Abstract
A distinctive feature of both type 1 and type 2 diabetes is the waning of insulin-secreting beta cells in the pancreas. New methods for direct and specific targeting of the beta cells could provide platforms for delivery of pharmaceutical reagents. Imaging techniques such as Positron Emission Tomography (PET) rely on the efficient and specific delivery of imaging reagents, and could greatly improve our understanding of diabetes etiology as well as providing biomarkers for viable beta-cell mass in tissue, in both pancreas and in islet grafts.
The DiGeorge Syndrome Critical Region Gene 2 (DGCR2) protein has been suggested as a beta-cell specific protein in the pancreas, but so far there has been a lack of available high-affinity binders suitable for targeted drug delivery or molecular imaging. Affibody molecules belong to a class of small affinity proteins with excellent properties for molecular imaging. Here, we further validate the presence of DGCR2 in pancreatic and stem cell (SC)-derived beta cells, and then describe the generation and selection of several Affibody molecules candidates that target human DGCR2. Using an in-house developed directed evolution method, new DGCR2-binding Affibody molecules were generated and evaluated for thermal stability and affinity. The Affibody molecules variants were further developed as targeting agents for delivering imaging reagents to beta cell. The Affibody molecule ZDGCR2:AM106 displayed nanomolar affinity, suitable stability and biodistribution, with negligible toxicity to islets, qualifying it as a suitable lead candidate for further development as a tool for specific delivery of drugs and imaging reagents to beta cells.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-84574-y.
Keywords: Affibody molecule, Beta cells, DGCR2, Drug delivery, Imaging reagents
Subject terms: Molecular imaging, Positron-emission tomography, Protein design
Introduction
Type 1 Diabetes (T1D) is a disease that is characterized by almost complete loss of beta cell mass and function, leading to insulin insufficiency. Curative treatments of T1D remain elusive1,2. Despite preclinical progress in the development of pharmaceutical agents that protect or even reverse the loss of beta-cell mass in T1D models, such successes have been challenging to translate into clinical fruition and meaningful patient benefit. A critical bottleneck in the drug development process and the major cause of failure, is the balance between sufficient drug exposure in the target tissue and the emergence of off-target effects during dose escalation. Additionally, the target of interest may be expressed in other tissues where drug interactions may generate undesirable effects.
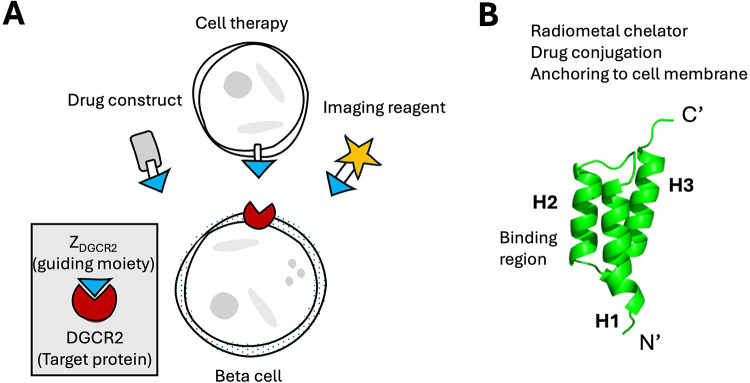
Thus, there is an urgent need for novel simple and flexible drug delivery systems to increase drug exposure in the pancreas, while minimizing the off-target and off-tissue effects and toxicity (Fig. 1A). Such delivery systems may enable re-assessment of existing and emerging drug targets and candidates, which previously have been associated with and discarded due to unacceptable side effects.
Fig. 1.
(A) Concept of using DGCR2 targeting for delivery of drugs and imaging reagents to beta cells. (B) Overview of the Affibody molecule scaffold and opportunities for modifications to enable drug delivery or radiolabeling.
Similarly, targeted delivery of imaging reagents to beta cells has been suggested as a tool for visualizing and quantifying the beta-cell mass (BCM) in a non-invasive manner, e.g. using molecular imaging technologies3. Accurate estimation of the BCM in either the pancreas or in transplanted islet grafts would greatly improve our understanding of diabetes etiology, as well as the ability to evaluate therapeutic efficacy, thus highlighting the importance of developing a reliable imaging marker for this purpose4.
The DiGeorge Syndrome Critical Region Gene 2 (DGCR2) is a surface protein which is strongly expressed in pancreatic beta cells5–7. Importantly, DGCR2 is absent in the exocrine pancreas. DGCR2 is only present in the beta cells of the islets of Langerhans, and not in e.g. the alpha cells5. Additionally, the expression of DGCR2 is drastically reduced in insulin deficient islets in T1D patients, further indicating its beta-cell restricted expression5. DGCR2 is located at the cell membrane, with a large extracellular region and thus accessible for binding by proteins. Recently, it was also shown that beta-cell derived extracellular vesicles (EVs) can be identified by their selective expression of DGCR2 at the surface8. DGCR2 is therefore a potential target for directed delivery of e.g. drug conjugates and imaging reagents in the pancreas. Finally, DGCR2 is not present in any other tissues in human adults, making it an ideal target protein for directed drug delivery, while minimizing unintended off-tissue and off-target exposure.
The exact function of DGCR2 is unknown, but deletion of the DGCR2 gene is responsible for the 22q11.2 deletion syndrome (22q11DS, previously known as DiGeorge Syndrome) and closely associated with both defects in the immune system, obesity and Type 2 Diabetes (T2D)9,10. Furthermore, DGCR2 is localized in a region on chromosome 22, which is home to several long non-coding RNA sequences that have been identified in pancreatic beta cells, as well as islet enhancer clusters11,12.
Currently, there is a lack of reported DGCR2 binders to be used as carriers for drug conjugates or imaging reagents in vivo. Polyclonal and monoclonal antibodies raised towards DGCR2 are available, but antibodies have slow clearance and high background signal13. With the rise of modern combinatorial library methods, novel protein scaffolds mimicking the binding properties of antibodies are rising in popularity. Affibody molecules constitute a class of small scaffold proteins with molecular weights of around 7 kDa (58 amino-acids)14. Affibody molecules consist of three alpha-helixes, where helix 1 and 2 are engineered to provide the binding surface by encompassing 13–15 variable amino acid residues, while the rest of the sequence as well as helix 3 are constant (Fig. 1B). Affibody molecules typically demonstrate both excellent stability and refolding characteristics, as well as flexible binding properties. Importantly, the C-terminal just after helix 3 is located away from the binding surface and thus can be site-specifically modified for e.g. addition of imaging reagents or drug conjugates without normally perturbing the molecular interactions at helixes 1 and 2 (Fig. 1B). Affibody molecules have been extensively investigated both in preclinical and clinical studies as tracers for molecular imaging applications with promising data in terms of imaging contrast, which is largely due to their small size and generally high affinity and specificity. The small sizes results in fast blood clearance through renal excretion, yielding a favorable pharmacokinetic profile and excellent contrast for molecular imaging14. Moreover, Affibody molecules extravasate rapidly from blood and efficiently penetrate tissues which is an advantageous feature for targeting diseased tissue15,16. Affibody molecules are currently also being investigated as therapeutics in late phase clinical trials for several indications including e.g. psoriasis and psoriatric arthriris17.
Here, we present the efforts to further validate DGCR2 as a selective and suitable marker of human beta cells, and also the generation of a first-in-class series of Affibody molecules with high affinity towards human DGCR2 which are optimized for delivery of imaging reagents to beta cells. A previously published lead candidate, ZDGCR2:AM106, which could be used to deliver imaging reagents to human stem cell (SC)-derived islet grafts in immunodeficient mice7, is here further characterized for binding efficiency, biodistribution and safety to support its further development as an agent for delivery of drugs or imaging reagents to beta cells.
Materials and methods
Generation of human SC-islets
Human embryonic stem cells (hESC; H1 cell line, Wicell®) were differentiated into SC-islets using a published differentiation protocol of seven stages18 with a few differences as described previously7: Briefly, H1 cells were propagated on treated human recombinant Laminin 521 (BioLamina; LN 521) in mTeSR-Plus medium (STEMCELL Technologies; #100–0274/100–275). To prepare for the differentiation step, cells were seeded at a density of 16 million cells/10 cm dish. The dishes were coated with Laminin 521 in mTeSR-Plus medium with the addition of 10 µM of ROCK inhibitor Y-27,632 (STEMCELL Technologies; 72,304). Differentiation was then triggered 24 h after the seeding process. During the first four stages (S1-4), cells were differentiated on adherent culture and a single cell suspension was prepared during S4:2 to generate uniform cluster formation. At stage 5:2, all SC-islets were moved to an ultra-low attachment 6-wells plate, on a rotating platform at 95 rpm. The last stages (S5-7) of the differentiation were carried out in suspension culture. During the stages S1-5, the medium was changed every day, while it was changed every second day over S6 and S7.
Immunofluorescent staining and confocal microscopy of SC-islets and primary human islets
SC-islets were fixed in 4% paraformaldehyde (PFA) at RT for 20 min and frozen. Cryosections of SC-islets were prepared with a thickness of 10 μm. Cryosections were blocked with 3% donkey serum (Jackson Immunoresearch Laboratories) in PBS for 30 min followed by staining with the primary polyclonal rabbit antibodies against DGCR2 (1:400, Thermo Fisher Scientific, #PA5-51,474), and polyclonal guinea pig antibodies against human insulin (1:400, Fitzgerald, Acton, #20-IP30) at 4 °C overnight. The next day, the following secondary antibodies diluted 1:300: Alexa Fluor 488-conjugated donkey anti-rabbit (Jackson Immunoresearch Laboratories).
and Alexa Fluor 594-conjugated donkey anti-guinea pig (Jackson Immunoresearch Laboratories) were incubated for 1 h at RT. The nuclei were stained with DAPI. All images were obtained using a laser scanning confocal microscope Zeiss LSM 780.
Staining and confocal microscopy of human primary islets was performed as described previously5 and is described in Supplementary Materials in detail.
Affinity maturation of first generation DGCR2 specific affibody molecules
A first-generation DGCR2 binder ZDGCR2:3D1 was previously discovered by E. coli display technology using the autotransporter Adhesin Involved in Diffuse Adherence (AIDA-I)19. Here, we report the characterization of ZDGCR2:3D1 as well as further affinity maturation of this initial DGCR2 binder. A similar technology was previously used for affinity maturation of a class of CD69 binding Affibody molecules20.
Subcloning of the affinity maturation library and electroporation into E.coli.
The randomized oligonucleotide sequences encoding helix 1 and 2 of the Affibody molecules (Twist Bioscience) were subcloned into the surface display vector. Ligation of the insert into the vector was done by adding a threefold molar excess of insert over vector and CloneSmart DNA ligase. Thirty electroporations were performed in total, divided into groups of 10. After electroporation, the cells were transferred to 10 ml of LB media and incubated for 1 h at 37 °C. The cell suspension was thereafter inoculated into 100 mL LB medium containing chloramphenicol and incubated overnight at 37 °C. A sample from the pooled cells was diluted and spread on agar plates containing chloramphenicol to determine the total library size. The following day, the culture was centrifuged at 5000 x g at 4 °C for 5 min, weighed and resuspended in 15% glycerol. The E. coli library was thereafter stored at -80 °C. 96 clones were picked at random and sequenced for evaluation of library diversity and functionality.
Selections of affinity matured variants by fluorescence activated cell sorting (FACS)
The recombinant E. coli displayed affinity maturation library was induced and cells were washed with PBSP buffer (10 mM PBS, pH 7.4 containing 0.1% Pluronic F127). Cells were then resuspended in PBSP containing biotinylated DGCR2 (Glu22-Ala349) purchased from R&D Systems. The mix was incubated on a rotamixer at room temperature (RT) for 1 h, followed by extended washes with ice cold PBSP before resuspending in 150 nM human serum albumin (HSA)-Alexa 647 conjugate and 0.5 µg/ml streptavidin conjugated with R-phycoerythrin (SAPE) (Invitrogen) or neutravidin conjugated with Oregon Green 488 (NAOG) (Life Technologies), followed by incubation on ice for 30 min. The cells were thereafter washed with ice-cold PBSP and resuspended in ice-cold PBSP for sorting in a MoFlo Astrios EQ flow cytometer (Beckman Coulter). The sort gate was set to sort out the top fraction of cells displaying Z variants (typically 0.1%) showing the highest R-phycoerythrin or Oregon Green 488 to Alexa Fluor 647 fluorescence intensity ratio. Bacteria were sorted into a 1.5 ml tube containing LB medium and chloramphenicol. The sorted cells were incubated for 1 h on rotamixer at 37 °C and thereafter inoculated to 50 ml LB medium with chloramphenicol for overnight cultivation.
Recombinant expression of the first generation and affinity-matured DGCR2-binding Affibody molecules
The gene encoding the first-generation DGCR2 binder ZDGCR2:3D119 and the affinity maturated variants were subcloned into an E. coli expression vector based on the pETsystem (GenScript Biotech Corp) under the control of a T7 promoter and a N-terminal hexahistidine tag. A unique cysteine residue was added at the C-terminal to enable site specific conjugations of imaging reagents or radiometal chelators using maleimide chemistry. The recombinantly produced peptide consisted of > 74 residues, incorporating both the Affibody molecule helices as well as the histidine tag, two restriction sites and in some cases a C-terminal cysteine. The recombinant proteins were purified using HisPur Cobalt Resin (#89,966, Thermo Scientific) according to the manufacturer’s instructions.
Circular dichroism spectroscopy
Thermal stability and refolding after heat induced denaturation were measured for each variant using circular dichroism spectroscopy. All measurements were performed on a Chirascan instrument (Applied Photophysics Ltd) and three spectra were run for each Affibody molecule. First, a wavelength spectrum was run to confirm the presence of a helical structure associated with the Affibody molecule scaffold. The ellipticity was measured at 20 °C from 195 nm to 260 nm in five technical replicates. The thermal stability was determined by following the ellipticity at 221 nm during variable temperature measurements (5 °C/min from 20 °C to 100 °C). After the heat induced denaturation, the samples were cooled to 20 °C before measuring ellipticity from 195 to 260 nm in five technical replicates to check for refolding.
Affinity analysis by surface plasmon resonance (SPR)
Recombinant human DGCR2 protein was diluted in 10 mM sodium acetate, pH 4.5 and immobilized on a CM5 chip surface using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC)/ N-hydroxysuccinimide (NHS) coupling chemistry for use as immobilized target in a Biacore 8 K instrument (Cytiva). The surfaces were inactivated using ethanolamine prior to binding studies. One surface was activated/inactivated for blank subtraction. A first screening was performed by injecting 100 nM of the respective variant over the respective immobilized target for 120 s, followed by running buffer for 300 s before regeneration of the surfaces. 10 mM glycine–HCl pH 2.5 was used for regeneration in the experiments. All the variants selected in the affinity maturation procedure expressed as described above were injected in concentrations ranging from 1 nM to 100 nM in triplicates over both surfaces.
Indium-111 radiolabeling of DGCR2 targeting affibody molecules
The DOTA-conjugated, recombinantly expressed Affibody molecules ZDGCR2:AM106, ZDGCR2:AM115, ZDGCR2:AM156 and ZDGCR2:3D1 were radiolabeled by Indium-111.
Indium-111 chloride (370 MBq/mL) was purchased from Curium. Ammonium acetate buffer (0.2 M, pH 5.5) was prepared in a plastic flask (200 mL, HDPE low metal resin from Nalgene) by dissolving 0.771 g ammonium acetate in 200 mL water. The pH was measured with a pH-meter (Mettler Toledo) and adjusted to 5.5 by adding glacial acetic acid (> 99%, TraceSELECT). Chelex 100 sodium form was added to the buffer and allowed to stand in a refrigerator (4 °C) overnight. NAP-5 column (from illustra) was conditioned and eluted with phosphate buffered saline (PBS). Protein LoBinding Eppendorf tubes (1.5 mL) were used for the reaction and elution collection.
Analytical high-performance liquid chromatography (HPLC) consisted of a VWR Hitachi Chromaster 5110 pump, a Knauer UV detector 40D, a Bioscan Flow count equipped with an Eckert & Ziegler extended range module 106, a Bioscan B-FC-3300 radioactivity probe, a VWR Hitachi Chromaster A/D Interface box, and a Vydac 214MS, 5 μm C4, 50 × 4.6 mm column. The eluents were: A = 0.1% trifluoroacetic acid (TFA) in water, B = 0.1% TFA in acetonitrile, 5–70% B over 15 min at a flow rate of 1.0 mL/min. The details of the radiolabeling procedures of the different Affibody molecule variants were continuously updated based on performance. Here are the details for each variant:
DOTA-ZDGCR2:3D1: 120 µL of 111InCl3 solution (89.5 MBq) was added to 240 µL of ammonium acetate buffer (pH 5.5) followed by 14 nmol of DOTA-ZDGCR2:3D1 (123 µL in PBS). The reaction mixture was heated at 80 °C for 1 h and was then purified by NAP-5 column.
DOTA-ZDGCR2:AM115: 80 µL of 111InCl3 solution (36.2 MBq) was added to 180 µL of ammonium acetate buffer (pH 5.5) followed by 2.8 nmol of DOTA-ZDGCR2:AM115 (20 µL in PBS). The reaction mixture was heated at 70 °C for 30 min and was purified by NAP-5 column.
DOTA-ZDGCR2:AM106: 30 µL of 111InCl3 solution (29.5 MBq) was added to 60 µL of ammonium acetate buffer (pH 5.5) followed by 3.3 nmol of DOTA-ZDGCR2:AM106 (15 µL in PBS). The reaction mixture was heated at 70 °C for 30 min and was purified by NAP-5 column.
DOTA-ZDGCR2:AM156: 40 µL of 111InCl3 solution (40.5 MBq) was added to 80 µL of ammonium acetate buffer (pH 5.5) followed by 2.4 nmol of DOTA-ZDGCR2:AM106 (20 µL in PBS). The reaction mixture was heated at 70 °C for 30 min and was purified by NAP-5 column.
In vivo SPECT/CT and organ biodistribution of the affibody molecule variants.
All the procedures performed were in accordance with the ARRIVE guidelines for animal research. The animal experiments were ethically reviewed, authorized by the Animal Ethics Committee of the Swedish Animal Welfare Agency and carried out in accordance with the European Directive 2010/63/EEC and the institutional guidelines at Uppsala University (UFV 2007/724). Sprague Dawley rats (n = 3 per Affibody molecule, n = 12 in total, weight = 250–300 g) were ordered and transported to the animal facility a week prior to each individual Affibody molecule experiment. The aim of the rat biodistribution was primarily to rank the candidates based on clearance (a faster clearance leads to low tissue background), off target non-specific binding (unsuitable as a drug delivery vector) and kidney uptake (high renal retention yields a poor dosimetry profile). The intent was not to evaluate binding in tissues as it is unknown if the Affibody molecules bind rodent DGCR2, nor has DGCR2 expression in rat islets been demonstrated previously.
Anesthesia was given through inhalation of 4% vaporized sevoflurane mixed with a 400 mL/min flow of medical air and oxygen. A computed tomography (CT) was first performed for structural observation (480 project; Semi Circ MultiFOV; 50 kVp/600 µA; Binning 1:4; Acqui time: 7’ 46’’; Voxel size: 251 × 251 × 251 μm). The CT was followed by the intravenous injection of 2 MBq (~ 7 kBq/g) of the Indium-111 labeled Affibody molecule (ZDGCR2:3D1, ZDGCR2:AM106, ZDGCR2:AM115 or ZDGCR2:AM156) dissolved in PBS through the tail vein of the rat. A Single Photon Emission Tomography (SPECT) scan was performed to obtain images from three time-points after injection of the radioactive compound (5 min, 30 min and 60 min) with the following acquisition parameters (Number of beds: 3; Acquisition time: 5’/25’/25’; Frame duration: 4’’/19’’/19’’; 245.35 keV/171.30 keV). Manual segmentation and quantification of the kidney were performed on sequential trans-axial projections using the PBAS modeling tool (pmod technologies LLC). The signal uptake in Bq/cc was converted to Standardized Uptake Values (SUV) by correcting for the amount and time of administered radioactivity (MBq) and the weight (g) of each rat. At the end of the scanning period, organs of interest were collected from each rat for gamma radiation counts measurements using a WIZARD Automatic Gamma Counter (PerkinElmer) and radioactive decay corrected.
Chemical synthesis of DOTA-ZDGCR2:AM106
Based on the results from the characterization of the Affibody molecule variants with regard to affinity, stability, radiolabeling and biodistribution, ZDGCR2:AM106 was selected as lead candidate. The recombinantly expressed Affibody molecules, including ZDGCR2:AM106, incorporated additional residues to allow purification e.g. histidine tags, bringing the total length to 75 amino acids which may impact biodistribution and non-specific binding. Thus, for further evaluation, the lead candidate was produced as minimized peptide by chemical synthesis, which only incorporated the Affibody molecule scaffold as well as a C-terminal cysteine (in total 59 amino acids: VDNKFNKEQTHARNEILQLPNLNRMQKSAFIRSLIDDPSQSANLLAEAKKLNDAQAPK[C/DOTA]-amide). This format of the peptide is expected to be the most relevant with regards to delivery of drugs or imaging reagents in vivo, and most comparable to other Affibody molecules being evaluated for similar purposes towards other targets.
DOTA-maleimide conjugated ZDGCR2:AM106 was manufactured by custom Solid Phase Peptide Synthesis (New England Peptide) in aliquots of 100 µg per vial. Purity was > 95% as assessed by HPLC. Separately, a similar ZDGCR2:AM106 construct also generated by chemical synthesis and conjugated with a Transcyclo-octene (TCO) group was used for radiolabeling with Fluorine-18 ([18F]ZDGCR2:AM106) and evaluated as a vector for delivery of imaging reagents to SC-islets7.
Gallium-68 radiolabeling of DOTA-ZDGCR2:AM106
The DOTA-conjugated Affibody molecule ZDGCR2:AM106, was radiolabeled with Gallium-68 as follows: The 68Ge/68Ga generator (Eckler Zigler) was eluted with 0.1 M metal-free 32-35% ultrapure HCl (Merck). Various fractions were collected, with the fraction containing the highest amount of radioactive material subsequently used for labeling DOTA-ZDGCR2:AM106. 30 µg of DOTA-ZDGCR2:AM106 was suspended in HEPES 2 M (pH 3.3) to reach a final volume of 40µL. 250µL of Gallium-68 containing eluate was mixed with the Affibody molecule before incubation at 60 °C for 5 min. At the end of the incubation time, the mixture was purified using a NAP-5 size-exclusion column (Cytiva) using PBS containing 10% Ethanol to obtain a radiochemical purity > 95%.
Copper coated plates ELISA-based binding of [68Ga]ZDGCR2:AM106
Binding of [68Ga]ZDGCR2:AM106 was evaluated against two sequences of the extracellular domain of human DGCR2 (based on the DGCR2 canonical structure, UNIPROT entry P98153, 550 residues). Recombinant human DGCR2 His-tag protein fragment (residues 21–349 of DGCR2, bio-techne, #10161-DG), as well as recombinant human DGCR2 protein fragment (residues 114–297 of DGCR2, Atlas Antibodies, PrEST HPA000873) was purchased and suspended in PBS. The recombinant human DGCR2 protein fragments were coated at 10 µg/mL onto a Pierce copper coated high capacity plate (Thermofisher) according to the manufacturer´s instruction using a 15 min blocking step with PBS containing 1% BSA. Pierce copper coated high capacity plate with PBS without the recombinant protein was used as negative control. Endogenous ligands towards DGCR2 are currently unknown, and thus there are limited options available for blocking studies to verify specificity in vitro. [68Ga]ZDGCR2:AM106 was incubated in each well at 5 MBq/mL for 60 min at RT with shaking. A series of 3 washes using PBS containing 0.05% Tween-20 was performed prior to measurement in a NaI well counter (Uppsala Imanet AB, Uppsala, Sweden).
In vitro autoradiography binding to human primary and SC-islets
The binding of [68Ga]ZDGCR2:AM106 was evaluated by in vitro autoradiography. Briefly, primary human islets or SC-islets were embedded in OCT and frozen at -80 °C. For the primary human islets, preparations of various islet purity were used (98% or 27% islet purity, the rest being exocrine tissue). The use of human tissue was approved by the Uppsala Ethical Review Board (Dnr 2015 − 401; #2011/473, #Ups 02-577). CHO cells with negligible normal expression of DGCR2 was use as comparison. Each preparation was processed into 4–20 μm slices using a cryotome, and mounted upon Superfrost Plus microscope slides (VWR).
Sections were first pre-incubated in assay buffer (PBS, pH 7.4, 150mL) for 10 min at RT. Next, radiolabeled Affibody molecule was added in a concentration at or slightly above the expected dissociation constant for each binder according to prior measurement using SPR (27 nM ZDGCR2:3D1, 2.1 nM ZDGCR2:AM115 or 6.1 nM for ZDGCR2:AM106).
The sections were incubated for 40 min at RT, and then washed 3 × 3 min in 150 mL of cold PBS (4 °C, pH 7.4) to remove excess unbound ligand. The sections were then airdried at 37 °C for 10 min before exposure to a digital phosphor-imager screen for 1 h. Readout of the phosphor-imager screen was performed using an Amersham Typhoon phosphorimager (GE Healthcare). A 10 µl of known radioactivity was included to allow for quantification of the results.
In vivo PET/CT and organ biodistribution of [68Ga]ZDGCR2:AM106
Sprague Dawley rats (n = 4, weight = 250–300 g) were ordered and transported to the animal facility prior to the experiment. N = 1 rat was anesthetized prior to a CT with similar parameters as described previously. Subsequently, around 15 kBq/g (total amount of 5 MBq) of [68Ga]ZDGCR2:AM106 dissolved in PBS and 10% ethanol were injected intravenously through the tail vein of each rat. A dynamic PET imaging of 60 min was then acquired on a nanoScan PET/MRI scanner (Mediso) using the following parameters: 3 bed positions x 5 frames of 2 × 1.4’, 2 × 3.3’, 1 × 10’, 0.4 × 0.4 × 0.4 mm voxels, 225 × 225 × 579 matrix size. The PET data were analyzed via manual segmentation using the PBAS image processing tool (PMOD technologies LLC). The tracer uptake in each tissue was expressed as SUV.
For the endpoint biodistribution, n = 3 rats were injected with 15 kBq/g [68Ga]ZDGCR2:AM106 (total amount of 5 MBq) through the lateral tail vein of n = 3 rats. Euthanasia of the rats was conducted 60 min post-injection and organs were collected for measurement in the well counter (Uppsala Imanet AB, Uppsala, Sweden).
Effect of in vitro ZDGCR2:AM106 exposure to isolated human islets
The function of DGCR2 expressed in human islets is so far unknown. Therefore, the potential effect on islet viability and e.g. insulin secretion was evaluated at different doses. Isolated islets from human donors (n = 3, normal HbA1c and no diagnosis of diabetes) were cultured in 6 well plates in 2 ml complete culture media (CMRL) with different doses of ZDGCR2:AM106 for at 37 °C for 48 h (75 islets per condition; 60µM [high dose], 60 nM [low dose], 0 nM [control]).
The islets were counted under the microscope before and after incubation. In order to evaluate the functional capacity of the islet cells, glucose-stimulated perifusion experiments were conducted. A dynamic perifusion system (Biorep Technologies, TP software Version 4.7, cRIO software Version 4.7. Miami Lakes, Florida, USA) was used for this purpose. After washing with low glucose for stabilization, the islets were stimulated with high glucose from 28 to 60 min at 37 °C. Each experimental setup was run as two technical replicates with 20 hand-picked islets placed in filter-closed chambers and perifused sequentially. Following perifusion, insulin ELISA was performed according to the manufacturer’s instructions (Mercodia, Uppsala, Sweden).
Results
Target validation of DGCR2 as a marker of beta cells
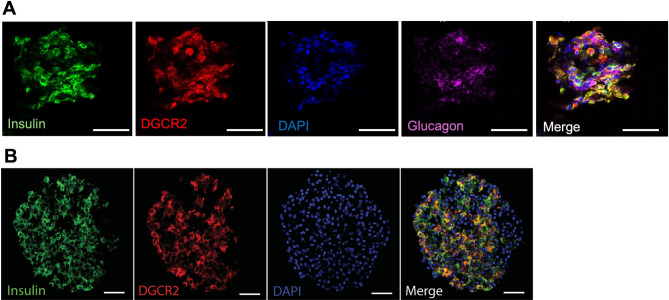
DGCR2 protein was detected in human primary islets (Fig. 2A). Importantly, there was overlap between staining for DGCR2 and insulin, but not DGCR2 and glucagon, indicating that DGCR2 is primarily present in beta cells in human islets. Similar patterns were also seen in SC-islets, where immunostaining for DGCR2 demonstrated overlap with insulin (Fig. 2B). Thus, SC-islets retained expression of DGCR2, especially in the beta cells.
Fig. 2.
(A) Confocal microscopy of primary human islets demonstrating DGCR2 expression in primary human islets, overlapping with insulin but not glucagon. Scale bar 50 μm. (B) Confocal microscopy of human stem cell-derived islet (SC-islet) showing DGCR2 positivity overlapping with insulin staining. Scale bar 50 μm.
Affinity maturation of first generation DGCR2 specific affibody molecules
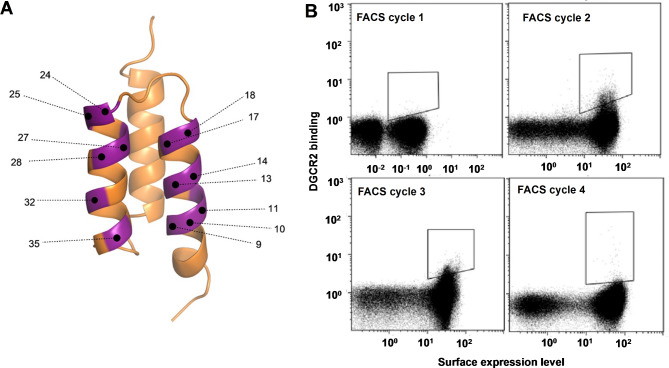
To obtain Affibody molecules with increased binding potential towards DGCR2, an affinity maturation library was designed by randomizing 13 positions on the binding surface of helix 1 and helix 2 (Fig. 3A). In each randomized position, the DNA library contained a combination of 70% of the original codon belonging to the first-generation DGCR2 binder ZDGCR2:3D119 and 30% of a mix of the other codons with the exceptions of cysteine, proline and glycine. The DNA library was subcloned into the E. coli display vector and electroporation of the plasmid library resulted in 2 × 107 transformants. From the transformed library, 96 clones were picked at random and sequenced. The sequencing showed that the library was highly functional, and no sequence occurred more than once. After verification, the library was expressed on the surface of E. coli and subjected to four rounds of FACS with alternating rounds of amplification by cell growth. Briefly, cells were incubated with biotinylated DGCR2 followed by extensive washes and then incubated with fluorescently labeled streptavidin for subsequent fluorescence-mediated detection of cell-bound DGCR2 as well as fluorescently labeled HSA for monitoring of surface expression levels. The incubation of secondary reagents and HSA was performed on ice in order to reduce the dissociation rate of bound DGCR2. The selection stringency (target concentration, sorting parameters and sorting gates) was increased with each sorting round. The top 0.1% of the library demonstrating the highest ratio of target binding to surface expression was gated and isolated for amplification and subsequent rounds of sorting. The advantage of a cell-based selection system is the straightforward monitoring of the obtained enrichment throughout the selection process. The visualization of the target-binding properties of the library in the flow cytometer revealed an enrichment of DGCR2 positive clones in each sorting round (Fig. 3B). After the third and fourth selection rounds, cells were spread on semi-solid medium and randomly picked variants were sequenced. Clones appearing more than once among the sequences were selected for further characterization.
Fig. 3.
(A) Schematic figure of the Affibody library design with randomized positions highlighted in purple. (B) Dot plots showing the four consecutive rounds of FACS of the E. coli displayed affinity maturation library. X-axes: fluorescence intensity corresponding to surface expression levels measured by fluorescently labeled HSA. Note that the x-axis for FACS cycle 1 has larger range than the other cycles. Y-axes: fluorescence intensity corresponding to labeled DGCR2 binding. Sorting gates are indicated in the dot plots.
Subcloning and expression of affibody molecules in soluble format
The new variants selected for further characterization (ZDGCR2:AM106, ZDGCR2:AM115 and ZDGCR2:AM156) and the original Affibody molecule ZDGCR2:3D1 were subjected to site-directed mutagenesis and introduction of the scaffold position mutations S42A, E43N, S46A and S54A. The genes encoding these four variants were each subcloned into an E. coli expression vector with an N-terminal hexahistidine tag. Additionally, the four genes were also subcloned to a format containing a C-terminal cysteine for site-specific conjugation of chelators for radiolabeling, resulting in eight different Affibody molecule constructs.
Circular dichroism spectroscopy
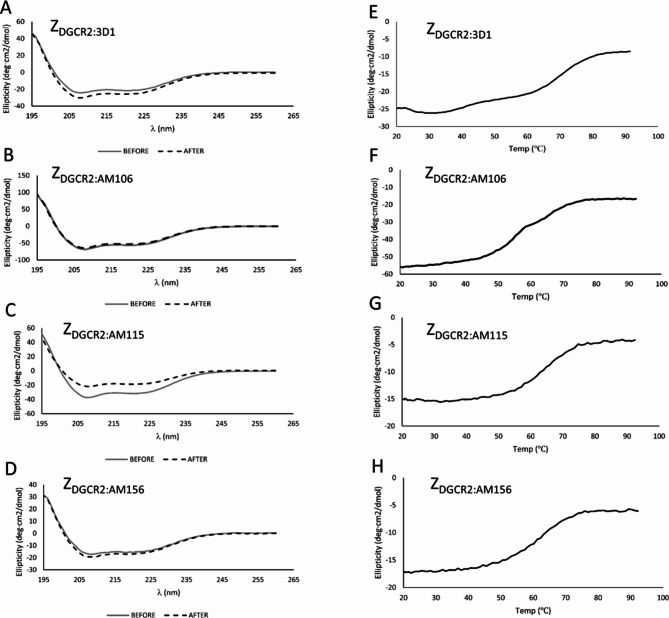
The melting temperature (Tm) for ZDGCR2:3D1, ZDGCR2:AM106, ZDGCR2:AM115 and ZDGCR2:AM156 without the C-terminal cysteine was determined by circular dichroism (CD) spectroscopy and variable temperature measurements (Table 1). Measuring the CD spectrum before and after heat induced denaturation demonstrated complete refolding for three of the variants (ZDGCR2:3D1, ZDGCR2:AM106 and ZDGCR2:AM156) with good thermal stability up to temperatures of around 60°C for all the evaluated Affibody molecules (Fig. 4).
Table 1.
Melting temperature as determined by CD and equilibrium dissociation constants as determined by SPR.
| Affibody | Tm (°C) | KD ± SD (nM) |
|---|---|---|
| ZDGCR2:3D1 | 69.0 | 26.7 ± 3.8 |
| ZDGCR2:AM106 | 56.4 | 2.4 ± 0.3 |
| ZDGCR2:AM115 | 64.0 | 2.1 ± 0.1 |
| ZDGCR2:AM156 | 60.0 | 8.5 ± 0.1 |
Fig. 4.
(A–D) Circular dichroism spectra of the affinity maturated Affibody molecules at 20 °C before and after heat induced denaturation. (E–H) Thermal stability evaluation by variable temperature measurement of ellipticity between 20 and 100 °C.
SPR characterization of DGCR2-binding Z variants
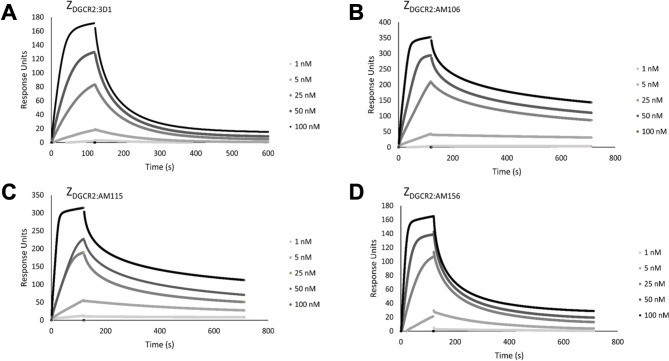
The interactions of the affibody molecules (ZDGCR2:3D1, ZDGCR2:AM106, ZDGCR2:AM115 and ZDGCR2:AM156) with DGCR2 were analyzed in SPR based biosensor assays and the three affinity-matured variants were found to have significantly increased affinity towards DGCR2 compared to the primary variant ZDGCR2:3D1 (Fig. 5). The calculated affinity values (KD) for the interaction with DGCR2 are given in Table 1.
Fig. 5.
Series of SPR sensorgrams showing the original (A) and the affinity maturated Affibody molecules (B–D) binding to surface-immobilized DGCR2 at different concentrations (1 nM, 5 nM, 25 nM, 50 nM and 100 nM).
Function and biodistribution of the Indium-111 labeled DOTA-conjugated affibody molecules
The four DGCR2 binding Affibody molecules were successfully conjugated with DOTA at the C-terminal (Supplementary Fig. 1) and radiolabeled with Indium-111 demonstrating acceptable yields, radiochemical purity and stability (Supplementary Table 1).
[111In]DOTA-ZDGCR2:3D1 and [111In]DOTA-ZDGCR2:AM115 retained binding to human primary islets in line with their respective affinity towards DGCR2, demonstrating that the ligands can carry imaging reagents without compromising their function (Supplementary Figure S2). [111In]DOTA-ZDGCR2:3D1 binding was low in 98% pure human primary islet preparations, but undetectable at 27% pure islets and negative in DGCR2-control cells. [111In]DOTA-ZDGCR2:AM115 which has 10-fold improved affinity according to SPR, demonstrated binding to 98% pure islets and weaker binding to 27% pure islets, while binding in negative cells was nearly undetectable (Supplementary Figure S2).
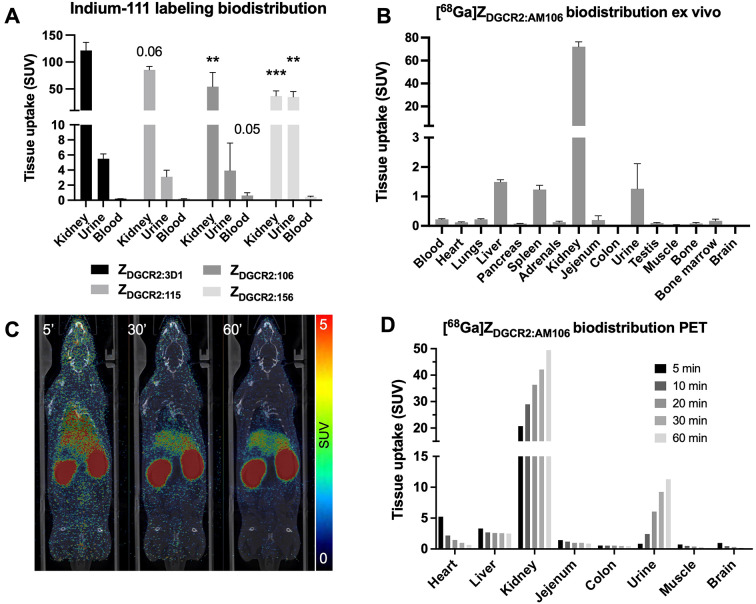
The Indium-111 labeled Affibody molecules were evaluated for biodistribution, background tissue uptake and clearance in rats by SPECT. All Affibody molecules demonstrated primarily renal clearance. [111In]DOTA-ZDGCR2:3D1 displayed the highest renal uptake, based on the ex vivo gamma counter measurement 60 min after injection (Fig. 6A). The renal uptake was decreased for [111In]DOTA-ZDGCR2:AM106, as well as for [111In]DOTA-ZDGCR2:AM156. [111In]DOTA-ZDGCR2:AM156 also exhibited distinct and strong pooling in the urine, potentially indicating partial instability (Fig. 6A). [111In]DOTA-ZDGCR2:AM106 demonstrated a tendency for longer circulatory half-life in blood, which is desirable for increased tissue exposure (Fig. 6A).
Fig. 6.
(A) Uptake of Indium-111 labeled Affibody molecules in kidney, urine and blood, based on ex vivo gamma counter measurements of excised tissues. (B) Biodistribution of [68Ga]ZDGCR2:AM106 by ex vivo organ distribution 60 min after injection. (C) Representative coronal PET/CT projections at 5 min, 30 min and 60 min after injection of [68Ga]ZDGCR2:AM106. (B) Biodistribution of [68Ga]ZDGCR2:AM106 in individual organs over time, from quantitative PET/CT scans.
All Affibody molecules demonstrated rapid clearance from blood and liver during the first hour after administration both assessed by in vivo SPECT/CT (Supplementary Figure S3) as well as ex vivo gamma counter measurement of tissues (Supplementary Figure S4). Background signal in peripheral tissues was subsequently low, including in the pancreas (SUV < 1) for all the Affibody molecules, indicating that high contrast delivery of imaging reagents could be accomplished given the presence of human DGCR2 in beta cells.
In general, [111In]DOTA-ZDGCR2:AM106 exhibited the most promising biodistribution profile, given its moderate kidney uptake (leading to a better dosimetry profile when delivering imaging reagents) combined with low pooling into urine as well as improved residency in the blood pool (important for a drug delivery vector to enable high tissue exposure).
Selection of lead candidate for further development
Based on an aggregate ranking of the in vitro and in vivo characterization of the four Affibody molecule candidates, the lead candidate ZDGCR2:AM106 was selected for further evaluation and development. ZDGCR2:AM106 combined excellent affinity towards human DGCR2 in the low nanomolar range, with acceptable thermal stability, high refolding after denaturation, as well as promising in vivo biodistribution profile. ZDGCR2:AM115 was also highly promising, with even better affinity and thermal stability, but crucially it did not exhibit complete refolding after denaturation (Fig. 4C), which could limit its utility in applications requiring harsh labeling or conjugation conditions.
ZDGCR2:AM106 was thus generated in a minimized format by chemical synthesis, for further evaluation for binding, delivery and toxicity. The successful performance of ZDGCR2:AM106 labeled with Fluoride-18 for in vivo delivery of imaging reagents to transplanted SC-islets was previously reported7.
Copper coated plates ELISA-based binding of [68Ga]ZDGCR2:AM106
DOTA-ZDGCR2:AM106 was successfully labeled with Gallium-68 to form [68Ga]ZDGCR2:AM106, which was tested for binding to DGCR2 protein and tissues.
The measured signal of [68Ga]ZDGCR2:AM106 was significantly higher in wells containing the a large fragment comprising the extracellular domain of recombinant human DGCR2 (residues 22–349) compared to both a smaller region of the extracellular domain (residues 114–297) of DGCR2 or the negative control (Fig. 7A).
Fig. 7.
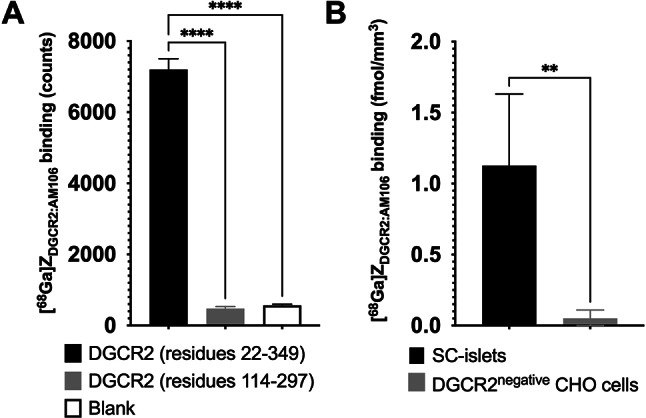
(A) Binding signal of [68Ga]ZDGCR2:AM106 to the large fragment of recombinant of human DGCR2 protein comprising almost the entire extracellular region (residues 22–349) was higher than the smaller region of extracellular DGCR2 (residues 114–297) and the negative control. (B) Binding of [68Ga]ZDGCR2:AM106 to human SC-islets was increased compared to non-transfected DGCR2 negative CHO cells.
In vitro autoradiography binding to human primary and SC-islets
[68Ga]ZDGCR2:AM106 exhibited binding to SC-islets, while binding to negative control cells was negligible (Fig. 7B). The binding intensity, approximately 1 fmol/mm3, corresponds to 1.4 pmol per islet (assuming an islet equivalent diameter of 150 μm), which equals in the range of 850.000 bound [68Ga]ZDGCR2:AM106 ligands per islet at this concentration.
In vivo PET/CT and organ biodistribution of [68Ga]ZDGCR2:AM106
The dynamic biodistribution of [68Ga]ZDGCR2:AM106 was evaluated in rat over 60 min with dynamic PET/CT, further confirmed by ex vivo organ measurement with a gamma counter. [68Ga]ZDGCR2:AM106 demonstrated mainly a renal clearance (Fig. 6B-D). The ex vivo biodistribution in the collected organs aligned with the dynamic PET/CT results with [68Ga]ZDGCR2:AM106 mainly being cleared through the kidney, combined with increased basal liver and spleen signal likely due to their high vascularization and perfusion. Background uptake in most tissue was otherwise low and decreased with time.
The in vitro plasma metabolite analysis of [68Ga]ZDGCR2:AM106 showed > 99% intact tracer after 60 min and 90% intact after 90 min incubation (Supplementary Figure S5A-B).
Effect ofin vitro ZDGCR2:AM106 exposure to isolated primary human islets
Given the unknown function of DGCR2 in human beta cells, it is important to assess any impact of exposure to ZDGCR2:AM106, either on function, insulin secretion or untoward effects on viability.
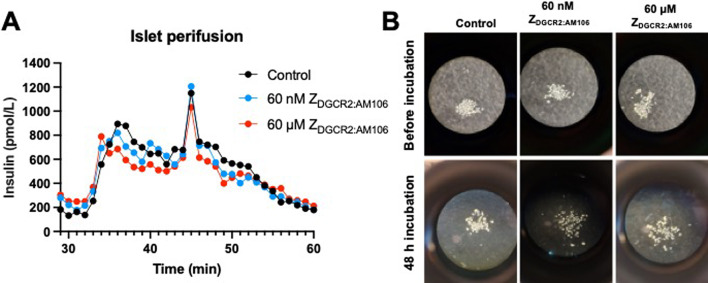
Islet perifusion experiments (n = 3) demonstrated a characteristic biphasic pattern of insulin release during stimulation with high glucose regardless of exposure to high doses of ZDGCR2:AM106 or not (Fig. 8A). Thus, exposure of human islets to ZDGCR2:AM106 did not inhibit or potentiate glucose induced insulin secretion, which could interfere with delivery drugs intending to affect the same or cause unwanted side effects such as hypoglycemia.
Fig. 8.
(A) Representative islet perfusion experiments showing biphasic insulin release by stimulation with high glucose. There was no apparent inhibition or stimulation of GSIS at either high or low dose of ZDGCR2:AM106. (B) Representative microscopy images of isolated islets before and after 48 h incubation with no (control), low dose (60 nM) or high dose (60 µM) ZDGCR2:AM106. There was no observable effect on islet number or viability.
Furthermore, incubation of human islets with ZDGCR2:AM106 at doses up to 60 µM did not have any apparent visible effect on islet number or viability (Fig. 8B).
Discussion
Straightforward and flexible systems for targeted delivery of drugs or reagents to the pancreatic beta cells would enable conceptually new anti-diabetic therapeutics and repurposing of earlier drug candidate failures due to off-target toxicity21. An obvious solution for achieving tissue specific delivery is to modify the drug with a guiding moiety, which in turn binds to a target exclusively expressed on the cells of interest. There are two critical parts of a successful delivery system, firstly the target protein and secondly the guiding moiety. The target protein must be restricted to the beta cell in the pancreas, as well absent from other tissues to ensure beta cell specific drug exposure and delivery. Then, a guiding moiety must be developed, with high affinity as well as specificity for the target protein. Furthermore, the guiding moiety must have certain properties, including suitable size, stability, molecular handles for flexible functionalization as well as low off-target interactions and rapid blood clearance.
There is an urgent need for drug delivery systems to increase drug exposure in the pancreas, while minimizing the off-target and off-tissue effects and toxicity. Such delivery systems may enable re-assessment of existing and emerging drug targets and candidates, which previously have been associated with and discarded due to unacceptable side effects. There exist a multitude of established or emerging drug platforms that usually are un-targeted, that would benefit greatly from delivery systems as proposed here, including but not limited to cell therapies, extracellular vesicles (EVs), RNA and gene therapy, and nanoparticles22–24.
Here, we present the development of a new class of Affibody molecules that target the beta-cell specific DCGR2 protein. We propose that the lead candidate identified here, ZDGCR2:AM106, could be a suitable targeting agent for beta cell specific delivery of therapeutic cargo or imaging reagents (Fig. 1A). Here we confirm via immunostaining the beta-cell specificity of DGCR2, with similar expression of DGCR2 in SC-islets. This is conceptually important, as SC-islet transplantation is an emerging therapeutic strategy for T1D and is expected to become an important supply of islets grafts in the future.
DGCR2 is interesting as a target for pharmaceutical delivery or imaging in diabetes. However, the only available ligands for human DGCR2 until now have been monoclonal or polyclonal antibodies, which have a biological half-life and clearance in the order of days due to their size and inherent recycling capabilities. In the context of the delivery of imaging reagent, antibodies are not an ideal scaffold. To enable molecular imaging with antibodies, they must be labeled with relatively long-lived positron emitters such as Zirconium-89 (3.3 days of radioactive half-life). This radiolabeling strategy will lead to a significant radioactive dose to the examined individual, making it unsuitable for T1D patients where one aims to image young adults or even children. Thus, for further progress it is imperative to develop smaller peptide binders towards DGCR2, with a clearance in the order of hours suitable for radiolabeling with e.g. Fluorine-18 (109 min radioactive half-life) or Gallium-68 (68 min radioactive half-life) given their lower associated radiation dose.
Affibody molecules are small proteins that overcome the challenges associated with larger scaffolds in the context of delivery of imaging reagents, while retaining excellent affinity and specificity. Affibody molecules are generally stable and can be modified both at the C- and N terminus without interfering with the binding to the target (mediated via a binding surface made up from amino acid residues on the alpha helices 1 and 2, Fig. 1B). This is an important feature for a targeting agent that is expected to be conjugated with many different kinds of molecular structures, from small imaging reagent chelators, to drug molecules, to EVs and nanoparticles or entire cell therapy products.
Here, we describe the generation of a new class of Affibody molecules with strong binding to recombinant DGCR2. The affinity of the original Affibody molecule ZDGCR2:3D1 was in the range of 30 nM19, which was an important achievement in itself, given the lack of other known small molecule or peptide-based ligands for DGCR2. To further improve the affinity, we designed an affinity maturation library based on the sequence of ZDGCR2:3D1 and used an in-house developed bacterial display method for selection of new binders. The selection was successful and the affinity towards DGCR2 was significantly improved (up to tenfold) for several new binders. Affinity is an important parameter for probes intended for delivery of drugs or imaging reagents. Here, affinity in the low nanomolar range was achieved for three of the candidates (ZDGCR2:AM106, ZDGCR2:AM115 and ZDGCR2:AM156). Furthermore, all presented Affibody molecules exhibit suitable thermal stability with a Tm of around 60°C, which is often sufficient for the majority of chemical conjugations. More importantly, three of the Affibody molecules (ZDGCR2:3D1, ZDGCR2:AM106, and ZDGCR2:AM156) exhibited complete refolding after denaturation. This is a critical advantage as it opens up the possibility of using an array of chemical modification strategies involving high temperatures without losing binding potential.
Based on the combined data of the four Affibody molecules presented here, ZDGCR2:AM106 was selected as the most suitable lead candidate for further development as a DGCR2 targeting ligand. ZDGCR2:AM106 combines excellent in vitro and in vivo properties including affinity, refolding, high plasma stability and appropriate biodistribution. As Affibody molecules usually are renally excreted and conjugated imaging reagents accumulate in the kidneys via reuptake and trapping (potentially causing unacceptable radioactive dosing), particular attention was focused on this excretory organ when comparing the different candidates.
Based on the data presented herein, ZDGCR2:AM106 was evaluated as a scaffold for the delivery of Fluoride-18 imaging reagents to SC-islets, in a previously presented study7. [18F]ZDGCR2:AM106 successfully targeted DCGR2 expressed on human SC-islets both in vitro and in vivo, after transplantation to immunodeficient mice.
[68Ga]ZDGCR2:AM106 was confirmed to bind to the extracellular domain of human recombinant DGCR2 by an ELISA-style assay. The DGCR2 protein consists of 550 amino acids (UNIPROT entry P98153), of which residues 1-349 form the extracellular domain, residues 350–367 comprise a transmembrane domain, and 368–550 make up the intracellular part. [68Ga]ZDGCR2:AM106 did not, however, bind to the extracellular region of residues 114–297 in this assay. Thus, important parts of the binding epitope of [68Ga]ZDGCR2:AM106 could likely be mapped to either residues 22–113 or residues 289–349 of the extracellular domain of human DGCR2.
Here, we further demonstrate that Indium-111 or Gallium-68 radiolabeled ZDGCR2:AM106 not only exhibit safe dosimetry in the kidney, but also that ZDGCR2:AM106 display negligible toxicity on human islets. No effects on insulin secretion in response to glucose was seen after 48 h incubation of islets with either 60 nM or 60 µM ZDGCR2:AM106. The lower of the two different doses (60 nM) was selected based on the microdose commonly associated with the delivery of imaging agents (e.g. PET scans). On the other hand, higher doses (e.g. 60 µM) might be required in the context of pharmaceutical delivery but also at this dose no toxicity was observed.
In conclusion, we have produced and characterized several high affinity binders to the beta-cell specific protein DGCR2 based on the Affibody molecule scaffold technology. ZDGCR2:AM106 in particular displayed excellent affinity, stability and refolding, a suitable biodistribution and safety profile, thus showing promise as a lead candidate for further development of DGCR2-directed delivery of therapeutics or imaging reagents.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The molecular imaging work in this publication was performed at the Preclinical PET-MRI Platform (PPP) at Uppsala University. Amina Khalil and Zhijun Huang (Uppsala University) are acknowledged for excellent technical assistance.
Author contributions
P.C. performed, analyzed and interpreted the in vitro and in vivo imaging experiments and wrote the manuscript. J.P designed, performed, analyzed, interpreted the studies and wrote the manuscript. B.Z. performed the radiochemistry and edited the manuscript. S.V. and J.La. performed and analyzed all experiments with SC-islets and interpreted the results, and edited the manuscript. S.I. planned, performed and analyzed all experiments around islet perifusion and islet viability. O.K. conceived, planned and interpreted the study. S.S. conceived, planned and interpreted the study. J.Lö. conceived, performed, analyzed, interpreted the study and edited the manuscript. O.E. conceived, planned and interpreted the study and wrote the manuscript. All co-authors read and approved the final version of the manuscript.
Funding
The study was funded by ExoDiab, Diabetes Wellness Sweden (#2409), Barndiabetesfonden, Diabetesfonden, Science for Life Laboratory, the Swedish Research Council (2020–02312 and 2024-03659; OE, CAN 20 1090 PjF and CAN 23 2717 Pj; JL, 2019 − 01415 and 2023-02221; OK), the Swedish Agency for Innovation VINNOVA (2019/00104, JL; CellNova 2017–02105, JL), the Knut and Alice Wallenberg Foundation through the Wallenberg Center for Protein Technology (KAW 2019.0341; SS), Cancerfonden (24 3754 Pj; OE) Novo Nordisk Foundation, an EFSD/Novo Nordisk grant, the Ernfors Family Fund, the Sten A Olssons Foundation, Helmsley Charitable Trust, JDRF (1-SRA-2020-973-S-B) and the Juvenile Diabetes Foundation International.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Declarations
Competing interests
O.E. is an employee of Antaros Tracer. O.E. and O.K. are minority shareholders of Antaros Tracer AB, which hold patents related to DGCR2 binding peptides. Otherwise, the authors declare no competing interests.
Ethics approval and consent to participate
All the performed procedures were in accordance with the ARRIVE guidelines for animal research. The animal experiments were ethically reviewed, authorized by the Animal Ethics Committee of the Swedish Animal Welfare Agency and carried out in accordance with the European Directive 2010/63/EEC and the institutional guidelines at Uppsala University (UFV 2007/724).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
John Löfblom and Olof Eriksson contributed equally to this work.
Contributor Information
John Löfblom, Email: lofblom@kth.se.
Olof Eriksson, Email: olof.eriksson@ilk.uu.se.
References
- 1.Grattoni, A. et al. Harnessing cellular therapeutics for type 1 diabetes mellitus: progress, challenges, and the road ahead. Nat. Rev. Endocrinol. Published Online September. 310.1038/s41574-024-01029-0 (2024). [DOI] [PubMed]
- 2.von Herrath, M. G., Korsgren, O. & Atkinson, M. A. Factors impeding the discovery of an intervention-based treatment for type 1 diabetes. Clin. Exp. Immunol.183 (1), 1–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung, P. & Eriksson, O. The current state of beta-cell-mass PET imaging for diabetes research and therapies. Biomedicines9 (12), 1824 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eriksson, O. et al. In vivo imaging of beta cells with radiotracers: State of the art, prospects and recommendations for development and use. Diabetologia [Internet]. 59 (7), 1340–1349 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Lindskog, C. et al. Novel pancreatic beta cell-specific proteins: Antibody-based proteomics for identification of new biomarker candidates. J. Proteom.75 (9), 2611–2620 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Bruin, J. E. et al. Accelerated maturation of human stem cell-derived pancreatic progenitor cells into insulin-secreting cells in immunodeficient rats relative to mice. Stem Cell. Rep.5 (6), 1081–1096 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, P. et al. Preclinical evaluation of affibody molecule for PET imaging of human pancreatic islets derived from stem cells. EJNMMI Res.13 (1), 107 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tesovnik, T. et al. Extracellular vesicles derived human-miRNAs modulate the immune system in type 1 diabetes. Front. Cell. Dev. Biol.8, 202 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voll, S. L. et al. Obesity in adults with 22q11.2 deletion syndrome. Genet. Med.19 (2), 204–208 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van, L. et al. 22q11.2 microdeletion and increased risk for type 2 diabetes. EClinicalMedicine26, 100528 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scoville, D. W., Gruzdev, A. & Jetten, A. M. Identification of a novel lncRNA (G3R1) regulated by GLIS3 in pancreatic β-cells. J. Mol. Endocrinol.65 (3), 59–67 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasquali, L. et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat. Genet.46 (2), 136–143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayer, A. T. et al. Practical immuno-PET radiotracer design considerations for human immune checkpoint imaging. J. Nucl. Med.58 (4), 538–546 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ståhl, S. et al. Affibody molecules in biotechnological and medical applications. Trends Biotechnol.35 (8), 691–712 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Feldwisch, J. & Tolmachev, V. Engineering of affibody molecules for therapy and diagnostics. Methods Mol. Biol.899, 103–126 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Schmidt, M. M. & Wittrup, K. D. A modeling analysis of the effects of molecular size and binding affinity on tumor targeting. Mol. Cancer Ther.8 (10), 2861–2871 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.https://clinicaltrials.gov/study/NCT05623345?term=izokibep&rank=2, accessed 09-OCT-2024.
- 18.Balboa, D. et al. Functional, metabolic and transcriptional maturation of human pancreatic islets derived from stem cells. Nat. Biotechnol.40 (7), 1042–1055 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersson, K. G., Persson, J., Ståhl, S. & Löfblom, J. Autotransporter-mediated display of a naïve affibody library on the outer membrane of Escherichia coli. Biotechnol. J.14 (4), 1–8 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Persson, J. et al. Discovery, optimization and biodistribution of an Affibody molecule for imaging of CD69. Sci. Rep.11 (1), 19151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Simaeys, D. et al. RNA aptamers specific for transmembrane p24 trafficking protein 6 and Clusterin for the targeted delivery of imaging reagents and RNA therapeutics to human β cells. Nat. Commun.13 (1), 1815 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlsson, P. O., Schwarcz, E., Korsgren, O. & Le Blanc, K. Preserved β-cell function in type 1 diabetes by mesenchymal stromal cells. Diabetes64 (2), 587–592. 10.2337/db14-0656 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Witwer, K. W. et al. Defining mesenchymal stromal cell (MSC)-derived small extracellular vesicles for therapeutic applications. J. Extracell. Vesicles. 8 (1). 10.1080/20013078.2019.1609206 (2019). [DOI] [PMC free article] [PubMed]
- 24.Solaro, R., Chiellini, F. & Battisti, A. Targeted delivery of protein drugs by nanocarriers. Materials3 (3), 1928–1980 (2010). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.