Abstract
Our study explores the complex dynamics of the integrated stress response (ISR) axis, highlighting PIM2 kinase’s critical role and its interaction with the BCL2 protein family, uncovering key mechanisms of cell survival and tumor progression. Elevated PIM2 expression, a marker of various cancers, often correlates with disease aggressiveness. Using a model of normal and malignant plasma cells, we show that inhibiting PIM2 kinase inhibits phosphorylated BAD production and activates ISR-mediated NOXA expression. This shift towards MCL1 dependence underscores the synergy achieved through combined PIM/MCL1 inhibition, driven largely by ISR-mediated NOXA expression. In mouse xenograft models, dual targeting of PIM2 and MCL1 effectively controls tumor growth—a response reversed by ISR-specific inhibition and upregulation of genes linked to tumor cell dissemination. This work elucidates the molecular intricacies of PIM2 inhibition and its implications for cancer therapy, especially in tumors with elevated PIM2 expression.
Subject terms: Molecular medicine, Apoptosis, Cancer therapy
The integrated stress response (ISR) allows cells to restore cellular homeostasis upon a wide range of stress stimuli. Here, the authors show that PIM2 inhibition activates the ISR and promotes NOXA expression, in turn making tumour cells vulnerable to MCL1 inhibition.
Introduction
The PIM serine/threonine kinase family comprises three members: PIM1, PIM2, and PIM3. Known for their oncogenic properties, these kinases are tightly regulated in normal tissues but often upregulated in various cancers1. Among them, PIM2 has garnered particular attention due to its association with advanced stages of cancers such as prostate cancer and multiple myeloma, highlighting its significance in oncology research2–4. While the precise mechanisms underlying PIM2’s role in cancer progression remain incompletely understood, its involvement in promoting cell survival has emerged as a focal point in oncological investigations5.
Initially recognized in the early 2000s as a crucial transcriptionally regulated serine/threonine kinase implicated in apoptosis inhibition through phosphorylating BAD6,7, PIM2 plays a pivotal role in promoting cell survival, especially within B cells destined to become antibody-secreting cells (ASCs)8. ASCs, including plasma cells (PCs), express elevated levels of PIM2, particularly during prolonged endoplasmic reticulum (ER) stress induced by robust immunoglobulin (Ig) secretion, which triggers the unfolded protein response (UPR) signaling cascade9–12. Given the longevity of PCs within their specialized niches, maintaining a delicate balance to prevent inadvertent apoptosis becomes paramount.
While the ER stress/UPR axis primarily safeguards cellular integrity, it can induce apoptosis under certain conditions13, with the mitochondrion acting as a central sensor organelle of cellular stress. The pro-apoptotic proteins BAX and BAK transition from latent monomers to toxic oligomers upon activation, disrupting the mitochondrial outer membrane potential (MOMP) and initiating the caspase cascade. Conversely, anti-apoptotic members like BCL2, BCL-XL, and MCL1 counteract mitochondrial damage by sequestering activated BAX/BAK monomers. BH3-only proteins function as cellular stress sensors and transmit pro-apoptotic signals to the core apoptotic machinery14. MCL1, characterized by its short half-life, undergoes regulation through proteasomal degradation via ubiquitination, as well as caspase cleavage15,16. Notably, while MCL1 binds BH3-only proteins like PUMA, BIM, and NOXA, only the latter binds MCL1 with high affinity and promotes its ubiquitination and degradation, culminating in apoptotic activation17–21.
In our study, we explore how PIM2 modulates mitochondrial apoptosis in primary human ASCs and multiple myeloma cell lines (MMCLs). We find that PIM2 inhibition activates the integrate stress response (ISR)-related ATF4/CHOP axis, leading to NOXA expression in parallel with pBAD suppression. This highlights PIM2’s role in the ISR pathway and its impact on cell survival in PCs. The resulting increase in BH3-only proteins binding to MCL1 amplifies apoptotic priming. We further investigate the critical role of ISR-mediated NOXA expression in the synergy observed with combined PIM and MCL1 inhibitors in MMCLs8, confirming this effect both in vitro and in vivo.
Results
Modulating cell viability in B cell differentiation to plasma cells: effects of BH3 mimetics and PIM kinase inhibition
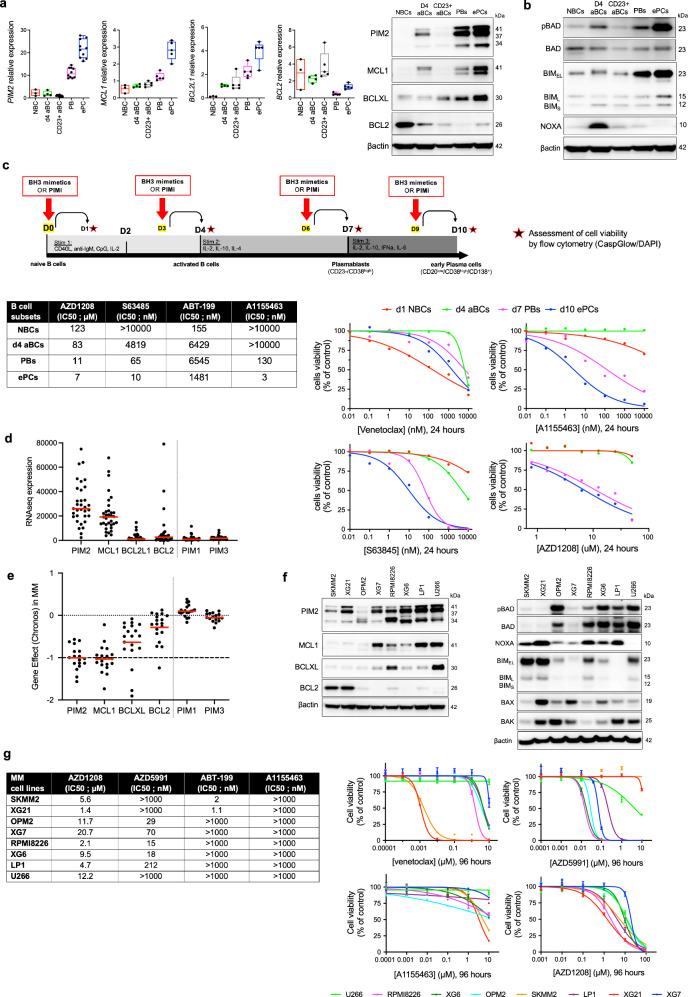
In vitro differentiation of human naïve B cells (NBCs) generates activated B cells (aBCs) by day 4, which can further differentiate into plasmablasts (PBs) by day 5-6, and mature into early PCs by day 10. However, some cells retain their B cell identity, becoming CD23+ aBCs22. We examined the expression of anti-apoptotic proteins BCL2, BCL-XL, MCL1, and PIM kinases in these populations. BCL2 expression declined rapidly, while BCL-XL, MCL1, and PIM2 levels increased as ASCs differentiated (Fig. 1a). PIM1 peaked in day 4 aBCs and decreased thereafter, whereas PIM3 appeared on day 4 but was not maintained later (Supplementary Fig. 1a, b). To further investigate PIM kinase expression, we analyzed RNAseq data from memory B cells, various PCs, 33 MMCLs, and a cohort of 97 myeloma patients previously investigated8. These data confirmed the predominant expression of PIM2 in PC populations, a significant expression of PIM1 in mature bone marrow PCs but not in myeloma patients, and generally low expression of PIM3 (Supplementary Fig. 1c).
Fig. 1. B Cell Differentiation and Response to BH3 Mimetics and PIM Kinase Inhibition.
a mRNA and protein levels during B cell differentiation across subsets and time points. Data are shown as floating bars (min to max, mean as line) for NBC (n = 3) and as box plots (median ± IQR, whiskers min to max) for d4 aBC (n = 4), CD23+ aBC (n = 5), PB (n = 5), and ePC (n = 5). b Protein levels across B cell and PC subsets. Samples derived from the same experiment with two different processed gels, including one for pBAD, BAD, and BIM (same gel of Fig. 1a), and another for NOXA. The blot is representative of 2 independent experiments. c Schematic of the assay design during B cell differentiation: cells were treated at specified time points with BH3-mimetic molecules and AZD1208. Data are representative of one experiment. Cell viability was assessed by flow cytometry. Percentage of cell viability were normalized to control condition and IC50 were calculated. d RNAseq expression of anti-apoptotic factors and PIM kinases in 33 MMCLs. Data are shown as mean (n = 33, ‘n’ denotes the number of distinct MMLs). e CRISPR screening data from DepMap in 17 MMCLs: dependency scores showing stronger dependency on PIM2 and MCL1 than on BCL-XL and BCL2. No dependency was observed for PIM1 or PIM3. Data are shown as mean (n = 17, ‘n’ denotes the number of distinct MMLs). f Protein levels of antiapoptotic and proapoptotic factors in MMCLs. Samples derived from the same experiment with different processed gels, including one for PIM2, MCL1, BCLXL, BCL2, p-BAD, BAD, BIM, NOXA and another for BAK, BAX. The blot is representative of 2 independent experiments. g Cell viability in MMCLs after treatment with BH3-mimetics and AZD1208, with calculated IC50 values. Data are shown as mean +/− SEM (Venetoclax, n = 3; AZD5991 and AZD1208, n = 4; A1155463, n = 2). NBC, naïve B cell; d4 aBC, day-4 activated B cell; CD23+ aBC, CD23-positive activated B cell; PB, plasmablast; ePC, early plasma cell; MMCLs, multiple myeloma cell lines; PIMi, pan-PIM inhibitor AZD1208. “n”, denotes the number of biological replicates except for Fig. 1d and Fig. 1e as indicated. Source data are provided as a Source Data file.
Additional protein detection revealed high levels of the proapoptotic protein BIM, while the MCL1 antagonist NOXA was absent. This absence may contribute to reduced initiation of apoptosis in ASCs compared to post-activation B cells8 (Fig. 1b). Furthermore, we evaluated the response of these cell populations to BH3 mimetics targeting BCL2, BCL-XL and MCL1 in parallel with the pan-PIM inhibitor AZD1208, revealing differential sensitivities based on the expression levels of these anti-apoptotic factors (Fig. 1c). The IC50 calculations and correlation with mRNA expression suggested that the expression profiles of BCL2 family proteins, particularly BCL-XL, MCL1, and PIM2 play a critical role in balancing the increase of pro-apoptotic BIM to maintain the viability and functionality of differentiated ASCs (Fig. 1c, Supplementary Fig. 1d).
In our study of MMCLs, we examined the expression of key apoptotic factors and their response to various drug inhibitors. RNA-seq data from 33 MMCLs revealed high levels of MCL1 and PIM2 expression, with lower levels of PIM1, PIM3, BCL2L1/BCL-XL, and BCL2 (Fig. 1d). Some MMCLs with the t(11;14) translocation showed particularly high levels of BCL2 gene expression. We also examined the RNA expression of other anti-apoptotic factors and found that BCL2A1/BFL1 and BCL2L2/BCL-W are expressed at very low levels in MMCLs, particularly compared to BCL2L1/BCL-XL and BCL2 (Supplementary Fig. 2a, b). Data from the CRISPR/Cas9 DepMap portal project (https://depmap.org/portal/) revealed the dual co-dependency of MMCL on PIM2 and MCL1, confirming MMCL reliance on PIM2 and MCL1 (Fig. 1e). In addition, RNA expression of BCL2 family pro-apoptotic molecules revealed that only BCL2L11/BIM, BAK1, BAX and to a lesser extent BMF genes had significant expression, in contrast to PMAIP1/NOXA and BCL2L8/BAD.
We then selected eight MMCLs based on their PIM2 protein expression (Fig. 1f, Supplementary Fig. 3a, b). BCL2-positive SKMM2 and XG21 cells exhibited low levels of BCL-XL and MCL1, but high levels of BIM and NOXA at both RNA and protein levels. (Fig. 1f, Supplementary Fig. 2b). An IC50 study showed that MCL1-positive MMCLs, except for U266 cells, were sensitive to the MCL1 inhibitor (Fig. 1g). Early PCs were sensitive to BCL-XL inhibition, while tumor PCs were resistant. SKMM2 and XG21 cells were sensitive to venetoclax, and all eight MMCLs were sensitive to the PIM inhibitor, albeit at higher doses than ePCs (Fig. 1g).
Our study highlights a significant shift in proapoptotic protein expression as B cells transition to ASCs, marked by reduced BCL2 and increased BCL-XL and MCL1, correlated with PIM2 presence. Notably, this alters sensitivity to BH3 mimetics, particularly showing remarkable susceptibility to MCL1 inhibition in both normal and malignant ASCs. These findings underscore the critical role of prosurvival BCL2 family proteins in regulating the survival and function of PCs during antibody production.
PIM2 inhibition elicits pBAD suppression and concurrent enhancement of NOXA expression
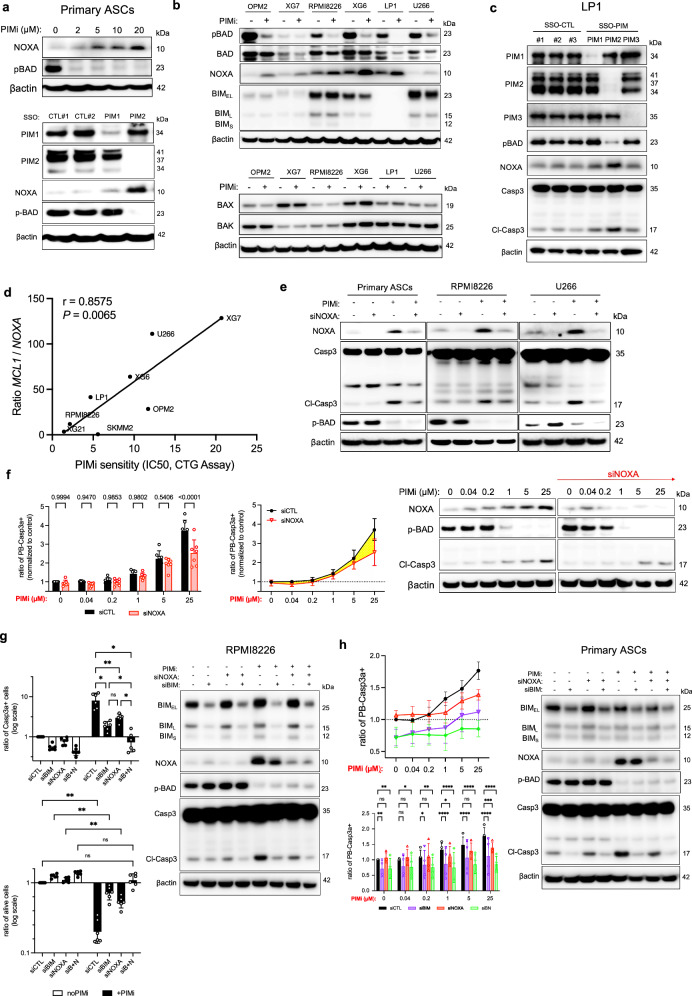
Our investigation revealed a notable pattern in NOXA expression, with a distinct increase observed during the transition to day 4 aBCs (Fig. 1b). Interestingly, as B cells differentiate into PCs, phosphorylation of BAD occurs, leading to its cytoplasmic sequestration, a process facilitated by PIM2 kinase activity8. This coincides with the complete loss of NOXA expression (Fig. 1b). To explore this relationship, we assessed the effects of escalating doses of the pan-PIM inhibitor AZD1208. Our results revealed a dose-dependent induction of NOXA expression in primary PBs and PCs, hereafter referred to as ASCs (Fig. 2a). Moreover, the reduction of pBAD and the increase in NOXA expression were particularly associated with PIM2 activity, rather than PIM1, as demonstrated by targeted morpholino antisense oligonucleotide experiments (Fig. 2a, Supplementary Fig. 4a).
Fig. 2. PIM2 Inhibition Modulates pBAD and NOXA Expression.
a ASCs were treated with increasing doses of AZD1208 for 6 h or with SSO-PIM for 24 h. Protein levels were assessed by immunoblotting. The blot is representative of 2 independent experiments. b MMCLs were treated with AZD1208 for 24 h. Protein levels were assessed by immunoblotting. Samples were from the same experiment, with different gels processed parallel. The blot is representative of 2 independent experiments. c Cells were treated with SSO-PIM for 24 h. Protein levels were assessed by immunoblotting with different processed gels, including one for PIM1, PIM2, another for PIM3 and pBAD, and another for NOXA and Caspase 3. The blot is representative of 2 independent experiments. d Correlation analysis between MCL1/PMAIP1 expression ratio with AZD1208 IC50 values. e Cells were transfected with NOXA or control siRNA, and then treated with AZD1208 for 6 h. Protein levels were assessed by immunoblotting. The blot is representative of 2 independent experiments. f ASCs were transfected with NOXA or control siRNA, and then treated with increasing doses of AZD1208 for 6 h. Caspase 3-active ASCs were analyzed by flow cytometry. Data were normalized to the control. Data are shown as mean ± SD (n = 7). Protein levels were assessed by immunoblotting. The blot is representative of 2 independent experiments. g Cells were transfected with NOXA, BIM, BIM + NOXA (BN) siRNA, or control siRNA, and then treated with AZD1208 for 24 h. Caspase 3-active cells and cell viability were analyzed by flow cytometry. Data are shown as mean ± SD (n = 6). Protein levels were assessed by immunoblotting. The blot is representative of 2 independent experiments. h ASCs were transfected with NOXA, BIM, BIM + NOXA (BN) or control siRNA, and then treated with AZD1208 for 6 h. Caspase 3-active ASCs were analyzed by flow cytometry. Data are shown as mean ± SD (n = 4). Protein levels were assessed by immunoblotting, with different processed gels, including one for NOXA, pBAD, and BIM, and another for Caspase 3. The blot is representative of 2 independent experiments. P-values were calculated using Pearson’s test (two-tailed) with linear regression for (d), two-way ANOVA with Holm-Sidak’s adjustment for (f), two-way ANOVA with Tukey adjustment for (f) and (g), and two-way ANOVA with Dunnett adjustment for (h). P < 0.05 is indicated as *P < 0.01 as **P < 0.001 as *** and P < 0.0001 as ****. Exact p-values are indicated in the Source Data file. “n”, denotes the number of biological replicates. Source data are provided as a Source Data file. ASCs, antibody-secreting cells; PIMi, pan-PIM inhibitor AZD1208; CTL, control; MMCLs, multiple myeloma cell lines; SSO, splice-switching oligonucleotide.
Similar trends were observed in MMCLs, where PIM2 inhibition, but not PIM1 or PIM3 inhibition, led to decreased pBAD levels and increased NOXA expression, while BIM, BAX, and BAK levels remained stable. This underscores PIM2’s specific role in regulating certain pro-apoptotic molecules (Fig. 2b, c, Supplementary Fig. 4b). Interestingly, an inverse correlation was found between MCL1 to PMAIP1/NOXA gene expression ratio and AZD1208 IC50 values in MMCLs, suggesting that high MCL1 and low NOXA expression correlate with reduced drug sensitivity in tumor cells (Fig. 2d).
To gain insights, we employed siRNA targeting PMAIP1/NOXA expression. In primary ASCs and RPMI8226 and U266 MMCLs, AZD1208 treatment in siNOXA-transfected cells effectively inhibited NOXA protein production. Interestingly, this led to a concurrent decrease in Caspase 3 cleavage in the absence of pBAD expression, indicating reduced apoptosis (Fig. 2e, Supplementary Fig. 4c). In PBs, NOXA deletion notably attenuated Caspase 3 activity at 25 µM AZD1208, highlighting its specific role in Caspase 3 activation following PIM kinase inhibition. These findings suggest that while pBAD inhibition alone initiates apoptosis at low AZD1208 doses, full Caspase 3 activation occurs above 5 µM when NOXA induction is achieved (Fig. 2f; Supplementary Fig. 5).
We next examined the impact of inhibiting both NOXA and BIM in the context of AZD1208 treatment to elucidate their individual contributions to apoptosis induced by PIM inhibition. In RPMI8226 cells, after 48 h of exposure to AZD1208, silencing NOXA (siNOXA) or BIM (siBIM) reduced Caspase 3 cleavage, with siBIM demonstrating greater efficacy than siNOXA alone. However, the most pronounced reduction in Caspase 3 cleavage was observed when both molecules were simultaneously inhibited, compared to each inhibition alone (Fig. 2g). Given our interest in understanding the distinct roles of NOXA and BIM in MMCLs, we extended this analysis to primary PBs. Our results showed that siBIM significantly inhibited apoptosis even in the absence of PIM inhibitor treatment, while siNOXA had no effect. This finding is consistent with the high expression of BIM and the minimal expression of NOXA in PBs under control conditions. Overall, siBIM was more effective than siNOXA in controlling Caspase 3 activation induced after PIM inhibition, with dual depletion proving significantly more efficient than either condition alone (Fig. 2h).
Our comprehensive findings provide compelling evidence that PIM2 inhibition induces cell death by suppressing pBAD levels and concomitantly inducing NOXA expression, which specifically contributes to apoptosis induced by the pan-PIM inhibitor. These results underscore the pivotal role of PIM2 in regulating the interplay between BH3-only proteins and BCL-XL/MCL1 at the mitochondrial level in ASCs, profoundly influencing cell survival and the delicate balance between survival and apoptosis.
Integrated stress response (ISR) activation, NOXA induction, and Caspase 3 activation in response to PIM2 inhibition
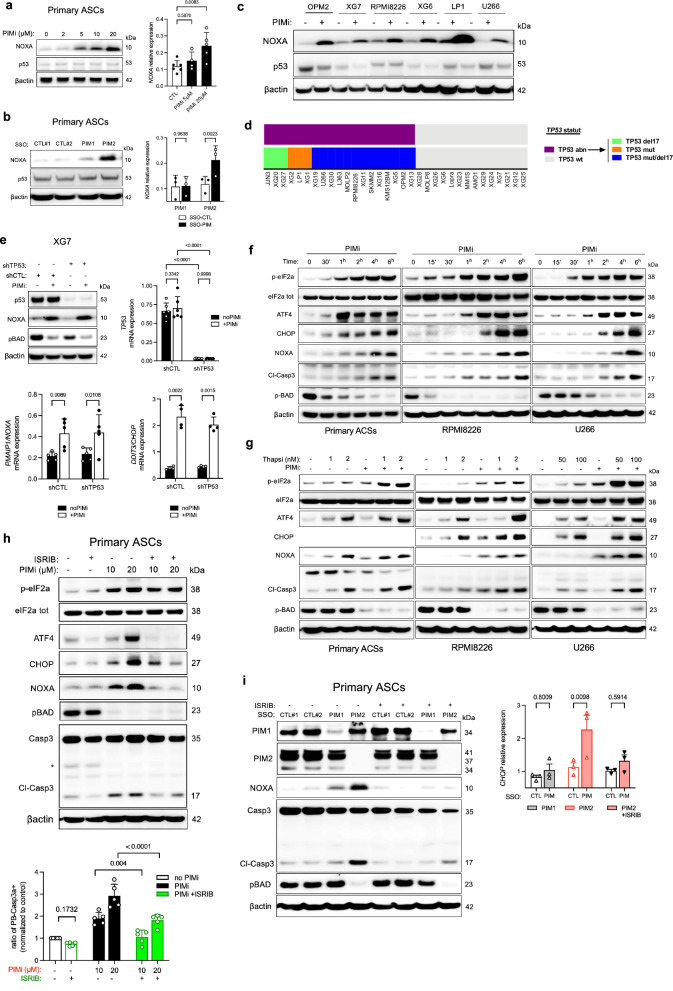
The induction of NOXA protein following PIM2 inhibition provides additional insight into B cell differentiation. Although NOXA was originally identified as a proapoptotic BH3-only protein induced by p53, our study found that PIM inhibition did not affect p53 expression, despite a concurrent increase in NOXA levels in both primary ASCs and selected MMCLs (Fig. 3a, Supplementary Fig. 6a). This pattern persisted even when using morpholinos targeting PIM2 or PIM1, indicating that NOXA induction by PIM2 inhibition is independent of the p53 pathway (Fig. 3b). Treatment with the pan-PIM inhibitor AZD1208 did not alter p53 protein expression in any of the six MMCLs tested (Fig. 3c). Additionally, four of the cell lines harbored TP53 mutations or deletions, further excluding p53 dependence. Notably, XG7 and XG6 cells are wild-type for the TP53 gene (Fig. 3 d) (23). To definitively rule out the involvement of p53 in NOXA production following AZD1208 treatment, we used the XG7 cell line and compared the drug response between cells depleted of p53 via shRNA (shTP53) and control cells (shCTL). As anticipated, p53 depletion did not affect NOXA production after PIM inhibition (Fig. 3e).
Fig. 3. Impact of PIM2 Inhibition on ISR Activation and Apoptosis.
a ASCs were treated with increasing doses of AZD1208 for 6 h. Protein levels were assessed by immunoblotting. mRNA expression was assessed by qPCR. Data are shown as mean ± SD (CTL, n = 6; PIMi 5 µM, n = 4; PIMi 20 µM, n = 5). b ASCs were treated with SSO-PIM for 24 h. mRNA expression was assessed by qPCR. Data are shown as mean ± SD (n = 3). c MMCLs were treated with AZD1208 for 24 h. Protein levels were assessed by immunoblotting. The blot is representative of 2 independent experiments. d TP53 mutational and deletion status in 33 MMCLs identifying three TP53 abnormal (TP53 abn) profiles. Cell lines without abnormalities were classified as wild type (TP53 wt). e shTP53 XG7 cells were treated with AZD1208 for 8 h. Protein expression was assessed by immunoblotting. mRNA expression assessed by qPCR. Data are shown as mean ± SD (TP53, n = 6; PMAIP1, n = 5; DDIT3, n = 4). f Cells were treated with AZD1208 and harvest at different time points. Protein levels were assessed by immunoblotting. All markers were processed on the same gel, except for p-BAD processed on a different gel. The blot is representative of 2 independent experiments. g Cells were treated with AZD1208 for 2 h, followed by thapsigargin for 4 h. Protein levels were assessed by immunoblotting. The blot is representative of 2 independent experiments. h ASCs were pre-treated with ISRIB before adding AZD1208 for 6 h. Protein levels were assessed by immunoblotting. Caspase 3-active ASCs were analyzed by flow cytometry. Data were normalized to the control condition. Data are shown mean ± SD (n = 5). i ASCs were pretreated with ISRIB for 2 h followed by SSO-PIM treatment. Protein levels were assessed by immunoblotting and mRNA expression was assessed by qPCR. Data are mean ± SEM (n = 3). P-values were calculated using one-way ANOVA with Dunnett’s adjustment for (a), and two-way ANOVA with Sidak’s adjustment for (b), (h), and (i), and for (e). “n”, denotes the number of biological replicates. Source data are provided as a Source Data file. ASCs, antibody-secreting cells; PIMi, pan-PIM inhibitor AZD1208; Thapsi, thapsigargin; MMCLs, multiple myeloma cell lines; SSO, splice-switching oligonucleotide.
The ATF4-CHOP signaling axis, activated by the ISR, transcriptionally upregulates PMAIP1/NOXA expression independently of p5323–25. AZD1208 treatment, and especially selective PIM2 inhibition, correlated with increased expression of PMAIP1/NOXA and DDIT3/CHOP genes (Fig. 3a, b, f, Supplementary Fig. 6b). In primary ASCs and MMCLs, this was mirrored by upregulated expression of upstream proteins such as p-eIF2α, ATF4, CHOP, and NOXA (Fig. 3f, g). Cellular stress induction with thapsigargin (Tg), an ER stress inducer, alongside AZD1208 treatment, revealed a sequential induction of ISR, NOXA expression, and Caspase 3 cleavage, potentiated in the presence of Tg in both ASCs and MMCLs (Fig. 3h, Supplementary Fig. 6c). These findings confirm that NOXA expression is influenced by ISR activation upon PIM inhibition, with heightened cellular response to AZD1208 under increased stress conditions.
For further validation, we used ISRIB, a specific ISR inhibitor, to assess its impact on NOXA expression and Caspase 3 activation during PIM inhibition. In ASCs, ISRIB effectively suppressed ATF4/CHOP expression, preventing NOXA induction and subsequent Caspase 3 activation independently of pBAD inhibition (Fig. 3i). Similarly, ISRIB treatment alongside targeted PIM2 depletion via morpholinos improved cell viability (Fig. 3j, Supplementary Fig. 6d). These observations were confirmed in all tested MMCLs, with ISRIB counteracting ISR activation, NOXA expression, and Caspase 3 activation triggered by AZD1208, again independently of pBAD (Supplementary Fig. 6e). Notably, the moderate or minimal Caspase 3 activation in specific MMCLs, particularly XG7 and U266, may be attributed to the short 6-hour exposure to AZD1208.
In summary, our investigation demonstrates a consistent response to PIM2 kinase inhibition in both ASCs and MMCLs, particularly under conditions of heightened cellular stress. This response involves activation of the ISR signaling pathway and subsequent upregulation of the proapoptotic NOXA protein. Notably, NOXA’s role in Caspase 3 activation remains significant regardless of p53 expression, emphasizing its pivotal role. Furthermore, this impact is distinct from the decline in pBAD levels, directly attributed to PIM kinase inhibition.
Augmented apoptotic priming via PIM inhibition: loading of pro-apoptotic BH3-only proteins onto MCL1
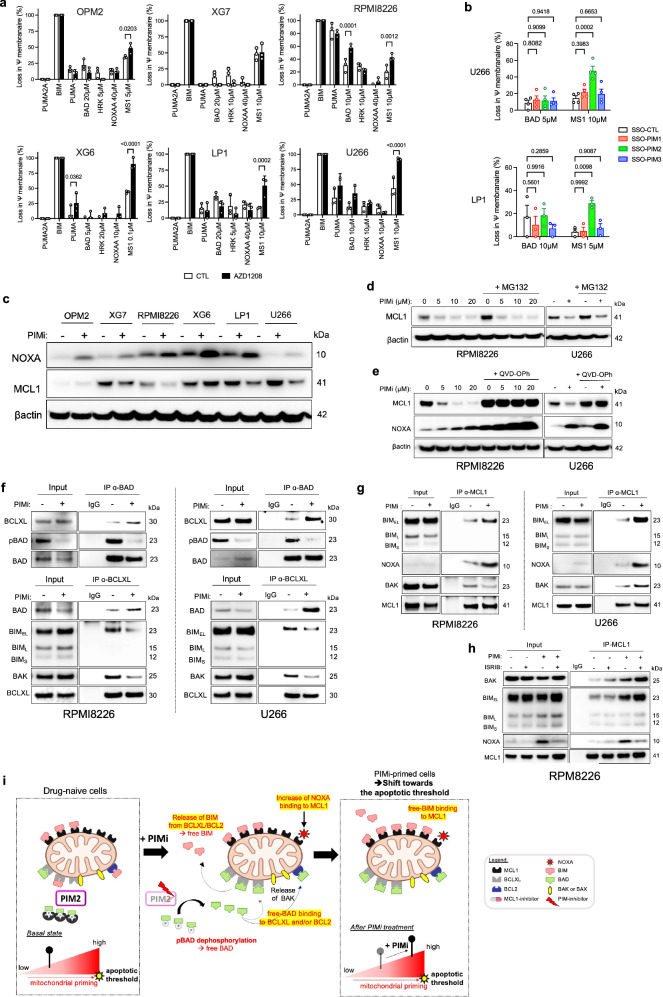
The analysis of ASCs under PIM2 inhibition reveals a complex interplay of molecular events reshaping the apoptotic landscape. Loss of pBAD liberates BAD, coinciding with increased PMAIP1 transcription and NOXA protein. This likely promotes proapoptotic protein influx to mitochondria, altering apoptotic priming and cellular dependence on pro-survival molecules. BH3 profiling of six MCL1-positive MMCLs treated with AZD1208 reveals consistent and significant mitochondrial depolarization when exposed to the MCL1-specific MS1 peptide following PIM inhibition. This effect is statistically significant for PIM2 depletion, but not for the depletion of PIM1 and PIM3, even in LP1 cells expressing highly all three kinases (Fig. 4a, b). Additionally, RPMI8226 cells show increased dependence on BCL-XL/BCL2, likely due to the high expression of BCL-XL (Fig. 1e, and Fig. 4a).
Fig. 4. Enhanced apoptotic priming through PIM inhibition-induced loading of pro-apoptotic BH3-only proteins onto MCL1.
a MMCLs were treated with AZD1208 for 16 h, followed by BH3-profiling. Data are shown as mean +/− SEM (n = 3). b Cells were treated with specific SSO-PIM for 24 h, followed by BH3 profiling. Data are shown as mean +/− SEM (LP1, n = 3; U266, n = 4). c MMCLs were treated with AZD1208 for 24 h. Protein levels were assessed by immunoblotting (the same gel as in Fig. 2b, β-actin was the same as in Fig. 2b). The blot is representative of 2 independent experiments. d Cells were treated with AZD1208 for 16 h, followed by MG132 addition for 3 h. Protein levels were assessed by immunoblotting. The blot is representative of 2 independent experiments. e Cells were treated with AZD1208 for 24 h with or without the pan-Caspase inhibitor QVD-OPH. Protein levels were assessed by immunoblotting. The blot is representative of 2 independent experiments. f Cells were treated with AZD1208 for 6 h. Immunoprecipitation of BAD and BCL-XL evaluated their interactions with BCL-XL, BIM, pBAD, and BAD. Samples of the IP-BCLXL were processed on different gels, one for BAD and other for BIM and BAK. The blot is representative of 1 independent experiment. g Cells were treated with AZD1208 for 6 h. Immunoprecipitation of MCL1 assessed its interaction with BAK, BIM, and NOXA. The blot is representative of 1 independent experiment. h Cells were treated with ISRIB for 2 h before addition of AZD1208 for 6 h. Immunoprecipitation of MCL1 evaluated its interaction with BAK, BIM, and NOXA. The blot is representative of 1 independent experiment. i Apoptotic Priming Pattern upon PIM Inhibition: plasma cells exhibit basal priming levels. PIM2 inhibition increases the pool of unphosphorylated BAD available for BCL-XL binding and BIM release. BIM and BAK bind to free MCL1 sites. NOXA induction further enhances MCL1 binding. Excess free pro-apoptotic molecules may cross the apoptotic threshold. Created with BioRender.com. P-values were calculated using ordinary two-way ANOVA with Sidak’s adjustment for a and b. “n”, denotes the number of biological replicates. Source data are provided as a Source Data file. PIMi, pan-PIM inhibitor AZD1208, MMCLs, multiple myeloma cell lines; SSO, splice-switching oligonucleotide.
At the protein level, no significant changes were observed for BCL2 and BCL-XL after PIM inhibition (Supplementary Fig. 7a). However, in some MMCLs, particularly U266 and RPMI8226, a decrease in MCL1 protein levels coinciding with increased NOXA expression was observed (Fig. 4c, Supplementary Fig. 7a). Despite previous findings linking NOXA binding to MCL1 degradation17, our investigations suggest an alternative mechanism for MCL1 decline, independent of NOXA binding. Indeed, addition of the proteasome inhibitor MG132 did not reverse the decrease in MCL1 protein levels, and ubiquitinated forms of MCL1 remained unaffected (Fig. 4d, Supplementary Fig. 7b). Interestingly, the pan-caspase inhibitor QVD-OPH prevented the decrease in MCL1 protein levels, even in the presence of high NOXA expression following PIM inhibition (Fig. 4e). Additionally, cycloheximide assays with QVD-OPH indicated that PIM inhibition does not directly impact MCL1 protein stability (Supplementary Fig. 7c). These results were consistent in primary ASCs (Supplementary Fig. 7d, e). Collectively, these findings highlight the complexity of molecular interactions following PIM inhibition in PCs, suggesting that PIM inhibition-induced NOXA expression may mediate MCL1 reduction through caspase activation16, independently of ubiquitin-mediated degradation observed in other contexts26–29.
The interplay between pro-apoptotic BH3-only proteins and their interactions with BCL-XL and MCL1 following PIM2 inhibition was explored, focusing on heightened dependence on MCL1. MMCLs were selected based on their BCL2-family protein profiles (Fig. 1e) and apoptotic priming levels, highlighted by endogenous co-immunoprecipitations (co-IPs) shedding light on binding dynamics of the proteins between pro and antiapoptotic factors (Supplementary Fig. 8a). RPMI8226 and U266, resembling in primary ASCs in terms of BCL-XL and MCL1 expression, were chosen. AZD1208 treatment led to shifts in binding patterns: an increase in BAD binding to BCL-XL, reduction in BIM and BAK binding to BCL-XL due to pBAD loss, followed by increased binding of BIM, BAK, and NOXA to MCL1 (Fig. 4f, g, Supplementary Fig. 8b, c). As expected, the addition of ISRIB, which suppressed NOXA production, resulted in a decrease in the amount of NOXA bound to MCL1 (Fig. 4h). In primary ASCs, PIM inhibition show similar shifts (Supplementary Fig. 8d, e).
These findings reveal a cascade of events triggered by PIM inhibition (Fig. 4i). Increased BAD binding to BCL-XL frees BIM and BAK, relocating them to the mitochondrial membrane where they engage with MCL1. Concurrently, NOXA binds to MCL1. These successive events saturate MCL1 with proapoptotic BH3-only proteins and accentuates apoptotic priming dependent on MCL1. These results align with previous BH3 profiling experiments indicating distinct priming propensity with the MCL1-specific MS1 peptide upon PIM2 inhibition (Fig. 4a).
PIM2 inhibition heightens MCL1 inhibitor sensitivity via NOXA expression
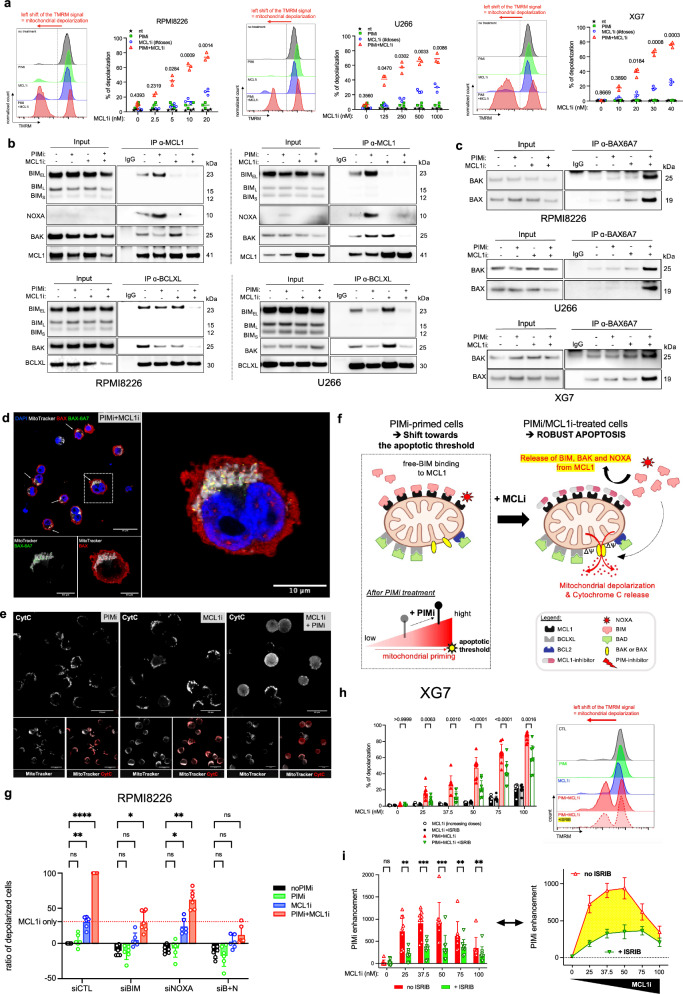
Next, our investigation focused on the role of NOXA induction in enhancing the efficacy of MCL1 inhibitors during PIM2 inhibition. We confirmed previous findings, the strong synergistic effect of inhibiting both PIM2 and MCL1 in six MMCLs using AZD1208 and AZD5991 (Supplementary Fig. 9a)8. Analysis with optimal doses of MCL1 inhibitor for each MMCL revealed a dose-dependent enhancement of MCL1 inhibitor effect by PIM inhibitor (Supplementary Fig. 9b). These results were validated by using PIM447 and S63845, two other pan-PIM and MCL1 inhibitors, respectively (Supplementary Fig. 9c). Functional studies on MMCLs demonstrated that PIM inhibition enhanced mitochondrial depolarization (TMRM signal) induced by AZD5991, especially in AZD5991-sensitive MMCLs (Fig. 5a, Supplementary Fig. 10a). Moreover, specific PIM inhibition in these cells via morpholinos amplified the response to AZD5991-induced mitochondrial depolarization after PIM2 depletion (Supplementary Fig. 10b). Although, MCL1 inhibitor-resistant U266 cells showed a delayed response, higher doses of AZD5991 yielded similar results after a 10-hour delay (Fig. 5a). Similar enhancement was observed in primary ASCs (Supplementary Fig. 11a, b, c).
Fig. 5. PIM2 Inhibition Heightens MCL1 Inhibitor Sensitivity via NOXA Expression.
a Cells were pretreated with AZD1208 before adding increasing doses of MCL1 inhibitor. Percentage of depolarized cells were analyzed by flow cytometry. Data are shown as mean (n = 3). b Cells were pretreated with AZD1208 before adding increasing doses of MCL1 inhibitor. Immunoprecipitation of MCL1 and BCL-XL assessed their interaction with BIM, NOXA, BAK, MCL1, and BCL-XL. The input for RPMI8226 is the same as for RPMI8226 in Fig. 4f, because the cells for IP were from the same experiments. The blot is representative of 1 independent experiment. c Cells were pretreated with AZD1208 before adding increasing doses of MCL1 inhibitor. Immunoprecipitation of BAX-6A7 (BAX active) evaluated its interaction with BAK. The blot is representative of 1 independent experiment. d Cells were pretreated with AZD1208 before adding increasing doses of MCL1 inhibitor. Immunofluorescence of full-length of BAX (red), activated BAX (BAX 6A7; green) and mitochondria (MitoTracker dye; white). Provided images are representative of 3 images. e Cells were pretreated with AZD1208 before adding increasing doses of MCL1 inhibitor. Immunofluorescence of Cytochrome c (red) and mitochondria (MitoTracker dye; white). Provided images are representative of 3 images. f Schematic Illustration: PIM inhibition prevents reuptake of BIM and BAK by BCL-XL after MCL1 inhibition, leading to BAX activation and mitochondrial depolarization. Created with BioRender.com. g RPMI8226 cells were transfected with NOXA, BIM, BIM + NOXA siRNA or control siRNA, then treated with AZD1208 before adding increasing doses of MCL1 inhibitor. The percentage of depolarized cells was analyzed by flow cytometry. Data are shown as mean +/− SEM (n = 6). h Cells were treated with AZD1208 overnight in the presence or absence of ISRIB before adding MCL1 inhibitor. Percentage of depolarized cells were analyzed by flow cytometry. Data are shown as mean +/− SEM (n = 7). i Enhancement of depolarization by PIM inhibitor was assessed by normalizing depolarization levels after combined PIMi/MCL1i treatment to those with PIM inhibitor alone, with or without ISRIB. Data are shown as mean ± SEM (n = 7). P-values were calculated using two-way ANOVA with Sidak’s adjustment for (a), (h), and (i), and two-way ANOVA with Tukey’s adjustment for (g). P < 0.05 is indicated as *P < 0.01 as **P < 0.001 as ***P < 0.0001 as ****. Exact p-values are indicated in the Source Data file. “n”, denotes the number of biological replicates. Source data are provided as a Source Data file. ASCs, antibody-secreting cells; PIMi, pan-PIM inhibitor AZD1208; MCL1i, MCL1 inhibitor AZD5991; BN, BIM + NOXA.
Co-IP assays were conducted following treatment with the synergistic dose of AZD5991 and AZD1208 in RPMI8226 and U266 cell lines. AZD5991 treatment released BIM and NOXA from MCL1 independently of AZD1208, indicating direct disruption of these interactions by AZD5991 (Fig. 5b). However, BAK remained bound to MCL1 in the presence of AZD5991 and required PIM inhibition for its release, highlighting the necessity of PIM kinase inhibition to free BAK from MCL1 (Fig. 5b). In MCL1 inhibitor-resistant U266 cells, AZD5991 treatment increased the binding of BIM and BAK to BCL-XL, a phenomenon prevented by PIM inhibitor pretreatment, highlighting the role of BCL-XL in resistance to MCL1 inhibition (Fig. 5b)30,31. Co-IPs of active BAX (BAX-6A7) revealed that only the combination of MCL1 and PIM inhibitors allowed the formation of BAX/BAX and BAX/BAK complexes, indicating activation of BAX and BAK and resulting in mitochondrial pore formation. These results were found in XG7, RPMI8226 and U266 cell lines (Fig. 5c). We then performed immunofluorescence staining to examine the subcellular localization of BAX and its relocalization to the mitochondria, using MitoTracker dye to identify mitochondria. Full-length BAX was broadly expressed in all conditions. However, the activated form of BAX, detected by BAX-6A7 antibody labeling, was mainly observed under PIM and MCL1 inhibitor treatment, and was restricted to the mitochondria, as shown by co-staining of BAX-6A7 labeling and MitoTracker dye. These results suggest that under the combined treatment, BAX relocalizes to the mitochondria, where it becomes activated, potentially forming homo-BAX/BAX or hetero-BAX/BAK dimers, which are indicative of mitochondrial pore formation (Fig. 5d, Supplementary Fig. 12a). Interestingly, these observations coincide with the cytoplasmic diffusion of Cytochrome c and the increase in Cytochrome c release (Fig. 5e and Supplementary Fig. 12b, c, d). Overall, these findings underscore the pivotal role of PIM inhibition in fully releasing BAK from MCL1, preventing recapture of BIM and BAK by BCL-XL, and activating BAX, processes leading to MOMP, eventual mitochondrial depolarization, and Cytochrome c release, even at lower doses of MCL1 inhibitor in susceptible MMCLs (Fig. 5f).
We then investigated the effects of inhibiting NOXA and/or BIM to determine their contributions to mitochondrial depolarization in the context of the synergistic effect between AZD1208 and AZD5991. In RPMI8226 cells, silencing either NOXA (siNOXA) or BIM (siBIM) led to a significant reduction in mitochondrial depolarization, with siBIM demonstrating greater efficacy than siNOXA alone. However, the most pronounced reduction was observed when both molecules were inhibited simultaneously, compared to each inhibition alone (Fig. 5g, Supplementary Fig. 13a, b).
Next, we examined the role of NOXA induction via the ISR pathway in the synergy between PIM and MCL1 inhibitors by using ISRIB in conjunction with NOXA depletion through siNOXA. In LP1 cells, the enhancement of MCL1 inhibitor-induced depolarization by PIM inhibition was significantly diminished in the presence of ISRIB (Fig. 5h, i, Supplementary Fig. 13c). Similarly, depleting NOXA with siRNA before PIM inhibition also reduced mitochondrial depolarization induced by the combination of PIM and MCL1 inhibitors (Supplementary Fig. 13d, e).
In summary, these findings highlight that in malignant PCs with inhibited PIM2 kinase activity, sensitivity to MCL1 inhibitors is heightened, partially mediated by ISR and NOXA expression. This sensitivity leads to pro-apoptotic protein shifts unique to each MMCL, culminating in the formation of active BAX/BAX and BAK/BAK complexes in all tested cell lines.
Enhanced myeloma xenograft regression via synergistic PIM kinase and MCL1 dual targeting
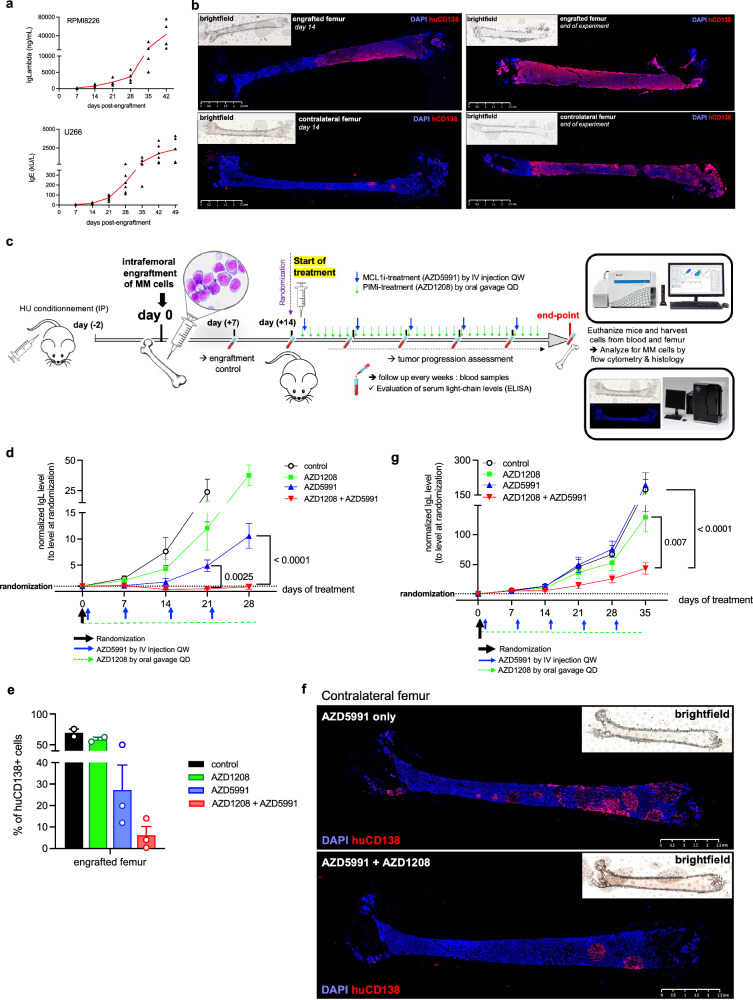
The study transitioned from in vitro investigations to an in vivo model using intrafemoral xenografts of the RPMI8226 and U266 cell lines. Optimization of the model resulted in progressive tumor growth and clear infiltration of human tumor cells in the mice’s femurs, confirmed by the presence of human monoclonal Ig in mice serum (Fig. 6a, b). Optimal doses of AZD1208 and AZD5991 were determined, showing significant and sustained effects on tumor progression and intra-femoral infiltration (Supplementary Fig. 14a, b, c). The model provided a platform for assessing the efficacy of the combination of PIM and MCL1 inhibitors for treating multiple myeloma (Fig. 6c). Combination treatment halted tumor progression (Fig. 6d), reduced infiltration of CD138-positive human cells in grafted femurs (Fig. 6e, Supplementary Fig. 15) and prevented dissemination to contralateral femurs (Fig. 6f). Animals receiving combination treatment did not show tumor progression or femoral tumor masses, indicating enhanced antitumor efficacy compared to single-agent treatments or no treatment (Supplementary Fig. 14c). The combination treatment also delayed tumor growth and prevented progression in U266 cells, overcoming primary resistance observed with single-agent MCL1 inhibition (Fig. 6g).
Fig. 6. Significant tumor growth delay achieved through effective synergistic dual targeting of PIM kinase and MCL1 in a myeloma xenograft model.
a Tumor progression was monitored weekly with measurement of mice serum human IgL and IgE levels after RPMI8226 and U266 cells engraftment, respectively. Data are shown as mean (n = 4). b Immunostaining of engrafted and contralateral femurs with human CD138 (hCD138) marker at the start and end of the experiment. c Female NSG mice received hydroxyurea prior to transplantation of tumor cells into the right femur. When mean human Ig levels reached 1000 ng/mL, mice were randomized to receive vehicle, AZD1208 alone (oral gavage, daily), AZD5991 only (IV injection, weekly), or both. Blood samples were taken weekly, and femurs were collected at the endpoint for analysis. Created with BioRender.com. d Serum IgL levels were tracked from baseline to 4 weeks following treatment with vehicle, AZD1208 alone, AZD5991 alone, or their combination, normalized to randomization values. Data are shown as mean ± SEM (n = 4). Statistical comparisons were made across all treatment groups. e Flow cytometry analysis of viable hCD138-positive cells in harvested femurs at the endpoint. Data are mean +/− SEM (n = 3). f hCD138 immunostaining of contralateral femurs from mice treated with AZD5991 alone or AZD5991/AZD1208 combination. g Serum IgL levels were tracked from baseline to 5 weeks of treatment with vehicle, AZD1208 alone, AZD5991 alone, or both, normalized to randomization. Data are shown as mean ± SEM (n = 6). Statistical analysis compared the AZD1208 + AZD5991 condition only to AZD1208 alone and AZD5991 alone. P-values were calculated using two-way ANOVA with Sidak’s adjustment for (d) and one-way ANOVA with Dunnett’s adjustment for (g). “n”, denotes the number of distinct mouse samples. Source data are provided as a Source Data file. IgL immunoglobulin light chain lambda.
In summary, these in vivo experiments confirmed the synergistic effect of combining PIM and MCL1 inhibitors, demonstrating improved outcomes compared to single-agent treatments.
Leverage ISR-mediated NOXA expression and migration reduction in dual PIM2/MCL1 targeting
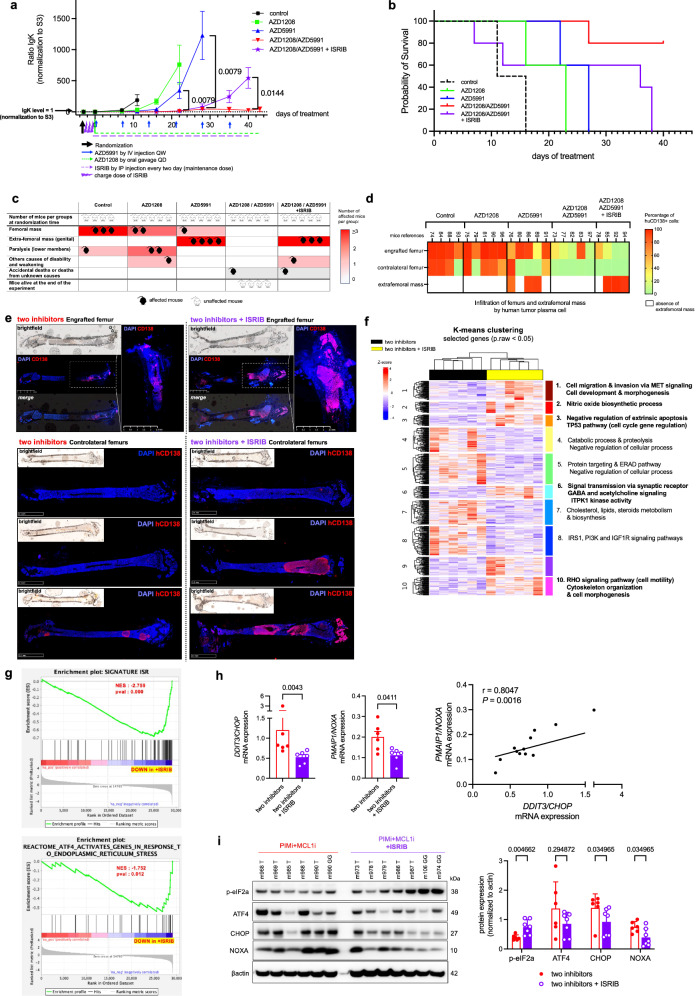
We investigated NOXA expression’s impact on combining PIM and MCL1 inhibitors to control tumor growth. Using XG7 cells, which responded well to AZD5991 combined with low doses of AZD1208 in vitro (Supplementary Fig. 9a), we observed significant tumor progression inhibition with the combined treatment compared to controls and single-agent therapies (Fig. 7a). Survival at day 40 was ensured only in mice receiving the combination, reflecting the absence of tumor progression as evidenced by the low levels of immunoglobulin kappa light chain (IgK) detected (Fig. 7b). Conversely, mice in the control and single-agent groups required premature euthanasia due to tumor progression, as evidenced by high levels of Ig Kappa (IgK) and confirmed by massive invasion of the femurs by hCD138+ PCs (Fig. 7a–d). These results highlight the significant efficacy enhancement of MCL1-targeted therapy with combined treatment, confirmed in another MMCL.
Fig. 7. Reduced tumor progression Control with ISR-Induced NOXA Inhibition in response to PIM/MCL1 inhibitors.
a Serum IgK levels were monitored from treatment initiation to the endpoint at 4 weeks, normalized to baseline levels. Data are shown as mean ± SEM. Statistical comparisons were made between the AZD1208 + AZD5991 group (red, n = 5) and the AZD5991 group (blue, n = 5) on days 22 and 28, and between AZD1208 + AZD5991 with ISRIB (purple, n = 3) and without ISRIB (red, n = 4) on day 40. b Animals were considered “dead” when euthanasia was required due to various reasons. Survival was monitored throughout the experiment. c Mice were categorized based on clinical signs such as visible masses, extrafemoral tumors, limb paralysis, and general weakness. Each group was qualitatively scored. d Flow cytometry analysis of viable hCD138 + XG7 cells harvested from engrafted and control femurs and extrafemoral masses at the endpoint. e hCD138 immunostaining of engrafted and contralateral femurs from different treatment groups. The bone architecture of the femurs, particularly in the ISRIB condition, is severely altered by tumor invasion. f Unsupervised analysis revealed enriched pathways between treatment groups. Ten enriched pathways and associated genes are shown. g GSEA was conducted independently (one-sided) using two distinct signatures on the RNAseq dataset comparing AZD5991 and AZD1208 + ISRIB conditions, with analysis performed using GSEA software. h Expression levels of PMAIP1/NOXA and DDIT3/CHOP genes in tumor cells across treatment conditions, with correlation analysis. Data are mean ± SEM (n = 6). i Protein levels in tumor cells across treatment conditions were assessed by immunoblotting and protein quantification was performed using ImageJ software. Data are mean +/− SEM (n = 7). The blot is representative of 1 independent experiment. P-values were calculated using Mann-Whitney or unpaired t-tests (two-tailed) as specified for (a); Mann-Whitney test (two-tailed) for (h, left); Pearson correlation test (two-tailed) with linear regression for (h, right); and Mann-Whitney tests (two-tailed) for (i). “n”, denotes the number of distinct mouse samples. Source data are provided as a Source Data file. Two inhibitors = AZD1208 + AZD5991; IgK, immunoglobulin kappa light chain; NES, Normalized Enrichment Score; pval, nominal p-value; GSEA, Gene Set Enrichment Analysis.
We further challenged the AZD5991/AZD1208 regimen by introducing ISRIB, known to be no toxic in rodent32, in a subset of mice. Following a 3-day loading dose prior to initiating AZD5991 and AZD1208, ISRIB was administered intraperitoneally every 2 days throughout the 40-day experiment (Supplementary Fig. 16a). While initial treatment weeks showed no significant differences in IgΚ levels between groups, mice in the ISRIB condition exhibited a significant increase in IgΚ levels in subsequent days, leading to euthanasia of all animals by day 40 (Fig. 7a, b). Tumor progression was notably rapid, evidenced by large extra-femoral tumors and femurs involvement, confirmed by cytometric assessment of hCD138+ PCs, necessitating euthanasia due to compromised physical condition (Fig. 7c, d, Supplementary Fig. 16b and Supplementary Fig. 17a, b). In contrast, AZD5991/AZD1208-only treated animals displayed limited tumor masses, suggesting ISR activation induced by PIM2 inhibition plays a pivotal role in tumor control (Fig. 7c, d).
Following these discoveries, subsequent experiments were conducted to deepen our understanding of the phenotype and transcriptomic alterations induced by ISRIB in transplanted XG7 cells. Animals were stratified into two groups based on their administration of ISRIB alongside the AZD5991/AZD1208 combination. Observations validated prior findings, confirming early tumor progression in the ISRIB-treated group, marked by pronounced disparities in extra-femoral tumor development and IgΚ levels; all ISRIB-treated mice required euthanasia prior to day 40 (Supplementary Fig. 17b). Histological examination of the engrafted and contralateral femurs revealed striking differences in tumor invasion, with significant infiltrations and cortical bone lysis evident in ISRIB-treated mice (Fig. 7e, Supplementary Fig. 17c). Viable hCD138+ tumor PCs were isolated from the extra-femoral tumor masses of the 6 AZD5991/AZD1208-only and 6 AZD5991/AZD1208 + ISRIB euthanized animals, with the latter exhibiting notably higher tumor infiltrations (Supplementary Fig. 17c).
Subsequent RNA-seq analysis identified 546 differentially expressed genes (DEGs) between the two conditions (Supplementary Fig. 7d). An unsupervised heatmap delineated 10 DEG modules linked to specific biological changes consequent to ISRIB’s addition to dual PIM2/MCL1 targeting (Fig. 7f). ISRIB yielded DEG signatures associated with signaling, cell cycle, negative apoptosis regulation, cell motility with the RhoA signaling pathway, and cell migration and invasion via the MET pathway, consistent with higher tumor burden and dissemination in mice treated by ISRIB. Conversely, AZD5991/AZD1208-only cells displayed specific features of lipid and steroids metabolism and biosynthesis, suggesting adaptive cellular changes following targeted therapy, including rewiring of protein synthesis after PIM2 inhibition-mediated ISR activation (Fig. 7f, supplementary Fig. 18).
Significant downregulation of ATF4 and ISR signatures in ISRIB-treated mice was confirmed by GSEA (Fig. 7g). Consistent with these observations, qPCR analysis demonstrated a significant reduction in DDIT3/CHOP and PMAIP/NOXA gene expression in the ISRIB condition, with a positive correlation observed between both gene expressions across the 12 samples (Fig. 7h). At the protein level, ISRIB supplementation led to a notable increase in p-eIF2a, as expected32, associated with a significant diminished expression of CHOP and NOXA, and a downward trend for ATF4 (Fig. 7i).
In conclusion, our findings strongly support that the synergistic effect observed upon combining PIM kinase inhibition with anti-MCL1 treatment predominantly stems from the induction of NOXA. Furthermore, ISR inhibition revealed gene expression signatures indicative of cell dissemination, consistent with observations in ISRIB-treated mice. Hence, the concurrent administration of PIM and MCL1 inhibitors holds promise in augmenting tumor cell apoptosis through ISR-mediated NOXA expression and thwarting tumor progression associated with protein translation rewiring.
Discussion
Our investigation delves into the intricate dynamics between PIM2 kinase activity, the ISR pathway, and MCL1 interactions within the BCL2 family, shedding light on mechanisms underlying cell survival. Elevated PIM2 expression often accompanies the progression of cancers, potentially exacerbating disease aggressiveness2,4. We focused on the B cell differentiation model into PCs due to PIM2’s pivotal role in this process8. The elevation in apoptotic priming within primary ASCs aligns with their susceptibility to spontaneous cell death, particularly in the absence of IL-6/STAT3/PIM2 signaling8. Inhibition of PIM2 in PCs triggers saturation of MCL1 binding sites, releasing and displacing BIM, ultimately activating BAK and BAX, followed by mitochondrial depolarization. Understanding the precise molecular events post-PIM2 inhibition holds significant biological relevance, especially in cancers characterized by heightened PIM2 expression.
The remarkable increase in PIM2 expression observed during B cell differentiation into ASCs prompted an exploration of concurrent changes in BCL2 family molecules. As PC differentiation progresses, there is a marked increase in BIM expression, indicative of the cellular stress associated with the heightened secretory demands and maturation into PCs33. This rise in proapoptotic signals is counteracted by the increased levels of BCL-XL and MCL1, alongside PIM2’s role in apoptosis resistance through BAD7. Notably, our findings reveal that only PIM2 inhibition, as opposed to PIM1 or PIM3, the former being previously implicated in myeloid cells34,35, maintains BAD in its unphosphorylated state. This permits the release of BIM, facilitating its displacement from BCL-XL to MCL1, while BAK is released from MCL1. The dynamic regulation of the BCL2 family is further emphasized by the induction of NOXA expression following PIM2 inhibition, aligning with prior studies in prostate cancer cells5. These results underscore the critical role of PIM2 as an apoptotic regulator in PCs, impacting key molecules such as BH3-only, BAD, and NOXA.
MCL1, distinct among anti-apoptotic BCL2 family members, plays in PCs a critical role in protecting cells against apoptotic stimuli36,37. While MCL1 degradation is facilitated by the ubiquitin-proteasome pathway, PIM inhibition in PCs reduces MCL1 levels through caspase-mediated degradation, as observed in myeloma cells under bortezomib treatment38. Additionally, MCL1 degradation is enhanced by the E3 ligase Mule, which promotes ubiquitination and subsequent degradation of MCL1, particularly in the presence of NOXA39,40. However, in our models, PIM2 inhibition leads to MCL1 degradation independent of NOXA binding and proteasomal degradation, which reinforce the mechanism involving caspase-mediated degradation16.
Our study demonstrates that PIM2 inhibition enhances apoptotic priming by inducing NOXA expression via the ISR pathway. This induction facilitates the translocation of proapoptotic BH3-only molecules to MCL1, thereby increasing the cells’ dependence on MCL1 for survival. Consistent with these findings, inhibition of MCL1 upon PIM inhibition disrupts the interaction between MCL1 and BH3-only molecules including BAK, thereby coupling ER stress with mitochondrial apoptosis41. Co-targeting PIM2 and MCL1, even at low doses of both inhibitors, enhances BAK and BIM availability, ultimately activating BAX and BAK and culminating in caspase 3 activation. Our experiments further highlight the critical role of this pathway, as BIM inhibition underscores the importance of this regulatory mechanism. Importantly, the synergy achieved through PIM2 inhibition is largely dependent on ISR-mediated NOXA expression. This synergy is significantly impaired by the ISR-specific inhibitor ISRIB, which blocks caspase 3 activation while maintaining BAD in its non-phosphorylated state.
Our mouse xenograft models demonstrate the evident synergy of dual targeting of PIM2 and MCL1, leading to rapid tumor growth control using RPMI8226 cells. Furthermore, inhibition of PIM2 in transplanted U266 cells reverses primary resistance to MCL1, aligning with previous findings describing synergistic combinations of MCL1 inhibitors with standard chemotherapy or targeted agents42. However, treatment with ISRIB compromises this synergy, leading to robust tumor progression compared to mice treated solely with the dual PIM/MCL1 inhibition. Notably, ISRIB treatment is associated with reduced PMAIP/NOXA gene expression, underscoring NOXA’s critical role in sustaining the long-term synergistic effect. RNAseq analysis confirms a reduction in ISR under ISRIB treatment, while concurrently revealing an upregulation of genes associated with tumor cell migration and dissemination, consistent with observed tumor progression in the animals. These findings shed light on the role of the ISR pathway in cancer43, where tumor cells treated with ISRIB show upregulation of the MET gene and its downstream signaling pathways, known for their association with metastatic potential and poor prognosis in cancer44. This observation is in line with previous studies implicating PIM1 in the regulation of MET expression and signaling in solid cancers45,46, suggesting a similar role for PIM2 in sustaining the MET signaling in myeloma cells by inhibiting ISR pathway activation.
Our findings suggest that PIM2-mediated ISR inhibition may involve the suppression of one of the four kinases that phosphorylate Ser51 in eIF2a to activate ISR, as demonstrated by the induction of both p-eIF2a and ATF4 following PIM2 inhibition13,43.
In summary, while our study unveils dynamic effects of PIM2 inhibition on BCL2 family members and reinstated ISR in tumor cells, it has certain limitations. Further investigation into MCL1 stability and the compensatory roles of other kinases in the absence of PIM2 activity is warranted. Additionally, ongoing efforts aim to elucidate how PIM2 intricately modulates the ISR pathway in PCs. Future studies will determine whether PIM2 elicits similar effects in other cancers and enhances responses to anti-MCL1 therapy under pan-PIM inhibition, holding promise as an adjuvant therapy.
Methods
Ethics
Human peripheral blood primary B cells were isolated from the buffy coats of healthy blood donors. All volunteers were recruited in accordance with the national French guidelines of the “Etablissement Français de Sang” (EFS) and provided informed consent for the use of their blood cells in research, in addition to donating blood for patient needs.
Mice experiments
All animal studies were performed in accordance with the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care International (ARRIVE guidelines), Reporting of In Vivo Experiments (ARRIVE guidelines) and with the approval of the Ethics Committee of the University of Rennes. Our mouse research program is registered with the French Ministry of Research and Universities under the number APAFiS #33438-2021100319339304.
Animals will be housed and cared for according to the standard guidelines, in social groups, in a protected, adapted and enriched environment, with free access to food and water. Experimental care and handling will always be carried out by the same experimenters and animal handlers to reduce animal stress. Daily monitoring of their state of health, regular monitoring of endpoints and recourse to euthanasia procedures as soon as necessary will be used to reduce, avoid and alleviate their suffering. The breeding and experimental conditions of these immunocompromised animals will help to avoid the expression of a damaging phenotype: housing in ventilated racks, autoclaved drinking water and bedding, manipulation under laminar flow hood, maintenance of a positive pressure differential in the experimental area.
The following methods and procedures were carried out
Animal model
Adult female NSG (NOD.Cg-Prkdc<scid>Il2rg<tm1Wjl > /SzJ) mice (6–9 weeks old) were used for the study. To prepare the mice for cell transplantation, hydroxyurea was administered via intraperitoneal injections at a dose of 1 mg/g. After 48 h, 0.5 × 10^6 viable multiple myeloma cell lines (MMCLs) were injected into the femur of the experimental mice.
Assessment of engraftment success
The success of grafting MMCL cells was assessed by measuring mice serum immunoglobulin (Ig) levels. Human Ig levels in the serum served as an indicator of graft success. A waiting period of 15 days was typically observed to allow for graft acceptance and establishment.
Drug administration
Mice were randomly assigned to different treatment groups. The PIM inhibitor AZD1208 (referred to as PIMi) was administered through daily oral gavage. The anti-MCL1 inhibitor AZD5991 (referred to as MCL1i) was administrated by intravenous (IV) injection once a week. The combination of AZD1208 and AZD5991 was also administered according to the experimental design.
Drug solutions
Drug solutions were prepared for administration. AZD1208 was dissolved in a mixture of 10% DMSO, 45% PEG300, 5% Tween80, and 40% NaCl 0.9. AZD5991 was dissolved in a mixture of 10% DMSO and 90% vehicle (20% HPBCD). For the administration of ISRIB (SML0843), a solution was prepared by dissolving the drug in 5% DMSO, 2% Tween20, 20% PEG400, and 73% Dextrose5%.
ISRIB treatment
In some experiments, an additional treatment involving the ISR inhibitor ISRIB was introduced. ISRIB was administered by intraperitoneal injection at a specific dose and schedule.
Follow-up and monitoring
Mice were followed up for a period of up to 6 weeks after cell transplantation, depending on the kinetics of tumor cell dissemination and progression. Mice were monitored weekly for tumor growth and treatment response by measurement of human Ig levels in mice sera.
Animals’ welfare
The humane endpoints were adapted to be sufficiently predictive and early to limit the animals’ pain to its minimum, without compromising the results of the project. To ensure animal welfare, the animals are monitored daily, with an evaluation of general condition (posture and behavior), followed by weight and mouse grimace scale (MGS score). If humane endpoints were achieved (MGS score>1.5, weight loss>20% of reference weight, or observation of abnormal behavior (isolation in cage, limited movement, abnormal appearance or behavior)), the animals were euthanized. In particular, the observation of paralysis or emergence of femoral masses (that could impair the animals’ movements) in certain animals immediately led to their euthanasia. During the course of the experiment, some animals died without showing any signs of pain or suffering on the previous day; the reasons for their death could not be identified (possibly related to undetected pain or suffering following blood sampling or administration of a treatment, or to another condition of unknown origin). In order to preserve full transparency of the data and not to exclude any points, these animals have been included in the results of the analysis. The different procedures performed on animals as well as the defined endpoints and measures to ensure animal welfare are detailed under our mouse research program registered with the French Ministry of Research and Universities the number APAFiS #33438-2021100319339304.
Flow cytometry analysis
At the end of the experiment, mice were sacrificed, and flow cytometric analysis was performed on bone marrow samples to determine the extent of tumor infiltration. Specifically, murine CD45-negative and human CD138-positive cells were identified and analyzed.
Immunohistofluorescence staining
To further investigate tumor infiltration, an immunohistofluorescence approach was used. Sagittal sections of both femurs from each mouse were labeled with antibodies specific to human CD138. This staining allowed visualization and quantification of the presence of human MM cells in the bone marrow.
These experimental procedures were used to evaluate the in vivo activity of the combination of PIMi and MCL1i in the MMCL xenograft model. Measurements of serum Ig levels, flow cytometry analysis, and immunohistofluorescence staining provided valuable insight into the growth, infiltration, and response of MMCL cells to the combination treatment, as well as the potential influence of ISR pathway inhibition on this response.
Multiple myeloma cell lines (MMCLs) culture and viability assays
Cell lines
Several MMCLs were used, including OPM2, SKMM2, RPMI8226, U266, LP1, and XG-6/7 MMCLs. OPM2 (ACC 50) and SKMM2 (ACC 430) were purchased from DSMZ (Braunsweig, Germany), RPMI8226 (CCL-155) and U266 (TIB-196) from ATCC (American Tissue Culture Collection, Rockville, MD, USA). XG MMCLs were provided by Jérôme Moreaux, Montpellier, France. All MMCLs used were characterized as previously described in ref. 47. The XG7 shCTL and shTP53 cell lines were provided by Jérôme Moreaux and used as reported in ref. 48. Control and TP53 shRNA sequences were cloned in the pLenti4-EZ-mIR plasmid as previously described in ref. 49.
Culture conditions
MMCLs were cultured in RPMI1640 Glutamax medium supplemented with 10% fetal calf serum (FCS) and antibiotics. For XG MMCLs, which are IL-6-dependent, 2 ng/mL of IL-6 was added to the culture medium. shCTL and shTP53 XG7 cells were maintained in culture with Zeocin (Thermo Scientific, R25001) at 3 µg/mL.
Compound treatment
Several compounds were tested for their effects on cell viability. These compounds included: AZD1208/PIMi (PIM kinase inhibitor); AZD5991/MCL1i (anti-MCL1 inhibitor); Venetoclax/VTX (BCL-2 inhibitor); A1155463/BCLXLi (BCL-XL inhibitor).
Viability assessment
To assess cell viability, MMCLs were plated at a density of 50,000 cells per well in 96-well flat-bottom microtiter plates. Cells were treated with the selected compounds for 96 h. Cell viability was assessed using the Promega CellTiter-Glo® Luminescence Assay. This assay measures cellular ATP levels as an indicator of viable cells. The assay was performed according to the manufacturer’s instructions.
Data analysis
Cell viability assay results were normalized to untreated samples (set to 100%). Dose-response curves were plotted using software such as GraphPad Prism. IC50 values (concentration required for 50% inhibition of cell viability) were calculated from these curves.
Synergy analysis
Synergy scores were calculated by Bliss method using the SynergyFinder web-application. This method evaluates the combined effects of two compounds and assesses whether the observed effect is greater than the sum of the individual effects.
By performing these viability assays, we were able to determine the effects of different compounds on the viability of MMCLs, calculate their IC50 values and assess any potential synergy between different inhibitors. These assays provide critical information about the effectiveness of compounds in inhibiting cell growth and how they interact when used in combination.
Primary B cells purification
Sample collection
Peripheral blood samples were obtained from healthy volunteers. The collection was performed according to ethical guidelines and with the approval from of the relevant authorities, such as the French Ministry of Higher Education and Research.
Isolation of PBMCs
Blood samples were processed to isolate peripheral blood mononuclear cells (PBMCs). This isolation process typically involves Ficoll density centrifugation, a technique used to separate blood cells based on their density.
Purification of B cells
We focused on isolating specific subsets of B cells. Two distinct types were isolated: Naive B cells (NBCs) (CD19 + /CD27-) and total B cells (CD19 + ). They were isolated using a negative selection approach, often with magnetic cell separation. Cells were enriched by depleting unwanted cell types using specific antibodies coupled to magnetic beads (Naive B Cell Isolation Kit II or B Cell Isolation Kit II). The AutoMACS depletion sensitive program from Miltenyi Biotech was used for this purpose.
Verification of purity
The isolated CD19 + /CD27- NBCs or CD19+ total B cells were routinely checked for purity. This is a critical step to ensure that the isolated cells are of the desired type. A purity level greater than 99% indicates that the isolated cells are highly enriched for the specific B cell subset of interest.
In vitro culture and differentiation of primary B cells and cell viability assays
In Vitro NBC differentiation Model
Purified human NBCs were labeled with 1 µM Violet Proliferation Dye (VPD) 450 (BD Biosciences). This dye allows you to track cell divisions and sort cell populations based on the dye intensity (VPD450hi/lo or D4hi/lo). Purified NBCs were cultured at a concentration of 0.75 × 10^6 cells/ml in 24-well plates. NBCs were stimulated with a cocktail (cocktail 1) composed of F(ab’)2 fragment goat anti-human IgM (H + L), recombinant human soluble CD40L, CpG oligodeoxynucleotide (ODN 2006), and recombinant IL-2. After 2 days, antigen-primed NBCs were stimulated with IL-10. On Day 4, activated B cells were washed and cultured at a concentration of 0.5 × 10^6 cells/ml with a different cocktail (cocktail 2) composed of IL-2, IL-10, and IL-4. At specific time points, activated B cells were recovered, washed, stained, and sorted based on fluorescence intensity or used for functional studies using flow cytometry.
In vitro total B-cell differentiation model
The general process of stimulation and differentiation for total B cells is like that of NBCs. The main differences are in the antibodies used for stimulation (F(ab’)2 anti IgM/IgG/IgA fragment) and the absence of VPD450 labeling. On Day 7, cells were recovered, and dead cells were removed. The cells were then cultured with a different cocktail (cocktail 3) composed of IL-2, IL-10, IFN-alpha, and IL-6. Similar to the NBC model, cells were recovered, washed, stained, and sorted based on fluorescence intensity or used for functional studies using flow cytometry.
Viability assays
For viability assays on Day 7, viable cells were recovered, and viable cells were plated in microtiter plates at a specific concentration. The viable cells were treated with a concentration range of selected compounds: AZD1208 (PIM inhibitor) and AZD5991 (MCL1 inhibitor) for 24 h. Cell viability was assessed using the CellTiter-Glo® Luminescent Assay. Results were normalized to samples without treatment (100%) and analyzed using GraphPad Prism software.
Immunophenotyping and cell sorting
At the appropriate time point, activated B cells were washed, stained with either CD38-APC (BD Biosciences; 555462) and CD23-PC.7 (BD Biosciences; 561167) for phase 2, either with CD20-FITC (Miltenyi Biotech; 130-113-429), CD38-APC (Miltenyi Biotech; 130-113-429) and CD138-PE (Miltenyi Biotech; 130-119-840) for the phase 3 and cell sorted using a FACS-ARIA cell sorter (BD Biosciences). For quantitative RT-PCR (RT-qPCR) and immunoblot assays, activated B cells were harvested and sorted as either CD23+ activated B cells (CD23+ aBCs; CD38lo/CD23 + ) and plasmablasts (PBs; CD38hi/CD23−) populations or as early plasma cells (ePCs; CD20lo/CD38hi/CD138+).
SSO experiments
Inhibition of PIM expression is achieved using a vivo-morpholino PIM splice-switching antisense oligonucleotide (SSO-PIM2, SSO-PIM1 and SSO-PIM3) from GeneTools, LLC (Philomath, OR, US) that binds to the exon 2 donor (5’) splice site. Hybridization of the SSO-PIM interferes with PIM pre-mRNA splicing and induces out-of-frame exon skipping, resulting in the appearance of a premature termination codon in exon 4. An irrelevant standard vivo morpholino oligonucleotide was used as a control (SSO-CTL; GeneTools, LLC). The efficiency of PIM2 mRNA knockdown was first evaluated by RT-PCR (forward: 5’-GGGGGTTCAGTGGGCTCAATCT (5’UTR); reverse: 5’-CTTCGAGTGGGCATGTGACT (exon 4)) to simultaneously identify full-length (286 pb) and alternatively spliced (176 bp) PIM2 mRNAs on a 2% agarose gel. Quantification of inhibition was also performed by RT-qPCR using exon 2 targeting primers (forward: 5’-CCTCATCAGGGAGGGTTAGG-3’; reverse: 5’-GCCCTCCTGTGTCTCAAACC-3’) and verified at the protein level by immunoblot. The efficiency of PIM1 mRNA knockdown was first evaluated by RT-PCR (forward: 5’-AAAATCAACTCGCTTGCCCA (5’UTR); reverse: 5’-AGTCCAGGAGCCTAATGACG (exon 4)) to simultaneously identify full-length (313 pb) and alternatively spliced (206 bp) PIM1 mRNAs on a 2% agarose gel. Quantification of inhibition was also performed by RT-qPCR using exon 2 targeting primers (forward: 5’-CTACTCAGGCATCCGCGTC-3’; reverse: 5’-CAGTCGGAAATCCGGTCCTT-3’) and verified at the protein level by immunoblot. The efficiency of PIM3 mRNA knockdown was first evaluated by RT-PCR (forward: 5’- GATGCTGCTCTCCAAGTTCG (5’UTR); reverse: 5’- GTCACCCGCTCCTTCACC (exon 3)) to simultaneously identify full-length (213 pb) and alternatively spliced (141 bp) PIM1 mRNAs on a 2% agarose gel. Quantification of inhibition was also performed by RT-qPCR using exon 2 targeting primers (forward: 5’- GATGCTGCTCTCCAAGTTCG-3’; reverse: 5’-GCTCTCCTTGTCCGCCTT-3’) and verified at the protein level by immunoblot. For each SSO and cell type, dose escalation was performed to determine the optimal dose for use.
For primary B-cell experiments, SSO-PIM1 and -PIM2 (1.25 and 2.5 µM, respectively) were added directly to the culture medium on D6 for 24 h. For MMCL experiments, cells were cultured at 0.5 × 10^6 cells/ml with IL-6 (2 ng/ml) and SSO-PIM1 and -PIM2, -PIM3 (1.5, 2 and 1.5 µM, for LP1 cells and 1.25, 4 and 1.5 µM for U266, respectively) were added directly to the culture medium for 24 h.
siRNA experiments
For all siRNA experiments, cells were washed in PBS, resuspended in 100 uL electroporation buffer (V cell line nucleofector; Lonza) to which 200 pmol siRNA was added, and finally transfected using the AMAXA system. A siRNA control (siCTL) (ON TARGET Plus, Control, non-targeting pool; Dharmacon) was used as a control. Inhibition of NOXA was achieved using a specific siRNA targeting the gene PMAIP1 (siNOXA) (ON TARGET Plus, SMART pool, human PMAIP1; Dharmacon). Inhibition of BIM was achieved using a specific siRNA targeting the gene BCL2L11 (siBIM) (ON TARGET Plus, SMART pool, human PMAIP1; Dharmacon). Cells were transfected on D5 and the efficiency of PMAIP1 mRNA and NOXA protein knockdown was evaluated after 24 h in ASCs on D6. For MMCLs experiments, PMAIP1 mRNA and NOXA protein knockdown efficiencies were evaluated after 24 h.
Chemical inhibitors and reagents
Pan-PIM kinase activity was inhibited with AZD1208 as PIMi (S7104; Selleckchem). For most experiments in MMCLs, PIMi was used at 20 µM, except for a few experiments where the dose used is indicated. MCL1 activity was inhibited with either AZD5991 (HY-101533; MedChemExpress) or S86485 (HY-100741; MedChemExpress). BCL-XL and BCL2 were inhibited with A1155463 (TA-T6748; Selleckchem) as BCLXLi and ABT-199/Venetoclax (S8048; Selleckchem) as VTX, respectively. The drug ISRIB (SML0843; Sigma-Aldrich) was used at 1 µM (except for one experiment where the dose used is indicated) to inhibit the ISR pathway. Thapsigargine, as Tg, (Sigma-Aldrich), a potent inhibitor of SERCA (sarco-endoplasmic reticulum Ca2+ ATPase), was used to induce an ER stress. For experiments, ISRIB was incubated for 2 h prior to the addition of PIMi or Tg. To evaluate the additive effect of PIMi and Tg, PIMi was incubated 2 h prior to the addition of Tg. The caspase pathway was inhibited by a pan-caspase inhibitor, QVD-OPH (OPH001; R&D) at 10 µM, which was added to the culture 30 min before the addition of AZD1208. The proteasome inhibitor MG-132 (474790; Sigma-Aldrich) was used at 20 µM to block proteasome degradation of proteins. Cycloheximide (CHX) (239764; Sigma-Aldrich) is used at 50 µg/mL to block protein translation.
BH3 profiling
For BH3 profiling, 75 × 10^3 cells (in triplicate) were washed with DTEB buffer (135 mM trehalose, 20 µM EDTA, 20 µM EGTA, 5 mM succinic acid, 0.1% BSA, 10 mM HEPES, 50 mM KCl, pH 7.5) and permeabilized with DTEB + 2.5 µg/mL digitonin. Loss of mitochondrial membrane potential (ψm) was assessed using JC-1 (200 nM) for 2 hours at 30 °C in the presence of different concentrations of each of the BH3 peptides listed below. Depolarized or inactive mitochondria have a reduced membrane potential and are unable to sequester JC-1, resulting in a loss of red fluorescence (Ex = 545 nm, Em = 590 nm). Fluorescence was monitored using a Biotek Synergy H1 microplate reader. The area under each curve was normalized to PUMA2A and BIM to obtain the final result. The peptides used are listed and characterized in the Supplementary Table 1: PUMA2A (negative control), BIM (activator; positive control) and PUMA (sensitizer) are used at 10 µM for all MMCLs. As sensitivities to the other peptides differed for each MMCL, assays were performed by testing increasing dose ranges of peptides to determine the best dose. Only the dose of interest selected is shown in the figures.
Caspase-3 active staining, mitochondrial membrane potential for the monitoring of the TMRM signal
For the in vitro total B cell differentiation model, cells were incubated with CD38-APC (BD Biosciences; 555462) and CD23-PC.7 (BD Biosciences; 561167) antibodies for phase 2, either with CD38-APC (BD Biosciences; 555462) and CD138-PE (Miltenyi Biotech; 130-119-840) for phase 3, and CaspGLOW-FITC (ThermoFisher) for 30 min at 37 °C. For MMCLs, cells were directly stained with CaspGLOW-FITC (ThermoFisher) for 30 min at 37 °C. If mitochondrial depolarization was required, samples were then incubated with 20 nM of TMRM for 30 min at 37 °C according to the MitoPROBE TMRM (ThermoFisher) kit protocol. After washing in PBS, cells were analyzed by flow cytometry after the addition of DAPI to exclude dead cells. The decrease in TMRM signal is characteristic of mitochondrial membrane depolarization.
Cytochrome c release
For Cytochrome c release analysis, MMCLs were initially resuspended thoroughly in 100 µL digitonin lysis buffer (50 mg/ml digitonin; 100 mM KCl; in 1x PBS) by pipetting the cells up and down. The cells were then incubated for 5 min at room temperature to permeabilize the plasma membrane. Following this, the cells were fixed with 100 µL of 3.5% paraformaldehyde (PFA) at room temperature for 30 min. After centrifugation to remove the PFA, the cells were washed once with 100 µL of 1x PBS. Next, the cells were incubated with 100 µL of blocking buffer (3% bovine serum albumin, 0.05% saponin in 1x PBS) for 30 min at room temperature. The cells were then incubated overnight at 4 °C with a mouse anti-Cytochrome c antibody (BioLegend; 612310) at a dilution of 1:100 in blocking buffer. After washing in 1x PBS, the cells were analyzed by flow cytometry. A decrease in the Cytochrome c signal indicates the release of Cytochrome c from the mitochondria.
Quantitative RT-PCR analysis
RNA was extracted using a NucleoSpin® RNA Plus XS kit (740984; Macherey-Nagel) and reverse transcribed to cDNA using a Superscript II kit (Invitrogen). RT-qPCR was performed using TaqMan® or Sybergreen® Gene Expression Master Mix. Gene expression levels were quantified using ABL1 as an endogenous control. The 2^(-ΔΔCt) method was used to determine the relative expression of each gene. See Supplementary Tables 2 for the sequence of all primers used.
Cell protein extractions and Immunoblotting
For total protein extraction, cell pellets were lysed in RIPA lysis buffer (89900; Pierce) in the presence of protease inhibitors (PIC, 04693116001; Roche). After centrifugation to remove debris, protein concentration was determined by the Bradford method (5000006; Biorad). Denatured protein extracts (at least 10 µg) were then separated on a NuPAGE 4–12% Bis-Tris acrylamide gel (Thermo Scientific) and transferred to nitrocellulose membranes using an iBlot 2 transfer device (Invitrogen). After incubation with specific primary antibodies, immunoblots were revealed with HRP-conjugated secondary mAb or, for immunoprecipitation, Clean-Blot IP Detection Reagent (Thermo Scientific; 21230) after addition of ECL plus (Thermo Scientific; 32132) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific; 34094) using the G:BOX Chemi imaging system (Syngene) and analyzed with GeneSys software. β-actin was used as loading control. Immunoblots shown in the figures were sometimes obtained with samples derived from the same experiment but processed on separate gels for different markers. Only one loading control was used in the figure, but all loading control were presented in the Source data file. The list of primary and secondary antibodies used for immunoblotting assays is provided in the Supplementary Table 3.
Co-immunoprecipitation assays
At least 10 × 10^6 D6-P1 cells were lysed in NP40 lysis buffer (50 mM Tris-HCl pH 7.5; 0.15 M NaCl; 0.5% deoxycholic acid; 0.5% NP40; 1 mM phenylmethylsulfonyl fluoride; 1 mM PIC). Protein concentration was determined by the Bradford method after centrifugation. Cell lysate proteins (approximately 1 mg protein extract) were immunoprecipitated with the appropriate antibodies and incubated overnight at 4 °C, followed by 1 h incubation with Dynabeads Protein A or Protein G (10001D, 10003D; Invitrogen) at 4 °C. Immunoprecipitates were washed four times with a DynaMagnet (Invitrogen) and NP40 wash buffer (50 mM Tris-HCl pH7.5; 0.4 M NaCl; 0.5% deoxycholic acid; 0.5% NP40; 1 mM phenylmethylsulfonyl fluoride; 1 mM PIC) according to the manufacturer’s instructions. Dynabead antibody-protein complexes were resuspended in loading buffer, denatured, and placed on a magnet. Supernatants were loaded on a gel for immunoblotting.
Ubiquitin enrichment assays
For MCL1 ubiquitination analysis, cells were incubated with MG-132 (10 μM) for 3 h after PIMi treatment. After cell lysis using BlastR lysis buffer (Cytoskeleton), ubiquitinated proteins were enriched using the Signal-Seeker Ubiquitination Detection Kit (Cytoskeleton). Briefly, after protein quantification, 300 μg of total protein was incubated with ubiquitination affinity beads or control beads for 2 h at 4 °C. After washes, the precipitates were eluted, separated from the beads using special spin columns, heated, and loaded onto a bis-tris gel. Proteins were transferred to PVDF membranes and detected with anti-MCL1 antibody. The presence of ubiquitinated proteins was checked with the supplied anti-ubiquitin HRP antibody.
ELISA
Mouse sera were tested for the presence of human Ig light chain kappa or lambda by ELISA. ELISAs were performed in polycarbonate 96-well plates coated overnight at 4 °C with Ig kappa (IgK) or Ig lambda (IgL) antibodies (goat anti-human kappa-UNLB, SouthernBiotech; 2060-01 and 2070-01, respectively) in PBS. After washing, sera were blocked with 3% bovine serum albumin (BSA) (Sigma-Aldrich) in PBS for 1 h at 37 °C. Sera were diluted into successive wells in 3% BSA/PBS buffer and incubated at 37 °C for 2 h. After washing, alkaline phosphatase (AP)-conjugated goat anti-human antibodies (Goat Anti-Human kappa-AP, SouthernBiotech; 2060-04 and 2070-04, respectively) were incubated in PBS for 1.5 h at 37 °C. AP activity was determined by adding the appropriate substrate for AP (SIGMAFAST™ p-nitrophenyl phosphate tablets, Sigma-Aldrich) for ~15 min, followed by blocking the reaction by adding 3 M NaOH (Sigma-Aldrich). Optical density was measured at 405 nm on a Multiskan FC microplate photometer (Varioskan LUX, Thermo Scientific) using Skanlt software.
Immunofluorescence assays
Cells were seeded on slides at a concentration of 2 × 10^6 cells per slide in complete medium containing MitoTracker (MitoTracker Deep Red FM; Thermo Scientific M22426) and incubated for one hour at 37 °C. The cells were then fixed with 4% paraformaldehyde (PFA) for 10 min before being incubated for 1 h in a blocking solution (PBS, 2% bovine serum albumin, 4% donkey serum, and 0.1% saponin). Following blocking, the slides were incubated overnight at 4 °C in a humidified chamber with primary antibodies: Cytochrome c (6H2.B4) (BD Pharmingen; 556432), full-length BAX (R&D Systems; NBP2-67285), or BAX-6A7 (BioLegend; 633802). After washing with PBS/0.1% saponin, the slides were incubated with the corresponding secondary antibodies for 1 h at room temperature. Finally, the slides were mounted using DAPI-Fluoromount-G® (SouthernBiotech; 0100-20) and analyzed by confocal microscopy using a Leica SP8 (Leica Microsystems). Image analysis was performed using ImageJ software (National Institutes of Health). A detailed list of the primary and secondary antibodies used for immunofluorescence assays is provided in the Supplementary Table 3.
Bone immunostaining
Femurs were isolated and incubated in PBS/4% paraformaldehyde (PFA) buffer for 24 h at 4 °C. Bones were then sequentially incubated for 1 h in 10%, 20%, and 30% sucrose solution in PBS at 4 °C, embedded in cryoprotectant gel (SCEM; Section-Lab), frozen, and stored at −80 °C. Sections of 20 µm were cut on a cryostat (Leica) using the Kawamoto transfer technique as described in (60). For immunofluorescence labeling, sections were fixed with 4% PFA for 10 min at 4 °C, rinsed three times with PBS, and then incubated in a saturation permeabilization solution (PBS-Triton 0.5%-donkey serum 20%) for 2 h at room temperature. For labeling, sections were incubated overnight at 4 °C in a humidity chamber with anti-CD138 antibody (R&D Systems; NB100-64980) in PBS/Triton 0.1%/donkey serum 2%. After washing in PBS, sections were incubated with secondary antibody Alexa Fluor 594 donkey anti-rabbit (Jackson Immuno Research; 711-586-152) for 1 h. Before mounting, sections were incubated with a solution of DAPI (1/1000 in PBS). Mounting was performed with commercial medium DAPI Fluoromount-G® (Southern Biotech; 0100-20). A drop of mounting medium is applied to each section and a coverslip is gently placed on top. The slides were then dried overnight at room temperature in the dark. Slides were scanned on the Nanozoomer 2.0 RS, and NPD.view-Plus software (Hamamatsu) was used for immunostaining analysis. Basic brightfield imaging has been performed and shown in the figures. For this procedure, a focused beam of white light is directed through the sample and the transmitted light is collected to produce an image in natural color. QuPath software was used for quantitative analysis of whole slide images (T. Kawamoto, M. Shimizu, A method for preparing 2- to 50 µm-thick fresh-frozen sections of large samples and undecalcified hard tissues. Histochem. Cell Biol. 113, 331–339 (2000).
RNAseq process & bioinformatic analysis
Six samples from mice of the two treatment combination groups (two inhibitor-only and two inhibitor+ISRIB) were selected for RNAseq analysis. RNA was extracted (as described above) and sent to Macrogen (Macrogene Europe, The Netherlands) for library preparation and sequencing. Reads were aligned to the hg19 human genome using TopHat2, and only reads with a single alignment were retained. Raw counts for each sample were obtained using HTSeq counts. Differential expression of filtered genes (HTSfilter, 29136 genes) was analyzed using DESeq228 in R. ISR pathway enrichment analysis was performed using GSEA software using the statistic (‘stat’) of the 20136 genes and by querying a curated ISR gene list from Liang Wong et al.50 and a known gene set from the Reactome database (R-HSA-380994); showing downregulation of ISR molecular signatures after ISRIB treatment. DEGs analysis was performed by selecting genes with p-value < 0.05 and performed on normalized read counts for the filtered genes. Functional implications (canonical pathways and biological functions) of these DEGs were generated using iDEP 2.0 software (South Dakota State University (SDSU)) (Ge, Son & Yao, iDEP, BMC Bioinformatics 19:1-24, 2018). Unsupervised analysis and K-means clustering into 10 clusters were performed to identify gene sets of enriched pathways. Gene Ontology (GO) and Reactome gene libraries were queried using Gene Ontology Resource software (https://geneontology.org) (Z. Xie, A. Bailey, M. V. Kuleshov, D. J. B. Clarke, J. E. Evangelista, S. L. Jenkins, A. Lachmann, M. L. Wojciechowicz, E. Kropiwnicki, K. M. Jagodnik, M. Jeon, A. Ma’ayan, Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 1, e90 (2021).
Statistics & reproducibility
Data are shown as bars, scatter plots and presented as mean+SD or mean + SEM depending on the results, or with box and whiskers. Data were analyzed using the nonparametric Mann-Whitney test, or exceptionally with unpaired Student t-test (parametric), to determine significance between population means when comparing two groups. When more than two groups were compared, Kruskal-Wallis’s test (for independent samples) with Dunn’s multiple comparisons was used to compare each group with a control group. A one-way ANOVA with Dunnett multiple comparisons or two-way ANOVA with adapted multiple comparisons (Sidak’s, Tukey or Fisher’s LSD) were used when appropriate for comparing each group together. Pearson’s correlation coefficient was calculated for correlation analysis. Statistical tests used for each experiment are indicated on figures and were always two-sided. One-way, Two-way ANOVA, Pearson correlation, Mann-Whitney tests and t-tests were performed using the GraphPad Prism software (GraphPad Software). Values of P < 0.05 were considered as statistical significance. P < 0.05 is indicated as *P < 0.01 as **P < 0.001 as ***P < 0.0001 as ****; ‘ns’ designates “not significant” on graphs. Gene Set Enrichment Analysis (GSEA) were performed using the GSEA software.
Immunoprecipitations were performed once, but always in different cell types (primary and MMCLs, or two distinct MMCLs). Immunoblottings were performed twice and mostly in different cell types (primary ASCs and/or two distinct MMCLs). All experiments with statistical analysis were repeated with, at least, three biological replicates. Biological replicates refer to distinct biological sources. For experiments involving MMCLs, these include independent experimental replicates and cells from different culture passages. For primary samples, biological replicates consist of cells from distinct donors, while for mouse experiments, they represent individual animals. In some case, experiments without statistical analysis were performed once (primary cells) or twice. See Source data file.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank all members of the Fest Laboratory for helpful discussion, J. Destin for technical assistance and F. Jouan for mouse colony management. Cell sorting was performed at the Biosit Flow Cytometry and the CytomeTRI cell sorting facilities (UMS6480 Biosit). Immunofluorescence studies were performed in the H2P2 facility, a member of the UMS 6480 Biosit (Rennes, France) and the French National France-BioImaging infrastructure network funded by the French Research Agency (ANR-10-INBS-04). We gratefully acknowledge funding from “Fondation ARC” the “Programmes labellisés” (PGA) 2022 N° ARCPGA2021120004244_4856, the Internal grant from the Hematology Laboratory, CHU de Rennes, the ANR Carnot-CALYM program funded the purchase of NGS mice. M.H. has received a doctoral fellowship from FHU CAMIn, Ligue Contre le Cancer/Comité d’Ile et Vilaine. M.H. is supported by a “CCA–Inserm-Bettencourt Schueller” Fund. A.J.’s team was supported by Inserm and by “Fondation ARC” Equipe labellisée 2022–2025.
Author contributions
Conceptualization: M.H., T.F. Methodology: M.H., A.J., G.C., L.D., P.A., E.D., T.M., T.F. Investigation: M.H., S.C., L.N., J.M., M.B. Funding acquisition: T.F. Project administration: T.F. Supervision: M.H., T.F. Writing – original draft: M.H., T.F. Writing – review & editing: M.H., T.F.
Peer review
Peer review information
Nature Communications thanks Frank Essmann, Stephen Nutt and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The RNAseq data generated for this study are available in GEO database: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE266112, under accession number GSE266112. Other dataset used for this study are available in GEO database under accession number GSE136990. The authors declare that all other source data supporting the findings of this study are available within the paper and its supplementary information files, including the Source Data file. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55572-5.
References
- 1.Santio, N. M. & Koskinen, P. J. PIM kinases_ From survival factors to regulators of cell motility. Int. J. Biochem. Cell Biol93, 74–85 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Dai, H. et al. Pim‐2 upregulation: Biological implications associated with disease progression and perinueral invasion in prostate cancer. Prostate65, 276–286 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Garcia, P. D. et al. Pan-PIM kinase inhibition provides a novel therapy for treating hematologic cancers. Clin. Cancer Res. J Am. Ass. Cancer Res.20, 1834–1845 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Wang, Y., Xiu, J., Ren, C. & Yu, Z. Protein kinase PIM2: A simple PIM family kinase with complex functions in cancer metabolism and therapeutics. J. Cancer12, 2570–2581 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song, J. H. & Kraft, A. S. Pim Kinase Inhibitors Sensitize Prostate Cancer Cells to Apoptosis Triggered by Bcl-2 Family Inhibitor ABT-737. Cancer Res.72, 294–303 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan, B. et al. The PIM-2 Kinase Phosphorylates BAD on Serine 112 and Reverses BAD-induced Cell Death. J. Biol. Chem.278, 45358–45367 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Fox, C. J. & Thompson, C. B. The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev.17, 1841–1854 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas, M. et al. Pivotal role of PIM2 kinase in plasmablast generation and plasma cell survival, opening new treatment options in myeloma. Blood (2022) 10.1182/blood.2021014011. [DOI] [PubMed]
- 9.Ron, D. & Walter, P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol.8, 519–529 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Iwakoshi, N. N. et al. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol.4, 321–329 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Shaffer, A. L. et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity21, 81–93 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Szegezdi, E., Logue, S. E., Gorman, A. M. & Samali, A. Mediators of endoplasmic reticulum stress‐induced apoptosis. EMBO Rep.7, 880–885 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walter, P. & Ron, D. The Unfolded Protein Response: From Stress Pathway to Homeostatic Regulation. Science334, 1081–1086 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Czabotar, P. E., Lessene, G., Strasser, A. & Adams, J. M. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell Biol.15, 49–63 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Senichkin, V. V., Streletskaia, A. Y., Gorbunova, A. S., Zhivotovsky, B. & Kopeina, G. S. Saga of Mcl-1: regulation from transcription to degradation. Cell Death Differ27, 405–419 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrant, M. et al. Cleavage of Mcl-1 by caspases impaired its ability to counteract Bim-induced apoptosis. Oncogene23, 7863–7873 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Willis, S. N. et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev.19, 1294–1305 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, K. et al. BH3-only proteins target BCL-xL/MCL-1, not BAX/BAK, to initiate apoptosis. Cell Res.29, 942–952 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Senft, D. et al. In non-transformed cells Bak activates upon loss of anti-apoptotic Bcl-XL and Mcl-1 but in the absence of active BH3-only proteins. Cell Death Dis.6, e1996–e1996 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Day, C. L. et al. Structure of the BH3 Domains from the p53-Inducible BH3-Only Proteins Noxa and Puma in Complex with Mcl-1. J. Mol. Biol.380, 958–971 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Certo, M. et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell9, 351–365 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Pignarre, A. et al. Plasmablasts derive from CD23– activated B cells after the extinction of IL-4/STAT6 signaling and IRF4 induction. Blood137, 1166–1180 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Núñez‐Vázquez, S. et al. NOXA upregulation by the prohibitin‐binding compound fluorizoline is transcriptionally regulated by integrated stress response‐induced ATF3 and ATF4. FEBS J.288, 1271–1285 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Sharma, K. et al. p53‐independent Noxa induction by cisplatin is regulated by ATF3/ATF4 in head and neck squamous cell carcinoma cells. Mol. Oncol.12, 788–798 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weller, S. et al. The BCL-2 inhibitor ABT-199/venetoclax synergizes with proteasome inhibition via transactivation of the MCL-1 antagonist NOXA. Cell Death Discov.8, 215 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Czabotar, P. E. et al. Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc Natl. Acad. Sci.104, 6217–6222 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Djajawi, T. M. et al. MARCH5 requires MTCH2 to coordinate proteasomal turnover of the MCL1:NOXA complex. Cell Death Differ.27, 2484–2499 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haschka, M. D. et al. MARCH5-dependent degradation of MCL1/NOXA complexes defines susceptibility to antimitotic drug treatment. Cell Death Differ.27, 2297–2312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arai, S. et al. MARCH5 mediates NOXA-dependent MCL1 degradation driven by kinase inhibitors and integrated stress response activation. Elife9, e54954 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez-Bougie & M, A. BH3-mimetic toolkit guides the respective use of BCL2 and MCL1 BH3-mimetics in myeloma treatment. Blood132, 2656–2669 (2018). [DOI] [PubMed]
- 31.Seiller, C. et al. Dual targeting of BCL2 and MCL1 rescues myeloma cells resistant to BCL2 and MCL1 inhibitors associated with the formation of BAX/BAK hetero-complexes. Cell Death Dis11, 316 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sidrauski, C. et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife2, e00498 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puthalakath, H. et al. ER Stress Triggers Apoptosis by Activating BH3-Only Protein Bim. Cell129, 1337–1349 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Aho, T. L. T. et al. Pim‐1 kinase promotes inactivation of the pro‐apoptotic Bad protein by phosphorylating it on the Ser112 gatekeeper site. FEBS Lett571, 43–49 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Didichenko, S. A., Spiegl, N., Brunner, T. & Dahinden, C. A. IL-3 induces a Pim1-dependent antiapoptotic pathway in primary human basophils. Blood112, 3949–3958 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Nijhawan, D. et al. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev17, 1475–1486 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maurer, U., Charvet, C., Wagman, A. S., Dejardin, E. & Green, D. R. Glycogen Synthase Kinase-3 Regulates Mitochondrial Outer Membrane Permeabilization and Apoptosis by Destabilization of MCL-1. Mol. Cell21, 749–760 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Gomez-Bougie, P. et al. Noxa Up-regulation and Mcl-1 Cleavage Are Associated to Apoptosis Induction by Bortezomib in Multiple Myeloma. Cancer Res67, 5418–5424 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Gomez-Bougie, P. et al. Noxa controls Mule-dependent Mcl-1 ubiquitination through the regulation of the Mcl-1/USP9X interaction. Biochem. Biophys. Res. Commun.413, 460–464 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Zhong, Q., Gao, W., Du, F. & Wang, X. Mule/ARF-BP1, a BH3-Only E3 Ubiquitin Ligase, Catalyzes the Polyubiquitination of Mcl-1 and Regulates Apoptosis. Cell121, 1085–1095 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Gomez-Bougie, P., Halliez, M., Moreau, P., Pellat-Deceunynck, C. & Amiot, M. Repression of Mcl-1 and disruption of the Mcl-1/Bak interaction in myeloma cells couple ER stress to mitochondrial apoptosis. Cancer Lett383, 204–211 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Tantawy, S. I., Timofeeva, N., Sarkar, A. & Gandhi, V. Targeting MCL-1 protein to treat cancer: opportunities and challenges. Front. Oncol.13, 1226289 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costa-Mattioli, M. & Walter, P. The integrated stress response: From mechanism to disease. Science368, eaat5314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raj, S. et al. Molecular mechanism(s) of regulation(s) of c-MET/HGF signaling in head and neck cancer. Mol. Cancer21, 31 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.An, N., Xiong, Y., LaRue, A. C., Kraft, A. S. & Cen, B. Activation of Pim Kinases Is Sufficient to Promote Resistance to MET Small-Molecule Inhibitors. Cancer Res75, 5318–5328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao, L. et al. PIM1 kinase promotes cell proliferation, metastasis and tumor growth of lung adenocarcinoma by potentiating the c-MET signaling pathway. Cancer Lett444, 116–126 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Vikova, V. et al. Comprehensive characterization of the mutational landscape in multiple myeloma cell lines reveals potential drivers and pathways associated with tumor progression and drug resistance. Theranostics9, 540–553 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herviou, L. et al. Targeting the methyltransferase SETD8 impairs tumor cell survival and overcomes drug resistance independently of p53 status in multiple myeloma. Clin. Epigenetics13, 174 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viziteu, E. et al. RECQ1 helicase is involved in replication stress survival and drug resistance in multiple myeloma. Leukemia31, 2104–2113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong, Y. L. et al. eIF2B activator prevents neurological defects caused by a chronic integrated stress response. Elife8, e42940 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNAseq data generated for this study are available in GEO database: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE266112, under accession number GSE266112. Other dataset used for this study are available in GEO database under accession number GSE136990. The authors declare that all other source data supporting the findings of this study are available within the paper and its supplementary information files, including the Source Data file. Source data are provided with this paper.