Abstract
To explore CRT gene family members and their responses to low-temperature stress, bioinformatics methods were used to identify the CRT gene family in pepper. In this study, a total of 4 CRT gene family members were identified by screening. The genes were found to be located on different chromosomes, and phylogenetic tree and collinearity analyses were performed. In addition, the cis-acting elements identified in the 2.0 kb upstream promoter regions of the 4 CaCRTs described in this study can be divided into three categories. The expression patterns of CRTs in different tissues and organs under low-temperature stress were analysed. Virus-induced gene silencing (VIGS) technology was used to verify the function of CaCRT1, and the results revealed that CaCRT1 participated in the pepper response to low-temperature stress and the accumulation of excessive reactive oxygen species (ROS) and malondialdehyde (MDA). In summary, this study systematically verified the regulatory role of CaCRT gene family members in pepper under low-temperature stress and provided a foundational understanding for further research on the biological functions of pepper CRT genes.
Keywords: Pepper, CaCRT, Low-temperature stress, Gene family
Subject terms: Biochemistry, Molecular biology
Introduction
Calreticulin (CRT) is a Ca2+-binding soluble protein widely present on the endoplasmic reticulum membranes in eukaryotes1 and is mainly localized to the endoplasmic reticulum (ER)2 and plasmodesmata (PD)3. CRT is the major Ca2+-binding protein in the endoplasmic reticulum and has a molecular chaperone functions4. CRT, which is ubiquitous in animals and plants, represents a class of calcium-binding proteins with extremely high Ca2+ affinity and highly conserved sequences5. It was first discovered on the endoplasmic reticulum membranes of skeletal muscle in mammals6 and was subsequently cloned from Rattus norvegicus7, Caenorhabditis elegans8, and Arabidopsis thaliana9. CRT is involved in the regulation of Ca2+ homeostasis10, signal transduction11, endoplasmic reticulum quality control (ERQC), growth and development, and the immune response, among other biological processes12.
Research on CRT in plants is relatively lacking13. CRT is known to respond extensively to biotic and abiotic stresses, such as drought14, low temperature15, salt stress16, fungi17, bacteria18, and viral infections19. Studies of transgenic plants revealed that the overexpression of CRT genes in plants can increase resistance traits. The CRT gene in Triticum aestivum L. responds to drought stress20, and the Arabidopsis thaliana CRT gene responds to water stress21. CRT gene was expressed in Oryza sativa L., and overexpression improved the endurance of rice plants under cold conditions22. A CRT gene was expressed in Nicotiana tabacum L., and a medium level of overexpression could increase the resistance against the movement of tobacco mosaic virus between cells, greatly improving the ability of Nicotiana tabacum L. to resist tobacco mosaic virus infection23. Other studies have shown that CRT genes may play important roles in the normal growth of plants. For example, mutation of the CRT gene in Brassica pekinensis affects the ability of plant cells to bind to calcium ions, thereby affecting the ability of plants to cope with calcium deficiency stress and ultimately causing dry burning heart disease in plants24. In addition, the CRT gene regulates plant tolerance to drought, high temperature, and cold stress25. Furthermore, the Castor CRT gene plays a role in the development of the microtubule systems of vegetative and flower organs, including in terms of seed formation. Petunia CRT is involved in pollen‒stigma interactions, and CRT2 regulates the immune response of Arabidopsis thaliana to pathogenic bacteria through a dual mechanism of action. Thus, CRT genes may play important roles in plant physiological metabolism.
Pepper is the main cultivated vegetable in China and is also an important model plant that is sensitive to the stress response and susceptible to low-temperature, drought, and salinity. Owing to the lack of water resources in the semiarid region of Northwest China, coupled with the unpredictable fertilization system and extreme weather, the production of pepper is often subjected to drought, salinity and low-temperature stress. In recent years, increasing attention has been given to the study of gene functions related to stress resistance in peppers. Current studies have investigated CRT genes in higher plants such as Arabidopsis thaliana and Triticum aestivum L., but information on the CRT gene family in pepper and their expression patterns at low temperatures are still scarce. Plants accumulate many reactive oxygen species in low-temperature environments, which leads to changes in the content of malondialdehyde. Moreover, damage to the cell membrane can be judged by the permeability of electrolytes. In addition, plants have low-temperature resistance mechanisms, and cold-related genes have been described in relevant reports. Therefore, in this work, a genome-wide identification of CRT family members in pepper was carried out, and their gene expression patterns under low-temperature stress were analysed to provide a reference for the subsequent study of the functions of CRT genes in pepper and their molecular mechanisms in response to stress.
Materials and methods
Plant material and treatments
This experiment was based on Shenghan 740, which is resistant to low temperatures and was donated by the Vegetable Research Center of the Beijing Academy of Agriculture and Forestry Sciences. Pepper seedlings were planted in a mixed substrate (peat: vermiculite: perlite = 3:1:1; V/V/V). The seedlings were transferred to a climate chamber with a temperature of 25/20°C (day/night), a photoperiod of 12/12 h (day/night), a relative humidity of 75 ± 5%, and a light intensity of 200 µmol m− 2 s− 1. During the cultivation process, pepper seedlings were watered with a 1/2 concentration of Hoagland nutrient solution when the cotyledons were fully expanded, and when two leaves and one heart formed, the plants were watered with a normal concentration of Hoagland nutrient solution every 2 days. When the plants’ 6–7 true leaves were fully developed, the low temperature treatment was started (treatment group: 4 °C; control group: 25 °C, relative humidity of 75 ± 5%, and light intensity of 200 µmol m− 2 s− 1).
The experimental treatment was carried out at 8:00 in the morning, and samples were taken at 0, 3, 6, 9, 12, 24, and 48 h after the treatment began. Three to six fully expanded true leaves above the cotyledons of the pepper were obtained after the experimental treatment, 0.1 g was accurately weighed, frozen with liquid nitrogen and placed in a − 80 °C freezer for later use, and each treatment was repeated three times.
qRT‒PCR analysis of CRT gene family members in pepper
The primers used were designed by Sangon Bioengineering (Shanghai) according to the CDSs of the CRT genes of pepper, and the actin internal reference gene was used as the control. Total RNA was extracted using TsingZol Total RNA Extraction Reagent (Qingke Biotechnology) and reverse transcribed into cDNA using a Goldenstar™ RT6 cDNA Synthesis Kit, and 2×TSINGKE Master Mix was used to verify the reliability of the RNA-Seq results with a qPCR Mix (SYBR Green I) kit for fluorescence quantification. The PCR program was as follows: predenaturation at 95 °C for 3 min; 40 cycles of extension at 95 °C for 10 s, 57 °C for 10 s, and 72 °C for 15 s; and 72 °C to acquire fluorescence signals. The relative expression levels of the genes were calculated using the 2−ΔΔCT method.
Gene identification and prediction of the physicochemical properties of CRT genes in pepper
The Capsicum annuum genome file (Zunla-1 version, https://solgenomics.net/) was downloaded from the Solanaceae Genomics Network website, and the protein sequence file of Capsicum annuum was extracted using TBtools. The CRT gene family characteristic domain (PF00262) of pepper was searched in the Pfam database (http://pfam.xfam.org/), and the amino acid sequence of CRT in pepper was screened using a Simple HMM search. The protein sequences of the genes identified using the two screening methods were extracted, and the conserved domains of pepper CRT were predicted using the online bioinformatics tools from the National Center for Biotechnology Information CDD Search Database (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) and the SMART website (https://smart.embl.de/). The sequences that were incomplete and did not contain conserved domains of CRT were removed.
The physicochemical characteristics of the CRT gene protein products in pepper were calculated using the ProtParam tool (https://web.ExPASy.org/protparam/), including amino acid sequence length, theoretical isoelectric point (pI), molecular weight (Mw), instability index, aliphatic index and average hydrophilicity index (GRAVY) index, and the proteins subcellular localization were predicted with the SubCELlular Localization Predictor Tool (http://cello.life.nctu.edu.tw/).
The three-dimensional structures of the CRT genes in pepper were predicted using the SWISS-MODEL website (https://swissmodel.expasy.org/).
Chromosomal mapping, phylogenetic analysis, gene structure and conserved motif analysis of the CRT genes in pepper
The chromosomal position information of the CRT genes in pepper was extracted from the genome annotation file of Capsicum annuum and visualized using the Gene Location Visualize from the GTF/GFF module in TBtools software. The CRT protein sequences of these species were retrieved using HMMER search, and MEGA11 was used to construct a maximum likelihood (ML) tree, with bootstrapping set to 1000. Finally, iTOL (https://itol.embl.de/) was used for phylogenetic tree visualization and beautification.
MEME software (http://www.OMIcsclass.com/article/67) was used to analyse the motif structures of CRT family members in pepper. The maximum number of motifs was set to 15, the minimum width of each motif was set to 6, and the maximum width of each motif was set to 50. The structure of the CRT genes in Capsicum, including the CDSs and UTRs, was extracted from the genome annotation file of Capsicum annuum. TBtools v2.057 was used for visual analysis.
Collinearity analysis of the CRT genes in pepper
The BLASTP program was used to identify homologous CRT genes in pepper, with an e-value threshold set to < e− 10. The collinearity between CRT genes in pepper was analysed using the default parameters of MCScanX. TBtools was used to visualize the resulting homologous CRT gene pairs. Collinearity analysis was performed at the same time, and the Arabidopsis genome sequence and annotation file were downloaded from the Phytozome v13 website (https://phytozome-next.jgi.doe.gov/), whereas the pepper genome sequence and annotation file were downloaded from the NCBI website. The MCScanX program was used to analyse collinearity between pepper and Arabidopsis thaliana.
Analysis of cis-acting elements and protein‒protein interactions in the promoters of the CRT genes in pepper
The 2000 bp region upstream of the start codon (ATG) of each pepper CRT gene was considered a promoter sequence (with the exception of the promoter sequence for CaCRT3, which was less than 2000 bp upstream). The number of cis-acting elements associated with the stress response, plant growth and development, and plant hormone reactivity was determined using Python, and a heatmap was generated. Online STRING software (http://string-db.org/) was used to analyse the CRT family interaction relationships in pepper, and the confidence of the interaction relationships was greater than 0.700. The results were then visualized using Cytoscape.
Construction and characterization of the silencing vector TRV2-CRT1
Using the reverse transcription product as a template, the CRT gene sequence information of pepper relatives was retrieved from NCBI, and a vector based on tobacco brittle virus (TRV) was constructed. The CRT target fragment and TRV2 plasmid were double-digested with AscI and KpnI restriction enzymes, transferred to DH5α competent cells after overnight ligation with T4 ligase, coated on LB plates (containing Gen 50 mg/L, Kan 50 mg/L, or Rif 50 mg/L), inverted and incubated overnight at 37 °C. Bacterial clones were picked, inoculated in cultures, and incubated with shaking, and the plasmids were then extracted following culture expansion. Each plasmid was verified by sequencing and transformed into Agrobacterium strain GV3101, and the transformants were subsequently coated on LB plates (containing Gen 50 mg/L, Kan 50 mg/L, and Rif 50 mg/L), which were inverted and incubated at 28 °C for 2 days to observe the growth of colonies. The CRT, TRV1 and TRV2 bacterial solutions were inoculated in 50 mL centrifuge tubes containing 20 mL of LB liquid medium, incubated at 28 °C with shaking, and then centrifuged to collect the bacteria. The bacteria were subsequently resuspended in infection buffer (containing 10 mmol L− 1 MES, and 10 mmol L− 1 MgCl2), which was repeated once, and the OD600 value was adjusted to 0.004. When the cotyledons were fully expanded, the TRV2:CRT and TRV2 bacteria were combined with TRV1 and TRV1 solution, respectively. The groups were mixed evenly, with TRV2:00 empty carrier as a blank control and TRV2:CaPDS plants to show photobleaching as a positive control, wait for pepper plants to grow to the five-leaf one-heart stage, sampling, quickly frozen in liquid nitrogen, and the samples were stored at − 80 °C for later use.
Measurement of antioxidant enzyme activity and ROS
The content of MDA in the leaves was determined by the thiobarbituric acid method. In accordance with the methods of Amin et al., the enzyme mixture was extracted, 0.4 g of the treated pepper leaf sample was weighed, 4 ml of 50 mmol/L phosphate buffer (pH = 7.8, containing 0.1 mmol of EDTA) was added, the mixture was fully ground in an ice bath, and the resulting crude enzyme mixture was used for the determination of superoxide dismutase (SOD), peroxidase (POD) and catalase (CAT) activity levels. In accordance with the methods of Stewart et al., SOD activity was determined by the nitrogen blue tetrazolium method, with 50% inhibition of the photochemical reduction of NBT considered one unit of enzyme activity. The POD activity was determined by the guaiacol method according to the methods of Polle et al. CAT activity was determined according to the method of Aebi and was modified to 1 enzyme activity unit for every 0.1 decrease at A240. DAB staining was used to detect the production of H2O2 in the leaves. Nitrotetrazolium blue chloride (NBT) staining was used to detect the production of O2− in leaves. Magenta sulfurous acid staining was used to detect the production of MDA in leaves. All of the above indices were measured in triplicate.
Statistical analysis
Origin 2022 statistical software was used for data analysis. One-way analysis of variance (ANOVA) was used to analyse the differences between treatments. According to Tukey’s test, significant differences were determined at p < 0.05. All the data were from at least three independent biological replicates.
Results
Identification, prediction of physicochemical properties and chromosome localization analysis of CRT family members in pepper
In this study, a total of 4 CRT gene family members were identified via screening, and they were named CaCRT1, CaCRT2, CaCRT3, and CaCRT4 according to their chromosomal order. Protein characterization revealed that the average amino acid number of the CaCRTs was 548 aa (range = 286–978 aa). Other characteristics of the CaCRTs are as follows: average molecular weight = 62.1 kDa (range = 32.9–109.7 kDa); mean fat coefficient = 73.2 (range = 58.8–80.2); mean isoelectric point = 6.0 (range = 4.6–8.9); mean instability index = 39.26 (range = 35.46–42.28); and mean GRAVY value for each protein = − 0.62. These results indicate that CaCRTs are mostly acidic, hydrophilic and unstable proteins. Subcellular localization prediction analysis revealed that the CaCRTs were all localized in the endoplasmic reticulum (Table 1).
Table 1.
Identification of physicochemical properties of CaCRT gene family in pepper.
| No. | Gene name | Gene ID | Number of amino acid (aa) | Molecular weight (KDa) | pI | Instability index | Aliphatic index | Grand average of hydropathicity | Subcellular localization prediction |
|---|---|---|---|---|---|---|---|---|---|
| 1 | CaCRT1 | Capana03g000601 | 978 | 109.65652 | 5.27 | 42.28 | 80.18 | −0.461 | Endoplasmic reticulum |
| 2 | CaCRT2 | Capana05g002540 | 286 | 32.89467 | 8.85 | 37.92 | 78.71 | −0.377 | Endoplasmic reticulum |
| 3 | CaCRT3 | Capana06g001088 | 537 | 61.2444 | 5.1 | 41.39 | 75.21 | −0.714 | Endoplasmic reticulum |
| 4 | CaCRT4 | Capana08g001603 | 390 | 44.59345 | 4.64 | 35.46 | 58.79 | −0.92 | Endoplasmic reticulum |
Number of Amino Acid (aa): Determine the various types of amino acids in the protein and their relative quantities. Molecular weight: also known as relative molecular weight, refers to the mass of protein molecules. Isoelectric point (PI): refers to the pH of a protein that has a neutral charge under specific conditions. Instability index (Instability lndex): The instability index of the protein, the higher the index, the more unstable the protein Decide. Aliphatic Ilndex: Describes the abundance and relative content of non-polar amino acids. Hydrophilic and hydrophobic (GRAVY): represents the relative ratio of hydrophilic and hydrophobic amino acids in the protein molecule, the lower the value, the lower the protein.
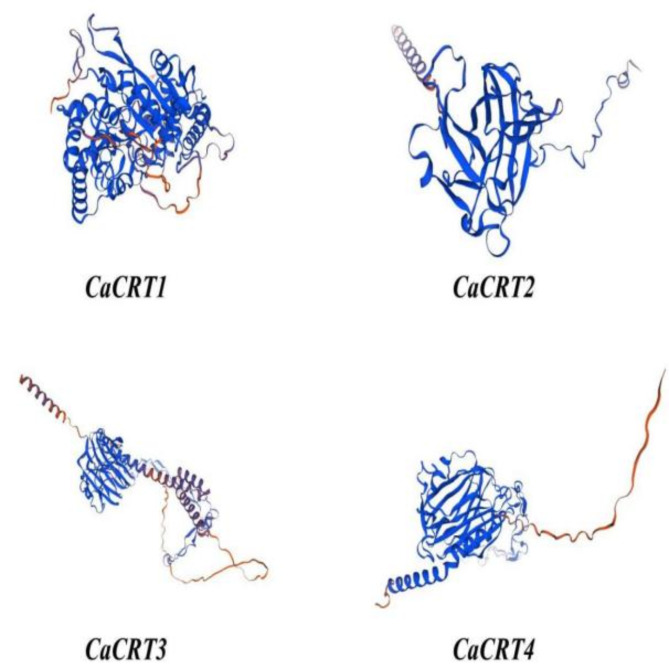
The results of 3D homology modelling revealed that the CaCRTs were structurally similar and were composed mainly of α-helices (21.79–37.01%), extension chains (13.41–18.8%), random coils (41.92–58.46%), and β-steering (3.35–6.34%) (Fig. 1).
Fig. 1.
The results of 3 D homology modeling of CaCRTs show that the four CRT genes are structurally similar, mainly composed of α helix, extension chain, random coil and β turn.
Chromosomal localization analysis of the CRT genes in pepper
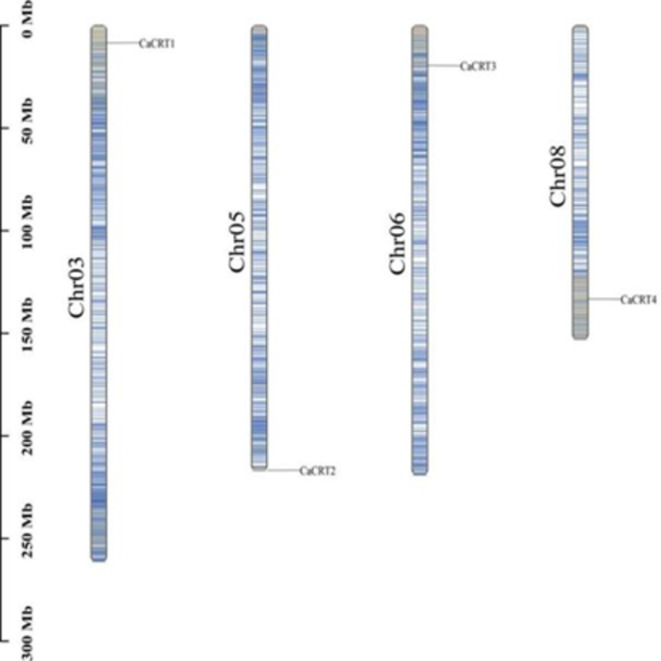
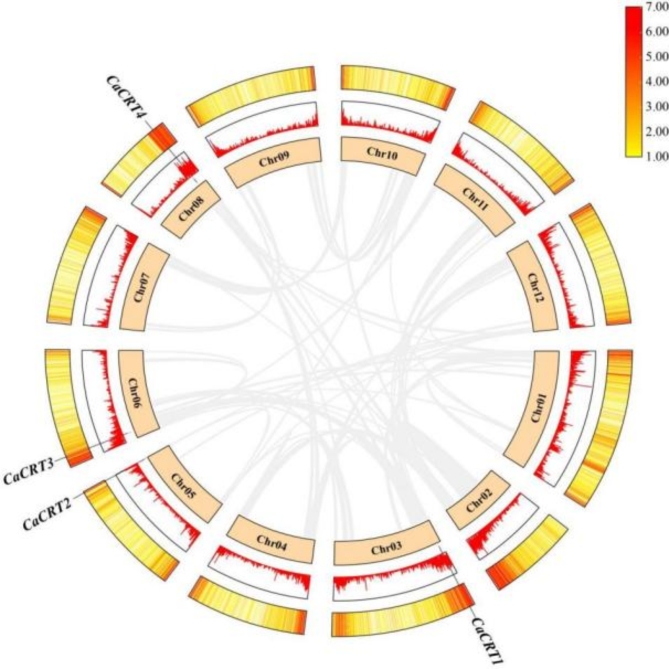
According to the gene structure annotation information of CaCRTs, TBtools was used to generate chromosome distribution maps. The 4 CaCRTs were evenly distributed across the 4 chromosomes: CaCRT1 was on Chr03, CaCRT2 was on Chr05, CaCRT3 was on Chr06, and CaCRT4 was on Chr08 (Fig. 2).
Fig. 2.
Chromosome distribution of CaCRTs. Chr stands for chromosome. The scale indicates the chromosome length (Mb).
Analysis of CRT family protein interactions in pepper
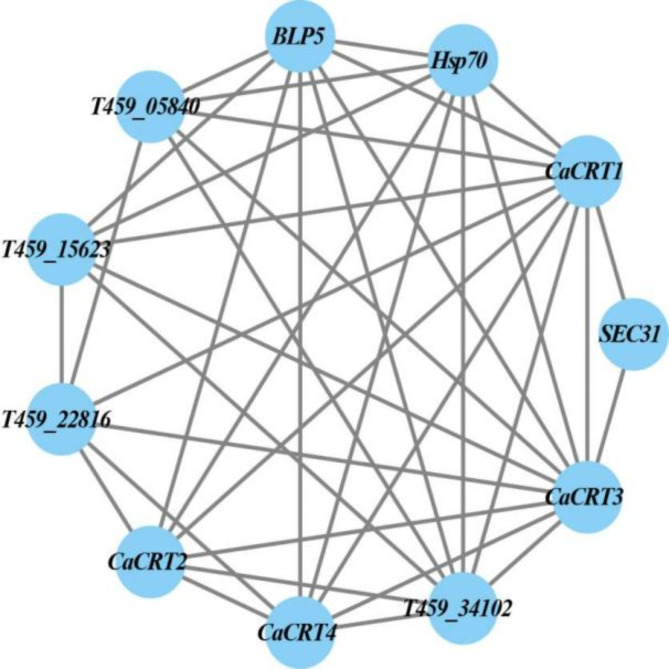
The prediction of potential proteins that interact with CRT family proteins in pepper revealed that all CRT family proteins could interact with each other. Among them, CaCRT1 and CaCRT4 could interact with the heat stress proteins Hsp70 and BLP5 and other proteins without clear annotation functions. In addition, only CaCRT1 and CaCRT3 interacted with the protein transporter Sect. 31 (Fig. 3).
Fig. 3.
Protein-protein interaction protein gene results showed that the CRT gene family proteins in pepper could interact with each other.
Phylogenetic analysis of the CRT gene family in pepper
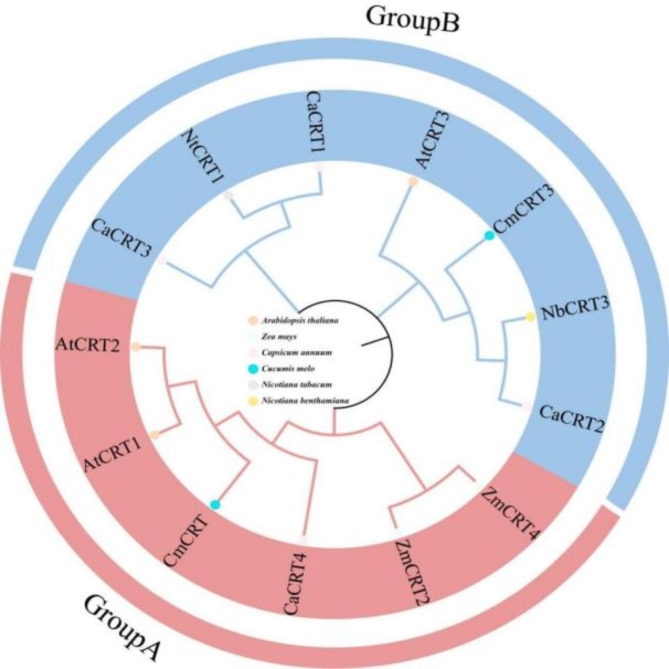
Through phylogenetic tree analysis, the CRT family can be divided into two subgroups: A and B. CaCRT1, CaCRT3, and CaCRT2 belonged to one subgroup, whereas CaCRT4 was part of a separate subgroup. Overall, CaCRT2 was close to AtCRT3 and AtCRT2 in Arabidopsis thaliana, and CaCRT4 was close to AtCRT1 and AtCRT2 in Arabidopsis thaliana (Fig. 4).
Fig. 4.
Phylogenetic tree of Capsicum. annuum, Arabidopsis thaliana, Zea mays, Cucumis melo, Nicotiana tabacum, Nicotiana benthamiana,. sativa CaCRTs. Different colored circles represent different species.
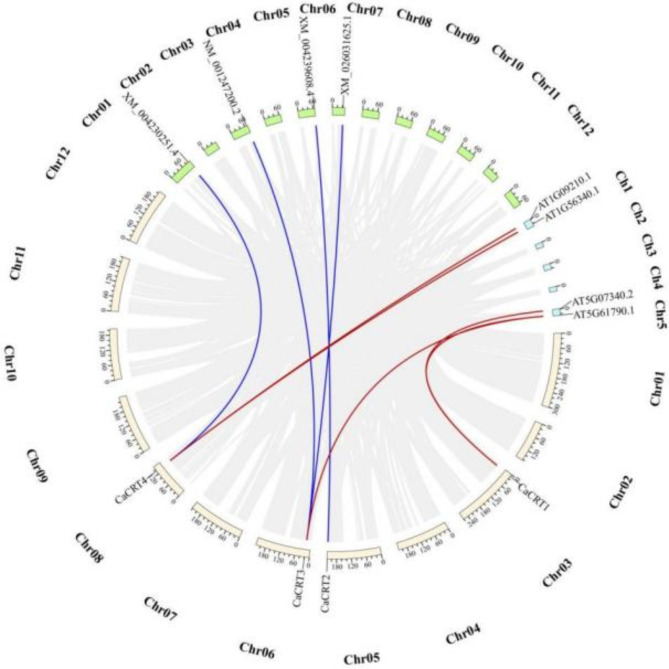
Intra- and interspecies collinearity analysis of the CRT genes in pepper
A collinearity analysis of the CRT genes in pepper revealed that there were no collinear gene pairs within the CRT gene family (Fig. 5). Four pairs of CRT genomic collinearity genes were identified in pepper and tomato and 5 pairs of genome collinearity genes were identified in pepper and Arabidopsis thaliana. However, while CaCRT1–CaCRT4 were included in the collinear gene pairs of pepper and tomato, CaCRT2 did not have a corresponding collinear gene pair in the pepper and Arabidopsis thaliana gene pairs, and CaCRT1 and CaCRT4 each had two collinear pairs (Fig. 6). These results suggest that peppers are evolutionarily close to tomatoes.
Fig. 5.
Intra-species collinearity analysis of the CRT gene family in pepper.
Fig. 6.
Collinear analysis of peppers, tomatoes, and Arabidopsis. The gray rectangular boxes represent chromosomes, and the colored lines represent collinearity pairs of the CaCRTs gene.
Analysis of the conserved domains, gene structures and conserved motifs of the CRT genes in pepper
The conserved domains of CaCRT indicate that all the CaCRTs contain calreticulin domains, which suggests that they are all CRT genes, and that CaCRT1 contains amidase superfamily domains in addition to its calreticulin domain, which indicates that CaCRT1 may have other functions. Based on the GFF3 gene structure annotation information, an exon/intron structure diagram of CaCRTs was generated using TBtools (Fig. 7a). As shown in the figure, all CaCRTs contain introns, with CaCRT1 having the greatest number of introns and CaCRT2 having the least number of introns (Fig. 7b). Conserved motif analysis revealed 15 motifs from CaCRTs. Among them, CaCRT2 has the fewest motifs, with only 8; CaCRT1 contains the largest number of motifs, with 15. Phylogenetic analysis revealed that more recent CaCRTs contained more similar motifs. In addition, domain analysis revealed that Motifs 1, 3, 5, 7, 9, and 13 constituted the key CRT functional domains, whereas the other motifs did not match the known key functional domains.
Fig. 7.
Gene structure and conserved motif analysis of CaCRTs in pepper (a) The basic composition of the 15 conserved motifs (b) Analysis of conserved motifs, conserved domains, and gene structures.
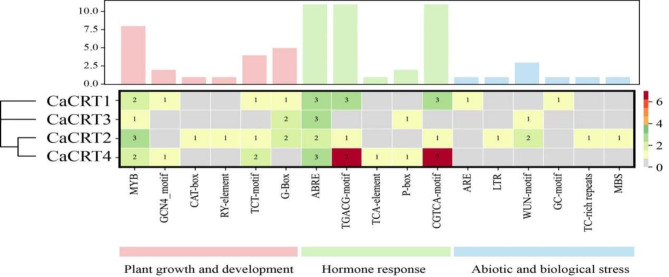
Analysis of cis-acting elements in the promoter of the CRT genes in pepper
Cis-regulatory elements are a class of DNA sequences located in transcription initiation regions and play key roles in regulating gene transcription. They function by binding to transcription factors and regulating the transcription process of genes. In this study, the cis-acting elements identified in the 2.0 kb upstream promoter regions of the four CaCRTs could be divided into three categories (Fig. 8): 6 elements related to plant growth and development, 5 elements related to hormonal responses, and 6 elements associated with abiotic and biotic stresses. We detected plant growth- and development-related response elements, including endosperm expression elements (GCN4_motif elements), meristem development elements (CAT-box elements), seed development elements (RY-element), and light response elements (TCT-motifs, G-box elements), in the CaCRT promoters. Gibberellin (P-box element), abscisic acid (ABRE), jasmonic acid (CGTCA-motif, TGACG-motif element) and salicylic acid (SARE, TCA-element) were the most common jasmonic acid reactive elements. In addition, biotic/abiotic stress response elements such as anaerobic inducible elements (AREs, GC motif elements), low-temperature response elements (LTR elements) and drought response elements (MBS elements) were also found, indicating that the expression of CaCRTs was induced by environmental stress.
Fig. 8.
Statistics of cis-acting elements and their numbers in the 2 kb region upstream of the CaCRTs gene family in pepper. Different elements in the promoter sequence of the CaCRTs gene are indicated by different colors.
Tissue-specific expression of the CRT genes in pepper and analysis of their expression under low-temperature stress
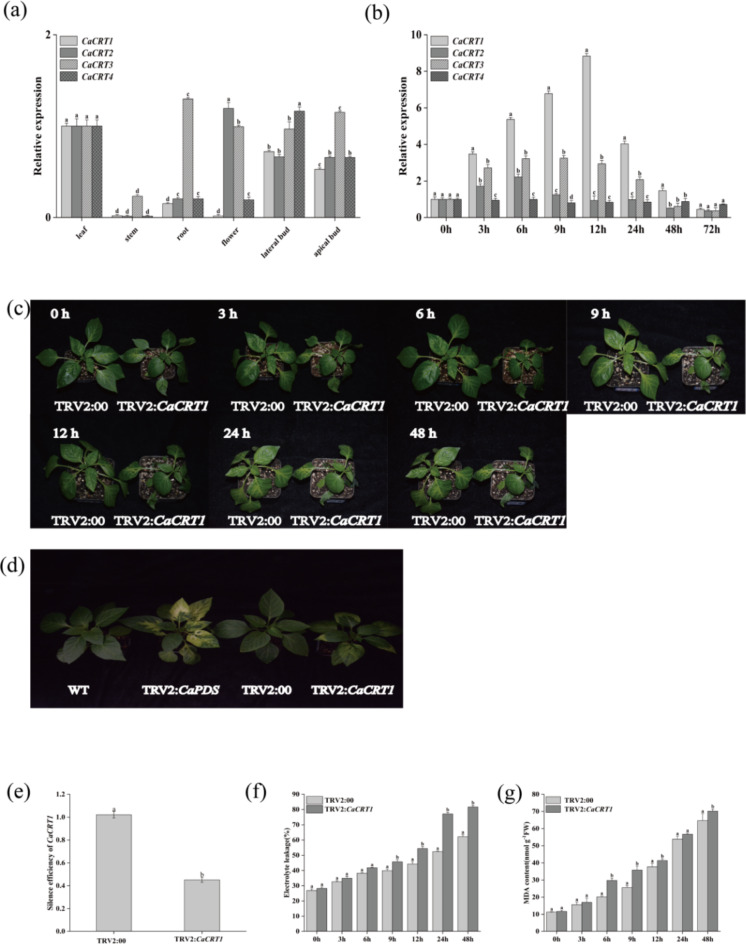
In this study, fluorescence quantification was used to detect the expression patterns of the CRT genes in different tissues of pepper. According to the results (Fig. 9a), the expression level of CRT1 in pepper was the highest in leaves and the lowest in flowers, and it was concluded that CaCRT gene family members presented specific differences in expression in the plant. Moreover, to explore the responses of the CRT gene family members of Capsicum under various stresses, five-leaf single-phase samples of Capsicum were treated, and the expression profiles of the CRT genes in Capsicum were detected by RT‒qPCR (Fig. 9b). After the onset of cold stress, the response patterns of CaCRT gene family members to cold stress differed. These results indicate that members of the CaCRT gene family respond specifically to low-temperature stress. These results indicate that CaCRT1 can be induced by low temperature and may be involved in the regulation of pepper plants under low-temperature stress.
Fig. 9.
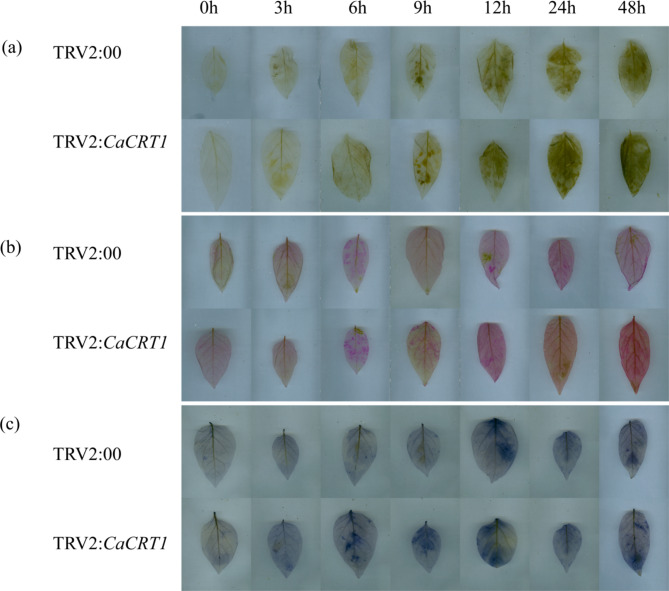
Tissue-specific expression, expression patterns under low temperature stress, and data changes after silencing of CaCRTs gene family members in pepper. (a) Changes in the expression content of CaCRTs in different parts of pepper plants. (b) Changes in gene expression of CaCRTs under low temperature stress at 4 °C. (c) Phenotypic changes of control and silent pepper plants under 4 °C low temperature stress. (d) Phenotypic control of control plants, silent plants, positive seedlings, and wild plants. (e) Gene silencing efficiency of CaCRT1. (f) Changes in the conductivity of CaCRT1 under 4 °C low temperature stress between control and silent plants. (g) Changes in malondialdehyde content of pepper control plants and CaCRT1-silenced plants under low temperature stress at 4 °C. The data represent three biological replicates, with significant differences determined according to tukey.
VIGS-mediated silencing of CaCRT1 affects pepper tolerance under low-temperature stress
In this study, VIGS was used to verify whether CaCRT1 is involved in the regulation of pepper plants under low-temperature stress and infected pepper seedlings growing to the two-cotyledon stage. After being cultured for a period of time, the phenotype of the positive control seedlings was observed (Fig. 9c). After the infected pepper seedlings grew to five leaves and one heart, they were subjected to low-temperature treatment. After 12 h of low-temperature treatment, the CaCRT1-silenced plants wilted significantly more than TRV2:00 plants did, and all the CaCRT1-silenced plants presented severe wilting by 48 h, with no significant difference (Fig. 9d). The silencing efficiency of CaCRT1 in pepper leaves was also verified (Fig. 9e). Moreover, CaCRT1-silenced plants presented greater REL (Fig. 9f) and MDA (Fig. 9g) levels than TRV2:00 control plants did. Three different experimental methods, DAB, NBT and magenta sulfurous acid, were used to stain the leaves of peppers picked from the same parts. Experimental comparisons revealed that TRV2:00 silenced plants accumulated more H2O2, O2− and MDA than the other groups (Fig. 10).
Fig. 10.
Detection of H2O2 production in leaves by diaminobenzidine DAB staining. Nitrotetrazolium blue chloride (NBT) staining was used to detect the production of O2− in leaves. Magenta sulfurous acid staining was used to detect the production of MDA in leaves. All of the above indexes were measured in 3 replicates.
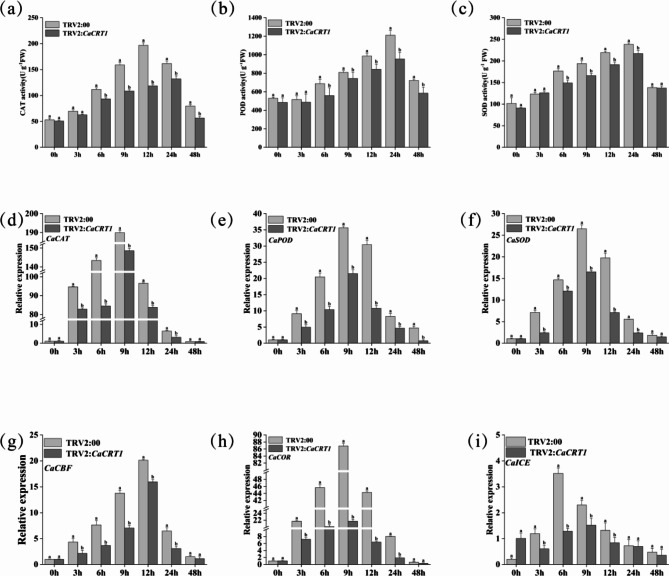
The enzyme activity levels of members of the antioxidant system, including CAT, POD, and SOD, were also determined by fluorescence quantification (Fig. 11a,b,c). The results revealed that the antioxidant capacity of the TRV2:00 control plants was significantly lower than that of the CaCRT1-silenced plants. There were also significant differences in the relative expression levels of antioxidant enzyme-encoding genes (CaCAT, CaPOD and CaSOD) (Fig. 11d,e,f). Finally, the expression levels of cold regulatory genes (CaCBF, CaCOR and CaICE) were significantly lower in the TRV2:00 plants than in the CaCRT1-silenced plants (Fig. 11g,h,i). In summary, silencing the CaCRT1 gene in pepper under low-temperature stress led to severe damage to the cell membrane, reduced antioxidant capacity, excessive accumulation of ROS, and the inability to induce cold-response regulatory genes. Our results suggest that CaCRT1 may play a role in the cold stress response in peppers.
Fig. 11.
Changes in antioxidant enzyme activity, antioxidant enzyme genes and cold regulatory genes under low temperature stress. (a–c) Changes in the activities of three enzymes, CAT, POD and SOD in silencing plants and control plants under low temperature stress. (d–f) Silent plants under low temperature stress showed changes in CaCRT1 and CaCAT, CaPOD and CaSOD enzyme activity regulatory genes in control plants. Changes of three cold-corresponding regulatory genes of CaCBF, CaCOR and CaICE in the silencing plant and control plants under low temperature stress (g–i). The data represent three biological replicates, with significant differences determined according to tukey.
Discussion
Pepper is a vegetable crop widely grown worldwide and has important economic value. The annual planting area of pepper accounts for approximately 8% of the vegetable planting area in China26. The growth, production and planting of pepper are very easily affected by various biological and abiotic stresses, such as cold damage, drought, light, disease and insect pests27. CRT gene family members play crucial roles in plant growth and development and in response to biotic and abiotic stresses28. Therefore, the expression characteristics of the CRT gene family of Capsicum under low-temperature stress should be comprehensively identified and analysed. The present study focuses on the mechanism of action of shenghan 740 under low-temperature stress, which may provide fundamental understanding of chili peppers under stress, especially shenghan 740 under low-temperature stress. These findings will provide a basic understanding of the response functions of these genes under low-temperature stress29.
In this study, we downloaded the Capsicum annuum genome file (Zunla-1 version, https://solgenomics.net/) from the Solanaceae Genomics Network website and extracted the protein sequence file of Capsicum annuum using TBtools. A total of 4 CRT genes were identified by bioinformatics by searching the CRT gene family characteristic domains (PF00262) of pepper from the Pfam database (http://pfam.xfam.org/), and the amino acid sequences of pepper CRT genes were screened in a Simple HMM search. The results of 3D homology modelling of CaCRTs revealed that the four CRT genes were structurally similar (Fig. 1). Through genome alignment, TBtools was used to generate chromosome distribution maps (Fig. 2). The four CaCRTs were evenly distributed on four chromosomes: CaCRT1 was located on Chr03, CaCRT2 was located on Chr05, CaCRT3 was located on Chr06, and CaCRT4 was located on Chr08. Analysis of the conserved domains of CaCRT revealed that all CaCRTs contained calreticulin domains, which proved that they were all CRT genes, and CaCRT1 contained amidase superfamily domains in addition to calreticulin domains, suggesting that CaCRT1 may have other functions (Fig. 3). Moreover, a phylogenetic tree was constructed (Fig. 4), and intraspecies (Fig. 5) and interspecies (Fig. 6) collinearities were analysed. Based on the GFF3 gene structure annotation information, TBtools was used to generate exon/intron structure diagrams of the CaCRTs (Fig. 7). All the CaCRTs contained introns, among which CaCRT1 had the greatest number of introns, and CaCRT2 had the lowest number of introns. Conserved motif analysis revealed 15 motifs from CaCRTs (Fig. 8). Among them, CaCRT2 had the fewest motifs, with only 8, and CaCRT1 contained the largest number of motifs, with 15. Phylogenetic analysis revealed that the more recent CaCRTs contained more similar motifs. In addition, domain analysis revealed that Motifs 1, 3, 5, 7, 9 and 13 constituted the key CRT functional domains (calreticulin), whereas the other motifs did not match the known key functional domains. The results of the cis-acting element analysis that these CRT genes may respond to multiple stresses.
As a conserved calcium-binding protein, CRT plays an extremely important role in cell signal transduction, protein sorting and biological metabolism by regulating intracellular calcium homeostasis and protein folding30. Research on CRT genes in plants has been based on cloning, protein expression or functional analysis of CRT genes in model plants such as Arabidopsis thaliana, rice and wheat31. In this study, calreticulin CaCRT genes were cloned from pepper. The gene families were identified, and their expression patterns were explored32. Existing evidence shows that CRTs may play important roles in plant growth and development, hormone responses, and physiological and immune responses to stress33. The molecular expression characteristics of CRT genes are closely related to the physiological and metabolic processes regulated by plants. TaCRT1 is expressed mainly in stems, leaves, pistils, and heads of wheat, and the expression levels are similar, indicating that the function of this gene is different during plant development34. The CRT gene of castor bean is expressed in whole plants, and its expression level is relatively high in young and tender tissues35. CRT gene treatment results in tissue-specific expression and can respond to various biological and abiotic stresses, including treatments with TaCRT136 and AtCRT237, by inducing or inhibiting the expression of target genes and participating in various physiological processes in plants. In this study, the expression patterns of the CRT genes in pepper were explored, and the CRT genes were found to be expressed in roots, stems, leaves, flowers, top buds and side buds; however, the expression levels were different, which indicated that the CRT genes exhibited functional differences during plant growth and development (Fig. 9a). Through protein interaction analysis, it was found that the Capsicum CRT genes may interact with a variety of molecular chaperones, which is consistent with the findings of previous studies, indicating that the Capsicum CRT genes can affect the normal folding and expression of proteins by interacting with other molecular chaperones in vivo.
In this study, the expression profiles of CRT genes in pepper under low-temperature stress were analysed by fluorescence quantification after different stresses were applied to pepper seedlings, and the results revealed that the expression levels of CRT1 and CRT3 were greater than that of CRT4 under low-temperature stress, indicating that CRT1 and CRT3 may respond specifically to low-temperature stress (Fig. 9b). Combining the tissue-specific expression of the identified members of the gene family in shenghan 740 and the response pattern under low-temperature stress and the fact that the object of our study was pepper leaves, CaCRT1 was identified as the object of study. Moreover, VIGS technology was used to obtain CaCRT1-silenced plants. After 12 h of low-temperature stress, the silenced plants began to wilt, and after 24 h, the silent plants had severely wilted (Fig. 9c). The vaccine efficacy in the control plants (Fig. 9d) and the silencing efficiency (Fig. 9e) were analysed. As useful as the VIGS technology is, the potential specificity and efficiency should also be taken into account, and we can’t deny that there will be deeper insights for future work on knockout or overexpression. The levels of REL (Fig. 9f) and MDA (Fig. 9g) were measured, and the results revealed that the levels in the TRV2:00 control plants were lower than those in the silenced plants after low-temperature treatment, indicating that the degree of cell membrane damage was lower. The activities of SOD, POD and CAT and the corresponding genes in the silenced plants and the control plants were measured (Fig. 11d,e,f), indicating that the ROS content was too high, which represented low degradation and affected the resistance of the plants to stress; the expression levels of several cold-responsive genes were also measured (Fig. 11g,h,i). The results revealed that the contents were lower than in the control plants, indicating that the silenced genes may be involved in the resistance of the plants to low-temperature stress. Several types of basic information concerning CRT genes in peppers can be understood from the results of this study. The information obtained may be of great significance for the use of CRT genes to improve plant stress resistance.
Conclusion
In this work, the Ca2+-bound soluble protein CRT, which is widely present in the endoplasmic reticulum membranes of eukaryotes, was identified, and a total of four pepper CRT genes were identified by bioinformatics methods. Different genes were specifically expressed in plant roots, stems, leaves, flowers, terminal buds and lateral buds, and the expression patterns of the four different genes under low-temperature stress were obtained through measurements of gene expression. Moreover, VIGS technology was used to verify the role of CaCRT1 in response to low-temperature stress, which provided a basic understanding for further exploration of the biological function of the CRT genes in pepper and provided a theoretical basis for the study of the CRT genes in pepper.
Author contributions
YZW processed and analyzed the experimental data and wrote the paper. JL (Jian Li) conducted the experiment. JL (Juan Li) conducted the experiment. KYZ conducted the experiment. MKP conducted the experiment. PY designed the trial and secured funding. JL (Jie Li) designed the trial and secured funding. HBD designed the trial and secured funding. All authors have read and agree to the published version of the manuscript.
Funding
This research was funded by Yunnan Fundamental Research Projects (202401AT070059) and Research on Key Technologies for High-yield and High-quality Cultivation of Vegetables in Huangsha Substrate Facilities (2021AB018), and Yunnan Young and Middle Aged Academic and Technical Leaders Reserve Talent Program (202205AC160056).
Data availability
Data Availability Statement: All datasets supporting the conclusions of this article are included within the article. The genome data and sequences of CaCRT genes used in the current study are available in the Solanaceae Genomics Network (https://solgenomics.net/). The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jie Li, Email: gsau23@126.com.
Hongbin Du, Email: dhbzky@163.com.
References
- 1.Joshi, R., Paul, M., Kumar, A. & Pandey, D. Role of calreticulin in biotic and abiotic stress signalling and tolerance mechanisms in plants. Gene714, 144004 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Michalak, M., Robert Parker, J. M. & Opas, M. Ca2+ signaling and calcium binding chaperones of the endoplasmic reticulum. Cell Calcium32, 269–278 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Gelebart, P., Opas, M. & Michalak, M. Calreticulin, a Ca2+-binding chaperone of the endoplasmic reticulum. Int. J. Biochem. Cell. Biol.37, 260–266 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Samaj, J., Napier, R. & Volkmann, D. Maize calreticulin localizes preferentially to plasmodesmata in root apex. Plant J.19, 481–488 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Chen, M. H., Tian, G. W., Gafni, Y. & Citovsky, V. Effects of calreticulin on viral cell-to-cell movement. Plant Physiol.138, 1866–1876 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ostwald, T. J. & MacLennan, D. H. Isolation of a high affinity calcium-binding protein from sarcoplasmic reticulum. J. Biol. Chem.249 (3), 974–979 (2017). [PubMed] [Google Scholar]
- 7.Smith, M. J. & Koch, G. L. E. Multiple zones in the sequence of calreticulin (CRP55, calregulin, HACBP), a major calcium binding ER/SR protein. EMBO J.8 (12), 3581–3586. (1990). [DOI] [PMC free article] [PubMed]
- 8.Smith, M. J. Elegans gene encodes a protein homologous to mammalian calreticulin. DNA Seq.2 (4), 235–240 (1992). [DOI] [PubMed] [Google Scholar]
- 9.Nelson, D. E., Glaunsinger, B. & Bohner, H. J. Abundant accumulation of the calcium-binding molecular chaperone calreticulin in specific floral tissues of Arabidopsis thaliana. Plant. Physiol.114 (1), 29–37 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michalak, M., Groenendyk, J., Szabo, E., Gold, L. I. & Opas, M. Calreticulin, a multi-process calcium-buffering chaperone of the endoplasmic reticulum. Biochem. J.417, 651–666 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Garg, G., Yadav, S. & Ruchi, Yadav, G. Key roles of calreticulin andcalnexin proteins in plant perception under stress conditions: a review. Adv. Life Sci.5, 18–26 (2015). [Google Scholar]
- 12.Krysko, D. V., Ravichandran, K. S. & Vandenabeele, P. Macrophages regulate the clearance of living cells by calreticulin. Nat. Commun.9, 4644 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menegazzi, P., Guzzo, F., Baldan, B., Mariani, P. & Treves, S. Purification of calreticulin-like protein(s) from spinach leaves. Biochem. Biophys. Res. Commun.190, 1130–1135 (1993). [DOI] [PubMed] [Google Scholar]
- 14.Onodera, H., Ugaki, M., Tanaka, H. & Komatsu, S. Characterization of calreticulin as a phosphoprotein interacting with cold-induced protein kinase in rice. Biol. Pharm. Bull.26, 256–261 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Xiang, Y. et al. Overexpression of a Triticum aestivum calreticulin gene (TaCRT1) improves salinity tolerance in tobacco. PLoS ONE10, e0140591 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pröbsting, M. et al. Loss of function of CRT1 (calreticulin) reduces plant susceptibility to verticillium longisporum in both Arabidopsis thaliana and oilseed rape (Brassica napus). Plant Biotechnol. J. (2020). [DOI] [PMC free article] [PubMed]
- 17.Tintor, N. et al. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J.28, 3439–3449 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen, W. T. et al. Helper component-proteinase (HC-Pro) protein of papaya ringspot virus interacts with papaya calreticulin. Mol. Plant. Pathol.11, 335–346 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia, X. Y. et al. Molecular cloning and characterization of wheat calreticulin (CRT) gene involved in droughtstressed responses. J. Exp. Bot.59 (4), 739–751 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Mamillapalli, P., Marathe, R., Anandalakshmi, R. & Dinesh-Kumar, S. P. Induced ER chaperones regulate a receptor-like kinase to mediate antiviral innate immune response in plants. Cell Host Microbe6, 457–469 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen, N. H., Hong, N. N. T., Lee, H. J. & S.W., and Loss of all three calreticulins, CRT1, CRT2 and CRT3, causes enhanced sensitivity to water stress in Arabidopsis. Plant Cell Rep.32 (12), 1843–1853 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Komatsu, S., Yamada, E. & Furukawa, K. Cold stress changes the concanavalin A-positive glycosylation pattern of proteins expressed in the basal parts of rice leaf sheaths. Amino Acids36 (1), 115–123 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Chen, M. H., Tian, G. W., Yedidya, G. & Vitaly, C. Effects of calreticulin on viral cell-to-cell movement. Plant Physiol.138 (4), 1866–1876 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su, T. B. et al. (2019).
- 25.Komatsu, S., Yamada, E. & Furukawa, K. Cold stress changes the concanavalin a positive glycosylation pattern of proteins expressed in the basal parts of rice leaf sheaths. J36, 115–123 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Zou, X. X. et al. Review and prospects of pepper breeding for the past 60 years in China. Acta Hortic. Sin.49, 2099–2118 (2022). [Google Scholar]
- 27.Han, X. W. et al. Genome-wide identification and expression analysis of pepper DUF966 gene family. J. South. Agric.54, 1341–1351 (2023). [Google Scholar]
- 28.Du, Y. L. et al. Genome-wide identification of the SWEET gene family in Phaseolus vulgaris L. and their patterns of expression under abiotic stress. J. Plant. Interact.17, 390–403 (2022). [Google Scholar]
- 29.Liu, J. H., Peng, T. & Dai, W. S. Critical cis-acting elements and interacting transcription factors. 19, 375–398 (2014). [Google Scholar]
- 30.Clapham, D. E. Calcium signaling. Cell134 (6), 1047–1058 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Michalak, M. et al. Calreticulin:one protein,one gene, many functions. Biochem. J. 281–292. (1999). [PMC free article] [PubMed]
- 32.Nelson, D. E. Glaunsinger B,bohner H J,abundant accumulation of the calcium-binding molecular chaperone calreticulin in specific floral tissues of Arabidopsis thaliana. Plant Physiol.114 29–37 (1997). [DOI] [PMC free article] [PubMed]
- 33.Jia, X. Y. et al. Molecular cloning and characterization of wheat calreticulin gene involved in drought-stressed responses. J. Exp. Bot. 739–751. (2008). [DOI] [PubMed]
- 34.Xiang, Y. et al. Overexpression of a Triticum aestivum calreticulin gene improves salinity tolerance in tobacco. PloS ONE 236–247. (2015). [DOI] [PMC free article] [PubMed]
- 35.Coughlans, S. J., Hastings, C. & Winfrey, R. J. R. Cloning and characterization of the calreticulin gene from Ricinuss communis. L Plant. Mol. Biology :897–911 (1997). [DOI] [PubMed]
- 36.Lenartowska, M. et al. Immunocy to chemical evidence of calireticulin-link protein in pollen tubes and styles of putunia hybrida hort. Protoplasma 23–30. (2022). [DOI] [PubMed]
- 37.Qiu, Y. et al. A dual regulatory role of Arabidopsis calretiaculin in plant innate immunity. Plant J. 489–500. (2012). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data Availability Statement: All datasets supporting the conclusions of this article are included within the article. The genome data and sequences of CaCRT genes used in the current study are available in the Solanaceae Genomics Network (https://solgenomics.net/). The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.