Abstract
A balance between stem cell self-renewal and differentiation is required to maintain concurrent proliferation and cellular diversification in organoids; however, this has proven difficult in homogeneous cultures devoid of in vivo spatial niche gradients for adult stem cell-derived organoids. In this study, we leverage a combination of small molecule pathway modulators to enhance the stemness of organoid stem cells, thereby amplifying their differentiation potential and subsequently increasing cellular diversity within human intestinal organoids without the need for artificial spatial or temporal signaling gradients. Moreover, we demonstrate that this balance between self-renewal and differentiation can be effectively and reversibly shifted from secretory cell differentiation to the enterocyte lineage with enhanced proliferation using BET inhibitors, or unidirectional differentiation towards specific intestinal cell types by manipulating in vivo niche signals such as Wnt, Notch, and BMP. As a result, we establish an optimized human small intestinal organoid (hSIO) system characterized by high proliferative capacity and increased cell diversity under a single culture condition. This optimization facilitates the scalability and utility of the organoid system in high-throughput applications.
Subject terms: Intestinal stem cells, Self-renewal, Stem-cell differentiation
Culturing adult stem cell-derived organoids requires a balance between stem cell self-renewal and differentiation, but this can be challenging to accomplish. Here, the authors present an optimized method for growing human intestinal organoids that achieve both rapid growth and diverse cell populations in a single culture system.
Introduction
Adult stem cell (ASC)-derived organoids are generated by mimicking the intricate processes of tissue development, homeostasis, and regeneration in vitro. They demonstrate a remarkable ability to recapitulate aspects of the tissue structure, cellular composition, and function, making them an attractive platform for studying development and disease in vitro1,2. Despite significant efforts, previous attempts to culture ASC-derived organoids have encountered significant challenges in replicating the complex and dynamic processes that occur in vivo. Conventional organoid culture systems for many tissues are optimized to maintain stem cell self-renewal for expansion, resulting in decreased cellular diversity as cells remain undifferentiated. Conversely, attempts to promote differentiation and maturation often lead to cellular heterogeneity but limited proliferative capacity, as seen in the liver, pancreas, lung, and other tissues3–8. Thus, separate expansion and differentiation steps are required for typical organoid cultures, which impedes their scalability and utility in high-throughput screening.
Mouse intestinal organoid, the first ASC-derived organoid system, demonstrates remarkable parallel self-renewal and multidirectional differentiation processes under the ENR condition9. However, achieving an equal balance in human intestinal organoids has been challenging. Despite efforts to improve culture conditions to induce cellular diversity while maintaining proliferation ability, achieving this balance remains elusive. For example, Paneth cells, an essential cell type in the intestinal epithelium, are absent or rare in the improved IF culture condition10 and require specific signals for their generation, such as IL22, which also results in decreased cell proliferation in the organoid11. Moreover, the signals that regulate the self-renewal and multidirectional differentiation of human intestinal stem cells (ISCs) have yet to be determined.
Intestinal epithelial cells exhibit remarkable plasticity, allowing for continuous self-renewal, differentiation, and dedifferentiation along the crypt-villi axis. For instance, ISCs maintain self-renewal by competing for niche signals at the base of crypts and starting to differentiate outside the crypt12–14. At the same time, multiple differentiated cell types could also revert to a stem cell state when re-entering the stem cell niche15–18. These processes occur in a highly dynamic manner in vivo and are tightly regulated by intrinsic processes and extrinsic niche signals.
Generating diverse and rapidly proliferating cells necessitates stem cells with the capacity to generate multiple cell types and orchestrate localized signaling gradients for spatially regulated self-renewal and differentiation19,20. Achieving this remains challenging in homogeneous organoid cultures lacking the spatial niche gradients found in vivo. In this study, we hypothesize that enhancing organoid stem cell stemness can amplify their differentiation potential, which would increase the cellular diversity in organoids without applying artificial spatiotemporal signaling gradients. Furthermore, recreating the dynamic modulation of cell fate observed in vivo in organoid systems by regulating niche-intrinsic and cell-intrinsic signals may facilitate this outcome. Here, we demonstrate that a combination of small molecule pathway modulators can facilitate a controlled shift in the equilibrium of cell fate towards a specific direction, leading to controlled self-renewal and differentiation of cells. Additionally, we have developed an optimized culture condition that captures the delicate balance of self-renewal and differentiation of cells, resulting in a highly proliferative organoid system with increased cellular diversity.
Results
Establishment of organoid system with enhanced stemness
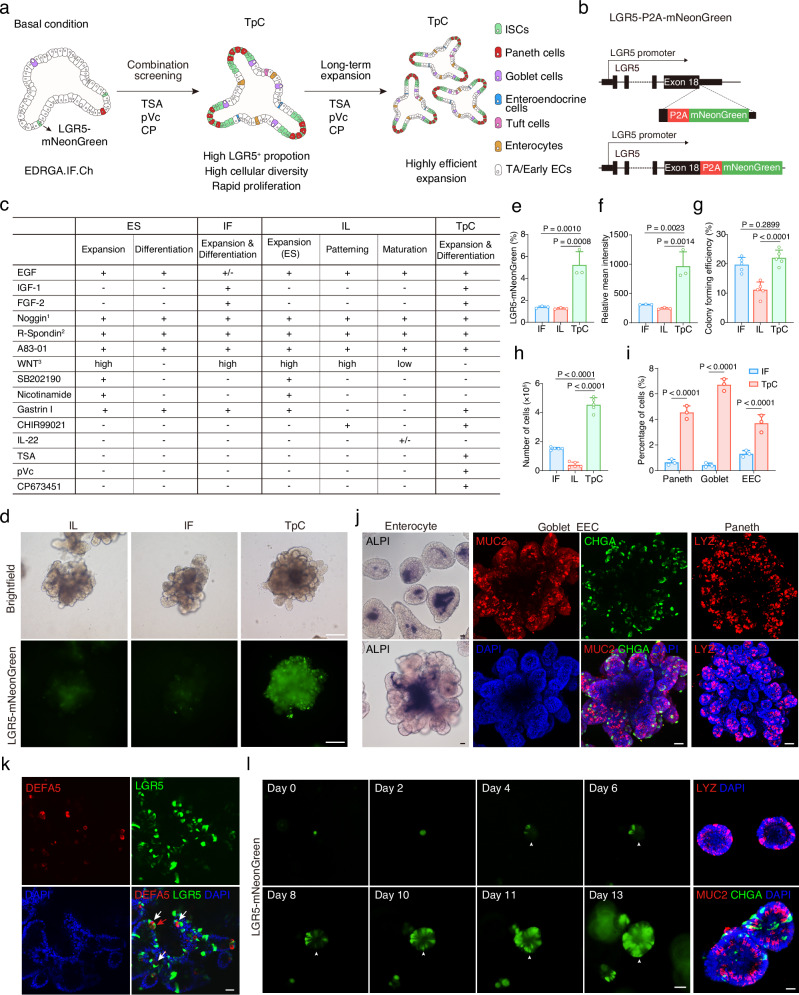
To enhance cellular diversity in human intestinal organoids, our initial goal was to increase the proportion of LGR5+ stem cells rather than driving differentiation directly. To achieve this, we sought to replicate an in vitro niche for stem cells using a combination of small molecules and growth factors (Fig. 1a). We generated an LGR5-mNeonGreen reporter system to visualize LGR5+ stem cells using CRISPR-Cas9 technology (Fig. 1b and Supplementary Fig. 1a–c). We compared the components used in previously identified culture conditions (Fig. 1c). The ES condition facilitated the expansion of progenitor cells but inhibited their differentiation, resulting in organoids devoid of secretory cell types21. Although the improved IF condition enabled multi-differentiation and self-renewal of the organoids, it was still associated with limited cellular diversity, as evidenced by the absence or rarity of mature enterocytes and Paneth cells10. The recently developed optimized condition employs IL-22 to induce the generation of Paneth cells based on the ES condition, but this comes at the cost of inhibited organoid growth11.
Fig. 1. Optimized human intestinal organoid demonstrates enhanced stemness and increased cell diversity.
a Schematic of screening strategy to optimize cultures of human intestinal organoids. b Schematic of the targeting strategy to generate LGR5-mNeonGreen reporter system. c Medium composition comparison among ES, IF, IL, and TpC culture systems. 1Noggin or BMP pathway inhibitor DMH1, 2R-Spondin1 conditioned media, 3WNT3a protein, WNT3a conditioned media or WNT surrogate. d Representative brightfield and fluorescence images of LGR5-mNeonGreen organoids cultured in IL patterning condition, IF condition or TpC condition. Representative images from three independent experiments. Scale bars, 200 μm. Quantification of LGR5-mNeonGreen proportion (e) and relative LGR5-mNeonGreen intensity (f) in IF, IL patterning and TpC organoids cultured for 4 weeks. n = 3 samples. Quantification of colony forming efficiency (g) and cell proliferation as indicated by the number of cells (h) in IF, IL patterning and TpC organoids cultured for ten days from single cells (8000 cells per well seeding). n = 4 samples. One-way ANOVA with Dunnett’s multiple comparisons test; data are presented as mean ± SD. i Proportion of secretory lineage cells in IF and TpC organoids quantified by positive staining of LYZ, MUC2, and CHGA, respectively. Two-tailed unpaired t-test; data are presented as mean ± SD; n = 3 samples. j Representative images of enterocytes (ALPI), goblet cells (MUC2), enteroendocrine (CHGA), and Paneth cells (LYZ) in TpC organoids. Representative images from five independent experiments. k Representative confocal images of LGR5-mNeonGreen and DEFA5 positive Paneth cells in TpC organoid. White arrowheads indicate Paneth cells adjacent to LGR5 intestinal stem cells, red arrowhead points out a double-positive cell. Representative images from four independent samples. l The growth and mNeonGreen expression of a single LGR5-mNeonGreen+ cell cultured in TpC condition over 13 days. Also showing Paneth (LYZ), goblet (MUC2), and enteroendocrine (CHGA) cells in the 13-day colonies detected by immunofluorescence staining. Representative images from three biological replicates. Scale bars, (j–l), 50 μm. Arrowheads indicate the emergence of mNeonGreen expression at the same location. Source data for this Figure are provided as a Source Data file.
Using the LGR5-mNeonGreen reporter, we find that stem cells under the IF and IL patterning conditions11 exhibit minimal expression of LGR5-mNeonGreen (Fig. 1d) while IF condition capable of generating multiple secretory cell types (Supplementary Fig. 2b), which is consistent with previous research. To promote the maintenance of LGR5+ stem cells while preserving their differentiation process, we incorporated key factors utilized in mouse intestinal culture, including EGF, the BMP inhibitor Noggin (or small molecule DMH1), and R-Spondin1. We eliminated factors such as SB202190, Nicotinamide, and PGE2, which have been demonstrated to impede the generation of secretory cell types21. Additionally, we combined previously identified niche factors, such as IGF-1 and FGF-210, and employed CHIR99021 as a replacement for Wnt proteins, as it promotes the self-renewal of ISCs22. We also included the ALK inhibitor A83-01, which has been shown to promote cell growth21. This basal condition was utilized during the screening process (Fig. 1a, c). A combination of 3 small molecules (TpC), including Trichostatin A (TSA or T, an HDAC inhibitor), 2-phospho-L-ascorbic acid (pVc or p, Vitamin C), and CP673451 (CP or C, a PDGFR inhibitor) was found to substantially increase the proportion of LGR5-mNeonGreen positive cells and their relative mNeonGreen expression in the culture (Fig. 1d-f). Moreover, the colony-forming efficiency of dissociated single cells significantly improved, and the total cell count in the culture also considerably increased (Fig. 1g, h and Supplementary Fig. 2a).
Generation of diverse and plastic cell types under the TpC condition
Under the TpC condition, organoids could be efficiently generated from dissociated single cells, wherein scattered LGR5-mNeonGreen expression was observed in each colony (Supplementary Fig. 2c). Prolonged culture led to extensive crypt-like budding structures within the organoids, with budding structures containing Paneth-like cells with dark granules (Supplementary Fig. 2d), indicating cell differentiation.
Interestingly, under the TpC condition, multiple intestinal lineage cells were readily generated, as evidenced by positive staining of markers including mature enterocytes (intestinal alkaline phosphatase, ALPI), goblet cells (mucin 2, MUC2), enteroendocrine cells (chromogranin A, CHGA), and Paneth cells (defensin alpha 5, DEFA5, and lysozyme, LYZ) (Fig. 1i-k). Notably, we observed widespread expression of LYZ mRNA in human intestinal organoids; however, at the protein level, lysozyme specifically marked Paneth cells, which co-expressed DEFA5, another Paneth cell marker (Supplementary Fig. 3a–c).
Within the organoids, interspersed DEFA5-/LGR5+ cells and DEFA5+ Paneth cells were observed, closely resembling their in vivo counterparts. DEFA5+/LGR5+ cells were also observed co-existing in the organoids, likely reflecting the retention of LGR5-mNeonGreen expression as LGR5+ stem cells differentiated into Paneth cells (Fig. 1k). The budding area also exhibited extensive OLFM4 expression, a specific stem cell marker for mouse intestinal stem cells23 (Supplementary Fig. 2e). Moreover, the organoids displayed EEC subtype markers, including somatostatin (SST) and glucagon (GCG) (Supplementary Fig. 2e).
Notably, LGR5+ stem cells and secretory progenies, such as Paneth cells, goblet cells, and enteroendocrine cells, were uniformly distributed in the organoids cultured under the TpC condition, both in short-term (7-10 days) and prolonged cultures (3-4 weeks) (Supplementary Fig. 2c, 2g, h and Supplementary Fig. 3d, e). The majority of TpC organoids were budding-structured, although a small number of round-shaped organoids also existed (Supplementary Fig. 3d, e). This indicates a high degree of homogeneity between TpC-generated organoids in terms of composition and structure, which is an advantage for downstream applications. Additionally, the TpC condition supported the generation and long-term maintenance of hSIOs from multiple donors (Supplementary Fig. 2f), indicating the robustness of this culture system.
To investigate cell growth dynamics, we longitudinally tracked individual LGR5+ stem cells under the TpC condition. Remarkably, we observed that a single LGR5+ stem cell could give rise to organoids comprising various secretory cell types, such as Paneth cells, goblet cells, and enteroendocrine cells (Fig. 1l). Intriguingly, we also detected the loss and re-emergence of LGR5-mNeonGreen expression in the organoids, indicative of dynamic differentiation and dedifferentiation processes (Fig. 1l).
Intestinal cells possess remarkable plasticity16,18,24. To test the ability of different cell types to initiate organoids, we sorted LGR5-negative, low, and high cells and cultured them under the TpC condition (Supplementary Fig. 4a, b). We found that LGR5-negative cells had the highest colony forming efficiency, with LGR5-positive cells re-emerging in these organoids (Supplementary Fig. 4c, d). We hypothesize that non-LGR5 cells, such as differentiating progenitor cells and alternative stem cells like isthmus progenitor cells, could serve as initiation cells for organoids25. These findings suggest that the TpC condition supports generating a dynamic and plastic population of intestinal cells in organoids, with the potential to give rise to multiple lineages and undergo cellular plasticity.
scRNA-seq analysis revealed cellular diversity and cell fate dynamics in TpC-organoids
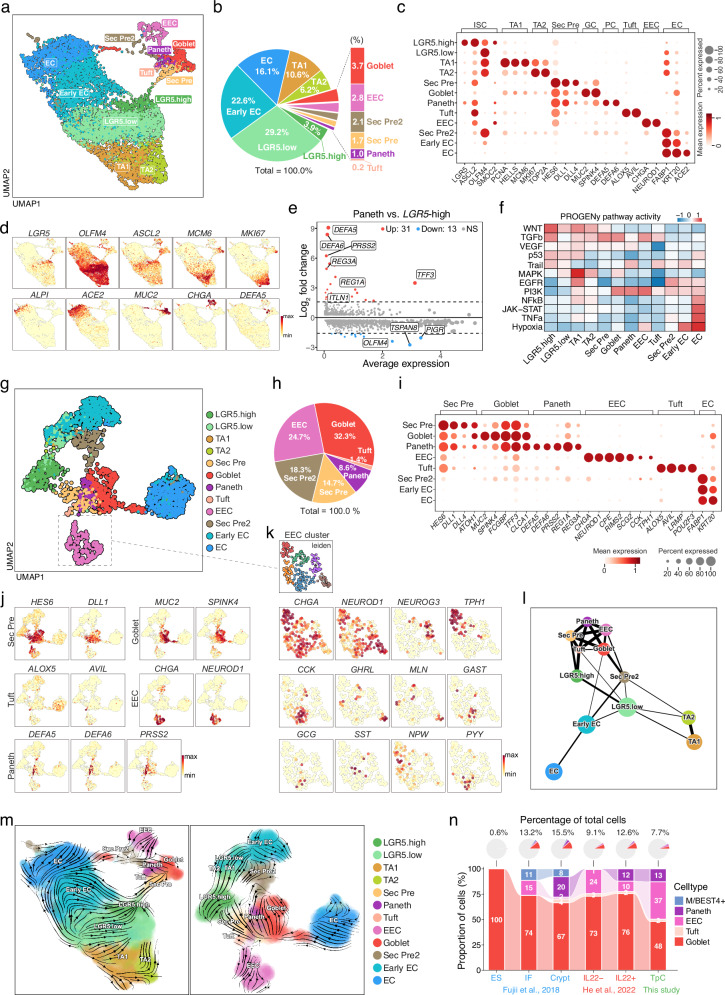
To gain insight into the cellular diversity of TpC-cultured organoids, we conducted single-cell RNA sequencing (scRNA-seq) on the cells. We annotated the cells based on the expression of well-established markers and identified 12 distinct clusters of cells (Fig. 2a, b). Among these clusters, we found that ISCs could be further divided into two subpopulations based on their expression levels of LGR5 and OLFM4 (Fig. 2a–d). Specifically, we identified LGR5-high and LGR5-low ISCs, two populations of cycling transit amplifying (TA) cells expressing low levels of LGR5 but high levels of cell cycle genes such as MKI67 and PCNA, a cluster of FABP1+ early enterocytes (early EC) lacking LGR5 expression, mature enterocytes (EC) expressing ALPI and ACE2, and multiple secretory cell types, including Paneth cells (DEFA5, DEFA6), goblet cells (MUC2, SPINK4), enteroendocrine cells (CHGA, NEUROD1), and tuft cells (ALOX5, AVIL) (Fig. 2c, d). Of particular interest, we identified two distinct secretory precursors. The first (Sec Pre) expressed classical secretory precursor markers such as HES6, DLL1, and DLL4, representing typical secretory precursors. The second (Sec Pre2) expressed OLFM4 as well as markers of both secretory (goblet, Paneth, and enteroendocrine cell markers) and enterocyte lineages (FABP1 and KRT20) (Fig. 2c). We inferred that these cells represent transdifferentiated cells from enterocyte lineages.
Fig. 2. scRNA-seq analysis reveals cellular diversity and cell fate dynamics in TpC-organoids.
a UMAP plot showing clustering of cells from scRNA-seq analysis of TpC organoids. Cluster labels indicate cell types. LGR5.high and LGR5.low indicate intestinal stem cells with different levels of LGR5 expression; TA1, transit-amplifying cell type 1; TA2, transit-amplifying cell type 2; Sec Pre, Secretory Precursor; Sec Pre2, Secretory Precursor Type 2; EC, enterocyte; EEC, enteroendocrine cell. b Proportion of each cell type from scRNA-seq sample in a. c Dot plot showing expression and percentage of cells expressing cell type-specific markers in clusters from scRNA-seq analysis. Dot size and color indicate normalized gene expression level. d UMAP plots showing expression of cell type-specific markers in TpC organoids. Color intensity indicates normalized gene expression level. e MA plot showing differentially expressed genes between Paneth cells and LGR5-high cells. f Heatmap showing predicted PROGENy pathway activity for each cell cluster. g UMAP plot showing subset analysis of secretory cells along with connected cell clusters. h Pie chart showing the proportion of each secretory cell type. Total percentages of all secretory cell types equal 100%. i Dot plot showing expression and percentage of cells expressing cell type-specific markers in clusters shown in g. Dot size and color indicate normalized gene expression level. j UMAP plots showing expression of cell type-specific markers in the secretory subset. Color intensity indicates normalized gene expression level. k UMAP plots highlighting EEC cells and depicting expression of EEC subtype-specific markers. Color intensity indicates normalized gene expression level. l PAGA trajectory analysis depicting cell connectivity. Circle size indicates cell number, and line thickness indicates connectivity strength between cell clusters. m RNA velocity analysis using Dynamo showing inferred transition direction between cell states. n Pie charts (top) showing proportions of indicated cell types among total cells, and bar charts (bottom) showing the composition of indicated cell types from scRNA-seq analysis of ES, IF, IL, TpC organoids, and crypt.
Differential expression analysis confirmed the upregulation of classical markers for each cell type relative to LGR5-high ISCs (Fig. 2e and Supplementary Fig. 5a). Notably, DEFA5, DEFA6, and PRSS2 exhibited the most marked fold change for Paneth cells. PROGENy pathway activity analysis revealed distinct pathway activities for each cell type. LGR5+ stem cells exhibited high Wnt activity, whereas TA cells showed high MAPK pathway activity (Fig. 2f). To better understand secretory lineage cells, we performed a subset analysis of secretory cells (Fig. 2g–k). Goblet cells were the most abundant, accounting for 32.3% of all secretory lineage cells, consistent with their composition in vivo. Paneth cells were less abundant, accounting for 8.6% of the secretory lineage cells, while a small number of tuft cells were also detected (1.4%) (Fig. 2h). These cells expressed classical markers (Fig. 2i–k). Enteroendocrine cells accounted for 24.7% of all secretory lineage cells and expressed multiple subtype markers, including NEUROG3 (progenitors). Among these subtypes, TPH1+ enterochromaffin and CCK+ I cells were the most abundant, consistent with their in vivo abundance26,27. We also observed similar expression patterns as previously reported, where GHRL+ cells co-expressed MLN10,28 and mutually exclusive expression for MLN and GAST, for GHRL and CHGA, and for TPH1 and GCG27. This suggests a high similarity of our organoid with the in vivo intestine.
Notably, the Uniform Manifold Approximation and Projection (UMAP) plot of TpC organoid-derived cells suggests a differentiation path of secretory cells from LGR5-high ISCs to secretory precursors (Sec Pre), partially differentiated LGR5-low/early ECs to the second population of secretory precursors (Sec Pre2), as well as more differentiated early ECs/mature ECs to goblet cells, in both the whole organoid dataset (Fig. 2a) and the secretory subset (Fig. 2g). These connections were also captured using the force atlas layout (Supplementary Fig. 5b) and PAGA trajectory analysis (Fig. 2l), where a major source of secretory lineage cells is a path from LGR5-high ISCs towards secretory precursors (Sec Pre), which was confirmed by RNA velocity analysis using the Dynamo package (Fig. 2m). This observation is consistent with scRNA-seq data from freshly isolated intestinal biopsies29, where Paneth cells and secretory lineages were found to descend from LGR5+ stem cells, and with ENR organoid data from mouse organoids30. These trajectories are also present in fresh crypt data, but not in IF10 or IL22 organoid datasets11 (Supplementary Fig. 5c), where a possible source of secretory cells is partially differentiated early ECs (Supplementary Fig. 5c). Notably, when we compared the pathway activity of ISCs from different samples, we found that ISCs in the TpC condition mostly resembled ISCs from freshly isolated crypt samples (Supplementary Fig. 5e). To compare the cell composition of organoids cultured under different conditions, we integrated scRNA-seq datasets of cells cultured under multiple organoid conditions using the Seurat package (Supplementary Fig. 5d). This analysis confirmed our previous observation that an LGR5-high population connects absorptive and secretory lineages, which was present in the Crypt data but mostly absent in the IF and IL22 samples (Supplementary Fig. 5d). Furthermore, the cell composition comparison confirmed that TpC organoids capture major secretory cell types, including Paneth, goblet, enteroendocrine, and tuft cells (Fig. 2n). We further compared organoid data with in vivo data (Supplementary Fig. 6a). Harmony analysis showed that most cell types were similar and co-clustered with in vivo data. EC lineage cells, including early and mature EC cells, formed distinct populations in all organoid conditions (ES, IF, IL22, and TpC) and in vivo enterocytes. However, only TpC organoids had significant numbers of EC cells that co-clustered with the in vivo EC cluster (Supplementary Fig. 6b, Supplementary Fig. 7, Supplementary Fig. 8). We believe this might be due to specific culture conditions, such as high Wnt and low BMP levels, which influence the properties of EC cells in organoid cultures. Additionally, cells in organoids exhibited similar marker gene expression to those in vivo cell types, with TpC organoids showing the highest similarity (Supplementary Fig. 9). These results demonstrate the fidelity of our organoid system in recapitulating the cellular diversity of the intestinal epithelium.
Small molecules play distinct roles in supporting stemness and differentiation
Our TpC condition maintains cellular diversity and self-renewal of LGR5+ stem cells, providing a platform to dissect the roles of individual chemicals in human intestinal cell fate regulation. The basal molecules in the TpC condition are well characterized, including EGF, IGF-1, FGF-2, the BMP inhibitor DMH1, R-Spondin 1, and A83-01, which have been shown to play essential roles in maintaining mouse and human intestinal organoids. Another molecule, CHIR99021, a GSK3 inhibitor, activates the Wnt pathway and promotes ISC self-renewal22.
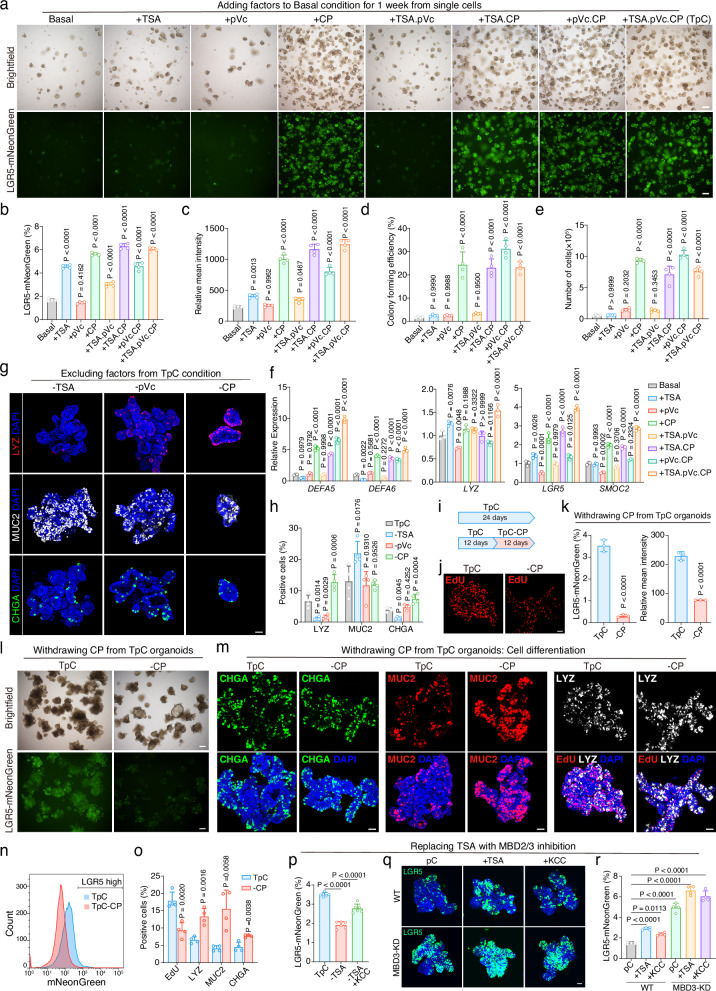
In our study, we focused on the roles of TSA, pVc, and CP. When TSA, pVc, and CP were added individually or in combination to the basal condition, TSA and CP significantly increased the LGR5-mNeonGreen percentages, with CP showing the most pronounced effect on cell proliferation, colony formation from single cells, and LGR5-mNeonGreen expression (Fig. 3a–e). TSA and CP also boosted the expression of Paneth cell markers (DEFA5, DEFA6, LYZ) and stem cell markers (LGR5, SMOC2), with the TpC condition showing the highest levels (Fig. 3f). CP also increased secretory cell diversity when added to the basal condition (Supplementary Fig. 10a), probably owing to increased stem cells in the organoids.
Fig. 3. The combination of TSA, pVc, and CP induces enhanced stemness and increased cellular diversity in organoids.
a Brightfield and fluorescence images of organoids cultured for 1 week in indicated conditions. Representative images from three independent experiments. b, c Quantification of the percentage of LGR5-mNeonGreen cells (b) and relative fluorescence intensity (c) in organoids cultured in indicated conditions, n = 4 samples. Colony forming efficiency (d) and the number of cells (e) of organoids cultured in indicated conditions, n = 4 samples. f RT-qPCR quantification of cell markers expression levels, n = 3 samples. g Representative confocal images of LYZ, MUC2, and CHGA in organoids cultured in indicated conditions. Representative images from three independent experiments. h Proportions of secretory lineages in organoids cultured in indicated conditions, n = 4 samples. b–f and h, one-way ANOVA with Dunnett’s multiple comparisons test; data are presented as mean ± SD. i Schematic of experiments in (j–o). j Confocal images of EdU staining of organoids. k Quantification of the percentage of LGR5-mNeonGreen cells and relative fluorescence intensity in organoids cultured under the indicated conditions, n = 3 samples. l Brightfield and fluorescence images of organoids cultured in indicated conditions. m EdU and immunofluorescence staining of CHGA, MUC2, and LYZ in organoid cultures as indicated. EdU was added for 1 hour before staining. Representative images of (j, l, m) were from three independent experiments. n FACS gating strategy of LGR5-mNeonGreen cells. o Quantification of EdU, LYZ, MUC2 and CHGA positive cell percentage as in (m), n = 4 samples. k and o, two-tailed unpaired t-test; data are presented as mean ± SD. p Quantification of the percentage of LGR5-mNeonGreen cells in organoids cultured under the indicated conditions, n = 6 samples. q LGR5-mNeonGreen fluorescence images of organoids cultured in indicated conditions. Representative images from four independent experiments. r Proportions of LGR5-mNeonGreen cells in organoids cultured in conditions as in (q), n = 4 samples. (p and r), one-way ANOVA with Dunnett’s multiple comparisons test; data are presented as mean ± SD. Scale bars, (a and l), 200 μm; (g, j, m and q), 50 μm. Source data are provided as a Source Data file.
We further investigated the effects of TpC factors by omitting each factor from the TpC combination when initiating organoids. Excluding any molecule from the TpC condition reduced ISC self-renewal, as evidenced by the percentage of LGR5-mNeonGreen high cells and their mean fluorescence intensity (Supplementary Fig. 10b, c). Although excluding TSA and pVc did not affect cell growth, the absence of CP significantly impacted organoid growth (Supplementary Fig. 10b). Excluding TSA from the TpC combination markedly decreased Paneth and EEC cells while increasing goblet cells. Excluding pVc decreased Paneth cells but had little effect on EEC and goblet cells. This could be attributed to the loss of ISCs capable of generating Paneth cells and EECs (but not goblet cells) under conditions lacking TSA or pVc. In contrast, excluding CP increased Paneth and EEC cells but reduced cell proliferation, possibly due to immediate differentiation of newly expanded ISCs in this condition (Fig. 3g-h). The combination of TpC demonstrated the best overall performance in organoid growth, colony formation, LGR5-mNeonGreen expression, and secretory cell diversity (Fig. 3a–h, and Supplementary Fig. 10b, c).
We then evaluated the effects of withdrawing factors from already established TpC organoids. Withdrawing CP in this context led to decreased cell proliferation (Fig. 3i, j, o), decreased LGR5 expression and LGR5 cell proportion (Fig. 3k, l, n). It also increased cell differentiation, confirmed by immunostaining for Paneth, goblet, and EEC markers in the organoids (Fig. 3m, o), suggesting a role for CP in preventing ISC differentiation, consistent with the effects of CP on organoid generation from single intestinal cells.
TSA enhances LGR5 stem cell maintenance by targeting HDAC-MBD-NuRD complex
To further dissect the mechanism of TSA, which is a pan-HDAC inhibitor, we tested multiple HDAC inhibitors and found they similarly promoted LGR5-mNeonGreen expression and increased cellular diversity, albeit with varying degrees of efficacy (Supplementary Fig. 11a–d). Another pan-HDAC inhibitor, VPA, also increased the LGR5-mNeonGreen proportion but significantly decreased organoid growth (Supplementary Fig. 11a). HDAC6 inhibitor Tubastatin A and HDAC2 inhibitor CAY10683 also significantly increased the LGR5-mNeonGreen proportion and intensity, similar to TSA (Supplementary Fig. 11a–d), while Tubastatin A did not affect Paneth cell generation (Supplementary Fig. 11e). These results suggest that TSA might promote stem cell maintenance and Paneth cell differentiation through HDAC1/2. To further investigate the target chromatin complex, we used small molecules to regulate complexes containing the HDAC1/2 subunit. We found that KCC-07 (KCC), an MBD2 inhibitor that prevents MBD2 from binding to methylated DNA, could rescue the budding ratio and LGR5-mNeonGreen expression in the absence of TSA (Fig. 3p and Supplementary Fig. 12a–d). Organoids treated with KCC-07 showed a greatly increased frequency of Paneth cells compared to the -TSA condition (Supplementary Fig. 12e, f). The expression of cell type-specific markers further confirmed the similar effects of KCC-07 and TSA (Supplementary Fig. 12g–k). MBD2 is a core subunit of the Nucleosome Remodeling and Deacetylation (NuRD) complex, which includes HDAC1/2 proteins. These results suggest that TSA targets the MBD2-NuRD complex to promote LGR5-mNeonGreen expression and subsequent Paneth cell differentiation.
The Methyl-CpG-binding domain (MBD) protein family includes two members, MBD2 and MBD3. Previous studies have implied the MBD3-NuRD repressor complex inhibits Lgr5 expression and progenitor proliferation in the mouse intestine31. We inferred that MBD3-NuRD also mediated TSA-induced gene expression in the human intestine. To test this, we used CRISPR-Cas9 to genetically interfere with MBD3 expression in human intestinal cells, using guide RNA targeting exon 3 of human MBD3 due to the lack of a specific MBD3 inhibitor (Supplementary Fig. 13a). We isolated an independent clone with reduced MBD3 expression, confirmed by Western blotting and qPCR (Supplementary Fig. 13b, c). Knockdown of MBD3 significantly increased LGR5 expression (Supplementary Fig. 13c) and the proportion of LGR5-mNeonGreen stem cells (Fig. 3q-r; Supplementary Fig. 13d, e). Notably, LGR5 expression and the proportion of LGR5 stem cells were significantly higher in MBD3 knockdown organoids than in WT organoids, even under TSA or KCC-07 conditions (Fig. 3q-r; Supplementary Fig. 13d, e). Simultaneous inhibition of both MBD2 and MBD3 (MBD3-KD + KCC-07) effectively enriched LGR5-mNeonGreen stem cells. In summary, these experiments confirmed that TSA, as an HDAC1/2 inhibitor in human intestinal organoids, led to MBD2 and MBD3 NuRD-mediated enrichment of LGR5 stem cells, consequently enhancing Paneth cell abundance.
We further performed single-cell RNA-seq to explore the effect of TSA on different cell types. The withdrawal of TSA from established TpC organoids (Supplementary Fig. 14a) induced premature differentiation and subsequent death of Paneth cells in the organoids, as indicated by the presence of a significant number of Paneth cell marker (DEFA5, DEFA6, and PRSS2) expressing cells with low UMI counts and feature numbers (Supplementary Fig. 14b, c). These cells also co-expressed markers of progenitors such as OLFM4, FABP2, and FGFBP1 (Supplementary Fig. 15a, b, e). Cell type priority analysis indicated that Paneth cells, secretory precursor and LGR5-high cells were most significantly affected by TSA withdrawal (Supplementary Fig. 15c, d). Pseudotime and trajectory analysis indicated a similar cellular differentiation path through LGR5 cells to secretory cells (Supplementary Fig. 15f, g), where Metacell analysis revealed the topology of cellular differentiation. LGR5 cells form a center that connects secretory cells with TA cells and enterocyte lineages (Supplementary Fig. 15h-i), which express typical cell type markers along pseudotime (Supplementary Fig. 15j). These results are consistent with the role of TSA in LGR5 cell maintenance and preventing the premature differentiation of ISCs into secretory cells.
iBET-151 reversibly promotes proliferation and inhibits secretory differentiation
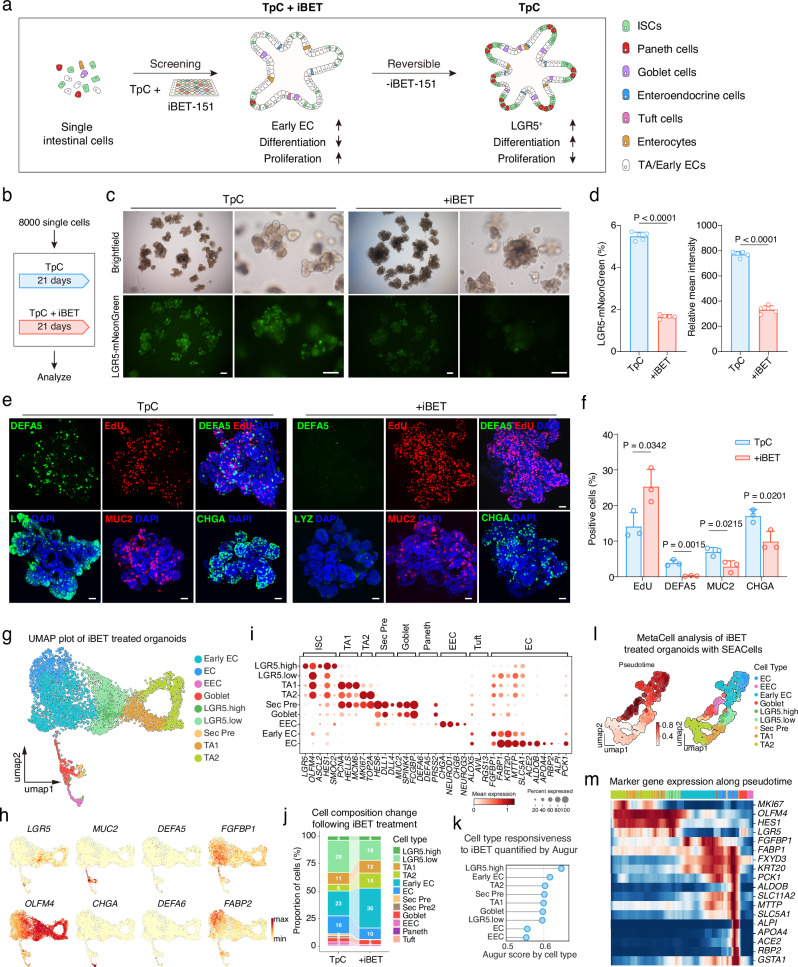
The cell fate conversion of intestinal epithelial cells in vivo is highly dynamic, involving various simultaneous cellular events, including self-renewal, differentiation, and dedifferentiation. We next asked whether these processes could be similarly regulated in our TpC organoids, which exhibit a balanced self-renewal and differentiation. To this end, we further evaluated the effects of small molecule pathway regulators based on the TpC condition (Fig. 4a). We discovered that iBET-151 (iBET or I), a BET bromodomain inhibitor, effectively enhanced cell proliferation while decreasing the number of LGR5-mNeonGreen cells (Fig. 4b–f). Concurrently, we observed reduced differentiation of secretory cells, including Paneth cells, goblet cells, and EECs (Fig. 4e, f and Supplementary Fig. 16a, b). Notably, Paneth cells were nearly absent in the organoids (Fig. 4e, f and Supplementary Fig. 16b, d). Additionally, the presence of iBET induced a distinct morphological change in the crypt domain of the organoids, resulting in a more homogeneous cell population rather than intermingled Paneth and ISC-like cells (Supplementary Fig. 16c). These findings imply that the TpCI combination maintains the organoids in a more progenitor-like state, partially by inhibiting differentiation towards secretory cell types.
Fig. 4. iBET-151 reversibly promotes proliferation and inhibits secretory differentiation.
a Schematic of the screening strategy for modulating early EC enrichment in organoids. b Schematic of organoid culture conditions for experiments in (c–f). c Representative brightfield and fluorescence images of organoids cultured in indicated conditions. Representative images from three independent experiments. Scale bars, 200 μm. d Proportion of LGR5-mNeonGreen cells in organoids shown in (c). n = 5 samples. Two-tailed unpaired t-test; data are presented as mean ± SD. e EdU and immunofluorescence staining of Paneth cells (LYZ and DEFA5), goblet cells (MUC2), and enteroendocrine cells (CHGA) in organoids cultured as in (b). Representative images from three independent experiments. Scale bars, 50 μm. f Quantification of positive staining as in (e). n = 3 samples. Two-tailed unpaired t-test; data are presented as mean ± SD. g UMAP plot showing clustering of cells from scRNA-seq analysis of TpCI (TpC+iBET) organoids. Cluster labels indicate distinct cell types. h UMAP plots displaying the expression levels of cell type-specific markers (LGR5, MUC2, DEFA5, FGFBP1, OLFM4, CHGA, DEFA6, FABP2) in TpCI organoids. Color intensity indicates normalized gene expression levels. i Dot plot illustrating the expression and percentage of cells expressing cell type-specific markers across the clusters identified in (g). Dot size represents the percentage of cells expressing the marker, while color intensity indicates normalized gene expression levels. j Bar charts depicting the composition of indicated cell types from scRNA-seq analysis, comparing TpCI organoids with TpC organoids. Proportions of cell types are shown as percentages. k Cell type responsiveness to iBET treatment quantified by Augur, comparing the TpCI dataset with the TpC dataset. Augur scores for each cell type are displayed, indicating the degree of responsiveness to iBET treatment. l UMAP plot demonstrating pseudotime analysis and cell type labeling of MetaCells in the iBET dataset. Pseudotime was calculated using the Stavia package, and MetaCells were identified using the SEACells package. m Heatmap of cell type-specific marker gene expression along the pseudotime trajectory as shown in (l). The color bar indicates cell type labels consistent with (l). Source data are provided as a Source Data file.
To examine the reversibility of iBET’s effects and assess the functionality of TpCI-cultured cells, we removed iBET from TpCI-cultured organoids (Supplementary Fig. 16e). After seven days of continued culture in TpC conditions, the LGR5-mNeonGreen expression increased to levels comparable to those observed in TpC organoids (Supplementary Fig. 16f, g), accompanied by the resurgence of secretory cells in the organoids (Supplementary Fig. 16h, i).
To investigate the impact of iBET on established organoids, we introduced iBET to TpC organoids (Supplementary Fig. 16j). After culturing for 12 days in the TpCI condition, we observed a notable enhancement in organoid proliferation as evidenced by the increased organoid size and the presence of EdU-positive proliferating cells (Supplementary Fig. 16j-k). In contrast, secretory cell differentiation was reduced in the organoids (Supplementary Fig. 16j-k), consistent with observations in organoids derived from single intestinal cells. These findings suggest that the effects of iBET are reversible, and the cell fate of cells in TpC or TpCI organoids is dynamic, adjusting in response to different culture conditions.
To further elucidate the cellular diversity and cell fate trajectory within TpCI-cultured organoids, we performed single-cell RNA sequencing (scRNA-seq). Cell type annotation revealed the presence of most cell types found in TpC organoids, with the notable exception of Paneth cells (Fig. 4g). This absence was further confirmed by the lack of expression of Paneth cell markers (Fig. 4h, i). Cell composition analysis indicated a lower abundance of secretory cells, such as enteroendocrine cells (EECs) and secretory precursor cells, while the abundance of goblet cells remained similar. However, there was a notable increase in enterocyte lineage cells and transient amplifying (TA) cells (Fig. 4j). This observation aligns with the cellular phenotype of TpCI organoids, which exhibit increased proliferation and reduced differentiation into secretory cells. Cell type priority analysis using Augur indicated that LGR5-high and early enterocyte (EC) cells were most affected by iBET treatment (Fig. 4k). This suggests that iBET treatment alters the cell differentiation trajectory, shifting it from secretory differentiation towards the enterocyte lineage. Further trajectory and MetaCell analysis revealed a major cell differentiation pathway from TA cells to enterocytes, mediated by LGR5 cells (Fig. 4l, m). This data supports the hypothesis that iBET treatment promotes the proliferation of enterocyte lineage cells at the expense of secretory cell differentiation.
Controlled lineage shifts in organoids via defined niche signals
Our optimized combination of chemical modulators effectively sustained a balance between stem cell self-renewal and differentiation in the organoids, as well as the controlled shift of this equilibrium decisively towards enhanced proliferation. Going forward, we aimed to identify conditions that can selectively and unidirectionally shift this equilibrium towards specific differentiated cell types, allowing us to generate organoids enriched in Paneth, goblet, enteroendocrine, or enterocyte cells. Although several protocols have been reported to induce Paneth cell or enteroendocrine cell differentiation, such as using IL-22 for Paneth cells11 or modulating the endocannabinoid receptor, c-Jun N-terminal kinase (JNK) or Forkhead box O1 (FOXO1) pathways for enteroendocrine cells32, we aimed to induce differentiation by recapitulating the physiologic cell fate dynamics directed by in vivo niche signals such as Wnt, Notch and BMP signaling.
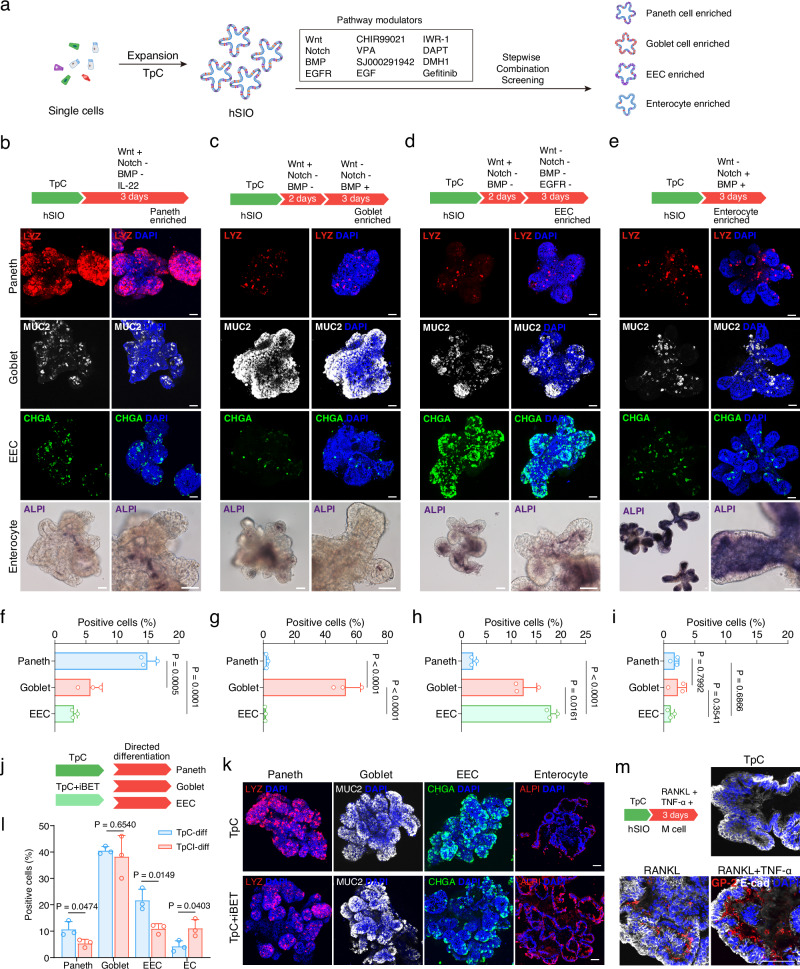
We systematically screened various combinations of Wnt, Notch, BMP, and EGFR pathway modulators in a stepwise fashion (Fig. 5a). We found that the effects of Wnt and Notch signaling on mouse ISCs also applied to human cells. Specifically, inhibiting Notch signaling with DAPT promoted differentiation towards secretory cells33–35. Additional modulation of Wnt activity further directed differentiation towards Paneth cells, goblet cells, or enteroendocrine cells. Sustained Wnt activation in the presence of DAPT enhanced Paneth cell differentiation. Further addition of IL-22 promoted Paneth cell differentiation (Supplementary Fig. 17a), resulting in a 15% enrichment of Paneth cells after a rapid 3-day induction protocol (Fig. 5b, f). In contrast, we found that an initial step of Wnt activation and Notch inhibition followed by Wnt inhibition and sustained Notch inhibition was required to enhance goblet cell and enteroendocrine cell differentiation. This may correspond to a secretory precursor stage in vivo. Moreover, switching BMP signaling from inhibition to activation was essential for goblet cell induction (Supplementary Fig. 17b), leading to nearly 50% goblet cell differentiation efficiency from TpC organoids (Fig. 5c, g). While CP mildly inhibited enteroendocrine cell generation in TpC organoids, removing CP did not further enhance its differentiation. Therefore, we included CP and the EGFR inhibitor Gefitinib in the protocol to prevent Paneth cell or goblet cell differentiation and induce enteroendocrine cell differentiation36. Under these conditions, CHGA-positive enteroendocrine cells comprised 17% of organoid cells (Fig. 5d, h). The duration of step 1 and step 2, primarily distinguished by the presence of Wnt signaling, significantly affected the differentiation efficiency of goblet cells and enteroendocrine cells (Supplementary Fig. 17b, c), similar to the differentiation of secretory cells in mouse organoid cells37. Furthermore, the combination of IWR-1, SJ000291942, and VPA rapidly and efficiently induced enterocyte differentiation in three days (Fig. 5e). Under these conditions, Paneth cells, goblet cells, and enteroendocrine cells were not enriched (Fig. 5i). Removing any single factor reduced enterocyte differentiation (Supplementary Fig. 17d).
Fig. 5. Directed differentiation of intestinal organoids via niche signaling modulation.
a Schematic of small molecules screening strategy for induction of differentiated cell types. Differentiation protocols for Paneth cells (b), goblet cells (c), enteroendocrine cells (d), and enterocytes (e) (upper panels) and representative confocal images of organoids after differentiation showing Paneth cells (LYZ), goblet cells (MUC2), enteroendocrine cells (CHGA) and enterocytes (ALPI) (lower panels). Representative images from three independent experiments. f–i Quantification of the percentage of differentiated cells as shown in (b–e) respectively. One-way ANOVA with Tukey’s multiple comparisons test; data are presented as mean ± SD; representative experiment showing n = 3 samples from each condition. j Schematic of differentiation conditions for organoids cultured in TpC and TpCI (iBET-treated) conditions for experiments (k and l). k Representative confocal images of differentiated organoids, showing Paneth cell (LYZ), goblet cell (MUC2), and enteroendocrine cells (CHGA) in TpC and TpCI organoids. Representative images from three independent experiments. l Quantification of the percentage of differentiated cells as in (k). Two-tailed unpaired t-test; data are presented as mean ± SD; representative experiment showing n = 3 samples from each condition. m Schematic and representative confocal images illustrating M cell differentiation in organoids cultured under TpC condition. Representative images from three independent experiments. Scale bars, 50 μm. Source data for this Figure are provided as a Source Data file.
We further tested these differentiation protocols on TpCI organoids, which are enriched for enterocyte lineage cells (Fig. 5j). As anticipated, these protocols induced differentiation with higher efficiency of EC cells in TpCI organoids compared to TpC organoids (Fig. 5k, l), while relatively lower differentiation efficiency of Paneth cell and EEC than TpC organoids. We also tested the Paneth cell differentiation protocol on IF organoids and detected a relatively low proportion of Paneth cells after a 3-days induction (Supplementary Fig. 17e). Additionally, we applied a previously published M cell differentiation protocol for mouse intestinal cells38,39 to human TpC organoids and observed high-efficiency differentiation (Fig. 5m and Supplementary Fig. 18a, b).
These results demonstrate that stem cells in TpC organoids remain multipotent. The equilibrium state of TpC organoids can be selectively shifted through defined signaling toward the major functional cell types of the human intestine (Supplementary Fig. 19a). By integrating our differentiation data with single-cell trajectory analysis, we propose a unified model of cell fate transitions in the human intestinal epithelium (Supplementary Fig. 19b). In this model, spatiotemporal gradients of Wnt, BMP, and Notch signaling direct ISC self-renewal and differentiation. ISC self-renewal at the crypt base results from high Wnt, low BMP, and activated Notch signaling and can be further enhanced by factors such as pVc and CP. Downregulation of Notch signaling at the crypt base gives rise to secretory precursors in a high Wnt, low BMP niche. Depending on the Wnt and BMP levels, these cells further differentiate into distinct secretory cell types. In contrast, loss of Wnt reduces ISC self-renewal and promotes their differentiation into enterocytes. During this enterocyte differentiation process, partially differentiated cells can further differentiate into secretory cells, especially goblet cells, upon Notch inactivation. This equilibrium can be further shifted by chemical or genetic perturbations. For instance, we showed that NuRD complex inhibition and BET inhibition have distinct effects on cell fate trajectory and consequently on cellular composition in the organoids.
Discussion
In this study, we demonstrate the ability to mimic complex cell fate dynamics observed in vivo by using growth factors and small molecules within an organoid system. By precisely controlling cell fate dynamics, our method fosters a balanced state of self-renewal and differentiation in organoids, which in turn allows for parallel proliferation and enhanced cellular diversity. Furthermore, by manipulating combinations of niche factors, we induced cell fate shifts towards different differentiation directions, significantly altering the cellular composition of the organoid.
Maintaining balanced self-renewal and differentiation of adult stem cells is crucial for tissue homeostasis in vivo. However, achieving this balance in vitro has been challenging in organoid culture, primarily due to the absence of signaling gradients regulating localized self-renewal and differentiation within specific niches12,40,41. Furthermore, while progenitor cells within organoids are sufficient for driving both proliferation and differentiation, their limited plasticity and stemness ultimately restrict the diversity of cell types in organoids. In this study, we hypothesize that maximal cellular diversity in organoids can be more effectively achieved by inducing enhanced stemness within organoids, thereby unlocking their potency and potential for multidirectional differentiation rather than relying on lineage-restricted progenitors or directly driving differentiation. We demonstrated that this “regress to progress” strategy applies to human intestinal organoids. By modulating the combinations of small molecules, we enhanced the stemness of intestinal stem/progenitor cells without hindering their differentiation, thereby achieving a balanced self-renewal and differentiation within the organoids. This approach led to both rapid growth and increased cellular diversity, offering advantages over existing protocols. The abundance of LGR5+ stem cells enables efficient organoid establishment and rapid expansion and, importantly, endows the capacity to generate a diverse range of intestinal cell types. Our protocol produced Paneth cells, enteroendocrine cells, goblet cells, tuft cells, and mature enterocytes without requiring a separate differentiation step. This substantially improves previous methods and provides an optimized human intestinal organoid system for applications such as drug development and disease modeling.
We have verified the specific roles of the three key components, TSA, pVc, and CP673451, in the TpC system and demonstrated that TSA, as an HDAC inhibitor, exerts its effects through the NuRD complex. Further investigation into the epigenetic mechanisms of HDAC inhibitors, using tools such as ATAC-seq, would provide valuable insights and represent a promising direction for future research.
While a signaling pathway regulation network for mouse ISC differentiation to specific cell types like Paneth, goblet, enteroendocrine, enterocytes, M cells, and Tuft cells has been established22,36,42,43, developing similar networks for human intestinal epithelial cells has been challenging due to the difficulty in maintaining human ISCs. Although obtaining high-purity hISC cultures has been difficult, we established an organoid system enriched in LGR5+ cells. Additionally, we showed that iBET-151 reversibly shifted cell fate from secretory cell differentiation to the enterocyte lineage while maintaining cells in a highly proliferative state, providing a tool to dissect the regulatory network for major cell types in the human intestinal epithelium. We found similar signaling mechanisms regulate Paneth, enteroendocrine, goblet cell, and enterocyte differentiation in both human and mouse intestinal epithelial cells, involving combinations and timing of Wnt, Notch, BMP, and MAPK pathways. Paneth cells can be obtained from human cells using high Wnt and Notch inhibitors, as in mouse ISCs22. We found an intermediate, high Wnt, and low BMP step upon Notch inhibition effectively increased secretory cell differentiation, representing secretory precursor differentiation at the crypt base in vivo upon Notch inactivation. These results suggest that basic cell fate regulation mechanisms are conserved in human and mouse intestinal cells, relying on niche signaling combinations.
While our scRNA-seq analysis and experiments suggest potential cellular plasticity in our organoid system, conclusively demonstrating direct transdifferentiation between cell types requires additional validation. Future studies using advanced lineage tracing techniques, such as CRISPR-mediated reporter knock-in organoids and unbiased cell barcoding approaches, will be essential to definitively map cell fate transitions and better understand the dynamics of cellular plasticity in human intestinal organoids. These studies will help elucidate the precise mechanisms underlying differentiation, dedifferentiation, and potential transdifferentiation events observed in our system.
Beyond these molecular and cellular dynamics, another key challenge remains in recreating the complex tissue architecture of the intestinal epithelium. Although our TpC system has increased cellular diversity in the organoids, it has not yet achieved the correct spatial structure and cell distribution as seen in vivo. Ultimately, creating an in vitro model that fully emulates in vivo physiology requires reproducing the capacity of stem cells to respond to complex spatiotemporal cues governing their self-renewal and differentiation, where a homogenous culture environment may limit the maturation of differentiated cells. Thus, we envision that artificially generating signaling gradients through bioengineering approaches could further improve the fidelity of organoid models.
Methods
The research reported here complies with all relevant ethical regulations and guidelines. Small intestine tissue biopsies used in this study were obtained from patients who provided written informed consent under the ethical committee of Shanghai East Hospital (approval number: 2023-064).
Genetic engineering of human small intestinal organoids
For the construction of a fluorescent reporter, an LGR5-P2A-mNeonGreen knock-in reporter was introduced into the last exon of LGR5 via electroporation, using previously published sgRNA (Supplementary Table 1) and homology arms (Supplementary Table 2)44. Two LGR5-targeting sgRNAs were cloned into SpCas9-equipped HP180 plasmids (a gift from Hui Yang lab), generating LGR5 sgRNA plasmids. HDR templates were made by replacing the CMV promoter in pBluescript II SK (+) (Addgene, #212205) with LGR5-P2A-mNeonGreen sequence flanked by LGR5 homology arms using the Fast-Fusion™ Cloning Kit (GeneCopoeia). The LGR5 sgRNA plasmids and HDR template were co-electroporated into human intestinal organoids using a NEPA21 electroporator (Nepa Gene) following published methods45. Organoid colonies derived from single cells were manually picked, expanded, and genotyped. To interfere with MBD3 expression, sgRNA sequences targeting MBD3 were cloned into SpCas9-equipped HP180 plasmids and electroporated into established LGR5-mNeonGreen reporter cells. Colonies from single cells were manually picked, expanded, and screened by Western blotting using an MBD3 antibody (1:200, Santa Cruz). The CRISPR target sites, homology arms, knock-in sequence and PCR primers are listed in Supplementary Table 1.
Crypt isolation and organoid culture
Tissues were transferred into Dulbecco’s phosphate-buffered saline (DPBS) and placed on ice until processing. Crypt isolation was performed as previously reported45 with modifications. Briefly, tissues were washed with DPBS to remove debris. The tissues were treated with 2.5 mM EDTA at 4 °C for 40 minutes to release crypts. The supernatant containing crypts was collected and embedded in Cultrex RGF BME (R&D Systems) droplets in 48-well plates (~50 crypts/well). After solidification, droplets were overlaid with culture medium containing 10 μM Y-27632 (TargetMol). The medium was changed every 3 days. Established organoids were dissociated into single cells with TrypLE (Gibco) and re-embedded in fresh BME at a density of approximately 8000 cells per well, in a total volume of 20 μL, within a 48-well plate. The BME Matrigel utilized in the culture system was pre-diluted with Advanced DMEM/F12 at a 5:3 ratio (Matrigel: Advanced DMEM/F12). Basal medium contained Advanced DMEM/F12 (Gibco) supplemented with B-27 (Gibco), 10 mM HEPES (Corning), 2 mM GlutaMAX (Gibco), Penicillin-Streptomycin (Gibco), and 1 mM N-Acetyl-L-cysteine (Sigma). For IL-hSIO culture, 50% Wnt3A conditioned medium, 10% R-spondin1 conditioned medium, 50 ng/mL EGF (Peprotech), 2 μM DMH1 (ApexBio), 0.5 μM A83-01 (ApexBio), and 3 μM CHIR (ApexBio) were added to the basal medium. For IF-hSIO culture, 10 nM Gastrin I (MCE), 100 ng/mL recombinant mouse Noggin (Peprotech), 50 ng/mL EGF (Peprotech), 100 ng/mL IGF-I (Peprotech), 50 ng/mL FGF-2 (Peprotech), 10% R-spondin1 conditioned medium, 0.5 μM A83-01 (ApexBio) and 50% Wnt3A conditioned medium were added to the basal medium as described10. TpC-hSIO culture medium was made by supplementing the basal medium with 50 ng/mL EGF(Peprotech), 100 ng/mL human IGF-I (Peprotech), 50 ng/mL FGF-2 (Peprotech), 2 μM DMH1 (ApexBio), 10% R-spondin1 conditioned medium, 10 nM Gastrin I (MCE), 0.5 μM A83-01 (ApexBio), 4 μM CHIR (ApexBio), 20 nM TSA (ApexBio), 100 μg/mL pVc (Sigma) and 0.5 μM CP673451 (TargetMol).
Direct differentiation in organoids
Organoids were induced to differentiate by directly changing the culture media to differentiation media. The differentiation protocols are as follows:
Paneth cells
EGF (50 ng/mL), IGF-I (100 ng/mL), FGF-2 (50 ng/mL), DMH1 (2 μM), R-spondin1 CM (10%), Gastrin I (10 nM), A83-01 (0.5 μM), CHIR (4 μM), DAPT (10 μM), pVc (100 μg/mL) and IL-22 (2 ng/mL). Culture for 3 days.
Goblet cells
SJ000291942 (10 μM), R-spondin1 CM (10%), Gastrin I (10 nM), A83-01 (0.5 μM), CHIR (4 μM) and DAPT (10 μM) for 2 days and then replace CHIR with IWR-1 (2 μM) for 3 days.
Enteroendocrine cells
DMH1 (2 μM), R-spondin1 CM (10%), Gastrin I (10 nM), A83-01 (0.5 μM), CHIR (4 μM), DAPT (10 μM) and CP673451 (0.5 μM) for 2 days, and then DMH1 (2 μM), Gastrin I (10 nM), A83-01 (0.5 μM), DAPT (10 μM), CP673451 (0.5 μM) and Gefitinib (5 μM) for 3 days.
Enterocytes
EGF (50 ng/mL), IGF-I (100 ng/mL), FGF-2 (50 ng/mL), SJ000291942 (10 μM), Gastrin I (10 nM), A83-01 (0.5 μM), IWR-1 (2 μM) and VPA (0.75 mM) for 3 days.
M cells
TNF-α (50 ng/mL) treated for the first 4 hours and RANKL (100 ng/mL) for total 3 days.
Proteins and small molecules used in this study are listed in Supplementary Table 2.
EdU cell proliferation assay
Organoid culture medium was supplemented with 10 µM EdU (5-ethynyl-2’-deoxyuridine, Beyotime) and incubated for 1 hour. Organoids were then collected, fixed, permeabilized, and EdU detection was performed according to the manufacturer’s instructions (Beyotime).
Flow cytometry
Organoids were randomly taken from whole wells without being segmented and dissociated from the BME with TrypLE and dissociated into single cells. Cells were washed with ice-cold DPBS and filtered into fluorescence-activated cell sorting (FACS) tubes through a 40 μm cell strainer (Falcon). Cells were stained with Hoechst (Beyotime) to identify live cells before FACS analysis. FACS was performed on a BD FACSVerse (BD Biosciences), and data were analyzed in FlowJo (V10). mNeonGreen fluorescence was quantified at the single-cell level across the entire organoid. The gating strategy for mNeonGreen fluorescent reporter was consistent throughout this study (Supplementary Fig. 20).
Immunostaining
Immunofluorescence was performed as previously described22. In brief, organoids were released from BME with DPBS, fixed with 4% paraformaldehyde (Sangon Biotech) for 30 minutes at room temperature, and permeabilized in DPBS containing 0.5% Triton X-100 (Sigma), 0.5% Tween 20 (Sigma), and 2% bovine serum albumin (BSA, Sigma). Samples were blocked and incubated in blocking buffer (0.1% BSA, 0.5% Tween 20 in DPBS) containing primary antibodies at 4 °C overnight. Primary antibodies used in this study were as follows: rabbit anti-Lysozyme (1:500, Invitrogen), mouse anti-Mucin 2 (1:100, Santa Cruz), mouse anti-Chr-A (1:100, Santa Cruz), mouse anti-α-defensin 5 (1:100, Santa Cruz), rabbit anti-OLFM4 (1:200, Cell Signaling Technology), rat anti-Somatostatin (1:200, Millipore), mouse anti-Glucagon (1:200, Santa Cruz), mouse anti-ALPI (1:50, Santa Cruz), mouse anti-GP2 (1:500, MBL), rabbit anti-E-cadherin (1:500, Proteintech). Samples were washed and incubated with secondary antibodies (1:1000, Invitrogen) for 1 hour at room temperature, counterstained with DAPI (1 µg/mL, ApexBio). Alkaline phosphatase activity (ALPI) was detected using a commercial kit (Beyotime) according to the manufacturer’s instructions. The Olympus IX73 fluorescence microscope with cellSens (2.3) was used to take phase-contrast or fluorescence images. Confocal images were taken by Zeiss LSM880 microscope with Zeiss Zen 3, all the images were analyzed in ImageJ. The antibodies used are listed in Supplementary Table 3.
RNA extraction and reverse transcription (RT)-quantitative PCR (RT-qPCR)
Organoids were collected and washed with ice-cold DPBS before lysis in Trizol reagent (TransGen Biotech). Total RNA was isolated according to manufacturer’s instructions. Up to 1 µg of total RNA was used for cDNA synthesis using the NovoScriptPlus Reverse Transcriptase kit (NovoProtein). qPCR was performed using the ChamQ Universal SYBR qPCR Master Mix (Vazyme) on a CFX384 Real-Time System (BioRad). Relative gene expression levels were calculated using the ΔΔCt method. A list of the primer sequences for RT-qPCR is provided in Supplementary Table 4.
Colony formation assay
To assess colony-forming efficiency, organoids were dissociated into single cells and replated at a density of 8000 cells/well BME in 48-well plates. After 7 days, colonies were fixed, stained with DAPI, and counted manually. Colony-forming efficiency was then calculated. Colony-forming efficiency = number of colonies/number of plated single cells (8000) X 100%.
Statistics and reproducibility
All statistical analyses were performed using GraphPad Prism (version 10.1.2, GraphPad software). Data are presented as means ± standard deviation (SD). Two-tailed unpaired t-test, one-way ANOVA with Dunnett’s multiple comparisons test, one-way ANOVA with Tukey’s multiple comparisons test, two-way ANOVA analysis with Sidak’s multiple comparisons test and two-way ANOVA with Dunnett’s multiple comparisons test were used to determine the statistical significance. The significance levels are ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05. For all analyses, P < 0.05 was considered statistically significant. The specific statistical test used and the number of experiments performed for each analysis are indicated in the corresponding figure legend. Normal distribution was assumed for all analyses, and no data points were excluded. All experiments were performed three or more times independently under identical or similar conditions.
Single-cell RNA-sequencing data processing
Cell and library preparation
Organoids cultured from single cells in TpC-hSIO medium for 28 days were dissociated into single-cell suspensions for single-cell RNA sequencing. Organoids were incubated in TrypLE at 37°C for 10 minutes. The resulting single-cell suspension was passed through a 40 μm cell strainer (Falcon) to remove any remaining cell aggregates. A cell counter (Countstar) was used for quality control. Single-cell RNA sequencing libraries were prepared from the 10,000 live cells using the 10x Genomics Chromium 3’ Gene Expression solution v3.1 following the manufacturer’s protocol and sequenced on a NovaSeq6000 (Illumina). For organoids with TSA withdrawal and iBET treatment, single-cell RNA sequencing libraries were prepared using the MobiNova platform and sequenced on a NovaSeq6000 (Illumina).
Clustering and annotation
Sequences from the hSIO sample were processed using Cellranger (v7.1.0, 10x Genomics). The generated filtered cell UMI count matrix was imported into Seurat (v4.3.0)46 for analysis in R. Doublets were removed using scDblFinder (v1.13.8)47 with default parameters. Data were normalized, scaled, and variance stabilized using SCTransform (V2) in Seurat. The first 30 principal components were used for dimensional reduction, shared Nearest Neighbor (SNN) graph construction, and clustering. Clusters with high mitochondrial content and low UMI/gene counts were excluded, retaining 7595 cells for downstream analysis. Following UMAP visualization, cells were annotated based on classical intestinal cell type markers, including stem cells (LGR5, OLFM4, ASCL2), TA1 (PCNA, HELLS, MCM6), TA2 (MKI67, TOP2A), early enterocytes (FABP1, KRT20), mature enterocytes (ALPI, ACE2), secretory precursors (HES6, DLL1, DLL4), goblet cells (MUC2, SPINK4, FCGBP), Paneth cells (DEFA5, DEFA6, PRSS2), enteroendocrine cells (CHGA, NEUROD1), and tuft cells (ALOX5, AVIL). To analyze secretory cell types in detail, secretory cells and connected clusters representing stem cells, early enterocytes, and mature enterocytes were selected to create a subset dataset and visualized using UMAP. Unsupervised graph-based clustering failed to separate Paneth cells, goblet cells, tuft cells, and secretory precursor cells, which were manually assigned based on their expression levels of cell type markers and further annotated accordingly in the full dataset. Differentially expressed genes between cell types were identified using FindMarkers in Seurat (p < 0.05, fold change >3).
Trajectory analysis
The annotated full dataset and secretory subset were further analyzed in Python using Scanpy (v1.9.3)48. Trajectory analysis was performed using Partition-based Graph Abstraction (PAGA) in Scanpy (tl.paga). RNA velocity analysis on the full dataset and secretory subset was performed using Dynamo (v1.2.0)49 following the published tutorial (https://dynamo-release.readthedocs.io/). Briefly, the top 3000 highly variable genes used in Seurat UMAP visualization and the top 2000 genes from Dynamo (recipe_monocle) were combined as top genes for velocity dynamics calculation. UMAP coordinates from Seurat were retained for visualization and streamline plotting in Dynamo. Cell trajectory analysis was performed using CytoTRACE250 following the published tutorial (https://github.com/digitalcytometry/cytotrace2). Pseudotime analysis was carried out using StaVia (v0.1.82)51 following the published tutorial (https://pyvia.readthedocs.io/en/latest/). Briefly, processed and annotated datasets were analyzed using pyVIA with default parameters. The root cell (root_user) was set as the cell with the maximum CytoTRACE score, representing the highest developmental potential predicted by CytoTRACE2. Pseudotime values were extracted following the trajectory construction.
Metacell analysis was performed using the SEACells package52 following the published tutorial (https://github.com/dpeerlab/SEACells). Briefly, the number of SEACells was set to 200 for each dataset. Soft assignments were used to assign cells to SEACells. Model weights were further used to calculate the weight-averaged mean for CytoTRACE2 scores and pseudotime values, which were added to the metacell object. This object was further processed in Scanpy following standard procedures. The OmicVerse package with CellFateGenie was then used to construct a heatmap for gene expression along pseudotime (https://omicverse.readthedocs.io/en/latest/)53.
Data integration and comparison
Publicly available scRNA-seq datasets for fresh human crypts (Crypt, GEO: GSE119969), and organoids cultured under the ES, IF (GEO: GSE119969), and IL22 (GEO: GSE189423) conditions were processed following the same procedure as the TpC dataset. Briefly, count matrices were loaded into Seurat, doublets removed with scDblFinder, and stressed/non-epithelial cells filtered according to the original filtering criteria as described in the original publications. The same annotation criteria as the TpC dataset were applied. TA2 cells in the IL22 dataset were renamed TA1 and TA2 based on their expression of TA markers as described above. TA1 cells were renamed early enterocytes for their expression of early EC markers but not TA markers. The ISC population was divided into LGR5-high and LGR5-low subpopulations. Note that for the IL22 dataset, the LGR5-high population was not identified, and all ISC cells were labeled as LGR5-low. Data were integrated using Seurat (IntegrateData, https://satijalab.org/seurat/). The TpC, IF, ES, IL22- IL22+, and Crypt datasets were normalized with SCTransform (v2) and then integrated using Pearson residuals (IntegrateData).
To integrate and compare organoid datasets with in vivo datasets, the following publicly available scRNA-seq datasets for human intestinal epithelial cells were used as reference: GSE125970, GSE185224, E-MTAB-9543. The original cell type annotations were preserved. Raw counts for all datasets were normalized, and PCA was calculated. Harmony54 was then employed to integrate all organoid datasets with in vivo datasets, resulting in a unified dataset. A split view of each dataset was plotted to compare cell cluster distributions within the integrated dataset.
For cell type harmonization, CellHint (v1.0.0)55 was utilized according to the published tutorials (https://cellhint.readthedocs.io/en/latest/). A tree plot was generated to compare cell type assignments, and a heatmap was produced to illustrate how cell types were cross-assigned across different studies. Notably, previously identified M cells in the IF and Crypt dataset, as well as Paneth cells in the Wang et al., 2019 dataset, clustered with BEST4 cells, but the original cell annotations were retained for consistency.
To predict cell types responsive to perturbation such as TSA withdrawal and iBET addition, the Augur and perpy packages56 (https://pertpy.readthedocs.io/en/latest/) were employed with default parameters.
Pathway activity analysis with PROGENy
The PROGENy R package (v1.17.3)57 was used to infer signaling pathway activity on a single-cell basis for the TpC and integrated datasets. For the TpC dataset, the top 200 most highly correlated genes for each pathway were used to generate the model matrix. For the integrated dataset, the top 1000 genes were used. A model matrix containing pathway activity scores was appended as a Seurat object assay, following the PROGENy scRNA-seq tutorial (https://saezlab.github.io/progeny/articles/ProgenySingleCell.html).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank all lab members for the helpful discussion of the paper, Chunying Li, Yongfei Hou, and Tianzeng Sun for R-Spondin1 conditioned medium production, Dr. Hui Yang for providing CRISPR plasmids, Dr. Rui Yue and Dr. Shan Bian for suggestions for the paper. This work was supported by the National Key R&D Program of China (2020YFA0112500, 2019YFA0110003) to X.Y., the National Natural Science Foundation of China (31970820) to X.Y., the Key Project of the Science and Technology of Shanghai Municipality (19JC1415300) to X.Y., and the Fundamental Research Funds for the Central Universities (22120230292) to X.Y.
Author contributions
X.Y. and L.Y. conceived the study, designed the experiments, and interpreted the results. L.Y. performed most experiments. X.W. and X.Z. performed preliminary screening. X.W. performed RT-qPCR and gene expression analysis. H.C. performed molecular cloning. X.Y. performed bioinformatics analysis. S.S. provided human samples. L.D. and Y.Y. assisted with sample preparation. X.Y. and L.Y. wrote the manuscript. All authors reviewed and approved the manuscript.
Peer review
Peer review information
Nature Communications thanks Prisca Liberali and Nobuo Sasaki for their contribution to the peer review of this work. A peer review file is available.
Data availability
The raw sequence data for scRNA-seq reported in this paper have been deposited in the Genome Sequence Archive58 in National Genomics Data Center59 (NGDC), China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences, under accession numbers: HRA004691 and HRA009421. The raw fastq data are available under restricted access for academic use only, in accordance with regulations on human genetic resources management by the Minister of Science and Technology. Access can be obtained by registering with the GSA database website (https://ngdc.cncb.ac.cn/gsa-human/) and submitting an application to the Data Access Committee (DAC) via the GSA-Human System. The typical response time for access requests is approximately 2 weeks. Additional scRNA-seq datasets for ES, IF, Crypt10, and IL2211 condition cultured human organoids, as well as in vivo intestinal epithelial cells, are available through published accessions GSE189423, GSE119969, GSE125970, GSE185224, and E-MTAB-9543. Source data are provided with this paper.
Competing interests
X.Y., L.Y., X.W., and X.Z. have submitted a patent application related to the culture of human intestinal organoids reported in this paper. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Li Yang, Xulei Wang.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55567-2.
References
- 1.Clevers, H. Modeling Development And Disease With Organoids. Cell165, 1586–1597 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Zhao, Z. et al. Organoids. Nat. Rev. Methods Prim.2, 94 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang, D. et al. Long-Term Expansion Of Pancreatic Islet Organoids From Resident Procr(+) progenitors. Cell180, 1198–1211.e1119 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Hu, H. et al. Long-Term Expansion Of Functional Mouse And Human Hepatocytes as 3D Organoids. Cell175, 1591–1606.e1519 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Huch, M. et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell160, 299–312 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huch, M. et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J.32, 2708–2721 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLean, W. J. et al. Clonal expansion of Lgr5-Positive Cells From Mammalian Cochlea aNd High-purity Generation Of Sensory Hair Cells. Cell Rep.18, 1917–1929 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim, K. et al. Organoid modeling of human fetal lung alveolar development reveals mechanisms of cell fate patterning and neonatal respiratory disease. Cell Stem Cell30, 20–37.e29 (2023). [DOI] [PubMed] [Google Scholar]
- 9.Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature459, 262–265 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Fujii, M. et al. Human Intestinal Organoids Maintain Self-renewal Capacity And Cellular Diversity In Niche-inspired Culture Condition. Cell Stem Cell23, 787–793.e786 (2018). [DOI] [PubMed] [Google Scholar]
- 11.He, G. W. et al. Optimized human intestinal organoid model reveals interleukin-22-dependency of paneth cell formation. Cell Stem Cell29, 1333–1345.e1336 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beumer, J. et al. BMP gradient along the intestinal villus axis controls zonated enterocyte and goblet cell states. Cell Rep.38, 110438 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Ootani, A. et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med.15, 701–706 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato, T. et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature469, 415–418 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jadhav, U. et al. Dynamic reorganization of chromatin accessibility signatures during dedifferentiation of secretory precursors into Lgr5+ intestinal stem cells. Cell Stem Cell21, 65–77.e65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tetteh, P. W. et al. Replacement of lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell18, 203–213 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Yan, K. S. et al. Intestinal Enteroendocrine lineage cells possess homeostatic and injury-inducible stem cell activity. Cell Stem Cell21, 78–90.e76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Es, J. H. et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat. Cell Biol.14, 1099–1104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin, X. et al. Engineering stem cell organoids. Cell Stem Cell18, 25–38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofer, M. & Lutolf, M. P. Engineering organoids. Nat. Rev. Mater.6, 402–420 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato, T. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology141, 1762–1772 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Yin, X. et al. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat. Methods11, 106–112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuijers, J., van der Flier, L. G., van Es, J. & Clevers, H. Robust cre-mediated recombination in small intestinal stem cells utilizing the olfm4 locus. Stem Cell Rep.3, 234–241 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buczacki, S. J. et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature495, 65–69 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Malagola, E. et al. Isthmus progenitor cells contribute to homeostatic cellular turnover and support regeneration following intestinal injury. Cell187, 3056–3071.e3017 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gehart, H. et al. Identification of Enteroendocrine regulators by real-time single-cell differentiation mapping. Cell176, 1158–1173.e1116 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Beumer, J. et al. High-resolution mRNA and Secretome Atlas of human enteroendocrine cells. Cell181, 1291–1306.e1219 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Wierup, N. et al. Ghrelin and motilin are cosecreted from a prominent endocrine cell population in the small intestine. J. Clin. Endocrinol. Metab.92, 3573–3581 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Triana, S. et al. Single-cell transcriptomics reveals immune response of intestinal cell types to viral infection. Mol. Syst. Biol.17, e9833 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qu, M. et al. Establishment of intestinal organoid cultures modeling injury-associated epithelial regeneration. Cell Res.31, 259–271 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aguilera, C. et al. c-Jun N-terminal phosphorylation antagonises recruitment of the Mbd3/NuRD repressor complex. Nature469, 231–235 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Zeve, D. et al. Robust differentiation of human enteroendocrine cells from intestinal stem cells. Nat. Commun.13, 261 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.VanDussen, K. L. et al. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut64, 911–920 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, Y. et al. Formation of human colonic crypt array by application of chemical gradients across a shaped epithelial monolayer. Cell Mol. Gastroenterol. Hepatol.5, 113–130 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung, P. et al. Isolation and in vitro expansion of human colonic stem cells. Nat. Med.17, 1225–1227 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Basak, O. et al. Induced quiescence of Lgr5+ stem cells in intestinal organoids enables differentiation of hormone-producing enteroendocrine cells. Cell Stem Cell20, 177–190.e174 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Yang, L., Wang, X., Zhao, G., Deng, L. & Yin, X. Leveraging temporal Wnt signal for efficient differentiation of intestinal stem cells in an organoid model. Stem Cells Dev.33, 11–26 (2024). [DOI] [PubMed] [Google Scholar]
- 38.Wood, M. B., Rios, D. & Williams, I. R. TNF-α augments RANKL-dependent intestinal M cell differentiation in enteroid cultures. Am. J. Physiol.-Cell Ph311, C498–C507 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knoop, K. A. et al. RANKL is necessary and sufficient to initiate development of antigen-sampling M cells in the intestinal epithelium. J. Immunol.183, 5738–5747 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malijauskaite, S., Connolly, S., Newport, D. & McGourty, K. Gradients in the in vivo intestinal stem cell compartment and their in vitro recapitulation in mimetic platforms. Cytokine Growth Factor Rev.60, 76–88 (2021). [DOI] [PubMed] [Google Scholar]
- 41.McCarthy, N. et al. Distinct mesenchymal cell populations generate the essential intestinal BMP signaling gradient. Cell Stem Cell26, 391–402.e395 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heuberger, J. et al. Shp2/MAPK signaling controls goblet/paneth cell fate decisions in the intestine. Proc. Natl Acad. Sci. USA111, 3472–3477 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Lau, W. et al. Peyer’s patch M cells derived from Lgr5(+) stem cells require SpiB and are induced by RankL in cultured “miniguts. Mol. Cell Biol.32, 3639–3647 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimokawa, M. et al. Visualization and targeting of LGR5(+) human colon cancer stem cells. Nature545, 187–192 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Fujii, M., Matano, M., Nanki, K. & Sato, T. Efficient genetic engineering of human intestinal organoids using electroporation. Nat. Protoc.10, 1474–1485 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Stuart, T. et al. Comprehensive integration of single-cell data. Cell177, 1888–1902.e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Germain, P. L., Lun, A., Garcia Meixide, C., Macnair, W. & Robinson, M. D. Doublet identification in single-cell sequencing data using scDblFinder. F1000Res.10, 979 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol.19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu, X. et al. Mapping transcriptomic vector fields of single cells. Cell185, 690–711.e645 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang, M. et al. Mapping single-cell developmental potential in health and disease with interpretable deep learning. BioRxiv10.1101/2024.03.19.585637 (2024).
- 51.Stassen, S. V., Yip, G. G. K., Wong, K. K. Y., Ho, J. W. K. & Tsia, K. K. Generalized and scalable trajectory inference in single-cell omics data with VIA. Nat. Commun.12, 5528 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Persad, S. et al. SEACells infers transcriptional and epigenomic cellular states from single-cell genomics data. Nat. Biotechnol.41, 1746 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeng, Z. et al. OmicVerse: a framework for bridging and deepening insights across bulk and single-cell sequencing. Nat. Commun.15, 5983 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu, C. et al. Automatic cell-type harmonization and integration across Human Cell Atlas datasets. Cell186, 5876–5891.e5820 (2023). [DOI] [PubMed] [Google Scholar]
- 56.Skinnider, M. A. et al. Cell type prioritization in single-cell data. Nat. Biotechnol.39, 30–34 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schubert, M. et al. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun.9, 20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen, T. et al. The genome sequence archive family: toward explosive data growth and diverse data types. Genomics Proteom. Bioinforma.19, 578–583 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Members, C.-N., Partners. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res.50, D27–D38 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data for scRNA-seq reported in this paper have been deposited in the Genome Sequence Archive58 in National Genomics Data Center59 (NGDC), China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences, under accession numbers: HRA004691 and HRA009421. The raw fastq data are available under restricted access for academic use only, in accordance with regulations on human genetic resources management by the Minister of Science and Technology. Access can be obtained by registering with the GSA database website (https://ngdc.cncb.ac.cn/gsa-human/) and submitting an application to the Data Access Committee (DAC) via the GSA-Human System. The typical response time for access requests is approximately 2 weeks. Additional scRNA-seq datasets for ES, IF, Crypt10, and IL2211 condition cultured human organoids, as well as in vivo intestinal epithelial cells, are available through published accessions GSE189423, GSE119969, GSE125970, GSE185224, and E-MTAB-9543. Source data are provided with this paper.