Abstract
Metabolic reprogramming fuels cancer cell metastasis and remodels the immunosuppressive tumor microenvironment (TME). We report here that circPETH, a circular RNA (circRNA) transported via extracellular vesicles (EVs) from tumor-associated macrophages (TAMs) to hepatocellular carcinoma (HCC) cells, facilitates glycolysis and metastasis in recipient HCC cells. Mechanistically, circPETH-147aa, encoded by circPETH in an m6A-driven manner, promotes PKM2-catalyzed ALDOA-S36 phosphorylation via the MEG pocket. Furthermore, circPETH-147aa impairs anti-HCC immunity by increasing HuR-dependent SLC43A2 mRNA stability and driving methionine and leucine deficiency in cytotoxic CD8+ T cells. Importantly, through virtual and experimental screening, we find that a small molecule, Norathyriol, is an effective inhibitor that targets the MEG pocket on the circPETH-147aa surface. Norathyriol reverses circPETH-147aa-facilitated acquisition of metabolic and metastatic phenotypes by HCC cells, increases anti-PD1 efficacy, and enhances cytotoxic CD8+ T-cell function. Here we show that Norathyriol is a promising anti-HCC agent that contributes to attenuating the resistance of advanced HCC to immune checkpoint blocker (ICB) therapies.
Subject terms: Cancer microenvironment, Hepatocellular carcinoma, Cancer metabolism
Tumor-associated macrophages (TAMs) are associated with poor prognosis and low responses to immunotherapy. Here, authors discover that a circular RNA, circPETH, produced by TAMs generates a metabolic reprogramming in hepatocellular carcinoma that promotes metastasis and impairs the activity of cytotoxic CD8 + T cells.
Introduction
Hepatocellular carcinoma (HCC) is the sixth most commonly diagnosed cancer and the third leading cause of cancer-related death worldwide1. Few effective treatments are available because most patients are diagnosed at an advanced disease stage2. Today, the predominant drugs used for the systematic treatment of late-stage HCC are multikinase inhibitors and immune checkpoint blockers (ICBs). However, oral multikinase inhibitors have shown limited benefit in the clinical setting3. ICBs targeting programmed cell death-1 (PD-1) block the binding of programmed cell death 1-ligand 1 (PD-L1) to PD-1, inhibiting antitumoral effector T-cell exhaustion, and have thus revolutionized the treatment of numerous advanced malignancies4,5. Unfortunately, despite initial success, the incidence of anti-PD1 resistance is 70–80%6; thus, HCC recurrence and response failure are common. Hence, investigating the underlying mechanisms of ICB resistance may contribute to the development of new treatment strategies and novel drugs for the purpose of overcoming the resistance of advanced HCC to ICB therapies.
To sustain their malignant biological properties, HCC cells tend to compete with adjacent normal cells for relatively limited nutrients via shifts in their metabolic phenotypes, which is termed metabolic reprogramming7. Aerobic glycolysis, known as the Warburg effect, is an essential form of glucose metabolism in tumor cells; through this process, large amounts of glucose are consumed, and large amounts of lactate are produced in the presence of sufficient oxygen8. Accumulating evidence has demonstrated that aerobic glycolysis exerts significant multifaceted effects on various biosynthesis pathways, biological processes and processes responsible for the generation of signaling metabolites that facilitate cancer cell metastasis and immune evasion, followed by the formation of an immunosuppressive tumor microenvironment (TME)9–11.
Effector T-cell depletion in the TME is regarded as the crucial factor for the development of ICB resistance in HCC12. As the most abundant inflammatory cells in the TME, tumor-associated macrophages (TAMs) are highly associated with poor prognosis and poor responses to immunotherapy in patients with various cancers13–15. TAMs can accelerate cancer progression by increasing the glycolytic capacity and metastatic potential of tumor cells16,17 and inhibiting the T-cell-mediated killing of tumor cells18. Nevertheless, the precise roles of TAMs in metabolic reprogramming and the immunosuppressive TME in HCC are not clear, and further research is needed.
Circular RNAs (circRNAs) constitute a class of covalently closed and single-stranded RNA transcripts. An increasing number of studies have described the importance of circRNAs in multiple biological regulatory processes, including organism development, cell differentiation and apoptosis, the immune response, tumorigenesis and metastasis13,19,20. Recently, circRNAs have been identified as translatable RNAs that carry putative open reading frames (ORFs) and internal ribosome entry sites (IRESs) that can recruit ribosomes and then initiate translation without a 5′ cap structure21, which results in the production of novel proteins with the ability to regulate the malignant biological behaviors of cancer cells. It has been reported that circRNAs participate in interactions between tumor and immune cells, affecting the resistance of HCC cells to immunotherapy22. Previous findings have shown that TAMs can release extracellular vesicles (EVs) that transmit different classes of RNAs to tumor cells to regulate cell phenotype acquisition16,23. However, whether EV-loaded circRNAs secreted by TAMs are critically involved in aerobic glycolysis and resistance to ICB therapies remains unclear. In this study, we found that circPETH (a circRNA derived from exons 13–15 of the low-density lipoprotein receptor gene) is packaged in EVs secreted by TAMs and taken up by HCC cells. Additionally, we identified a protein encoded by circPETH (circPETH-147aa) and clarified its role in metabolic reprogramming and the immunosuppressive TME in HCC. Ultimately, we discovered a small-molecule circPETH-147aa inhibitor, norathyriol, via high-throughput screening and evaluated its ability to reduce HCC resistance to ICBs.
Results
TAMs promote the HCC cell invasion, migration and aerobic glycolysis via EV-loaded circPETH
To explore the biological function of TAMs in regulating the acquisition of metastatic and metabolic phenotypes by HCC cells, we first isolated TAMs from human HCC tissues (Supplementary Fig. 1a) and monocyte-derived macrophages (MDMs) from human peripheral blood. As determined via flow cytometry, these two kinds of macrophages highly expressed CD11b on the cell membrane (Supplementary Fig. 1b, left). TAMs expressed the mannose receptor CD206 at a significantly greater level than MDMs (Supplementary Fig. 1b, right), and TAMs mainly undergo M2 polarization24. TAMs and MDMs were subsequently cocultured with HCC cells, but prevented from physically interacting. After 72 h, Hep3B and Huh-7 cells cocultured with TAMs exhibited a spindle-shaped appearance (Supplementary Fig. 1c), suggesting that these TAMs might promote the acquisition of a metastatic phenotype by HCC cells. Indeed, Hep3B and Huh-7 cells cocultured with TAMs presented significantly greater invasion and migration capacities than cells cultured alone or those cocultured with MDMs did (Supplementary Fig. 1d–f). Importantly, HCC cells cocultured with TAMs presented significantly increased extracellular acidification rates (ECARs) (Supplementary Fig. 1g–j), glucose consumption (Supplementary Fig. 1k, l) and lactate production (Supplementary Fig. 1m, n). Collectively, these data demonstrate that TAMs facilitate HCC cell invasion, migration, and aerobic glycolysis via soluble mediators.
As the effects of TAMs on the acquisition of metastatic and metabolic phenotypes by HCC cells were found to be independent of direct cell‒cell contact, we investigated the communicators critical to the related signaling pathways. Numerous studies have emphasized the importance of EVs in cell‒cell communication in the TME25,26. To confirm that EVs mediate the interaction between TAMs and HCC cells, EVs from the culture medium (CM) of TAMs and MDMs were isolated via ultracentrifugation. The morphology and size distribution of these EVs and the expression of biomarkers (CD9 and CD81) were analysed via transmission electron microscopy, nanoparticle tracking and nanoparticle flow cytometry (nanoFCM) (Supplementary Fig. 2a–c). Moreover, the dynamics of TAM-secreted EV uptake by Hep3B and Huh-7 cells was visualized via confocal microscopy (Supplementary Fig. 2d), which revealed that EVs markedly promoted the invasion and migration capacities of HCC cells (Supplementary Fig. 2e–g) and increased aerobic glycolysis in HCC cells (Fig. 1a–h). Notably, eliminating EVs from the CM of TAMs via the addition of an anti-CD81 neutralizing antibody markedly reversed the impacts of EVs on HCC cell invasion, migration and aerobic glycolysis (Supplementary Fig. 2e and Fig. 1a–h). Taken together, these findings indicate that EVs mediate the effects of TAMs by enhancing the acquisition of metastatic and metabolic phenotypes by HCC cells.
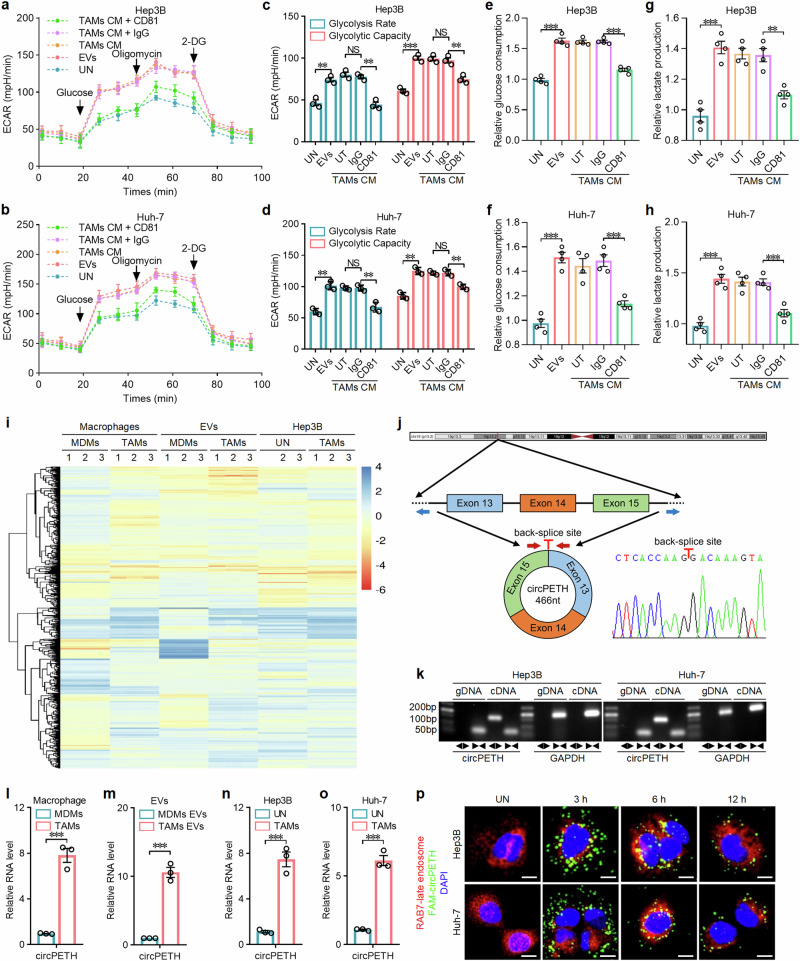
Fig. 1. TAMs promote the invasion, migration and aerobic glycolysis of HCC cells via the action of EV-loaded circPETH.
a, b ECARs of Hep3B and Huh-7 cells with the indicated treatments. n = 3 independent experiments. Data are shown as mean ± SD. c, d Glycolysis rates and glycolysis capacities of Hep3B and Huh-7 cells with the indicated treatments. n = 3 independent experiments. Data are shown as mean ± SEM. Exact P values from left to right (c): 0.002350, 0.001789, 0.000133, 0.009653. Exact P values from left to right (d): 0.002215, 0.008657, 0.001573, 0.003177. e, f Glucose consumption of Hep3B and Huh-7 cells with the indicated treatments. n = 4 independent experiments. Data are shown as mean ± SEM. Exact P values from left to right (e): 1.56E − 05, 9.65E − 06. Exact P values from left to right (f): 6.38E − 05, 0.000543. g, h Lactate production of Hep3B and Huh-7 cells with the indicated treatments. n = 4 independent experiments. Data are shown as mean ± SEM. Exact P values from left to right (g): 0.000244, 0.002215. Exact P values from left to right (h): 0.000123, 0.000772. i Heatmap of microarrays. j Scheme illustrating the genomic locus and production of circPETH. The special primers used for detection of circPETH by qRT‒PCR are indicated by red arrows and validated by Sanger sequencing. k qRT‒PCR products with divergent primers showing the circularization of circPETH. GAPDH used as control. l circRNA expression in TAMs and MDMs by qRT‒PCR. n = 3 independent experiments. Data are shown as mean ± SEM. Exact P value: 0.000369. m circRNA expression in EVs from TAMs and MDMs by qRT‒PCR. n = 3 independent experiments. Data are shown as mean ± SEM. Exact P value: 0.000182. n, o circRNA expression in Hep3B and Huh-7 cells cocultured with TAMs by qRT‒PCR. n = 3 independent experiments. Data are shown as mean ± SEM. Exact P value (n): 0.000622. Exact P value (o): 0.000163. p Representative confocal microscopy images of the collected Hep3B and Huh-7 cells assessing the colocalization of FAM-labeled circPETH and late endosomes (denoted by RAB7). Scale bars, 10 μm. NS, not significant; **P < 0.01; ***P < 0.001 by two-tailed Student’s T-test. Data are representative of at least three independent experiments. Source data are provided as a Source Data file.
Emerging evidence has highlighted the contribution of EV-loaded circRNAs to the progression of various human diseases27. Therefore, microarrays were employed to identify circRNAs packaged in EVs and transported from TAMs to HCC cells. The differentially expressed circRNAs for the following comparisons were intersected and analysed: TAMs versus MDMs; EVs from TAMs versus those from MDMs; and Hep3B cells cocultured with TAMs versus those cultured alone (Fig. 1i). For the intersection analysis, 18 circRNAs whose expression levels were significantly altered were identified (Supplementary Fig. 3a). Among these circRNAs, 5 were found to be significantly upregulated for the three comparison groups (Supplementary Fig. 3a). Among the candidate circRNAs, hsa_circ_0049398, derived from exons 13–15 of the low-density lipoprotein receptor gene on chromosome 19p13 (Fig. 1j), was found to be the most highly upregulated in EVs from TAMs versus EVs from MDMs (Supplementary Fig. 3b) and the most highly upregulated in Hep3B cells cocultured with TAMs versus Hep3B cells cultured alone (Supplementary Fig. 3c). Therefore, hsa_circ_0049398 was preliminarily identified as the key circRNA transported via EVs from TAMs to HCC cells (circPETH). The back-splice junction site was determined via Sanger sequencing of the PCR products (Fig. 1j). Divergent and convergent primers, RNase R, random hexamer primers, oligo (dT)18 primers and actinomycin D were used to validate the circular characteristics of circPETH (Fig. 1k and Supplementary Fig. 3d–i). As expected, quantitative real-time PCR (qRT‒PCR) verified that the expression of circPETH in TAMs was approximately 8-fold greater than that in MDMs (Fig. 1l), and it was elevated by approximately 10-fold in EVs from TAMs (Fig. 1m) and by at least 6-fold in HCC cells cocultured with TAMs (Fig. 1n, o). Furthermore, subcellular RNA fractionation and fluorescence in situ hybridization (FISH) revealed that circPETH was predominantly located in the cytoplasm of both HCC cells and macrophages (Supplementary Fig. 3j–l), and its level was markedly elevated in HCC cells cocultured with TAMs (Supplementary Fig. 3l). To visualize the direct transmission of circPETH, we transfected artificially synthesized FAM-labeled circPETH into TAMs via electroporation and then isolated EVs. Dynamic confocal microscopy revealed that FAM-labeled circPETH was largely incorporated into late endosomes 6 h after EV treatment, which was followed 12 h later by a marked increase in the secretion of FAM-labeled circPETH from late endosomes in Hep3B and Huh-7 cells (Fig. 1p). Overall, these observations indicate that the increased expression of circPETH in HCC cells cocultured with TAMs is attributable to EV packaging and transmission.
Next, the relevance of circPETH expression to clinical characteristics and patient prognosis was determined by analysing 72 matched HCC and adjacent normal samples. Sex and gender were not considered in the study design. The results revealed that circPETH was considerably overexpressed in HCC tissues compared with adjacent normal tissues (Supplementary Fig. 3m). The upregulation of circPETH was statistically associated with a larger tumor size, a larger tumor number, the occurrence of microvascular invasion, a more advanced TNM stage and a more advanced Barcelona Clinic Liver Cancer (BCLC) stage (Supplementary Table 1). Gender ratio was not significantly correlated with circPETH expression level in HCC samples. Kaplan‒Meier analysis revealed that high circPETH expression was significantly associated with poor overall survival (OS) and recurrence-free survival (RFS) following surgical resection (Supplementary Fig. 3n, o). In addition, Cox proportional hazards regression analysis led to the identification of circPETH as an independent prognostic indicator for OS and RFS (Supplementary Table 2 and Supplementary Table 3), suggesting a critical role for circPETH in promoting HCC progression.
ADAR1 inhibits the circularization of circPETH, and the loss of introns leads to the knockout of circPETH
Numerous RNA-binding proteins (RBPs) bind complementary sequences in long flanking introns, and their binding impedes the back splicing of certain exons, explaining the biogenetic formation of circRNAs20,28,29. Since circPETH originates primarily from TAMs, we analysed the expression profiles of 10 previously reported RBPs in microarray datasets of TAMs and MDMs to identify the RBPs involved in the biogenesis of circPETH. Only 3 RBPs (ADAR1, QKI and PTBP1) were associated with circPETH biogenesis, as indicated by adjusted P values less than 0.05 (Supplementary Fig. 4a), and among these RBPs, adenosine deaminase RNA specific 1 (ADAR1) showed the most significant change in expression (Supplementary Fig. 4b), suggesting that ADAR1 might participate in the production of circPETH. Indeed, knocking down ADAR1 in Hep3B and Huh-7 cells resulted in substantially increased expression of circPETH but did not affect the expression of its linear messenger RNA counterpart, LDLR (mLDLR), or precursor messenger RNA, LDLR (pLDLR) (Supplementary Fig. 4c–e). Consistent with the microarray data, the protein level of ADAR1 was markedly lower in TAMs than in MDMs (Supplementary Fig. 4f). More importantly, overexpression of ADAR1 in TAMs efficiently abolished the upregulation of circPETH in Hep3B and Huh-7 cells induced by coculture with vector-TAMs (Supplementary Fig. 4g–i). In addition, we compared the sequences in the introns flanking exons 13–15 and observed highly complementary reverse sequences between introns 12 and 15 (77% identity in 618 nucleotides; Supplementary Fig. 4j), which were named I12RC (reverse complementary sequence in intron 12) and I15RC (reverse complementary sequence in intron 15), respectively. To examine whether the circularization of circPETH depends on I12RC or I15RC, the wild-type sequence (spanning from intron 12 to intron 15, # 2) and a series of plasmids with deletion mutations (plasmids # 3–5) were constructed (Supplementary Fig. 4k). The qRT‒PCR and Northern blot results demonstrated that circPETH was overexpressed in cells transfected with the wild-type-expressing plasmid but not in cells transfected with the deletion mutant-expressing plasmids, indicating that the coexistence of I12RC and I15RC is indispensable for the formation of circPETH (Supplementary Fig. 4l, m). Afterwards, to determine the binding affinity of ADAR1 for I12RC and I15RC, RNA immunoprecipitation (RIP) was performed using an endogenous anti-ADAR1 antibody. As expected, I12RC and I15RC, but not exon 13, were markedly enriched among the ADAR1-immunoprecipitated RNAs from HEK293T cells and TAMs (Supplementary Fig. 4n, o). Moreover, silencing and overexpressing ADAR1 in cells transfected with the plasmids (# 1–5) led to pervasive increases and decreases in circPETH levels, respectively (Supplementary Fig. 5a, b). Considering this evidence, we deduced that the high expression of circPETH in TAMs, but not in MDMs, can be attributed to the relatively low level of ADAR1, which inhibits the pairing of the I12RC and I15RC sequences within pLDLR and thereby inhibits the circularization of circPETH.
To clarify the role of circPETH in the acquisition of metastatic and metabolic phenotypes by HCC cells, we initially knocked down circPETH in Hep3B and Huh-7 cells via small interfering RNA (siRNA) targeting the back-spliced sequence without significantly reducing the expression of mLDLR or pLDLR (Supplementary Fig. 5c, d). The increase in circPETH levels in HCC cells induced by TAMs was subsequently abrogated by transfection of a short hairpin RNA (shRNA) against circPETH into TAMs (Supplementary Fig. 5e, f). Notably, the effect of depleting circPETH in TAMs was similar to that of suppressing the release of EVs by silencing RAB27 in TAMs30 (Supplementary Fig. 5g, h). Loss of circPETH in TAMs clearly reversed the TAM-mediated facilitation of HCC invasion, migration and aerobic glycolysis (Supplementary Figs. 5i and 6a–j). We designed two small guide RNAs (sgRNAs) targeting the sequences flanking I15RC (Supplementary Fig. 6k) and successfully knocked out I15RC via the CRISPR‒Cas9 technique, as verified by Sanger sequencing (Supplementary Fig. 6l). The deletion of circPETH did not affect the expression of mLDLR or pLDLR in HCCLM9 cells (Supplementary Fig. 6m). Knockout (KO) of circPETH markedly decreased the invasion and migration capacities of HCC cells (Supplementary Fig. 6n, o) and aerobic glycolysis in these cells (Supplementary Figs. 6p and 7a–c). Next, we assessed the impacts of circPETH on HCC metastasis in vivo by establishing lung metastasis and liver orthotopic intrahepatic metastasis models via implantation of HCC cells. After HCC cell implantation, the circPETH KO group presented a markedly reduced bioluminescence intensity, a considerably decreased fluorescence intensity and fewer histopathologically detectable metastatic foci in the lungs (Supplementary Fig. 7d). Similarly, the intrahepatic fluorescence intensity and the number of histopathologically detectable metastatic foci in the liver were reduced after circPETH was completely knocked out (Supplementary Fig. 7e). Together, these data show that TAMs strengthen the metastatic and metabolic phenotypes of HCC cells via EV-loaded circPETH.
Lactate functions as a stimulating factor that can drive the polarization of MDMs into TAMs in the TME16,31. Because TAMs but not MDMs exhibited a substantial decrease in ADAR1 levels, we speculated that lactate might exert inhibitory effects on the expression of ADAR1 in TAMs. As anticipated, lactate treatment reduced the RNA and protein levels of ADAR1 in TAMs in a concentration-dependent manner (Supplementary Fig. 7f, g). Additionally, given that circPETH transport via TAM-derived EVs obviously elevated lactate production in HCC cells, it can be inferred that circPETH in HCC cells conversely modulates the expression of ADAR1 in TAMs. To test this hypothesis, TAMs were cocultured with circPETH knockdown HCC cells or control HCC cells. The results revealed that the RNA and protein levels of ADAR1 in TAMs were significantly greater after circPETH silencing in Hep3B and Huh-7 cells (Supplementary Fig. 7h–j). Overall, it is reasonable to propose that circPETH, ADAR1 and lactate form a positive feedback loop between HCC cells and TAMs that sustains the increase in circPETH expression.
A 147-acid protein encoded by circPETH associated with N6-methyladenosine (m6A)-driven translation enhances the metastatic and metabolic phenotypes of HCC cells
Some circRNAs have been reported to be translated into bioactive proteins that are prominently involved in cancer progression32,33. According to circRNADb34, a cross-junction ORF encoding a predicted protein consisting of 147 amino acids was found in the circPETH sequence (Fig. 2a), and this predicted protein was termed circPETH-147aa. Moreover, via dual-luciferase assays, we identified a putative IRES and validated its ability to recruit ribosomes and trigger translation (Fig. 2b, c). Confirming the protein-coding potential of circPETH, circPETH-147aa was highly overexpressed in Hep3B and Huh-7 cells, whereas neither mLDLR nor pLDLR expression was significantly changed (Supplementary Fig. 7k, l). Notably, we observed an evident band slightly larger than the 15 kD band in the circPETH-overexpressing group but not in the control vector group on an SDS‒PAGE gel (Fig. 2d). The amino acid sequence of the 15 kD band was determined and aligned to the predicted sequence of circPETH-147aa obtained by mass spectrometry (MS) (Fig. 2e). Thereafter, a haemagglutinin (HA)-tag sequence was inserted into a circPETH-overexpressing plasmid just before the stop codon, and this plasmid was transfected into Hep3B and Huh-7 cells. The HA-tagged protein was detected as a band slightly larger the 15 kD band following western blotting with an anti-HA antibody (Fig. 2f). Subcellular protein fractionation and immunofluorescence (IF) revealed that the expression of this HA-tagged protein was greater in the nucleus than in the cytoplasm (Fig. 2g, h).
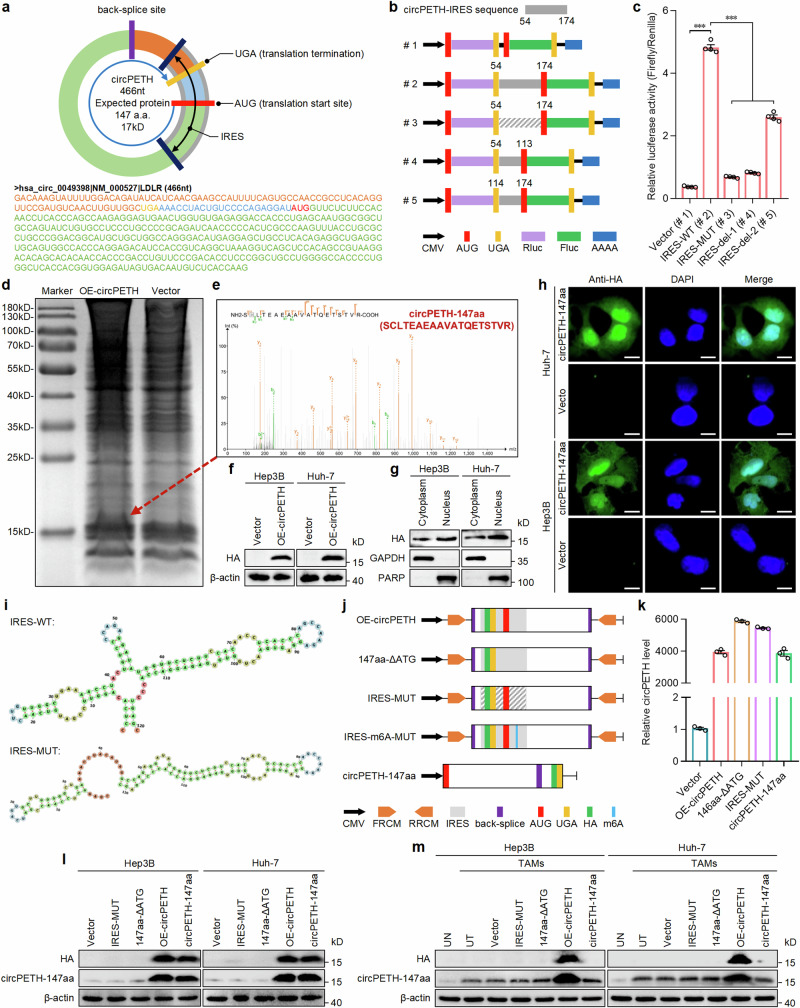
Fig. 2. circPETH encodes a protein termed as circRNA-147aa.
a The putative ORF and its location in the genomic region of circPETH. b Schematic of five different vectors for luciferase reporter assays. Wild-type, mutated IRES sequence or a series of deletions of IRES sequences were inserted into vectors (#1 to #5). c The relative luciferase activity of the indicated five vectors. Data are shown as the relative ratio of Firefly luciferase activity to Renilla luciferase activity. n = 4 independent experiments. Data are shown as mean ± SEM. Exact P values from left to right: 3.72E − 09, 5.90E − 09, 7.35E − 09, 8.83E − 07. d Total proteins from Hep3B cells transfected with circPETH overexpression or control plasmid were separated through SDS-PAGE. e The evident band, a little above 15 kD, was cut and subjected to MS. The circPETH-147aa was identified. f HA-tagged circRNA-147aa expression in Hep3B and Huh-7 cells with circPETH overexpression by western blot. The samples derive from the same experiment but different gels for HA, and another for β-actin were processed in parallel. g, h Subcellular location of HA-tagged circRNA-147aa by subcellular protein fractionation and IF. Scale bar, 10 μm. The GAPDH and PARP were applied as positive controls in the cytoplasm and nucleus. i The predicted wild-type and mutated secondary structures of IRES. j Schematic of five different vectors for circPETH overexpression. k circPETH expression of the indicated five vectors by qRT‒PCR. n = 3 independent experiments. Data are shown as mean ± SEM. l The expressions of HA and circPETH-147aa in Hep3B and Huh-7 cells transfected with these five vectors by western blot. The samples derive from the same experiment but different gels for HA, another for circPETH-147aa and another for β-actin were processed in parallel. m The expressions of HA and circPETH-147aa in Hep3B and Huh-7 cells cocultured with indicated TAMs by western blot. ***P < 0.001 by two-tailed Student’s T-test. Data are representative of at least three independent experiments. Source data are provided as a Source Data file.
Owing to a natural frameshift that overlaps the back-splice junction ORF, circPETH-147aa is equipped with a unique 28-aa tail (GQSILDRYHQRSHFQCQPPHRFRCQLVG) at its carboxy terminus that is not homologous to the tail of any known protein. To precisely detect circPETH-147aa, we produced a highly specific antibody against the 28-aa tail and identified a specific protein that had a mass slightly greater than 15 kD in Hep3B, Huh-7 and HCCLM9 cells (Supplementary Fig. 7m, n). Notably, after circPETH knockdown or knockout, circPETH-147aa was undetectable, whereas the expression of LDLR remained stable (Supplementary Fig. 7m, n). Consistent with the variable trend of circPETH expression, the circPETH-147aa level was markedly elevated in HCC cells cocultured with TAMs before circPETH or RAB27 was knocked down in TAMs (Supplementary Fig. 7o, p).
To determine the importance of general coding elements in the translation of circPETH, a series of mutant vectors based on the HA-tagged wild-type circPETH-overexpressing plasmid (OE-circPETH), including a vector expressing circPETH lacking the initiation codon (ATG) (147aa-ΔATG), a vector expressing circPETH with a mutation in the IRES (IRES-MUT), causing it to lack the tRNA-like structure of the wild-type IRES (Fig. 2i), and a linear ORF vector expressing circPETH without the back-splice site (circPETH-147aa) (Fig. 2j), were constructed and successfully expressed with high efficiency (Fig. 2k). Western blot analysis revealed that the expression levels of HA and circPETH-147aa were markedly greater in both the OE-circPETH group and circPETH-147aa group than in the control vector group, whereas circPETH-147aa levels were unchanged and HA was undetectable in the IRES-MUT group and 147aa-ΔATG group (Fig. 2l). In addition, we transfected these plasmids into TAMs and cocultured the transfected TAMs with HCC cells. Intriguingly, in contrast to the previously obtained data, HA and circPETH-147aa levels were increased in the OE-circPETH group but not in the other vector groups (Fig. 2m), indicating that only circular circPETH, not linear circPETH-147aa, was loaded into the EVs secreted by TAMs. Collectively, our findings clarify that IRES activity, the initiation codon (ATG) and the circular structure are essential for circPETH protein translation in HCC cells. Notably, upregulation of circPETH or circPETH-147aa expression, but not that of IRES-MUT or 147aa-ΔATG, significantly promoted metastasis and aerobic glycolysis in Hep3B and Huh-7 cells (Supplementary Figs. 7q, r, 8a–i and 9a–h), demonstrating that like circPETH, circPETH-147aa completely mediated the re-establishment of HCC metastasis and aerobic glycolysis.
Although TAMs in the TME presented the highest circPETH level, we unexpectedly observed that circPETH-147aa was undetectable in TAMs and MDMs but not in Hep3B or Huh-7 cells (Supplementary Fig. 9i). In other words, circPETH was expressed mainly in TAMs, but it was predominantly translated in HCC cells, suggesting that TAMs might lack certain elements that play crucial roles in the translation of circPETH. Notably, m6A, the most common modification of RNA, drives the efficient initiation of circRNA translation in human cells35–37. Hence, we explored whether the lack of circPETH translation in TAMs was caused by a deficiency in m6A methylation. First, neither METTL3 nor METTL14, as vital N6-adenosine methyltransferases, was expressed at lower levels in TAMs or MDMs than in Hep3B or Huh-7 cells (Supplementary Fig. 9i). Second, circPETH did not undergo substantially m6A modification in macrophages (Supplementary Fig. 9j). In contrast, circPETH underwent significantly m6A modification in HCC cells, and the m6A level was considerably decreased in cells transfected with siRNA targeting METTL3 or METTL14 (Supplementary Fig. 9k, l). We subsequently identified a potential m6A site in the IRES of circPETH via m6A motif sequence alignment analysis38 and constructed a vector expressing circPETH with a mutation in the IRES m6A site (IRES-m6A-MUT) (Supplementary Fig. 10a). As expected, the expression levels of HA and circPETH-147aa in the IRES-m6A-MUT group were substantially lower than those in the OE-circPETH group (Supplementary Fig. 10b, c). Moreover, the expression levels of HA and circPETH-147aa in the OE-circPETH group were reduced to almost the same levels as those in the IRES-m6A-MUT group by METTL3 and METTL14 silencing, which had no influence on the level of linear circPETH-147aa (Supplementary Fig. 10c, d). In contrast to canonical cap-dependent mRNA translation mediated by EIF4G1, circRNA translation is mediated by EIF4G2, which recognizes the IRES and thus initiates translation complex assembly and recruits the activated 40S ribosomal subunit39,40. As expected, knockdown of EIF4G2, but not EIF4G1, led to the downregulation of circPETH-147aa protein expression in Hep3B and Huh-7 cells (Supplementary Fig. 10e). In contrast, EIF4G1, but not EIF4G2, was responsible for the protein expression of LDLR (Supplementary Fig. 10f). Notably, YTHDF3, an m6A reader, has been reported to bind the m6A site in the IRES and simultaneously interact with EIF4G2, serving as a pivotal connector in the recognition of the IRES by EIF4G236. Consistently, a RIP assay revealed that circPETH was highly enriched among YTHDF3-immunoprecipitated RNAs, and the level of circPETH was markedly reduced after the loss of METTL3 and METTL14 (Supplementary Fig. 10g, h). Reciprocal coimmunoprecipitation (co-IP) assays verified that the levels of YTHDF3 and EIF4G2 were obviously higher in the EIF4G2 and YTHDF3 immunoprecipitates, respectively, than in the IgG pellets (Supplementary Fig. 10i). In addition, the expression of circPETH-147aa and the enrichment of circPETH in EIF4G2-immunoprecipitated RNAs were markedly decreased by YTHDF3 depletion (Supplementary Fig. 10j–l). Finally, western blotting revealed that the coexpression of YTHDF3 and EIF4G2 was absolutely necessary for the translation of circPETH in HCC cells (Supplementary Fig. 10m). In summary, the m6A modification of the circPETH IRES, which requires the binding of the initiation factor EIF4G2 and the m6A reader YTHDF3 in a cap-independent manner, triggers the translation of circPETH into circPETH-147aa in HCC cells but not in macrophages.
circPETH-147aa functions as a guide that enhances the phosphorylation of ALDOA catalyzed by PKM2
Research has shown that aerobic glycolysis endows tumor cells with particular metabolic properties and endows transcription factors, such as YAP, with potent metastasis-promoting properties41. Because circPETH increased the glycolytic and metastatic capacities of HCC cells, we first evaluated whether the expression or subcellular location of YAP was regulated by circPETH. As anticipated, YAP translocated into the cytoplasm after circPETH was deleted in HCCLM9 cells. In contrast, circPETH overexpression induced the nuclear translocation of YAP in Hep3B and Huh-7 cells, as validated by IF (Supplementary Fig. 11a, b) and subcellular protein fractionation (Supplementary Fig. 11c–e) assays. Furthermore, circPETH was shown to attenuate the ubiquitination of YAP (Supplementary Fig. 11f), thus inactivating the Hippo signaling pathway. In summary, glycolysis is the primary bioenergetic pathway that drives cytoskeletal remodeling in diverse cancer cells42, generally by modulating the Hippo kinase cascade43. Specifically, excessive filamentous actin (F-actin), formed by the subunit globular actin (G-actin), binds with LATS1 and suppresses its phosphorylation, reducing the degradation rate of YAP44. Notably, LATS1 exclusively interacts with F-actin, not G-actin, although both actin types are categorized as β-actin, the main actin isoform in nonmuscle cells44. Therefore, to elucidate whether β-actin polymerization mediates the regulation of YAP by circPETH, co-IP assays were performed to assess the association between F-actin and LATS1 in HCC cells with or without altered circPETH expression. After knockout of circPETH, we observed a considerable downwards trend in the β-actin level in the LATS1 immunoprecipitates (Supplementary Fig. 11g); the β-actin level was evidently elevated by the upregulation of circPETH (Supplementary Fig. 11h, i), and this change was abrogated by treatment with Lat. B, which depolymerizes F-actin (Supplementary Fig. 11g–i). Ultimately, the effects of circPETH on cytoskeletal reorganization and Hippo signaling were confirmed via IF assays (Supplementary Figs. 11j, k and 12a, b).
To gain insight into the regulatory mechanism underlying the functions of circPETH through comprehensive analysis of gene expression, RNA sequencing (RNA-seq) was performed, and differences in expression for the following two comparisons were analysed: the OE-circPETH group versus the vector group and the circPETH KO group versus the NTC group (Supplementary Fig. 12c). The differentially expressed genes were most significantly enriched in the DNA-template transcription gene set according to Gene Ontology (GO) analysis (Supplementary Fig. 12d). More importantly, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed that circPETH most significantly altered the expression of genes enriched in metabolic pathways (Supplementary Fig. 12e), indicating the prominent role of circPETH in the metabolic reprogramming of HCC cells. Afterwards, after HA-tagged circPETH-147aa was transfected into Hep3B cells, we performed co-IP using an antibody against HA, and the potential binding proteins were stained with Coomassie Brilliant Blue (Supplementary Fig. 13a), followed by identification via MS. Pyruvate kinase M2 (PKM2), fructose-bisphosphate aldolase A (ALDOA) and ELAV-like protein 1 (ELAVL1/HuR) were found to bind circhPETH-147aa (Supplementary Fig. 13b). Subsequent co-IP assays confirmed the interaction among circPETH-147aa, PKM2 and ALDOA, as well as the mutual interaction between circPETH-147aa and HuR (Supplementary Fig. 13c).
In addition to regulating the final rate-limiting step of glycolysis by catalyzing the transfer of a phosphate group from phosphoenolpyruvate (PEP) to ADP to produce pyruvate and ATP, PKM2 has also been identified as a protein kinase that phosphorylates a variety of signaling protein substrates involved in cell growth, division, differentiation, adhesion, motility, and death45,46. A recent high-quality study involving the integrated proteogenomic characterization of HCC described the critical role of ALDOA S36 phosphorylation in driving HCC glycolytic metabolism47. Therefore, we hypothesized that PKM2 might bind to ALDOA and catalyze its phosphorylation, with circPETH-147aa possibly acting as a guide for catalysis. To test this hypothesis, a series of reciprocal co-IP assays were performed. In accordance with the trend of the change in circPETH-147aa expression, the interaction between PKM2 and ALDOA was markedly increased in HCC cells cocultured with TAMs but abolished by circPETH depletion in TAMs (Supplementary Fig. 13d); this finding is consistent with was observed after the deletion of circPETH in HCCLM9 cells (Supplementary Fig. 13e). In addition, the binding of PKM2 to ALDOA was substantially strengthened by the upregulation of circPETH-147aa but not by circPETH itself or any of the mutants in HCC cells (Supplementary Fig. 13f, g), revealing that circPETH-147aa specifically facilitated the interaction between PKM2 and ALDOA. Next, we examined the effects of circPETH-147aa and PKM2 on ALDOA phosphorylation. As predicted, the knockout and knockdown of circPETH markedly repressed ALDOA phosphorylation, which was additionally inhibited by silencing PKM2 (Supplementary Fig. 14a, b). The phosphorylation level of ALDOA clearly increased only after circPETH-147aa was overexpressed, and this change was subsequently largely reversed after PKM2 was downregulated (Supplementary Fig. 14c, d). Overall, these observations illustrate that circPETH-147aa guides PKM2 to preferentially catalyze the phosphorylation of ALDOA by simultaneously interacting with PKM2 and ALDOA.
circPETH-147aa promotes PKM2-dependent ALDOA-S36 phosphorylation via the MEG pocket
To identify the binding domain of ALDOA responsible for its interaction with PKM2, a series of FLAG-tagged ALDOA deletion mutants were established and transfected into HEK293T cells expressing HA-tagged circPETH-147aa (Supplementary Fig. 15a). Co-IP assays revealed that ALDOA, through the 1–155 aa region and 240–364 aa region, interacted with PKM2 (Supplementary Fig. 15b). Notably, ALDOA has been shown to bind to cellular proteins via a surface hydrophobic pocket created by the side chains of two amino acids, R43 and R30448, which are located in the 1-155-aa region and 240-364-aa region, respectively. To explore whether the R43 and R304 sites are critical for the interaction of ALDOA with PKM2, these two sites were mutated to alanine (A) separately or in combination, yielding R43A, R304A and RR/2 A (Supplementary Fig. 15c). The RR/2 A mutation eliminated the binding of ALDOA to PKM2 (Supplementary Fig. 15d). Moreover, binding behavior was assessed using a sensor chip immobilized with recombinant wild-type ALDOA (WT) or mutant ALDOA (RR/2 A). Surface plasmon resonance (SPR) assays revealed no interaction between ALDOA (RR/2 A) and PKM2 (Supplementary Fig. 15e). The dissociation constant (KD) for ALDOA (WT) was 382.1 nM (Supplementary Fig. 15f), highlighting that the hydrophobic pocket is indispensable in mediating the binding of ALDOA with PKM2. Additionally, we investigated the active sites of PKM2 that may fit into the hydrophobic pocket of ALDOA. Since PKM2 differs from PKM1 in the 388-433 aa region46 (Supplementary Fig. 15g), we initially hypothesized that PKM1 interacts with ALODA. As expected, ALDOA was not enriched in the green fluorescent protein (GFP)-PKM1 immunoprecipitate, even after circPETH-147aa was overexpressed (Supplementary Fig. 15h), indicating that PKM2 bound to ALDOA via the 388--433 aa region. We constructed a battery of GFP-tagged PKM2 variants with deletion mutations in the 388-433 aa region (Supplementary Fig. 15g) and found that, among the mutants, only PKM2 Δ404-418 failed to interact with ALDOA (Supplementary Fig. 15i). Hydrophobic pockets are likely to accommodate the side chains of hydrophobic amino acids, forming hydrophobic bonds to permit interactions48. All four hydrophobic amino acid sites in the 404-418 aa region of PKM2 were individually mutated to A to generate I404A, P408A, V414A and V417A (Supplementary Fig. 15j). The binding of PKM2 to ALDOA was abrogated only by the I404A mutation (Supplementary Fig. 16a), even when the other three sites were simultaneously mutated (Supplementary Fig. 16b). To confirm the binding site that mediates this interaction, molecular docking based on ZDOCK calculations was performed with the I404 residue of PKM2 loosely docked into the hydrophobic pocket of the ALDOA surface through hydrophobic bonds (Supplementary Fig. 16c). Finally, the binding of ALDOA to recombinant wild-type PKM2 (WT) or mutant PKM2 (I404A) was analysed via SPR. The dissociation constant (KD) for the ALDOA-PKM2 (WT) interaction was 322.5 nM (Supplementary Fig. 16d), whereas no response was observed for the ALDOA-PKM2 (I404A) interaction (Supplementary Fig. 16e). Collectively, these findings show that the hydrophobic bonds generated by the I404 residue of PKM2 and the hydrophobic pocket on the ALDOA surface clearly provide a foundation for the PKM2‒ALDOA interaction.
Therefore, we clarified that PKM2 interacted with circPETH-147aa through the 388--433 aa region (Supplementary Fig. 16f) and that this interaction was abolished by the combination of PKM2 Δ389-403 and PKM2 Δ419-433 mutations (Supplementary Fig. 16g). To predict the specific binding sites involved, we performed structural prediction and modeled circPETH-147aa (Supplementary Fig. 16h) and then carried out molecular docking studies on the basis of ZDOCK calculations, which revealed that two pockets on the PKM2 surface, which were individually created by the side chains of two sets of amino acids, accommodated the P96 and P106 residues of circPETH-147aa (Supplementary Fig. 17a). Two sets of amino acids in PKM2 were mutated to glycine (G) separately or in combination (Supplementary Fig. 17b), whereas the P96 and P106 sites in circPETH-147aa were individually or simultaneously mutated to A (Supplementary Fig. 17c). Co-IP assays revealed that both of these combinations of mutations, PKM2 ERRAKC/6 G and circPETH-147aa PP/2 A, resulted in almost no interaction between PKM2 and circPETH-147aa (Supplementary Fig. 17d, e), which was verified by SPR (Supplementary Fig. 17f–i). Thereafter, molecular docking based on ZDOCK calculations was implemented to investigate the binding sites between ALDOA and circPETH-147aa, and the findings revealed that three sets of amino acid residues in ALDOA were able to bind a pocket on the circPETH-147aa surface, which was generated primarily by the side chains of three amino acids, M1, E65 and G147 (Supplementary Fig. 18a). All three sets of amino acid sites in ALDOA were mutated to G individually or in combination (Supplementary Fig. 18b). The interaction between ALDOA and circPETH-147aa was abrogated by the KHVNHA/6 G mutation (Supplementary Fig. 18c), which was validated by SPR (Supplementary Fig. 18d, e). The M1, E65 and G147 residues in circPETH-147aa were subsequently mutated to A individually or simultaneously (Supplementary Fig. 18f). As indicated by co-IP and SPR, the MEG/3 A mutation led to the most significant decrease in the enrichment of ALDOA in HA-tagged circPETH-147aa immunoprecipitates (Supplementary Fig. 18g) and a large increase in the dissociation constant (KD) for the ALDOA-circPETH-147aa interaction, from 42.38 nM to 1407 nM (Fig. 3a, b).
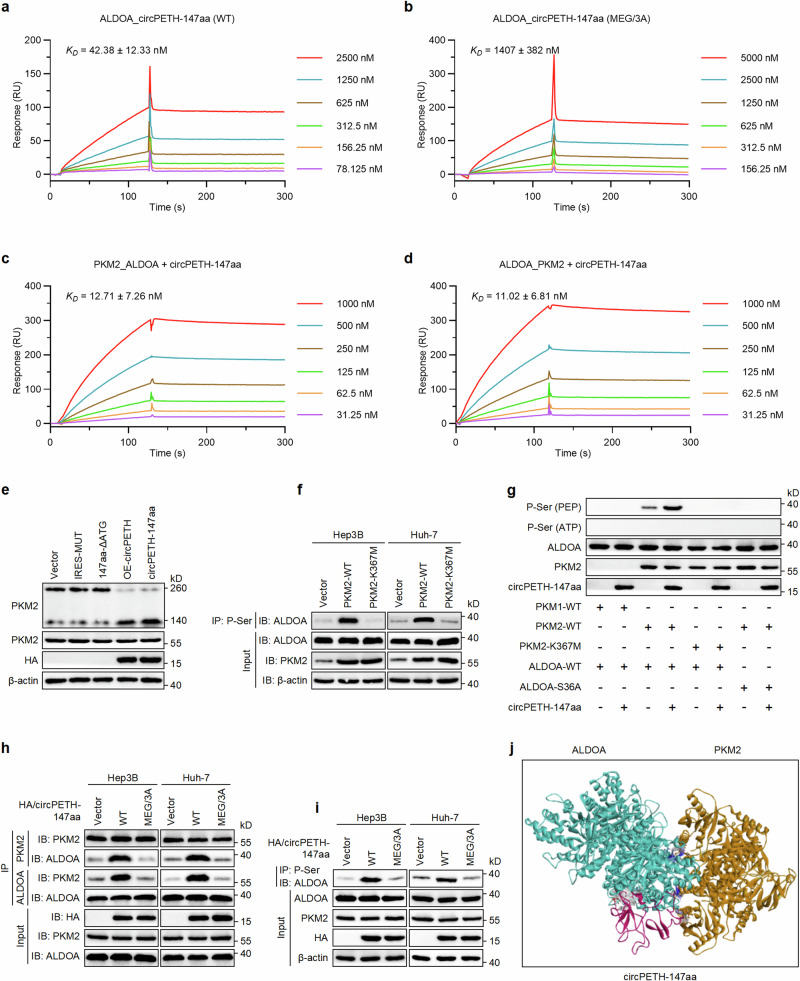
Fig. 3. circPETH-147aa promotes PKM2-catalyzed ALDOA-S36 phosphorylation via the MEG pocket.
a SPR sensorgrams and binding affinity between wild-type circPETH-147aa and ALDOA. b SPR sensorgrams and binding affinity between site-mutated circPETH-147aa and ALDOA. c SPR sensorgrams of ALDOA-PKM2 binding in the presence of circPETH-147aa in solution. d SPR sensorgrams of PKM2-ALDOA binding in the presence of circPETH-147aa in solution. e PKM2 dimer and tetramer formation analyzed by BN-PAGE and total PKM2 detected by western blot. The samples derive from the same experiment but different gels for PKM2 dimer and tetramer, another for PKM2 and another for HA and β-actin were processed in parallel. f The phosphorylation level of ALDOA in Hep3B and Huh-7 cells transfected with PKM2 wild-type or K367M-mutated plasmid by co-IP assays. The samples derive from the same experiment but different gels for ALDOA and PKM2, and another for ALDOA and β-actin were processed in parallel. g In vitro kinases assays were performed by mixing recombinant PKM2-WT, PKM2-K367M, or PKM1 with purified recombinant ALDOA-WT or ALDOA-S36A in the presence of circPETH-147aa and PEP or ATP. The samples derive from the same experiment but different gels for ALDOA, and another for PKM2, ALDOA and circPETH-147aa were processed in parallel. h The interaction between PKM2 and ALDOA in Hep3B and Huh-7 cells with overexpression of wild-type or MEG/3 A mutated circPETH-147aa by co-IP assays. The samples derive from the same experiment but different gels for ALDOA and PKM2, and another for PKM2, ALDOA and HA were processed in parallel. i The phosphorylation level of ALDOA in Hep3B and Huh-7 cells with overexpression of wild-type or MEG/3 A mutated circPETH-147aa by co-IP assays. The samples derive from the same experiment but different gels for ALDOA, and another for PKM2, β-actin and HA were processed in parallel. j Molecular docking of a complex consisting of PKM2, ALDOA and circPETH-147aa. Data are representative of at least three independent experiments. Source data are provided as a Source Data file.
Finally, we evaluated whether circPETH-147aa directly increases the phosphorylation of ALDOA catalyzed by PKM2. As predicted, the dissociation constants (KD) for the PKM2-ALDOA and ALDOA-PKM2 interactions were markedly reduced, from 382.1 nM and 322.5 nM to 12.71 nM and 11.02 nM, respectively, in the presence of circPETH-147aa (Fig. 3c, d), which demonstrated that circPETH-147aa considerably increased the affinity between PKM2 and ALDOA. Notably, R399 formed stable charge‒charge interactions with residues E418 and E396 of PKM2 located on the other dimer of the PKM2 tetramer49. As R399 and E396 are located in the binding sites formed between PKM2 and circPETH-147aa, we surmised that circPETH-147aa might disrupt the assembly of dimeric PKM2 into tetrameric PKM2. The results of blue native PAGE (BN-PAGE) suggested that upregulation of circPETH-147aa facilitated the conversion of PKM2 tetramers to dimers (Fig. 3e), increasing the protein kinase activity of PKM2 because dimeric PKM2 is the activated form of the protein kinase, whereas tetrameric PKM2 exhibited pyruvate kinase activity49. Furthermore, the protein kinase activity of PKM2 was abolished by the K367M mutation (Fig. 3f), which was in line with previous reports50. Next, in vitro kinase assays were conducted to evaluate whether ALDOA is directly phosphorylated via the catalytic activity of PKM2. No ALDOA phosphorylation was observed when ATP was the phosphate group donor and recombinant PKM2 was the protein kinase. However, incubation of PKM2 with ALDOA in the presence of PEP revealed that PKM2 (WT), but not kinase-dead PKM2 (K367M) or PKM1, phosphorylated ALDOA (WT) but not ALDOA (S36A), and the catalytic reaction was consolidated by circPETH-147aa (Fig. 3g). On the other hand, the binding of circPETH-147aa to ALDOA revealed that circPETH-147 was the substrate for PKM2-catalyzed phosphorylation, which depended on the pocket formed by three amino acids (M1, E65 and G147), termed the MEG pocket, on the circPETH-147aa surface. Indeed, the strengthening of the PKM2‒ALDOA interaction and increase in ALDOA phosphorylation, both caused by the overexpression of circPETH-147aa (WT), were abolished by the MEG/3 A mutation (Fig. 3h, i). Therefore, we propose the existence of a complex consisting of PKM2, ALDOA and circPETH-147aa according to molecular docking via ZDOCK calculations (Fig. 3j). Specifically, circPETH-147aa promotes the protein kinase activity of PKM2 by driving its dissociation into the dimeric form and strengthening targeted ALDOA phosphorylation at S36, which is mediated by the MEG pocket on the circPETH-147aa surface.
We preliminarily assess whether the interaction of circPETH-147aa with ALDOA mediated by the MEG pocket contributes to the biological functions of circPETH-147aa. Mutation of the MEG pocket abolished the ability of circPETH-147aa to promote F-actin overgrowth in HCC cells, YAP nuclear translocation in HCC cells (Supplementary Fig. 19a, b), and HCC cell invasion and migration (Supplementary Fig. 19c, d).
circPETH-147aa impairs anti-HCC immunity by increasing HuR-dependent SLC43A2 mRNA stability and stimulating methionine and leucine deficiency in cytotoxic CD8+ T cells
Considering that metabolic pathways are the pathways most significantly regulated by circPETH, metabolomics was performed after liquid chromatography‒tandem mass spectrometry (LC‒MS) to identify differentially abundant metabolites after circPETH expression was altered (Supplementary Fig. 20a), and the differentially abundant metabolites were found to be significantly enriched in the following terms: metabolic pathways, biosynthesis of amino acids, protein digestion and absorption, multiple amino acid metabolism and central carbon metabolism in cancer (Supplementary Fig. 20b). Through combined metabolome and transcriptome analysis, we constructed a correlation network, which revealed strong correlations between changes in SLC43A2 expression and changes in methionine and leucine levels induced by circPETH-147aa (Fig. 4a). We aimed to clarify the molecular mechanism underlying the modulatory effects of circPETH-147aa on SLC43A2 levels and intriguingly found that SLC43A2 was one of the HuR-bound mRNAs identified via RIP microarray analysis51. The RNA-binding protein HuR, which regulates the stability of many target mRNAs, can interact with circPETH-147aa according to our previous co-IP data (Supplementary Fig. 13c). Hence, it can be deduced that HuR might be responsible for the regulation of SLC43A2 expression by circPETH-147aa. To test this possibility, RIP was first performed using an endogenous anti-HuR antibody. Indeed, SLC43A2 mRNA was clearly enriched among the HuR-immunoprecipitated RNAs from Hep3B and Huh-7 cells (Supplementary Fig. 20c, d). The RNA and protein levels of SLC43A2 markedly decreased upon downregulation of HuR expression in HCC cells (Supplementary Fig. 20e–g), suggesting that HuR bound to SLC43A2 mRNA and increased its stability. More importantly, consistent with the ability of circPETH-147aa to alter gene expression, the enrichment of SLC43A2 mRNA among HuR-immunoprecipitated RNAs from HCC cells increased when HCC cells were cocultured with TAMs, and this increase was abolished by circPETH knockdown (Supplementary Fig. 20h, i), which was confirmed by knocking out circPETH in HCCLM9 cells (Fig. 4b). The binding of HuR to SLC43A2 mRNA was markedly enhanced by increasing exclusively translatable circPETH levels (Supplementary Figs. 20j, k and 21a, b), implying that circPETH-147aa strengthened the association of HuR with SLC43A2 mRNA. Next, we assessed the joint effects of circPETH-147aa and HuR on SLC43A2 expression. As expected, both deleting and silencing circPETH resulted in obvious decreases in the RNA and protein levels of SLC43A2, which were further reduced by downregulating HuR (Supplementary Fig. 21c–f and Fig. 4c). The RNA and protein levels of SLC43A2 increased only when circPETH-147aa was overexpressed, and this effect was reversed by HuR depletion (Supplementary Figs. 21g–k and 22a).
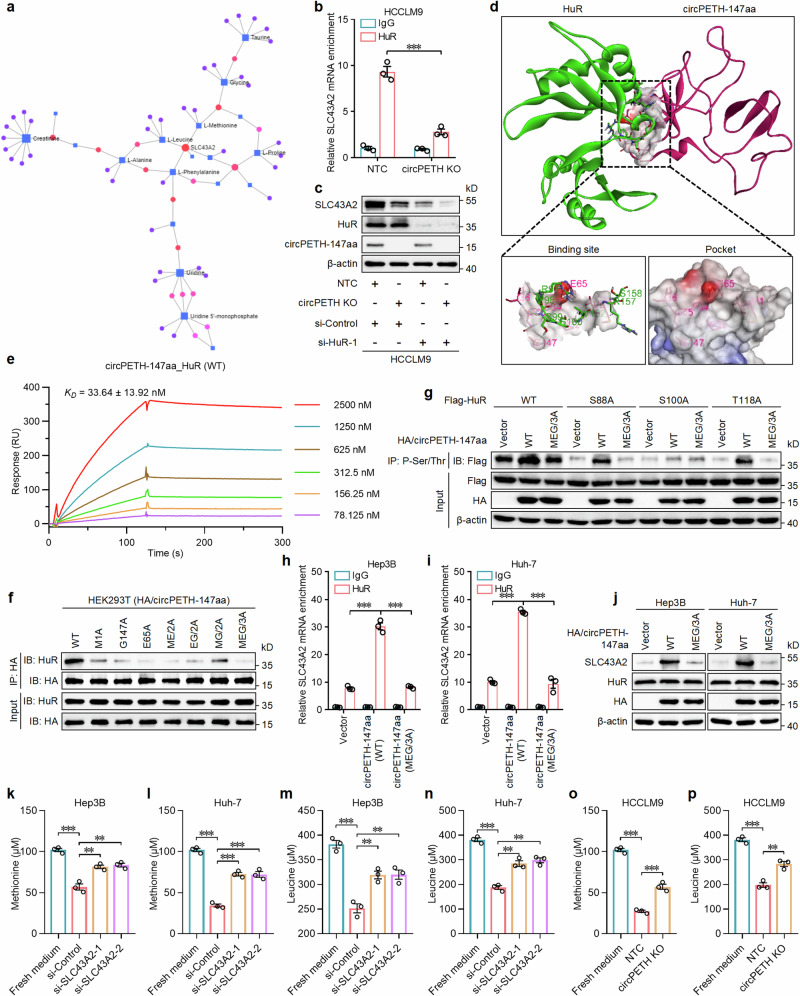
Fig. 4. circPETH-147aa stimulates methionine and leucine consumption in HCC cells by elevating HuR-dependent SLC43 A2 mRNA stability.
a The conjoint analysis of metabolomics and transcriptomics. b The enrichment of SLC43 A2 mRNA in HuR-immunoprecipitated RNAs for HCCLM9 cells with circPETH KO by RIP assays. n = 3 independent experiments. Data are shown as mean ± SEM. Exact P value: 0.000543. c The expressions of SLC43 A2, HuR and circPETH-147aa in HCCLM9 cells with circPETH KO and HuR knockdown by western blot. The samples derive from the same experiment but different gels for SLC43A2, HuR and circPETH-147aa, and another for β-actin were processed in parallel. d Molecular docking. e SPR sensorgrams and binding affinity between wild-type HuR and circPETH-147aa. f The interactions of HuR and HA-tagged circPETH-147aa site-directed mutants in HEK293T cells by co-IP assays. The samples derive from the same experiment but different gels for HuR and HA, and another for HuR and HA were processed in parallel. g The phosphorylation levels of total HuR, HuR-S88, HuR-S100 and HuR-T118 upon overexpression of wild-type or MEG/3A mutated circPETH-147aa by co-IP assays. The samples derive from the same experiment but different gels for Flag, and another for Flag, HA and β-actin were processed in parallel. h, i The enrichment of SLC43A2 mRNA in HuR-immunoprecipitated RNAs for Hep3B and Huh-7 cells with overexpression of wild-type or MEG/3A mutated circPETH-147aa by RIP assays. n = 3 independent experiments. Data are shown as mean ± SEM. Exact P values from left to right (h): 7.39E − 05, 7.15E − 05. Exact P values from left to right (i): 3.08E − 06, 9.87E − 05. j The expressions of SLC43A2, HuR and HA in Hep3B and Huh-7 cells with overexpression of wild-type or MEG/3A mutated circPETH-147aa by western blot. The samples derive from the same experiment but different gels for SLC43A2, and another for HuR, HA and β-actin were processed in parallel. k–n Effects of Hep3B and Huh-7 cells with SLC43A2 knockdown on methionine and leucine consumption for 24 h. n = 3 independent experiments. Data are shown as mean ± SEM. Exact P values from top to bottom (k): 0.000458, 0.003639, 0.004687. Exact P values from top to bottom (l): 1.63E − 05, 0.000597, 0.000183. Exact P values from top to bottom (m): 0.000406, 0.006196, 0.004932. Exact P values from top to bottom (n): 6.31E − 05, 0.002213, 0.001020. o, p Effects of HCCLM9 cells with circPETH KO on methionine and leucine consumption for 24 h. n = 3 independent experiments. Data are shown as mean ± SEM. Exact P values from top to bottom (o): 7.20E − 06, 0.000830. Exact P values from top to bottom (p): 0.000111, 0.003735. **P < 0.01; ***P < 0.001 by two-tailed Student’s T-test. Data are representative of at least three independent experiments. Source data are provided as a Source Data file.
To explore the binding sites between circPETH-147aa and HuR, molecular docking based on ZDOCK calculations was performed, and two sets of amino acid residues in HuR were found to precisely fit into the MEG pocket on the circPETH-147aa surface (Fig. 4d). These two sets of amino acid sites in HuR were mutated to A separately or simultaneously (Supplementary Fig. 22b). Co-IP assays revealed that the interaction between HuR and circPETH-147aa was abrogated by the combined RPSSVRS/7 A mutation (Supplementary Fig. 22c), which was verified by SPR (Fig. 4e and Supplementary Fig. 22d). Mutation of the MEG pocket led to negligible enrichment of HuR in the HA-tagged circPETH-147aa immunoprecipitate (Fig. 4f), which was in accordance with the ZDOCK results.
It has been reported that HuR phosphorylation profoundly affects the association of HuR with target mRNAs51. In particular, HuR is generally phosphorylated at S88, S100 and T118; the phosphorylation of HuR at S100 enhances its binding to mRNAs, whereas the phosphorylation of HuR at S88 and T118 reduces this interaction52. Given that the S100 residue precisely fit in the MEG pocket of circPETH-147aa, we investigated whether HuR phosphorylation at S100 is influenced by the MEG pocket. The results revealed that the MEG pocket largely sustained HuR phosphorylation at S100 (Fig. 4g). As predicted, the increases in the HuR–SLC43A2 mRNA interaction and SLC43A2 expression induced by increasing the circPETH-147aa (WT) level were abrogated after the loss of the MEG pocket in Hep3B and Huh-7 cells (Fig. 4h–j). Since SLC43A2 has been identified as an amino acid transporter responsible for methionine and leucine consumption53, we measured amino acid levels in fresh medium and the supernatant of HCC cells and observed that the consumption of methionine and leucine was significantly decreased by SLC43A2 knockdown in Hep3B and Huh-7 cells (Fig. 4k–n), an effect that was similar to that of circPETH knockout in HCCLM9 cells (Fig. 4o, p). In contrast, the consumption of methionine and leucine was markedly enhanced via the upregulation of circPETH-147aa or translatable circPETH before large expression changes were induced by silencing SLC43A2 (Supplementary Fig. 22e–h).
Emerging high-quality research has revealed that cancer cells avidly consume and outcompete CD8+ T cells for methionine via high expression of SLC43A2, impairing spontaneous and checkpoint-induced tumor immunity54. In addition, leucine has been described as an important essential amino acid and a nutrient signal that activates complex 1 of the mammalian target of rapamycin (mTORC1), which is a critical regulator of T-cell proliferation, differentiation, and function55. Therefore, we evaluated whether circPETH-147aa weakens the killing effects of CD8+ T cells on HCC cells. First, T cells with high expression of CD8 on the cell membrane were isolated from human peripheral blood via fluorescence-activated cell sorting (FACS) (Supplementary Fig. 23a) and were subjected to activation and amplification by anti-IL-2, anti-CD3 and anti-CD28 antibodies (Supplementary Fig. 23b). Then, we cocultured circPETH-altered HCC cells with CD8+ T cells (Supplementary Fig. 23c) and measured the efficiency of CD8+ T cells in killing the HCC cells by flow cytometry. The data demonstrated that the apoptosis rates of Hep3B and Huh-7 cells were markedly increased after these cells were cocultured with CD8+ T cells, and the number of these cells was further elevated by circPETH silencing (Supplementary Fig. 23d–f). In contrast, upregulation of circPETH-147aa or translatable circPETH considerably inhibited the apoptosis of HCC cells cocultured with CD8+ T cells, and HCC cells were significantly rescued from apoptosis by methionine and leucine supplementation (Supplementary Fig. 23g–i). Taken together, these findings reveal that circPETH-147aa sustains HuR phosphorylation at S100 via the MEG pocket on the circPETH-147aa surface, which accommodates the S100 residue of HuR, increasing the association of HuR with SLC43A2 mRNA. Moreover, the stability of SLC43A2 mRNA and its expression are increased, and the uptake of methionine and leucine by HCC cells is increased, thereby attenuating the tumor-killing effects of CD8+ T cells.
To assess the specific antitumour response, we engineered Hepa1-6 cells, well-characterized H-2Kb+ mouse HCC cells, to express a truncated form of the tumor antigen ovalbumin257-264 (OVA257-264) (Supplementary Fig. 24a). To evaluate the expression of antigen-specific pMHC-I, we used a unique mouse TCR-mimicking antibody that binds the immunogenic OVA257-264 peptide presented by the H-2Kb molecule (H-2Kb:OVA257-264, hereafter referred to as H-2Kb:OVA)56 to verify successful antigen presentation (Supplementary Fig. 24b). Correspondingly, OT-I T cells, a type of primary mouse CD8+ T cells expressing a unique T-cell receptor (TCR) that specifically recognizes H-2Kb:OVA, were isolated from the spleens of OT-I TCR transgenic mice and subsequently cocultured with Hepa1-6 OVA cells (Supplementary Fig. 24a); the results revealed that circPETH-deficient Hepa1-6 OVA cells were more susceptible to OT-I T-cell-mediated killing (Supplementary Fig. 24c). Conversely, Hepa1-6 OVA cells were more resistant to OT-I T-cell-mediated killing in the presence of circPETH-147aa, and this effect was reversed by methionine and leucine supplementation (Supplementary Fig. 24d).
Finally, we assessed the relationship between circPETH expression and the response to ICBs in HCC patients, as well as immune cell infiltration in tumor tissues. The results revealed that patients with high circPETH levels exhibited increased infiltration of CD8+ T cells and decreased infiltration of TAMs in HCC tissues (Supplementary Fig. 24e–h). Importantly, 38 patients received ICB therapy before hepatectomy, with 14 patients achieving a partial response (PR) and 24 patients displaying progressive disease (PD). We found that patients with low circPETH levels exhibited a significantly greater response to ICB therapies (Supplementary Fig. 24i), which preliminarily implies a crucial role for circPETH in immunotherapy for HCC in patients.
Norathyriol reverses the circPETH-147aa-facilitated acquisition of metabolic and metastatic phenotypes by HCC cells by specifically occupying the MEG pocket
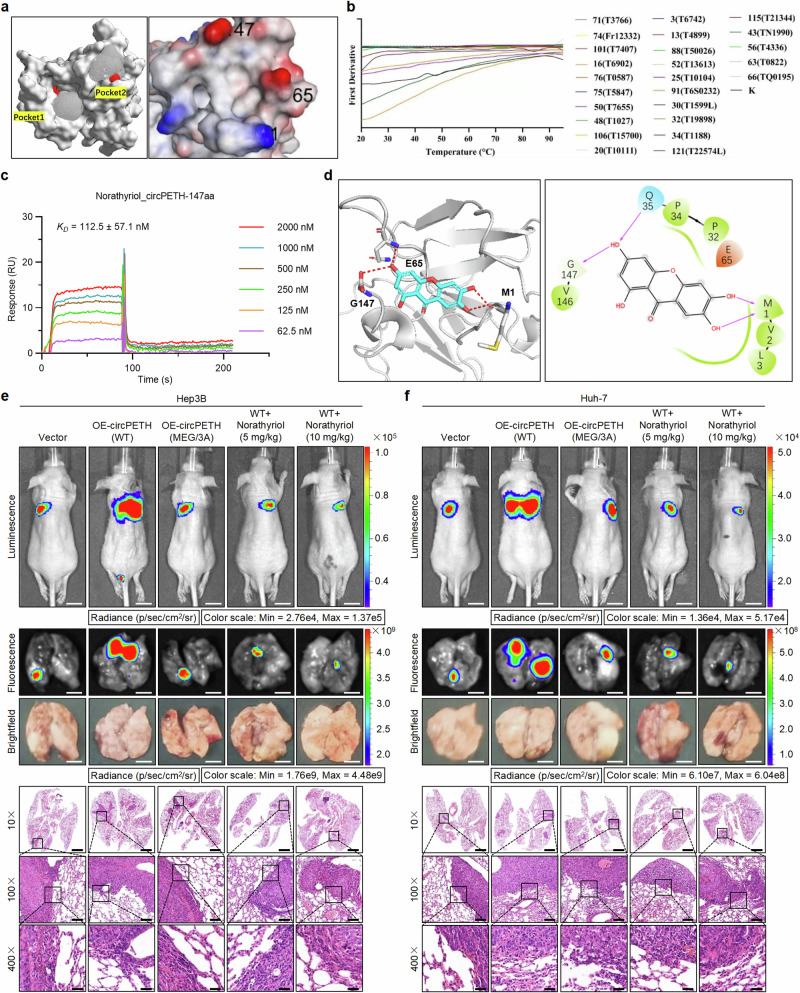
Considering the clear importance of the MEG pocket in the biological functions of circPETH-147aa, we explored small-molecule compounds that might inhibit circPETH-147aa–protein interactions by targeting the MEG pocket. Initially, we carried out molecular operating environment (MOE) analysis with the site Finder module and identified two pockets on the circPETH-147aa surface (Fig. 5a), among which Pocket 2 was the MEG pocket. High-throughput virtual screening (HTVS) was subsequently performed with the small-molecule library TargetMol to identify potential circPETH-147aa inhibitors. A total of 122 candidate chemicals and their docking scores are listed in Supplementary Data 1; preliminary experimental validation of these candidates was subsequently performed via nanodifferential scanning fluorimetry (nanoDSF), which indicated that 25 molecules significantly affected the stability of the circPETH-147aa protein (Fig. 5b and Supplementary Data 2), possibly by interacting with it. A series of SPR assays were subsequently performed, and the results accurately revealed the binding behavior of circPETH-147aa. Norathyriol, a natural product, presented the highest affinity for circPETH-147aa, with a dissociation constant (KD) of 112.5 nM (Fig. 5c). The precise interaction between norathyriol and circPETH-147aa was delineated by molecular docking analysis (Fig. 5d). Therefore, we selected norathyriol as a potential inhibitor of circPETH-147aa.
Fig. 5. Norathyriol reverses circPETH-147aa-facilitated lung metastasis of HCC cells by specifically occupying the MEG pocket.
a Two pockets identified on circPETH-147aa surface by site Finder module of MOE. b 25 compounds significantly affected the stability of circPETH-147aa protein by nanoDSF. c SPR sensorgrams and binding affinity between circPETH-147aa and Norathyriol. d Molecular docking of 3-dimensional structures shows the occupation of Norathyriol on MEG pocket of circPETH-147aa. e, f Top: Representative bioluminescence photographs of the murine tail vein injection lung metastasis models with indicated Hep3B and Huh-7 cells. Norathyriol was intraperitoneally injected (5 mg/kg or 10 mg/kg, once daily for 2 weeks). Middle: Representative fluorescence and brightfield photographs of pulmonary metastatic nodules in lung metastasis models with indicated Hep3B and Huh-7 cells. Bottom: Representative HE staining of metastatic foci in the lung. Scale bars from top to bottom: 1 cm, 4 mm, 4 mm, 2.8 mm, 280 μm, 70 μm. n = 5 for each group. Data are representative of at least three independent experiments. Source data are provided as a Source Data file.
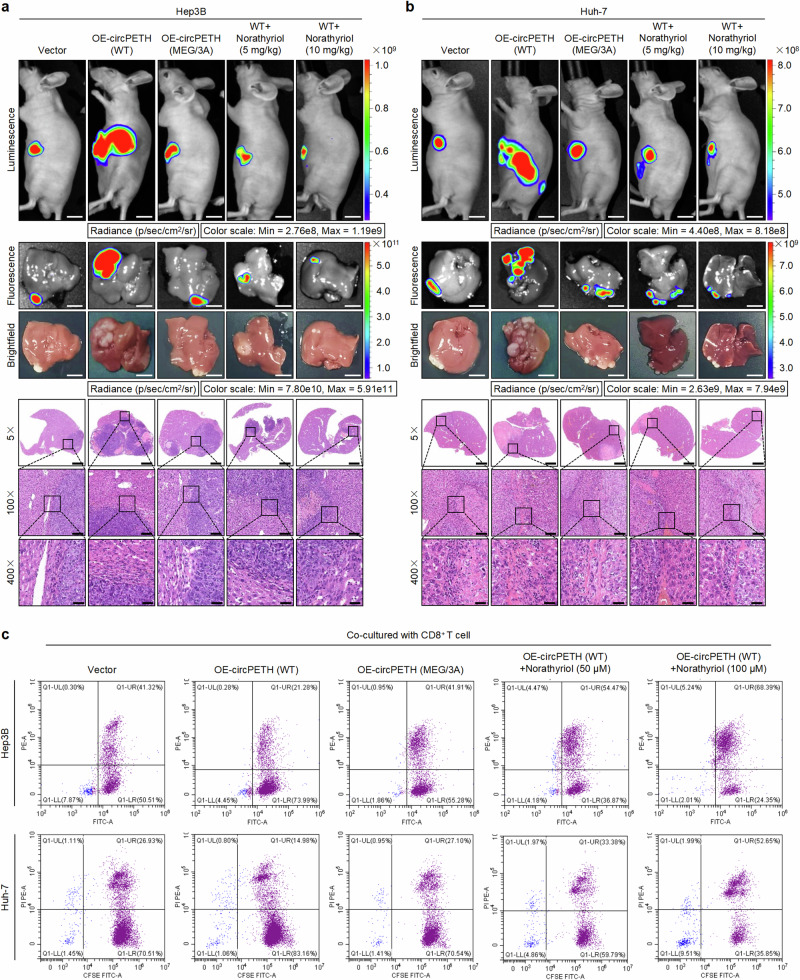
CCK-8 assays revealed that the half maximal inhibitory concentrations (IC50s) for Hep3B, Huh-7 and HCCLM9 cells were 43.86 μM, 59.29 μM and 175.5 μM, respectively (Supplementary Fig. 25a). We found that mutation of the MEG pocket presumably rescued the promoting effects of circPETH-147aa on HCC cell aerobic glycolysis and metastasis, and norathyriol treatment substantially reversed these effects in a dose-dependent manner (Supplementary Fig. 25b–l and Figs. 5e, f, 6a, b and Supplementary Fig. 26a–d), indicating that norathyriol is an effective suppressor of the malignant biological properties of HCC cells. Notably, it has been reported that norathyriol has multiple anticancer activities57. Indeed, in addition to suppressing the function of the MEG pocket of circPETH-147aa, norathyriol had nonspecific effects on HCC metastasis (Supplementary Fig. 26e–h). However, the inhibitory effects of norathyriol in the OE-circPETH MEG/3 A group were considerably weaker than the inhibitory effects of norathyriol in the OE-circPETH WT group. Overall, the inhibitory effect of norathyriol on HCC cells was dependent mainly on circPETH-147.
Fig. 6. Norathyriol inverts circPETH-147aa-facilitated intrahepatic metastasis of HCC cells and enhances tumor-killing effects of CD8+ T cells.
a, b Top: Representative bioluminescence photographs of liver orthotopic implanted intrahepatic metastasis models with indicated Hep3B and Huh-7 cells. Norathyriol was intraperitoneally injected (5 mg/kg or 10 mg/kg, once daily for 2 weeks). Middle: Representative fluorescence and brightfield photographs of intrahepatic metastatic nodules in liver orthotopic implanted intrahepatic metastasis models with indicated Hep3B and Huh-7 cells. Bottom: Representative HE staining of metastatic foci in the liver. Scale bars from top to bottom: 1.2 cm, 7.5 mm, 7.5 mm, 2.8 mm, 280 μm, 70 μm. n = 5 for each group. c Flow cytometry analysis of indicated Hep3B and Huh-7 cells cocultured with CD8+ T cells and treated with Norathyriol at different concentrations for 24 h. Data are representative of at least three independent experiments. Source data are provided as a Source Data file.
Norathyriol increases anti-PD1 efficacy and enhances cytotoxic CD8+ T-cell function
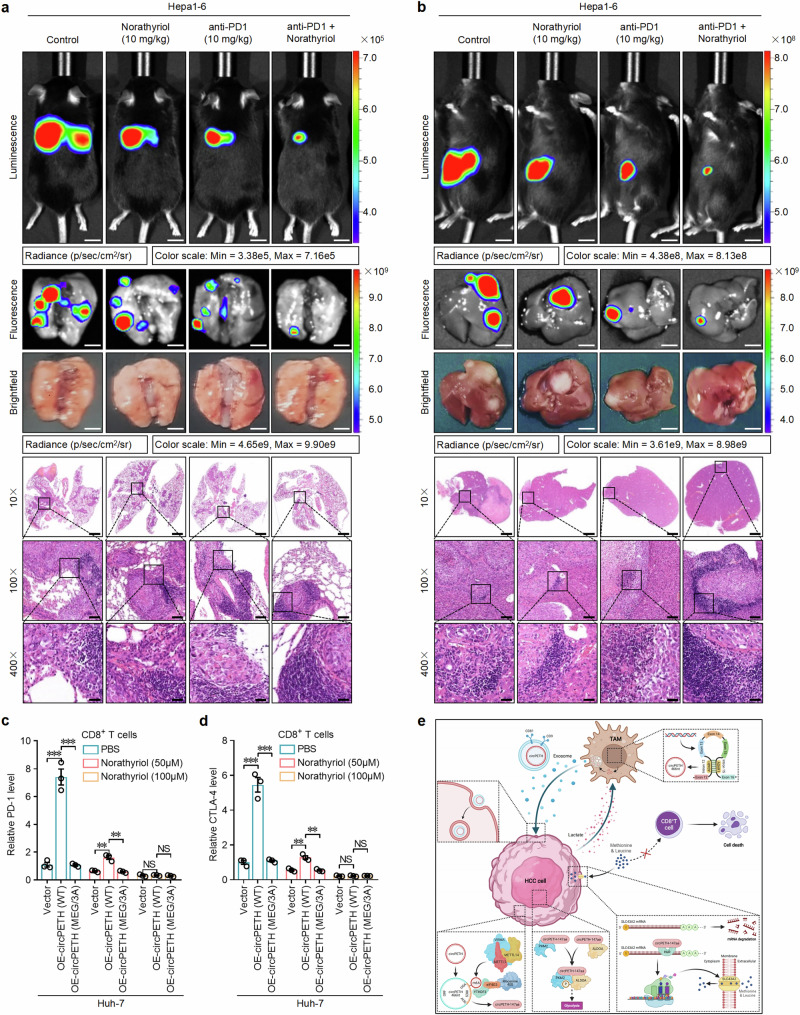
Importantly, the weakened tumor-killing effects of CD8+ T cells caused by circPETH-147aa overexpression were reversed by mutation of the MEG pocket and further increased by norathyriol treatment in a dose-dependent manner (Fig. 6c and Supplementary Fig. 26i, j). Accordingly, we hypothesized that norathyriol might improve the efficacy of ICB treatment. To test this possibility, C57BL/6J mouse models of lung metastasis and liver orthotopic intrahepatic metastasis were established via the implantation of Hepa1‒6 mouse HCC cells. Afterwards, norathyriol and anti-PD1 antibodies were administered individually or in combination. As expected, although the pulmonary and hepatic bioluminescence intensity, fluorescence intensity and number of histopathologically detectable metastatic foci were significantly lower after norathyriol or anti-PD1 monotherapy than in the control group to some extent, anti-PD1 plus norathyriol was markedly more effective than anti-PD1 monotherapy was (Fig. 7a, b and Supplementary Fig. 26k, l). Notably, an remarkably high increase in the proportion of lymphocytes accumulated in or around HCC foci in lung and liver tissue sections was detected in the combination therapy group. Consequently, we analysed the infiltration of CD8+ T cells and the expression levels of representative markers (PD-1, CTLA-4 and CD69) in HCC tissues via IF and IHC to determine the effects of norathyriol on tumor-infiltrating CD8+ T cells in vivo. The results revealed that norathyriol treatment was associated with significantly increased infiltration of CD8+ T cells, increased expression of CD69 and decreased levels of PD-1 and CTLA-4 (Supplementary Fig. 27a–f), indicating that norathyriol strengthened the function of tumor-infiltrating CD8+ T cells.
Fig. 7. Norathyriol increases anti-PD1 efficacy and boosts cytotoxic CD8+ T-cell function.
a Top: Representative bioluminescence photographs of the murine tail vein injection lung metastasis models with Hepa1-6 cells. Norathyriol was intraperitoneally injected (10 mg/kg, once daily for two weeks). Anti-PD1 was intraperitoneally injected (10 mg/kg, three times every week, for two weeks). Middle: Representative fluorescence and brightfield photographs of pulmonary metastatic nodules in lung metastasis models with Hepa1-6 cells. Bottom: Representative HE staining metastatic foci in the lung. Scale bars from top to bottom: 1.2 cm, 3.6 mm, 3.6 mm, 2.4 mm, 240 μm, 60 μm. n = 5 for each group. b Top: Representative bioluminescence photographs of liver orthotopic implanted intrahepatic metastasis models with Hepa1-6 cells. Norathyriol was intraperitoneally injected (10 mg/kg, once daily for two weeks). Anti-PD1 was intraperitoneally injected (10 mg/kg, three times every week, for two weeks). Middle: Representative fluorescence and brightfield photographs of intrahepatic metastatic nodules in liver orthotopic implanted intrahepatic metastasis models with Hepa1-6 cells. Bottom: Representative HE staining of metastatic foci in the liver. Scale bars from top to bottom: 1.2 cm, 8.5 mm, 8.5 mm, 2.4 mm, 240 μm, 60 μm. n = 5 for each group. c PD-1 expression in CD8+ T cells with indicated Huh-7 cells coculture and Norathyriol treatment at different concentrations for 24 h. n = 3 independent experiments. Data are shown as mean ± SEM. Exact P values from left to right: 0.000471, 0.000410, 0.002400, 0.002130. d CTLA-4 expression in CD8+ T cells with indicated Huh-7 cells coculture and Norathyriol treatment at different concentrations for 24 h. n = 3 independent experiments. Data are shown as mean ± SEM. Exact P values from left to right: 0.000501, 0.000486, 0.002863, 0.002195. e Summary figure of the mechanism of circPETH in TME [Created in BioRender. Cai, Y. (2024) https://BioRender.com/v84x844]. NS, not significant; **P < 0.01; ***P < 0.001 by two-tailed Student’s T-test. Data are representative of at least three independent experiments. Source data are provided as a Source Data file.
To further understand the mechanistic association of norathyriol with CD8+ T cells, RNA-seq was performed to determine the expression profiles of CD8+ T cells in norathyriol- and PBS-treated coculture systems (Supplementary Fig. 27g). The differentially expressed genes were enriched mainly in T-cell-related functions and T-cell-related pathways according to GO and KEGG analyses, respectively (Supplementary Fig. 27h, i), and the expression of CXCL13, reported to affect CD8+ T-cell immune function58, was significantly altered by norathyriol treatment. Next, we cocultured circPETH-altered Huh-7 cells with CD8+ T cells and treated them with different concentrations of norathyriol. Indeed, the expression of CXCL13 on CD8+ T-cell membranes was markedly elevated by upregulation of circPETH-147aa, and this effect was abrogated by mutation of the MEG pocket and markedly reversed by norathyriol treatment in a dose-dependent manner (Supplementary Fig. 28a). Methionine supplementation repairs T-cell immunity via the STAT5 pathway54. Consistent with this, mutation of the MEG pocket rescued the suppressive effects of circPETH-147aa on the expression of STAT5A and STAT5B in CD8+ T cells, and this effect was markedly reversed by norathyriol or methionine treatment in a dose-dependent manner (Supplementary Fig. 28b, c). Finally, we examined the influence of norathyriol on the immune function of CD8+ T cells in a norathyriol-treated coculture system. As expected, norathyriol decreased the levels of immune checkpoints, namely, exhaustion markers, including PD-1, CTLA-4, Tim-3 and TIGHT in CD8+ T cells (Fig. 7c, d and Supplementary Figs. 28d, e and 29a–c and Supplementary Fig. 30a), in a dose-dependent manner. In contrast, norathyriol treatment considerably elevated the activation of molecules (IFN-γ and TNF-α), the levels of cytotoxic molecules (GZMB and PRF-1), degranulation capability (CD107a) and proliferation ability (Ki67) in CD8+ T cells in a dose-dependent manner (Supplementary Fig. 28f–k and Supplementary Fig. 30b, c). These findings suggest that norathyriol increases the therapeutic efficacy of anti-PD1 against HCC by restoring the function of CD8+ T cells in the immunosuppressive TME.
Discussion
As the predominant facilitator of malignant biological features, metabolic reprogramming fuels cancer cell metastasis and remodels the immunosuppressive TME11. TAMs can promote the acquisition of metabolic and metastatic phenotypes by cancer cells16. Nevertheless, how TAMs regulate metabolic reprogramming and the immunosuppressive TME in the context of HCC is largely unknown. Our evidence confirmed that TAMs are oncogenic drivers of invasion, migration and aerobic glycolysis in HCC. EVs are pivotal communicators involved in noncontact cell‒cell interactions in the TME25. Accordingly, we found that EVs mediated the functions of TAMs in accelerating liver cancer progression. The mechanism underlying EV shuttling, which is required for the transportation of various bioactive molecules involved in the development of diverse human diseases, is unknown16. Our work has expanded on current research by illustrating that circRNAs loaded in EVs can be transported from inflammatory cells to cancer cells, in which they regulate biological behaviors. Specifically, circPETH, a circRNA loaded in EVs secreted by TAMs and taken up by HCC cells, reprogrammed metabolism and promoted the metastasis of recipient HCC cells, and this effect was significantly associated with worse clinical symptoms and prognosis in HCC patients.
The biogenesis of many circRNAs relies on the binding of some RBPs to inverted complementary sequences flanking the back-splice junction28. In this study, we discovered that ADAR1 repressed the circularization of circPETH by suppressing the back splicing of exons 13–15, which was consistent with the previously described function of ADAR159. Some studies have revealed that biomolecules from TAMs and lactate from cancer cells can form positive feed-forward loops to promote malignancy31,60. Similarly, our results revealed a positive feedback loop composed of circPETH encapsulated in EVs, lactate produced by HCC cells and inhibition of ADAR1 in lactate-polarized TAMs (Fig. 7e).
Numerous circRNAs have been demonstrated to be translatable RNAs61. Herein, we revealed that the protein, circPETH-147aa, with a unique 28-aa tail, is encoded by circPETH and that this protein represents an exceptional target for the effective detection and treatment of HCC. More importantly, circPETH-147aa, but not circPETH, was found to have a cancer-promoting function. In addition, TAMs presented high circPETH levels but not circPETH-147aa levels because circPETH did not undergo significant m6A modification, which triggers cap-independent protein translation of circRNAs via EIF4G2 and YTHDF336,40.
PKM2 tends to function as a dimer and exhibits high protein kinase activity in cancer cells; however, the tetrameric form with pyruvate kinase function is not the predominant form in normal cells49. The charge‒charge interaction between residues R399 and E396 is indispensable for the association of dimeric PKM2 with tetrameric PKM262 and was proven to be disrupted by circPETH-147aa, which competed for binding sites in our study. Furthermore, ALDOA was identified as the substrate phosphorylated by PKM2 at S36 via the MEG pocket-dependent interaction with circPETH-147aa (Fig. 7e). High-quality research has revealed the strong stimulatory effects of ALDOA S36 phosphorylation on metabolic reprogramming47, suggesting that the glycolysis-enhancing function of circPETH-147aa might be reflected by PKM2-catalyzed ALDOA S36 phosphorylation.
Metabolites in the TME are crucial modulators of antitumoral immunity63. Here, through a combined analysis of metabolomic and transcriptomic data, we revealed that circPETH-147aa increased the consumption of methionine and leucine. Specifically, the expression of SLC43A2, a transporter for methionine and leucine absorption53, was regulated by the MEG pocket-mediated phosphorylation of HuR, which increases the stability of SLC43A2 mRNA51. Previous studies have revealed that methionine or leucine deficiency weakens the killing effects of CD8+ T cells on HCC cells54,55. Indeed, circPETH-147aa impaired anti-HCC immunity by inducing methionine and leucine insufficiency in cytotoxic CD8+ T cells (Fig. 7e).
Finally, we successfully identified norathyriol by virtual and experimental screening as a small-molecule inhibitor that targets the MEG pocket on the circPETH-147aa surface. It has been reported that norathyriol exhibits potent chemopreventive activity through the inhibition of the transcription factors AP-1 and NF-κB by targeting ERKs in the context of ultraviolet-induced skin carcinogenesis57. In HCC cells, aerobic glycolysis and metastasis were suppressed by norathyriol in a dose-dependent manner. More importantly, the tumor-killing effects of CD8+ T cells and the expression of functional markers of CD8+ T cells were increased by norathyriol treatment in a dose-dependent manner. Accordingly, we deduced that norathyriol might increase the efficacy of ICB treatment. As expected, anti-PD1 plus norathyriol showed markedly greater effectiveness and induced greater lymphocyte infiltration than did anti-PD1 monotherapy. Therefore, this is a comprehensive work confirming that norathyriol is a promising effective agent, potentially opening an avenue to increase treatment efficacy in HCC patients. We hope to develop clinical protocols involving the use of anti-PD1 antibodies combined with norathyriol for treating patients with HCC and ICB resistance.
Methods
Ethics statement
All animal studies were approved by the Animal Ethics Committee of West China Hospital of Sichuan University (IACUC Number: 2021572 A), and conducted according to the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) guidelines. This paper describes studies involving human tissue samples that were approved by the Ethics Committee of West China Hospital of Sichuan University. All tissue samples were collected and used in compliance with informed consent policy. Written informed consent was obtained by human participants. All studies were performed in accordance with the Declaration of Helsinki.
Primary human TAMs isolation from HCC tissues and MDMs isolation from peripheral blood
HCC tissues and adjacent normal tissues were acquired from HCC patients undergoing curative surgical resections in the West China Hospital (Sichuan University, Chengdu, China). TAMs were isolated from fresh HCC tissues as previously described64. To be brief, the fresh tissues were minced into small pieces (1 mm3) and digested with 1 mg/ml collagenase I and 2 mg/ml collagenase IV (Sigma) at 37 °C for 40 min, which was terminated by 10% fetal bovine serum RPMI-1640 medium (Gibco). Then, cells were sequentially filtered through 70-μm and 40-μm cell strainers for two times, and subjected to Ficoll density gradient centrifugation as previously described16. CD14+ macrophages were isolated using MACS separation method based on CD14 Microbeads (Miltenyi Biotec) according to the manufacturer’s instructions. MDMs from peripheral blood of patients with HCC or healthy donors were also isolated by Ficoll density gradient centrifugation. Macrophages were grown in RPMI-1640 medium (Gibco) supplemented with 10% fetal bovine serum, 50 U/ml penicillin (Gibco) and 50 μg/ml streptomycin (Gibco).
Isolation, activation and amplification of primary human CD8+ T cells from peripheral blood
Lymphocytes from peripheral blood of healthy donors were isolated by Ficoll density gradient centrifugation, followed by an incubation with antibody against CD8 (555369, BD), after which CD8+ T cells were isolated by FACS. 100,000 CD8+ T cells per well were seeded into 96-well plates at 37 °C. CD8+ T cells were activated and amplified in RPMI-1640 medium (Gibco) containing 20 IU/ml IL-2 (PeproTech), 1:100 T Cell TransAct (Miltenyi Biotec), 10% fetal bovine serum, 50 U/ml penicillin (Gibco) and 50 μg/ml streptomycin (Gibco).
HCC cells and co-culture
Three human HCC cell lines, SK-Hep1 (TCHu109), Huh-7 (TCHu182) and Hep3B (TCHu106), and Hepa1-6 (TCM39) mouse HCC cells were purchased from the Shanghai Cell Bank Type Culture Collection Committee (CBTCCC, Shanghai, China). HEK293T (CRL-11268) cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, Virginia, USA). HCCLM9 cells were provided by Prof. Yufeng Yuan from Zhongnan Hospital of Wuhan University, Wuhan, China. All cell lines were characterized by short tandem repeat (STR) analysis by third-party biology services (Feiouer Biology Co., Ltd, Chengdu, China). Nunc carrier plate system (Thermo), consisted of 6-well carrier plate and culture insert with 0.4-μm pore size, was used for co-culture with macrophages. Human HCC cells (1 × 105) were added to the lower chamber, while macrophages (1 × 106) were added to the upper chamber. After 72 h of co-culture, the macrophages were withdrawn and human HCC cells were washed by PBS and grown in fresh medium. As for the co-culture with CD8+ T cells, human HCC cells (1 × 105) were labeled with CFSE (TargetMol) and seeded into 24-well plates. CD8+ T cells (2 × 106) were seeded into the same chambers with direct physical contact. The apoptotic rates of human HCC cells were analyzed by CytoFLEX Research Flow Cytometer (Beckman Coulter).
Isolation, activation and amplification of primary mouse CD8+ T cells from spleens
OT-I TCR transgenic mice were purchased from Cyagen Biosciences Inc. CD8+ T cells were isolated from spleens from OT-I TCR transgenic mice using the EasySep mouse CD8+ T cell isolation kit (STEMCELL) according to the manufacturer’s protocol. CD8+ T cells were activated and amplified in RPMI-1640 medium (Gibco) containing 20 ng/ml IL-2 (Biolegend), anti-CD3/CD28 beads (Thermo Fisher Scientific), 10% fetal bovine serum, 50 U/ml penicillin (Gibco) and 50 μg/ml streptomycin (Gibco).
OT-I T cell killing assay
Hepa1-6 OVA cells (1 × 106) with circPETH knockdown or overexpression were plated with activated OT-I T cells (2 × 106) in a 6-well plate in 2 ml of complete RPMI-1640 media. Flow cytometric analyses were applied to assess viable cell numbers for the tumor cells using AccuCheck counting beads (Invitrogen) at 48 h post-coculture. DAPI (Sigma) was used to exclude dead cells. “OT-I T cell killing (%)” was calculated as: 1 – (cell numbers in the OT-I group/cell numbers in the untreated group without coculture with OT-I T cells).
In vitro invasion and migration assays
Transwell chambers ((8.0 μm pore size, Corning Costar, Kennebunk, USA)) were put into 24-well plates and the membranes were precoated with Matrigel. DMEM medium without 10% FBS was added to the upper chamber, while DMEM medium containing 10% FBS was added to the lower chamber. Then, 20,000 cells were plated in the upper chamber, followed by the incubation for 24 h, before Transwell chambers were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. The number of cells was counted in five randomly selected regions using Image J (1.8.0).
Seahorse assays
Seahorse XFe24 analyzer (Agilent Technologies) was used to measure the ECARs. 10,000 HCC cells per well were seeded into 24-well Seahorse XF24 culture plates in the absence of CO2 overnight, followed by incubation with base medium with 2 mM L-glutamine at 37 °C for 1 h, after which final pH was adjusted to 7.4 and culture plates were kept at 37 °C. ECARs measurements were performed according to the manufacturer’s instructions. After every 3 measurements in 8-min intervals, glucose, oligomycin or 2-DG was added into the wells at the indicated time points to a final concentration of 10 mM, 10 μM or 50 mM, respectively. Glycolysis rate was calculated as the difference between the ECARs before the injection of oligomycin and the basal ECARs values. Glycolytic capacity was calculated as the difference between the ECARs following the injection of oligomycin and the basal ECARs values.
Glucose consumption and lactate production assays
EonTM Microplate Reader (BioTek) was used to measure the glucose consumption and lactate production. 20,000 HCC cells per well were seeded into 96-well plates at 37 °C for 6 h. Glucose consumption and lactate production were detected using glucose uptake assay kit (Abcam) and L-Lactate assay kit (Abcam) according to the manufacturer’s instructions. The glucose and lactate levels in the medium of negative controls were used for comparison in order to calculate the relative glucose uptake and relative lactate secretion.
EVs experiments
EVs from culture medium were isolated by ultracentrifugation. Briefly, culture medium was collected and centrifuged at 2000 × g for 30 min at 4 °C, after which the supernatants were centrifuged at 10,000 × g for 45 min at 4 °C, followed by a filtration using 0.22-μm filter and a centrifugation step of 100,000 × g for 70 min at 4 °C, before the pellets were centrifuged at 100,000 × g for 70 min at 4 °C. EVs were finally resuspended and preserved in PBS. EVs were analyzed by transmission electron microscopy, nanoparticle tracking system and nanoFCM. Primary antibodies against CD9 (1:50; 555371, BD) and CD81 (1:50; 551108, BD) were used for the identification of EVs. For the visualization of EVs incorporation, EVs were labeled with Mem Dye-Green using ExoSparkler Exosome Membrane Labeling Kit-Green (EX01, Dojindo) according to the manufacturer’s instructions. Labeled EVs were added to HCC cells and visualized by A1R + MP confocal laser microscope system (Nikon). For EVs elimination, culture medium of TAMs was collected and filtered through a 0.22-μm filter, followed by an incubation with antibody against CD81 (1:100; MAB4615, R&D), labeled with biotin using Zenon Biotin-XX Mouse IgG2b Labeling kit (Z25252, Molecular Probes), overnight at 4 °C, before the supplementation with Dynabeads MyOne Streptavidin T1 (65601, Invitrogen) and incubation for 30 min. The EV-antibody complex was depleted using a magnet. For the visualization of EVs-encapsulated RNA internalization, linear circPETH, synthesized by in vitro transcription, was labeled with 488-X-UTP and subsequently cyclized using circular RNA synthesis kit (Beyotime) according to the manufacturer’s instructions, followed by transfection into TAMs using NEPA21 Super Electroporator (NEPA GENE) according to the manufacturer’s instructions. EVs from transfected TAMs were collected after 48 h and added to HCC cells. Images were obtained by A1R + MP confocal laser microscope system (Nikon).
Microarray analysis
Total RNA of macrophages and HCC cells was extracted and purified using a RNeasy Maxi kit (Qiagen) according to the manufacturer’s instructions. EVs-derived RNA extraction was performed using exoRNeasy Maxi kit (Qiagen) according to the manufacturer’s instructions. The purity, concentration and integrity were analyzed using NanoDrop ND-2000 (Thermo) and Agilent Bioanalyzer 2100 system (Agilent). circRNA expression profiles were examined using Human (4 × 180 K) RNA microarray (LC-Bio). Raw images were obtained using Agilent Scanner G5761A (Agilent) and raw data were extracted using Feature Extraction (Agilent), followed by normalization using the quantile algorithm, Genespring (Agilent). Limma package in R (v.3.2.2) was employed to compare the differential expressions of circRNAs and mRNAs between matched profiles, followed by false discovery rate (FDR) correction using Benjamini and Hochberg’s approach. Genes with Fold change (FC) > 1.5 and P < 0.05 were considered to be differentially expressed.
qRT-PCR assays
qRT-PCR was performed with a ChamQTM SYBR@ qPCR Master Mix (Vazyme). All reactions were run in triplicate on the CFX96TM Touch Real-Time PCR System (Bio-Rad). The primer sequences are listed in Supplementary Data 3. β-actin or U6 were applied as endogenous control. The relative expression was calculated using the comparative Ct (2-ΔΔCT) method.
RNase R digestion
Total RNA was extracted using Trizol reagent (Invitrogen). Approximately 3 μg RNA was subjected to 10 U RNase R digestion at 37 °C for 30 min, followed by incubation at 75 °C for 10 min to deactivate the RNase R. Circ7, an already known circRNA, was used for control. The relative RNA expression levels of circPETH and mLDLR were analyzed by qRT-PCR and normalized to the values of mock group (without RNase R treatment).
Actinomycin D assays
HCC cells were treated with actinomycin D (1 μg/ml) and harvested after 0, 4, 8, 12, and 24 h, respectively. The relative RNA expression levels of circPETH and mLDLR were analyzed by qRT-PCR and normalized to the values at 0 h.
Subcellular RNA fractionation
Cytoplasmic and nuclear RNA fractions were collected according to the instructions of the PARISTM Kit (Invitrogen), followed by qRT-PCR analysis. U3 small nuclear RNA was used as the nuclear endogenous control. β-actin mRNA was used as the cytoplasmic endogenous control.
Subcellular protein fractionation
Cytoplasmic and nuclear protein fractions were extracted according to the protocol of the NE-PERTM Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, CA, USA), after which the samples were separated on 10% SDS-PAGE gels and analyzed by Western blot. Histone H3 was used as the nuclear endogenous control. β-Tubulin was used as the cytoplasmic endogenous control.
Fluorescence in situ hybridization (FISH)
FISH was performed to determine the subcellular location of circPETH. Fluorescein-labeled probes against circPETH were designed and synthesized by RiboBio. FISH was conducted using the Fluorescent in Situ Hybridization Kit (RiboBio), according to the manufacturer’s protocol. Fluorescence signals were scanned using the A1R + MP confocal laser microscope system (Nikon).
Generation of circPETH-147aa antibody
6-week-old female BALB/c mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. and were subcutaneously immunized 3 times at 10-d intervals with 100 μg purified recombinant polypeptides in PBS and Freund’s complete adjuvant, which was made up of the 28-aa tail of circPETH-147aa. The serum titers were detected by enzyme-linked immunosorbant assay (ELISA). Next, mouse spleen cells were isolated and subsequently fused with an equivalent number of vital mouse myeloma SP2/0 cells by polyethylene glycol (PEG) at a ratio of 10:1, followed by the suspension in HAT selection medium supplemented with 20% FCS (Gibco, Grand Island, NY) and the seeding into 96-well plates, before aminopterin was added for 14 days to select the melted cells, after which Hypoxanthine and thymidine were added to the selection medium for a further period of 7 days. At last, the remaining hybridoma cells were cultivated in RPMI1640/glutamine medium supplemented with 10% FCS and then subjected to selection with the purified recombinant 28-aa polypeptides. After the acquirement of three clones of hybridoma with positive activity of 28-aa polypeptides binding, 1 × 106 hybridoma cells were intraperitoneally injected into a BALB/c mouse for monoclonal antibody enrichment of antibody production. Ascites was collected after 7 days.
RNA interference and CRISPR‒Cas9 techniques
The transfection of siRNAs were performed using GenMuteTM Reagent (SignaGen Laboratories) following the manufacturer’s protocol. The oligonucleotide sequences of siRNAs and shRNAs were listed in Supplementary Data 4. Two sgRNAs targeting the flanking sequences of I15RC were designed with 5’-GCCACTGAAGGGACCCGTCTC-3’ and 5’-GCAGTTTGGGCGCCCTCTGG-3’ oligonucleotides using the online tool at https://www.zlab.bio/resources and inserted into pCRISPR-LvSG03 vector with mcherry expression, which was transfected into HCCLM9 cells with overexpression of Cas9 protein and GFP. Therefore, the interest cells successfully transfected with two sgRNAs and Cas9 showed co-expressions of mcherry and GFP, followed by the screening of interest single cell using FACS with excitation wavelengths of 587 nm and 488 nm. KO stable HCCLM9 cells were collected and characterized by Sanger sequencing.
Northern blot
The NB assay was performed using a Northern blot Kit (Thermo) according to the manufacturer’s instructions. In brief, a total of 15 μg RNA was treated with RNase Rand denatured in formaldehyde, followed by electrophoresing in a 1% agarose-formaldehyde gel. Then the RNA was transferred onto a Hybond-N+nylon membrane (GE Healthcare) and subsequently hybridized with digoxin-labeled circPETH DNA probe. Thereafter, a DIG oligonucleotide 3’-end labeling kit (Roche) was applied for development of the bound RNA. The intensity of signals was scanned by X-ray films with chemiluminescence substrate CSPD (Roche). 18S rRNA was used as control. The DNA probe was listed in Supplementary Table 4.
Western blot
Total protein was extracted from the cells using RIPA lysis buffer supplemented with protease inhibitor cocktail (Beyotime). Protein concentration was detected using the BCA protein assay kit (Beyotime), separated by SDS-PAGE, and transferred to PVDF membranes (Millipore). After incubation with primary antibodies, the membranes were incubated with HRP-conjugated secondary antibodies. The information of primary antibodies was listed in Supplementary Data 5. Immunoreactivity was visualized using enhanced chemiluminescent (ECL) chromogenic substrate (Beyotime). The intensity of signals was scanned by ChemiDoc MP Imager System (Bio-Rad) and analyzed by Image Lab 5.2 software (Bio-Rad).
Luciferase reporter assays
The sequences of Renilla luciferase (Rluc) and Firefly luciferase were respectively inserted into the ORF of pCDNA3.1 vector. The potential IRES sequence of circPETH and mutants were amplified and inserted in the middle of Rluc and Fluc. Dual luciferase reporter assay kit was used to detect luciferase activity 48 h after transfection using the Synergy Mx Multi-Mode Microplate Reader (BioTek). Firefly luciferase activity was normalized to Renilla luciferase activity and was presented as the relative luciferase activity.
Immunofluorescence (IF)
For IF analysis, cultured cells were fixed with 4% formaldehyde for 20 min and then blocked with 5% normal goat serum with or without 0.1% Triton X-100 in PBS for 60 min at room temperature. Immunostaining was performed using primary antibodies against RAB7 (1:100; No. 9367, CST), HA (1:400; No. 66006-2-Ig, Proteintech) and YAP1 (1:200; No. 66900-1-Ig, Proteintech), and Alexa Fluor secondary antibodies (Invitrogen). Nuclei were counterstained with Hoechst. Fluorescence signals were scanned using the A1R + MP confocal laser microscope system (Nikon).
Immunohistochemistry (IHC)
FFPE tissues underwent dewaxing, antigen repair and blocking before incubating with primary antibodies overnight at 4 °C. After being washed with PBST three times, slides were incubated with HRP-conjugated secondary antibodies for 1 h at RT. The slides were incubated with DAB chromogenic Kit (ZSGB-Bio). The slides were stained with hematoxylin, dehydrated and sealed for observation and scanning. Antibodies: CD8 (1:100; No. ab316778, Abcam), CD68 (1:1000; No. ab303565, Abcam). For immune cells (CD8, CD68), the counts of all positive cells by immunostaining were changed into density as cells/mm2 by three pathologists independently.
Co-immunoprecipitation (co-IP), RNA immunoprecipitation (RIP) and methylated RNA immunoprecipitation (MeRIP)
The co-IP, RIP and MeRIP assay were performed using Pierce Crosslink Magnetic IP/co-IP Kit (Thermo), Magna RIPTM RNA-binding Protein Immunoprecipitation Kit (Millipore) and Magna MeRIPTM m6A Kit (Millipore) according to the manufacturer’s instructions, respectively. As for the F-actin immunoprecipitation assays, cells were collected and lysed with IP lysis buffer (100 mM NaCl, 0.5 mM EDTA, 20 mM Tris-HCl, pH7.4, 0.5% NP-40), and immunoprecipitated with 1 μg anti-LATS antibody (Cell Signaling Technology, cat# 3477) at 4 °C overnight. Then lysates were incubated with protein A Sepharose (GE Healthcare, cat# 17-0963-03) or protein G agarose (Millipore, cat# 16-266) for 2 h followed by 3 washes using binding buffer (150 mM NaCl, 0.5 mM EDTA, 20 mM Tris-HCl, pH7.4, 0.5% NP-40). Finally, the beads were eluted with SDS-PAGE loading buffer and the resultant elutes were subjected to immunoblotting analysis.
Methionine quantification
The methionine concentration in culture medium was measured using Methionine Assay Kit (Abcam, ab234041). The medium was collected and diluted to 100 μl with PBS, and then 2 µl of Sample Clean-up mix was added and incubated at 37 °C for 30 min. Samples were filtered through a 10 kDa spin column (10,000 × g, 4 °C, 10 min), and the ultrafiltrate was retained. For each sample, two parallel wells were prepared, one for the determination of methionine and one for a sample background control. Totally, 5 µl ultrafiltrate and 20 µl detection buffer were added to each well. Then 25 µl reaction mix and 25 µl background control mix were added to their parallel sample wells, respectively. Meanwhile, 100 µM methionine standard was used to prepare the standard curve. Fluorescence was read in endpoint mode (Ex/Em = 535/587 nm) after 30 min of incubation of the plate at 37 °C. The absolute methionine concentration was calculated for each sample using the standard curve.
Leucine quantification
The quantification of leucine in culture medium was conducted using liquid chromatography-tandem mass spectrometry (LC-MS/MS) (API3200, AB Sciex). A mass spectrometry conjugated with an electrospray ionization (ESI) interface was used to produce positive ions. Optimized mass parameters of leucine were determined by infusing each solution of the compounds at 1 ng/mL into the mass spectrometer in the positive ion mode at 10 μL/min. The chromatographic separation of leucine in culture medium was conducted with Gemini-NX 3 μ C18 110 A New column 50 × 2.0 mm (656065-14) using an isocratic elution condition of 0.1% formic acid in water and methanol (50:50, v/v) at 0.3 mL/min and 40 °C using the Agilent 1200 series HPLC system (Agilent Technologies).
Mass spectrometry (MS)
For MS analysis, the gels were stained with Coomassie brilliant blue (Beyotime). The number of biological replicates was 3. Thus, the gel bands of interest in total 6 samples (3 for circPETH-overexpressing group and 3 for vector group) were manually cut into small particles, followed by acetonitrile dehydration and trypsin digestion. After that, the digested peptides were desalted and collected by a Q-Exactive Plus mass spectrometer (Thermo) in series EASY-nLC 1200 liquid phase LMS system. The peptide samples were dissolved by the loading buffer, and then combined with the analytical column (50 μm × 15 cm, C18, 2 μm, 100 A) after inhalation by the automatic injector for separation. Two mobile phases (mobile phase A: 0.1% formic acid and mobile phase B: 0.1% formic acid, 80% ACN) were used to establish a 100 min analysis gradient. The liquid phase flow rate was set to 300 nL/min. Mass spectrometry data acquisition in DDA mode, each scan cycle contains a MS full scan (R = 70 K, AGC = 3e6, max IT = 20 ms, scan range = 350–1800 m/z), And then 15 MS/MS scans (R = 17.5 K, AGC = 2e5, max IT = 50 ms). The HCD collision energy is set to 28. The screening window of the four-level bar is set to 1.6 Da. The dynamic exclusion time of repeated ion collection was set to 35 s. The mass spectrum data were retrieved by MaxQuant (V1.6.6) software, and the database retrieval algorithm was Andromeda. The database used for the search was the Human proteome reference database in Uniprot. The main search parameters are as follows: variable modification selected Oxidation (M), Acetyl (Protein N-term); Fixed modification Carbamidomethyl (C); Trypsin/P; The primary mass spectrum matching tolerance was set to 20 ppm in the primary search and 4.5 ppm in the main search. The secondary mass spectrum matching tolerance is set to 20 ppm. The search results were screened based on the 1% FDR level of proteins and peptides, and the entries of anti-library proteins, polluting proteins, and proteins with only one modified peptide segment were deleted. The remaining identification information was used for subsequent analysis.
In vivo metastasis assays
The animal studies were authorized by the Animal Ethic Review Committees of the West China Hospital. 5-week-old male athymic BALB/c nude mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. and were housed in a specific-pathogen-free condition with a dark/light cycle of 12-h of light/12-h of darkness, ambient temperature of 20–26 °C and humidity of 40–70%. The lung metastases were visualized using the IVIS@ Lumina II system (Caliper) 15 min after intraperitoneal injection of 3.0 mg of D-Luciferin (ABP) at 6 weeks after the establishment of tail intravenous injection models. The liver metastases were visualized using the IVIS@ Lumina II system (Caliper) 15 min after intraperitoneal injection of 3.0 mg of D-Luciferin (ABP) at 6 weeks after the construction of orthotopic implanted models. The number of metastatic foci was counted in HE staining in tissue sections of lungs and livers under the AX10 imager A2/AX10 cam HRC microscope (Zeiss). 6-week-old male C57BL/6J mice were also purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. 5 × 105 Hepa1-6 mouse HCC cells were injected into the tail veins to construct lung metastasis models and 1 × 106 Hepa1-6 mouse HCC cells were injected into the livers to construct orthotopic implanted intrahepatic metastasis models. Treatment was performed 2 weeks after mice model construction. Anti-mouse PD1 (HRP00262-022, provided by Hengrui Medicine Com., Jiangsu, China) was intraperitoneally injected (10 mg/kg, three times every week, for 2 weeks). Norathyriol (C13H8O6, CAS No.: 3542-72-1, 260.20 Da, purity: 99.83%) was purchased form MedChemExpress (MCE, Catalog No.: HY-N1029). Norathyriol was intraperitoneally injected (5 mg/kg and 10 mg/kg for two different concentration groups, respectively, once daily for two weeks). The metastases were visualized using the IVIS@ Lumina II system (Caliper) 15 min after intraperitoneal injection of 3.0 mg of D-Luciferin (ABP) in 200 µl of sterile PBS without magnesium or calcium. The number of metastatic foci was counted in HE staining in tissue sections of lungs and livers under the AX10 imager A2/AX10 cam HRC microscope (Zeiss). All animal experiments were strictly implemented in compliance with the NIH Guide for the Care and Use of Laboratory Animals. The maximal tumor size permitted by the Animal Ethics Committee was 15 mm in diameter. Mice were sacrificed when they developed tumors larger than 15 mm in diameter. In this study, we confirm that at no point during the study did any animal exceed the maximal permitted tumor size.
Molecular docking
The 3D structures of the ALODA protein (PDB ID: 1ALD), PKM2 protein (PDB ID: 1T5A) and HuR protein (PDB ID: 4ED5) were downloaded from RCSB Protein Data Bank. For the prediction of interactions, 3D structures were submitted to ZDOCK calculation via ZDOCK protocol in Discovery Studio v3.1 software. The resultant protein poses were ranked by ZDOCK scoring and the image of top-ranked protein pose was futher processed and presented.
Virtual screening and nano differential scanning fluorimetry (nanoDSF)
Molecular operating environment (MOE) through site Finder module was used to find and score the pockets on circPETH-147aa surface. The Schördinger Maestro package was employed to perform dock analysis. The 2D structures of small molecular chemicals in a TargetMol library (bioactive compound library, 40,000 small molecules, Target Molecule Corp) were prepared using LigPrep (Epik). The circPETH-147aa structure was constructed following the Protein Prepare Wizard workflow in the Maestro package. The property of docking pocket was calculated by the Receptor Grid Generation module. Three-step screening was performed by taking a step-wise strategy, including HTVS, general (SP) precision and extra (XP) precision utilizing Glide, to gradually increase the filtration precision. Small-molecule flexible docking was employed in each step, followed by energy optimization, after which the top 20% molecules were selected according to the docking score and subjected to the next screening. Next, these selected molecules were subjected to Protein-ligand Interaction Fingerprint (PLIF) analysis in MOE to investigate the binding residues of amino acids. The chemical candidates with docking scores from -8 to -5 were finally determined and were subsequently subjected to nanoDSF using a protein stability analytic system Prometheus NT.48 in the final concentration of 100 μg/ml. The molecule with Tm difference value above 5 was regarded to possibly interact with interest protein.
Surface plasmon resonance (SPR)
The binding behaviors were measured using SPR on a BIAcore X100 system. Interest recombinant protein was immobilized on a CM5 chip through its amine groups using EDC/NHS according to the standard amine coupling protocol, after which different dilutions of analyte were orderly injected over the flow cell. Running buffer was passed over the sensor surface for a 1.5 min period for dissociation. The response was determined as a function of time (sensorgram) at 25 °C. Data were analyzed using Biacore Evaluation software, version 2.0.2 (GE Healthcare).
In vitro kinase assays
The kinase reactions were performed as described previously65. In brief, the recombinant PKM2 (200 ng) were incubated with ALDOA (100 ng) with kinase buffer (50 mM Tris-HCl pH = 7.5, 100 mM KCl, 50 mM MgCl2, 1 mM Na3VO4, 1 mM PMSF, 1 mM DT, 5% glycerol, 0.5 mM PEP, 0.05 mM FBP) in 100 μl at RT for 1 h. The reactions were terminated by addition of SDS-PAGE loading buffer and heated to 100 °C. The reaction mixtures were then subjected to 10% SDS-PAGE analyses.
Statistics and reproducibility
Statistical analysis was performed using SPSS v.25 or Prism GraphPad 8. For most of the in vitro and animal experiments, independent sample t-tests were used to calculate the P values. Survival curves were plotted using the Kaplan–Meier method and compared using log-rank tests. All statistical analyses were performed using two-tailed P values. Statistical details and methods used are indicated in the figure legends, text or methods. No statistical method was used to pre-determine the sample size. No data were excluded from the analyses. Unless stated otherwise, the experiments were not randomized and the investigators were not blinded to treatment allocation during experiments or to the outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank Yan Wang from the Core Facilities of West China Hospital, Sichuan University for technical assistance in primary human cells isolation experiments, and Jingyao Zhang from the Core Facilities of West China Hospital, Sichuan University for helping us perform SPR experiments, and Xiaoting Chen and Yang Yang from the Animal Experimental Center of West China Hospital for technical assistance in animal experiments. We are most grateful for West China Biobanks, Department of Clinical Research Management, West China Hospital, Sichuan University for their support of human tissue samples. The study was supported by grants from the National Natural Science Foundation of China (82103533, 82372791, 82173124 and 82303220), the National Key Research and Development Program of China (2022YFC2407604), the Science and Technology Support Program of Sichuan Province (2023ZYD0171 and 2024NSFSC0745), and the Postdoctoral Science the fellowship of China National Postdoctoral Program for Innative Talents (BX20200227).
Author contributions
Conceptualization, T.L., F.G., Y.C., and H.W.; Methodology, T.L., Y.C., Y.L., J.Z., H.L., and S.X.; Investigation, T.L., Y.C., Y.L., J.Z., and H.W.; Resources, T.L., Y.C., H.H., K.X., C.L., and H.W.; Writing-Original Draft, T.L.; Writing-Review & Editing, F.G., Y.C., C.L., and H.W.; Supervision, T.L. and H.W.; Founding Acquisition, T.L. and H.W.
Peer review
Peer review information
Nature Communications thanks Chunxiao Liu, Wei Zhao, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The microarray datasets, RNA-seq dataset of HCC cells and RNA-seq dataset of CD8+ T cells have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE241868, GSE242016 and GSE242130. Metabolomics data have been deposited in the MetaboLights under accession code MTBLS11254. This study analyzes existing, publicly available anti-HuR RIP microarray data, which is listed in the Supplementary Table I of a previous study51. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Tian Lan, Fengwei Gao, Yunshi Cai.
Contributor Information
Tian Lan, Email: blue_sky_land@163.com.
Hong Wu, Email: wuhong@scu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55577-0.
References
- 1.Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: Cancer J. Clin.71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Vogel, A., Meyer, T., Sapisochin, G., Salem, R. & Saborowski, A. Hepatocellular carcinoma. Lancet400, 1345–1362 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Yang, C. et al. Evolving therapeutic landscape of advanced hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol.20, 203–222 (2023). [DOI] [PubMed] [Google Scholar]
- 4.Havel, J. J., Chowell, D. & Chan, T. A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer19, 133–150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dual Immunotherapy Makes Strides against HCC. Cancer Discov.12, Of1 (2022). [DOI] [PubMed]
- 6.Donne, R. & Lujambio, A. The liver cancer immune microenvironment: Therapeutic implications for hepatocellular carcinoma. Hepatology77, 1773–1796 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai, W. et al. OGDHL silencing promotes hepatocellular carcinoma by reprogramming glutamine metabolism. J. Hepatol.72, 909–923 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Paul, S., Ghosh, S. & Kumar, S. Tumor glycolysis, an essential sweet tooth of tumor cells. Semin. Cancer Biol.86, 1216–1230 (2022). [DOI] [PubMed] [Google Scholar]
- 9.Li, J. et al. CircRPN2 inhibits aerobic glycolysis and metastasis in hepatocellular carcinoma. Cancer Res.82, 1055–1069 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Guo, D. et al. Aerobic glycolysis promotes tumor immune evasion by hexokinase2-mediated phosphorylation of IκBα. Cell Metab.34, 1312–1324.e1316 (2022). [DOI] [PubMed] [Google Scholar]
- 11.He, H., Xiao, L., Wang, J., Guo, D. & Lu, Z. Aerobic glycolysis promotes tumor immune evasion and tumor cell stemness through the noncanonical function of hexokinase 2. Cancer Commun.43, 387–390 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng, C. et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell169, 1342–1356.e1316 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Zhang, Y. et al. Macrophage-associated PGK1 phosphorylation promotes aerobic glycolysis and tumorigenesis. Mol. Cell71, 201–215.e207 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Vitale, I., Manic, G., Coussens, L. M., Kroemer, G. & Galluzzi, L. Macrophages and metabolism in the tumor microenvironment. Cell Metab.30, 36–50 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Gordon, S. R. et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature545, 495–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, F. et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat. Cell Biol.21, 498–510 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Qian, B. Z. & Pollard, J. W. Macrophage diversity enhances tumor progression and metastasis. Cell141, 39–51 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, C. et al. Macrophage-derived CCL5 facilitates immune escape of colorectal cancer cells via the p65/STAT3-CSN5-PD-L1 pathway. Cell Death Differ.27, 1765–1781 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, H. et al. Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR. Theranostics11, 1396–1411 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, H. et al. IL-6-induced cGGNBP2 encodes a protein to promote cell growth and metastasis in intrahepatic cholangiocarcinoma. Hepatology75, 1402–1419 (2021). [DOI] [PMC free article] [PubMed]
- 21.Gao, X. et al. Circular RNA-encoded oncogenic E-cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR-STAT3 signalling. Nat. Cell Biol.23, 278–291 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Huang, X. Y. et al. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol. Cancer19, 92 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang, L. et al. Exosomal hsa_circ_0004658 derived from RBPJ overexpressed-macrophages inhibits hepatocellular carcinoma progression via miR-499b-5p/JAM3. Cell Death Dis.13, 32 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei, Y. T. et al. Thymosin α-1 reverses M2 polarization of tumor-associated macrophages during efferocytosis. Cancer Res.82, 1991–2002 (2022). [DOI] [PubMed] [Google Scholar]
- 25.Yang, E. et al. Exosome-mediated metabolic reprogramming: the emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduct. Target. Ther.5, 242 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He, G. et al. Exosomes in the hypoxic TME: from release, uptake and biofunctions to clinical applications. Mol. Cancer21, 19 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, C. et al. Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transduct. Target. Ther.6, 383 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu, J. et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J. Hepatol.68, 1214–1227 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Zhao, W. et al. Splicing factor derived circular RNA circUHRF1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. Cell Death Differ.27, 919–933 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ostrowski, M. et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol.12, 19–30 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Su, S. et al. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell25, 605–620 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Wu, X. et al. A novel protein encoded by circular SMO RNA is essential for Hedgehog signaling activation and glioblastoma tumorigenicity. Genome Biol.22, 33 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen, S. Y., Qadir, J. & Yang, B. B. Circular RNA translation: novel protein isoforms and clinical significance. Trends Mol. Med.28, 405–420 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Chen, X. et al. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci. Rep.6, 34985 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, Y. et al. Expanding uncapped translation and emerging function of circular RNA in carcinomas and noncarcinomas. Mol. Cancer21, 13 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang, Y. et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res.27, 626–641 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao, W. et al. Epigenetic regulation of m(6)A modifications in human cancer. Mol. Ther. Nucleic Acids19, 405–412 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lan, T. et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer18, 186 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liberman, N. et al. DAP5 associates with eIF2β and eIF4AI to promote internal ribosome entry site driven translation. Nucleic Acids Res.43, 3764–3775 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sonenberg, N. & Hinnebusch, A. G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell136, 731–745 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Enzo, E. et al. Aerobic glycolysis tunes YAP/TAZ transcriptional activity. EMBO J.34, 1349–1370 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiraishi, T. et al. Glycolysis is the primary bioenergetic pathway for cell motility and cytoskeletal remodeling in human prostate and breast cancer cells. Oncotarget6, 130–143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun, S. & Irvine, K. D. Cellular organization and cytoskeletal regulation of the hippo signaling network. Trends Cell Biol.26, 694–704 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang, Z. et al. PDLIM1 inhibits tumor metastasis through activating hippo signaling in hepatocellular carcinoma. Hepatology71, 1643–1659 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Lu, Z. & Hunter, T. Metabolic kinases moonlighting as protein kinases. Trends Biochem. Sci.43, 301–310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang, J. et al. Mitochondrial PKM2 regulates oxidative stress-induced apoptosis by stabilizing Bcl2. Cell Res.27, 329–351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao, Q. et al. Integrated proteogenomic characterization of HBV-Related hepatocellular carcinoma. Cell179, 561–577.e522 (2019). [DOI] [PubMed] [Google Scholar]
- 48.St-Jean, M., Izard, T. & Sygusch, J. A hydrophobic pocket in the active site of glycolytic aldolase mediates interactions with Wiskott-Aldrich syndrome protein. J. Biol. Chem.282, 14309–14315 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Gao, X., Wang, H., Yang, J. J., Liu, X. & Liu, Z. R. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol. Cell45, 598–609 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, W. et al. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature480, 118–122 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abdelmohsen, K. et al. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J.28, 1271–1282 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu, T. X. et al. Chk2-dependent HuR phosphorylation regulates occludin mRNA translation and epithelial barrier function. Nucleic Acids Res.39, 8472–8487 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bodoy, S. et al. Identification of LAT4, a novel amino acid transporter with system L activity. J. Biol. Chem.280, 12002–12011 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Bian, Y. et al. Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. Nature585, 277–282 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ananieva, E. A., Powell, J. D. & Hutson, S. M. Leucine metabolism in T cell activation: mTOR signaling and beyond. Adv. Nutr.7, 798s–805s (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen, X. et al. A membrane-associated MHC-I inhibitory axis for cancer immune evasion. Cell186, 3903–3920.e3921 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li, J. et al. Norathyriol suppresses skin cancers induced by solar ultraviolet radiation by targeting ERK kinases. Cancer Res.72, 260–270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dai, S. et al. Intratumoral CXCL13(+)CD8(+)T cell infiltration determines poor clinical outcomes and immunoevasive contexture in patients with clear cell renal cell carcinoma. J. Immunother. Cancer9, e001823 (2021). [DOI] [PMC free article] [PubMed]
- 59.Omata, Y. et al. RNA editing enzyme ADAR1 controls miR-381-3p-mediated expression of multidrug resistance protein MRP4 via regulation of circRNA in human renal cells. J. Biol. Chem.298, 102184 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hernandez, L. et al. The EGF/CSF-1 paracrine invasion loop can be triggered by heregulin beta1 and CXCL12. Cancer Res.69, 3221–3227 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kristensen, L. S. et al. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet.20, 675–691 (2019). [DOI] [PubMed] [Google Scholar]
- 62.Wei, Y. et al. Pyridoxine induces glutathione synthesis via PKM2-mediated Nrf2 transactivation and confers neuroprotection. Nat. Commun.11, 941 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elia, I. & Haigis, M. C. Metabolites and the tumour microenvironment: from cellular mechanisms to systemic metabolism. Nat. Metab.3, 21–32 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu, Y. et al. The XOR-IDH3α axis controls macrophage polarization in hepatocellular carcinoma. J. Hepatol.79, 1172–1184 (2023). [DOI] [PubMed]
- 65.Yang, W. et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell150, 685–696 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The microarray datasets, RNA-seq dataset of HCC cells and RNA-seq dataset of CD8+ T cells have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE241868, GSE242016 and GSE242130. Metabolomics data have been deposited in the MetaboLights under accession code MTBLS11254. This study analyzes existing, publicly available anti-HuR RIP microarray data, which is listed in the Supplementary Table I of a previous study51. Source data are provided with this paper.