Abstract
Detection and teeth segmentation from X-rays, aiding healthcare professionals in accurately determining the shape and growth trends of teeth. However, small dataset sizes due to patient privacy, high noise, and blurred boundaries between periodontal tissue and teeth pose challenges to the models’ transportability and generalizability, making them prone to overfitting. To address these issues, we propose a novel model, named Grouped Attention and Cross-Layer Fusion Network (GCNet). GCNet effectively handles numerous noise points and significant individual differences in the data, achieving stable and precise segmentation on small-scale datasets. The model comprises two core modules: Grouped Global Attention (GGA) modules and Cross-Layer Fusion (CLF) modules. The GGA modules capture and group texture and contour features, while the CLF modules combine these features with deep semantic information to improve prediction. Experimental results on the Children’s Dental Panoramic Radiographs dataset show that our model outperformed existing models such as GT-U-Net and Teeth U-Net, with a Dice coefficient of 0.9338, sensitivity of 0.9426, and specificity of 0.9821. The GCNet model also demonstrates clearer segmentation boundaries compared to other models.
Subject terms: Computational models, Machine learning
Introduction
Deep learning has been widely used in medical image analysis, such as object detection1,2, classification3,4, segmentation5,6, and registration7,8. Compared to traditional methods, Convolutional Neural Networks (CNNs) have unique advantages9. CNNs can automatically learn and extract high-level features from images, which is particularly crucial in medical image processing since medical images often contain complex histological and anatomical structures. By capturing these information, CNNs can perform certain tasks at the level of professional healthcare providers10. The deep structure of CNNs allows for a thorough understanding of the contextual information in images, which is vital for accurately interpreting the structures and lesion areas in medical images11. Additionally, CNNs have the capability for end-to-end learning, directly outputting the final segmentation results from raw images. This makes CNNs more efficient and accurate compared to traditional methods1.
With the improvement of living standards, people’s pursuit of dental aesthetics has also increased. They are not only concerned about the health of their teeth but also their appearance and alignment. Accurate teeth segmentation technology plays a crucial role in assisting dentists with clinical diagnosis12. Consequently, convolutional networks are widely used in the segmentation of dental X-ray images13. Numerous studies14,15 have attempted to use modified convolutional modules to achieve precise segmentation of teeth and periodontal tissue from X-ray images. However, this process faces multiple challenges.
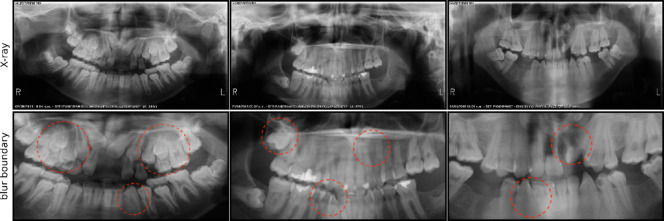
Firstly, privacy concerns make it challenging to obtain large-scale datasets, and X-ray images often contain noise points. Convolutional Neural Networks (CNNs) are highly sensitive to noise, which can significantly affect the model’s stability and cause it to focus on irrelevant areas. Secondly, the contrast between teeth and periodontal tissue in X-ray images is low, with blurred boundaries and individual differences (Fig. 1). This issue is more pronounced in children’s X-ray images, where permanent dentition is not fully developed, creating complex structures. These challenges make diagnosis and treatment difficult, requiring doctors to carefully observe and interpret the images for accurate assessment and treatment planning. As a result, even improved CNN modules struggle with clear boundary segmentation. Additionally, the reliance of CNNs on large datasets is problematic due to the limited availability of patient X-ray images, which impacts the stability and generalization capabilities of models trained on small datasets.
Fig. 1.
Three examples for teeth Segmentation because the quality of X-ray scan. First column shows the X-ray of teeth and the red dots in second column shows the blur boundaries between teeth and teeth, and also between teeth and periodontal tissue.
To address the aforementioned issues, we integrated CNNs with attention mechanisms to specifically address the challenges posed by datasets like tooth segmentation from X-ray, which are characterized by noise, significant individual variation, burr boundary and limited dataset size. By leveraging the strengths of both CNNs and attention mechanisms, our method ensures precise feature extraction and robust handling of complex data variability, providing a valuable tool to support professionals in making accurate diagnoses.
In summary, this paper contributes in the following ways:
We propose a U-shaped network to clearly segment periodontal tissue and teeth in X-ray images. This model includes two main modules: the Grouped Global Attention Module and the Cross-Layer Fusion Modules. The Grouped Global Attention Module not only captures global information from shallow inputs but also guides high-level features from the Cross-Layer Fusion Modules to selectively focus on key regions. Finally, a Dual-output Decoder combines texture-rich and position-rich features from the global attention-guided module with high-level features from the cross-layer attention fusion module to achieve accurate segmentation.
Grouped Global Attention Module: Since low-level features contain more texture and contour information16. To capture important positional information, we designed the Grouped Global Attention Module, which includes two Grouped Uni-directional Attention Modules and a Global Feature Fusion module. Initially, input features are extracted using various convolution kernels and dilation coefficients. Then, two parallel attention modules extract global position and texture information. These features are merged through the Global Feature Fusion module to create the Grouped Global Map, guiding high-level features to focus on important image areas. The output is also fed into the Dual-output Decoder for feature fusion.
Cross-Layer Fusion Modules: High-level features contain richer semantic information than low-level features17. To maximize the use of this information, the Cross-Layer Fusion Modules employ the Merge-and-Share Module for sharing information across layers. Guided by the Grouped Global Map from the Grouped Global Attention Module, the Cross-Layer Attention directs high-level features to focus on key contour and texture information. This fusion of low-level and high-level features creates cross-layer features. These features, combined with the Grouped Global Map, are then input into the Dual-output Decoder to produce edge prediction maps and final prediction images.
In Sect. 2, we will present the applications of deep learning in semantic segmentation and analyze its advantages in detail. Subsequently, this paper will elaborate on our research approach and corresponding solutions. Section 3 provides a comprehensive and detailed description of our proposed network architecture and in-depth explanations of key modules. Section 4 outlines the specific details of experiments conducted using this network to validate the effectiveness of the network structure and its core components. The final research conclusions are clearly presented in Sect. 5.
Related work
In recent years, convolutional networks have revolutionized medical imaging. Since the introduction of U-net in 201518, Various networks based on improvements to U-net have been developed to handle a wide range of medical image segmentation tasks, such as Vnet19, UnetPlusPlus17, and SegNet20.
While convolutional networks excel at feature extraction, guiding them to focus on critical information and ignore noise is crucial. Initially, the attention mechanism was proposed in21 and widely applied to various sequence modeling tasks, becoming the foundation for many subsequent studies, including the well-known “Transformer” model22. These mechanisms are now used in image23 and video24 domains to improve performance by focusing on key information. Similarly, combining attention with CNNs in feature channels is also common. For example, Hu et al. proposed the SENet (Squeeze-and-Excitation Networks) model25, which significantly enhances network performance by re-weighting the outputs of convolutional layers. Attention maps clearly show which parts of the image the model focused on during decision-making, providing practical value for understanding the model’s behavior and verifying if it operates as expected26.
Given the limited dataset size of dental X-ray images and the presence of noise, models with a large number of parameters often overfit to random noise or sample-specific features, reducing generalizability. To address this, Howard et al.27 proposed depthwise separable convolutions, which decompose standard convolutions into depthwise and pointwise operations, significantly reducing parameters and computational costs while maintaining comparable performance. This lightweight architecture is particularly effective for small datasets, reducing the risk of overfitting and enabling real-time performance in resource-constrained environments. To further capture broader contextual information without increasing computational complexity, dilated convolutions, as introduced by Yu et al.25, expand the receptive field by inserting spaces within convolution filters. Similarly, large kernels (e.g., 7 × 7) enhance semantic understanding, as shown by Peng et al.28, who used global convolutional networks to improve segmentation tasks. However, large kernels increase computational complexity, prompting Szegedy et al.29 to combine multiple kernel sizes and dilation rates for efficiency, albeit at the cost of slower inference. To tackle this, Zhang et al.30 introduced a multi-branch training structure that simplifies into a single convolutional layer during inference, significantly improving efficiency while maintaining performance.
In the field of tooth root segmentation, Li et al.31 proposed GT U-Net, a novel network architecture combining the U-Net framework with group Transformer modules to address the limitations of convolutional networks in capturing global features. The GT U-Net incorporates a hybrid structure of convolution and Transformer, allowing it to model long-range dependencies without relying on pre-trained weights. To reduce the high computational cost associated with Transformers, the architecture employs a grouping and bottleneck structure, significantly enhancing efficiency.
In parallel, the Teeth U-Net model was introduced by Senbao Hou et al. to address challenges such as blurred tooth boundaries and low contrast between teeth and alveolar bone in dental panoramic X-ray images. This model incorporates several key innovations: a dense skip connection mechanism between the encoder and decoder to retain teeth detail information and reduce semantic gaps, a multi-scale aggregation attention block (MAB) to adaptively extract and fuse multi-scale features, and a dilated hybrid self-attentive block (DHAB) to suppress irrelevant background information while enhancing contextual information.
Senbao Hou et al.32 introduced the Inf-Net model which is an innovative edge-focused approach to address challenges associated with low-contrast and blurred infection boundaries in CT images. The model integrates an edge attention module (EA), which explicitly enhances the representation of infection region boundaries by learning edge-aware features derived from low-level image features. This process utilizes a convolutional layer to generate an edge map, which is further refined using a Binary Cross-Entropy (BCE) loss function to guide the model in capturing accurate boundary information. This component collectively improves the segmentation of boundaries of lesions.
X-ray images in medical segmentation often have low contrast at boundaries, making accurate segmentation challenging. Adding boundary supervision signals enhances the model’s focus on hard-to-learn edge pixels, ensuring the model focuses more on these difficult-to-capture boundary details during training. This method ensures the model prioritizes difficult boundary details during training, improving segmentation accuracy and providing a more reliable basis for medical diagnosis and treatment.
Methodology
In this section, we will explain the proposed network architecture, including its key module components and specific implementation details.
Model overview
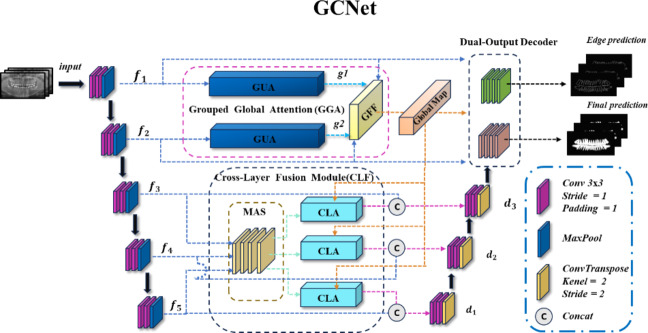
The overall structure of the model, as shown in Fig. 2, exhibits a U-shaped architecture. The model consists of five encoders, with the first two encoders,  and
and 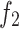 , being shallow encoders, while
, being shallow encoders, while 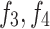 , and
, and 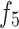 are deep encoders. Unlike many medical image segmentation models, this model employs three decoders (labeled
are deep encoders. Unlike many medical image segmentation models, this model employs three decoders (labeled 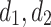 and
and 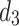 ) and features a Dual-output Decoder (DOD). The DOD merges high-level features from
) and features a Dual-output Decoder (DOD). The DOD merges high-level features from 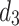 with the Grouped Global Map from the Grouped Global Attention Module (GGA) to produce edge and final prediction results. In the following sections, we will provide a detailed introduction and analysis of each proposed module.
with the Grouped Global Map from the Grouped Global Attention Module (GGA) to produce edge and final prediction results. In the following sections, we will provide a detailed introduction and analysis of each proposed module.
Fig. 2.
Overview of the GCNet model architecture.
Grouped global attention module (GGA)
Given that low-level features contain high-resolution and rich texture information16, we specifically designed the GGA to capture the critical texture and contour information contained in these low-level features. This global guidance module includes two Grouped Uni-directional Attention Modules (GUA) and a Global Feature Fusion module (GFF). Through the processing of the GUA, global texture and positional features are captured, resulting in the Grouped Global Map, which is then passed to the Cross-Layer Fusion Modules (CLF) to guide the precise extraction of global features and edge contours. Additionally, to ensure that the details and textures of the input features are accurately learned and preserved, the Grouped Global Map also transmits the captured feature information to the DOD.
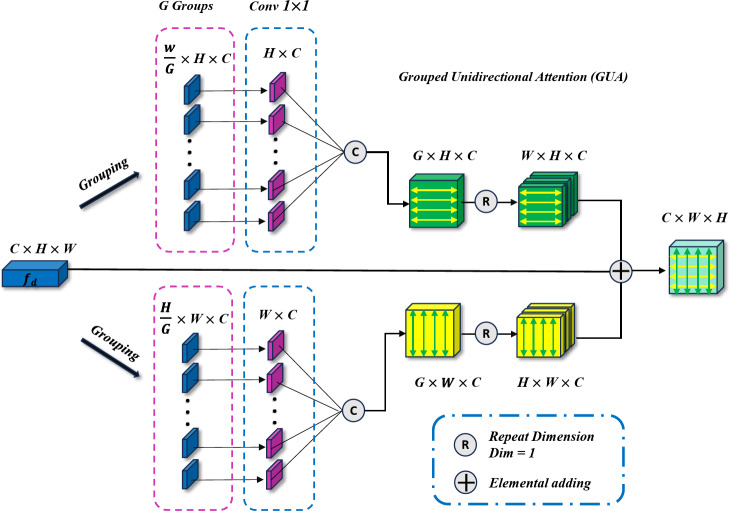
Grouped uni-directional attention module (GUA)
The Grouped Uni-directional Attention Module (GUA) employs a parallel structure. To extract as much texture and dental contour information as possible from the high-resolution features 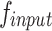 provided by the encoder, convolutional kernels of different scales and dilation rates are used for feature extraction. We use two 5 × 5 convolutional kernels with dilation rates of 1 and 2, and three 3 × 3 convolutional kernels with dilation rates of 3, 4, and 5 respectively. In the shallow feature layers, which are rich in texture and structural information, we utilize larger convolutional kernels (5 × 5) with smaller dilation rates (1 and 2) to capture finer details around the current pixel. Conversely, smaller convolutional kernels (3 × 3) with larger dilation rates (3, 4, and 5) are employed to extract structural information from the surrounding area, resulting in the extracted features denoted as
provided by the encoder, convolutional kernels of different scales and dilation rates are used for feature extraction. We use two 5 × 5 convolutional kernels with dilation rates of 1 and 2, and three 3 × 3 convolutional kernels with dilation rates of 3, 4, and 5 respectively. In the shallow feature layers, which are rich in texture and structural information, we utilize larger convolutional kernels (5 × 5) with smaller dilation rates (1 and 2) to capture finer details around the current pixel. Conversely, smaller convolutional kernels (3 × 3) with larger dilation rates (3, 4, and 5) are employed to extract structural information from the surrounding area, resulting in the extracted features denoted as 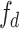 . To complement the use of dilated convolutions, we employ depthwise separable convolutions27, aiming to reduce the model’s parameter count and enable learning of general features that capture different receptive fields on small-scale datasets, thus enhancing the model’s resistance to noise interference. The entire GUA process is illustrated in Fig. 3.
. To complement the use of dilated convolutions, we employ depthwise separable convolutions27, aiming to reduce the model’s parameter count and enable learning of general features that capture different receptive fields on small-scale datasets, thus enhancing the model’s resistance to noise interference. The entire GUA process is illustrated in Fig. 3.
Fig. 3.
Grouped Uni-directional attention module (GUA) model architecture. The input features are first grouped and then fed into parallel uni-directional attention mechanisms.
Next, to guide the model in learning the correlation of image pixels independently in the horizontal and vertical directions and to reduce computational complexity to avoid overfitting, we perform parallel vertical and horizontal attention extraction. For example, in the vertical branch, we first adjust the input feature dimensions from 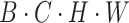 to
to 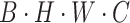 . Then, we divide the captured features
. Then, we divide the captured features 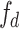 into
into 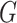 groups, each group having a height dimension of
groups, each group having a height dimension of 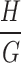 . Then, we use a 1 × 1 convolution to compress the channel number of each group to 1, representing the features of that group, thus transforming the feature dimensions to
. Then, we use a 1 × 1 convolution to compress the channel number of each group to 1, representing the features of that group, thus transforming the feature dimensions to 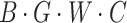 . We then extract vertical attention from these features, finally adjusting the dimensions back to
. We then extract vertical attention from these features, finally adjusting the dimensions back to 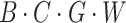 . Similarly, in the horizontal attention branch, we process the height dimension
. Similarly, in the horizontal attention branch, we process the height dimension 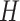 in a similar manner, resulting in a feature matrix with dimensions
in a similar manner, resulting in a feature matrix with dimensions 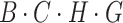 . Finally, using broadcasting and addition operations, we fuse the results of vertical and horizontal attention to generate an output with dimensions
. Finally, using broadcasting and addition operations, we fuse the results of vertical and horizontal attention to generate an output with dimensions 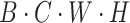 . The attention mechanism process is detailed in Formulae 1, demonstrating how our parallel attention mechanism effectively captures feature information from different dimensions.
. The attention mechanism process is detailed in Formulae 1, demonstrating how our parallel attention mechanism effectively captures feature information from different dimensions.
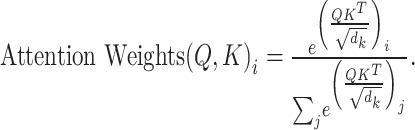 |
1 |
By using the grouping and uni-directional mechanism, the overall computational complexity is significantly reduced. Specifically, the complexity decreases from 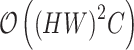 to
to 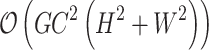 where
where 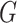 is the number of groups (set to 16 in this paper), and the width
is the number of groups (set to 16 in this paper), and the width 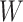 of the input image is twice the height
of the input image is twice the height 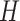 . Substituting
. Substituting 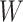 with 2H in the formula, the resulting complexity is shown in Formulae 2. Our proposed grouping and uni-directional attention mechanism significantly saves computational costs (in
with 2H in the formula, the resulting complexity is shown in Formulae 2. Our proposed grouping and uni-directional attention mechanism significantly saves computational costs (in 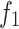 ,
, 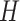 is set to 256 and
is set to 256 and 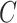 to 64; in
to 64; in 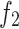 , H is set to 128 and
, H is set to 128 and 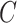 is increased to 128), thus conserving a substantial amount of computational resources.
is increased to 128), thus conserving a substantial amount of computational resources.
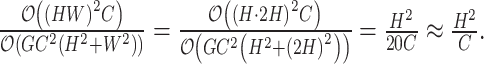 |
2 |
Compared to existing models, the use of a uni-directional attention mechanism not only reduces computational complexity but also enables the model to learn more generalized features. In contrast, self-attention often causes the model to focus excessively on noisy points, leading to overfitting. Consequently, the uni-directional attention mechanism improves the model’s performance while significantly enhancing its generalization ability, as demonstrated by the experimental results in Sect. 4.
The processing procedure is illustrated in Formula 3. The initial feature dimension is set to 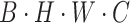 . The operation
. The operation 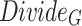 splits the input into a dimension of
splits the input into a dimension of 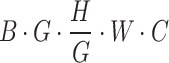 . After applying
. After applying 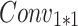 , the feature dimension changes to
, the feature dimension changes to 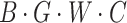 . Finally, through the
. Finally, through the 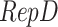 operation, which stands for “Repeating Dimension,” the feature dimension is restored to
operation, which stands for “Repeating Dimension,” the feature dimension is restored to 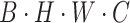 .
.
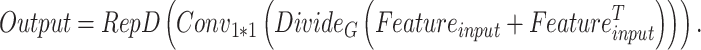 |
3 |
Global feature fusion module (GFF)
The Global Feature Fusion Module (GFF) is designed to effectively fuse the Grouped Global Maps generated by the two GUAs at different resolutions and to pass the fused information to the Cross-Layer Fusion Modules (CLF). This process ensures combination of deep and shallow information. Additionally, this module outputs the processed features to the DOD for further integration with the features from the third decoder, 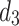 . This fusion plays a critical role in edge prediction, especially in handling challenging edge regions, serving as a supervisory signal to achieve more precise edge prediction results19. Compared to existing modules, the output of GFF not only provides information for edge supervision but also offers global guidance for deep features. The process of this module is illustrated in Fig. 4.
. This fusion plays a critical role in edge prediction, especially in handling challenging edge regions, serving as a supervisory signal to achieve more precise edge prediction results19. Compared to existing modules, the output of GFF not only provides information for edge supervision but also offers global guidance for deep features. The process of this module is illustrated in Fig. 4.
Fig. 4.
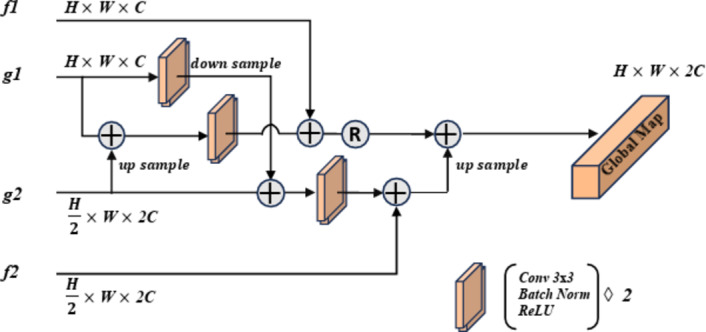
Diagram of the Global Feature Fusion module (GFF) process. This diagram illustrates how to combine features from the Grouped Uni-directional Attention Module (GUA) and the encoder modules to produce a global feature map.
Cross-layer fusion modules (CLF)
The purpose of the Cross-Layer Fusion Modules (CLF) is to effectively combine the Grouped Global Map from the GGA with the features from the deep level encoders ,
,  , and
, and . Through this fusion, high-level features can capture critical position, contour, and other information from the global feature map, resulting in features that are rich in semantics, texture, and contour information, thereby enhancing their expressive power and accuracy. This module comprises the Merge-And-Share Modules (MAS) and Cross-Layer Attention.
. Through this fusion, high-level features can capture critical position, contour, and other information from the global feature map, resulting in features that are rich in semantics, texture, and contour information, thereby enhancing their expressive power and accuracy. This module comprises the Merge-And-Share Modules (MAS) and Cross-Layer Attention.
Merge-and-share module (MAS)
Unlike existing models such as Teeth U-Net33, which lack the ability to integrate deep features and effectively extract hierarchical features from other deep-level decoders, we specifically designed the Merge-and-Share Module (MAS) to address this limitation. Positioned before the cross-layer feature fusion module, as shown in Fig. 5, MAS is tailored to handle high-level features, which have numerous channels and rich semantic information34. Its unique role is to facilitate seamless information exchange among hierarchical features, ensuring that each cross-layer feature fusion module extracts the most critical information from the three high-level features (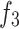 ,
, 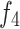 , and
, and 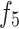 ). This design sets our model apart by enabling more precise and effective feature fusion across layers.
). This design sets our model apart by enabling more precise and effective feature fusion across layers.
Fig. 5.
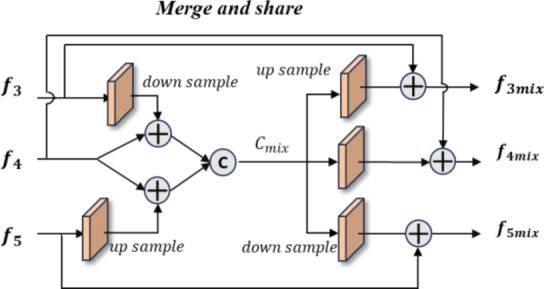
Merge-and-share module. This module integrates and extracts information from encoders 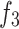 ,
, 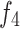 , and
, and 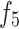 to enable the sharing of features among the three deep features.
to enable the sharing of features among the three deep features.
In the operation process, we first apply downsampling and upsampling using bilinear interpolation to align the channels and spatial dimensions of 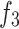 and
and 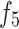 with
with 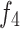 , thereby eliminating mismatches in channel and spatial dimensions. Next, an element-wise addition operation is performed, followed by concatenation to generate a fused channel,
, thereby eliminating mismatches in channel and spatial dimensions. Next, an element-wise addition operation is performed, followed by concatenation to generate a fused channel, 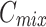 . Convolution operations, along with downsampling and upsampling, are employed to adjust the number of channels and spatial dimensions. Finally, shortcuts from the initial features are used to extract the required features for each layer (
. Convolution operations, along with downsampling and upsampling, are employed to adjust the number of channels and spatial dimensions. Finally, shortcuts from the initial features are used to extract the required features for each layer (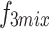 ,
, 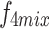 and
and 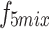 ) from
) from 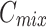 , ensuring that all high-level feature information is comprehensively captured at a single layer, achieving cross-layer information sharing.
, ensuring that all high-level feature information is comprehensively captured at a single layer, achieving cross-layer information sharing.
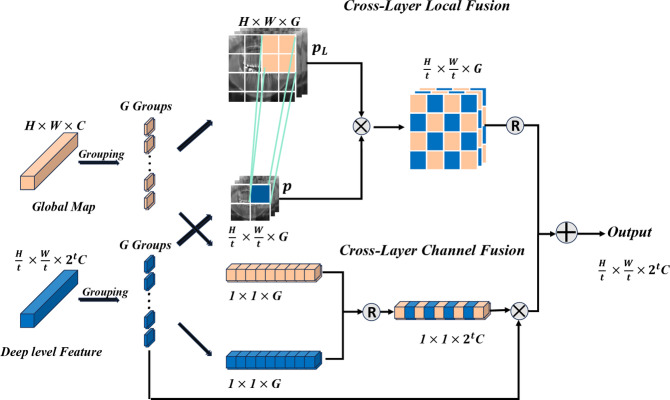
Cross-layer attention (CLA)
We pass 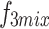 ,
, 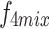 , and
, and 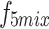 into their respective Cross-Layer Attention (CLA) modules to extract global information from the Grouped Global Map. Each CLA module includes Cross-Layer Local Fusion and Cross-Layer Channel Fusion operations, as detailed in Fig. 6. Low-level features, compared to high-level features, have higher resolution and are rich in texture and edge information16. However, during multiple downsampling processes, low-level features compress the information of a certain area into a single pixel in the high-level features, inevitably leading to the loss of some contour and texture information. To maximize the effective information in low-level features, we adopt the Cross-Layer Local Fusion strategy. This strategy extracts information using the corresponding regions on the feature guidance map for each pixel in the high-level features. First, we expand the length and width of the high-level features to match the feature guidance map so that each pixel p corresponds to the local features
into their respective Cross-Layer Attention (CLA) modules to extract global information from the Grouped Global Map. Each CLA module includes Cross-Layer Local Fusion and Cross-Layer Channel Fusion operations, as detailed in Fig. 6. Low-level features, compared to high-level features, have higher resolution and are rich in texture and edge information16. However, during multiple downsampling processes, low-level features compress the information of a certain area into a single pixel in the high-level features, inevitably leading to the loss of some contour and texture information. To maximize the effective information in low-level features, we adopt the Cross-Layer Local Fusion strategy. This strategy extracts information using the corresponding regions on the feature guidance map for each pixel in the high-level features. First, we expand the length and width of the high-level features to match the feature guidance map so that each pixel p corresponds to the local features 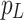 in the Grouped Global Map. Then, we use the same grouping method as in the GUA to divide both the high-level features and the feature guidance map into
in the Grouped Global Map. Then, we use the same grouping method as in the GUA to divide both the high-level features and the feature guidance map into 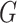 groups. Finally, we perform a convolution operation, fusing pixel p with the corresponding region
groups. Finally, we perform a convolution operation, fusing pixel p with the corresponding region 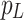 in the Grouped Global Map. This approach allows us to effectively capture key contour and positional information from the Grouped Global Map, as illustrated in the Cross-Layer Local Fusion process in Fig. 6.
in the Grouped Global Map. This approach allows us to effectively capture key contour and positional information from the Grouped Global Map, as illustrated in the Cross-Layer Local Fusion process in Fig. 6.
Fig. 6.
Diagram of the Cross Layer Attention module process, where t represents the number of downsampling times. The upper branch represents the Cross-Layer Local Fusion operation, aimed at extracting local information from high-resolution global features. The lower branch represents the Cross-Layer Channel operation, aimed at integrating channel information from deep features and global features.
To extract the rich semantic features contained in the high-level features, we incorporate the SE mechanism to capture the channel information of the high-level features35. We innovatively improve this by also capturing the channel information of the Global Feature Map and effectively fusing them through the Cross-Layer Channel Fusion process, as shown in Fig. 6. We use two different fusion strategies: this dual approach not only fully utilizes the critical contour and positional information from the Global Feature Map but also successfully integrates channel information, achieving cross-layer information extraction.
Dual-output decoder (DOD)
The DOD leverages the Grouped Global Map and decodes features merged from various layers in the CLF using the decoder 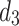 , as shown in Fig. 7. This process generates an edge prediction result, serving as a supervisory signal, and a final segmentation result as the output prediction map. Compared to the edge supervision module in existing models like Inf-Net, our approach integrates semantic information from deep-level decoders into the edge feature fusion process, enhancing the overall performance.
, as shown in Fig. 7. This process generates an edge prediction result, serving as a supervisory signal, and a final segmentation result as the output prediction map. Compared to the edge supervision module in existing models like Inf-Net, our approach integrates semantic information from deep-level decoders into the edge feature fusion process, enhancing the overall performance.
Fig. 7.
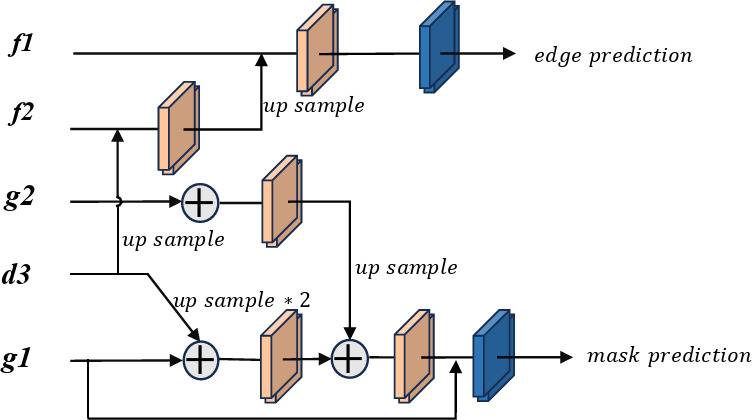
Diagram of the dual-output decoder process. Edge prediction maps for edge supervision, while mask prediction represents the prediction results.
Loss function
For the edge prediction results, we use a combination of weighted and from36,37 as the loss function 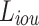 , and
, and 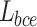 . The advantage of this loss function is that it assigns higher weights to pixels that are difficult to predict, thereby allowing the model to focus more on these pixels and achieve better prediction results. This is particularly effective for edge prediction, as the boundaries between teeth and the background in the input X-ray scans are blurred and difficult to segment. This loss function is detailed in Formulae 4.
. The advantage of this loss function is that it assigns higher weights to pixels that are difficult to predict, thereby allowing the model to focus more on these pixels and achieve better prediction results. This is particularly effective for edge prediction, as the boundaries between teeth and the background in the input X-ray scans are blurred and difficult to segment. This loss function is detailed in Formulae 4.
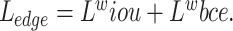 |
4 |
For the final prediction results, we use the loss function 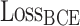 as the supervisory signal. The loss function is defined in Formulae 5,
as the supervisory signal. The loss function is defined in Formulae 5,
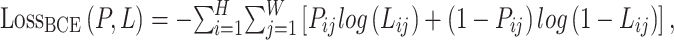 |
5 |
Where 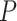 represents the model’s prediction, and
represents the model’s prediction, and 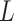 represents the label.
represents the label. 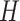 and
and 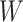 denote the width and height of the output image, respectively.
denote the width and height of the output image, respectively.
Finally, our total loss function 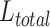 is defined in Formulae 6, where
is defined in Formulae 6, where 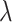 a hyperparameter, set to 0.8 in this paper to achieve the best results.
a hyperparameter, set to 0.8 in this paper to achieve the best results.
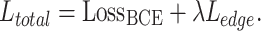 |
6 |
Experiments
Experimental setup
Our model was deployed in an environment equipped with PyTorch 1.10.0 and CUDA 11.8, and trained on a single RTX 4090 GPU with 24GB of VRAM. The training process consisted of 200 epochs with a batch size of 2. We utilized the Adam optimizer with a learning rate set to 1e-3. To maximize the preservation of the original image quality, we finely processed the X-ray images with an original size of 1991 × 1227 pixels: the width was mirror-padded to 2048 pixels, and the height was cropped to 1024 pixels. Subsequently, we applied bi-linear interpolation to downsample the adjusted X-ray images, obtaining X-ray images with widths and heights of 1024 and 512 pixels, respectively.
Dataset
Dental Panoramic Radiographs Dataset: This dataset was first introduced in38 as the first dataset specifically for caries segmentation and dental disease detection in pediatric panoramic radiographs. It includes X-ray scan images of children, with panoramic dental radiographs and related caries segmentation and dental disease detection cases from 106 child patients aged between 2 and 13 years. Additionally, the dataset incorporates three previously released adult dental datasets13,38,39 (2692 images). In total, this dataset comprises 3187 pairs of panoramic dental radiographs and corresponding MASKs, with each image being three-channel and having dimensions of 1991 × 1227 pixels. For edge ground truth, we utilize the Sobel operator in two directions (horizontal and vertical). Gradients are computed for the masks using the respective filters, resulting in the extracted edges.
Evaluation metrics
We adopted five widely used evaluation metrics: Accuracy, Mean IoU, Dice coefficient, Sensitivity (Sen), and Specificity (Spec).
Additionally, to more accurately measure the average error between pixels and visually represent the model’s average prediction error, we selected the Mean Absolute Error (MAE), which is widely used in the field of object detection, as an evaluation metric. The formula for MAE is as follows:
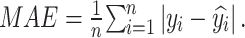 |
7 |
Each 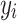 represents the value of each pixel in the predicted image, and
represents the value of each pixel in the predicted image, and 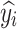 represents the corresponding pixel value
represents the corresponding pixel value 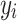 in the label image.
in the label image.
To further and comprehensively evaluate the differences in contrast, brightness, and structural quality between the predicted image and the label image, we specifically selected the image quality evaluation metric—Enhanced Alignment Measure (E-measure)40. The formula is expressed as follows:
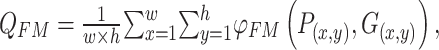 |
8 |
where 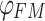 represents the enhanced alignment matrix, which applies a quadratic function to the enhanced alignment matrix. This metric combines precision and recall to calculate the global alignment measure. Additionally, it employs a weighted method by considering the different importance of foreground and background regions, thereby enhancing the accuracy of the alignment evaluation. The advantage of this method is that it takes into account the spatial distribution of the foreground and background, leading to more accurate evaluation results. It balances the impact of different regions, which reduces the sensitivity to imbalanced datasets.
represents the enhanced alignment matrix, which applies a quadratic function to the enhanced alignment matrix. This metric combines precision and recall to calculate the global alignment measure. Additionally, it employs a weighted method by considering the different importance of foreground and background regions, thereby enhancing the accuracy of the alignment evaluation. The advantage of this method is that it takes into account the spatial distribution of the foreground and background, leading to more accurate evaluation results. It balances the impact of different regions, which reduces the sensitivity to imbalanced datasets.
Experimental results and analysis
Comparative experiments
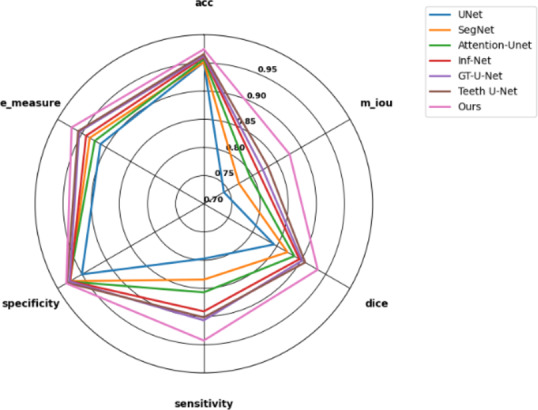
To comprehensively assess the segmentation performance of our model, we conducted extensive comparative experiments against several classical and improved segmentation models. These models include the classic U-Net18, Attention U-Net41, and SegNet20, as well as advanced models specifically designed for dental segmentation tasks such as GT-U-Net31, Teeth U-Net33, and Inf-Net32, which combines boundary and global guidance. The quantitative analysis results based on the dataset are detailed in Table 1.
Table 1.
Performance data of each model on each indicator.
| acc | m_iou | Dice | Sensitivity | Specificity | e_measure | MAE | |
|---|---|---|---|---|---|---|---|
| Unet | 0.9511 | 0.7390 | 0.8427 | 0.7943 | 0.9695 | 0.9115 | 0.0490 |
| SegNet | 0.9512 | 0.7719 | 0.8710 | 0.8332 | 0.9723 | 0.9337 | 0.0489 |
| Attention-Unet | 0.9566 | 0.7951 | 0.8857 | 0.8571 | 0.9759 | 0.9237 | 0.0435 |
| Inf-Net | 0.9604 | 0.8123 | 0.8963 | 0.8909 | 0.9744 | 0.9415 | 0.0411 |
| GT-U-Net | 0.9619 | 0.8212 | 0.9017 | 0.9067 | 0.9754 | 0.9558 | 0.0382 |
| Teeth U-Net | 0.9655 | 0.8318 | 0.9080 | 0.9015 | 0.9805 | 0.9579 | 0.0353 |
| Ours | 0.9742 | 0.8761 | 0.9338 | 0.9426 | 0.9821 | 0.9712 | 0.0259 |
The experimental results clearly indicate that traditional medical image segmentation models like U-Net18 and SegNet20 showed relatively limited performance. However, when the Attention U-Net41 incorporated an attention module into the U-Net architecture, the model gained stronger selective focus, thereby improving its performance. Models such as Inf-Net32, which integrates global information guidance, and GT-U-Net31 and Teeth U-Net33, designed specifically for dental segmentation, also achieved significant performance improvements, as evidenced by the results in Table 1. Notably, our proposed model exhibited outstanding performance across all evaluation metrics. It achieved the best results in the E-measure evaluation, surpassing all other models. This experimental outcome demonstrates that our model’s segmentation results closely align with the ground truth in terms of shape and position, proving its excellent segmentation quality. It also indicates that our model achieved the best alignment between the predicted results and the ground truth, ensuring optimal segmentation accuracy. Furthermore, our model exhibited superior performance in distinguishing between the foreground and background. It not only improved the accuracy of foreground and background recognition but also ensured that the predicted results closely matched the labels in terms of shape, position, and structure. This significant achievement in segmentation accuracy can be largely attributed to the introduction of additional edge supervision signals, which effectively enhanced edge prediction accuracy and maintained high consistency with the ground truth.
According to the evaluation standards defined in Table 1, our model achieved the best performance across various metrics on the dataset. This not only demonstrates its ability to maintain excellent performance when handling noisy and highly individualized datasets but also highlights its strong robustness and stability in dealing with fuzzy boundary features.
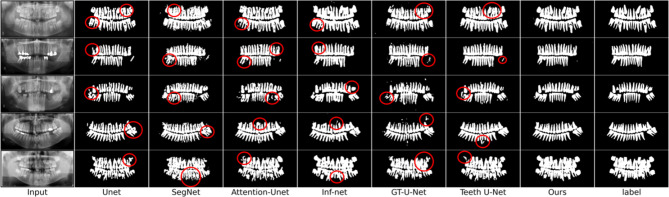
Visual comparison
As shown in Fig. 8, we visually compared the segmentation results of our model with other models. The results clearly illustrate that our model outperformed the others in segmentation effectiveness. Specifically, the segmentation results of GCNet were closer to the ground truth, significantly reducing missegmentation and more accurately depicting edges and contours.
Fig. 8.
Visualization comparison of prediction results for models. The red circles highlight the areas where the segmentation results are inaccurate.
In contrast, the performance of U-Net18 and SegNet20 was unsatisfactory. These models simply extracted features of different levels and resolutions without incorporating improved modules to capture common features across input images. The blurred boundaries in the input images further hindered their performance.
Attention U-Net42 showed some improvement due to its attention mechanism, which focuses on important parts of the input, optimizing feature extraction and information transfer. However, a single attention module was insufficient to fully extract effective information. GT U-Net31 combined the structural advantages of U-Net with the self-attention mechanism from Transformer22, using a grouping method to handle spatial dependencies of features. While this improved understanding of complex patterns, the high computational cost and loss of useful information due to convolutional dimensionality reduction, along with the lack of a global guidance module, limited its boundary segmentation performance.
Inf-Net30, using global guidance and edges as supervision signals, performed better than the previous models. However, its simple edge guidance module limited its ability to capture edge details effectively. Teeth U-Net33, designed for dental segmentation, demonstrated better results by using multi-scale attention blocks to capture irregular shapes and fuse features, aiding in low-contrast, overlapping panoramic X-ray images. However, it struggled with complex edges.
In contrast, GCNet generates the Grouped Global Map through the Grouped Global Attention Module to guide high-level features and uses edge labels as supervision signals. This mechanism effectively extracts comprehensive edge and texture information from the images. Additionally, our model employs Cross-Layer Fusion Modules to fuse features at deep layers and extracts and merges texture and contour information from low-level features in the Grouped Global Map. This method captures global contour details and fully leverages the correlation between local information, significantly enhancing segmentation performance.
Ablation experiment
In this section, we conduct ablation experiments on the core modules of our model, GGA, CLA, and DOD, to verify their effectiveness. The detailed performance is shown in Table 2.
Table 2.
Performance data of ablation experiments.
| DOD | CLF | GGA | acc | m_iou | Dice | Sensitivity | Specificity | e_measure | MAE |
|---|---|---|---|---|---|---|---|---|---|
| ✓ | 0.9503 | 0.7846 | 0.8922 | 0.8434 | 0.9735 | 0.9334 | 0.0461 | ||
| ✓ | 0.9591 | 0.8032 | 0.8928 | 0.8592 | 0.9767 | 0.9394 | 0.0429 | ||
| ✓ | 0.9631 | 0.8272 | 0.9039 | 0.9071 | 0.9764 | 0.9547 | 0.0376 | ||
| ✓ | ✓ | 0.9613 | 0.8168 | 0.9004 | 0.8934 | 0.9776 | 0.9458 | 0.0410 | |
| ✓ | ✓ | 0.9684 | 0.8518 | 0.9225 | 0.9304 | 0.9805 | 0.9619 | 0.0324 | |
| ✓ | ✓ | 0.9714 | 0.8618 | 0.9225 | 0.9304 | 0.9805 | 0.9619 | 0.0324 | |
| ✓ | ✓ | ✓ | 0.9742 | 0.8761 | 0.9338 | 0.9426 | 0.9821 | 0.9712 | 0.0259 |
Verifying the Effectiveness of DOD: In the baseline of Table 2, we replaced the top two decoders of Unet18 with the DOD module and used low-level features to decode an edge prediction result. This aimed to achieve better fusion of shallow and high-level features and accurate extraction of edge features. The experimental results clearly demonstrate that DOD, by outputting an additional edge prediction map as a supervision signal, plays a crucial role in improving model performance.
Verifying the Effectiveness of CLF: The effectiveness of CFM is evident in Table 2. By using MAS to efficiently fuse high-level features and share this information with each CLA, the model significantly enhances its performance in image segmentation tasks. This is achieved by accurately extracting local features from high-resolution features and fusing channel information from both high and low-resolution features.
Verifying the Effectiveness of GGA: According to the third row of data in Table 2, the model incorporating GGA shows significant improvement in all evaluation metrics. GGA, particularly GUA, efficiently learns the position and boundary features of the target regions in the samples. This module also serves as a supervision signal, guiding the targeted fusion of high-level features to ensure that both texture and semantic information are present in the features. This demonstrates that guiding the fusion of high-level features with shallow information can achieve superior segmentation results.
Verifying the Combined Use of DOD, CLF, and GGA: We conducted multiple combination experiments to explore the effects of different combinations of these three modules (as shown in rows 4, 5, and 6 of Table 2). The experimental results clearly indicate that the combined use of all three modules optimizes the model’s performance. Furthermore, when CLF and GGA are used together, the performance on various evaluation metrics is close to that of using all three modules simultaneously. This further emphasizes the importance of the combined use of shallow and high-level features and highlights their critical role in enhancing model performance.
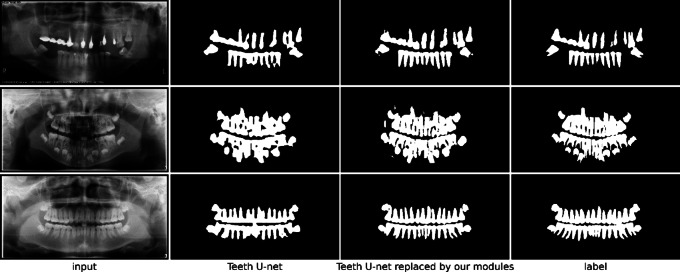
Module generalization experiment
To verify the generalization capabilities of our proposed modules, we conducted a series of experiments. In these experiments, we replaced the corresponding modules in Teeth U-Net33 with our designed modules. The experimental results showed significant performance improvements with the replaced modules (see Table 3), effectively demonstrating the good generalization and learning capabilities of our modules.
Table 3.
Performance data of replacing Teeth U-net modules by our modules.
| DSM replaced by CLA | DHAB replaced by GGA | acc | m_iou | Dice | Sensitivity | Specificity | e_measure | MAE |
|---|---|---|---|---|---|---|---|---|
| 0.9655 | 0.8318 | 0.9080 | 0.9015 | 0.9805 | 0.9579 | 0.0353 | ||
| ✓ | 0.9679 | 0.8432 | 0.9134 | 0.9249 | 0.9803 | 0.9591 | 0.0307 | |
| ✓ | 0.9694 | 0.8522 | 0.9243 | 0.9308 | 0.9809 | 0.9645 | 0.0289 | |
| ✓ | ✓ | 0.9742 | 0.8641 | 0.9274 | 0.9365 | 0.9814 | 0.9655 | 0.0273 |
In the experiments, we replaced the DSM and DHAB modules in Teeth U-Net33 with our CFM and GGA modules, respectively. The results indicated that the modified model achieved better performance across all evaluation metrics compared to the original model. Additionally, the visualizations of the prediction results (as shown in the Fig. 9) showed clear improvements. This finding strongly supports the effectiveness and generalizability of the CLF and GGA modules.
Fig. 9.
Visualization of prediction results before and after replacing the modules in Teeth U-net.
Experiment on the grouped global attention module
We conducted comparative experiments between the Grouped Uni-directional Attention (GUA) module and the self-attention(SA) method. The experimental results on the training and test sets are shown in Tables 4 and 5.
Table 4.
Performance data on training and testing set using GUA.
| GUA | acc | m_iou | Dice | Sensitivity | Specificity | e_measure | MAE |
|---|---|---|---|---|---|---|---|
| Training set | 0.9782 | 0.8934 | 0.9536 | 0.9571 | 0.9857 | 0.9757 | 0.0257 |
| Testing set | 0.9742 | 0.8761 | 0.9338 | 0.9426 | 0.9821 | 0.9712 | 0.0259 |
Table 5.
Performance data on training and testing set using SA.
| Self attention | acc | m_iou | Dice | Sensitivity | Specificity | e_measure | MAE |
|---|---|---|---|---|---|---|---|
| Training set | 0.9874 | 0.9434 | 0.9846 | 0.9618 | 0.9931 | 0.9874 | 0.0183 |
| Testing set | 0.9581 | 0.7995 | 0.8863 | 0.8568 | 0.9762 | 0.9233 | 0.0434 |
The loss values from the experiments (Figs. 10 and 11) indicate that using GUA in our model yields better results on the validation set compared to using self-attention method (Table 5). It is evident that self-attention for all pixels performs significantly worse on the validation set than on the training set. This suggests that self-attention is affected by noise points in the input images during training, causing the model to learn specific features of the input images rather than general features across all images. In contrast, using GUA allows the model to learn general features in small datasets with high individual variability and significantly reduces computational complexity, thereby saving computational costs.
Fig. 10.
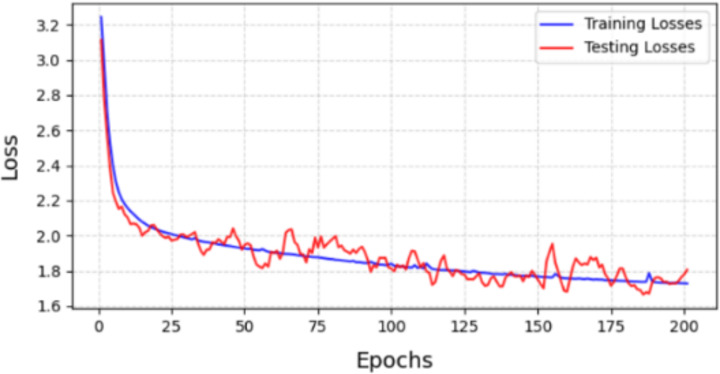
Loss of module between training set and testing set with GUA.
Fig. 11.
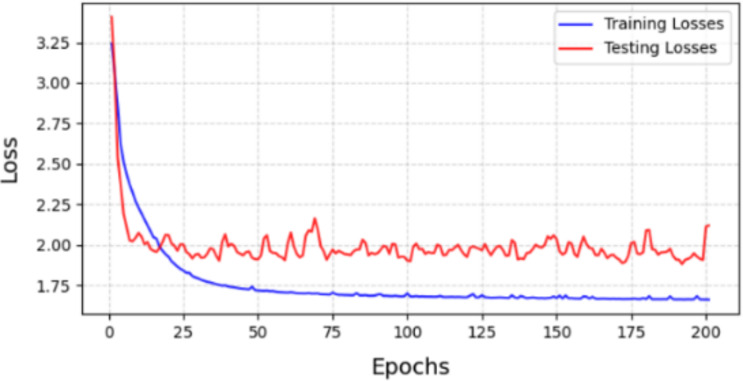
Loss of module between training set and testing set with SA.
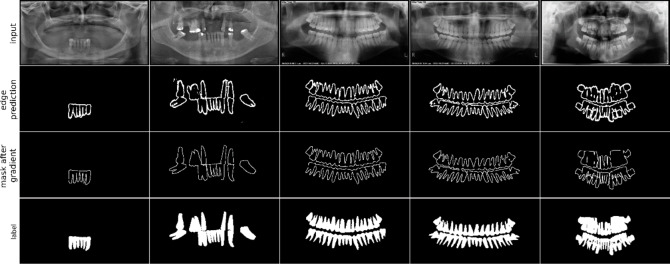
Experiment of edge supervision
We used FFA for edge prediction, enabling the model to better fit segmentation edges in noisy and low-quality inputs. The output results are shown in Fig. 12.
Fig. 12.
Visualization of edge prediction map.
Experimental validation revealed that combining DOD and GGA significantly improved the accuracy of edge prediction. This enhancement allows the model to fit blurred boundaries more precisely. The improvement is mainly due to the clever fusion of shallow and high-level features and the guidance of edge supervision signals, enabling the model to effectively infer those blurred or even barely discernible boundaries. Additionally, guiding high-level features to selectively acquire information through edge features results in the most accurate output.
Model performance in imbalanced data
Medical image data is often imbalanced due to patient privacy protection. In this dataset, for example, there are 3187 pairs of X-ray images and labels, with a larger proportion of adult data (2692 pairs) compared to pediatric data (492 pairs). This imbalance can bias the model’s predictions. When processing pediatric dental X-ray, the incomplete development of children’s teeth, with many hidden under the periodontal tissue or overlapping, increases prediction difficulty. The model may favor the more frequent adult data, neglecting the pediatric data. Consequently, while overall performance may be adequate, the model’s performance on pediatric data suffers significantly42. Recall and precision for the pediatric category may be noticeably lower, leading to missed samples (low recall) or prediction errors (low precision). High-frequency categories dominate gradient updates, making it difficult for the model to learn the minority category features adequately, affecting generalization.
In our experiments, we tested multiple models on both adult and pediatric X-ray data. The results for adult data are shown in Fig. 13, while the results for pediatric data are presented in Fig. 14.
Fig. 13.
Results of various models on adult data in the test set.
Fig. 14.
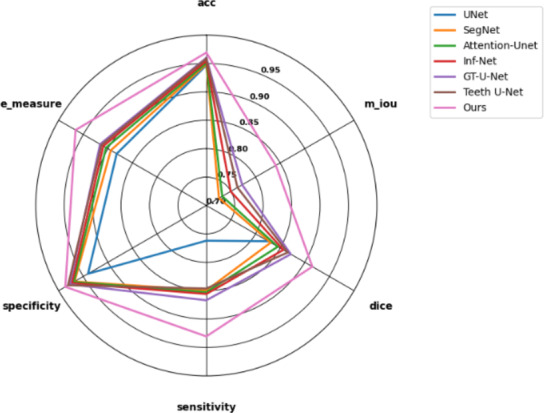
Results of various models on children data in the test set.
After thorough experimental validation, we found that GCNet significantly outperforms other similar models when processing pediatric X-ray datasets. More notably, compared to other models’ performance on adult X-ray datasets, GCNet showed the smallest performance gap. This significant result indicates that even in the presence of data imbalance, GCNet can effectively extract and learn general features from the data. This fully demonstrates GCNet’s strong anti-interference capability in complex data environments, further proving its robustness and reliability. Therefore, GCNet not only has theoretical superiority but also exhibits excellent segmentation capabilities in practical applications.
Future work
When dealing with small-scale X-ray datasets that have low contrast, significant individual variance, and noise interference, the model’s prediction results are often adversely affected. To optimize the model’s prediction performance, transfer learning strategies can be considered in the future. Specifically, the model can be initially trained on a large X-ray dataset to enhance its noise resistance and improve the accuracy of foreground and background segmentation. Subsequently, fine-tuning on the smaller dataset can be performed to improve the model’s performance in specific downstream tasks. Additionally, for input images with blurred edges and low contrast, image enhancement techniques such as contrast enhancement and denoising can be applied during the preprocessing stage to significantly improve segmentation outcomes. To address the limitations of traditional edge detection algorithms that rely on fixed parameters such as thresholds, future research can focus on developing adaptive edge detection algorithms. These algorithms can dynamically adjust parameters based on the specific content and quality of the images, thereby better accommodating datasets with significant individual variability.
Conclusion
This paper presents an innovative dental segmentation network framework—GCNet. By utilizing the Grouped Global Attention Module (GGA) and Cross-Layer Fusion Modules (CLF) in concert, this framework can accurately extract general features from dental X-ray images, resulting in precise segmentation outcomes. The experimental results demonstrate that the Grouped Uni-directional Attention Module mechanism within the GGA excels at capturing texture and positional information from low-level features, generating a Grouped Global Map that guides high-level features to focus on key areas. Additionally, the results confirm that the CLF effectively integrates features from three deep encoders, precisely extracting positional and texture information from low-level features under the guidance of the Grouped Global Map.
We also employed an efficient feature fusion strategy, combining features from both modules and outputting an additional edge prediction result as a supervisory signal to further enhance edge prediction accuracy. Experimental validation shows that the GUA mechanism outperforms self-attention when handling small datasets. This is primarily due to the significant individual variance in the input images, where complex networks and excessive parameters make models more susceptible to noise and increase computational complexity. The GUA mechanism effectively addresses this by extracting common information from the input, enhancing model stability and accuracy while reducing computational costs.
Our proposed strategy is not only applicable to dental segmentation tasks but can also be broadly applied to other medical image segmentation tasks, particularly those involving noisy images with blurred foreground and background boundaries. Considering the time-consuming nature of manual dental X-ray segmentation, GCNet is poised to become a valuable tool for medical professionals, significantly improving diagnostic efficiency and reducing their workload.
Acknowledgements
We are grateful to the Centre for Health Informatics, Australian Institute of Health Innovation, Macquarie University for their guidance and assistance with the experiments, and for their valuable suggestions on the manuscript.
Author contributions
W.Z. developed the model and conducted the experiments. W.Z. and X.R. drafted the main manuscript. H.Z. collected the dataset and prepared the figures. All authors reviewed and approved the manuscript.
Data availability
The code for this project will be publicly available at: https://github.com/ZJohnWenjin/GCNet-Automatic-X-ray-teeth-segmentation-with-Grouped-Attention.git upon acceptance.
Declarations
Competing interests
The authors declare no competing interests.
Ethical approval and informed consent
The data utilized in this study were obtained from publicly accessible databases that exclude any personally identifiable information. Hence, the question of informed consent does not apply. All procedures involving data handling adhered strictly to the ethical guidelines for research.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Litjens, G. et al. A survey on deep learning in medical image analysis. Med. Image Anal.42, 60–88 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Shen, D., Wu, G. & Suk, H-I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng.19, 221–248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim, H. E. et al. Transfer learning for medical image classification: a literature review. BMC Med. Imaging22, 69 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai, L., Gao, J. & Zhao, D. A review of the application of deep learning in medical image classification and segmentation. Ann. Transl. Med.8, 11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu, X. et al. A review of deep-learning-based medical image segmentation methods. Sustainability13, 1224 (2021). [Google Scholar]
- 6.Wang, R. et al. Medical image segmentation using deep learning: a survey. IET Image Proc.16, 1243–1267 (2022). [Google Scholar]
- 7.Fu, Y. et al. Deep learning in medical image registration: a review. Phys. Med. Biol.65, 20: 20TR01 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, J. et al. Transmorph: Transformer for unsupervised medical image registration. Med. Image Anal.82, 102615 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krizhevsky, A., Sutskever, I. & Hinton, G. E. ImageNet Classification with Deep Convolutional Neural Networks (2012).
- 10.Esteva, A. et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature542 (7639), 115–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajpurkar, P. et al. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. Preprint at http://arXiv.org/1711.05225 (2017).
- 12.Lin, P. L. et al. Teeth segmentation of dental periapical radiographs based on local singularity analysis. Comput. Methods Progr. Biomed.113 (2), 433–445 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Silva, G., Oliveira, L. & Pithon, M. Automatic segmenting teeth in X-ray images: Trends, a novel data set, benchmarking and future perspectives. Expert Syst. Appl.107, 15–31 (2018). [Google Scholar]
- 14.Mohammad-Rahimi, H. et al. Deep learning for caries detection: a systematic review. J. Dent.122, 104115 (2022). [DOI] [PubMed] [Google Scholar]
- 15.Wirtz, A. et al. Automatic teeth segmentation in panoramic X-ray images using a coupled shape model in combination with a neural network. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, September 16–20, Proceedings, Part IV 11 (Springer, 2018).
- 16.Liu, Y. et al. Richer convolutional features for edge detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2017).
- 17.Zhou, Z. et al. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIAand 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 20, 2018, Proceedings 4 (Springer, 2018). [DOI] [PMC free article] [PubMed]
- 18.Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5–9, Proceedings, Part III 18 (Springer, 2015).
- 19.Abdollahi, A., Pradhan, B. & Alamri, A. VNet: an end-to-end fully convolutional neural network for road extraction from high-resolution remote sensing data. IEEE Access8, 179424–179436 (2020). [Google Scholar]
- 20.Badrinarayanan, V., Kendall, A. & Cipolla, R., Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell.39, 2481–2495 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Bahdanau, D., Cho, K. & Bengio, Y. Neural machine translation by jointly learning to align and translate. Preprint at http://arXiv.org/1409.0473 (2014).
- 22.Vaswani, A. et al. Attention is all you need. Adv. Neural. Inf. Process. Syst.30, 1 (2017). [Google Scholar]
- 23.Dosovitskiy, A. et al. An image is worth 16x16 words: Transformers for image recognition at scale. Preprint at http://arXiv.org/2010.11929 (2020).
- 24.Arnab, A. et al. Vivit: A video vision transformer. In Proceedings of the IEEE/CVF International Conference on Computer Vision (2021).
- 25.Yu, F. & Koltun, V. Multi-scale context aggregation by dilated convolutions. Preprint at http://arXiv.org/1511.07122 (2015).
- 26.Woo, S. et al. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV) (2018).
- 27.Howard, A. G. et al. Mobilenets: Efficient convolutional neural networks for mobile vision applications. Preprint at http://arXiv.org/1704.04861 (2017).
- 28.Peng, C. et al. Large kernel matters–improve semantic segmentation by global convolutional network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2017).
- 29.Szegedy, C. et al. Going deeper with convolutions. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2015).
- 30.Ding, X. et al. Repvgg: Making vgg-style convnets great again. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (2021).
- 31.Li, Y. et al. Gt u-net: A u-net like group transformer network for tooth root segmentation. In Machine Learning in Medical Imaging: 12th International Workshop, MLMI 2021, Held in Conjunction with MICCAI 2021, Strasbourg, France, September 27, Proceedings 12 (Springer, 2021).
- 32.Fan, D-P. et al. Inf-net: automatic covid-19 lung infection segmentation from X-ray images. IEEE Trans. Med. Imaging39, 2626–2637 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Hou, S. et al. Teeth U-Net: a segmentation model of dental panoramic X-ray images for context semantics and contrast enhancement. Comput. Biol. Med.152, 106296 (2023). [DOI] [PubMed] [Google Scholar]
- 34.Zhe, W., Su, L. & Huang, Q. Cascaded partial decoder for fast and accurate salient object detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (2019).
- 35.Hu, J., Shen, J. & Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2018).
- 36.Qin, X. et al. Basnet: Boundary-aware salient object detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (2019).
- 37.Wei, J., Wang, S. & Huang, Q. F3Net: fusion, feedback and focus for salient object detection. Proc. AAAI Conf. Artif. Intell.34, 7 (2020).
- 38.Zhang, Y. et al. Children’s dental panoramic radiographs dataset for caries segmentation and dental disease detection. Sci. Data10, 380 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdi, A. & Kasaei, S. Panoramic dental X-rays with segmented mandibles. Mendeley Data2, 1 (2020). [Google Scholar]
- 40.Fan, D. P. et al. Enhanced-alignment measure for binary foreground map evaluation. IJCAI1, 698–704 (2018). [Google Scholar]
- 41.Oktay, O. et al. Attention u-net: Learning where to look for the pancreas. Preprint at http://arXiv.org/1804.03999 (2018).
- 42.Cong, C. et al. Adaptive unified contrastive learning with graph-based feature aggregator for imbalanced medical image classification. Expert Syst. Appl.251, 123783 (2024). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The code for this project will be publicly available at: https://github.com/ZJohnWenjin/GCNet-Automatic-X-ray-teeth-segmentation-with-Grouped-Attention.git upon acceptance.