Abstract
High mortality and low response rates in lung cancer patients call for novel therapeutic targets. Data mining of whole-genome genetic dependency screens suggest Cell Division Cycle 40 (CDC40) to be an essential protein for lung cancer cell survival. We characterized CDC40 knockdown effects in multiple lung cancer cell lines, revealing induced cell cycle defects that resulted in strong growth inhibition and activation of apoptosis. Global transcriptional and splicing changes were also investigated, where CDC40 knockdown resulted in perturbation of splicing- and translation-related genes as well as more transcripts with intron retention. In the transcript of the cell cycle regulatory protein CDCA5, CDC40 knockdown was shown to induce retention of the first intron, leading to an increase in the unspliced CDCA5 transcript and subsequent decrease in CDCA5 protein expression. Additionally, protein–protein interactions of CDC40 were explored and spliceosome components were found to be its main binding partners, further highlighting the role of CDC40 in splicing. CDC40 mutation analysis suggests that it may be difficult to disrupt key interactions using small molecules within a large complex. Our results demonstrate that CDC40 is essential for lung cancer cell growth, and that its inhibition may represent a viable therapeutic strategy for lung cancer.
Subject terms: Cancer, Cell biology, Molecular biology, Oncology
Introduction
Worldwide, lung cancer was the second most diagnosed and the leading cause of cancer mortality. In the United States, 0.2 million new cases and 0.1 million deaths are projected in 20231. Options for treatment include surgery, chemotherapy, and radiotherapy; however, these therapies lack specificity, leading to damaging side effects. Answering the call for novel targeted therapies are genetic studies of lung cancer dependencies, which open new opportunities for investigation and drug development.
Recently, results from large, pancancer shRNA screens were published, evaluating the genome-wide dependencies of cancer cell lines2–6. Cell Division Cycle 40 (CDC40), a WD40 repeat (WDR) protein involved in pre-mRNA splicing7, showed a significant essentiality in lung lines. In yeast, CDC40 participates in pre-mRNA splicing in the spliceosomal C complex, catalyzing intron/exon separation7. Here, CDC40 mutation induces splicing defects and G2/M cell cycle arrest, suggesting participation in both processes7. Interestingly, structural studies of yeast and mammalian spliceosomes have localized CDC40 within spliceosome Bact, B* (active), C, C* (active), P and ILS complexes, suggesting broad functionality8–11. However, CDC40 spliceosome interactions were not characterized and the mammalian role of CDC40 remains to be fully elucidated.
Pre-mRNA splicing is a fundamental cellular process essential to the regulation of gene expression which may support carcinogenesis when altered12. Changes in expression or regulation of several splicing factors can induce cell migration, proliferation, and apoptosis resistance12. Alternative splicing is essential for transcriptome and proteome diversity; however, aberrant alternative splicing favors the generation of lung cancer-associated splicing variants of Bcl-x and Cyclin D1, which contributes to disease progression12,13. Thus, therapeutic modulation of cancer-supporting splicing patterns could be an effective anti-cancer strategy.
Here we investigated the effects of CDC40 reduction in a panel of lung cancer cells, demonstrating that knockdown induces cell cycle defects, strong growth inhibition, and apoptosis in all tested lines. We investigated global transcriptional and splicing changes and found that CDC40 knockdown resulted in a perturbation of genes related to splicing and translation, together with an increase in intron retention events in transcripts. In the CDCA5 transcript, a potential prognostic marker for cancer, there was increased retention in the first intron, leading to decreased CDCA5 protein. Finally, we explored the protein interaction network of CDC40 in lung cancer and found spliceosome components to be its predominant partners. Mutated CDC40 showed no disruption in its protein–protein interactions. Furthermore, mutated CDC40 fully rescued CDC40-knockdown induced phenotypes in the same manner as wildtype. Overall, our results demonstrate the essentiality of CDC40 for lung cancer cell growth and its potential as a target for cancer therapeutics.
Results
CDC40 knockdown suppresses lung cancer cell growth
Analysis of genome-wide shRNA dropout screens revealed that approximately 20% of lung lines were highly dependent on CDC404,14. Although useful in providing information about pan-genome genetic vulnerabilities across 700 pan-cancer lines, these data lack concordance. Differences in shRNA efficiency, gene expression, and shRNA off-target activities may contribute to artifacts that compromise conclusions. Moreover, these screens employ different data analysis algorithms, adding complexity to result interpretation. These challenges support additional gene and disease-focused target validation. We used a doxycycline (DOX)-inducible lentivirus-based delivery system for shRNA transduction into H460 lung cancer cells. A pool of 4 non-targeting shRNAs (shCTR) and a toxic control (shPLK1) were used as negative and positive controls respectively for cell dependency within our studies.
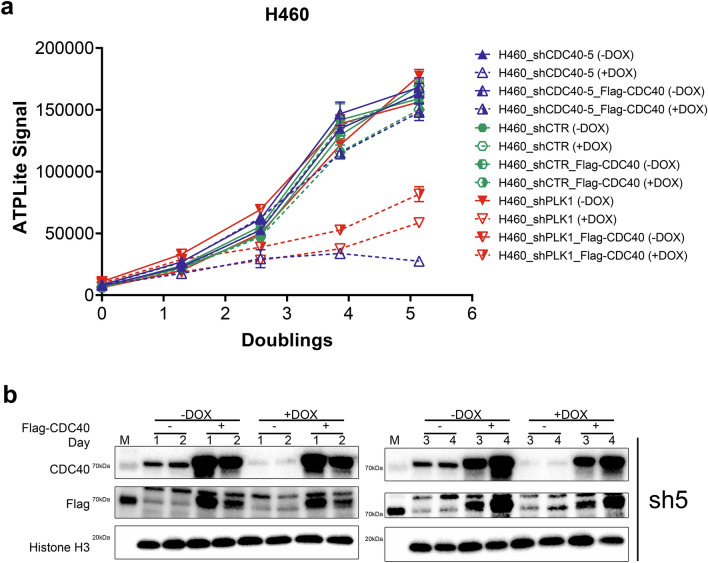
Upon DOX-induction in H460 cells (Fig. 1a, b), efficient knockdown of CDC40 mRNA (Figure S1a) and protein (Figure S1b and S20a) were observed after 8 and 24 h respectively, followed by reduced cell proliferation (Fig. 1a, Figure S20b) compared to shCTR. To rule out off-target activities, rescue experiments with overexpressed, shCDC40-resistant, Flag-tagged CDC40 (termed Flag-CDC40) were performed. High CDC40 levels (Fig. 1b) were maintained following knockdown, confirming successful over-expression (Fig. 1b) and normal proliferation was restored with 3 different shCDC40s in H460 and H1299 cells (Fig. 1a, Figure S2b and S20b). These results demonstrate that the observed anti-proliferative effect was caused by CDC40 knockdown.
Fig. 1.
Knockdown of CDC40 reduces proliferation of H460 lung cancer cells. (a) Assessing the effect of CDC40 knockdown on H460 cell proliferation after 4 days of shCDC40 induction. (n = 3, ± SD, Student’s T-test) (b) Western blot analysis showing the effect of Flag-CDC40 overexpression on the expression of Flag and CDC40 after 4 days of DOX induction (see Supplementary Fig. 12 for source blot images). Results are representative of at least two independent experiments.
Next, we assessed the effect of CDC40 knockdown in 5 additional lung lines (H2170, H1975, A549, H322, and H1915) representing different sub-types of non-small cell lung cancer (NSCLC; adenocarcinoma, large cell, and squamous cell). Within 24 h of CDC40 shRNA knockdown, protein expression was reduced in all lines (Figure S3 and S24a) as was cell proliferation (Figure S4 and S24b). Since CDC40 knockdown is detrimental to multiple lung cancer lines, we also assessed its essentiality in the non-tumorigenic, breast epithelial MCF10A line. CDC40 knockdown was shown to slow down MCF10A proliferation after 4 days of induction, albeit at a lesser magnitude. Compared to lung cancer cells, which had almost no cells remaining, the DOX-treated MCF10A cell number continues to climb with a shallower slope over 4 days, suggesting textural sensitivity between cancer and non-tumorigenic cell lines. (Figure S4 and S24b). Taken together, our results demonstrate that CDC40 knockdown induces strong growth suppressive phenotypes in multiple cell lines, although there is some texture in the severity of the phenotype.
CDC40 knockdown induces apoptosis in lung cancer cells
To investigate the growth suppression mechanism behind CDC40 knockdown, apoptosis activation was evaluated using a caspase-3/7 assay. Within the same time frame as the anti-proliferative phenotype, increased caspase-3/7 activity was detected at varying levels in all cell lines (Figure S5 and S25a), suggesting that suppression of proliferation is at least partially attributable to apoptotic induction. Notably, compared to knockdown of the toxic control PLK1, sh5-induced MCF10A cells displayed less caspase-3/7 activation than sh5-induced lung cancer cells, indicating reduced dependence on CDC40 for cell proliferation. This corroborates the previous observation of CDC40 knockdown leading to milder anti-proliferative phenotype in MCF10A cells than lung cancer cells. Furthermore, Flag-CDC40 rescued the apoptotic phenotype in H460 and H1299 cells over 4 days with three different shCDC40 hairpins (Figure S6 and S25b). These results demonstrate that CDC40 knockdown activates late-stage apoptosis markers in multiple lung cancer cells.
CDC40 knockdown alters cell cycle distribution
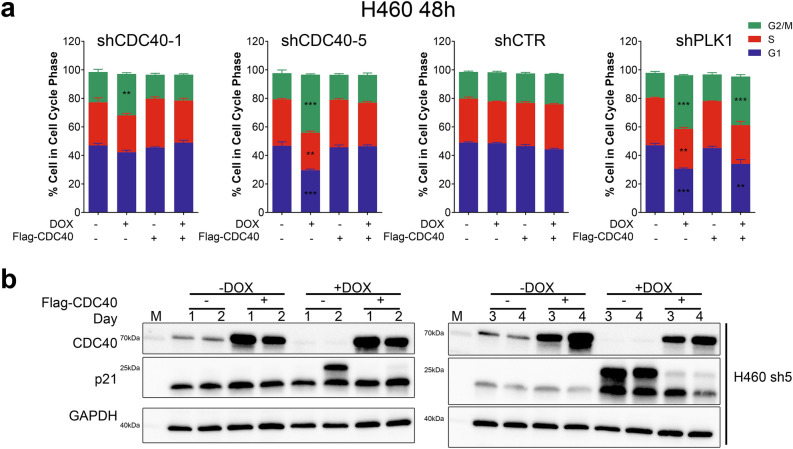
Previous studies have shown that CDC40 loss alters cell cycle dynamics, with G2/M arrest in yeast cells7. Consequently, we assessed the cell cycle effect of knockdown in lung cancer cells. Unsynchronized cells post-knockdown were collected for analysis via flow cytometry (using Propidium Iodide (PI)) to examine cell cycle distribution. After 48 h of shCDC40-5 (sh5) induction, there was increased G2/M in H460, H322, H1915, H1299 and H2170 cells (Fig. 2a, Figure S7a and S7c) which was reversed with over-expressing Flag-CDC40 (Fig. 2a). In A549 cells, a different pattern emerged with a more prominent decrease in S phase with redistribution into both G1 and G2 (Figure S7c). In MCF10A cells, cells redistributed from both S and G2/M phases into G1 (Figure S7c). To mechanistically explore the observed cell cycle defects, we checked the expression levels of p21, a core cell cycle checkpoint inhibitor. We found p21 significantly increased in H460 cells (p53 wild type) after knockdown which was restored to baseline levels by CDC40 over-expression (Fig. 2b). Interestingly, a higher molecular weight band appeared in p53 wild-type H460 cells and was rescued by over-expressed Flag-CDC40 (Fig. 2b and S9). Regarding the p21 alteration timeframe, a significant increase in the p21 protein level was observed in A549 and H460 cells as early as 48 h following DOX induction, and within 72 h in MCF10A and H1299 cells. In contrast, p21 protein level reduction was observed in H322 cells after 72 h of shCDC40-5 induction. The molecular weight shift in the p21 band was observed in A549, H322, MCF10A, H460 and H1299 cells (Figure S9 and S10). Our results revealed CDC40 knockdown alters cell cycle distribution: namely G1/S and G2/M accumulation and p21 levels alteration.
Fig. 2.
CDC40 knockdown leads to cell cycle distribution changes in multiple lung cancer cell lines. (a) The effect of Flag-CDC40 over-expression on cell cycle distribution of H460 cells after 48 h of shCDC40-1 and -5 induction in comparison to shCTR and shPLK1. (*, p < 0.05, **, p < 0.01, ***, p < 0.001, ****, p < 0.0001; n = 3, ± SD, Student’s T-test) (b) Western blot analysis showing p21 expression levels after 4 days of CDC40 knockdown using shCDC40-5 (representative) in H460 cells. (see Supplementary Fig. 14 for source blot images).
CDC40 knockdown upregulates splicing and translation pathway genes and downregulates genes in general pathways
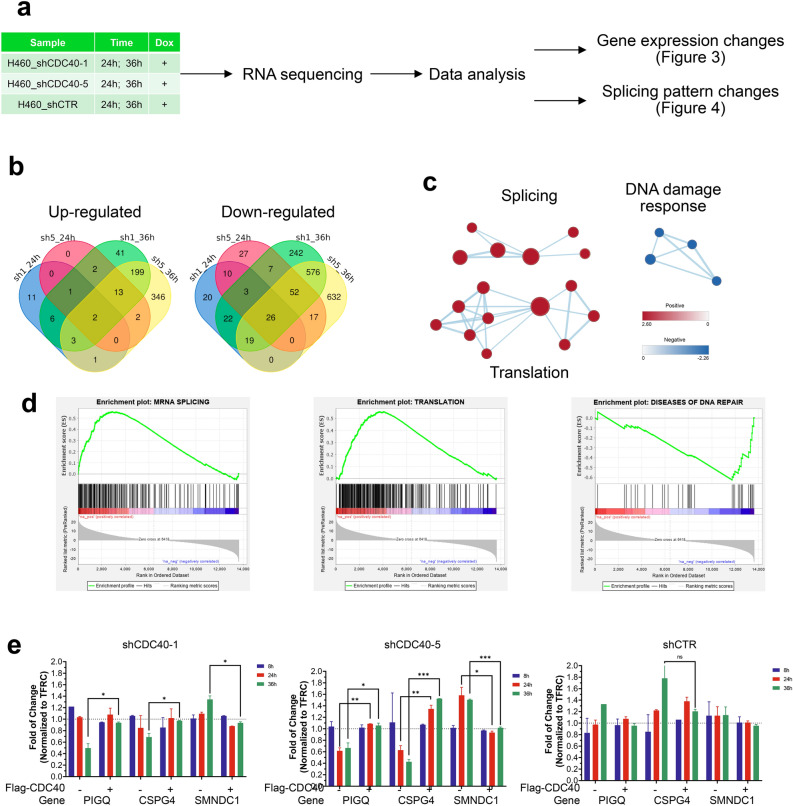
To better understand the splicing and transcriptional consequences of CDC40 knockdown, a RNA-sequencing (RNA-seq) study was conducted using total RNA extracted from H460 cells at 24 and 36 h after shCDC40-1 (sh1), sh5 and shCTR induction (Fig. 3a). Between the sh1- and sh5-induced cells, 3 genes were found to be upregulated after 24 h increasing to 217 genes after 36 h (Fig. 3b), and 39 and 673 genes were commonly downregulated after 24 h and 36 h, respectively (Fig. 3b). Following this, Gene Set Enrichment Analysis (GSEA) identified the top 30 up- and down-regulated pathways at 36 h (Figure S21). Noticeably, up-regulated pathways were enriched in splicing- and translation-related functions (Fig. 3c), among which splicing (NES = 2.26; ranked 21st) and translation (NES = 2.54; ranked 3rd) related genes were elevated (Fig. 3d, Figure S21). This suggests that CDC40 is implicated in splicing and translation-related cellular processes, as cells appear to compensate for the loss of CDC40 by upregulating splicing and translation pathways. On the other hand, although DNA damage response genes (NES = -1.99; ranked 4th) were down after 36 h (Fig. 3d, Figure S21), downregulated pathways were not enriched in any particular function, indicating that CDC40 knockdown induces general dysfunction of mRNA transcription leading to decreased expression of multiple transcripts. Three genes (SMNDC1, CSPG4 and PIGQ) were selected for validation using real-time qPCR, and we confirmed the increase in SMNDC1 (splicing-related)15 and decreases in CSPG4 (proteoglycan-, proliferation-related)16 and PIGQ (glycolipid biosynthesis-, proliferation-related)17 (Fig. 3e). These changes were prevented by over-expressing Flag-CDC40. Together, these results indicate that CDC40 knockdown results in upregulation of genes involved in splicing and downregulation of genes involved in proliferation.
Fig. 3.
CDC40 knockdown results in up-regulation of splicing and translation related genes and down-regulation of DNA damage response and proliferation related genes. (a) RNA-seq workflow using H460 cells. (b) Gene expression level changes after knockdown compared to shCTR. Left: Number of genes up-regulated after induction of shCDC40-1 and -5 for 24 h and 36 h in comparison to shCTR. Right: Number of genes down-regulated after induction of shCDC40-1 and -5 for 24 and 36 h in comparison to shCTR. (d) Pathway enrichment via GSEA using genes whose expression levels changed with both shCDC40-1 and -5 after 36-h induction and (c) visualization using a Cytoscape Enrichment Map. (e) RT-qPCR validation of RNA-Seq up-regulated gene (SMNDC1) and down-regulated genes (PIGQ and CSPG4) in response to shCDC40-1, shCDC40-5 and shCTR induction after 8, 24 and 36 h. (*, p < 0.05, **, p < 0.01, ***, p < 0.001, ****, p < 0.0001; n = 2, ± SD, Student’s T-test).
Our RNA-seq results revealed the upregulation of many translation-related genes, including the cap-dependent initiation pathway (Figure S21). We thus examined protein expression of cap binding factors from the eIF4 family and eIF2α, a critical factor involved in initiator tRNA recognition18–20. CDC40 knockdown increased eIF2α p-Ser51 (a marker for translation halt)20, and decreased phospho-eIF4B and phospho-eIF4E (Figure S8) which would also inhibit translation, pointing to a translation shutdown. Taken together, these results suggest that translation pathways were inhibited after losing CDC40, and the upregulation of genes involved in these pathways may act as a compensatory mechanism. The inhibitory effects on translation may result from accumulated improperly spliced transcripts due to CDC40 knockdown.
CDC40 knockdown induces intron retention
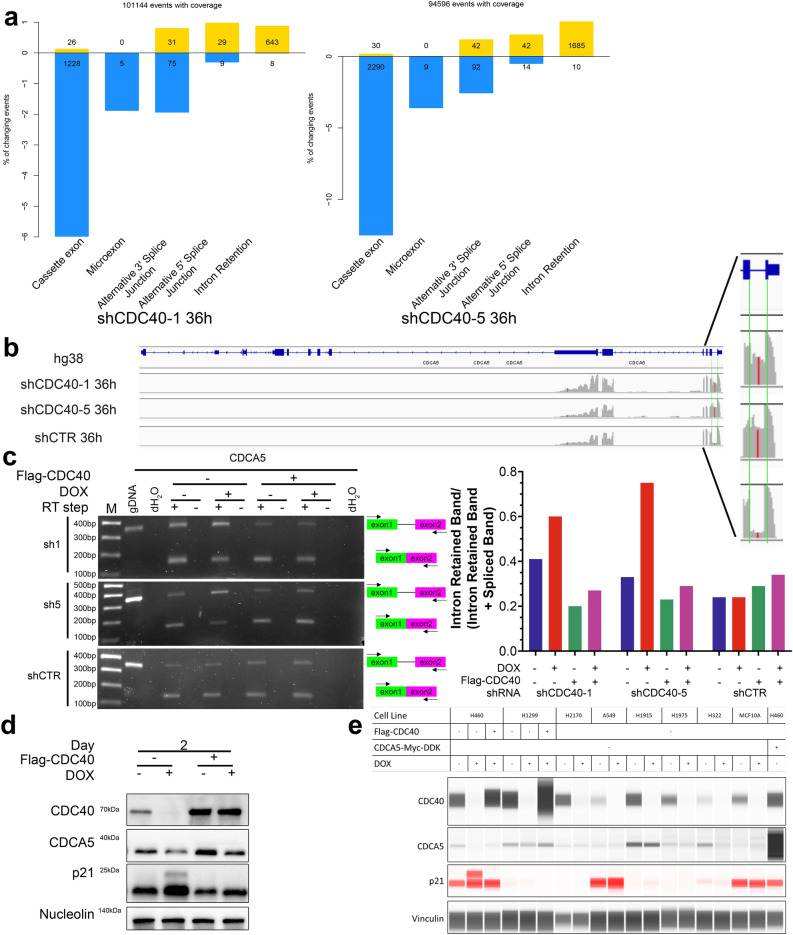
As CDC40 is implicated in pre-mRNA splicing7, we calculated the percent spliced in (PSI) index, which indicates how efficiently a sequence of interest is spliced into transcripts21. Our analysis revealed a significant increase in intron retention (IR) in many genes upon CDC40 knockdown (Fig. 4a). A total of 1478 common alternative splicing events, including 397 intron retention events, were identified in H460 cells after 36 h of sh1 and sh5 induction. Among the top hits, the first intron of CDCA5 was found to be the most retained (Fig. 4b).
Fig. 4.
Knockdown of CDC40 results in increased intron retention events. (a) Up-regulation (yellow) and down-regulation (blue) of different splicing events in H460 cells after 36 h induction of shCDC40-1 and shCDC40-5. (b) RNA-seq results of CDCA5 transcripts (grey) mapped to the CDCA5 human locus (in blue, genome assembly GRCh38, hg38) showing mapping to exonic (rectangle) and intronic (line) regions visualized using the Integrative Genomics Viewer (IGV). (c) Left: DNA gels of RT-PCR products amplified using CDCA5 primers. Right: Intron retention ratio of CDCA5 intron 1 after 36 h of shCDC40-1 and shCDC40-5 induction in comparison to shCTR. (d) Western blot analysis shows the effect of CDC40 knockdown using shCDC40-5 on CDCA5 and p21 expression after 48 h of DOX induction in H460 cells. (see Supplementary Fig. 15 for source blot images) (e) Jess analysis shows the effect of CDC40 knockdown using shCDC40-5 on CDC40, CDCA5 and p21 expression after 48 h of DOX induction in various cell lines. (see Supplementary Fig. 15 for source blot images).
Reverse transcription PCR was used to validate the intron (HsaINT0031855) retention increase by using forward and reverse primers within the flanking exons to generate amplicons of different sizes depending on intron retention status. Following sh5 induction in H460 cells, the proportion of the intron retained band increased, which was not seen when Flag-CDC40 was over-expressed (Fig. 4c). The expression of CDCA5 protein, a cycle regulator, was evaluated in H460 cells over a 4-day period, as the in-frame pre-mature stop codon within the retained intron of CDCA5 transcript should result in protein reduction. A rescuable decrease in CDCA5 protein level was seen after 2 days with sh5 induction in most lines tested (Fig. 4d, e). This decrease took place at the same timepoint as the p21 increase (Fig. 4d, e). Across all lines, CDCA5 protein was decreased except for H1915, while p21 increase was observed in H460, H1299, A549, and MCF10A; a molecular weight shift also occurred in all lines except for H2170, H1915 and H1975 (Figure S9 and S10). The consequences of simultaneous CDCA5 decrease and p21 increase, with varied intensities across different lines, could contribute to the various cell cycle defects observed.
CDC40 primarily interacts with components of the NineTeen complex (NTC)
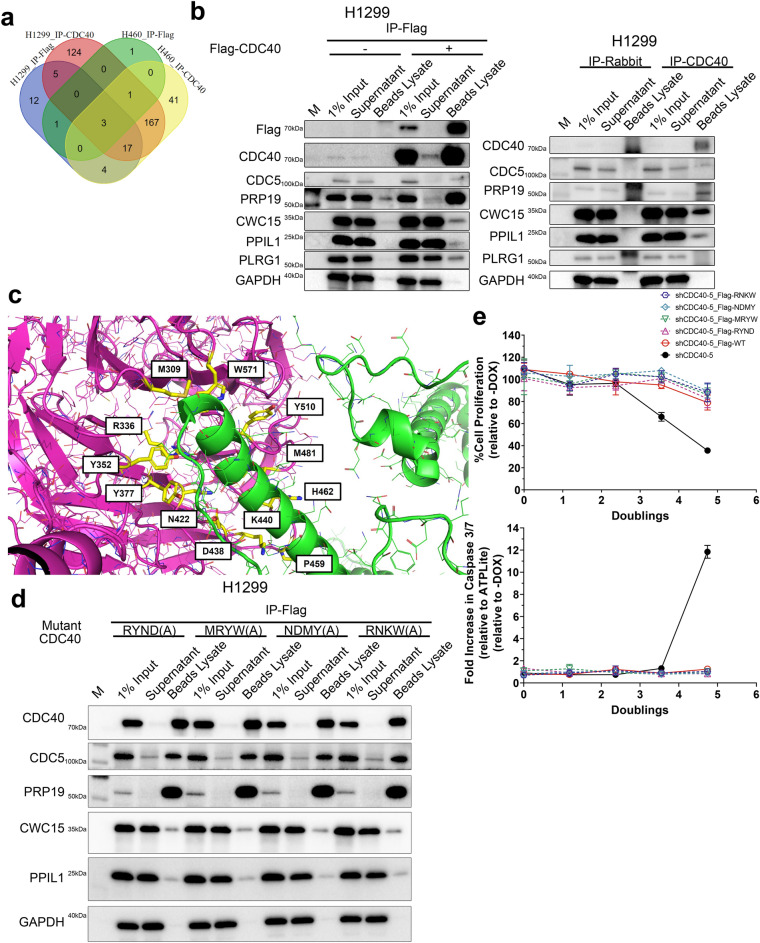
Understanding the protein–protein interaction network of CDC40 facilitates the discovery of small molecule effectors. We used immunoprecipitation-mass spectrometry (IP-MS) proteomics to identify interacting partners of Flag-CDC40 and endogenous CDC40. In H1299 cells, we identified 42 proteins enriched by Flag-CDC40 IP-MS, and 317 proteins enriched with endogenous CDC40 IP-MS. Similar experiments were conducted in H460 cells with 6 and 233 proteins enriched with Flag-CDC40 and endogenous CDC40 IPs, respectively. (Fig. 5a).
Fig. 5.
CDC40 primarily interacts with spliceosome components. (a) IP-MS hits identified after IP-Flag and IP-CDC40 in H1299 and H460 cells. (b) co-IP validation of the IP-MS results using western blot post pull-down with anti-Flag beads in shCTR containing H1299 cells expressing Flag-CDC40 (Left) and with anti-CDC40 (Right) beads in H1299 cells. (see Supplementary Fig. 16, 17 for source blot images) (c) Twelve residues for CDC40-CDC5 interaction were identified in silico for CDC40 mutation analysis. (d) coIP-WB with anti-Flag beads in shCTR H1299 cells transduced with Flag-CDC40 quadruple point mutants. (See Supplementary Fig. 23 for source blot) (e) Assessment of Flag-CDC40 quadruple-point mutant overexpression on the proliferation of H1299 cells after 4 days shCDC40-5 induction. Results were normalized to the doubling time of H1299 cells. (*, p < 0.05, **, p < 0.01, ***, p < 0.001, ****, p < 0.0001; n = 3, ± SD, Student’s T-test).
Using co-IP followed by western blot, five proteins (CDC5, PRP19, CWC15, PPIL1 and PLRG1) were confirmed to interact with Flag-CDC40 while PLRG1 fell off the list in H1299 cells with endogenous CDC40 (Fig. 5b). Other non-spliceosomal proteins identified from the IP-MS analysis, such as NOP2, RPLP2, and CCT2, were not confirmed to interact with CDC40 during co-IP (data not shown). All confirmed CDC40 interactors are known spliceosome components with most (CDC5, PRP19, CWC15 and PLRG1) being part of the NineTeen Complex (NTC).
Designed CDC40 mutants did not disrupt CDC40-CDC5 protein–protein interaction
To better understand protein–protein interactions of CDC40 within the spliceosome, point mutation analysis targeting the CDC40-CDC5 interaction was conducted to characterize its implication in future drug discovery efforts. Twelve residues for CDC40-CDC5 interaction were identified in silico (Fig. 5c) based on their occurrence in available Protein Data Bank (PDB) structures, the resolution of interactions, and the interaction strength, among which 4 single-point mutants of CDC40 were introduced into H1299 cells for interaction disruption analysis and phenotypic assessments (Figure S22a). Based on coIP-WB, CDC5 and PRP19 remained bound to Flag-CDC40 mutants in a similar manner to Flag-CDC40 wildtype, suggesting that none of the mutations affect the CDC40-CDC5 interaction (Figure S22c). Moreover, phenotypic assessments (ATPLite and caspase 3/7 assays) suggest that like Flag-CDC40 wildtype, proliferative phenotypes inhibited by CDC40 knockdown were fully rescued, in addition to blocking apoptosis induction (Figure S22d) after over-expressing these mutants. These results indicate that these 4 single-point mutants were not able to disrupt the CDC40-CDC5 interaction, which explains why their effects in H1299 cells were similar to Flag-CDC40 wildtype.
As the previously designed single-point mutants were not able to disrupt the CDC40-CDC5 interaction as expected, four CDC40 quadruple-point mutants were designed (Figure S22b) to disrupt the CDC40-CDC5 interface, and ultimately to prevent the association of both partners. Based on the co-IP WB, all five confirmed CDC40 interactors (CDC5, PRP19, CWC15, PPIL1 and PLRG1) remained bound to Flag-CDC40 quadruple-point mutants (Fig. 5d), which indicates that none of the quadruple mutations affect CDC40 interactions with spliceosomal components. Furthermore, proliferative phenotypes and reduced apoptosis were restored with overexpression of Flag-CDC40 quadruple mutants in a similar manner as Flag-CDC40 wild type (Fig. 5e). Overall, these results suggest that the protein–protein interactions of CDC40 within the spliceosome is extremely resistant to disruption even with quadruple-point mutations.
Discussion
CDC40 is part of the WD40 repeat (WDR) family of highly conserved scaffolding proteins. With previous success in targeting WDR members, as well as the growing demand to identify novel cancer targets, interest was extended to evaluate the feasibility of targeting CDC40 as a potential therapy for lung cancer. In this work, we characterized the effects of CDC40 knockdown in a panel of lung cancer cells and demonstrated that CDC40 knockdown induces cell cycle defects with strong growth inhibition and the activation of late-stage apoptotic markers. To further understand the consequences of CDC40 suppression, we investigated global transcriptional and splicing changes and found an alteration in splicing-related and translation-related gene expression in addition to a large increase in intron retention. We explored one specific intron retention event in the cell cycle regulator CDCA5 and validated that CDC40 knockdown leads to retention of its first intron and decreased CDCA5 protein expression. Interestingly, CDC40 knockdown induced a large increase in splicing pathway genes, which could be a compensatory mechanism. In addition, when exploring the protein interaction network of CDC40 in lung cancer, spliceosome components were the predominant binding partners for CDC40. Mutation analysis of the CDC40-CDC5 interaction implies that small molecule targeting strategies may not be optimal to disrupt protein–protein interactions within this large complex. Taken together, these findings support the important role that CDC40 plays in splicing.
Our results suggest that CDC40 is essential and may be a viable therapeutic target for lung cancer, as all analyzed lines displayed growth suppression upon knockdown. However, the therapeutic index will need to be carefully evaluated, as immortalized (but non-transformed) MCF10A cells also displayed anti-proliferative effects. MCF10A cells while not fully normal were a compromise to provide a non tumorigenic epithelial line that tolerated long term culturing to make stable lines. Supportively, while viable lung cancer cells were non-detectable after 4 to 7 days of CDC40 knockdown, MCF10A cells reached a lower plateau at about 20% viable after 7 days. Additionally, CDC40 knockdown in MCF10A cells displayed less caspase activation in comparison to knocking down PLK1, while the caspase increase in lung cancer cells after shCDC40 induction surpasses the magnitude of shPLK1 induction. If a reversible growth arrest predominates in normal cells vs. cell death in cancer cells, there may be a chance for an in vivo therapeutic window with appropriate scheduling. These findings point to the need to identify a chemical probe to further investigate the therapeutic potential of small molecule CDC40 inhibitors.
As CDC40 was previously linked to cell cycle progression in yeast studies7, we examined this possibility in human lung cancer cells. Cell cycle analysis of 6 lung cancer lines as well as a non-tumorigenic cell line with CDC40 knockdown showed a decrease in S phase with a re-distribution into G1 (MCF10A), G2 (H460, H322, H1915, H2170 and H1299) or both G1 and G2(A549) phases. Temporally, the onset of the cell cycle defects in H2170, A549, H460, H322 and MCF10A occurred 48 h after CDC40 knockdown while the dysregulation was observed later at 72 h in H1915 and H1299 cells. This timing coincides with our observed anti-proliferative profiles. While CDC40 knockdown induced a variety of cell cycle defects in our tested lung cancer lines, the direct cell cycle regulating kinase PLK122 induced a much more profound and rapid G2/M arrest as early as 24 h post-induction (data not shown).
The observed increase of p21 after CDC40 KD, though the underlying mechanism was not explored further, was confirmed to be p53-independent. Among all eight cell lines tested, three are p53 wild type (H460, A549 and MCF10A), four are homozygous p53 point mutants (H2170, H322, H1975 and H1915) and one is p53-null (H1299) based on the Cellosaurus knowledgebase23, while p21 increase was observed in H460, MCF10A, A549 and H1299 cell lines. These findings indicate that CDC40 KD induced p21 upregulation is not always regulated through p53. While we cannot rule out a direct role for CDC40 in cell cycle regulation, our studies have only linked CDC40 to the spliceosome in human cells, and we expect that CDC40 only indirectly contributes to cell cycle progression through the splicing of critical cell cycle genes.
This study offers the first characterization of gene expression alterations in lung cancer after knocking down CDC40. By RNA-seq we found that the expression of many splicing-related and translation-related genes was increased upon CDC40 knockdown, including our more complete validation of SMNDC1, a protein involved in pan-cancer intron retention15. This suggests a cellular compensatory mechanism in response to CDC40 loss.
The number of down-regulated genes after CDC40 knockdown is much higher than those up-regulated, which indicates that lung cancer cells lose their ability to maintain global transcriptome levels. These results further support our previous findings that CDC40 is essential for lung cancer cell survival. The two most down-regulated genes, PIGQ and CSPG4, are involved in cell proliferation and survival pathways16,17,24,25; however, analysis of the down-regulated pathways did not reveal many enriched biological functions (unlike our findings with the up-regulated gene set), suggesting that suppression is not biologically specific as it involves a multitude of cellular functions. Based on these results, we also analyzed CDC40 expression profile across different lung cancer pathological types using the public databases DepMap (cell line data) and TCGA (clinical data). The DepMap results suggest that small cell lung cancer exhibits the highest CDC40 expression. The TCGA data suggest that squamous cell carcinoma has the highest CDC40 expression due to the unavailability of small cell lung cancer samples, whereas typical lung adenocarcinoma does not show elevated expression (Supplementary Fig. 27).
We are the first to describe the consequences of CDC40 knockdown on splicing in lung cancer cells. Our deep RNA-seq experiment revealed that there was a large increase in aberrant intron retention (IR) (Fig. 4a). This increased IR may be explained by the dynamic stages of spliceosome assembly when CDC40 is present. CDC40 acts as an active site stabilizer during the two known steps of intron splicing8–11 . As a result, decreased CDC40 levels could lead to spliceosome malfunction resulting in intron removal inefficiency and ultimately cellular stress. Pathway analysis of upregulated genes upon CDC40 knockdown revealed nonsense-mediated decay (NMD) (NES = 2.43; ranked 9th) (Figure S21) as one of the top pathways, which may indicate an increase of premature stop codons caused by compromised intron removal. We also discovered that CDC40 knockdown will induce alterations in cap-dependent translation initiation factors including phospho-eIF4B, phospho-eIF4E and phospho-eIF2α, which generates inhibitory signals to translation pathways. As phospho-eIF4B is a known downstream effector of oncogenes26 while phospho-eIF4E is recognized as a driver of carcinogenesis27,these translational regulatory changes leading to reduced protein production show the potential of targeting CDC40 for cancer therapy.
RT-PCR was used to validate the CDCA5 IR (first intron) seen by RNA-seq after CDC40 knockdown (Fig. 4c). CDCA5 codes for a protein (also termed sororin) that is involved in sister chromatid cohesion regulation in mitosis28,29. It has been reported that the protein expression of CDCA5 decreases when its first intron is retained, which in turn leads to mitotic progression perturbation30,31. Similar decrease in CDCA5 protein expression was observed in our hands after 2 days of CDC40 knockdown (Fig. 4d) and may contribute to the cell cycle arrest seen with CDC40 knockdown. Rather than playing an active role in cell cycle control, CDC40 more likely acts indirectly by contributing to the splicing of cell cycle related factors. CDCA5 was identified to be upregulated in lung cancer and is described as an independent prognostic factor for lung cancer patients30.
CDC40 is part of the WD40 repeat (WDR) family of highly conserved scaffolding proteins. Their typical structure is a circular seven bladed β-propeller, with each blade containing approximately 40 amino acids, and with up to three major interaction sites on the top, bottom and/or side31. WDR proteins are involved in diverse cellular functions mediating protein–protein interactions within multi-protein complexes31. So far, at least nine WDRs (CDC40, PRPF4, PRPF19, CSTF1, SMU1, PLRG1, WDR33, PPWD1, snRNP40) have been implicated in the splicing pathway with the majority as scaffolds within different spliceosome complexes. Although all nine WDRs are involved in splicing function, CDC40 was identified to be the most essential splicing WD protein according to the public databases2–6,14. Recent studies, including from our group, have demonstrated that antagonism of WDR interactions with WDR5 and EED can inhibit cancer cell growth by disrupting large protein complexes essential for cancer cell survival32–37. As we have shown that CDC40 plays an essential role in lung cancer growth and survival there may be potential to target CDC40 as a cancer therapy.
Small molecule splicing factor inhibitors have demonstrated some in vitro success. E7107, a SF3B1 inhibitor of constitutive splicing, entered phase I clinical trials, although development was later halted due to toxicity38. Currently, the majority of splicing inhibitors in development target alternative splicing39. Targeting late spliceosome assembly and/or scaffolding proteins remains relatively under-explored. Additionally, identifying biomarkers to select patient populations that would most benefit from CDC40 targeting would help increase the therapeutic index. Studies have suggested that cancer cells with existing splicing factor mutations were more sensitive to splicing modulator intervention, such as cancer cells bearing Srsf2 and U2AF1 mutations39,40.
So far, structural information on CDC40 protein is limited to fifteen available Cryo-EM structures of human spliceosome complexes with resolution ranging from 2.9 Å to 8.0 Å. Although CDC40 is predicted to contain druggable sites, missing interface information as well as unresolved domain of proteins in some published structures further complicates in silico analysis of protein–protein interactions between CDC40 and other spliceosome components. Our study was the first to identify potential interactors of CDC40 in physiological conditions via IP-MS. In our study, CDC5 and PRP19 were identified as two primary interactors of CDC40 together with other three interactors CWC15, PRLG1 and PPIL1. These findings provide candidates to screen for small molecules targeting CDC40 that can interfere with these protein–protein interactions. Our findings encourage future drug discovery efforts with protein–protein interaction disruptors aimed at the CDC40 interfaces involved in CDC5, PRP19, CWC15 and PPIL1 binding. Inhibiting CDC40 interactions may not exactly mimic knockdown but destabilizing important scaffolding functions will impact spliceosome formation and function, and will likely lead to similar downstream consequences.
Many factors could contribute to the unsuccessful disruption of the CDC40-CDC5 interaction observed in our mutational analysis. One possible reason is that the currently available Cryo-EM structures of the human spliceosome only visualize a small part of CDC5 protein instead of the full-length version. The unresolved part of CDC5 may also interact with CDC40. Another potential explanation is that the interactions between spliceosome components in cells are stronger than expected. The presence of interlinked spliceosome components may strengthen the complex, making the CDC40-CDC5 interaction resistant to localized mutations. Due to this challenge targeting CDC40 with a proteolysis targeting chimera (PROTAC) approach could serve as an alternative strategy if a small molecule interactor can be identified.
This study seeks to investigate the role of mammalian CDC40 in splicing and cell cycle control while also attempting to validate CDC40 as a potential lung cancer target, and to characterize CDC40 interactions supporting oncogenesis. So far, we have discovered that CDC40 is essential for lung cancer cell growth and survival. By knocking down CDC40, lung cancer cells exhibit cell cycle alterations followed by anti-proliferative phenotypes and eventually undergo apoptosis. Our RNA-seq analysis suggests that in the absence of CDC40, splicing related genes are upregulated as a potential compensatory mechanism, indicating the importance of CDC40 in splicing pathway. Additionally, many genes are downregulated including proliferation related genes, which further explains the essentiality of CDC40. Furthermore, the changes in splicing patterns, particularly the intron retention events, after CDC40 knockdown reinforces the important role of CDC40 in splicing. Taken together, our results demonstrate that lung cancer is strongly dependent on the presence of CDC40 and CDC40 is vital for cancer cell splicing functions. Our protein–protein interaction analysis with CDC40 identified several protein binding partners within the spliceosome, providing potential candidates for future pharmacological interruption. Mutational analysis of a key CDC40 interaction interface implies that a PROTAC may serve as a better strategy for future CDC40 targeting. This study improves our understanding of the relationship(s) between mRNA splicing, cell cycle control, proliferation, and apoptosis in lung cancer cells, along with providing insight into the potential of CDC40 as a novel therapeutic target for lung cancer treatment. Ultimately, this could lead to the identification and development of drug-like small molecule binders to CDC40.
Online methods
Cell line and culture
HEK293EMT, NCI-H322, NCI-H460, NCI-H1299, A549, NCI-H1915, NCI-H2170, NCI-H1975 and MCF10A cells were obtained from American Type Culture Collection (ATCC). Cell lines were cultured in media including RPMI (Life Tech, 11875–119), DMEM (Life Tech, 12430–062) and F12 (Thermo Fisher, 11320082) supplemented with fetal bovine serum (FBS; Life Tech, A4736201) or horse serum (Thermo Fisher, 16050130) according to the ATCC recommendations (Table 1) and maintained in a humidified incubator at 37 °C supplemented with 5% CO2.
Table 1.
Cell culture media for selected cell lines. (Complete media = media + serum + 100 μg/mL Normocin (Invivogen, ANT-NR-2) + 50 μg/mL Gentamicin (Gibco, 15750078); Puro = puromycin (Invivogen, ANT-PR-1); Gene = Geneticin/G418 (Invivogen, ANT-GN-5); Hygro = Hygromycin B (Invivogen, ANT-HG-5); Zeo = Zeocin (Invivogen, ANT-ZN-1); EGF = Epithelial growth factor (Thermo, PHG0315); Hydrocortisone (Sigma, H0888-1G); Insulin (Sigma, I1882-100MG); Isoproterenol (Sigma, I2760-100MG)).
| Cell Line | Media | Serum | Selection | Additives |
|---|---|---|---|---|
| H2170 | RPMI | 10% FBS | Puro (2.5 µg/mL) | NA |
| H1915 | RPMI | 10% FBS | Puro (2.5 µg/mL) | NA |
| A549 | DMEM | 10% FBS | Puro (1.25 µg/mL) | NA |
| H322 | RPMI | 10% FBS | Puro (1 µg/mL) | NA |
| H1975 | RPMI | 10% FBS | Puro (1 µg/mL) | NA |
| MCF10A | F12 | 5% horse serum | Puro (1 µg/mL) | EGF (20 ng/mL); Hydrocortisone (0.5ug/mL); Insulin (10ug/mL); Isoproterenol (0.25ug/mL) |
| H460 | RPMI | 10% FBS | Puro (1 µg/mL) | NA |
| H460-OvE | RPMI | 10% FBS | Puro (1 µg/mL); Gene (0.75 mg/mL) | NA |
| H1299 | RPMI | 10% FBS | Puro (1 µg/mL) | NA |
| H1299-OvE | RPMI | 10% FBS | Puro (1 µg/mL); Gene (0.75 mg/mL) | NA |
| HEK293EMT | DMEM | 10% FBS | Gene (0.75 mg/mL); Hygro (0.2 mg/mL); Zeo (0.25 mg/mL) | NA |
shRNA construct generation
CDC40 shRNAs were designed using BLOCK-iT RNAi designer (Invitrogen). The trigger sequences of the 3 short hairpin constructs were 5’-GGAGGTGGCAGTTAAGGAAGA-3’ (shCDC40-1) and 5’- GGTGTTAATCTACGGTCAACT-3’ (shCDC40-5) targeting the coding sequence (CDS) as well as 5’-GCAGCTGTGATTACTGAAATC-3’ (shCDC40-8) targeting the 3’ untranslated region (UTR). Corresponding shCDC40 forward and reverse oligos were obtained from Integrated DNA Technology (IDT). Inserts were constructed to sequentially contain the attB1 site, tH1 promoter (H1 promoter for Tet-ON expression), shCDC40 hairpin, poly-thymine (polyT) and attB2 site and cloned into the pDONR vector using gateway cloning for each shCDC40. These vectors were sent to Advanced Center for Genome Technology (ACGT) for sequence validation. Validated vectors were cloned to a pLV series lentiviral transfer plasmid with a puromycin selectable marker.
shRNA-resistant Flag-CDC40 construct generation
shRNA-resistant Flag-CDC40 cDNA was designed using GeneArt Gene Synthesis (Invitrogen) software and cloned into pDONR 221 vector. Flag-CDC40 mutants were generated using wildtype shRNA-resistant Flag-CDC40 cDNA as template and cloned into pDONR 221 vector. These vectors were cloned to a pLV series lentiviral transfer plasmid with geneticin selectable marker.
Lentivirus generation
Lipofectamine®2000 (Invitrogen) was added to Opti-MEM media (Gibco) at a 1:50 ratio. 1.0 µg pBaculo-p35 plasmid, 1.0 µg pVSV-G plasmid, 2.5 µg pCMV-ΔR8.2 plasmid and 1.5 µg of the pLV shCDC40 construct were added to 2.5 mL of Opti-MEM media (Gibco) for each shCDC40. The Lipofectamine/Opti-MEM mix was added to each of the plasmid mixtures at a 1:1 ratio. Mixed solutions were added to 90% confluent HEK293EMT cells with 3.5 mL of Opti-MEM media (Gibco) for transfection overnight in the incubator (37 °C, 5% CO2). The next day, the Opti-MEM media was replaced with 8.5 mL of DMEM media (Gibco) and 24 and 48 h later, DMEM media containing the generated lentivirus was harvested and concentrated 30-fold using an Amicon Ultra-15 centrifugal filter. The concentrated lentivirus was stored at -80 °C until needed for transductions.
Generation of stable DOX-inducible cell lines
NCI-H322, NCI-H460, NCI-H1299, A549, NCI-H1915, NCI-H2170, NCI-H1975 and MCF10A cells were seeded in 6-well plates at 0.025–0.05 million cells/mL in duplicate for each shCDC40. 10 μL of lentivirus containing shCDC40 constructs were added into each well the following day. Puromycin (Sigma) at 1–2.5 μg/mL concentration was added to each well to select a population of resistant cells.
Generation of stable cell line constitutively expressing shCDC40 resistant Flag-CDC40
shCDC40-transduced NCI-H460 cells and NCI-H1299 cells were seeded at 0.025–0.05 million cells/ml in duplicates for each shCDC40 in 6-well plates. 10 μL of lentivirus containing Flag-CDC40 construct were added into each well the following day. Geneticin (Sigma) at the appropriate concentration was added to each well to select a population of resistant cells.
Real-time polymerase chain reaction (RT-qPCR)
Cells grown under equivalent conditions were collected in 1.5 mL microtubes and rinsed with ice-cold PBS (Gibco) before total RNA extraction following the RNeasy Plus Kit protocol (Qiagen). Each well of the RT-qPCR reaction contained: 10 μL of EXPRESS qPCR SuperMix Universal (Life Tech), 20 μL of 20X TaqMan Gene Expression Assay (Thermo Scientific), 0.04 μL of ROX Reference Dye (25 μM, Life Tech), 2 μL of RNase-free water, 2 μL of EXPRESS SuperScript Mix for One-Step qPCR (Life Tech) and 5 μL of RNA sample (20 ng/μL). RT-qPCR was performed on an Applied Biosystem’s ViiA 7 Real Time PCR machine using a 96-well standard block with the following protocol: 1) 50 °C for 15 min; 2) 95 °C for 3 min; 3) 40 cycles of 95 °C for 15 s + 60 °C for 1 min. Taqman probes used in the RT-qPCR were: CDC40 (Hs00210705_m1), HPRT1 (Hs99999909_m1), TFRC(Hs00961083_m1), COL8A1 (Hs00156669_m1), SCHP1 (Hs01053118_m1), SMNDC1 (Hs00195343_m1), CSPG4 (Hs00361541_g1) and PIGQ (Hs00188370_m1).
Western blot
Cells grown under equivalent conditions were collected in 1.5 mL microtubes and rinsed with 1 mL of ice-cold PBS (Gibco) before adding M-PER lysis buffer (Thermo Scientific) supplemented with 0.1% SDS and Halt protease and phosphatase inhibitor cocktail (Thermo Scientific) for cell lysis. Lysates were incubated on ice for 10 min and were then centrifuged at 21,100 rpm at 4 °C for 5 min. Supernatants were transferred into a new 1.5 mL microtubes for protein quantification performed using the DC Protein Assay Reagents (Bio-Rad) following the manufacturer’s protocol. Samples were then mixed with water and Laemmli sample buffer (Bio-Rad) and were heated at 95 °C for 5 min. Western blot analyses were performed after electrophoresis over Any kD™ Mini-PROTEAN® TGX Stain-Free™ protein gels (Bio-Rad) in 1X Tris/Glycine/SDS buffer (Bio-Rad) at 200 V for 30 min and transferred to 0.45 µm polyvinylidene difluoride (PVDF) membrane (Invitrogen) in 1X Tris/Glycine buffer (Bio-Rad) supplemented with 20% ethanol at 100 V for 1 h. Equal loading amounts were confirmed using Ponceau S staining (Sigma) and the membranes were cut in strips based on the molecular weight markers prior to antibody incubation as multiple proteins were detected from the same gel. Membrane strips were then blocked with Intercept™ (PBS) Blocking buffer (Mandel) for 30 min. After blocking, the membranes were incubated with primary antibodies at 4 °C on a shaker. The next day, the membranes were washed with 1X Tris Buffered Saline (Bio-Rad) supplemented with 0.1% Tween-20 (Bio-Rad) prior to being incubated with secondary antibodies in SuperBlock™ (PBS) blocking buffer (Thermo Scientific) for 50 min at room temperature on a shaker. Bands were illuminated using Immobilon Western Chemiluminescent HRP Substrate (Thermo Scientific) and imaged with a ChemiDoc XRS + imager (Bio-Rad). ImageLab software was used to generate the blot images using Auto Exposure with the “intense bands – optimized for all bands” setting to balance the exposure. No additional contrast controls were used. Primary antibodies used for western blot analysis were: anti-CDC40 (Abcam, ab175924, 1:1,000), anti-H3 (Cell Signaling Technology, 14269S,1:1,000), anti-GAPDH (Cell Signaling Technology, 2118S, 1:1,000), anti-CDC5 (Abcam, ab51320, 1:1,000), anti-DYKDDDDK (Sigma, F1804, 1:2,000), anti-PRP19 (Abcam, ab126776, 1:1000), anti-CWC15 (Abcam, ab188345, 1:1000), anti-PPIL1 (Abcam, ab154188, 1:1000), anti-PLRG1 (Abcam, ab86050, 1:1000), anti-EGFR (Abcam, ab52894, 1:5,000), anti-p21 (Cell Signaling Technology, 2947S, 1:5,000), Translation Initiation Complex Antibody Sampler Kit (Cell signalling Technology, 4763 T, 1:5000), anti-eIF2α (Cell Signaling Technology, 5324S, 1:5000), anti-Phospho-eIF2α (Ser51) (Cell Signaling Technology, 9721S, 1:5000), anti-Nucleolin (Cell Signaling Technology, 14574S, 1:1000). Secondary antibodies used were anti-Rabbit (Bio-Rad, 1,706,515, 1:10,000) and anti-Mouse (Bio-Rad, 1,721,011, 1:10,000).
Automated western blot analysis (Jess ProteinSimple)
Samples were prepared, loaded into the plate following manufacture’s guidance and read on a JESS machine (ProteinSimple, 004–650). 40 μL of dH2O was added into the clear tube from EZ Standard Pack to make 400 mM DTT solution. 20 μL of 10X sample buffer and 20 μL of 400 mM DTT solution were added into the pink tube from EZ Standard Pack to make 5X fluorescent master mix. 20 μL of dH2O were added into the green tube from EZ Standard Pack to make biotinylated ladder. Protein samples were diluted to 1 mg/mL with dH2O. One part of the 5X fluorescent master mix was mixed with 4 parts of the protein sample. The mixture was heated to 95 oC for 5 min and stored back on ice. 200 μL of Luminol-S were mixed with 200 μL of peroxide in a microcentrifuge tube for chemiluminescence detection assay. Antibodies were diluted to desired concentration with antibody diluent. Ladder, samples, antibody diluent, antibodies and luminol-peroxide mix were loaded onto the prepared microplate as instructed. The microplate was centrifuged at 1000 g for 5 min at room temperature. The Jess (ProteinSimple, JS-4296), which automates traditional western blot, was used to detect, and quantify CDCA5, CDC40, p21 and Vinculin protein expression levels in lung cancer cells.
Propidium iodine staining
Cells grown under appropriate conditions were collected in 1.5 mL microtubes and rinsed with 1 mL of ice-cold PBS (Gibco) before being fixed with 800 μL of ice-cold 80% ethanol and stored at -20 °C for less than two weeks. During the day of the experiment, cells were centrifuged at 5000 rpm for 5 min at 4 °C prior to being washed twice with 200 µL/tube of cold HBSS/BSA buffer (1X HBSS (Gibco), 10 mM HEPES (Gibco), 0.25% BSA (Gibco), 0.03% Sodium bicarbonate (Gibco)). Propidium iodine (Sigma) and RNAse (Sigma) were mixed in HBSS/BSA buffer to reach a final concentration of 0.1 mg/mL and 2 mg/mL respectively. 200 µL/tube of the mixture were added to the samples which were then incubated on ice in the dark for 30 min. 200 µL of HBSS/BSA buffer were added after samples were filtered through strainer cap tubes. Cell cycle analysis was performed on a FACS Canto II. Plots were analyzed by FlowJo software (BD Biosciences) to construct cell cycle profiles of singlets.
Cell proliferation and apoptosis assays
Cells were seeded at 1000 cells/well in 96-well plates. ATPLite assays (PerkinElmer, 6,016,739) and Caspase 3/7 assays (Promega, G8092) were performed following the manufactures’ protocols 4–7 days after 0.1 μg/mL doxycycline was added. Luminescence was measured using a Cytation 3 plate reader (Biotek) and their Gen5 software.
RNA-sequencing (RNA-seq)
Cells grown under described conditions were collected in 1.5 mL microtubes and rinsed with ice-cold PBS (Gibco) prior to total RNA extraction following the RNeasy Plus Kit protocol (Qiagen). Total RNA samples were sent to the Donnelly Center (Toronto, Canada) for mRNA sequencing using Illumina NovaSeq 6000 with paired-end reads of length 150 base pairs in each read and a minimum library depth of 40 million reads per sample.
FastQC41 was used for quality control of raw demultiplexed fastq files. Sequence alignment was performed using the STAR42 RNA-seq aligner and the human genome assembly from Genome Reference Consortium (Release 30 GRCh38.p12)43. A minimum of 80% uniquely mapped reads were observed for all samples sequencing data. Additionally, RseqC44 was used for quality check of the alignment results and the newly detected splicing junctions.
Gene expression analysis
Raw counts per gene from STAR42 alignment results were used for gene expression analysis. Genes with average values lower than 10 counts per million (cpm) were removed from the data to guarantee the quality of sequencing counts per gene for further analysis. Normalization of raw counts and differential gene expression analysis was done using Empirical Analysis of Digital Gene Expression Data in R 3.6 (edgeR45–47 Bioconductor package 3.24.3). Multidimensional scaling (MDS) was performed using edgeR on all samples to confirm the quality of the data and similarity among all replicates (data not shown). The edgeR’s quasi-likelihood test was applied to identify differentially expressed genes between control samples and DOX-induced samples at each time point (data not shown). To correct for multiple hypothesis testing and reducing false positive adjusted p-values were calculated using False Discovery Rate method (FDR, Benjamini-Hochberg). |Log2(Fold Change)| higher than 1 in comparison to shCTR and False Discovery Rate less than 5% were considered significant.
Functional analysis
To perform gene set enrichment analysis by GSEA48,49 (4.2.1) a ranked gene list was generated using the sign of log-scale fold changes multiplied by log10 p-values, and the human pathways gene set from the Reactome database50 was downloaded and used.
Cytoscape (3.9.1)51 was used for visualization of results from gene set enrichment analysis. GSEA results files were uploaded to the EnrichmentMap (3.3.3) application52 to cluster the enriched pathways.
Splicing pattern analysis
Splicing analysis between shCDC40-induced and non-induced conditions over time were measured using the VAST-TOOL pipeline53,54. diff function of VAST-TOOL utilizing Bayesian statistics was used for this analysis. Data were filtered with quality scores after running the files through diff. The scores account for coverage of the splicing event and the balance between the reads upstream and downstream of the alternative splicing site. Upon filtering the data, events with a change in per splice in (PSI (dPSI) > 0.1) were considered to be biologically significant. The events are divided into cassette exons, microexons, alternative 3’ and 5’ events, and intron retention events. The y-axis is the proportion of events and the numbers in each bar represent the total number of either increasing or decreasing splicing events.
Integrated genome viewer (IGV)
RNA data after QC, trimming and alignment were exported into .bam files. Human hg38 (human genome assembly G4Ch38) was loaded and set as the reference genome. BAM files were loaded and visualized using IGV. Genes of interest were selected and zoomed using the “search” function of IGV. RNA-seq readings were represented as grey bars located on the top of each sample.
Immunoprecipitation and co-immunoprecipitation
Cells grown under described conditions were collected in 1.5 mL microtubes and rinsed with ice-cold PBS (Gibco) prior to adding lysis buffer (0.1% Triton X-100 (Sigma) in PBS) supplemented with Halt protease and phosphatase inhibitor cocktails (Thermo Scientific). Lysates were vortexed, sonicated and centrifuged at 21,100 rpm at 4 °C for 5 min. Supernatants were transferred into new 1.5 mL microtubes.
For Flag pulldown experiments, Pierce™ Anti-DYKDDDDK Magnetic Agarose beads (Thermo Scientific) were added to the tube after two washes with 1X Tris Buffered Saline (Bio-Rad) supplemented with 0.05% Tween (Bio-Rad). Samples with beads were incubated overnight on a rotator at 4 °C. The next day, supernatants were separated from the beads by centrifugation and saved in new 1.5 mL microtubes.
For CDC40 pulldown experiments, anti-CDC40 antibody (Cell Signaling Technology, 99086S, 1:50) was added to samples at a 1:50 dilution. Samples were incubated overnight at 4 °C on a rotator. The next day, Protein A beads (Thermo Scientific) were added to the tube after first being washed twice with 1X Tris Buffered Saline (Bio-Rad) supplemented with 0.05% Tween (Bio-Rad). Samples were then incubated on a rotator at room temperature for 1 h. Supernatants were separated from the beads by centrifugation and saved in new 1.5 mL microtubes.
Beads were washed three times with corresponding lysis buffer without detergent followed by once with water before Laemmli sample buffer (Bio-Rad) was added into the beads and boiled at 95 °C for 5 min. Beads were then discarded. Boiled samples together with input samples and supernatant samples were loaded for western blot analysis.
Immunoprecipitation-mass spectrometry analysis
Raw data were exported as peptide readings per sample. Scaffold 5 software (5.1.1)55 was used to visualize the protein identification and readings. Protein identification threshold was set to be 99%, minimum number of unique peptides were set to be 2 and peptide identification threshold was set to be 95%. Afterwards, data was trimmed if both readings in samples and negative controls were less than 5. |Log2(Fold Change)| higher than 1 in comparison to negative control and False Discovery Rate less than 5% were considered significant. Cellular compartment enrichment analysis was performed using g:Profiler56.
One-step reverse transcription polymerase chain reaction (RT-PCR)
Cells grown under described conditions were collected in 1.5 mL microtubes and rinsed with ice-cold PBS (Gibco) prior to total RNA extraction following the RNeasy Plus Kit protocol (Qiagen). The master mix was prepared following the OneStep RT-PCR Kit protocol (Qiagen) with 0.5 µM CDCA5 primer per reaction.15 µL of reaction master mix was aliquoted into strip tubes. 5 µL of 2 ng/µL genomic DNA or total RNA template was added to each strip tube with pre-aliquoted reaction mixes. The mixture then underwent PCR as recommended in the QIAGEN OneStep RT-PCR Kit protocol. The annealing temperature was 53 °C with 32 amplification cycles. 10 µL of the total 20 µL reaction was loaded and resolved on a 1.5% agarose gel stained with SYBR™ safe (Invitrogen) DNA dye followed by imaging using a BioRad Chemdoc Imager. ImageLab software was used to measure the relative intensity of bands representing the intron retained (IR) and spliced isoforms in comparison to genomic DNA. The IR ratio was calculated as the amount of intron retained bands divided by the sum of both retained and spliced isoforms.
Supplementary Information
Acknowledgements
This research was supported by a grant from the Cancer Research Society (CRS). Ontario Institute for Cancer Research is funded by the government of Ontario.
Author contributions
D.H. designed the study, performed the experiments, and wrote the manuscript. B. L T., P.S., E.L-F., R.M., R.A., designed the study and wrote the manuscript. V. T., J. L., B. J. B., analyzed RNA-seq data. J. O., L. H., performed in silico protein structure analysis.
Funding
This project was supported by grants from Cancer Research Society and the Government of Ontario, Canada.
Data availability
The authors declare that all data supporting the findings presented in this study are available within the paper and its supplementary information files. The RNA-seq datasets generated and analyzed during the current study are available in the Sequence Read Archive (SRA) repository under the reference number PRJNA1097204.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-83337-z.
References
- 1.Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin.73, 17–48 (2023). [DOI] [PubMed] [Google Scholar]
- 2.Barretina, J. et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barretina, J. et al. Addendum: the cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature565, E5–E6 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Cowley, G. S. et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci. Data1, 140035 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsherniak, A. et al. Defining a cancer dependency map. Cell170(564–576), e516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McFarland, J. M. et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat. Commun.9, 4610 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben Yehuda, S. et al. Identification and functional analysis of hPRP17, the human homologue of the PRP17/CDC40 yeast gene involved in splicing and cell cycle control. RNA4, 1304–1312 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertram, K. et al. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature542, 318–323 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Zhan, X., Yan, C., Zhang, X., Lei, J. & Shi, Y. Structure of a human catalytic step I spliceosome. Science359, 537–545 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Zhang, X. et al. An atomic structure of the human spliceosome. Cell169, 918-929.e914 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Zhang, X. et al. Structure of the human activated spliceosome in three conformational states. Cell Res.28, 307–322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.da Silva, M. R. et al. Splicing regulators and their roles in cancer biology and therapy. Biomed. Res. Int.10.1155/2015/150514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pio, R. & Montuenga, L. M. Alternative splicing in lung cancer. J. Thorac. Oncol.4, 674–678 (2009). [DOI] [PubMed] [Google Scholar]
- 14.McDonald, E. R. 3rd. et al. Project DRIVE: A compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell170(577–592), e510 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Thomas, J. D. et al. RNA isoform screens uncover the essentiality and tumor-suppressor activity of ultraconserved poison exons. Nat. Genet.52, 84–94 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rolih, V. et al. CSPG4: a prototype oncoantigen for translational immunotherapy studies. J. Transl. Med.15, 151 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tiede, A. et al. Human and mouse Gpi1p homologues restore glycosylphosphatidylinositol membrane anchor biosynthesis in yeast mutants. Biochem. J.334(Pt 3), 609–616 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merrick, W. C. eIF4F: a retrospective. J. Biol. Chem.290, 24091–24099 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gingras, A. C., Raught, B. & Sonenberg, N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem.68, 913–963 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Fernandez, J., Yaman, I., Sarnow, P., Snider, M. D. & Hatzoglou, M. Regulation of internal ribosomal entry site-mediated translation by phosphorylation of the translation initiation factor eIF2alpha. J. Biol. Chem.277, 19198–19205 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Schafer, S. et al. Alternative splicing signatures in RNA-seq data: percent spliced in (PSI). Curr. Protoc. Hum. Genet.10.1002/0471142905.hg1116s87 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Roshak, A. K. et al. The human polo-like kinase, PLK, regulates cdc2/cyclin B through phosphorylation and activation of the cdc25C phosphatase. Cell Signal12, 405–411 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Cellosaurus knowledgebase, vol. 2023.
- 24.Price, M. A. et al. CSPG4, a potential therapeutic target, facilitates malignant progression of melanoma. Pigment Cell Melanoma Res24, 1148–1157 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Starr, L. J., Spranger, J. W., Rao, V. K., Lutz, R. & Yetman, A. T. PIGQ glycosylphosphatidylinositol-anchored protein deficiency: Characterizing the phenotype. Am. J. Med. Genet. A.179, 1270–1275 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Yang, J. et al. eIF4B phosphorylation by pim kinases plays a critical role in cellular transformation by Abl oncogenes. Cancer Res.73, 4898–4908 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Yang, X., Zhong, W. & Cao, R. Phosphorylation of the mRNA cap-binding protein eIF4E and cancer. Cell Signal73, 109689 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oka, Y. et al. UBL5 is essential for pre-mRNA splicing and sister chromatid cohesion in human cells. EMBO Rep.15, 956–964 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watrin, E., Demidova, M., Watrin, T., Hu, Z. & Prigent, C. Sororin pre-mRNA splicing is required for proper sister chromatid cohesion in human cells. EMBO Rep.15, 948–955 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen, M. H. et al. Phosphorylation and activation of cell division cycle associated 5 by mitogen-activated protein kinase play a crucial role in human lung carcinogenesis. Cancer Res.70, 5337–5347 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Stirnimann, C. U., Petsalaki, E., Russell, R. B. & Muller, C. W. WD40 proteins propel cellular networks. Trends Biochem. Sci.35, 565–574 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Grebien, F. et al. Erratum: Pharmacological targeting of the Wdr5-MLL interaction in C/EBPalpha N-terminal leukemia. Nat. Chem. Biol.11, 815 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Grebien, F. et al. Pharmacological targeting of the Wdr5-MLL interaction in C/EBPalpha N-terminal leukemia. Nat. Chem. Biol.11, 571–578 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He, Y. et al. The EED protein-protein interaction inhibitor A-395 inactivates the PRC2 complex. Nat. Chem. Biol.13, 389–395 (2017). [DOI] [PubMed] [Google Scholar]
- 35.He, Y. et al. Erratum: The EED protein-protein interaction inhibitor A-395 inactivates the PRC2 complex. Nat. Chem. Biol.13, 922 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Qi, W. et al. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat. Chem. Biol.13, 381–388 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Schapira, M., Tyers, M., Torrent, M. & Arrowsmith, C. H. WD40 repeat domain proteins: a novel target class?. Nat. Rev. Drug. Discov.16, 773–786 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seiler, M. et al. H3B–8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nat. Med.24, 497–504 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salton, M. & Misteli, T. Small molecule modulators of pre-mRNA splicing in cancer therapy. Trends Mol. Med.22, 28–37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, S. C. & Abdel-Wahab, O. Therapeutic targeting of splicing in cancer. Nat. Med.22, 976–986 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andrews S. FastQC: a quality control tool for high throughput sequence data., vol. 2019, 2010.
- 42.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frankish, A. et al. GENCODE reference annotation for the human and mouse genomes. Nucl. Acids Res47, D766–D773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, L., Wang, S. & Li, W. RSeQC: quality control of RNA-seq experiments. Bioinformatics28, 2184–2185 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCarthy, D. J., Chen, Y. & Smyth, G. K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucl. Acids Res40, 4288–4297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen, Y., Lun, A. T. & Smyth, G. K. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res.5, 1438 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A.102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mootha, V. K. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Jassal, B. et al. The reactome pathway knowledgebase. Nucl. Acids Res.48, D498–D503 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Merico, D., Isserlin, R., Stueker, O., Emili, A. & Bader, G. D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One5, e13984 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tapial, J. et al. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res.27, 1759–1768 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.VAST-TOOLS vol. 2019.
- 55.Proteome Software, vol. 2021. Proteome Software, Inc.: Portland, Oregon.
- 56.Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucl. Acids Res.47, W191–W198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings presented in this study are available within the paper and its supplementary information files. The RNA-seq datasets generated and analyzed during the current study are available in the Sequence Read Archive (SRA) repository under the reference number PRJNA1097204.