Abstract
SNARE (soluble N-ethylmaleimide sensitive factor attachment protein receptor) proteins are the minimal machinery required for vesicle fusion in eukaryotes. Formation of a highly stable four-helix bundle consisting of SNARE motif of these proteins, drives vesicle/membrane fusion involved in several physiological processes such as neurotransmission. Recycling/disassembly of the protein machinery involved in membrane fusion is essential and is facilitated by an AAA+ ATPase, N-ethylmaleimide sensitive factor (NSF) in the presence of an adapter protein, α-SNAP. Here we use single-molecule fluorescence spectroscopy approaches to elucidate the chain of events that occur during the disassembly of SNARE complex by NSF. Our observations indicate two major pathways leading to the sequential disassembly of the SNARE complex: one where a syntaxin separated intermediate state is observed before syntaxin disassembles first, and a second where Vamp disassembles from the other proteins first. These studies uncover two parallel sequential pathways for the SNARE disassembly by NSF along with a syntaxin separated intermediate that couldn’t be observed otherwise.
Subject terms: Single-molecule biophysics, SNARE, Microscopy, Fluorescence resonance energy transfer
SNARE proteins are the minimal machinery required for the vesicle fusion in eukaryotes. Here the authors use single-molecule fluorescence spectroscopy to elucidate the chain of events during the disassembly of SNARE complex by NSF, showing two parallel sequential pathways for SNARE disassembly.
Introduction
In eukaryotic cells, vesicular trafficking is essential in regulating many physiological processes, including neurotransmission1. Membrane fusion between the vesicles and the target membrane is an important, ubiquitous and highly orchestrated process in the trafficking2,3. A highly conserved family of proteins called SNAREs (soluble N-ethylmaleimide sensitive factor attachment protein receptor) regulate the membrane fusion and hence the vesicular transport4. Formation of a highly stable four-helix bundle between vesicular SNARE (v-SNARE) protein called Vamp (vesicle attached membrane protein or synaptobravin) and target SNARE (t-SNARE) (syntaxin (Stx) and synaptosome associated protein 25 (SN25)) mediates the membrane fusion in synaptic exocytosis during neurotransmission3,5–9. Upon the completion of vesicle fusion, the SNARE is called a cis-SNARE, with both v- and t-SNAREs anchored on the same membrane, which is disassembled by a clade 3, type II AAA + (ATPase associated with diverse cellular activities)10–12 called NSF (N-ethylmaleimide sensitive factor) with the help of an adaptor protein, α-SNAP (α- soluble NSF attachment protein). This momentary complex of cis-SNARE with several α-SNAPs and an NSF is termed as 20S complex, which is the fundamental unit for the disassembly of SNARE. NSF utilises energy from ATP hydrolysis and with the help of α-SNAP, disassembles the cis-SNARE complex into individual proteins, which will be used for next rounds of synaptic vesicle formation and fusion13–19.
Biochemical and structural studies on the individual components of the SNARE complex have provided valuable details of the assembly and its disassembly by NSF. Crystal structures of the cis-SNARE complex reveal a highly conserved central ionic layer termed a ‘zero-layer’, surrounded by 15 layers of hydrophobic amino acids that form a highly stable helical bundle5,20–25. In recent years, structural and single-molecule Förster resonance energy transfer (FRET) studies of the 20S complex have revealed a typical left-handed SNARE bundle wrapped in a right-handed symmetrical fashion by two to four α-SNAPs providing a tight physical grip useful for the disassembly26–32. Single-molecule studies have shown that NSF utilises only one round of ATP hydrolysis and proposed that SNARE disassembles in a single burst in a ‘spring loaded’ fashion during a global disassembly31,33,34. Structural analysis has revealed direct interactions between NSF D1 ring with the SNARE complex, and they also revealed that the N-terminal chain of SN25 interaction and intercalation into the D1 ring pore of NSF indicates loading and partial translocation of SNARE during disassembly29,30. Biochemical analysis of the residues involved in the specific interaction between SNARE and α-SNAP revealed that the latter may exert a different extent of interaction/force on the SNARE layer that may result in a sequential order of disassembly among different chains of SNARE30,35. The fastest disassembled chain should be responsible for the unwinding of the SNARE complex.
Here, we perform two- and three-colour single-molecule FRET (smFRET) to analyse the disassembly of a single SNARE complex by NSF in order to shed light on the disassembly pathway of the SNARE and its disassembly intermediates. With the ability to examine the movements of all three chains simultaneously, we reveal two parallel sequential pathways.
Results
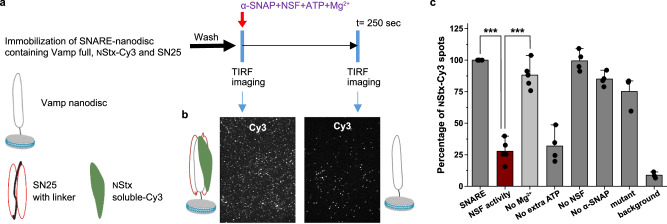
Single-molecule fluorescence assay of SNARE-nanodisc containing Vamp full, SN25 and Stx-Cy3 by NSF
We have developed a single-molecule assay for the disassembly of SNARE complex reconstituted into the nanodiscs, by NSF. One main difference over the previous single-molecule assays33 was that the SNARE complex was formed, purified and reconstituted into the nanodiscs, which was further used for the disassembly assay (details in methods). The experimental approach is shown in Fig. 1a. We have prepared a SNARE complex with syntaxin soluble-Cy3 (syntaxin soluble (2–262) with S200C, NStx linked to Cy3), Vamp with transmembrane domain and SN25 with linker, which is further purified using affinity chromatography. The obtained SNARE complex is reconstituted into nanodiscs (see methods for more details) and immobilised onto the microscope slide using biotin-streptavidin-biotin linkage. After washing out unbound SNARE, a mixture of NSF, α-SNAP, and 10 mM Mg2+, with or without 2 mM ATP, were added and incubated for 250 s, and the movies were collected to quantify the number of Cy3 spots per imaging field. The decrease in the number of spots after adding the NSF mixture represents the percent disassembly of the SNARE complex. From the data obtained, we observed that under our experimental conditions, > 75% of spots have disappeared, indicating high NSF activity (Fig. 1b). The reaction specificity was tested using various controls, as shown in Fig. 1c. The disappearance of the spots was observed only in the presence of all the components- NSF, α-SNAP, 10 mM Mg2+ and 2 mM ATP. Lack of any one of these components hindered the spot disappearance indicating the essential of α-SNAP, NSF binding to SNARE and the ATP hydrolysis of NSF.
Fig. 1. Single-molecule fluorescence assay of SNARE-nanodisc containing Vamp full, SN25 and Stx-Cy3 by NSF.
a Procedure of the single-molecule SNARE disassembly assay on total internal reflection fluorescence (TIRF) microscope. b TIRFM images of Cy3 fluorescence channel containing SNARE-nanodisc after immobilisation along with a pictorial representation of nanodisc present on the glass slide before and after the disassembly by NSF. c Number of Stx-Cy3 nanodiscs present under various experimental conditions. Disassembly reaction in the presence of all components (brown), without the addition of Mg2+ (light grey), only with ATP already bound to the NSF (No extra ATP) and other controls. p-values calculated using a two-tailed paired t test and mean ± SD values are given, ***= p < 0.005. (n = 3-6 independent SNARE disassembly experiments).
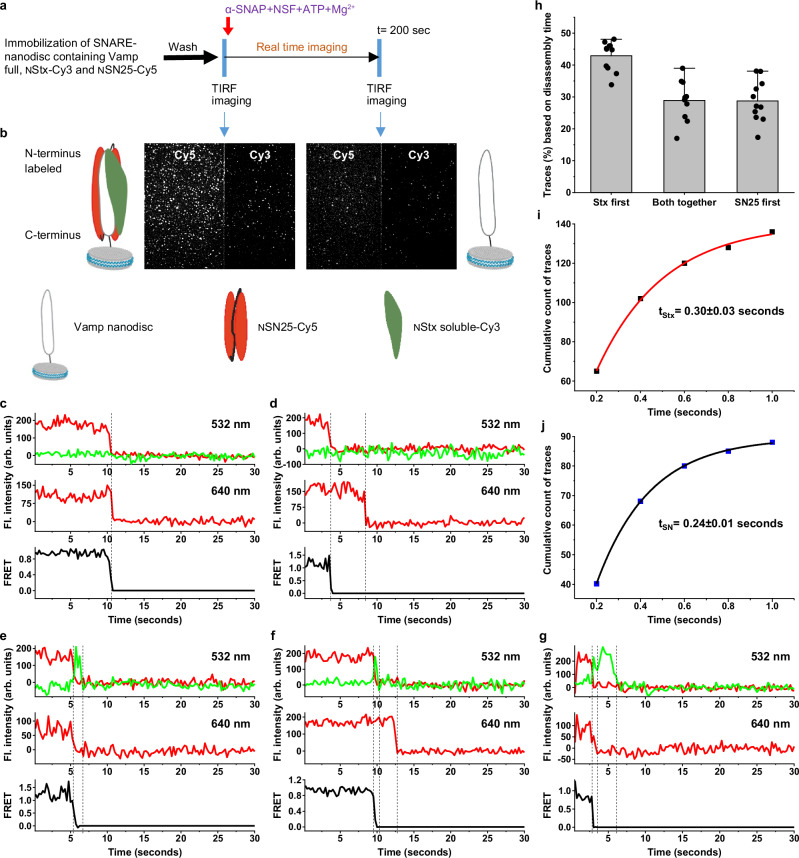
SNARE disassembly by NSF monitored using single-molecule FRET
Further, to examine the kinetics of disassembly of individual chains of the SNARE, single cysteine mutants of Syntaxin (Stx) and SNAP25 in the same layer of SNARE at the N-terminus, namely Stx S200C (NStx) and SN25 E27C (NSN25) were labelled with dyes of a FRET pair- Cy3 and Cy5, respectively (Supplementary Fig. 1). SNARE complex was formed with Vamp-full, NStx-Cy3 and NSN25-Cy5 and reconstituted into nanodiscs and the NSF disassembly assay was carried as described above (Fig. 2a). High FRET between Stx-Cy3 and SN25-Cy5 was observed as the mutations in Stx S200 and SN25 E27 are present in the same SNARE layer within 1-2 nm from each other (Supplementary Fig. 1). Addition of NSF, α-SNAP, 10 mM Mg2+ and 2 mM ATP resulted in loss of >70% of the individual spots and > 80% of the FRET pairs indicating high NSF activity (Fig. 2b and Supplementary Fig. 2a–c). For individual single-molecule fluorescence trajectories under Alternative Laser Excitation (ALEX) mode (see methods for details) between 532 nm and 640 nm lasers, the sudden decrease of Cy3 and FRET signal under 532 nm laser excitation indicates dissociation of NStx-Cy3 and the sudden decrease of Cy5 signals under 640 nm laser excitation indicates dissociation of NSN25-Cy5 (Fig. 2c–g, n = 743 traces). Under our experimental conditions, photobleaching rates were ~ 8-10 fold slower than disassembly, which would not affect our analysis (Supplementary Fig. 2g–i). Only ~28% of the traces had NStx-Cy3 and NSN25-Cy5 dissociated from Vamp at the same time within our time resolution (Fig. 2c, h and Supplementary Fig. 2d, n = 205 traces). For ~45% of the traces, NStx-Cy3 disassembles before NSN25-Cy5 (Fig. 2d, n = 336 traces), and for ~27% of the traces, NSN25-Cy5 disassembles before NStx-Cy3 (Fig. 2e, n = 202 traces). These results are surprising as both chains were proposed to leave together under previously hypothesised global disassembly33. The dwell times from injection of NSF mixture to initiate disassembly until dissociation of NStx-Cy3 and NSN25-Cy5 were quantified for each group of traces and shown in Supplementary Fig. 1e, f.
Fig. 2. SNARE disassembly by NSF monitored using single-molecule FRET.
a Procedure of the smFRET assay of SNARE disassembly by NSF using TIRF microscope. b TIRF microscopy images FRET pair, Cy3 and Cy5 fluorescence channels representing SNARE-nanodisc after immobilisation along with a pictorial representation of nanodisc present on the glass slide before and after the disassembly by NSF. c–g Single-molecule trajectories of the NStx-Cy3 (green), NSN25-Cy5 (red) and FRET (dark grey) collected under the ALEX cycle between 532 nm and 640 nm lasers during the disassemble. Fl., fluorescence, Arb., arbitrary. c a representative trace showing both the NStx-Cy3 chain and NSN25-Cy5 chain disassembling at the same time. d a representative trace where the NStx-Cy3 chain disassembles first, followed by the NSN25-Cy5 chain. e a representative trace where the NSN25-Cy5 chain disassembles first, followed by the NStxCy3 chain. f a representative trace where a separation of the NStx-Cy3 chain is observed, followed by NStx-Cy3 disassemble first and then the NSN25-Cy5 chain. g a representative trace where a separation of the NSN25-Cy5 chain is observed, followed by NSN25-Cy5 disassemble first and then the NStx-Cy3 chain. h Classification of the single molecule traces based on the chain disassembly time (n = 11, each n represents an independent SNARE disassembly assay), data shown as mean ± SD. i, j Graphs showing the average lifetime of the chain separated state before the chain disassembly, 0.30 ± 0.03 seconds in case of traces (f) and 0.24 ± 0.01 s in case of traces (g). The dotted lines show the FRET separation and chain disassembly in traces (c–g).
Further, upon careful examination of individual traces, where NStx and NSN25 chains did not dissociate at the same time, we have observed a short-lived zero-FRET state before either of them dissociates, indicated by the sudden disappearance of the FRET signal and simultaneously raising of Cy3 signal (Fig. 2f, g). The dwell times of the short-lived zero-FRET state were 0.30 ± 0.03 seconds in the case of NStx disassembling first (n = 148 traces) and 0.24 ± 0.01 seconds in case of NSN25 disassembling first (n = 95 traces) (Fig. 2i. j). Taken together these results indicate a transient intermediate state in which NStx and NSN25 chains were separated before their disassembly. In order to double-check the transient state, we performed a Hidden-Markov analysis (Supplementary Fig. 3)36. Upon analysing the traces, where a transient intermediate of NStx separates to a zero-FRET state before the disassembly, a transient state with a dwell time of ~ 0.38 ± 0.02 seconds was observed (Supplementary Fig. 3). Since the transient state is very short-lived compared to our time resolution (~ 1-1.5 ×), we carried out similar disassembly assay at higher resolution. NSF disassembly at 50 ms resolution also shown similar behaviour of disassembly as mentioned above, along with an observable transient state before the chain disassembly. Analysis has revealed a transient state with dwell time of 0.24 ± 0.02 seconds for NStx (n = 106) separated zero-FRET state and 0.32 ± 0.03 seconds for NSN25 (n = 103) separated zero-FRET states before their disassembly (Supplementary Fig. 4). Since, in our experiments we added α-SNAP, NSF, ATP and Mg2+ to the SNARE-nanodisc and monitored the disassembly, we tested if there is any delay in the formation of 20S complex that could affect the kinetics of the disassembly. So, we tested the ATP hydrolysis of the 20S complex, by forming the 20S complex under non-hydrolysing conditions on the microscope chamber by incubating SNARE-nanodisc with α-SNAP, NSF-EDTA, and ATP (methods). 20S complex disassembly was initiated by the addition of 10 mM Mg2+, and the dynamics of NStx-Cy3 disassembly were monitored. We observed a slightly faster time scale of NStx-Cy3 disassembly (4.1 ± 0.5 seconds) compared to our methodology (4.5 ± 0.2 seconds). This minimal difference indicates the formation of the 20S complex in our experiments is very fast (< 0.5 s) (Supplementary Fig. 5). Since the transient state is not observed before 1–2 s in our traces, we can conclude that the transient state observed is formed during the SNARE disassembly, not during the 20S complex formation.
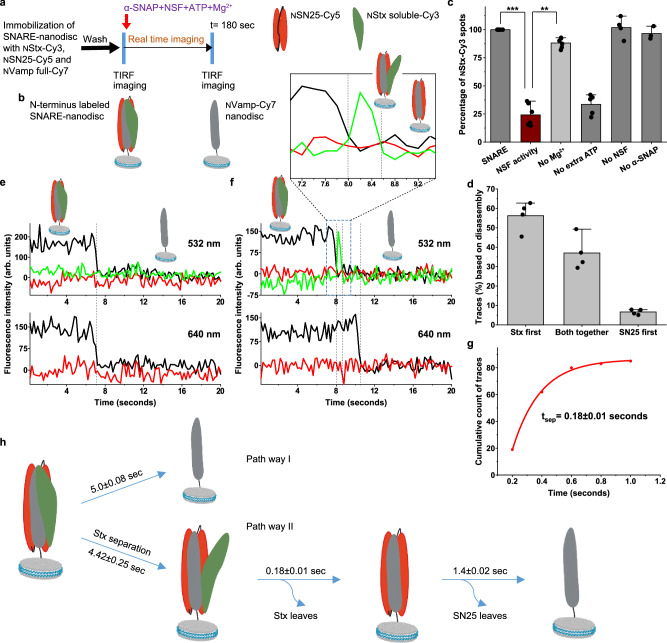
Vamp anchored SNARE-nanodisc disassembly by NSF monitored using three colour single-molecule FRET
In order to gain more insights into the separation between Stx and SN25 and to investigate the interaction between Vamp-Stx and Vamp-SN25, we have carried out three-colour FRET experiments on the SNARE complex by labelling Vamp with Cy7 in the same layer as the other two (Fig. 3a, band Supplementary Fig. 1). Based on our design, high FRET efficiencies between all the three FRET pairs have been observed: NStx-Cy3 to NSN25-Cy5, NStx-Cy3 to NVamp-Cy7 (Vamp R30C-Cy7) and NSN25-Cy5 to NVamp-Cy7. Hence, when we excite with either 532 nm laser or 640 nm laser, we only capture a predominantly Cy7 signal with almost no Cy3 and Cy5 signals. NSF disassembly of the three-colour labelled SNARE was checked based on the disappearance of the fluorescence spots of both NStx-Cy3 and NSN25-Cy5 (Fig. 3c). Individually, single trace analysis of NSF disassembly of the three-colour SNARE complex reveals two major classes of traces and one minor class (Fig. 3d). ~ 40% traces show a single signal transition where both NStx-Cy3 and NSN25-Cy5 chains disassemble from NVamp-Cy7, which remains attached to the surface (n = 98 traces) (Fig. 3e) and ~ 50% of the traces shows NStx-Cy3 leaving before NSN25-Cy5 (n = 131 traces). The dwell time of NStx-Cy3 dissociation after injecting the NSF mixture is 4–5 seconds (Supplementary Fig. 6a, b). When NStx-Cy3 disassembles before NSN25-Cy5, a uniform Cy5/Cy7 intensity is observed under 640 nm excitation before NSN25-Cy5 dissociation, whereas a sudden loss of the NVamp-Cy7 intensity with a concomitant rise in the NStx-Cy3 intensity is captured under 532 nm excitation (Fig. 3f). Such phenomenon indicates that the NStx-Cy3 chain is transiently separated from the NVamp and NSN25 complex before it leaves (Fig. 3f, more traces shown in Supplementary Fig. 7) with the lifetime of the separated state being 0.18 ± 0.01 s (Fig. 3g). Even though NStx separates and subsequently disassembles, Cy5/Cy7 signals under 640 nm excitation were almost unaffected, indicating that a Stx separation and chain leaving has little effect on the Vamp-SN25 binary complex, which has a lifetime of 1.4 ± 0.02 s, after which SN25 disassembles indicating complete disassembly of the SNARE (Fig. 3h and Supplementary 6c). Since previously no studies have been reported that show a stable binary complex between Vamp and SN25 that is observed transiently in our study, we verified the Vamp-SN25 binary complex formation. Upon 1:1 incubation > 2 h, a stable Vamp-SN25 binary complex has been formed (Supplementary Fig. 8). Together, consistent with our two-colour SNARE disassembly results, we were able to observe parallel pathways of SNARE disassembly and a short-lived intermediate state containing separated Stx chain when Stx dissociates before SN25 (Fig. 3h).
Fig. 3. Vamp anchored SNARE-nanodisc disassembly by NSF monitored using three-colour single-molecule FRET.
a Procedure of the smFRET assay of SNARE disassembly by NSF using TIRF microscope. b Pictorial representation of complexes on the glass slide before and after the disassembly by NSF. c Relative percentage of spots detected in the Cy3 channel under various experimental conditions. SNARE, NVamp-anchored SNARE containing NStx-Cy3, NSN25-Cy5 and NVamp-Cy7 immobilised on to the TIRFM surface; NSF activity, After the addition of α-SNAP, NSF along with 2 mM ATP and 2 mM MgCl2; No Mg2+, After the addition of α-SNAP, NSF along with 2 mM ATP, without MgCl2; No extra ATP, After the addition of α-SNAP, NSF complexed with ATP along with 2 mM MgCl2; No NSF, After the addition of α-SNAP, 2 mM ATP and 2 mM MgCl2; No α-SNAP, After the addition of NSF, 2 mM ATP and 2 mM MgCl2. The data shows mean ± SD from multiple experiments from 4 independent sample preparations with multiple (n = 3-6) SNARE disassembly experiments. p = ***< 0.005 and **< 0.01, two-sided paired t test. d Percentage of the classified traces based on the chain disassembly time. n = 4, each n is data from an individual sample preparation, with 2–4 experiments from each sample. Values are mean ± SD. e, f Single-molecule trajectories of the NStx-Cy3 (green), NSN25-Cy5 (red) and NVamp-Cy7 (dark grey) during the disassembly. e a representative trace showing both NStx-Cy3 and NSN25-Cy5 chains disassembling from NVAMP-Cy7 at the same time. f a representative trace where NStx-Cy3 separation is observed before it disassembles first, followed by NSN25-Cy5 dissociation. g Cumulative distribution of the lifetime of the transient NStx-Cy3 separated state (n = 84 traces). h Pictorial representation of two major pathways of SNARE disassembly by NSF under our experimental design. Pathway I, showing both NStx and NSN25 disassembly together from NVamp or vice versa (40%) and Pathway II, with an NStx separated transient state observed during NStx disassembly first, followed by NSN25 (50.5%).
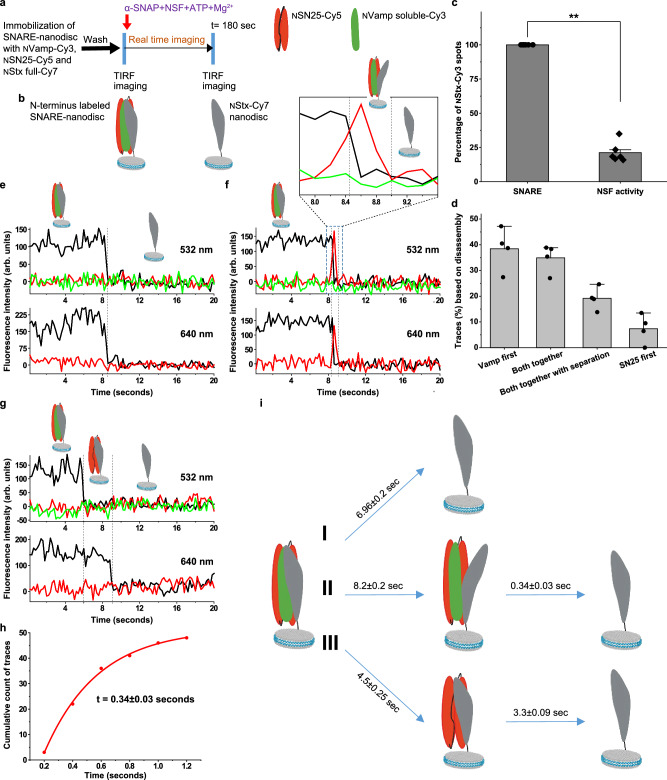
Syntaxin anchored SNARE-nanodisc disassembly by NSF monitored using three-colour single-molecule FRET
The pathway I observed in Fig. 3h, can be interpreted in two ways. It could be a global disassembly pathway, with all three proteins disassemble together or Vamp disassembling first from the Stx and SN25 or a combination of both. In order to investigate this process in detail along with the details on syntaxin separated state, a Stx-anchored SNARE was prepared with the same mutations as that of Vamp anchored SNARE on the N-terminus of the SNARE helix bundle (Supplementary Fig. 1). However, NStx-full was labelled with Cy7 and NVamp was labelled with Cy3 to form SNARE with NSN25-Cy5. A similar NSF disassembly assay, as described before, was performed (Fig. 4a–c). Single-molecule traces have revealed three major classes of traces and one minor class (Fig. 4d). ~ 36% traces show that both NVamp soluble-Cy3 and NSN25-Cy5 dissociate simultaneously from NStx-full-Cy7 without any detectable intermediate state (Fig. 4e, n = 73 traces). Interestingly, ~ 21% of traces display a transient intermediate state, indicated by the sudden disappearance of NStx-full-Cy7 signal accompanied by the appearance of NSN25-Cy5 signal under both 532 nm and 640 nm excitation before NVamp-Cy3 and NSN25-Cy5 dissociate from NStx-full-Cy7 (Fig. 4f and Supplementary Fig. 9) with a lifetime of 0.34 ± 0.03 s (Fig. 4h, n = 43 traces). Such pattern of fluorescence signals suggests that, in the transient intermediate state, NVamp-Cy3 and NSN25-Cy5 remain closely bound and NStx-Cy7 is transiently separated from them before its dissociation (Fig. 4f and Supplementary Fig. 9). In our experiments, each ALEX cycle between 532 nm and 640 nm lasers spends 0.2 s, and a transient state lasting shorter than two ALEX cycles (~ 0.4 s) might be missed in our state assignment. The average lifetime of the Stx-separated state is 0.34 s, indicating a significant amount of traces containing short Stx-separated states beyond our detection limit would be identified as the ones without detectable intermediate state. Together, we speculated that these two major classes (pathways I and II in Fig. 4i) should be assigned as the same pathway, with Stx dissociating first with a transient separated state. Their total percentage (~ 57%) is very similar to that of the Stx-separated pathway in Fig. 3h (~ 50.5%, pathway II). Furthermore, ~ 38% of traces have Vamp-Cy3 disassembles first, followed by NSN25-Cy5 (Fig. 4g, n = 78 traces), which matches well with the pathway I of Fig. 3h observed with Vamp-anchored SNARE. The average time spent in each state under different pathways was extracted from single-molecule trajectories (Fig. 4h and Supplementary Fig. 10) and summarised in Fig. 4i. These results clearly show the presence of a Stx separated intermediate state previously observed in the two-colour and Vamp anchored SNARE. These results also reveal a pathway where Vamp disassembles first from the Stx and SN25 (pathway III in Fig. 4i), which could be the same pathway observed with Vamp anchored SNARE (Pathway I in Fig. 3h).
Fig. 4. Syntaxin anchored SNARE-nanodisc disassembly by NSF monitored using three-colour single-molecule FRET.
a Procedure of the smFRET assay of SNARE disassembly by NSF using TIRF microscope. b Pictorial representation of complexes on the glass slide before and after the disassembly by NSF. c Percentage of spots detected in the NVamp soluble-Cy3 channel before and after the disassembly. p = **< 0.01, two-sided paired t test, n = 5, individual experiments from multiple sample preparations. Values are mean ± SD. d Relative percentage of the classified of the traces based on the chain disassembly time and separation. n = 4, each n is data from an independent sample preparation with 2-3 experiments from each sample. Data is shown as mean ± SD. e–g Single-molecule trajectories of the NVamp-Cy3 (green), NSN25-Cy5 (red) and NStx-Cy7 (dark grey) during the disassembly. e a representative trace showing NVamp and NSN25 disassemble from NStx together at the same time. f a representative trace where Stx separation from NVamp-Cy3 and NSN25-Cy5 is observed before NVamp and NSN25 disassemble from NStx. g a representative trace where NVamp-Cy3 disassembles first, followed by NSN25-Cy5. h Cumulative distribution of the lifetime of the NVamp-Cy3 and NSN25-Cy5 separated state (n = 43 traces). i Pictorial representation of three major pathways of SNARE disassembly by NSF under our experimental design. I- Pathway showing NStx disassemble from NVamp and NSN25 first and vice versa (36%, n = 73 traces) and II-Pathway where a transient state with NStx separated from NVamp and NSN25 is observed before NStx disassembles (20.5%, n = 43 traces). III- Pathway showing NVamp disassembles first, followed by NSN25 (38.5%, n = 78 traces).
Vamp anchored C-SNARE-nanodisc disassembly by NSF monitored using three-colour single-molecule FRET
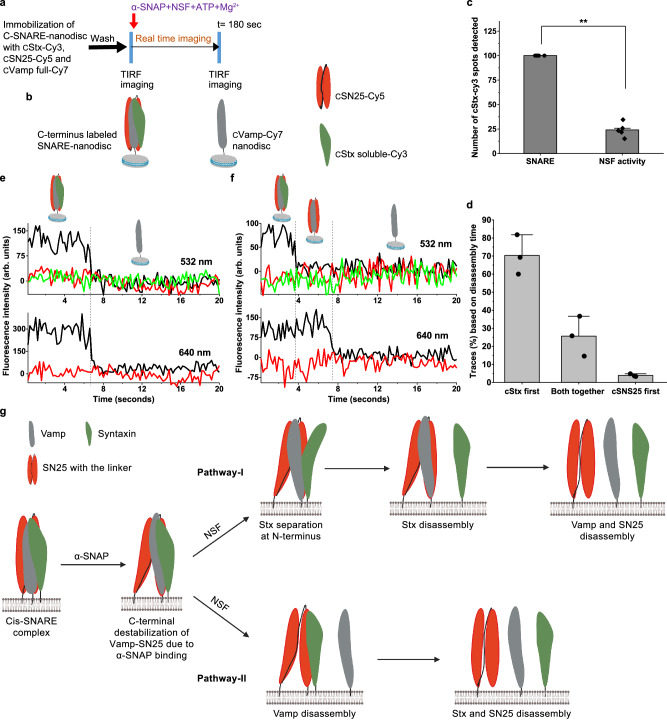
Further, we investigated if this separation of Stx persists all along the cylindrical SNARE complex or only restricted to the N-terminus, so we labelled the dyes on the C-terminal of the Vamp-anchored SNARE with the same dyes as that of the N-terminus and performed the NSF disassembly assay (Fig. 5a–d and Supplementary Figs. 1, 11a). In this case, we also observed two major classes of traces, one where cStx (C-terminal labelled Stx) and cSN25 (C-terminal labelled SN25) disassembles together from cVamp (C-terminal labelled Vamp) and another where cStx-Cy3 leaves first followed by cSN25-Cy5. No transient intermediate state, as shown in Figs. 2, 3, and 4, has been observed in these traces (Fig. 5e, f, n = 64 traces). These results indicate that during the disassembly, the transient separation between Stx and Vamp-SN25 chains is observed only at the N-terminal end of the SNARE bundle. The parallel disassembly pathways and the dwell times of each step were extracted from single-molecule trajectories (Supplementary Fig. 11b–d) and summarised in Supplementary Fig. 11e. Interestingly, the high FRET between the cVamp-Cy7 and cSN25-Cy5 has been changed to a medium or low FRET after the addition of α-SNAP and NSF (Supplementary Fig. 12a, n = 51 traces). This indicates the destabilization of the C-terminus of the SNARE complex. Further, we have added only α-SNAP separately to check their individual effect on the destabilization and found that the binding of α-SNAP alone is causing this destabilization and making the C-terminal of the SNARE complex flexible, as described earlier33 (Supplementary Fig. 12b). In all our dynamic experiments, a mixture of α-SNAP, NSF, ATP and Mg2+ was added while collecting the data. While it is true that we do not know when exactly the 20S complex has formed, the observed changes in the intensity of the C-terminal FRET pairs that correspond to the unwinding of the SNARE complex upon binding to the α-SNAP can be observed immediately. Since the association of α-SNAP with NSF is presumed to be instantaneous, we believe that this change in the intensity and FRET signal indicating structural perturbation upon binding to α-SNAP can be an initial step in the formation of the 20S complex.
Fig. 5. Vamp anchored C-SNARE-nanodisc disassembly by NSF monitored using three-colour single-molecule FRET.
a Procedure of the smFRET assay of C-SNARE disassembly by NSF on TIRF microscope. b Pictorial representation of SNARE-nanodisc with N- and C-terminal end labelling. c Relative percentage of spots detected in the cStx-Cy3 channel before and after the disassembly. p = **< 0.01, two-tailed paired t test, n = 5, individual experiments from multiple sample preparations. Values are mean ± SD. d Percentage of the classified traces based on the chain disassembly time, n = 3, each n is data from an independent sample preparation with 2-3 experiments from each sample. Values are mean ± SD. e, f Single-molecule trajectories of the cStx-Cy3 (green), cSN25-Cy5 (red) and cVamp-Cy7 (dark grey) during the C-SNARE disassembly. e a representative trace showing both cStx-Cy3 and cSN25-Cy5 chains disassemble at the same time from cVamp-Cy7. f a representative trace where cStx-Cy3 disassembles first, followed by cSN25-Cy5. g Pictorial representation of plausible major pathways of SNARE disassembly by NSF (Created in BioRender. Cheppali, S. (2024) https://BioRender.com/f62r845). Binding of α-SNAP itself can induce destabilization at the C-terminal end of the cis-SNARE complex. The attachment of NSF and the ATP hydrolysis leads to SNARE disassembly in two major sequential pathways. Pathway I shows sequential disassembly with Stx separated intermediate and proceeds through Stx disassembles first and Pathway II, where sequential disassembly occurs with Vamp disassembles first.
Discussion
Our present work also indicates the previous observation of one round of ATP hydrolysis for one round of SNARE disassembly by NSF, as additional ATP was not required for the disassembly33 (Fig. 1c). Due to our design of the SNARE complex, we were able to capture and obtain information regarding different pathways to that of reported earlier33. Two major pathways of sequential disassembly of the SNARE complex were observed from our design, with one pathway where Stx disassembles first with an observable transient state where it is separated from the rest of the complex at the N-terminus as evident from our two and three-colour FRET studies (Fig. 5g, pathway I). Another major pathway observed is where Vamp disassembles first from Stx and SN25. The percentage of traces following this route, in all the three-colour SNARE complexes matches well (pathway I in Fig. 3h and Supplementary Fig. 11e, and pathway III in Fig. 4i) with each other to consider them the same pathway (40, 42 and 38.5%) (Fig. 5g, pathway II). We cannot fully rule out the possibility that a minor global disassembly pathway might contribute to the percentages observed in pathway I in Fig. 3h and Supplementary Fig. 11e.
We observed that in all of our experiments with Stx N- or C-terminal labelled Vamp anchored SNARE or Stx anchored SNARE, the observed chain leaving time is Vamp ~ Syntaxin ≤ SN25 as shown in Figs. 3h, 4i and Supplementary Fig. 11e exhibiting delayed SN25 leaving in all the pathways. Thus, during the disassembly, either the force might be applied to Vamp or Syntaxin to break the SNARE complex while the SN25 N-terminus partially intercalate into the NSF D1 pore as supported by the results of the chemical cross-linking of proteins coupled with mass spectrometry analysis30. Therefore, the SN25 chain will remain bound until the whole 20S complex breaks down. This process explains the main feature of the sequential disassembly of the SNARE complex that the chain leaving time is Vamp ~ Syntaxin ≤ SN25. In addition, the interaction between the NSF D1 pore loop and SN25 is said to serve as loading of the SNARE complex onto NSF that triggers ATP hydrolysis35. Upon hydrolysis, this state with the SNARE complex positioned deeper in the pore can cause additional shearing force on the SNARE complex driving it to complete disassembly.
Considering Vamp disassembly, according to our previous observation from cryo-EM and mutational analysis based on the α-SNAP interaction with individual SNARE proteins, the Vamp residues E62 and D68 are very important for the SNARE disassembly, and Vamp could be the chain through which NSF and α-SNAP applies its force. Due to the dynamic nature of the N-terminal domains of NSF, a clear role has not been attributed to them in terms of application of force even though their interaction and orientation with α-SNAP has been proposed to exert force on the SNARE bundle to disassemble27,29,30. Among the interactions of α-SNAP with individual SNAREs, the interaction of Vamp with α-SNAP has been shown to be essential30. Such an interaction can cause resistance to the movement of the SNARE complex through the NSF pore upon ATP hydrolysis. This resistance, in turn, generates strain on the system, resulting in disassembly. Therefore, the force applied predominantly on Vamp could be a pivotal step for the disassembly of the SNARE complex and overcome the Vamp-SN25 interaction to facilitate early Vamp disassembly, thus possibly acting as a rate-limiting step. Hence, we observe another major pathway in which Vamp disassembles first. The two parallel disassembly pathways exhibiting similar proportions (Fig. 5g) suggest that the disassembly of Vamp and Stx have similar energetic barriers, which are determined by interactions among the adjacent helix chains of the SNARE complex and the disruption force applied to them.
Another major important observation in our studies is the presence of a Stx separated transient state. From the studies on the interaction of α-SNAP with individual SNARE proteins, it is clear that the interaction of α-SNAP with Stx is non-specific and weaker in nature, implies less force being applied to Stx, indicating a slower disassembly, hence our observation of a Stx-separated state at the N-terminus within our time resolution. As the interaction of α-SNAP with Vamp is stronger and due to the high force applied to Vamp30, the disassembly of Vamp may proceed abruptly without any separated state or the lifetime of such state was too small to be observed under our time resolution. Overall, from our observations, along with previous studies on the SNARE disassembly by NSF, the disassembly most likely proceeds in the following sequence. An ATP hydrolysis independent of SNARE loading on the NSF with a possible interaction of SN25 with pore loops of NSF35, the ATP hydrolysis resulting major structural changes in the NSF D1 rings27,30, this causes structure incompatibility and tension within the 20S complex, with NSF N-domains and α-SNAP exerting its force on the SNARE complex29,30,33 and thermal fluctuations drive the already strained 20S complex to unstability, resulting in the disassembly of SNARE complex. We cannot completely rule out the possibility of an SN25-separated state or any other state that could not be observed due to the time resolution and experimental setup/conditions we used. Nevertheless, several other questions remain regarding the process, such as the movement of the NSF N domains and α-SNAP with respect to the NSF D1 D2 rings and also the SNARE cylinder. Their relative motion may determine the exact mechano-model of the SNARE disassembly by NSF.
Methods
Protein expression and purification
NSF from Chinese Hamster ovary, α-SNAP from Bos Taurus and SNARE proteins from Rattus norvegicus Syntaxin 2–253, Vamp 1–94, SNAP25 1–206 were expressed and purified as described earlier. Full-length Vamp 1–106 was purified with 1% octyl glucopyranoside (OG) in all the buffers used. Full-length Syntaxin 2–284 was purified with 2% sodium cholate in the extraction buffer and 1% sodium cholate in the elution buffer. All the Cysteine residues in the SNARE proteins were mutated to serine (SNARE protein sequences are given in Supplementary Table S1), and single Cysteine mutants of the proteins were generated with overlap PCR using designed primers (Supplementary Table S2), using quick-site mutagenesis kit from Qiangen (Beijing) and the sequences were confirmed by DNA sequencing.
Protein labelling, SNARE complex formation and reconstitution into nanodisc
Single Cysteine variants of the SNARE proteins, Vamp R30C, Vamp A82C, Syntaxin S200C, Syntaxin K252C, SNAP25 E27C and SNAP25 K79C, were labelled with maleimide variants of cyanine dyes (Lumiprobe) based on our experimental design using a protein/dye ratio of 1:5. Dyes were labelled in buffer containing 50 mM HEPES, 100 mM NaCl, 2 mM TCEP, pH 7.4. 1% OG and 1% sodium cholate were included in the buffer for the labelling of Vamp full and Syntaxin full, respectively. Excess dye was removed using gel filtration and the protein fractions with labelling efficiency > 70% were used for further experiments (Supplementary Fig. 13A).
SNARE complex formation was initiated by adding syntaxin to SNAP25 followed by Vamp, and the final concentration of the detergent was maintained at 1% OG for Vamp full SNARE and 1% sodium cholate for Syntaxin full SNARE. The reaction was allowed to proceed up to 16 hr at 4 °C and thrombin enzyme was added to remove the His-tag. An additional urea wash step was added in the purification of SNARE to remove most of the higher-order and/or antiparallel SNARE complexes. The formation of the SNARE complex was assessed by SDS-PAGE and confirmed by SNARE dissociation upon heating in the SDS buffer. SNARE-nanodisc was prepared essentially in the same manner as described earlier30 with biotinylated-phosphatidylethanolamine (18:1 Biotinyl phosphatidylethanolamine (Avanti)) used instead of phoshatidylethanolamine in reconstituting lipids. Excess detergent was removed using bio-beeds, and the SNARE-nanodisc were purified using gel filtration using Superdex 200 (GE Healthcare) in buffer A (50 mM HEPES, 100 mM NaCl, pH 7.6) (Supplementary Fig. 13).
Acquisition and analysis of the single-molecule experiments
All the single-molecule experiments were performed at 37 °C in buffer A containing an oxygen scavenging system consisting of 3 mg/mL glucose, 100 μg/mL glucose oxidase (Sigma-Aldrich), 40 μg/mL catalase (Roche), 1 mM cyclooctatetraene (COT, Sigma-Aldrich), 1 mM 4-nitrobenzylalcohol (NBA, Sigma-Aldrich), 1.5 mM 6-hydroxy-2,5,7,8-tetramethyl-chromane-2-carboxylic acid (Trolox, Sigma-Aldrich, added from a concentrated DMSO stock solution). 1% BSA was included in the buffer to reduce the non-specific binding onto the slides. Streptavidin was added to the slide and incubated for 2 min before the addition of SNARE-nanodiscs for immobilisation. SNARE-nanodiscs containing biotin-PE were attached to the slides coated with polyethylene glycol and biotin using biotin-strapavidin-biotin linkage. The concentration of SNARE-nanodiscs were adjusted such that an average of 800–1200 nanodiscs was immobilised per field view such that the distance between the spots were much greater than the optical diffraction limit. Photobleaching of the dyes were carried out to investigate the stability (dwell times) of the dyes in the SNARE-nanodiscs (Supplementary Fig. 14). NSF disassembly assay was carried out by adding NSF, α-SNAP, 10 mM Mg2+ and 2 mM ATP in oxygen scavenging buffer. For the dynamic experiments, all these were added while the data was being collected. For the 20S complex disassembly assay, first SNARE-nanodiscs were immobilised, followed by the addition of NSF-EDTA, α-SNAP, 0.5 mM EDTA and 2 mM ATP in oxygen scavenging buffer and incubated for 10 min. The extra NSF and α-SNAP were washed off by oxygen scavenging buffer containing 0.5 mM EDTA. Disassembly was initiated by the addition of 10 mM Mg2+ and 2 mM ATP in oxygen scavenging buffer.
All the single-molecule experiments were performed on a home-built objective-type total internal reflection microscope based on a Nikon Eclipse Ti-E with an EMCCD camera equipped with 488, 532 and 640 nm solid state lasers37. All the movies were collected in Alternative Laser EXcitation (ALEX) mode with alternating laser excitation between 532 nm and 640 nm lasers, which enabled us to simultaneously capture Cy3-Cy5 and Cy3-Cy7 FRET under 532 nm excitation and Cy5-Cy7 FRET under 640 nm excitation. ALEX mode was achieved by synchronising the on-off switch with EMCCD frames. Fluorescence emission from the spots were spectrally separated and filtered by the interference dichroic (T635lpxr, Chroma) and bandpass filters ET585/65 m (Chroma, Cy3) and ET700/70 m (Chroma, Cy5) in a Dual-view spectral splitter for two-colour SNARE complexes. For three-colour experiments, an additional interference dichroic (T720lpxr, Chroma) and an additional bandpass filter (FF01-715LP-25, Semrock, Cy7) were included in a Quad-view spectral filter (Photometrics, Inc., Tucson, AZ, USA). All the movies were collected by Cell Vision software (Beijing Coolight Technology).
Collected movies were analysed by custom-made software programmes developed as an ImageJ plugin for both two-colour and three-colour FRET experiments. The fluorescence spots were fitted by a 2-D Gaussian function, matching the donor and acceptor spots using a variant of Hough transform38. The background-subtracted total volume of the 2-D Gaussian peak was used as raw fluorescence intensity I. Single-molecule trajectories were extracted from the movies using home-written Matlab scripts. The FRET values were calculated from the corrected intensities as described earlier FRET = Iacceptor/(Idonor+ Iacceptor)39,40. Two-colour smFRET traces were further analysed by Hidden Markov Model based programme for kinetics information36.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank Tsinghua-Peking Centre for Lifesciences for excellent postdoctoral fellowship funding to S.K.C. This work was supported by grants from the National Natural Science Foundation of China (92254306 to S.S., 22425701, 22061160466 and 22277063 to C.C., 32071192 to S.-F.S.), the National Basic Research Programme to S.-F.S. (2017YFA0504600).
Author contributions
S.-F.S. and S.S. conceived the project. S.K.C. and C.C. designed the experiments. S.K.C. purified most of the recombinant proteins, performed most of the single-molecule experiments and analysed all the data. C.L. purified some of the recombinant proteins and analysed the interaction between Vamp and SN25. W.X. and R.S. performed some of the single-molecule experiments. M.Y. helped to carry out the single-molecule experiments. Y.X. performed the computation of the binding free energy. S.-Y.L. and J.Y. provided advice regarding the manuscript review. C.C. provided analysis tools and advice regarding data processing. S.K.C. wrote the initial draft. S.S., C.C. and S.-F.S. edited the manuscript.
Peer review
Peer review information
Nature Communications thanks Tae-Young Yoon, and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The data that supports the reported results of the study are available in open repository Zenodo (10.5281/zenodo.14060740). Source data are provided in this paper.
Code availability
Custom written MATLAB codes used for the analysis of the smFRET traces are provided with this paper (Supplementary Data 1).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sudheer K. Cheppali, Chang Li, Wenjing Xing.
Contributor Information
Shan Sun, Email: shansun@mail.tsinghua.edu.cn.
Chunlai Chen, Email: chunlai@mail.tsinghua.edu.cn.
Sen-Fang Sui, Email: suisf@mail.tsinghua.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55531-0.
References
- 1.Südhof, T. C. The synaptic vesicle cycle. Annu. Rev. Neurosci.27, 509–547 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Bonifacino, J. S. & Glick, B. S. The mechanisms of vesicle budding and fusion. Cell116, 153–166 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Brunger, A. T., Choi, U. B., Lai, Y., Leitz, J. & Zhou, Q. Molecular mechanisms of fast neurotransmitter release. Annu. Rev. Biophys.47, 469–497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang, T., Li, L. & Hong, W. SNARE proteins in membrane trafficking. Traffic18, 767–775 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Sutton, R. B., Fasshauer, D., Jahn, R. & Brunger, A. T. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature395, 347–353 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Weber, T. et al. SNAREpins: minimal machinery for membrane fusion. Cell92, 759–772 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Jahn, R. & Scheller, R. H. SNAREs–engines for membrane fusion. Nat. Rev. Mol. Cell Biol.7, 631–643 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Ungar, D. & Hughson, F. M. SNARE protein structure and function. Annu. Rev. Cell Dev. Biol.19, 493–517 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Brunger, A. T. et al. The pre-synaptic fusion machinery. Curr. Opin. Struct. Biol.54, 179–188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erzberger, J. P. & Berger, J. M. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct.35, 93–114 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Khan, Y. A., White, K. I. & Brunger, A. T. The AAA+ superfamily: a review of the structural and mechanistic principles of these molecular machines. Crit. Rev. Biochem. Mol. Biol. 57, 156–187 (2021). [DOI] [PubMed]
- 12.Hanson, P. I. & Whiteheart, S. W. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol.6, 519–529 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Wilson, D. W., Whiteheart, S. W., Wiedmann, M., Brunner, M. & Rothman, J. E. A multisubunit particle implicated in membrane fusion. J. Cell Biol.117, 531–538 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Söllner, T., Bennett, M. K., Whiteheart, S. W., Scheller, R. H. & Rothman, J. E. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell75, 409–418 (1993). [DOI] [PubMed] [Google Scholar]
- 15.Hayashi, T., Yamasaki, S., Nauenburg, S., Binz, T. & Niemann, H. Disassembly of the reconstituted synaptic vesicle membrane-fusion complex in-vitro. EMBO J.14, 2317–2325 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayer, A., Wickner, W. & Haas, A. Sec18p (NSF)-driven release of Sec17p (alpha-SNAP) can precede docking and fusion of yeast vacuoles. Cell85, 83–94 (1996). [DOI] [PubMed] [Google Scholar]
- 17.May, A. P., Whiteheart, S. W. & Weis, W. I. Unraveling the mechanism of the vesicle transport ATPase NSF, the N-ethylmaleimide-sensitive factor. J. Biol. Chem.276, 21991–21994 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Baker, R. W. & Hughson, F. M. Chaperoning SNARE assembly and disassembly. Nat. Rev. Mol. Cell Biol.17, 465–479 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon, T.-Y. & Munson, M. SNARE complex assembly and disassembly. Curr. Biol.28, R397–R401 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Antonin, W., Fasshauer, D., Becker, S., Jahn, R. & Schneider, T. R. Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nat. Struct. Biol.9, 107–111 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Ernst, J. A. & Brunger, A. T. High resolution structure, stability, and synaptotagmin binding of a truncated neuronal SNARE complex. J. Biol. Chem.278, 8630–8636 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Zwilling, D. et al. Early endosomal SNAREs form a structurally conserved SNARE complex and fuse liposomes with multiple topologies. EMBO J.26, 9–18 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strop, P., Kaiser, S. E., Vrljic, M. & Brunger, A. T. The structure of the yeast plasma membrane SNARE complex reveals destabilizing water-filled cavities. J. Biol. Chem.283, 1113–1119 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Stein, A., Weber, G., Wahl, M. C. & Jahn, R. Helical extension of the neuronal SNARE complex into the membrane. Nature460, 525–528 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diao, J. et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature520, 563–566 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang, L. F. et al. Structural characterization of full-length NSF and 20S particles. Nat. Struct. Mol. Biol.19, 268–275 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Zhao, M. et al. Mechanistic insights into the recycling machine of the SNARE complex. Nature518, 61–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou, Q. et al. Cryo-EM structure of SNAP-SNARE assembly in 20S particle. Cell Res.25, 551–560 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White, K. I., Zhao, M., Choi, U. B., Pfuetzner, R. A. & Brunger, A. T. Structural principles of SNARE complex recognition by the AAA+ protein NSF. Elife7, 213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang, X. et al. Mechanistic insights into the SNARE complex disassembly. Sci. Adv.5, eaau8164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, C. et al. Extreme parsimony in ATP consumption by 20S complexes in the global disassembly of single SNARE complexes. Nat. Commun.12, 3206–3215 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi, U. B. et al. NSF-mediated disassembly of on- and off-pathway SNARE complexes and inhibition by complexin. Elife7, 10.7554/elife.36497 (2018). [DOI] [PMC free article] [PubMed]
- 33.Ryu, J.-K. et al. Spring-loaded unraveling of a single SNARE complex by NSF in one round of ATP turnover. Science347, 1485–1489 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryu, J.-K., Jahn, R. & Yoon, T.-Y. Progresses in understanding NSF-mediated disassembly of SNARE complexes. Biopolymers105, 518–531 (2016). [DOI] [PubMed]
- 35.Cipriano, D. J. et al. Processive ATP-driven substrate disassembly by the N-Ethylmaleimide-sensitive factor (NSF) molecular machine. J. Biol. Chem.288, 23436–23445 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKinney, S. A., Joo, C. & Ha, T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J.91, 1941–1951 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xing, W., Li, D., Wang, W., Liu, J.-J. G. & Chen, C. Conformational dynamics of CasX (Cas12e) in mediating DNA cleavage revealed by single-molecule FRET. Nucleic Acids Res.52, 9014–9027 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Illingworth, J. & Kittler, J. A survey of the hough transform. Comput. Vis. Graph. Image Process.44, 87–116 (1988). [Google Scholar]
- 39.Munro, J. B., Altman, R. B., Tung, C.-S., Sanbonmatsu, K. Y. & Blanchard, S. C. A fast dynamic mode of the EF-G-bound ribosome. EMBO J.29, 770–781 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, J. et al. Single-molecule four-color FRET. Angew. Chem. Int. Ed. Engl.49, 9922–9925 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The data that supports the reported results of the study are available in open repository Zenodo (10.5281/zenodo.14060740). Source data are provided in this paper.
Custom written MATLAB codes used for the analysis of the smFRET traces are provided with this paper (Supplementary Data 1).