Abstract
Microsatellite markers are cost-effective, rapid, efficient, and show great advantages in in large-sample kinship analysis and population structure studies. However, microsatellite loci are seriously underdeveloped in non-model organisms. The plateau zokor (Eospalax baileyi) is a key species living underground in the Tibetan Plateau, the effective management of which has long been challenging. In this study, we analyzed the distribution characteristics and functions of microsatellites in the genome of plateau zokors, and their polymorphic sites. The mononucleotide and dinucleotide types being the most abundant in the genome. The largest number of microsatellites and their abundance in the intergenic region whereas the smallest number of microsatellites and their abundance in the coding region. The coding sequences containing microsatellites were annotated to 52 major functional genes and assigned 19,358 Gene Ontology entries. The Kyoto Encyclopedia of Genes and Genomes pathway was the most enriched in the signal transduction pathway. Thirteen pairs of polymorphic loci were successfully amplified, with the number of alleles ranging from 3 to 8, observed heterozygosity ranging from 0.059 to 0.810, and expected heterozygosity ranging from 0.469 to 0.854. These microsatellite markers provide a cornerstone for studies on the identification of parentage and population genetics of plateau zokors.
Keywords: Plateau zokor, Genome-wide, Microsatellites, Polymorphism
Subject terms: Computational biology and bioinformatics, Genetics, Molecular biology
Introduction
The development of next-generation sequencing (NGS) has had a profound impact on the field of genomics and biomedical research, providing detailed genetic information and enabling the detection of small differences, such as single nucleotide polymorphisms (SNPs) and small insertions/deletions, among individuals1,2. Currently, the cost of genome sequencing is decreasing, and third-generation molecular markers, such as SNPs, have become a new research trend. However, at the population level, for kinship analysis and population structure studies of large-scale samples, microsatellites are more cost-effective and irreplaceable than whole-genome sequencing 3. Microsatellites, also known as simple sequence repeats (SSRs), usually consist of one to six nucleotide repeats, with length within 200 bp. Most of them are located in the non-coding regions of the genome, with a few CDS and exons4. Slip-strand mismatches occur during DNA replication, resulting in the addition or deletion of microsatellite motifs, forming different alleles that can be inherited by the next generation5,6. Moreover, the mutation rate of long microsatellite motifs is higher than that of short microsatellite motifs7. Compared with other molecular markers, microsatellites are co-dominant, multi-allele, homozygous, stable, inheritable, and easy to amplify with polymerase chain reaction (PCR), making them widely used as genetic markers6. Microsatellites are suitable for use in studies, such as kinship analysis, population genetic structure, and genetic diversity, but are not suitable for phylogenetic analyses of closely related species because of their high degree of polymorphism and homozygosity3.
Traditional methods, such as magnetic bead enrichment, selective hybridization, and expressed sequence tags, are usually used to identify and select microsatellites. These methods are effective but limited in number, time consuming, and expensive8. NGS has facilitated the development of microsatellites and availability of a much larger number of microsatellites9. Currently, microsatellites can be filtered from genome-wide databases using various computer software packages, such as McroSAtellite (MISA)10, Krait11, TBtools12, lobSTR13, and MegaSSR14. These research methods provide new opportunities for genome-wide studies using microsatellites and have been applied to a variety of animals, such as bats (Chiroptera)15, blackhead seabream (Acanthopagrus schlegelii)16, and spotted tail gobies (Acanthogobius ommaturus)17.
The plateau zokor (Eospalax baileyi) is a subterranean rodent endemic to the Qinghai-Tibet Plateau, distributed within the altitude range of 2800–4200 m18. It lives in a stable underground environment that is dark, with high carbon dioxide levels and low oxygen content19. At an appropriate population density, its behavior of piling up soil to form mounds has a positive impact on the grasslands, and it has a good reputation as an ecosystem engineer20,21. When the population density is high, it reduces the productivity of grasslands and causes grassland degradation, and is thus controlled as pests by local governments. However, the population control of plateau zokors always leads to an increase in their numbers instead, and the population recovers within two or three years. Currently, the management of plateau zokors is a vicious circle. Plateau zokor is a strictly subterranean species, and we can only visually observe the mounds on the ground. Its unique habits make it difficult to directly observe its behaviors, mating systems, and life history traits. Molecular markers can provide detailed data that are crucial for understanding population genetic structure, population dynamics, and disaster mechanisms.
Plateau zokors have been living in a stable underground environment since a long time, and there is little gene flow between populations in different regions, forming their own unique haplotypes and resulting in poor generalizability of microsatellites among different geographic populations. Therefore, the development of microsatellite primers suitable for different geographic populations must be addressed immediately. Su et al.22 developed 11 pairs of microsatellite primers with polymorphisms based on genomic data from Gansu zokor (E. cansus), of which 5 pairs were available in plateau zokors. Kang et al.23 developed 27 pairs of microsatellite loci for the Spalacidae family and 60 pairs of microsatellite loci for the closely related species Gansu zokor and then amplified the loci with PCR to detect polymorphisms in plateau zokors. Nine pairs of microsatellite loci were common in plateau zokors. Liu et al.24 developed 12 pairs of polymorphic microsatellite loci based on the transcriptome sequencing of plateau zokor brain. Microsatellite loci with 3–5 repeats are typically used for parentage determination. Currently, only 13 pairs of loci in plateau zokors meet these requirements. The whole genome of plateau zokor is available online, and microsatellite loci can be selected and optimized from the perspective of whole genome. In view of this, this study made use of the publicly available whole genome data of plateau zokor to select microsatellite markers with polymorphisms and performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses on the genes of coding sequences (CDS) containing microsatellites. The results of this study not only help deepen our understanding of the distribution pattern of microsatellites in the whole genome of plateau zokors but also provide additional molecular markers for the study of dispersal, population genetic structure, and mating systems of plateau zokors.
Material and methods
Species data sources
Plateau zokor whole genome data were downloaded from the National Genomics Science Data Centre (a part of the China National Center for Bioinformation), and the genome was assembled at the scaffold level (https://ngdc.cncb.ac.cn/gwh/Assembly/941/show). The CDS and exon regions of plateau zokors were extracted using TBtools12, and the intergenic and intronic regions of plateau zokors were extracted using self-written Perl scripts.
Microsatellite classification methods
The identification tool software MISA10 was used to search for plateau zokor microsatellites with 1–6 base repeats. When MISA was executed, the ini file was set to 1–12, 2–6, 3–5, 4–5, 5–5, and 6–4, and the detection criteria were restricted to identifying perfect microsatellites of 1–6 bp, where the minimum number of repetitions for mono-, di-, tri-, tetra-, penta-, and hexanucleotide microsatellites were 12, 6, 5, 5, 5, and 4 times, respectively. When the distance between two microsatellites was less than 100 bases, it was considered a composite microsatellite. Meanwhile, repeat units consisting of the same set of bases (e.g., TAC, ACT, and CTA) were considered the same motif.
Annotation analysis of CDS regions containing microsatellites
The CDS of microsatellite-containing genes was enriched, and the screened CDS were compared with the NR protein database using Blastx (Blast, version 2.10.1)25 with the parameters “evaluate E-5, num alignments 50, max hsps 50, num threads 10”. The released proteins were then imported into Blast2GO (version 5.2.5)26 for GO functional categorization, including cellular components, molecular functions, and biological processes. KEGG analysis was performed by blasting (Blast, version 2.10.1) to the KEGG database27, with the parameters set to “evaluate E-5, num alignments 50, max hsps 50, num threads 10” to obtain the KEGG pathway annotation results. Functional enrichment analysis was performed using Fisher’s exact test.
DNA extraction and detection
Sixty-seven plateau zokors with leg muscle tissue (preserved in 95% alcohol) were selected as test samples. Among them, 24 were from Tianzhu Tibetan Autonomous County, Wuwei City, Gansu Province (TZ) and 43 were from Haiyan County, Haibei Tibetan Autonomous Prefecture, Qinghai Province (HY). As part of pest control, all captured plateau zokors were euthanized under isoflurane inhalation anesthesia. The Animal Ethics Committee of Gansu Agricultural University (GAU-LC-2020-014) approved all animal experiments, and the experiments adhered to the ARRIVE guidelines. All methods were done in accordance with relevant guidelines and regulations. Extraction of plateau zokor muscle was performed using the phenol–chloroform extraction method. The concentration and purity of DNA were detected using UV spectrophotometry, and the quality of DNA was detected with 1% agarose gel electrophoresis and stored at – 20 ℃.
Primer synthesis and PCR amplification
Based on the sequences obtained from whole genome sequencing, Primer 3.0 software28 was used to design primers. Seventy pairs of primers with a GC content of 40–60% and annealing temperature of 50–60 ℃ were randomly selected, and the primers were synthesized by Shanghai Sangong Bioengineering Co. The total volume of the PCR reaction system was 25 µL, which consisted of 12.5 μL of 2× Taq PCR Master Mix, 1 µL each of upstream and downstream primers (10 μM), 1 µL of template DNA (20–50 ng/μL), and 9.5 µL of ddH2O. The reaction conditions were pre-denaturation at 94 ℃ for 5 min, denaturation at 94 ℃ for 30 s, annealing at 53.5 ℃ for 30 s, 35 cycles of extension at 72 ℃ for 30 s each, and extension at 72 ℃ for 10 min; the mixture was then stored at 4 ℃.
Microsatellite data analysis
Microsatellite typing was performed based on the results of polyacrylamide gel electrophoresis (PAGE) at a concentration of 10%, and the number of alleles (Na), observed heterozygosity (Ho), expected heterozygosity (He), and polymorphism information content (PIC) were counted based on the typing results using Cervus 3.0.729. Hardy–Weinberg equilibrium (HWE) was tested using Genepop 4.730.
Results
Microsatellite repeat types and abundance in the whole genome
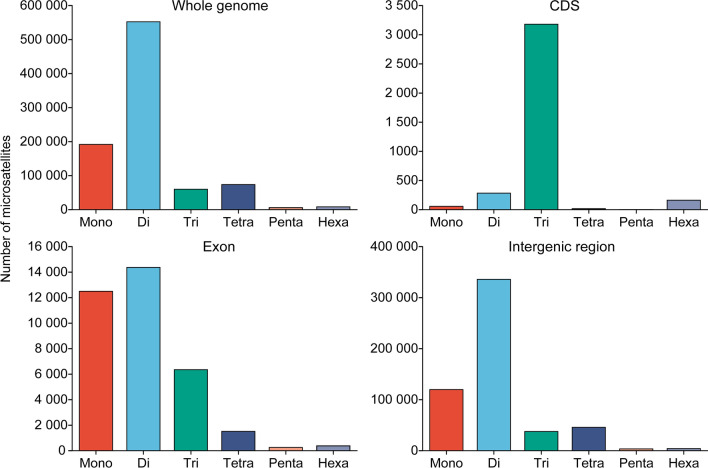
The whole genome of plateau zokor was 2.41 Gb. Screening the microsatellite data of the whole genome of plateau zokor revealed that perfect microsatellites were the most common type (891,748) followed by composite microsatellites (132,409), imperfect microsatellites were the least abundant (1001). In this study, only 1–6 base repeats of perfect-type microsatellites in the whole genome of plateau zokor were analyzed. The total number of the six types of microsatellites was 891,748, with a length of 20,649,725 bp, which accounted for 0.81% of the total genome sequence length, and the relative abundance was 352.81 loci/Mb (Table 1). Among them, the largest number and proportion of microsatellite types were mono- and dinucleotides, with 191,806 (21.51%) and 552,245 (61.93%), respectively, and a relative abundance of 75.89 loci/Mb and 218.49 loci/Mb, respectively. The remaining microsatellite types accounted for less than 10% of the total, in the order of 73,790 tetranucleotides (8.27%), 59,916 trinucleotides (6.72%), 8205 hexanucleotides (0.92%), and 5786 pentanucleotides (0.65%, with a relative abundance of 2.29 loci/Mb).
Table 1.
Distribution of microsatellites in the whole genome of plateau zokor.
| Type | Counts | Length (bp) | Percent (%) | Relative abundance (loci/Mb) |
|---|---|---|---|---|
| Mono | 191,806 | 3,127,384 | 21.51 | 75.89 |
| Di | 552,245 | 13,309,930 | 61.93 | 218.49 |
| Tri | 59,916 | 1,233,321 | 6.72 | 23.71 |
| Tetra | 73,790 | 2,535,012 | 8.27 | 29.19 |
| Penta | 5786 | 207,150 | 0.65 | 2.29 |
| Hexa | 8205 | 236,928 | 0.92 | 3.25 |
| Whole genome length (bp) | 2,569,336,452 | |||
| Microsatellite content of genome (%) | 0.81 |
Microsatellite dominant motif in the genome
A total of 283 motifs were found in 891,748 microsatellite loci in the plateau zokor genome, of which 2, 4, 9, 27, 71, and 170 were mono-, di-, tri-, tetra-, penta-, and hexanucleotide motifs, respectively, and the 6-basic motif was the most abundant. From the mononucleotide to hexanucleotide microsatellites, the most repetitive units were (A)n, (AC)n, (AGG)n, (ATAG)n, (AAAAC)n, and (AACCCT)n. The 15 most abundant microsatellite types in the whole genome of plateau zokor were (AC)n, (A)n, (AG)n, (AT)n, (AGG)n, and (ATAG)n, (AAT)n, (AAGG)n, (AAAT)n, (AAC)n, (AAAG)n, (AGGG)n, (AAAC)n, (ACC)n, and (ATC)n, accounting for 95.02% of the total number of all microsatellites (Fig. 1). The distribution of the number of repeats of different types of microsatellites in plateau zokors varied widely, with the number of mononucleotide repeats ranging from 12 to 35, with 12 being the most numerous, amounting to 43,766. The number of dinucleotide repeats ranged from 6 to 35, with 6 being the most numerous, with 120,017 dinucleotides. The number of repeats for the other four types is relatively low, mainly ranging from 4 to 20 (Fig. 2).
Fig. 1.
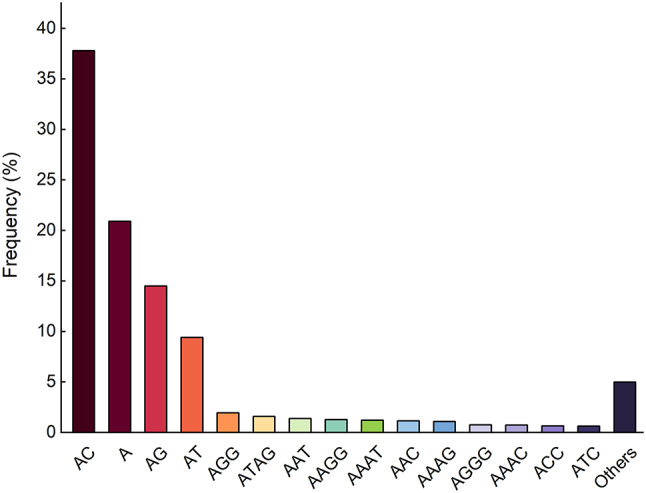
Most abundant motifs of plateau zokors.
Fig. 2.
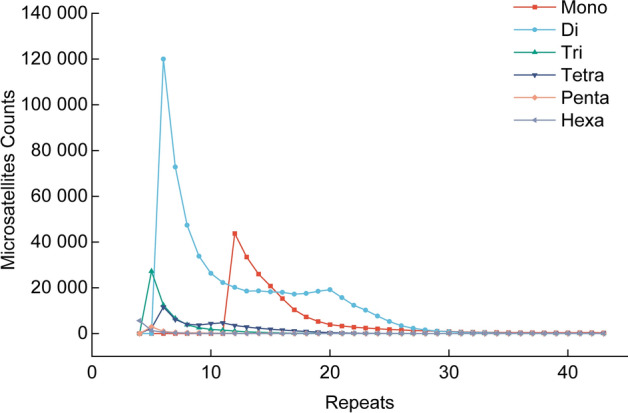
Distribution of microsatellite repeats in plateau zokors.
Distribution characteristics of microsatellites in different regions of the whole genome of plateau zokors
A comparison of the number and abundance of microsatellites in different genetic and intergenic regions of plateau zokors is shown in Table 2, in which the intergenic region had the largest number of microsatellites with an abundance of 547,279 and 367.02 loci/Mb and the CDS had the smallest number of microsatellites with an abundance of 3701 and 43.47 loci/Mb. The number (relative abundance) of microsatellites in the untranslated region, exons, and introns were 31,706 (258.97 loci/Mb), 35,379 (170.44 loci/Mb), and 138,422 (74.28 loci/Mb), respectively.
Table 2.
Comparison of the number and abundance of microsatellites in different genetic and intergenic regions of plateau zokors.
| Index | Genetic region | Intergenic region | |||
|---|---|---|---|---|---|
| CDS | Untranslated | Exon | Intron | ||
| Number | 3701 | 31,706 | 35,379 | 138,422 | 547,279 |
| Abundance (loci/Mb) | 43.47 | 258.97 | 170.44 | 74.28 | 367.02 |
A comparison of the number of microsatellite types in different regions of the whole genome of plateau zokor is shown in Fig. 3. The distributions of microsatellites in the whole genome and intergenic region were similar, with mononucleotides and dinucleotides being the most abundant. The most abundant microsatellites in CDS were trinucleotides and the most abundant type in the exons were dinucleotides.
Fig. 3.
Comparison of the distribution of microsatellite types in different genomic regions of plateau zokors.
Gene sequence annotation analysis of the CDS with microsatellites
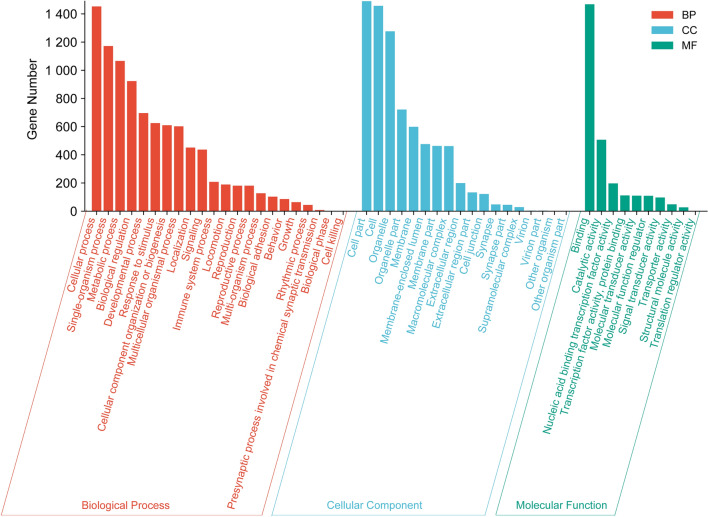
The CDS containing microsatellites from the whole genome of the plateau zokor were extracted and compared with the NR database using Blastx, and 1196 genes were annotated. GO annotations were made for 52 major functional genes, and 19,358 GO entries were assigned to them. Blast2GO analyses revealed 15,122 attributed to biological processes, 8885 to cellular components, and 2432 to molecular functions (Fig. 4). Among these, the cellular process (1453, GO:0009987), single-organism process (1172, GO:0044699), and metabolic process (1066, GO:0008152) in biological process ontology were assigned the highest numbers of CDS. The cellular component ontology had the highest number of CDS assigned to cell parts (1490, GO:0044464), cells (1457, GO:0005623), organelles (1276, GO:0043226), and binding (1468, GO:0005488) in molecular function ontology, which was the highest number of assigned CDS.
Fig. 4.
Gene ontology functional classification of microsatellite-containing genes in the coding region of plateau zokors.
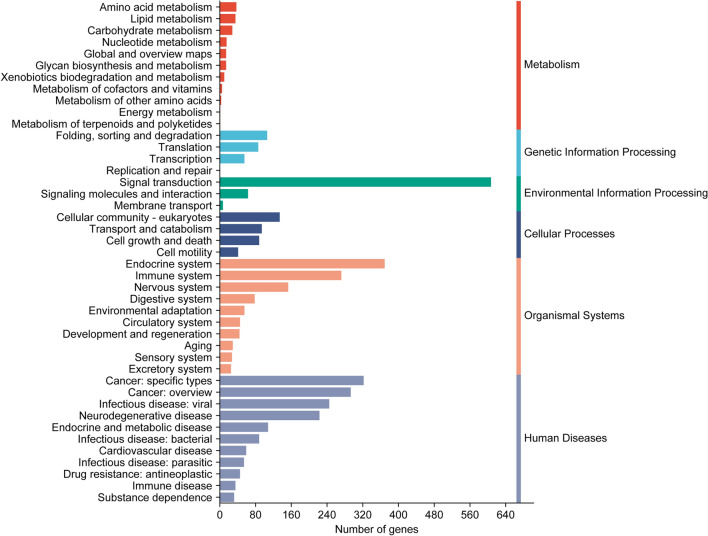
KEGG annotation of microsatellite-containing genes in plateau zokor exons produced 271 KO numbers, which were enriched into 43 pathways via enrichment analysis (Fig. 5). Of these pathways, signal transduction was the most significantly enriched with 607 genes. The pathways were functionally classified into metabolism, environmental information processing, genetic information processing, cellular processes, organismal systems, and human diseases. Among them, the pathways related to human diseases are the most numerous, with a total of 11 pathways containing 1504 genes. In contrast, the pathways related to environmental information processing are the fewest, with only 3 pathways containing 667 genes.
Fig. 5.
KEGG enrichment analysis of microsatellite-containing genes in the coding region of plateau zokor.
Screening of polymorphic microsatellite loci
From the analyzed microsatellites, 70 that fit the requirements of primer design were randomly selected for primer synthesis, of which 40 were trinucleotide and 30 were tetranucleotide microsatellites. PCR amplification and PAGE were performed using 24 TZ plateau zokors. The results showed that 43 pairs of primers could be successfully amplified in the muscle DNA of individual plateau zokors, which showed specific amplification products of the same size. Fifteen pairs of primers were successfully amplified and more than two alleles were found in the amplification products of these individuals. Subsequently, PCR amplification and PAGE were carried out using the genomic DNA of 43 HY plateau zokors as templates, and three pairs of primers that showed polymorphism in the TZ population did not show polymorphism in the HY population. The genetic characteristics of 13 pairs of microsatellite loci in the HY plateau zokor population were analyzed. The number of alleles ranged from 3 to 8; the observed heterozygosity (Ho) ranged from 0.059 to 0.810, with a mean value of 0.332; and the expected heterozygosity (He) ranged from 0.469 to 0.854, with a mean value of 0.734. Using the Bonferroni correction, only Z2357 microsatellite locus met HWE, whereas the others significantly deviated from HWE (P < 0.01). The PIC ranged from 0.389 to 0.824; all loci were moderately polymorphic (0.250 < PIC < 0.500), and 11 microsatellite loci showed high polymorphism (PIC > 0.500) (Table 3).
Table 3.
Genetic characteristics of 13 pairs of microsatellite loci in plateau zokors of HY population.
| Locus | Primer sequence | Repeat unit | Length/bp | Number of alleles | Ho | He | PIC | HWE |
|---|---|---|---|---|---|---|---|---|
| Z2305 |
F: ACTAGGGATACTGTGAGGCC R: TGTAGCATGCAGATTCCAGC |
(AAG)20 | 167 | 4 | 0.242 | 0.692 | 0.620 | 0.000 |
| Z2309 |
F: AGAAAGGCAAACACAGAGGG R: AAGAGCAATTGAAGGGTCTGG |
(AAT)9 | 208 | 3 | 0.810 | 0.542 | 0.450 | 0.000 |
| Z2319 |
F: GCCTGTGAAAGTGCTTTGC R: ACAGTAACCACTCCTTGTAGC |
(AGC)22 | 193 | 3 | 0.059 | 0.469 | 0.389 | 0.000 |
| Z2322 |
F: AGCAGGTCAGTTGAAGG R: AGAGATGAGAATTGTAAGGG |
(AGC)18 | 188 | 8 | 0.214 | 0.854 | 0.824 | 0.000 |
| Z2323 |
F: CCCCACAGAATATTAATGTTGC R: AATTCCTGCACTGTCAAACC |
(AGC)16 | 188 | 6 | 0.342 | 0.782 | 0.734 | 0.000 |
| Z2328 |
F: GGACAACTGGGACTACAGGG R: ACATCACCCTCATGCATAGC |
(CAG)18 | 156 | 7 | 0.400 | 0.828 | 0.794 | 0.000 |
| Z2330 |
F: AAGAGGAAGCTGTGAGACG R: AGTCCTTAGTCCTCAGAAAGC |
(AGC)16 | 169 | 6 | 0.333 | 0.749 | 0.694 | 0.000 |
| Z2345 |
F: TCAAGAAGATCCACACACACC R: CCACCAAACCATTATCACTTGC |
(AAAG)16 | 181 | 6 | 0.256 | 0.762 | 0.713 | 0.000 |
| Z2347 |
F: AGGTGGATGGAGATAGAAGGG R: CACCTCTGGTCACATTGCC |
(AAAG)16 | 239 | 5 | 0.172 | 0.747 | 0.691 | 0.000 |
| Z2356 |
F: CCCTGGGGTAAATCACTAGC R: TGTTTTCCTTTTCAGTTCCCC |
(AAGG)19 | 150 | 7 | 0.400 | 0.695 | 0.642 | 0.000 |
| Z2357 |
F: GTCTAGACTGCTGCTTCAGG R: GTTCCACAGATTCTTCCCGC |
(AAGG)16 | 156 | 7 | 0.561 | 0.823 | 0.789 | 0.002 |
| Z2359 |
F: AGAAAAAGGATGAGGGGAGG R: CACACTTGGAAATGGAGCC |
(AAGG)16 | 159 | 6 | 0.275 | 0.800 | 0.759 | 0.000 |
| Z2366 |
F: CCTGATCCAAATGAATGCTGC R: ATACCGTTCAAATGCTCCCG |
(AGCC)23 | 185 | 8 | 0.256 | 0.800 | 0.761 | 0.000 |
Discussion
The identification and classification of microsatellite sequences in plateau zokors using genome-wide data can provide useful information for molecular marker and population genetic diversity studies. In the present study, the whole genome size of the plateau zokor was 2.41 Gb. A total of 891,748 microsatellites were identified, with a total length of 20,649,725 bp. These microsatellites accounted for 0.81% of the total genome sequence length, and the relative abundance was 352.81 loci/Mb. Srivastava et al.31 analyzed the distribution of microsatellites in 15 taxonomic subgroups ranging from protozoa to mammals and found that the total microsatellite abundance was correlated with the genome size, whereas there was no correlation between the density of microsatellites (i.e. bp covered by microsatellites per Mb of the genome) and genome size. In mammals, microsatellite densities vary little, with the difference between the highest and lowest densities being approximately three-fold31.
The largest proportion of vertebrate genomes contains mononucleotide and dinucleotide repeats32. In plateau zokor genome, there were 191,806 (21.51%) monobasic and 552,245 (61.93%) dinucleotide repeats with relative abundances of 75.89 loci/Mb and 218.49 loci/Mb, respectively. Further, there were variations in the abundance of microsatellites among rodents. For example, in the vole genus (Microtus), dinucleotide repeats dominated in the field vole (M. agrestis), prairie voles (M. ochrogaster), and root voles (M. oeconomus), whereas in the common vole (M. arvalis), the percentage of mononucleotide repeats was 1.05% higher than that of dinucleotide repeats33. The most abundant repeat type in the entire genome of plateau zokors was (AC)n. It has been shown that most rodents have the most (AC)n repeats in their genomes; for example, (AC)n repeats account for 45% of the total genome of rats (Rattus norvegicus)34,35. In plateau zokors, (A)n repeats were the most abundant, accounting for 97.01% of the mononucleotide repeats. The high frequency of (A)n in the genome is due to an evident poly-A bias in mammals31.
The distribution of microsatellites in the genome is not random. Most of them are in the intergenic and non-coding regions of the genome, with a small portion in CDS6,35,36. The number and abundance of microsatellites in different genetic and intergenic regions varied, with the largest number of microsatellites (547,279) and their abundance (367.02 loci/Mb) in the intergenic region of plateau zokor, and the smallest number of microsatellites (3701) and their abundance (43.47 loci/Mb) in CDS. The distribution of microsatellites in the whole genome and intergenic regions was similar in character, and the numbers of both mononucleotide and dinucleotide repeats were the highest. The most abundant in CDS were trinucleotide repeats, and the most abundant exons were dinucleotides. Song et al.33 analyzed the microsatellite characteristics of CDS, exons, and introns of 57 genera of the primate order Euarchontoglires, and the most abundant types of CDS were trinucleotide repeats. Most trinucleotide repeat sequences in the CDS do not change in length37. When there is an increase in the number of trinucleotide repeats in CDS, it may increase the diversity of traits, thereby benefiting adaptive changes in the species during the evolutionary process38. Species differences exist in the dominant types of exons in rodents, and most of the dominant types are monobasic repeats, with Dipodomys ordii, Neotoma lepida, Mus musculus, and Mesocricetus auratus having the same dinucleotide repeats as plateau zokors33.
In this study, we investigated the potential functions of CDS containing microsatellites in the genome of plateau zokor using GO and KEGG pathway enrichment analyses. Many CDS with microsatellites were associated with environmental interactions, such as metabolic processes (GO:0008152), cellular processes (GO:0009987), signaling (GO:0023052), and responses to stimulus (GO:0050896), which had the highest number of genes in these entries. The same distribution pattern was found in 29 species of beetles39 and Pteropus vampyrus and Miniopterus natalensis in the order Chiroptera15. GO entries related to environmental interactions might be related to the evolutionary adaptation of plateau zokors to high-altitude and low-oxygen subterranean environments. The results of KEGG enrichment analyses also reflected the adaptation of plateau zokors to low oxygen levels. KEGG enrichment analysis of genes containing microsatellites revealed the most significant enrichment in signal transduction. Moreover, the MAPK signaling pathway was the most enriched signal transduction pathway, with 49 significantly enriched genes. MAPK signaling can promote the expression of hypoxia-inducible factor α, which indirectly regulates the HIF-1 signaling pathway. When an organism is in a hypoxic state, it regulates oxygen utilization by inducing a series of responses and thus regulating oxygen utilization40.
The length and sequence of repeat units affect the mutation rate of microsatellites, with the repeat sequences of shorter repeat units being more variable than those of longer ones. It has been observed that AT repeat sequences mutate more than other dinucleotide microsatellites, and AAAG and AAGG repeats are most likely to amplify polymorphisms compared to other tetranucleotide chains37. Consistent with this pattern, most of the tetranucleotide repeats amplified with polymorphisms in plateau zokors were AAAG and AAGG repeats. PIC can reflect genetic polymorphisms and have a direct linear relationship with gene diversity; this is a reliable means of determining the suitability of microsatellite loci for genetic analysis41.
In this study, 13 pairs of polymorphic loci were successfully amplified, of which 11 pairs were highly polymorphic, providing usable molecular markers for subsequent genetic studies of plateau zokor populations. Liu et al.24 designed 102 pairs of microsatellites based on transcriptome data and successfully amplified 12 pairs of polymorphic primers. Polymorphic screening has shown that microsatellites in the genome are more polymorphic than those in the transcriptome42. Transcriptomic microsatellites are conserved among species. Thus, microsatellites developed on the basis of transcriptomes have a higher success rate than those developed based on genomes in cross-species amplification42,43, and therefore, they can be considered a choice for cross-species amplification of near-origin species of plateau zokors. Of the 13 pairs of polymorphic loci amplified in this experiment, 12 significantly deviated from HWE due to heterozygous deletions, which might be responsible for the limited sample size for testing microsatellite polymorphisms. Microsatellite polymorphism screening revealed three microsatellite loci that showed polymorphisms in the TZ population but not in the HY population. Tang et al.44 reported severely restricted gene flow among plateau zokor populations. In this study, plateau zokors showed different microsatellite loci polymorphisms in different geographic populations, which may be related to the unique underground biology of plateau zokors44. It is also possible that different selective pressures in different regions contributed to this result.
Microsatellite typing can be performed using PAGE and capillary electrophoresis. In this experiment, PAGE was used to screen the microsatellite polymorphism primers. This method is simple and easy to perform; however, there may be errors in the interpretation of results, and it is suitable for the initial screening of polymorphic sites during microsatellite development. Capillary electrophoresis is accurate for microsatellite typing, but it is expensive and should be chosen when analyzing the genetic diversity of populations, phylogeny, and calculating kinship between individuals, which requires highly accurate results. With an abundance of genomic data, the development of microsatellite loci with polymorphisms by comparing genomic data from multiple individuals has become a new trend45. Luo et al.16 aligned the whole genome sequences (10×) of 42 blackhead seabreams with a reference genome and used the HipSTR tool to genotype the genes by comparing and counting the changes in the number of repeats at the microsatellite loci. Brandt et al.46 compared the sequences of two sumatran rhinoceros (Dicerorhinus sumatrensis), identified the read lengths of microsatellites corresponding to the same locus, and directly designed primers for microsatellite loci exhibiting polymorphisms. This method of developing microsatellite loci with polymorphisms, based on genome-wide data from multiple individuals, is more efficient and less labor-intensive. Given that plateau zokors do not have genome-wide data available for multiple individuals, only traditional methods of microsatellite development could be adopted.
The population of plateau zokor, a dominant subterranean rodent species on the Tibetan Plateau, is closely related to its reproduction and dispersal. Because basic information on population regulation has not been thoroughly researched, the effective management of plateau zokor populations remains difficult. Qinghai and Gansu provinces are the main distribution areas of plateau zokors47. In this study, we selected plateau zokor populations located in these two provinces, examined the polymorphisms and generality of microsatellites, and developed 13 microsatellite loci. The development of these loci provides a method for subsequent study of the mating system, population genetic structure, and dispersal of plateau zokors.
Supplementary Information
Acknowledgements
This work was supported by the open competition projects to select the best candidates for leading key initiatives of the key laboratory of grassland ecosystems, Gansu Agricultural University, Ministry of Education (KLGE-2024-02), the National Natural Science Foundation of China (32272566), the Industrial Support Program Project (2022CYZC-47) of Gansu Provincial Education Department, and the High-end Foreign Experts Recruitment Plan (G2022042008L).
Author contributions
Qiqi Hou: Investigation, Writing–original draft, Formal analysis. Weihong Ji: Supervision, Writing–review and editing. Kang An: Investigation, Writing–review and editing. Yuchen Tan: Investigation, Methodology. Penghui Liu: Investigation. Junhu Su: Conceptualization, Writing–review and editing, Funding acquisition, Supervision.
Data availability
All data generated or analyzed during this study are included in the supplementary information file. Plateau zokor genome downloaded from https://ngdc.cncb.ac.cn/gwh/Assembly/941/show, BioProject PRJCA002092, BioSample SAMC126955.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-84631-6.
References
- 1.Sachidanandam, R. et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature409, 928–933. 10.1038/35057149 (2001). [DOI] [PubMed]
- 2.He, Z., Xu, B., Buxbaum, J. & Ionita-Laza, I. A genome-wide scan statistic framework for whole-genome sequence data analysis. Nat. Commun.10, 3018. 10.1038/s41467-019-11023-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodel, R. G. J. et al. The report of my death was an exaggeration: a review for researchers using microsatellites in the 21st century. Appl. Plant Sci.4, 1600025. 10.3732/apps.1600025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellegren, H. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet.5, 435–445. 10.1038/nrg1348 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Fan, H. & Chu, J. Y. A brief review of short tandem repeat mutation. Genom. Proteom. Bioinf.5, 7–14. 10.1016/s1672-0229(07)60009-6 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vieira, M. L. C., Santini, L., Diniz, A. L. & Munhoz, C. D. Microsatellite markers: what they mean and why they are so useful. Genet. Mol. Biol.39, 312–328. 10.1590/1678-4685-Gmb-2016-0027 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber, J. L. & Wong, C. Mutation of human short tandem repeats. Hum. Mol. Genet.2, 1123–1128. 10.1093/hmg/2.8.1123 (1993). [DOI] [PubMed] [Google Scholar]
- 8.Zane, L., Bargelloni, L. & Patarnello, T. Strategies for microsatellite isolation: a review. Mol. Ecol.11, 1–16. 10.1046/j.0962-1083.2001.01418.x (2002). [DOI] [PubMed] [Google Scholar]
- 9.Satpathy, R. Application of bioinformatics resources for mining of simple sequence repeats (SSRs) marker in plant genomes: an overview. Res. J. Biotechnol.17, 136–143. 10.25303/1708rjbt1360143 (2022). [Google Scholar]
- 10.Beier, S., Thiel, T., Münch, T., Scholz, U. & Mascher, M. MISA-web: a web server for microsatellite prediction. Bioinformatics33, 2583–2585. 10.1093/bioinformatics/btx198 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du, L., Zhang, C., Liu, Q., Zhang, X. & Yue, B. Krait: an ultrafast tool for genome-wide survey of microsatellites and primer design. Bioinformatics34, 681–683. 10.1093/bioinformatics/btx665 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Chen, C. et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant13, 1194–1202. 10.1016/j.molp.2020.06.009 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Gymrek, M., Golan, D., Rosset, S. & Erlich, Y. lobSTR: a short tandem repeat profiler for personal genomes. Genome Res.22, 1154–1162. 10.1101/gr.135780.111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mokhtar, M. M., Alsamman, A. M. & El Allali, A. MegaSSR: a web server for large scale microsatellite identification, classification, and marker development. Front. Plant Sci.14, 1219055. 10.3389/fpls.2023.1219055 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao, W., Cai, W., Qiao, F., Lin, Z. & Wei, L. Comparison of microsatellite distribution in the genomes of Pteropus vampyrus and Miniopterus natalensis (Chiroptera). BMC Genom. Data24, 5. 10.1186/s12863-023-01108-7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo, X., Zhang, L. & Chen, S. Microsatellite genome-wide database development for the commercial blackhead seabream (Acanthopagrus schlegelii). Genes14, 620. 10.3390/genes14030620 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, B., Sun, Z., Lou, F., Gao, T. X. & Song, N. Genomic characteristics and profile of microsatellite primers for Acanthogobius ommaturus by genome survey sequencing. Biosci. Rep.40, BSR20201295. 10.1042/BSR20201295 (2020). [DOI] [PMC free article] [PubMed]
- 18.Wang, Z. et al. Impacts of climate change and human activities on three Glires pests of the Qinghai-Tibet Plateau. Pest Manag. Sci.80, 5233–5243. 10.1002/ps.8250 (2024). [DOI] [PubMed] [Google Scholar]
- 19.Kang, Y. et al. Introgression drives adaptation to the plateau environment in a subterranean rodent. BMC Biol.22, 187. 10.1186/s12915-024-01986-y (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang, Y. The biology and ecology of plateau zokors (Eospalax fontanierii). In Subterranean Rodents: News from Underground (eds. Begall, S., Burda, H. & Schleich, C. E.) 237–249 (Springer, 2007). 10.1007/978-3-540-69276-8_17.
- 21.Zhang, Y., Peng, S., Chen, X. & Chen, H. Y. H. Plant diversity increases the abundance and diversity of soil fauna: a meta-analysis. Geoderma411, 115694. 10.1016/j.geoderma.2022.115694 (2022). [Google Scholar]
- 22.Su, J. et al. Novel microsatellite markers obtained from Gansu zokor (Eospalax cansus) and cross-species amplification in Plateau zokor (Eospalax baileyi). Biochem. Syst. Ecol.57, 128–132. 10.1016/j.bse.2014.07.017 (2014). [Google Scholar]
- 23.Kang, Y. et al. Isolation of microsatellite markers by cross-amplification and transfer ability analysis in Eospalax baileyi. Grassl. Turf38, 56–60. 10.13817/j.cnki.cyycp.2018.02.009 (2018). [Google Scholar]
- 24.Liu, Q., Tan, Y., Yao, B., Kang, Y. & Su, J. Screening of polymorphic microsatellite markers in the plateau zokor based on transcriptome sequencing. Pratac. Sci.38, 2481–2489. 10.11829/j.issn.1001-0629.2021-0052 (2021). [Google Scholar]
- 25.Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res.25, 3389–3402. 10.1093/nar/25.17.3389 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conesa, A. et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics21, 3674–3676. 10.1093/bioinformatics/bti610 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Ogata, H. et al. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res.27, 29–34. 10.1093/nar/27.1.29 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koressaar, T. & Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics23, 1289–1291. 10.1093/bioinformatics/btm091 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Kalinowski, S. T., Taper, M. L. & Marshall, T. C. Revising how the computer program cervus accommodates genotyping error increases success in paternity assignment. Mol. Ecol.16, 1099–1106. 10.1111/j.1365-294X.2007.03089.x (2007). [DOI] [PubMed] [Google Scholar]
- 30.Rousset, F. genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour.8, 103–106. 10.1111/j.1471-8286.2007.01931.x (2008). [DOI] [PubMed] [Google Scholar]
- 31.Srivastava, S., Avvaru, A. K., Sowpati, D. T. & Mishra, R. K. Patterns of microsatellite distribution across eukaryotic genomes. BMC Genom.20, 153. 10.1186/s12864-019-5516-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arabfard, M. et al. Global abundance of short tandem repeats is non-random in rodents and primates. BMC Genom. Data23, 77. 10.1186/s12863-022-01092-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song, X. et al. Comparison of the microsatellite distribution patterns in the genomes of Euarchontoglires at the taxonomic level. Front. Genet.12, 622724. 10.3389/fgene.2021.622724 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walder, R. Y. et al. Short tandem repeat polymorphic markers for the rat genome from marker-selected libraries. Mamm. Genome9, 1013–1021. 10.1007/s003359900917 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Tóth, G., Gáspári, Z. & Jurka, J. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res.10, 967–981. 10.1101/gr.10.7.967 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, Y. C., Korol, A. B., Fahima, T. & Nevo, E. Microsatellites within genes: structure, function, and evolution. Mol. Biol. Evol.21, 991–1007. 10.1093/molbev/msh073 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Verbiest, M. et al. Mutation and selection processes regulating short tandem repeats give rise to genetic and phenotypic diversity across species. J. Evol. Biol.36, 321–336. 10.1111/jeb.14106 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loire, É., Higuet, D., Netter, P. & Achaz, G. Evolution of coding microsatellites in primate genomes. Genome Biol. Evol.5, 283–295. 10.1093/gbe/evt003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song, X. et al. Comparison of microsatellite distribution patterns in twenty-nine beetle genomes. Gene757, 144919. 10.1016/j.gene.2020.144919 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Teng, M. et al. Microtubular stability affects pVHL-mediated regulation of HIF-1alpha via the p38/MAPK pathway in hypoxic cardiomyocytes. PLoS One7, e35017. 10.1371/journal.pone.0035017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serrote, C. M. L., Reiniger, L. R. S., Silva, K. B., dos Santos Rabaiolli, S. M. & Stefanel, C. M. Determining the polymorphism information content of a molecular marker. Gene726, 144175. 10.1016/j.gene.2019.144175 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Xia, Y., Luo, W., Yuan, S., Zheng, Y. & Zeng, X. Microsatellite development from genome skimming and transcriptome sequencing: comparison of strategies and lessons from frog species. BMC Genom.19, 886. 10.1186/s12864-018-5329-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Postolache, D. et al. Transcriptome versus genomic microsatellite markers: Highly informative multiplexes for genotyping Abies alba Mill. and congeneric species. Plant. Mol. Biol. Rep.32, 750–760. 10.1007/s11105-013-0688-7 (2014).
- 44.Tang, L. et al. Gene flows of Eospalax baileyi geographical populations. J. Anhui Agric. Sci.38, 5123–5124. 10.13989/j.cnki.0517-6611.2010.10.033 (2010). [Google Scholar]
- 45.Trede, F. et al. A refined panel of 42 microsatellite loci to universally genotype catarrhine primates. Ecol. Evol.11, 498–505. 10.1002/ece3.7069 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brandt, J. R. et al. Characterization of 29 polymorphic microsatellite markers developed by genomic screening of Sumatran rhinoceros (Dicerorhinus sumatrensis). BMC Res. Notes14, 119. 10.1186/s13104-021-05522-x (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, T. et al. Phenotypic and genomic adaptations to the extremely high elevation in plateau zokor (Myospalax baileyi). Mol. Ecol.30, 5765–5779. 10.1111/mec.16174 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in the supplementary information file. Plateau zokor genome downloaded from https://ngdc.cncb.ac.cn/gwh/Assembly/941/show, BioProject PRJCA002092, BioSample SAMC126955.