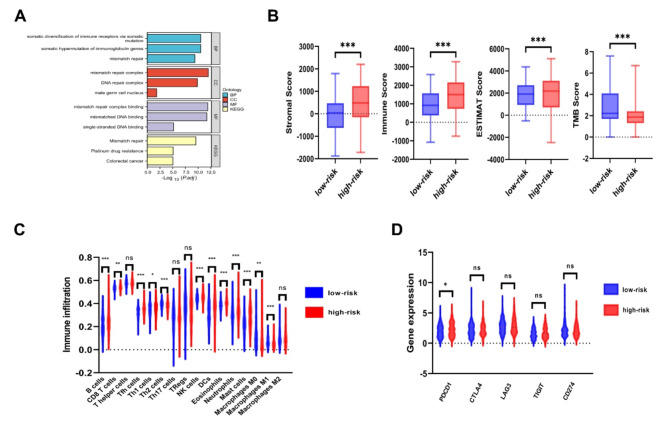
GO and KEGG enrichment analysis indicated that the four MMR genes were primarily associated with the somatic diversification of immune receptors via somatic mutation, somatic hypermutation of immunoglobulin genes, and mismatch repair (Fig. 6A). These data suggested that abnormal expression of four MMR genes may disrupt immune function in STAD patients. Subsequently, we compared the immune status between different risk groups of STAD. It was observed that the stromal score, immune score and ESTIMATE score were significantly higher in the high-risk group, whereas TMB score was significantly elevated in the low-risk group (Fig. 6B). Moreover, we found that the immune infiltration levels of B cells, CD8 + T cells, Tfh cells, Th1 cells, NK cells, DCs, Eosinophils, Neutrophils and Mast cells were higher in the high-risk group than in the low-risk group. The immune infiltration levels of Th2 cells, Macrophages M0 and Macrophages M1 were higher in the low-risk group than in the high-risk group (Fig. 6C). Notably, the expression of PDCD1 was upregulated in the high-risk group among the five known immune checkpoint genes (Fig. 6D).
Fig. 6.
Immune infiltration in high and low risk groups of STAD. A : Four MMR genes were enriched in somatic diversification of immune receptors via somatic mutation, somatic hypermutation of immunoglobulin genes, mismatch repair (GO and KEGG enrichment analysis). B : Stromal score, immune score and ESTIMATE scores in high and low risk groups of STAD. C : The levels of immune cells infiltration in high and low risk groups of STAD. D : The expression of five immune checkpoint genes in high and low risk groups of STAD. * p < 0.05, ** p < 0.01 and *** p < 0.001.
Potential role of MLH1 in STAD
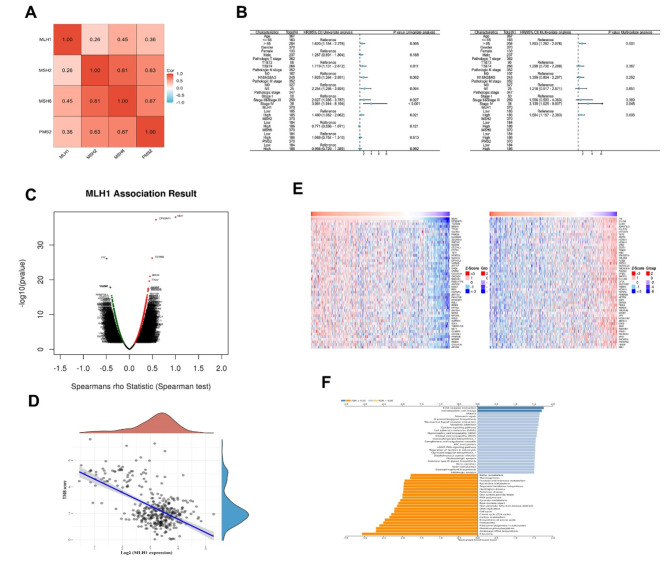
As shown in Fig. 7A, the expression of the four MMR genes was positively correlated with each other. Univariate and multivariate cox regression analyses indicated that age, clinical stage and MLH1 were independent predictors of STAD prognosis (Fig. 7B). Next, we investigated the MLH1-related genes in STAD patients on the LinkedOmics website (Fig. 7C). A negative correlation coefficient was obtained between MLH1 and TMB (Fig. 7D). The top 50 genes that were positively and negatively related with MLH1 are displayed in a heat map (Fig. 7E). KEGG enrichment analysis results indicated that MLH1-related genes participated in several aspects of cell physiological functions such as growth regulation, cell metabolism and apoptosis (Fig. 7F).
Fig. 7.
MLH1 co-expression genes in STAD. A: Correlations of the four MMR genes with each other in STAD. B: Univariate and multivariate Cox regression analysis for the MMR genes and clinical data (Forest plot). C: Correlation between MLH1 and genes differentially expressed in STAD. D: Correlation between MLH1 and TMB score in STAD. E: Heat maps showing the top 50 genes that were positively and negatively associated with MLH1 in STAD. F: KEGG enrichment analysis of MLH1 co-expression genes in STAD.
Discussion
Although the incidence and mortality rates of gastric cancer have been declining over the past decade in China, the burden of this disease remains significant. The pathogenesis of gastric cancer is complicated, requiring the development of precision medicine to improve treatment outcomes and survival of patients. The deoxyribonucleic acid (DNA) is the main carrier of genetic information and its molecular structure is highly conserved to maintain body homeostasis14. The MMR system plays a crucial role in maintaining genomic integrity under normal physiologic condition. MMR deficiency may lead to the activation of oncogenes and silencing of tumor suppressor genes, promoting tumor formation15. Gastric cancer is a heterogeneous disease composed of various subtypes with MSI-H accounting for around 22% of these cases9,16. Previous studies showed MSI-H gastric cancer was more prevalent in elderly women, primarily occurring in the gastric antrum and characterized by rare serosa invasion and relatively favorable prognosis9,17–19. MSI progresses from precancerous lesions to gastric cancer, and early detection of MSI may help to identify high-risk patients20–22. MSI-H/dMMR has been used to detect various types of advanced solid tumors, including stomach, colorectal and endometrial cancers, and is one of the most important biomarkers for predicting immunotherapy efficacy. However, about 30% MSI-H/dMMR patients with gastrointestinal cancer show resistance to immunotherapy23. This calls for deeper investigations into the MMR molecular mechanism to uncover new strategies for controlling gastric cancer progression.
MLH1, MSH2, MSH6 and PMS2 are the dominant members of the MMR system, regulating the DNA replication integrity. First discovered in hereditary nonpolyposis colorectal cancer, MLH1 was implicated in the initiation of the MMR system24,25. Aberrant expression of MLH1 has been linked to the occurrence of gene mutations and methylation, which can potentially cause MMR deficiency and subsequent cell malignant transform. Previous studies have reported that approximately 90% of MSI-H gastric cancers are caused by abnormal downregulation of MLH1 gene expression26,27. MSH2 was the first MMR gene isolated from hereditary non-polyposis colon cancer by Fishel et al. in 1993 . MSH2 can decrease the occurrence of spontaneous mutations by correcting base mismatches and insertion-deletion ring mismatches during DNA replication29. MSH6 also participates in the initiation of DNA repair through binding with MSH2 to form dimer complexes, which involves mismatch recognition, mismatch DNA strands degradation and correction30. PMS2 mainly corrects single base pair mismatch and small fragment mismatch by binding to MLH1 as a dimeric form to maintain genome stability31. The MMR deficiency may be attributed to mismatch genes mutations as well as aberrant epigenetic regulation of some genes such as POLD1 and POLE gene. These genes serve as important biomarkers for predicting the efficacy of immunotherapy in improving the of survival outcome32. In this study, we analyzed the mRNA expression profiles of the four MMR proteins in STAD. The results indicated that the mRNA expressions of MSH2, MSH6 and PMS2 were significantly increased in STAD tissues. However, only MLH1 influenced the prognosis of STAD. Besides, no significant relationships were obtained between MMR gene expression and clinical stage of STAD. Previously, Carolina et al. found no co-relationship between MSI-H gastric cancer and TNM stage based on the pathologic analysis of specimens33. In this study, we included the four MMR genes into LASSO Cox regression analysis, and found that the MLH1 and MSH2 could establish a risk model of STAD. The risk models revealed a worse prognosis in the high-risk patients than that in the low-risk patients. The AUC obtained by the model did not indicate the precision and accuracy of the signature which may be ascribed to the limitations of the sample size and follow-up duration.
The tumor microenvironment has been reported to be a complex and dynamic ecosystem, which is mainly composed of tumor cells, immune cells and extracellular matrix. It may promote or suppress tumor progression, and harbors several therapeutic targets34. Gastric cancer is a highly heterogeneous malignant tumor. Cell dataset revealed that the expression levels of the four MMR genes varied in different gastric cancer cell lines, which further confirmed the molecular heterogeneity of gastric cancer19. B cells can inhibit tumor development by producing tumor-specific antibodies or activating T cells through antigen presentation in most situations. However, some B cell subsets and specific antibodies may inhibit anti-tumor immunity which enhances tumor growth35. CD4 + T cells, known as T helper cells, are divided into multiple subtypes including T helper 1 (Th1), T helper 2 (Th2), T helper 17 (Th17), regulatory T cells (Tregs), follicular helper T cells (Tfh) and so on. These lymphocytes subsets have distinct roles in tumor progression36–38. For instance, Th1 cells not only exert anti-tumor immunity by releasing cytokines, but also directly stimulate the proliferation and activation of CD8 + T cells to kill tumor cells. On the other hand, Th2 cells were originally thought to secrete anti-inflammatory cytokines to promote oncogenesis. However, evidence from recent studies has shown that Th2 cells can recruit other immune cells to suppress tumor cells proliferation. Treg cells mediate immunosuppressive effects in the tumor microenvironment, thereby weakening the anti-tumor specific immune response and promoting tumor evasion from immune surveillance. Th17 cells can secrete cytokines which accelerate tumor cell proliferation and also enhances the expression of vascular endothelial growth factor to induce tumor neovascularization. Tfh cells promote the formation of tertiary lymphoid structure, stimulating tumor immune infiltration and inhibiting tumor growth. In addition, Tfh cells can indirectly enhance anti-tumor immunity mediated by CD8 + T cells by secreting IL-21. CD8 + T cells are the main cytotoxic effector cells involved in the generation of antitumor immune response. CD8 + T cells stimulate apoptosis of tumor cells via the perforin-granase and Fas/FasL pathways39. NK cells are natural immune cells that directly recognize and kill target cells without specific antigen stimulation. Therefore, NK cells are generally considered as the first line of defense against tumor development40. Dendritic cells (DCs) are described as the most powerful professional antigen presenting cells, which efficiently internalize, process and present antigens to immune cells to mediate anti-tumor effect. However, tumor microenvironment may also constrain the function of DCs, resulting in the suppression of anti-tumor immunity41. Macrophages can differentiate into M1 or M2 type depending on their microenvironment. M1 type is mainly involved in inflammatory response and anti-tumor process. Conversely, the M2 type exerts pro-tumor effects and accelerates the proliferation of tumor cells42. Mast cells, eosinophils and neutrophils are important members of body’s natural immune system, which influence tumor progression depending on the state of tumor microenvironment. They may inhibit tumor cell proliferation directly or indirectly, but may also induce neovascularization and matrix remodeling to promote tumor growth43–45. Immune checkpoint molecules have been shown to regulate the activation or inhibition of immune cells in the tumor microenvironment, which may ultimately affect tumor biology behavior46,47. PD-1/PD-L1 immune checkpoint pathways contribute to the occurrence of tumor immune escape by inducing immune cell apoptosis, incapacitation and depletion48. The TMB is defined as the number of somatic nonsynonymous mutations per million base pairs within a specific genomic region, which can indirectly reflect the ability and degree of tumor neoantigen production. TMB-H cancers generate more neoantigens which enhances immunogenicity, activates immune response and ultimately suppress tumor development49,50. In this study, we found that immune infiltration levels of most immune cells were increased in high-risk group with upregulated PDCD1 levels, suggesting that PDCD1 overexpression may suppress anti-tumor effect of immune cells and induce tumor immune escape to stimulates tumor growth. Besides, we observed that the low-risk group had higher TMB score implying stronger anti-tumor immunity and better prognosis. These data indicated that PDCD1 and TMB may influence the immune status in the tumor microenvironment to affect tumor development. Regression analysis showed that MLH1 was the only independent predictor of STAD prognosis among the four MMR genes.
Currently, the NGS method has been shown to simultaneously detect panel-covered driver gene variants, including MMR gene germline and somatic mutations, and even molecular tags such as TMB. NGS can detect DNA MMR gene somatic mutations accompanied by LOH and structural mutations in genes leading to dMMR. However, there is no consensus recommendation on the MSI/MMR gene combinations currently used for NGS detection, with findings from previous studies showing inconsistencies or false negatives. Moreover, there is no evidence-based medicine for NGS-microsatellite instability in relation to clinical immunotherapy32. In this study, we conducted preliminary bioinformatics analyses of the four MMR proteins at mRNA expression levels in stomach adenocarcinoma to determine patients who are most likely to benefit from immunotherapy. Despite the significant findings of this study, there are some limitations. Firstly, we did not validate the results of the MMR-related prognostic model of gastric cancer through experiments or actual patients. In future, real-world clinical data should be used to evaluate the feasibility of the prognostic model. We aim to conduct a retrospective review of patients with stomach adenocarcinoma receiving immunotherapy to further verify the feasibility and validity of this risk score model. In summary, the present findings present new research directions to improve the diagnosis and management of gastric cancer.
Conclusion
Our study constructed a risk model based on MLH1 and MSH2 for STAD at genetic level. In the MMR system, MLH1 may contribute to the STAD development. Besides, PDCD1 and TMB affect the cellular immune function in the tumor microenvironment. These findings reveal important strategies for the diagnosis and treatment of STAD patients.
Acknowledgements
The authors thank public database for the availability of the data and online tools for the bioinformatic analysis.
Author contributions
Authors’ contributionsZ.T. designed experiments, performed data analysis and drafted the manuscript; Z.T. and L.Y. collected the data and performed data analysis. R.Y and W.Y. supervised experiments. All authors read and approved the manuscript.
Data availability
The complete clinical data generated and analyzed during the current study are downloaded from the TCGA database (https://portal.gdc.cancer.gov/). And these datasets are included in supplementary information files. Thanks!
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Fund
No.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhihui Tian, Email: dtianzhihui@163.com.
Rong Yang, Email: wbbyr2005@163.com.
Wenhui Yang, Email: yangwenhui10012@163.com.
References
- 1.Sung, H. et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249. 10.3322/caac.21660 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Zeng, H. et al. Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries. Lancet Glob Health. 6, e555–e567. 10.1016/S2214-109X(18)30127-X (2018). [DOI] [PubMed] [Google Scholar]
- 3.Smyth, E. C., Nilsson, M., Grabsch, H. I., van Grieken, N. C. & Lordick, F. Gastric cancer. Lancet396, 635–648. 10.1016/S0140-6736(20)31288-5 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Ellegren, H. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet.5, 435–445. 10.1038/nrg1348 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Committee of Colorectal, Cancer, C. S. & o., C. O. Genetics Group of the Committee of Colorectal Cancer, C. A.-C. A. & Genetics Committee of the Committee of Colorectal Cancer, C. M. D. A. [Consensus on the detection of microsatellite instability in colorectal cancer and other related solid tumors in China]. Zhonghua Zhong Liu Za Zhi. 41, 734–741. 10.3760/cma.j.issn.0253-3766.2019.10.003 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Hechtman, J. F. et al. Retained mismatch repair protein expression occurs in approximately 6% of microsatellite instability-high cancers and is associated with missense mutations in mismatch repair genes. Mod. Pathol.33, 871–879. 10.1038/s41379-019-0414-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aaltonen, L. A. et al. Clues to the pathogenesis of familial colorectal cancer. Science260, 812–816. 10.1126/science.8484121 (1993). [DOI] [PubMed] [Google Scholar]
- 8.Haraldsdottir, S. Microsatellite instability testing using next-generation sequencing data and therapy implications. JCO Precis Oncol.1, 1–4. 10.1200/PO.17.00189 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research, N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature513, 202–209. 10.1038/nature13480 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marabelle, A. et al. Efficacy of Pembrolizumab in patients with Noncolorectal high microsatellite Instability/Mismatch repair-deficient Cancer: results from the phase II KEYNOTE-158 study. J. Clin. Oncol.38, 1–10. 10.1200/JCO.19.02105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res.51, D587–D592. 10.1093/nar/gkac963 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res.28, 27–30. 10.1093/nar/28.1.27 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci.28, 1947–1951. 10.1002/pro.3715 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watson, J. D. & Crick, F. H. Genetical implications of the structure of deoxyribonucleic acid. 1953. JAMA269, 1967–1969 (1993). [PubMed] [Google Scholar]
- 15.Bielska, A. A. et al. Tumor Mutational Burden and Mismatch Repair Deficiency Discordance as a mechanism of Immunotherapy Resistance. J. Natl. Compr. Canc Netw.19, 130–133. 10.6004/jnccn.2020.7680 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai, J. H. et al. An integrative morphomolecular classification system of gastric carcinoma with distinct clinical outcomes. Am. J. Surg. Pathol.44, 1017–1030. 10.1097/PAS.0000000000001521 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Polom, K. et al. Meta-analysis of microsatellite instability in relation to clinicopathological characteristics and overall survival in gastric cancer. Br. J. Surg.105, 159–167. 10.1002/bjs.10663 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Puliga, E., Corso, S., Pietrantonio, F. & Giordano, S. Microsatellite instability in gastric Cancer: between lights and shadows. Cancer Treat. Rev.95, 102175. 10.1016/j.ctrv.2021.102175 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Cristescu, R. et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med.21, 449–456. 10.1038/nm.3850 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Semba, S., Yokozaki, H., Yamamoto, S., Yasui, W. & Tahara, E. Microsatellite instability in precancerous lesions and adenocarcinomas of the stomach. Cancer77, 1620–1627. https://doi.org/10.1002/(SICI)1097-0142(19960415)77:8<1620::AID-CNCR30>3.0.CO;2-# (1996). [DOI] [PubMed]
- 21.Halling, K. C. et al. Origin of microsatellite instability in gastric cancer. Am. J. Pathol.155, 205–211. 10.1016/S0002-9440(10)65114-0 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, P., Zhang, X. Y., Shao, Y. & Zhang, D. F. Microsatellite instability in gastric cancer and pre-cancerous lesions. World J. Gastroenterol.11, 4904–4907. 10.3748/wjg.v11.i31.4904 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng, S. et al. Multi-omics of the gut microbial ecosystem in patients with microsatellite-instability-high gastrointestinal cancer resistant to immunotherapy. Cell. Rep. Med.5, 101355. 10.1016/j.xcrm.2023.101355 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bronner, C. E. et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature368, 258–261. 10.1038/368258a0 (1994). [DOI] [PubMed] [Google Scholar]
- 25.Hsieh, P. & Zhang, Y. The Devil is in the details for DNA mismatch repair. Proc. Natl. Acad. Sci. U S A. 114, 3552–3554. 10.1073/pnas.1702747114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi, Y. Y. et al. Is microsatellite instability a prognostic marker in gastric cancer? A systematic review with meta-analysis. J. Surg. Oncol.110, 129–135. 10.1002/jso.23618 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Setia, N. et al. A protein and mRNA expression-based classification of gastric cancer. Mod. Pathol.29, 772–784. 10.1038/modpathol.2016.55 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Fishel, R. et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell77, 1pfollowing166 (1994). [PubMed] [Google Scholar]
- 29.Nielsen, S. V., Hartmann-Petersen, R., Stein, A. & Lindorff-Larsen, K. Multiplexed assays reveal effects of missense variants in MSH2 and cancer predisposition. PLoS Genet.17, e1009496. 10.1371/journal.pgen.1009496 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edelmann, W. et al. Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell91, 467–477. 10.1016/s0092-8674(00)80433-x (1997). [DOI] [PubMed] [Google Scholar]
- 31.Mohd, A. B. et al. Truncation of the C-terminus of human MLH1 blocks intracellular stabilization of PMS2 and disrupts DNA mismatch repair. DNA Repair. (Amst). 5, 347–361. 10.1016/j.dnarep.2005.11.001 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Lee, V., Murphy, A., Le, D. T. & Diaz, L. A. Jr. Mismatch Repair Deficiency and Response to Immune Checkpoint Blockade. Oncologist 21, 1200–1211, (2016). 10.1634/theoncologist.2016-0046 [DOI] [PMC free article] [PubMed]
- 33.Kim, K. J. et al. Deletion in HSP110 T(17): correlation with wild-type HSP110 expression and prognostic significance in microsatellite-unstable advanced gastric cancers. Hum. Pathol.67, 109–118. 10.1016/j.humpath.2017.08.001 (2017). [DOI] [PubMed] [Google Scholar]
- 34.de Visser, K. E. & Joyce, J. A. The evolving tumor microenvironment: from cancer initiation to metastatic outgrowth. Cancer Cell.41, 374–403. 10.1016/j.ccell.2023.02.016 (2023). [DOI] [PubMed] [Google Scholar]
- 35.Sharonov, G. V., Serebrovskaya, E. O., Yuzhakova, D. V., Britanova, O. V. & Chudakov, D. M. B cells, plasma cells and antibody repertoires in the tumour microenvironment. Nat. Rev. Immunol.20, 294–307. 10.1038/s41577-019-0257-x (2020). [DOI] [PubMed] [Google Scholar]
- 36.Zhu, J. & Paul, W. E. CD4 T cells: fates, functions, and faults. Blood112, 1557–1569. 10.1182/blood-2008-05-078154 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chraa, D., Naim, A., Olive, D. & Badou, A. T lymphocyte subsets in cancer immunity: friends or foes. J. Leukoc. Biol.105, 243–255. 10.1002/JLB.MR0318-097R (2019). [DOI] [PubMed] [Google Scholar]
- 38.Speiser, D. E., Chijioke, O., Schaeuble, K. & Munz, C. CD4(+) T cells in cancer. Nat. Cancer. 4, 317–329. 10.1038/s43018-023-00521-2 (2023). [DOI] [PubMed] [Google Scholar]
- 39.van der Leun, A. M., Thommen, D. S. & Schumacher, T. N. CD8(+) T cell states in human cancer: insights from single-cell analysis. Nat. Rev. Cancer. 20, 218–232. 10.1038/s41568-019-0235-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, S. Y., Fu, T., Jiang, Y. Z. & Shao, Z. M. Natural killer cells in cancer biology and therapy. Mol. Cancer. 19, 120. 10.1186/s12943-020-01238-x (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Del Prete, A. et al. Dendritic cell subsets in cancer immunity and tumor antigen sensing. Cell. Mol. Immunol.20, 432–447. 10.1038/s41423-023-00990-6 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeNardo, D. G. & Ruffell, B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol.19, 369–382. 10.1038/s41577-019-0127-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grisaru-Tal, S., Itan, M., Klion, A. D. & Munitz, A. A new dawn for eosinophils in the tumour microenvironment. Nat. Rev. Cancer. 20, 594–607. 10.1038/s41568-020-0283-9 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Gungabeesoon, J. et al. A neutrophil response linked to tumor control in immunotherapy. Cell 186, 1448–1464 e1420, (2023). 10.1016/j.cell.2023.02.032 [DOI] [PMC free article] [PubMed]
- 45.Lichterman, J. N. & Reddy, S. M. Mast cells: a New Frontier for Cancer Immunotherapy. Cells1010.3390/cells10061270 (2021). [DOI] [PMC free article] [PubMed]
- 46.Geraud, A. et al. Clinical pharmacology and interplay of Immune Checkpoint agents: a Yin-Yang Balance. Annu. Rev. Pharmacol. Toxicol.61, 85–112. 10.1146/annurev-pharmtox-022820-093805 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Sanmamed, M. F. & Chen, L. A paradigm shift in Cancer Immunotherapy: from enhancement to normalization. Cell175, 313–326. 10.1016/j.cell.2018.09.035 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chamoto, K., Yaguchi, T., Tajima, M. & Honjo, T. Insights from a 30-year journey: function, regulation and therapeutic modulation of PD1. Nat. Rev. Immunol.23, 682–695. 10.1038/s41577-023-00867-9 (2023). [DOI] [PubMed] [Google Scholar]
- 49.Havel, J. J., Chowell, D. & Chan, T. A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer. 19, 133–150. 10.1038/s41568-019-0116-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghatalia, P. & Plimack, E. Biomarkers for neoadjuvant checkpoint blockade response in urothelial cancer. Nat. Med.25, 1650–1651. 10.1038/s41591-019-0645-6 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete clinical data generated and analyzed during the current study are downloaded from the TCGA database (https://portal.gdc.cancer.gov/). And these datasets are included in supplementary information files. Thanks!