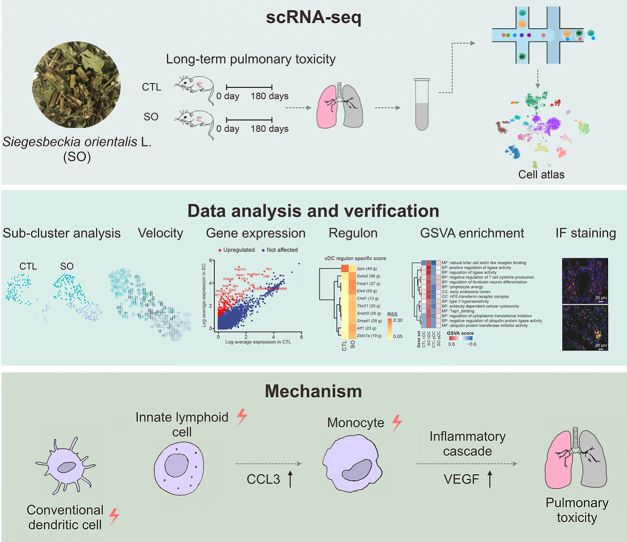
Graphical abstract
Highlights
-
•
scRNA-seq is a advanced technology to dissect the immune microenvironment of SO induced long-term pulmonary toxicity.
-
•
cDC, ILCs, and monocytes activation may play crucial roles in SO-induced pulmonary toxicity.
-
•
CCL3 signal released by ILCs may activate the inflammatory cascade of monocytes in lung tissues of SO-treated rat.
-
•
VEGF activation in monocytes participated in the inflammatory cascade caused by SO.
Siegesbeckia orientalis L. (SO) is traditionally used in Chinese medicine for joint health and is a common herbal remedy in Asian countries. The metabolic alterations and long-term pulmonary toxicity of the water extract of SO have been observed in our previous study [1]. Upon six months of SO treatment, the lung tissues of rat displayed signs of cellular edema, inflammation, and infiltration of inflammatory cells, as evidenced by hematoxylin-eosin (H&E) staining. These pathological alterations were most pronounced in the lungs when compared to other organs, suggesting that SO has long-term pulmonary toxicity. However, the underlying molecular mechanisms contributing to this pulmonary toxicity is limited. In this study, single-cell RNA sequencing (scRNA-seq) was employed to delineate the cellular diversity within lung tissues and to investigate the alterations in the immune environment and cellular functions following prolonged administration of SO.
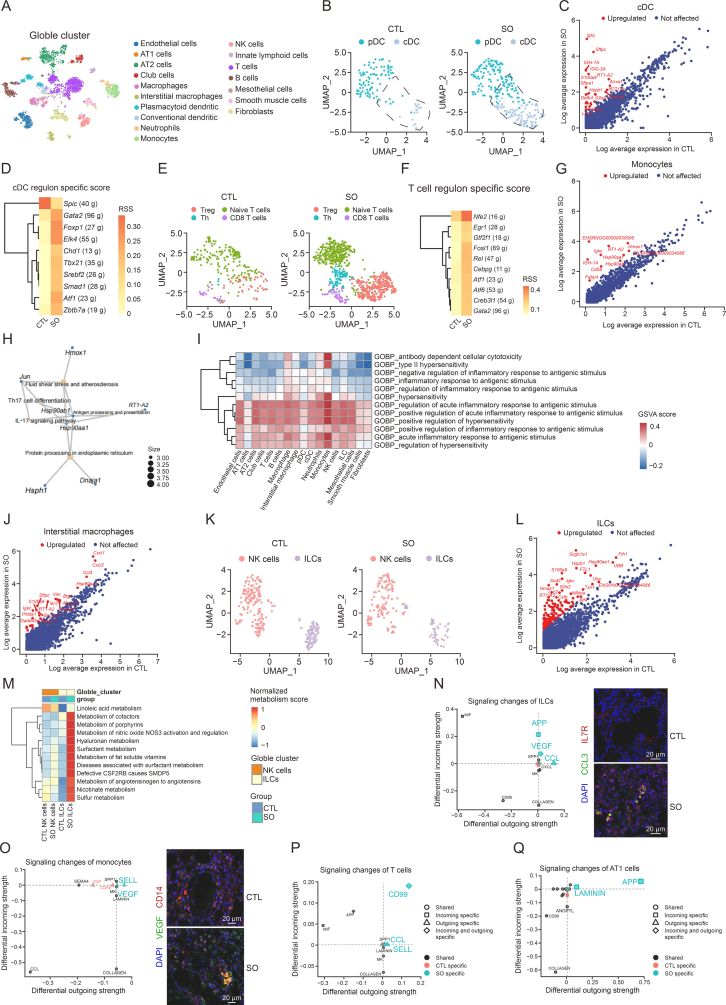
scRNA-seq analysis was performed with lung samples dissected from SO-treated and control (CTL) rats. After quality control and cell filtering following the protocol of Seurat, we obtained a total of 5,232 cells and 26,404 transcripts. Principal component analysis (PCA) and unsupervised clustering analysis revealed 17 major cell types, including myeloid, lymphoid, and stromal cells (Fig. 1A). The cell clusters exhibited high purity and homogeneity, as evidenced by their high entropy scores (Supplementary data). It was found that the treatment group had a high proportion of AT2 cells, conventional dendritic cells (cDCs), and T cells, and low proportions of interstitial macrophages, neutrophils, monocytes, and fibroblasts (Fig. S1A). Subsequently, we performed a comprehensive analysis of each immune cell type. Dendritic cells (DCs) subcluster analysis results showed that cDCs were significantly increased in treatment group, with elevated expressions of Igkc, Sftpc, and S100a9 (Figs. 1B and C). However, plasmacytoid dendritic cells (pDCs) did not exhibit substantial differentially expressed genes (DEGs), suggesting that SO has little effect on pDCs. Gene set variation analysis (GSVA) revealed that the ribosome pathway, natural killer cell lectin-like receptor binding, and positive regulation of ligase activity were significantly upregulated in DCs after treated with SO (Fig. S1B).
Fig. 1.
Single-cell RNA sequencing (scRNA-seq) was used to identify diverse cellular change in the lungs of the water extract of Siegesbeckia orientalis L. (SO)-treated rats. (A) t-distributed Stochastic neighbor embedding (t-SNE) plot showing the lung cells cluster from control (CTL) and SO groups. (B) Uniform manifold approximation and projection (UMAP) plot showing the dendritic cells (DCs) integration from two groups. (C) Conventional dendritic cells (cDCs) differentially expression genes (DEGs) analysis between CTL and SO groups. (D) cDCs regulon specific score reveals the differential regulon activation between CTL and SO. (E) Unsupervised clustering of T cells. (F) T cells regulon specific score showing the differential regulon activation between CTL and SO groups. (G) Monocytes DEGs analysis between CTL and SO groups. (H) Monocyte Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis based on the SO-upregulated DEGs. (I) Gene set variation analysis (GSVA) heatmap illustrating the expression of the inflammatory-related biological processes in all cell types within the SO group. (J) Interstitial macrophages DEGs analysis between CTL and SO groups. (K) UMAP plot showing innate lymphoid cells (ILCs) and natural killer (NK) cells from two groups. DEGs related to NK cells and ILCs based on the integrated dataset. (L) ILCs DEGs analysis between CTL and SO groups. (M) Metabolism heatmap to compare the metabolic changes in T cells and B cells between the CTL and SO groups. (N, O) Differential outgoing or incoming signaling pathways of ILCs and monocytes between CTL and SO groups from cell communication analysis. The expression of C−C motif chemokine ligand 3 (CCL3) in ILCs (N) and the expression of vascular endothelial growth factor (VEGF) in monocytes (O) were validated by immunofluorescence staining, respectively. (P, Q) Differential outgoing or incoming signaling pathways of T cells (P) and AT1 cells (Q) between CTL and SO groups from cell communication analysis. In Figs. 1C, G, J, and L, red dots represent the upregulated genes in SO when compared with CTL group and blue dots represent those not altered genes in both CTL and SO groups. pDC: plasmacytoid dendritic cells; RSS: regulon specific score; Th: T helper cell; IL-17: interleukin-17; NOS3: nitric oxide synthase 3; CSF2RB: colony stimulating factor 2 receptor subunit beta; SMDP5: pulmonary surfactant metabolism dysfunction-5; APP: amyloid precursor protein; SPP1: secreted phosphoprotein 1; CXCL: chemokine (C−X−C motif) ligand; MK: midkine; DAPI: 4’,6-diamidino-2’-phenylindole; IL-7R: IL-7 receptor; SEMA4: semaphorin 4; CSF: colony-stimulating factor; MIF: macrophage migration inhibitory factor; ANGPTL: angiopoietin like.
By integrating RNA velocity analysis (Supplementary data) with dynamic modeling using scVelo, Tcf4 was identified as a key driver in the vector field of the development trajectories of SO-treated cDCs (Figs. S1C and D). Single-cell regulatory network inference and clustering (SCENIC) analysis [2] revealed that the regulons Foxp1 and Elk4 were specifically activated in cDCs, whereas the Spic regulon was inactivated after treatments (Fig. 1D). Interestingly, SO treatment did not impact B cells but increased T cells population (Fig. 1E). GSVA showed that cellular cytotoxicity, hypersensitivity, and antigenic response were enhanced (Fig. S1E), and Nfe2 regulon was also activated in T cells after treatments (Fig. 1F). Regarding neutrophils and monocytes, SO has little effect on the neutrophils, while RT1-A2, Hmox1, and Hsp90aa1 were upregulated in monocytes, in which DEGs Jun, Hsp90aa1, and Hsp90ab1 were enriched in the interleukin-17 (IL-17) signaling pathway (Figs. 1G and H). GSVA enrichment showed that lymphocyte anergy was specifically upregulated in monocytes, while the regulation of complement-dependent cytotoxicity and anchored components of the external side of the plasma membrane were upregulated in both monocytes and neutrophils after treatments (Fig. S1F). Monocytes displayed the most significant activation in pathways associated with inflammation compared to other immune cell types, indicating their pro-inflammatory roles in mediating lung toxicity (Fig. 1I). Interestingly, SO treatment increased interstitial macrophages but not macrophages, which is also suggested by the differential expression tests that revealed a profound impact of SO on interstitial macrophages. The treatments induced the upregulation of Cxcl1, Cxcl2, Ccl3, Vim, and S100a9 in interstitial macrophages (Fig. 1J). The treatments also increased the acute inflammatory response to antigenic stimuli in macrophages (Fig. S1G). Glycosphingolipid metabolism and polyamine metabolism were significantly inhibited in interstitial macrophages. The defective colony stimulating factor 2 receptor subunit beta (CSF2RB)-caused surfactant metabolism dysfunction may play an important role in mediating the long-term pulmonary toxicity induced by SO (Fig. S1H). The SO treatments exerted a limited effect on natural killer (NK) cells but a profound effect on innate lymphoid cells (ILCs) (Figs. 1K and L). Although SO treatments reduced the number of ILCs, a significant amount of genes were upregulated in ILCs, including RT1-A2, Hsp90aa1, and Nfkbia (Fig. 1L). These genes were likely associated with antigen processing and presentation pathways (Fig. S1I). A defective CSF2RB-caused surfactant metabolism dysfunction was observed in ILCs (Fig. 1M). It also showed an activation in Myc, Sox17, and Cebpd regulons (Fig. S1J). The effects of SO treatments were more pronounced in AT2 cells when compared to AT1 cells, fibroblasts, smooth muscle cells, and endothelial cells as indicated by DEGs analysis (Fig. S1K). Nr3c1 and Irf7 regulons, as well as the inflammatory pathways were specifically activated in AT2 cells after the treatments (Figs. S1L and M).
Next, we employed CellChat [3] to investigate intercellular communications. Specifically, we found that both outgoing and incoming signals were attenuated after SO treatment in most clusters (Fig. S2A). However, there was an increase in the strength of cell interactions in certain clusters, such as ILCs and monocytes (Fig. S2B). C−C motif chemokine ligand (CCL) signaling was essential for monocytes recruitment during inflammation [4]. Here, we also observed that the ILCs may activate the inflammatory cascade of monocytes and macrophages through the CCL3-C−C motif chemokine receptor 1 (CCR1) signaling (Figs. 1N and S2C). The activation of CCL3 in ILCs also was validated by immunofluorescence staining (Fig. 1N). Reports has proposed that monocytes recruitment and vascular endothelial growth factor (VEGF) production play a critical role during lung injury [5]. Both the tissue immunofluorescence staining and CellChat results showed that monocytes specifically upregulated VEGF and SELL outgoing signals, which could affect the endothelial cells, pDCs, and ILCs via Vegfa-Vegfr2 and Sell-Podxl interactions (Figs. 1O and S2D). Therefore, the interaction between ILCs and monocytes may significantly contribute to the SO-induced chronic pulmonary toxicity. In T cells, the treatments increased SELL, CCL, and CD99 signaling that affected endothelial cells, macrophages, and fibroblasts (Fig. 1P). Enhanced interaction between AT1 cells and ILCs with reduced outgoing signals from smooth muscle cells were also observed. Amyloid precursor protein (APP) incoming signaling was enhanced in AT1 cells (Fig. 1Q).
In conclusion, our data suggests that the activation of cDC, ILCs, and monocytes play important roles in SO-altered pulmonary immune environment in rat. Notably, CCL3 signaling activated in ILCs was found to be crucial for the subsequent VEGF activation in monocytes, which emerged as a pivotal element in the inflammatory cascade induced by SO. These findings provide scientific justification for the rational and safe use of SO.
Ethical statement
All animal experiments were conducted in accordance with the guidelines of the Ethics Review Boards of the International Institute for Translational Chinese Medicine, Guangzhou University of Chinese Medicine, China (Approval No.: 2020W0088).
Data availability statement
The scRNA-Seq data were public available in China National Center for Bioinformation GSA database under accession CRA016051. The detailed methods were shown in Supplementary data.
CRediT author statement
Fang Zhang: Data curation, Visualization, Software, Writing - Reviewing and Editing; Shu Gan, Jingjing Liao, Ting Jiang, Zhiqiang Shi, and Xueying Fan: Methodology, Validation; Hiu-Yee Kwan: Supervision, Writing - Reviewing and Editing; Zhongqiu Liu: Project administration, Supervision, Writing - Reviewing and Editing; Tao Su: Project administration, Supervision, Writing - Original draft preparation, Reviewing and Editing.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (Grant No.: 2023YFC3502800), the National Natural Science Foundation of China (Grant Nos.: 82274158 and 82074019), the Characteristic Innovation Projects of Universities in Guangdong Province, China (Grant No.: 2023KTSCX023), the Administration of Traditional Chinese Medicine of Guangdong Province, China (Grant Nos.: 20222042 and 20241074), the Young Elite Scientists Sponsorship Program by China Association of Chinese Medicine (Grant No.: 2021-QNRC2-B15), Guangdong Basic and Applied Basic Research Foundation, China (Grant No.: 2020B1515130005), the Special Research Project of Traditional Chinese Medicine Science and Technology of Guangdong Provincial Hospital of Traditional Chinese Medicine, China (Grant No.: YN2020MS04), and the Special Program for Top Talents of Guangdong Provincial Hospital of Traditional Chinese Medicine, China (Grant No.: BJ2022YL03).
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2024.101035.
Contributor Information
Hiu-Yee Kwan, Email: hykwan@hkbu.edu.hk.
Zhongqiu Liu, Email: liuzq@gzucm.edu.cn.
Tao Su, Email: sutao@gzucm.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Jiang T., Liu L., Zhang M., et al. Metabolomics reveals the mechanisms for the pulmonary toxicity of Siegesbeckia orientalis L. and the toxicity-reducing effect of processing. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.630319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van de Sande B., Flerin C., Davie K., et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc. 2020;15:2247–2276. doi: 10.1038/s41596-020-0336-2. [DOI] [PubMed] [Google Scholar]
- 3.Jin S., Guerrero-Juarez C.F., Zhang L., et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi C., Pamer E.G. Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 2011;11:762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang T.-H., Fang P.-H., Li J.-M., et al. Cyclooxygenase-2 activity regulates recruitment of VEGF-secreting Ly6Chigh monocytes in ventilator-induced lung injury. Int J. Mol. Sci. 2019;20:1771. doi: 10.3390/ijms20071771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-Seq data were public available in China National Center for Bioinformation GSA database under accession CRA016051. The detailed methods were shown in Supplementary data.