Abstract
Background/Aim
α1-Acid glycoprotein (AGP), also known as orosomucoid, is an acute-phase protein that has been found increased in plasma of cancer patients. This study investigates the role of AGP expression in clear cell renal cell carcinoma (ccRCC) and its association with clinical outcomes.
Materials and Methods
We investigated the correlation between AGP levels and the prognosis of ccRCC through an analysis of The Cancer Genome Atlas (TCGA) database. To examine AGP expression and its clinico-pathological associations, immunostaining was performed on paraffin-embedded tissue samples of 92 ccRCC cases.
Results
AGP expression was found to be higher in RCC cell lines compared to normal renal epithelial cells. Analysis of the TCGA dataset showed that patients with AGP gene expression had significantly worse overall survival. However, AGP expression was not correlated with age, sex, or cancer stage. A mouse monoclonal antibody against AGP was generated. This antibody reacted with human and mouse hepatocytes, but not in AGP-deficient mice. From 92 examined ccRCC cases, AGP protein expression was detected in 89 cases, with only 3 being negative. AGP expression levels did not correlate with clinicopathological factors, such as age, tumor size, or nuclear grade. CD14, a receptor of AGP, was found to be expressed in Iba1-positive monocytes and tumor-associated macrophages (TAMs) but not in other cell types like lymphocytes or cancer cells. No significant correlation was found between AGP expression and the number of Iba1-positive cells in ccRCC tissues. Iba1-positive cells were correlated with Fuhrman grade, and patients with ≥30% Iba1-positive cells were, on average, significantly younger and had more aggressive tumor.
Conclusion
AGP expression is linked to poorer survival in ccRCC, but its association with immune cell infiltration (via Iba1-positive cells) is unclear.
Keywords: AGP, ORM1, clear cell renal cell carcinoma, macrophages
Kidney cancer is the most common malignant tumor affecting the kidneys in adults. Renal cell carcinoma (RCC) has various histological subtypes, with clear cell renal cell carcinoma (ccRCC) being the most frequent (1,2). Although the clinical outcomes for patients with early-stage ccRCC are generally favorable, those with metastatic ccRCC have a worse prognosis, with 5-year survival rates below 20% (3). Over the past few decades, immunotherapies, including cytokine-based treatments, have been explored for advanced ccRCC. Recent scientific advances have highlighted the effectiveness of immune checkpoint inhibitors. Clinical trials have shown that combination immunotherapy targeting tyrosine kinase inhibitors and immune checkpoint molecules, such as programmed cell death protein 1 (PD-1) and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), improves the clinical outcomes of patients with metastatic ccRCC (4-6).
Over the past decade, the relationship between the tumor microenvironment and patient prognosis has been increasingly recognized. It is well established that numerous immune cells infiltrate the ccRCC microenvironment, and higher densities of lymphocytes and macrophages have been associated with poorer clinical outcomes (7,8). Conversely, increased lymphocyte infiltration with tertiary lymphoid structure-like phenotypes has been linked to improved clinical outcomes (9). Tumor-associated macrophages (TAMs), which infiltrate tumor tissues, are associated with poor prognosis and treatment resistance in many cancers (10). The pro-tumor and immunosuppressive functions of TAMs have been implicated in resistance to immunotherapy. Recent studies suggest that targeting TREM2-positive TAMs with anti-TREM antibodies can reverse anti-PD-1 resistance in animal models (11). A clinical trial of combination therapy using anti-TREM and anti-PD-1 antibodies demonstrated clinical benefits in 29% of anti-PD-1-resistant ccRCC cases (12). Therefore, targeting TAMs may be beneficial for patients with immunotherapy-resistant ccRCC.
Several factors, such as C-reactive protein (CRP) and the neutrophil-to-lymphocyte ratio (NLR), have been identified as prognostic markers in various cancers. Elevated CRP and NLR have been suggested as biomarkers for predicting resistance to anti-PD-1 therapy in RCC patients (12,13). CRP, an acute-phase protein produced by the liver and released into the bloodstream, is considered a clinicopathological marker of systemic inflammation and immune activation (14). These proteins may show increased or decreased levels in the blood. Elevated levels of acute-phase proteins are commonly observed in patients with inflammatory diseases and malignancies (15). α1-Acid glycoprotein (AGP), also known as orosomucoid, is another acute-phase protein present in plasma. AGP levels can increase 2-5 times (1-2.5 mg/ml compared to the normal 0.5 mg/ml) in response to inflammation and various tumors, including lung cancer, hepatocellular carcinoma, and melanoma (16).
Although there exist some reports on the relationship between CRP and ccRCC, no studies have demonstrated a link between AGP and ccRCC. In the present study, we identified AGP gene expression in ccRCC tissues and found that AGP expression was associated with worse clinical outcomes based on statistical analysis using the Human Protein Atlas database (https://www.proteinatlas.org/). We subsequently developed a novel anti-AGP mouse monoclonal antibody and tested AGP expression in 92 cases of ccRCC.
Materials and Methods
Animals. C57BL/6N (wild type, CLEA JAPAN) and AGP-deficient mice [C57BL/6N-Orm2tm1(KOMP)Vlcg/Mmucd, RRID: MMRRC_048914-UCD] sperm was purchased from Knockout Mouse Project (KOMP) Repository, UC Davis, USA. Mice were kept in a 12 h light/dark cycle (light from 07:00 to 19:00) at a room temperature of 22˚C±2˚C, with free access to food and water. The Animal Care and Use Committee of Kumamoto University approved our protocols for animal experiments (approval No.: #A2019-015) and all methods were performed in accordance with the relevant guidelines and regulations.
The Cancer Genome Atlas (TCGA) database analysis. A total of 526 full RNAseq matrices and clinical data for ccRCC were downloaded from the TCGA database. Data analyses were performed using Prism (GraphPad Software, San Diego, CA, USA).
Cell lines and real-time PCR analysis of AGP. We utilized RCC cell lines (786-O, ACHN, and MAMIYA) as well as immortalized renal proximal tubular epithelial cells (RPTEC). Total RNA was extracted from the cells using RNAiso Plus (Takara Bio, Shiga, Japan). Quantitative real-time PCR (qRT-PCR) was performed using TB Green Premix Ex Taq II (Takara Bio) on an Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems, Tokyo, Japan). At least three biological replicates were analyzed, with expression levels calculated from a minimum of two technical replicates. mRNA expression levels were quantified using the 2ΔΔCt method and normalized to β-actin mRNA levels. All primers were pre-designed and obtained from Takara Bio (Shiga, Japan).
Generation of mouse monoclonal antibody against AGP. Human AGP protein (100 μg, WAKO, Tokyo, Japan), mixed with TiterMax Gold (TiterMax, Norcross, GA, USA), was injected intraperitoneally into AGP-deficient mice. Splenocytes were subsequently fused with NS-1 myeloma cells using PEG1500 (Roche, Munich, Germany). Hybridoma cells were selected in RPMI medium containing 10% FBS, HAT (Sigma, St. Louis, MO, USA), and 1% BM-Condimed (Roche). Antibody screening was performed via ELISA using human AGP-coated multi-well plates and immunohistochemistry on human liver tissue. Mouse liver tissue from wild-type and AGP-deficient mice was also used in the screening process. Clone No. 45 was selected as the anti-AGP monoclonal antibody. The immunoglobulin class of this clone was determined to be IgG1 with a ĸ chain (Immunoglobulin Isotyping Kit, Antagen Biosciences, Inc., Canton, MA, USA).
Immunohistochemical (IHC) analysis. Paraffin-embedded tissue samples from 92 ccRCC patients, diagnosed at Kumamoto University Hospital between 2019 and 2022, were collected and reviewed by two experienced pathologists. Specimens were sectioned into 3-μm slices. The primary antibodies used were anti-AGP, anti-Iba1 (clone NSNP27; Wako, Tokyo, Japan), and anti-CD14 (clone 4B4F12; Abcam, Cambridge, UK). Samples were incubated with peroxidase-labeled goat anti-mouse or rabbit secondary antibodies (anti-mouse, #424132, and anti-rabbit, #424142; Histofine, Nichirei Biosciences, Tokyo, Japan). Immunoreactions were visualized using a diaminobenzidine substrate kit (#425011; Nichirei Biosciences) and Simple Stain AEC Solution (#415182; Nichirei Biosciences).
AGP positivity in cancer cells was categorized into four groups based on the area of positive staining: score 0 (none), score 1 (<10%), score 2 (10%-50%), and score 3 (>50% positive) [proportion score]. Additionally, staining intensity was classified into four groups: score 0 (negative), score 1 (weak), score 2 (intermediate), and score 3 (strong) [intensity score]. These two scores were combined to obtain the total AGP score. Iba1-positive cells were counted in three randomly selected, non-overlapping high-power fields (200× magnification) within tumor areas free from necrosis and hemorrhage, using Halo software (Visualix, Hyogo, Japan). Two experienced investigators (T.A. and Y.K.), blinded to the patients' clinical characteristics and outcomes, independently evaluated all immunostained sections.
Single cell RNA-sequence. The analysis of scRNA-seq data was conducted in R using the Seurat package. Data from 30 primary ccRCC cases were obtained from the following datasets: GSE159115 (n=7) (17), GSE152938 (n=2) (18), GSE171306 (n=2) (19), and GSE207493 (n=19) (20). The analytical methods followed those described in a previous report (21).
Statistical analyses. Prism (GraphPad Software, San Diego, CA, USA) was used to perform the statistical analyses.
Ethical approval. The study was approved by the Institutional Review Board of Kumamoto University (#2059) and conducted in accordance with the Declaration of Helsinki.
Consent to participate. Patient consent for inclusion in this study was waived by the Institutional Review Board of Kumamoto University (#2059, #1169).
Approval for animal experiments. All procedures were carried out in accordance with the relevant guidelines and regulations. All animal procedures were planned according to the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines and were approved by the Animal Research Committee at Kumamoto University (#A2019-015).
Consent for publication. Informed consent for publication was obtained from the relevant participants.
Results
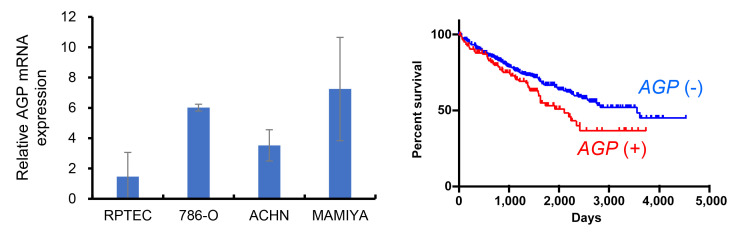
AGP expression was associated to worse clinical course in ccRCC. First, we assessed AGP gene expression in three RCC cell lines (786-O, ACHN, and MAMIYA) and in immortalized renal proximal tubular epithelial cells (RPTEC). AGP gene expression was significantly higher in the RCC cell lines compared to its expression in RPTEC (Figure 1A). Next, we examined the relationship between AGP expression and clinicopathological factors using the TCGA dataset obtained from the ProteinAtlas database (https://www.proteinatlas.org/). The analysis revealed that overall survival was significantly worse in ccRCC cases with AGP gene expression compared to cases without AGP gene expression (Figure 1B). However, no significant correlation was found between AGP gene expression and other factors, such as age, sex, or clinical stage.
Figure 1.
Gene expression of AGP in RCC. (A) AGP mRNA expression was tested by real-time PCR in immortalized renal tubular cell line and RCC cell lines. (B) The Kaplan-Meier survival analysis of The Cancer Genome Atlas (TCGA) database compared the overall survival between AGP mRNA (+) ccRCC cases and AGP mRNA(–) ccRCC cases. AGP: α1-Acid glycoprotein; RCC: renal cell carcinoma.
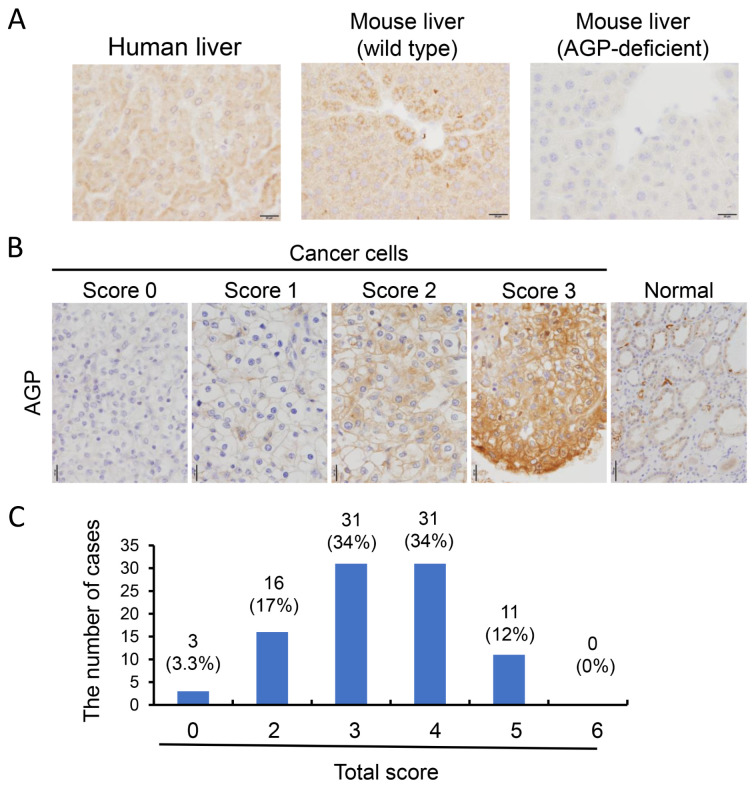
Generation of anti-AGP mouse monoclonal antibody, and the expression of AGP in ccRCC. Next, we aimed to develop a monoclonal antibody against AGP to evaluate its protein expression in ccRCC. Human recombinant AGP was used to immunize mice, as detailed in the materials and methods section. Several clones were generated using an ELISA assay with a human AGP-coated plate, and one clone was selected based on immunohistochemistry (IHC) performed on human liver tissue. This clone demonstrated reactivity with both human and mouse hepatocytes in paraffin-embedded liver tissue, while no reactivity was observed in the hepatocytes of AGP-deficient mice (Figure 2A).
Figure 2.
AGP protein expression in ccRCC. (A) To examine AGP protein expression in ccRCC, we generated an anti-AGP antibody. Positive reactivity to human and murine hepatocytes was presented. (B) AGP IHC was carried out on surgically resected ccRCC specimens. Given the heterogeneous expression of AGP, the IHC score was determined by assessing both the positive staining area and staining intensity. (C) AGP expression was detected in 89 out of 92 cases, with only 3 cases showing no AGP expression. AGP: α1-Acid glycoprotein; ccRCC: clear cell renal cell carcinoma; IHC: immunohistochemical analysis.
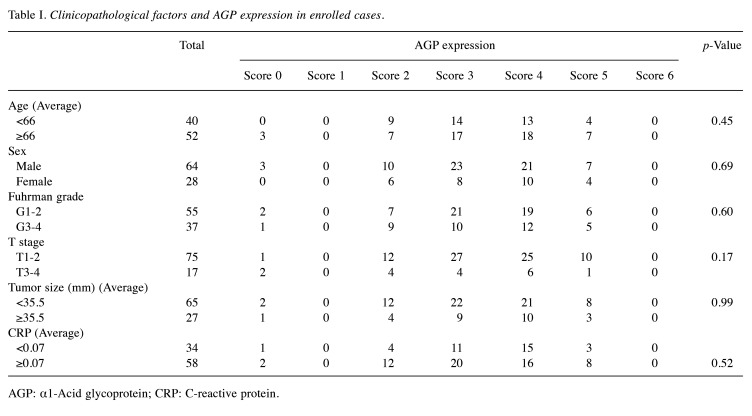
IHC for AGP was subsequently performed on surgically resected ccRCC specimens. Given the heterogeneous nature of AGP expression, the IHC score was calculated based on both the positive staining area and staining intensity, as described in the materials and methods section. Representative intensity scores for AGP are presented in Figure 2B. Of the 92 cases, only 3 were negative for AGP, while AGP expression was detected in the remaining 89 cases (Figure 2C). The correlation between AGP expression scores and clinicopathological factors was assessed; however, no significant association was found between AGP scores and factors, such as age, sex, nuclear grade, T stage, tumor size, or serum CRP levels (Table I).
Table I. Clinicopathological factors and AGP expression in enrolled cases.
AGP: α1-Acid glycoprotein; CRP: C-reactive protein.
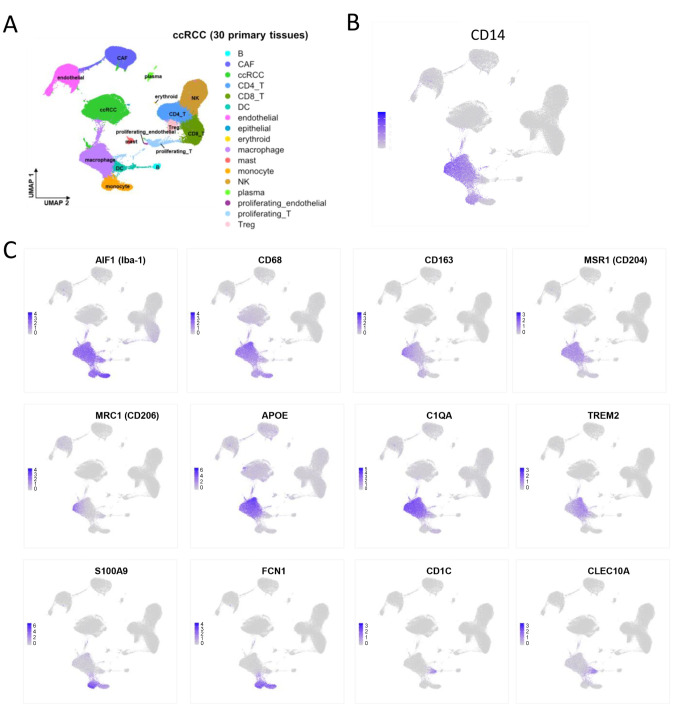
CD14, a receptor of AGP, was expressed in Iba1-positive monocyte and TAMs. It is well established that CD14 functions as a receptor for AGP, and AGP-CD14 signaling facilitates the protumor activation of tumor-associated macrophages (TAMs) (16). Consequently, CD14 gene expression was analyzed using published single-cell RNA sequencing data, as detailed in the materials and methods section. CD14 expression was identified in AIF1 (Iba-1) and CD68-expressing monocytes and TAMs, whereas no CD14 expression was detected in other cell lineages, including lymphocytes, dendritic cells, cancer cells, endothelial cells, or fibroblasts (Figure 3A-C).
Figure 3.
Single-cell RNA-sequence data of ccRCC cases. (A) UMAP plot of the 137,233 single cells from 30 primary ccRCC tissues. The cells are clustered into 17 cell types as indicated in the legend. (B) UMAP plot of the expression of CD14 was presented. (C) UMAP plots of the expression of AIF1(Iba-1), CD68, CD163, MSR1(CD204), MRC1(CD206), APOE, C1QA, TREM2, S100A9, FCN1, CD1C, CLEC10A were showed. ccRCC: Clear cell renal cell carcinoma.
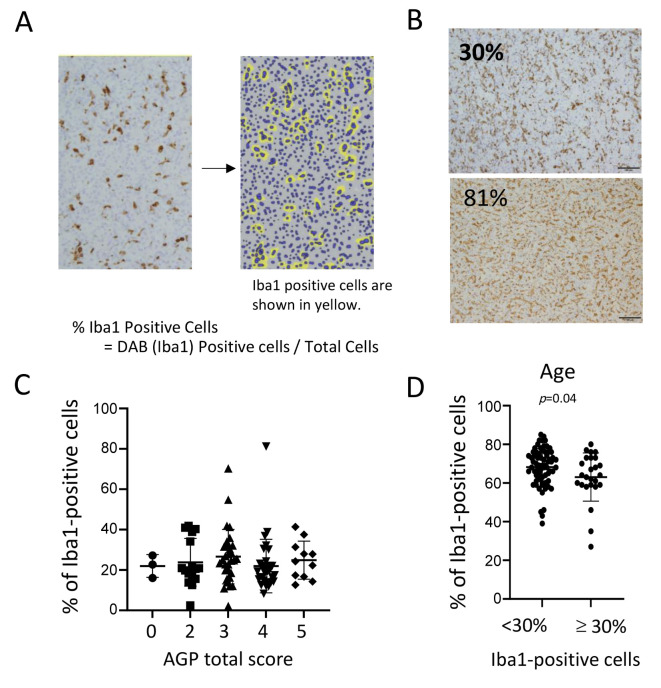
There was no significant correlation between the AGP expressions and Iba1-positive cells. In the final phase, immunohistochemistry (IHC) for Iba-1 was performed on ccRCC tissue samples, and the percentage of Iba1-positive cells relative to the total cell population was quantified using HALO software (Figure 4A and B). We subsequently analyzed the correlation between the density of Iba1-expressing cells and AGP expression in ccRCC. However, no significant association was identified between the percentage of Iba1-expressing cells and AGP expression (Figure 4C).
Figure 4.
The association between Iba-1-positive macrophages and AGP in ccRCC cases. (A) IHC for Iba-1 was conducted on ccRCC tissue, and the percentage of Iba1-positive cells among the total cell population was calculated using HALO software. In the HALO software, Iba1-positive cells are highlighted in yellow. Representative two cases of IHC for Iba-1 and analysis using HALO software were presented. (C) The relationship between the density of Iba1-expressing cells and AGP expression in ccRCC was analyzed. (D) Patients with more than 30% Iba1-positive cells have a significantly younger average age. AGP: α1-Acid glycoprotein; ccRCC: clear cell renal cell carcinoma; IHC: immunohistochemical analysis.
Next, the relationship between Iba1-positive cells and clinicopathological factors was assessed (Table II). Iba1-positive cells were found to correlate with Fuhrman grade, and patients with ≥30% Iba1-positive cells were, on average, significantly younger compared to those with <30% Iba1-positive cells (Figure 4D, Table II).
Table II. Clinicopathological factors and Iba1 expressions in enrolled cases.
AGP: α1-Acid glycoprotein; CRP: C-reactive protein.
Discussion
Recent studies have demonstrated that cancer development and progression are influenced not only by genetic mutations within cancer cells, but also by interactions between cells within the tumor microenvironment. Among these cell types, tumor-associated macrophages (TAMs) are highly abundant in tumor tissues, and their numbers increase with tumor growth and severity. Previous research has shown that α1-acid glycoprotein (AGP) can induce the expression of PD-L1, an immune checkpoint molecule, in macrophages, thus highlighting its role in TAM-mediated tumor progression within the tumor microenvironment (16).
AGP is primarily produced by hepatocytes but is also expressed in various other cell types. Analysis of ccRCC cases from the TCGA database revealed a correlation between elevated AGP expression and poor prognosis. Consequently, we developed an anti-AGP antibody and performed immunohistochemical staining, confirming its efficacy. Although AGP expression in renal cell carcinoma is generally low, our findings revealed that AGP is frequently present in human ccRCC specimens. However, due to the recent nature of the cases examined in this study, we were unable to assess the relationship between AGP expression levels and patient survival rates.
Previous studies have indicated that AGP-mediated induction of PD-L1 and secretion of IL-6 are regulated through its binding to CD14, a known co-receptor of TLR4. It has been reported that elevated AGP levels in cancer promote tumor progression by inducing immune suppression and enhancing cancer cell proliferation via CD14/TLR4 signaling in TAMs. This study further demonstrated that AGP contributes to the tumor-promoting activity of TAMs through CD14 signaling. AGP receptors include CD14, a marker for monocytes and macrophages, as well as TLR4 co-receptor and CCR5, a chemokine receptor (16,22). CD14-neutralizing antibodies significantly inhibited STAT1 activation and PD-L1 expression in AGP-induced macrophages, as well as STAT3 activation and IL-6 production, indicating that CD14/TLR4 serves as a receptor for AGP and plays a critical role in macrophage function. In patients with RCC, CD14 was detected on nearly all Iba1/CD68-positive TAMs and monocytes. Although no correlation was observed between the density of Iba1/CD68-positive cells and AGP expression, this does not account for their activation state, warranting further analysis to explore this relationship.
RCC is a unique cancer compared to other carcinomas, as it appears capable of absorbing various molecules from the serum into cells. It has been reported that the soluble form of CD163 is elevated in the cancer microenvironment and can be taken up by cancer cells via an unidentified receptor, with sCD163 subsequently being observed within cancer cells (23). Therefore, it is possible that the AGP-positive findings in renal cell carcinoma result from the uptake of AGP from the serum, and we plan to investigate the relationship between serum AGP levels and tumor AGP expression in future research.
In summary, we successfully developed an anti-AGP antibody suitable for immunohistochemistry in paraffin-embedded sections and evaluated AGP expression in ccRCC.
Conflicts of Interest
The Authors declare that they have no conflicts of interest.
Authors’ Contributions
Ayano Ezaki, Hiromu Yano, and Yoshihiro Komohara conducted the experiments and contributed to writing the manuscript. Yukio Fujiwara, Toshiki Anami, Yuki Ibe, Youjiro Ozaki, Hidekazu Nishizawa, Takanobu Motoshima, and Junji Yatsuda prepared the human tissue samples. Hiroshi Watanabe, Toru Maruyama, and Toru Takeo developed gene-deficient mice. Yoshihiro Komohara and Tomomi Kamba managed and coordinated the research activity planning. Cheng Pan analyzed the single cell-RNA sequence data set.
Acknowledgements
We thank Mr. Takenobu Nakagawa and Ms. Yuka Watanabe for the technical assistance.
Funding
This work was supported by Grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (Nos. 20H03459).
References
- 1.Grigolo S, Filgueira L. Immunotherapy of clear-cell renal-cell carcinoma. Cancers (Basel) 2024;16(11):2092. doi: 10.3390/cancers16112092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373(9669):1119–1132. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- 3.Feng X, Zhang L, Tu W, Cang S. Frequency, incidence and survival outcomes of clear cell renal cell carcinoma in the United States from 1973 to 2014: A SEER-based analysis. Medicine (Baltimore) 2019;98(31):e16684. doi: 10.1097/MD.0000000000016684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah NJ, Sura SD, Shinde R, Shi J, Singhal PK, Robert NJ, Vogelzang NJ, Perini RF, Motzer RJ. Real-world treatment patterns and clinical outcomes for metastatic renal cell carcinoma in the current treatment era. Eur Urol Open Sci. 2023;49:110–118. doi: 10.1016/j.euros.2022.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rotte A, Sahasranaman S, Budha N. Targeting TIGIT for immunotherapy of cancer: update on clinical development. Biomedicines. 2021;9(9):1277. doi: 10.3390/biomedicines9091277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apolo AB, Girardi DM, Niglio SA, Nadal R, Kydd AR, Simon N, Ley L, Cordes LM, Chandran E, Steinberg SM, Lee S, Lee MJ, Rastogi S, Sato N, Cao L, Banday AR, Boudjadi S, Merino MJ, Toubaji A, Akbulut D, Redd B, Bagheri H, Costello R, Gurram S, Agarwal PK, Chalfin HJ, Valera V, Streicher H, Wright JJ, Sharon E, Figg WD, Parnes HL, Gulley JL, Saraiya B, Pal SK, Quinn D, Stein MN, Lara PN, Bottaro DP, Mortazavi A. Final results from a phase I trial and expansion cohorts of cabozantinib and nivolumab alone or with ipilimumab for advanced/metastatic genitourinary tumors. J Clin Oncol. 2024;42(25):3033–3046. doi: 10.1200/JCO.23.02233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komohara Y, Hasita H, Ohnishi K, Fujiwara Y, Suzu S, Eto M, Takeya M. Macrophage infiltration and its prognostic relevance in clear cell renal cell carcinoma. Cancer Sci. 2011;102(7):1424–1431. doi: 10.1111/j.1349-7006.2011.01945.x. [DOI] [PubMed] [Google Scholar]
- 8.Nakano O, Sato M, Naito Y, Suzuki K, Orikasa S, Aizawa M, Suzuki Y, Shintaku I, Nagura H, Ohtani H. Proliferative activity of intratumoral CD8(+) T-lymphocytes as a prognostic factor in human renal cell carcinoma: clinicopathologic demonstration of antitumor immunity. Cancer Res. 2001;61(13):5132–5136. [PubMed] [Google Scholar]
- 9.Zhang D, Ni Y, Wang Y, Feng J, Zhuang N, Li J, Liu L, Shen W, Zheng J, Zheng W, Qian C, Shan J, Zhou Z. Spatial heterogeneity of tumor microenvironment influences the prognosis of clear cell renal cell carcinoma. J Transl Med. 2023;21(1):489. doi: 10.1186/s12967-023-04336-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chanmee T, Ontong P, Konno K, Itano N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers (Basel) 2014;6(3):1670–1690. doi: 10.3390/cancers6031670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Binnewies M, Pollack JL, Rudolph J, Dash S, Abushawish M, Lee T, Jahchan NS, Canaday P, Lu E, Norng M, Mankikar S, Liu VM, Du X, Chen A, Mehta R, Palmer R, Juric V, Liang L, Baker KP, Reyno L, Krummel MF, Streuli M, Sriram V. Targeting TREM2 on tumor-associated macrophages enhances immunotherapy. Cell Rep. 2021;37(3):109844. doi: 10.1016/j.celrep.2021.109844. [DOI] [PubMed] [Google Scholar]
- 12.Beckermann KE, Patnaik A, Winer I, Tan W, Bashir B, Kyriakopoulos CE, Sweis RF, Chamberlain M, Rini BI. A phase 1b open-label study to evaluate the safety, tolerability, pharmacokinetics, and pharmacodynamics of py314 in combination with pembrolizumab in patients with advanced renal cell carcinoma. Invest New Drugs. 2024;42(2):179–184. doi: 10.1007/s10637-024-01419-1. [DOI] [PubMed] [Google Scholar]
- 13.Young M, Tapia JC, Szabados B, Jovaisaite A, Jackson-Spence F, Nally E, Powles T. NLR outperforms low hemoglobin and high platelet count as predictive and prognostic biomarker in metastatic renal cell carcinoma treated with immune checkpoint inhibitors. Clin Genitourin Cancer. 2024;22(3):102072. doi: 10.1016/j.clgc.2024.102072. [DOI] [PubMed] [Google Scholar]
- 14.Tuttle CS, Thang LA, Maier AB. Markers of inflammation and their association with muscle strength and mass: A systematic review and meta-analysis. Ageing Res Rev. 2020;64:101185. doi: 10.1016/j.arr.2020.101185. [DOI] [PubMed] [Google Scholar]
- 15.Baumann H, Prowse KR, Marinković S, Won K, Jahreis GP. Stimulation of hepatic acute phase response by cytokines and glucocorticoids. Ann N Y Acad Sci. 1989;557(1):280–296. doi: 10.1111/j.1749-6632.1989.tb24021.x. [DOI] [PubMed] [Google Scholar]
- 16.Matsusaka K, Fujiwara Y, Pan C, Esumi S, Saito Y, Bi J, Nakamura Y, Mukunoki A, Takeo T, Nakagata N, Yoshii D, Fukuda R, Nagasaki T, Tanaka R, Komori H, Maeda H, Watanabe H, Tamada K, Komohara Y, Maruyama T. α1-acid glycoprotein enhances the immunosuppressive and protumor functions of tumor-associated macrophages. Cancer Res. 2021;81(17):4545–4559. doi: 10.1158/0008-5472.CAN-20-3471. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Narayanan SP, Mannan R, Raskind G, Wang X, Vats P, Su F, Hosseini N, Cao X, Kumar-Sinha C, Ellison SJ, Giordano TJ, Morgan TM, Pitchiaya S, Alva A, Mehra R, Cieslik M, Dhanasekaran SM, Chinnaiyan AM. Single-cell analyses of renal cell cancers reveal insights into tumor microenvironment, cell of origin, and therapy response. Proc Natl Acad Sci USA. 2021;118(24):e2103240118. doi: 10.1073/pnas.2103240118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su C, Lv Y, Lu W, Yu Z, Ye Y, Guo B, Liu D, Yan H, Li T, Zhang Q, Cheng J, Mo Z. Single-cell RNA sequencing in multiple pathologic types of renal cell carcinoma revealed novel potential tumor-specific markers. Front Oncol. 2021;11:719564. doi: 10.3389/fonc.2021.719564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Z, Lu W, Su C, Lv Y, Ye Y, Guo B, Liu D, Yan H, Mi H, Li T, Zhang Q, Cheng J, Mo Z. Single-cell RNA-seq identification of the cellular molecular characteristics of sporadic bilateral clear cell renal cell carcinoma. Front Oncol. 2021;11:659251. doi: 10.3389/fonc.2021.659251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Z, Lv Y, Su C, Lu W, Zhang R, Li J, Guo B, Yan H, Liu D, Yang Z, Mi H, Mo L, Guo Y, Feng W, Xu H, Peng W, Cheng J, Nan A, Mo Z. Integrative single-cell analysis reveals transcriptional and epigenetic regulatory features of clear cell renal cell carcinoma. Cancer Res. 2023;83(5):700–719. doi: 10.1158/0008-5472.CAN-22-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ezaki A, Yano H, Pan C, Fujiwara Y, Anami T, Ibe Y, Motoshima T, Yatsuda J, Esumi S, Miura Y, Kamba T, Komohara Y. Potential protumor function of CD74 in clear cell renal cell carcinoma. Hum Cell. 2024;37(5):1535–1543. doi: 10.1007/s13577-024-01110-w. [DOI] [PubMed] [Google Scholar]
- 22.Ceciliani F, Lecchi C. The immune functions of α1acid glycoprotein. Curr Protein Pept Sci. 2019;20(6):505–524. doi: 10.2174/1389203720666190405101138. [DOI] [PubMed] [Google Scholar]
- 23.Ma C, Horlad H, Ohnishi K, Nakagawa T, Yamada S, Kitada S, Motoshima T, Kamba T, Nakayama T, Fujimoto N, Takeya M, Komohara Y. CD163-positive cancer cells are potentially associated with high malignant potential in clear cell renal cell carcinoma. Med Mol Morphol. 2018;51(1):13–20. doi: 10.1007/s00795-017-0165-8. [DOI] [PubMed] [Google Scholar]