Abstract
Chromosome instability is a prevalent vulnerability of cancer cells that has yet to be fully exploited therapeutically. To identify genes uniquely essential to chromosomally unstable cells, we mined the Cancer Dependency Map for genes essential in tumor cells with high levels of copy number aberrations. We identify and validate KIF18A, a mitotic kinesin, as a vulnerability of chromosomally unstable cancer cells. Knockdown of KIF18A leads to mitotic defects and reduction of tumor growth. Screening of a chemical library for inhibitors of KIF18A enzymatic activity identified a hit that was optimized to yield VLS-1272, which is orally bioavailable, potent, ATP non-competitive, microtubule-dependent, and highly selective for KIF18A versus other kinesins. Inhibition of KIF18A’s ATPase activity prevents KIF18A translocation across the mitotic spindle, resulting in chromosome congression defects, mitotic cell accumulation, and cell death. Profiling VLS-1272 across >100 cancer cell lines demonstrates that the specificity towards cancer cells with chromosome instability differentiates KIF18A inhibition from other clinically tested anti-mitotic drugs. Treatment of tumor xenografts with VLS-1272 results in mitotic defects leading to substantial, dose-dependent inhibition of tumor growth. The strong biological rationale, robust preclinical data, and optimized compound properties enable the clinical development of a KIF18A inhibitor in cancers with high chromosomal instability.
Subject terms: Drug development, Cancer, Mechanisms of disease
Chromosomal instability occurs frequently in cancer, making it an attractive therapeutic target. Here, the authors identify KIF18A as a targetable vulnerability of cancer cells with chromosomal instability and target this using VLS-1272, a selective KIF18A inhibitor.
Introduction
Precise segregation of genetic material during cell division is crucial for organismal survival. However, instances arise where errors in chromosome segregation result in unequal distribution of genetic contents. These errors can result in parts of, or entire, chromosomes not properly segregated. Chromosomal instability (CIN) is a state in which these mis-segregation errors occur frequently over time. CIN is prevalent in a large percentage of human cancers and occurs across a wide range of human solid and liquid tumors1. Poor prognosis is associated with CIN, and high levels of CIN correlate with reduced survival in diffuse large B-cell lymphoma2, lung cancer3, breast cancer4 and colorectal cancer5. Beyond overall survival, CIN provides an avenue to permit tumor heterogeneity and evolution through acquisition of advantageous karyotypes6. This increased heterogeneity caused by CIN can result in resistance to standard of care therapies such as kinase inhibitors7,8. Finally, CIN can also drive metastatic disease9. Thus, therapies that target tumors with CIN could have widespread clinical benefit through extending patient survival and combating drug resistance.
As CIN is a result of ongoing mitotic errors, targeting mitotic machinery may be a strategy to combat CIN. Increased understanding of cell cycle regulators led to the development of targeted agents of the mitotic machinery such as inhibitors of Aurora Kinase10,11, PLK112 and TTK/Mps113. However, these drugs had limited clinical success due to dose-limiting toxicities, often in proliferative compartments such as the bone marrow and gut14. Thus, the need exists for therapies that specifically target mitotic cancer cells while sparing normal proliferating cells. Herein, we describe a therapeutic strategy targeting a component of the mitotic machinery, KIF18A, which preferentially impacts CIN tumors while allowing normal cells to proliferate.
KIF18A is a microtubule plus-end directed kinesin whose ATPase function powers its depolymerase activity15. During mitosis, KIF18A ensures that kinetochore-microtubule interactions are under proper tension, allowing chromosome congression and progression to metaphase16. In the absence of KIF18A, cells exhibit increased chromosome oscillations and reduced chromosome congression, resulting in cells with a wider metaphase plate17. However, normal, diploid cells tolerate loss of KIF18A and progress through mitosis18–20. The permissibility of KIF18A loss in normal cells is illustrated by the phenotype of mice with KIF18A loss of function, as they are viable and survive to adulthood21,22.
Although KIF18A is dispensable in normal cells, studies have demonstrated a requirement for KIF18A in certain cancer contexts. KIF18A loss in cancer cells that exhibit aneuploidy, whole genome doubling (WGD) or CIN results in mitotic defects and eventual cell death18–20. Cancer cells that are aneuploid and/or chromosomally unstable tend to have abnormal mitotic phenotypes19,20 such as longer spindles, altered chromosome oscillations and microtubule dynamics, increased mitotic duration and increased errors, and thus are more dependent on KIF18A to enable a successful mitosis.
In this work, we describe the generation and characterization of a potent and selective KIF18A inhibitor, VLS-1272. Pharmacological inhibition of KIF18A with VLS-1272 results in reduced proliferation of chromosomally unstable cancer cells due to defective chromosome congression and mitotic arrest, phenotypes similar to those observed with KIF18A knockdown. In contrast, normal cells and cancer cells with low levels of CIN can proliferate in the presence of VLS-1272. VLS-1272 treatment results in robust inhibition of tumor growth with evidence of KIF18A relocalization and mitotic defects in the tumor. Taken together, the development of KIF18A inhibitors is a promising therapeutic strategy with potential for a favorable therapeutic index.
Results
Identification of KIF18A as a vulnerability of CIN-high tumors
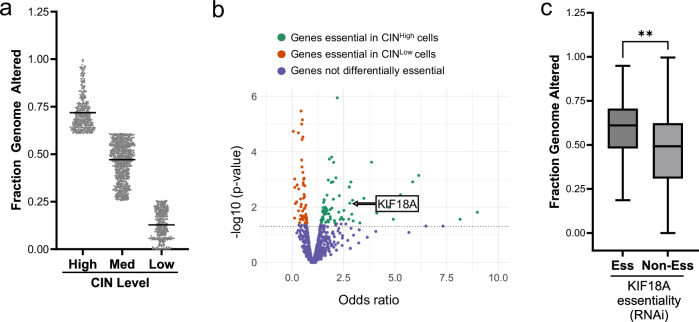
To identify unique dependencies of chromosomally unstable cells, we utilized the Cancer Dependency Map (DEPMAP)23–28, a database with gene essentiality assessments for hundreds of human cancer cell lines. CIN was calculated for each cell line using fraction genome altered (FGA). FGA measures the fraction of the genome with copy number alterations (CNA) using whole exome sequencing and has been correlated with experimental measures of CIN29. Cell lines were stratified to high, medium, and low levels of chromosome instability (CINHigh, CINMed, CINLow) based on even distribution of FGA scores ranging from 0-1 (Fig. 1a).
Fig. 1. Identification of KIF18A as a vulnerability of CINHigh tumors.
DEPMAP23 gene essentiality data was used to identify genes required for proliferation of CINHigh cells. a Categorization of CIN across 1660 cell lines as CINHigh (top quartile), CINMed or CINLow (bottom quartile) as measured by fraction genome altered (FGA). Data presented as mean values. Each dot represents 1 cell line. b Volcano plot visualizing the odds ratio from a Fisher’s Exact test to determine the association between RNAi essentiality scores and CIN, which identified genes that are significantly essential in CINHigh cell lines (FDR < 0.25). Each dot represents one gene. Genes in green are more essential in CINHigh cells, genes in red are more essential in CINLow cells, and genes in blue did not show differential essentiality. KIF18A p value = 0.0072 with a Fisher’s Exact test. c Cell lines requiring KIF18A for proliferation (Ess) exhibit higher median FGA compared to cell lines not requiring KIF18A for proliferation (Non-Ess) based on RNAi essentiality scores across 748 cell lines. Data presented as median (center) with 25th to 75th percentile bounds (boxes), minimum and maximum (whiskers). **p = 0.006 by unpaired, 2-tailed t test. Source data are provided in the Source Data file.
Using the DEPMAP gene essentiality data23, we identified genes differentially required in the CINHigh vs. CINLow groups across all tumor types (Fig. 1b). KIF18A was among the top-scoring genes more essential in CINHigh cells compared to CINLow cells. We further observed that essentiality for KIF18A correlated with higher overall FGA across all cell lines listed in the DEPMAP database (Fig. 1c). These initial computational analyses established KIF18A as a potential therapeutic target for cancers with CIN. Our internal findings were further corroborated by publications identifying KIF18A essentiality in models of aneuploidy, whole genome doubling, and CIN breast and colon cancer cells18–20. Based on these internal analyses, the independent identification of KIF18A by other groups, and the potential for therapeutic targeting of KIF18A by pharmacological inhibition, we focused on KIF18A in follow-up studies.
Genetic validation of KIF18A dependence in vitro and in vivo
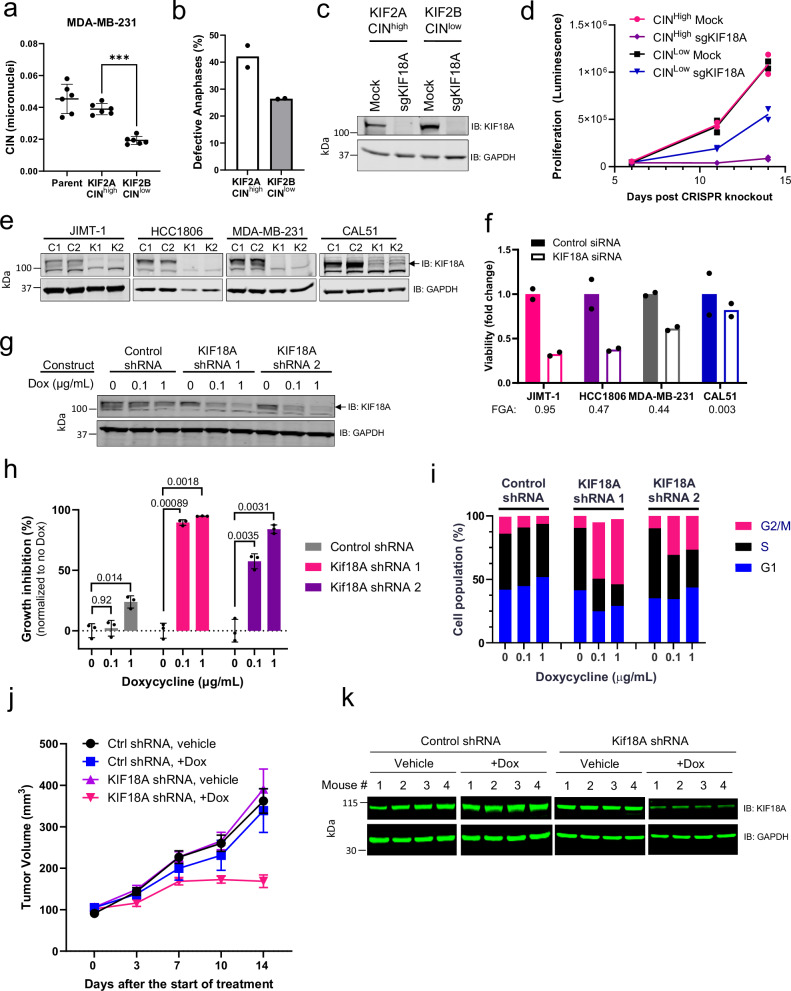
To extend the findings from our computational analyses, we re-created a model system where CIN is suppressed through alteration of kinetochore-microtubule dynamics30–32. In this system, expression of KIF2B reduces CIN whereas expression of the related KIF2A does not alter CIN. We confirmed that overexpression of KIF2A maintains the CIN levels of the parental MDA-MB-231, and overexpression of KIF2B reduces overall CIN levels (Fig. 2a, b). We used this paired cell line model of CIN to assess KIF18A dependency. Following the loss of KIF18A by CRISPR-Cas9 (Fig. 2c), proliferation of the CINHigh (KIF2A-overexpressing) cells was reduced to a greater extent compared to the CINLow (KIF2B-overexpressing) cells (Fig. 2d), confirming the DEPMAP analysis that CINHigh cells have increased vulnerability to KIF18A loss.
Fig. 2. Differential requirement of CINHigh vs. CINLow cells for KIF18A.
a Micronuclei levels (ratio of total micronuclei to total primary nuclei) in MDA-MB-231 cells over-expressing KIF2B or KIF2A compared to the parental line. Data presented as mean values -/+ SD from n = 6 biological replicates, ***p = 1.01 x 10−4 using one-way ANOVA with Dunnett’s multiple comparisons test. b Defective anaphases (as percentage of total anaphases) in MDA-MB-231 cells overexpressing KIF2A or KIF2B. Data presented as mean values from n = 2 biological replicates. c Western blot confirming CRISPR/Cas9-mediated KIF18A knockout. One representative experiment of 2 biological replicates is shown. d Proliferation (luminescence measured by CellTiter-Glo assay) upon KIF18A knockout. Data presented as individual values from n = 2 biological replicates. (e) Western blot of KIF18A knockdown (K1 and K2) compared to control siRNA (C1 and C2) 72 h after replating. Top panel: arrow indicates KIF18A (the highest band), bottom band is non-specific. One representative experiment of 2 biological replicates is shown. f Viability of cells shown in (e) 72 h after replating following siRNA transfection. Computed CIN values (FGA) of cell lines are indicated below the name. Data are presented as mean values from 2 biological replicates. g Western blot of KIF18A expression in engineered Dox-inducible KIF18A (shRNA 1 and 2) and Control (non-targeting) shRNA in JIMT-1 cells after 7 days Dox treatment. One representative experiment of 2 biological replicates is shown. h Proliferation of engineered Dox-inducible JIMT-1 cells measured after 10 days Dox treatment. Data presented as mean values +/- SD from n = 3 biological replicates. p values are indicated above the bars and determined by 2-way ANOVA with Tukey’s multiple comparisons test. i Cell cycle population frequencies in Dox-inducible KIF18A and Control shRNA JIMT-1 cells after 72 h of Dox treatment. Data presented as mean values from n = 2 biological replicates. j Tumor volume of Dox-inducible shRNA JIMT-1 xenografts implanted in SCID Beige mice (n = 4 mice per group). Data presented as mean values -/+ SEM. k Western blot of KIF18A expression from JIMT-1 xenograft tumors excised 4 days after initiation of Dox treatments. Source data are provided in the Source Data file.
We extended these findings to a small panel of breast cancer cell lines with varied levels of CIN, determined by FGA for each cell line. In this cell line panel, reduction of KIF18A with siRNA (Fig. 2e) decreased proliferation to an extent correlating with the baseline level of chromosome instability (Fig. 2f). In the CINHigh breast cancer cell line JIMT-1, similar anti-proliferative results were obtained with Doxycycline (Dox)-inducible shRNA-mediated knockdown of KIF18A (Fig. 2g, h). Cell cycle analysis (Supplementary Fig. 1a–c) after KIF18A knockdown displayed an increase in the G2/M population (Fig. 2i, Supplementary Fig. 1d). These data indicate that cells respond to KIF18A knockdown by arresting in mitosis, consistent with published data18,33,34.
To assess the requirement of KIF18A on tumor growth, we implanted cells expressing inducible control and KIF18A shRNA into immune-compromised mice, followed by doxycycline (Dox) treatment to induce knockdown and monitored tumor growth. In mice harboring tumors expressing inducible KIF18A shRNA, Dox treatment resulted in delayed tumor growth compared to tumors with control shRNA or mice not treated with Dox (Fig. 2j). In tumor samples taken four days after the introduction of Dox, western blot analysis confirmed knockdown of KIF18A in the tumor samples (Fig. 2k). These data support the dependence of chromosomally unstable cells on KIF18A for tumor growth in vivo.
Development of KIF18A small molecule inhibitors
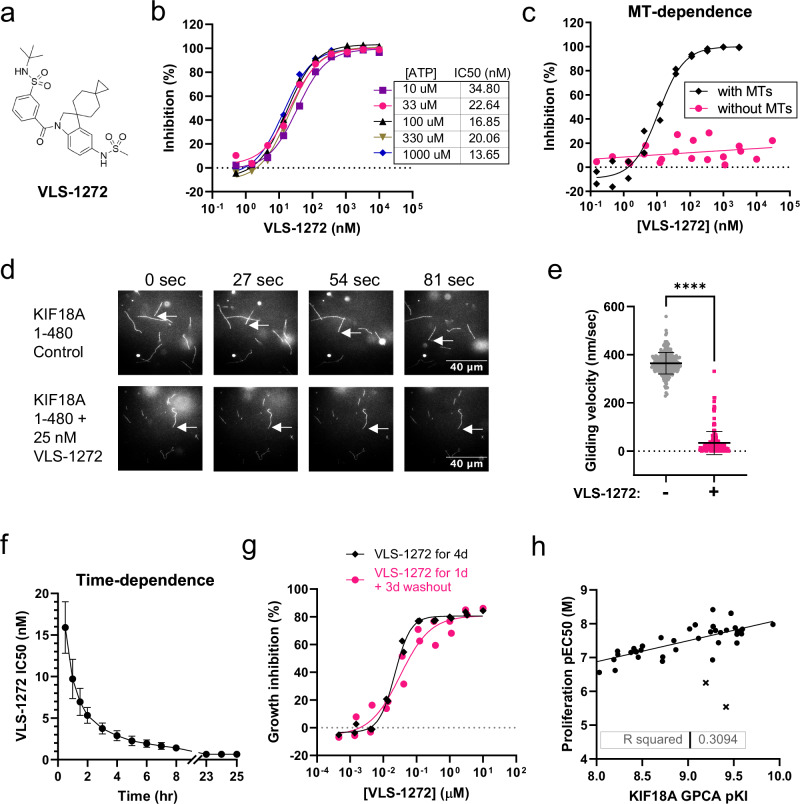
To further assess the potential of KIF18A as a therapeutic target, we developed and ran a high throughput screen to identify small molecule inhibitors of KIF18A ATPase activity. VLS-1272 (Fig. 3a) was derived from a high-throughput screening campaign hit from a library of 50,000 lead-like compounds for inhibition of microtubule-dependent ATPase activity of the motor domain of KIF18A (aa 1-374). Optimization of the hit for potency and pharmacokinetic (PK) properties resulted in VLS-1272, a compound with an IC50 in a 60 min KIF18A ATPase inhibition assay of 41 ×/ ÷ 1.1 nM when ATP is at its KM of 0.1 mM (Table 1).
Fig. 3. Development of KIF18A inhibitors.
a Structure of VLS-1272. b Potency of VLS-1272 in a KIF18A (1-374) biochemical ADP-Glo assay to measure ATPase activity with varied concentrations of ATP. Data presented as mean values from n = 2 biological replicates. c Inhibition of KIF18A (1-374) by VLS-1272 as measured by ADP-Glo assay in the absence or presence of 0.1 mg/ml microtubules. Data presented as individual values from n = 2 biological replicates. d Representative still images from microtubule gliding filament assays with purified KIF18A 1-480-GFP linked to the glass surface in absence (control) or presence of VLS-1272. Arrows indicate the ends of microtubules that move directionally (control) or remain stationary (VLS-1272). e Plot of microtubule velocities measured in gliding filament assays in the presence or absence of VLS-1272. n = 249 (control) and 177 (VLS-1272) microtubules from 3 independent experiments. Data were compared using an unpaired, two-tailed t-test. ****p < 1.0 x 10-15. Data points represent individual microtubule velocities, bars indicate mean and standard deviation. f Potency of VLS-1272 at various timepoints from a global progress curve analysis (GPCA). Data presented as mean values -/+ SD from n = 11 biological replicates. g Inhibition of HCC15 cell proliferation after 4-day treatment with VLS-1272 (black) versus 1 day of VLS-1272 treatment followed by a washout and replacement in media without inhibitor for an additional 3 days (pink). Data presented as individual values from n = 2 biological replicates. h Comparison of the KI calculated by GPCA vs. the EC50 from a 2.5-day CellTiter-Glo proliferation assay in JIMT-1 cells from n = 40 compounds. Each dot represents one compound. Compounds with poor permeability are denoted by “x”. Source data are provided in the Source Data file.
Table 1.
Selectivity summary for VLS-1272 against a panel of human and mouse kinesins
| Kinesin | ATP (mM) | IC50 (nM), 30–60 min | IC50 (nM), 24 h |
|---|---|---|---|
| Human KIF18A | 0.10 (1 x KM) | 41 ×/ ÷ 1.1 (n = 3) | |
| 1.0 (10 x KM) | 9.3 ×/ ÷ 1.4 (n = 11) | 0.58 ×/ ÷ 1.4 (n = 13) | |
| Murine KIF18A | 0.10 (1 x KM) | 8.8 ×/ ÷ 1.2 (n = 3) | |
| Human KIF19 | 0.030 (1.5 x KM) | 280 (n = 2) | 150 (n = 1) |
| Human KIF18B | 0.030 (1.5 x KM) | >100,000 (n = 1) | |
| Human KIF11/Eg5 | 0.030 (6 x KM) | >100,000 (n = 1) | |
| Human KIFC1 | 0.010 (1 x KM) | >100,000 (n = 1) |
Inhibitory activity of VLS-1272 against the proteins listed using ADP-Glo assay to measure ATPase activity.
To understand the inhibitory mechanism of VLS-1272, potency was measured with increasing ATP concentrations. VLS-1272 is non-competitive with ATP, with no change in IC50 values at ATP concentrations ranging from 10 µM to 1 mM, (Fig. 3b), demonstrating that VLS-1272 is an allosteric inhibitor of KIF18A. We compared the ATP dependence of VLS-1272 to previously reported small molecule inhibitors of KIF18A, BTB-135. and AM-188236. With increasing ATP concentrations, the IC50 of BTB-1 increases, reaching an IC50 of 6.7 ×/ ÷ 1.3 µM at 1 mM ATP compared to 2.2 ×/ ÷ 1.2 µM at 0.1 mM ATP, confirming the ATP-competitive nature of inhibition reported for BTB-1 (Supplementary Fig. 2a). Similar to VLS-1272 and consistent with published data36, AM-1882 demonstrated ATP non-competitiveness, and the potencies of ~20–50 nM across a range of ATP concentrations (Supplementary Fig. 2b) were similar to those observed with VLS-1272 (Fig. 3b).
As the ATPase activity of KIF18A drives translocation along microtubules, we assessed the requirement of microtubules for KIF18A inhibitory activity. VLS-1272 activity was dependent on the presence of microtubules (MTs), as potency significantly decreased when MTs were omitted from the ATPase assay (Fig. 3c), which suggests that VLS-1272 binds to the KIF18A-microtubule complex. To further probe the microtubule dependence of VLS-1272, a microtubule gliding assay was performed by immobilizing KIF18A (aa 1-480) to a surface, adding microtubules, and measuring the velocity of microtubule movement across the kinesin surface. In the absence of VLS-1272, microtubule gliding was evident in the presence of KIF18A (Supplementary Movie 1, Fig. 3d). However, with addition of VLS-1272, microtubule motility ceased (Fig. 3d, e; Supplementary Movie 2), and the average MT velocity decreased from 363.9 nm/sec to 33.73 nm/sec in the presence of VLS-1272 (Fig. 3e). Thus, by inhibiting the ATPase activity of KIF18A, VLS-1272 prevents the translocation function of the kinesin, resulting in the immobilization of KIF18A onto microtubules.
We discovered that VLS-1272 exhibits slow binding kinetics and did not achieve steady-state during the 60-minute assay time. Analysis of binding kinetics using a global progress-curve analysis (GPCA)37 of both time and dose dependent inhibition allowed us to determine the kon of 0.037 ± 0.010 nM−1h−1, and a koff of ≤0.02 h−1, resulting in a KI of ≤0.5 nM. The slow koff is notable and translates into a dissociation half-life of about 35 h (Fig. 3f). The slow dissociation was further observed in a compound washout experiment, where HCC15 cells showed similar growth inhibition after 96 h, whether compound was present only for the initial 24 h of the experiment or for the entire duration (Fig. 3g). The long target residence time of VLS-1272 contributed to the cellular efficacy of the drug, as evidenced by the correlation between KI and cellular anti-proliferative EC50 (Fig. 3h), with a few outliers due to lack of cell permeability. Long target residence time has been associated with improved in vivo efficacy and durable pharmacological responses38,39. The binding kinetics leading to sustained target engagement may be a beneficial property in an anti-mitotic agent, effectively arresting sensitive cells in mitosis and increasing the probability that arrested cells will die rather than reverse in mitosis or slip through cell division.
GPCA analysis demonstrated that AM-1882 is also a slow binding inhibitor of KIF18A with potency improving with increasing incubation time. Direct comparison of AM-1882 and VLS-1272 reveal that both inhibitors exhibit very similar binding kinetics (Supplementary Fig. 2c). Given the similarities between VLS-1272 and AM-1882, both compounds were docked into a KIF18A homology model40 to further understand the mechanism of inhibition. The binding of VLS-1272 and AM-1882 were compared to the published binding mode of Compound 2640 (Supplementary Fig. 3a–d). The docking of VLS-1272 and AM-1882 shows binding to the same allosteric site as described for Compound 26, supporting the conclusion from the ATP, MT and time-dependence studies that VLS-1272 and AM-1882 have similar modes of inhibition.
VLS-1272 is selective for KIF18A over other kinesins, with no inhibition of KIF11/Eg5, KIF18B, or KIFC1 at 100 µM, and an IC50 of 280 nM against KIF19 (Table 1). The selectivity over KIF19 (7x) is similar to that observed with AM-1882 (8x), and the selectivity profile of VLS-1272 closely matches that of AM-188236 as summarized in Supplementary Table 1. The VLS-1272 binding kinetics for KIF19 inhibition is fast, and too rapid to quantify with the GPCA method, indicating the biochemical KIF19 inhibition assay is at equilibrium binding conditions. KIF19 is a kinesin that controls cilia length41, and is not required for proliferation of >1000 cancer cell lines tested according to DEPMAP23. Thus, given the short residence time of VLS-1272 for KIF19, the biological function of KIF19, and lack of dependence of cancer cell lines on KIF19, it is unlikely that inhibition of KIF19 would drive cellular phenotypes observed with VLS-1272. Given the favorable properties of VLS-1272, we used the inhibitor to probe the therapeutic potential of KIF18A inhibition in cancer cell line models.
Differentiation of KIF18A inhibitors from other anti-mitotic therapies
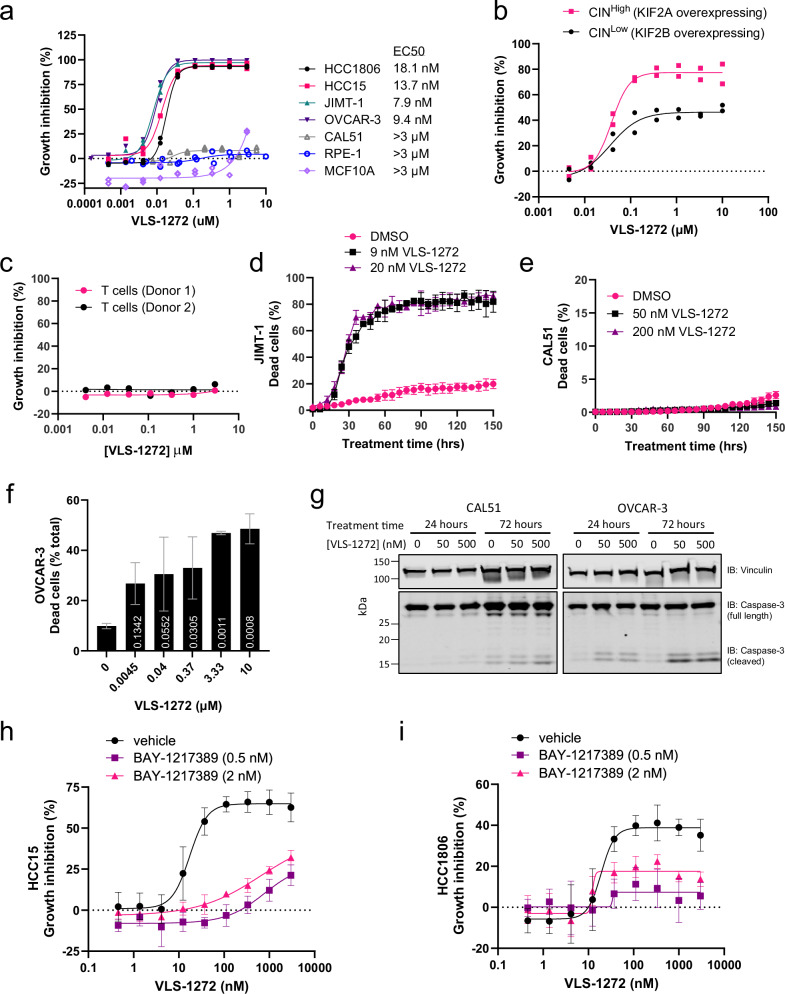
VLS-1272 was assessed for its anti-proliferative effect in a panel of CINHigh and CINLow cell lines (Fig. 4a). Inhibition of KIF18A was effective in preventing the growth of CINHigh tumor cells, with limited efficacy in the CINLow cell line CAL51 (Fig. 4a), phenocopying our previous findings in the genetic validation experiments (Fig. 2f). VLS-1272 was also tested in the MDA-MB-231 paired CIN cell line used in Fig. 2d. KIF18A inhibition with VLS-1272 prevented cell proliferation to a greater degree in the CINHigh (KIF2A over-expressing) vs. CINLow (KIF2B over-expressing) MDA-MB-231 cell line pair (Fig. 4b), consistent with our previous data with KIF18A knockout (Fig. 2d) and further supporting the greater dependence of CINHigh cells on KIF18A function. These results contrasted with those observed with BTB-1, where cell growth inhibition was only observed at the highest dose tested (50 µM), and without differential effects between CINHigh and CINLow cells (Supplementary Fig. 2d).
Fig. 4. Differential effects of KIF18A inhibition in cancer vs. normal cells.
a Growth inhibition as measured by CellTiter-Glo of indicated cancer and non-transformed epithelial cell lines after treatment with VLS-1272 for 168 h. Data presented as individual values from n = 2 biological replicates. b Proliferation of MDA-MB-231 cells over-expressing KIF2A (CINHigh) or KIF2B (CINLow) treated with VLS-1272 and assessed by CellTiter-Glo after 168 h of treatment. Data presented as individual values from n = 2 biological replicates. c Growth inhibition of primary T cells isolated from healthy donors after stimulation with αCD3/CD28 beads, treated with VLS-1272 and assessed by CellTiter-Glo after 72 h. Data presented as individual values from n = 2 donors. Dead cell (propidium iodide-positive cells) enumeration by live cell imaging in the presence of vehicle or indicated VLS-1272 concentration for 150 h in JIMT-1 cells (d) or CAL51 cells (e). Data presented as mean -/+ SD from n = 3 biological replicates. f Dead cell quantification by acridine orange/propidium iodide staining following 3-day treatment with the indicated concentrations of VLS-1272 in OVCAR-3 cells. Data presented as mean -/+ SD from n = 3 biological replicates. P values are calculated compared to untreated control and labeled on the corresponding bar, using one-way ANOVA with Dunnett’s multiple comparisons test. g Western blot of cleaved Caspase 3 (MW: 17 kDa) versus full length Caspase 3 (MW: 35 kDa) after 24- and 72 h treatment of CAL51 and OVCAR-3 cells with VLS-1272. One representative experiment of 2 biological replicates is shown. Growth inhibition as measured by CellTiter-Glo of HCC15 (h) and HCC1806 cells (i) after treatment with the indicated concentration of VLS-1272 with or without 0.5 or 2 nM BAY-1217389 (MPS1 inhibitor) for 3 days. Data presented as mean values -/+ SD from n = 4 biological replicates. Source data are provided in the Source Data file.
To further confirm that the anti-proliferative effect observed with KIF18A inhibition was specific to chromosomally unstable cancer cells, we tested the effect of VLS-1272 on several normal cell types, including proliferating T-cells isolated from two healthy donors, and the non-transformed epithelial cell lines MCF10A and RPE-1. Importantly, we observed little to no effect on proliferation of any of these non-tumorigenic cells following treatment with VLS-1272 (Fig. 4a, c). This critical finding demonstrates that KIF18A inhibition is selective for chromosomally unstable tumor cells while sparing proliferation of normal cell types.
Given the anti-proliferative effect observed with KIF18A inhibition, cell death was evaluated, and a dose-dependent increase in non-viable cells following KIF18A inhibition was observed within 48 h in both kinetic (Fig. 4d) and endpoint (Fig. 4f) analyses in CINHigh cell lines, but not in the CAL51 (CINLow) cell line (Fig. 4e). VLS-1272 also induced Caspase-3 cleavage in the sensitive OVCAR-3 cell line but not in the insensitive CAL51 cell line (Fig. 4g). Taken together, these data suggest that KIF18A inhibition in sensitive cancer cells results in the initiation of programed cell death, but cell death is absent in insensitive cells.
To further probe potential determinants of sensitivity beyond CIN, we assessed whether inhibition of the spindle assembly checkpoint (SAC) would alter responses to KIF18Ai. The reduction in kinetochore tension due to KIF18A loss can activate the SAC42, and an intact SAC is required for mitotic arrest induced by KIF18A loss18. Inhibition of the SAC permits cell division in the presence of erroneously connected chromosomes and leads to the formation of micronuclei43,44. Attenuation of the SAC can be accomplished by inhibiting MPS113, and we identified doses of the MPS1 inhibitor BAY-1217389 that promote micronuclei formation while maintaining cell viability (Supplementary Fig. 4a, b). Weakening of the SAC with low dose MPS1 inhibition was sufficient to robustly diminish the anti-proliferative response to VLS-1272 (Fig. 4h, i). These data with pharmacological KIF18A inhibition are consistent with published genetic data demonstrating that SAC inhibition through Mad2 loss can restore mitotic progression in cells depleted of KIF18A18,42 and with published data demonstrating reduction of KIF18A inhibitor toxicity with low doses of reversine45. Thus, activation of the SAC is a key requirement for KIF18A inhibition to result in proliferative defects.
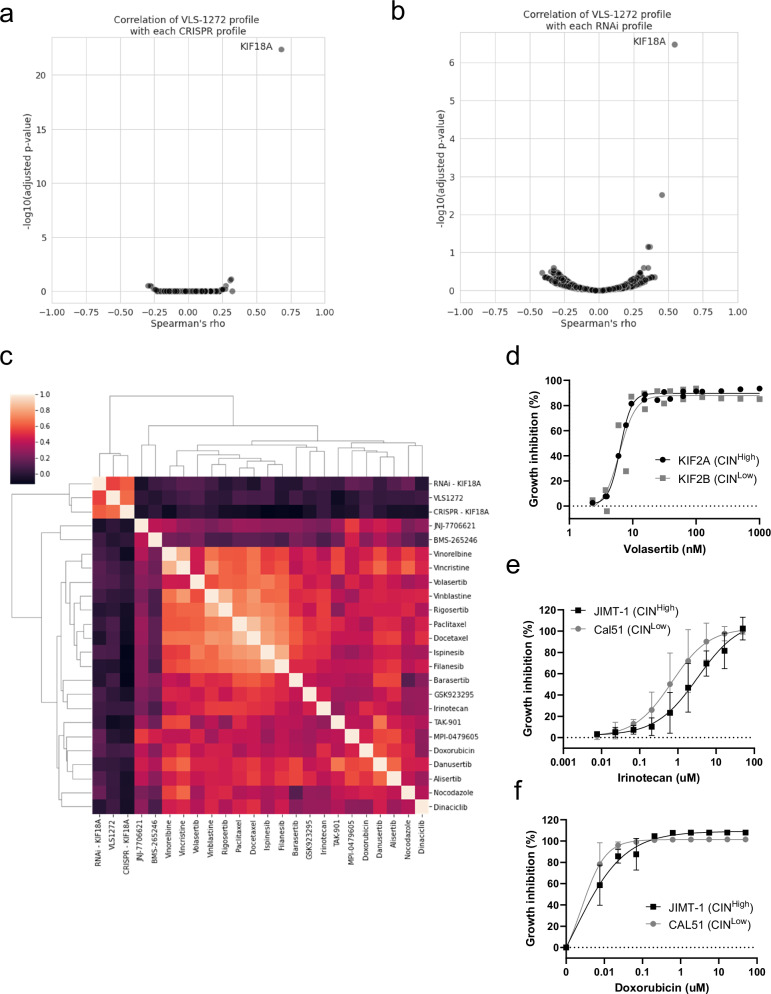
The differential effects of KIF18A inhibition on CINHigh, CINLow, and normal cells suggests that KIF18A inhibitors might differentiate from traditional anti-mitotic therapies that have been evaluated for clinical use. Anti-mitotic drugs suppress most proliferating cells, including CINLow and normal cells46–54 in contrast to KIF18A perturbation by genetic knockdown, knockout, and chemical inhibition with VLS-1272. To further probe differential effects of KIF18A inhibition vs. anti-mitotic drugs, we profiled the sensitivity to VLS-1272 across a > 100 cell line panel (Supplementary Data 1). We first compared the cell line sensitivity to VLS-1272 with sensitivity to genetic depletion of KIF8A. VLS-1272 response is highly correlated with the effect of KIF18A perturbation by either CRISPR (Fig. 5a) or RNAi (Fig. 5b) as determined in Depmap23. KIF18A was the gene most significantly correlated with VLS-1272 cell line sensitivity, providing further support that the effect of VLS-1272 on cells is through on-target inhibition of KIF18A. Comparison of VLS-1272 cell line sensitivities to those published with AM-188236 showed high correlation demonstrating that the proliferative defects observed were likely specific to KIF18A inhibition (Supplementary Fig. 2e). We then compared the VLS-1272 sensitivity profile to the reported sensitivity profiles of other anti-mitotic agents55. These agents included drugs targeting mitotic regulators such as AURK, PLK1, Eg5 and CENP-E, and MT-targeting therapies such as docetaxel and vinblastine (Table 2). In contrast to the correlation with KIF18A genetic dependency, the VLS-1272 response profile did not correlate with the response to each of 21 anti-mitotic compounds included in the analysis (Fig. 5c). These results confirm that while KIF18A functions in mitosis, KIF18A inhibitors display a unique profile compared to previously developed anti-mitotic therapies.
Fig. 5. Comparison of VLS-1272 to genetic perturbation of KIF18A and other anti-mitotics.
Volcano plot depicting the correlation (Spearman’s rho value) versus significance (p value computed with Spearman’s rho) of each gene effect profile in the DepMap CRISPR (a) or RNAi (b) dataset with the VLS-1272 response profile across up to 192 cell lines. c Heatmap of the Spearman’s rho values for each pairwise comparison of treatment response across up to 128 cell lines. The labels on each axis indicate the treatment, and the shading in each box indicates the computed rho value for the comparison of perturbation response. Area under the curve (AUC) values were computed from the VLS-1272 treatment response for use in the analyses in (a–c). d Growth inhibition as measured by CellTiter-Glo of MDA-MB-231 cells expressing KIF2A or KIF2B after treatment with the indicated concentrations of Volastertib for 168 h. Data presented as individual values from n = 2 biological replicates. Growth inhibition as measured by CellTiter-Glo of CAL51 and JIMT-1 cells after treatment with the indicated concentrations of Irinotecan (e) or Doxorubicin (f) for 168 h. Data presented as mean values -/+ SD from n = 4 (e) or n = 3 (f) biological replicates. Source data are provided in the Source Data file.
Table 2.
Compounds used in comparative analysis of anti-mitotic efficacies compared to VLS-1272
| Compound name | Mechanism of action | Target(s) |
|---|---|---|
| Docetaxel | Tubulin polymerization inhibitor | BCL2, MAP2, MAP4, MAPT, NR1I2, TUBB, TUBB1 |
| Vinorelbine | Tubulin polymerization inhibitor | TUBA1A, TUBA1B, TUBA1C, TUBA3C, TUBA3D, TUBA3E, TUBA4A, TUBB, TUBB1, TUBB2A, TUBB2B, TUBB3, TUBB4A, TUBB4B, TUBB6, TUBB8 |
| Paclitaxel | Tubulin polymerization inhibitor | BCL2, MAP2, MAP4, MAPT, NR1I2, TLR4, TUBB, TUBB1 |
| Vinblastine | Microtubule inhibitor, tubulin polymerization inhibitor | JUN, TUBA1A, TUBB, TUBD1, TUBE1, TUBG1 |
| Vincristine | Tubulin polymerization inhibitor | TUBA4A, TUBB |
| Nocodazole | Tubulin polymerization inhibitor | HPGDS |
| Danusertib | Aurora kinase inhibitor, growth factor receptor inhibitor | AURKA, AURKB, AURKC, FGFR1, NTRK1, RET, SLK |
| Barasertib | Aurora kinase inhibitor | AURKA, AURKB |
| Alisertib | Aurora kinase inhibitor | AURKA |
| TAK-901 | Aurora kinase inhibitor | AURKB |
| MPI-0479605 | Mitotic kinase inhibitor | TTK |
| Irinotecan | Topoisomerase inhibitor | TOP1, TOP1MT |
| Doxorubicin | Topoisomerase inhibitor | TOP2A |
| Rigosertib | Cell cycle inhibitor, PLK inhibitor | PLK1 |
| Volasertib | PLK inhibitor | PLK1 |
| Ispinesib | Kinesin inhibitor | KIF11/Eg5 |
| Filanesib | Kinesin inhibitor, kinesin-like spindle protein inhibitor | KIF11/Eg5 |
| GSK923295 | Centromere associated protein inhibitor | CENPE |
| JNJ-7706621 | CDK inhibitor | AURKA, AURKB, CDK1, CDK2 |
| BMS-265246 | CDK inhibitor | CDK1, CDK2 |
| Dinaciclib | CDK inhibitor | CDK1, CDK2, CDK5, CDK9 |
Mechanism of action and annotated targets as listed from DepMap used for the analysis in Fig. 5C.
To further probe the comparison to drugs targeting mitosis, we directly tested the sensitivity of CINHigh/Low cells to select anti-mitotic agents analyzed in Fig. 5c. In the paired CIN cell line expressing KIF2A or KIF2B, there was no difference in sensitivity to the PLK1 inhibitor Volasertib (Fig. 5d), in contrast to the differential response observed with VLS-1272 (Fig. 4b). Doxorubicin and Irinotecan were tested in the CINHigh JIMT-1 and CINLow CAL51 cells. Differential sensitivity was not observed between these cell lines (Fig. 5e, f), again in contrast to the results observed with VLS-1272 (Fig. 4a). These data further corroborate that VLS-1272 stands out amongst anti-mitotic agents as exhibiting differential sensitivity in chromosomally unstable cancer cell lines.
Cellular readouts of KIF18A inhibition
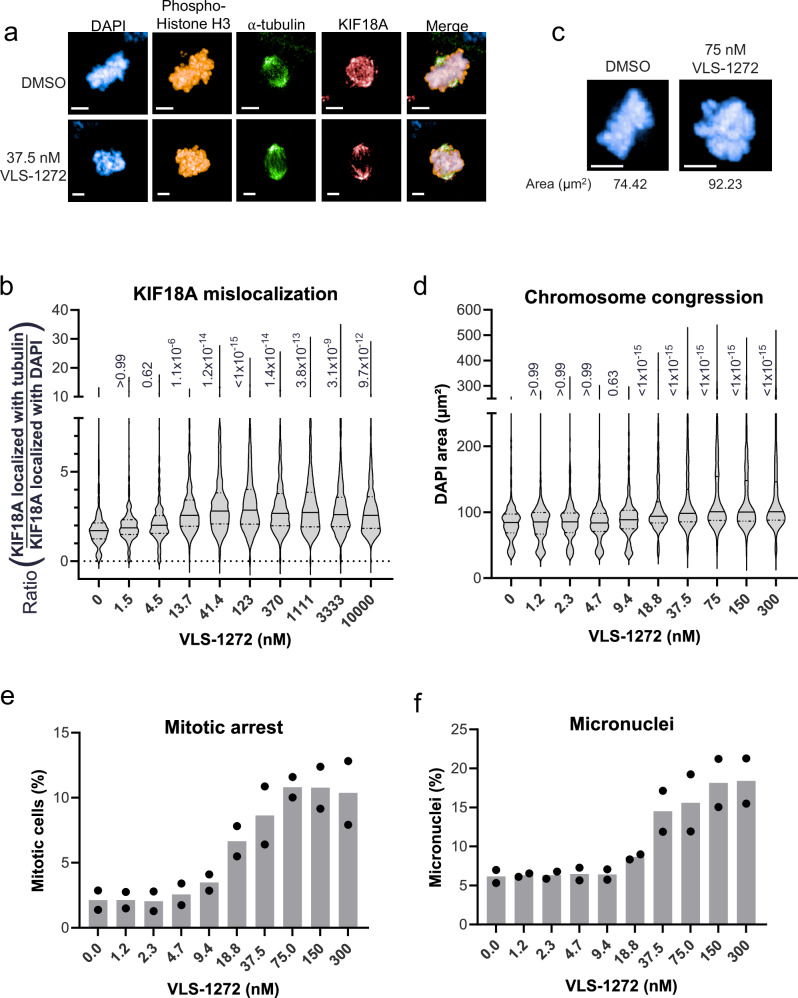
We next assessed the impact of KIF18A inhibition on KIF18A localization and mitotic progression. KIF18A expression increases during mitosis and localizes at the mitotic spindle, where it subsequently translocates towards the microtubule plus-ends to stabilize chromosome oscillations at the metaphase plate allowing progression from metaphase to anaphase17,56. To assess if inhibition of KIF18A ATPase activity impacts its translocation ability, we evaluated KIF18A localization following VLS-1272 treatment. Consistent with published data showing KIF18A localization near the plus ends of kinetochore-microtubules17,56, we observe KIF18A colocalization with DNA across the metaphase plate in vehicle treated cells (Fig. 6a, top). With VLS-1272 treatment, KIF18A still localizes to microtubules but is no longer localized along the metaphase plate. Instead, inhibited KIF18A is observed on microtubules near the spindle poles (Fig. 6a, bottom). The shift is KIF18A localization can be quantified by measuring the ratio of KIF18A colocalized with α-tubulin versus KIF18A colocalized with DAPI. This ratio increases, with KIF18A co-localization shifting away from DNA towards α-tubulin, as VLS-1272 inhibitor concentration increases above the IC50 concentration of the HCC1806 cell line (Fig. 6b). The impact of VLS-1272 on KIF18A spindle localization aligns with published data demonstrating the need for KIF18A motor activity for its plus-end localization17, and is consistent with relocalization of KIF18A towards the spindle poles observed with AM-188236. The KIF18A localization data, combined with the MT-dependence of VLS-1272 activity and impact on KIF18A motility, support the model that VLS-1272 binds to the KIF18A-microtubule complex, immobilizing KIF18A on microtubules and preventing its procession towards the plus end.
Fig. 6. Mitotic consequences of KIF18A inhibition in sensitive cells with VLS-1272.
a Representative immunofluorescence images of HCC1806 cells treated with DMSO (top) or 37.5 nM VLS-1272 (bottom). Scale bar = 5 µm. One representative image from the treatment and control group from two biological replicates is shown. b Quantification of KIF18A mis-localization in mitotic HCC1806 cells measured as the ratio of KIF18A colocalizing with the α-tubulin spindle to KIF18A colocalizing with DNA (DAPI). Colocalization masks were generated in Harmony high-content analysis software. Data presented as violin plots with the mean (solid line) and quartiles (dashed lines) from n = 233-803 mitotic cells. Adjusted p value is calculated compared to untreated control and labeled next to each condition, using one-way ANOVA with Bonferroni’s multiple comparisons test. c Representative fluorescent images of DAPI-stained mitotic DNA in HCC1806 cells treated with DMSO or VLS-1272. Scale bar = 5 µm. One representative image from the treatment and control group from two biological replicates is shown. d Quantification of the area of DAPI staining (µm2) in pHH3-positive cells treated with increasing doses of VLS-1272. The area of the DAPI-stained DNA was calculated in Harmony high-content analysis software. Data presented as violin plots with the mean (solid line) and quartiles (dashed lines) from n = 652–1434 mitotic cells. Adjusted p value is calculated compared to untreated control and labeled next to each condition, using one-way ANOVA with Bonferroni’s multiple comparisons test. e Quantification of mitotic HCC1806 cells identified by positive staining for pHH3 compared to the number of total DAPI-stained nuclei. The mitotic cell population was calculated in Harmony high-content analysis software. f Percentage of total micronuclei to primary nuclei in cells treated with VLS-1272, calculated in Harmony high-content analysis software. e, f Data presented as mean values with individual data points shown from n = 2 biological replicates. Source data are provided in the Source Data file.
Previous studies have demonstrated that KIF18A knockdown results in a disordered mitotic spindle with reduced chromosome congression due to increased chromosome oscillations17,56. To assess whether VLS-1272 would similarly impact chromosome congression, mitotic cells were identified with phospho-histone H3 (pHH3) staining and the DNA-stained area of those mitotic cells was quantified after treatment with VLS-1272. In the sensitive HCC1806 cell line, an increase in the surface area of mitotic chromosomes was observed due to the lack of chromosome congression with VLS-1272 treatment, providing evidence that KIF18A inhibition results in defective chromosome alignment (Fig. 6c, d), similar to published results using genetic loss-of-function. To determine the impact of KIF18A inhibition on mitotic progression, we quantified the number of mitotic cells by pHH3 detection. KIF18A inhibition increased the percentage of cells in mitosis (Fig. 6e), suggesting a deficiency in mitotic progression. Additionally, increased micronuclei levels were observed at concentrations above the IC50 of 18.1 nM in HCC1806 cells (Fig. 6f). This data suggests mitotic abnormalities prevent proper progression through mitosis, resulting in a greater percentage of cells transitioning to programmed cell death (as shown in Fig. 4d–g). These data affirm our hypothesis that KIF18A inhibitor-sensitive cells treated with VLS-1272 arrest in mitosis because KIF18A is no longer able to effectively align chromosomes at the metaphase plate.
To understand how cellular responses to VLS-1272 correlate with drug sensitivity, we evaluated the CINLow KIF18Ai-insensitive CAL51 cell line. KIF18A mis-localization after treatment was observed in CAL51 cells (Supplementary Fig. 5a, b), demonstrating that the effect on KIF18A localization is a result of target engagement and not a determinant of sensitivity to KIF18A inhibition. In contrast, VLS-1272 did not alter chromosome congression (Supplementary Fig. 5c) and did not arrest CAL51 cells in mitosis (Supplementary Fig. 5d), consistent with the lack of cell death in this cell line (Fig. 4e, g). An increase in micronuclei was observed in CAL51 cells (Supplementary Fig. 5e), although not to the extent seen in sensitive cells (compare to Fig. 6f). The micronuclei increase in CINLow cells aligns with published data of increased micronuclei in KIF18A-deficient normal tissues57, and identifies micronuclei formation as a readout of KIF18A inhibition separate from KIF18A dependency.
Tumor growth inhibition with KIF18A inhibitors
Our data demonstrate that inhibition of KIF18A ATPase activity leads to an inability of KIF18A to translocate along spindle MTs, resulting in dysfunctional chromosome congression, mitotic arrest, and finally programmed cell death. To translate the impact of these findings to a more disease-relevant setting, we evaluated the response of human tumor xenografts to KIF18A inhibition. We analyzed the pharmacokinetic properties of VLS-1272 and found that oral formulation and delivery resulted in drug exposures necessary to maintain target engagement (Supplementary Fig. 6a, b).
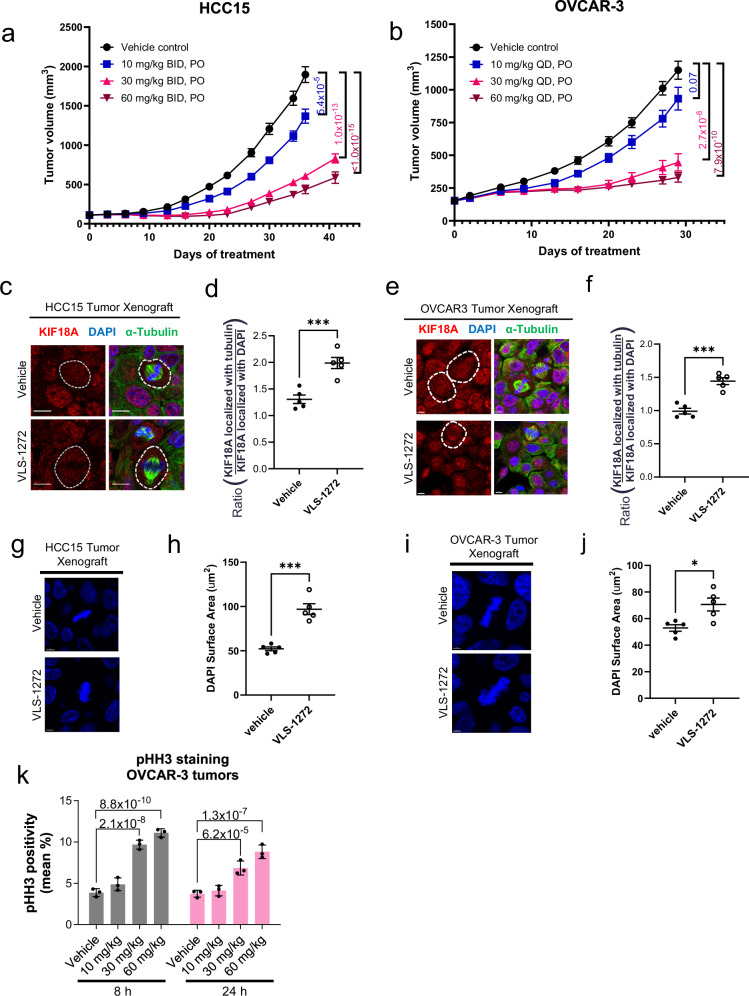
HCC15 and OVCAR-3 tumor models were selected for in vivo evaluation as they represented diverse tumor types and were among the most sensitive cell lines to KIF18A inhibition (Fig. 4a). Mice bearing these human tumor xenografts were randomized and dosed once (QD) or twice (BID) daily with increasing dosages of VLS-1272. Body weights and tumor sizes were measured twice a week. Mice treated with VLS-1272 exhibited robust dose-dependent inhibition of tumor growth in these two distinct tumor models (Fig. 7a, b). Body weight was maintained throughout the course of the study for the daily dosing groups, and with the aid of nutrient gel for all groups (including vehicle) with twice daily dosing (Supplementary Fig. 7a, b). Further evidence of tolerability to VLS-1272 was observed in a separate study of non-tumor bearing mice dosed daily with VLS-1272. After 7 days of dosing, complete blood count (CBC) analysis was performed, and no significant alterations of hematological lineages were observed when compared to strain reference data58 (Supplementary Table 2).
Fig. 7. Tumor growth inhibition with KIF18A inhibitor treatment.
Tumor volume measurements after administering vehicle or VLS-1272-SDD to (a) female SCID Beige mice bearing HCC15 tumor xenografts or (b) female Balb/c nude mice bearing OVCAR-3 tumor xenografts (n = 10 mice per treatment group). Data presented as mean values -/+ SEM. p values of VLS-1272-treated groups compared to vehicle at Day 36 (HCC15) or Day 29 (OVCAR-3) are indicated and determined using one-way ANOVA with Dunnett’s multiple comparison test. Representative fluorescence images of KIF18A alone or together with α-tubulin and DAPI in tumors harvested from mice treated with vehicle or (c) 30 mg/kg VLS-1272 in HCC15 or (e) 60 mg/kg VLS-1272 in OVCAR-3. Scale bar = 10 µm (c) and scale bar = 5 µm (e). Quantification of KIF18A mislocalization measured as the ratio of KIF18A fluorescence colocalizing with α-tubulin over the amount of KIF18A colocalizing with DAPI in HCC15 (d) and OVCAR-3 (f) xenograft tumor sections. Data presented as mean values -/+ SEM from n = 5 mice per treatment group, (d) ***p = 0.00082 and (f) ***p = 0.00015 using two-tailed unpaired t test. Representative fluorescence micrographs and quantification of DAPI staining of tumors harvested from mice treated with vehicle or (g, h) 30 mg/kg VLS-1272 in HCC15 or (i, j) 60 mg/kg VLS-1272 in OVCAR-3. Scale bar = 5 µm (g, i). Data presented as mean values -/+ SEM from n = 5 mice per treatment group, (h) ***p = 0.00012 and (j) *p = 0.012 from two-tailed unpaired t-test. k Mitotic cell counts by pHH3 staining of tumor sections from excised OVCAR-3 xenografts. Data presented as mean -/+ SD from n = 3 mice per group. p values are indicated above the bars and determined using one-way ANOVA with Dunnett’s multiple comparison test. Source data are provided in the Source Data file.
KIF18A target engagement and mechanistic impacts of VLS-1272 treatment in tumors were assessed using multiple readouts. Consistent with our earlier results in cell culture studies (Fig. 6), VLS-1272 treatment caused mis-localization of KIF18A from the spindle midbody to the spindle poles (Fig. 7c–f), as well as an increased mitotic DNA-stained area indicative of chromosome congression defects (Fig. 7g–j) in tumor sections from both tumor models tested. Additionally, we observed an increase in mitotic cells (pHH3-positive) in OVCAR-3 tumors sampled 8 and 24 h after initial treatments (Fig. 7k). These results confirm that VLS-1272 has pharmacokinetic properties to robustly engage KIF18A in vivo and prevent KIF18A translocation, resulting in decreased chromosome congression, mitotic arrest, and ultimately inhibition of tumor growth.
Discussion
A high-throughput screen followed by chemical optimization led to the discovery of VLS-1272, a potent inhibitor of KIF18A ATPase activity that is non-competitive with ATP and dependent on microtubules. VLS-1272, by interfering with KIF18A’s motor activity, causes mitotic KIF18A relocalization away from microtubule plus-ends, resulting in reduced chromosome congression and mitotic arrest. Ongoing chromosome instability and the presence of an active mitotic checkpoint both contribute to VLS-1272’s anti-proliferative effect. The phenotypes and dependencies observed with KIF18A inhibition are strikingly similar to those observed with KIF18A knockdown or knockout, described in this manuscript and previously published17,18,40,56. Finally, we demonstrate that both genetic knockdown and pharmacological inhibition of KIF18A results in tumor growth inhibition, illustrating the potential for clinical benefit of KIF18A inhibitors.
Previously discovered KIF18A inhibitors include BTB-135 and those developed by Amgen36,40. VLS-1272 differs from BTB-1 through its greater potency, ATP-uncompetitive mode of inhibition, and cellular differentiation between CINHigh and CINLow cells. The inhibitors described by Amgen, despite being a different structural class, display greater similarity with VLS-1272 with regards to potency, mode of inhibition, cellular activity, and demonstration of mitotic arrest in vivo (Supplementary Table 1). The similarity in phenotypes between these inhibitor classes provides additional confirmation that biological responses observed with KIF18A genetic manipulation can be translated to pharmacological inhibition. Furthermore, we extend previous findings by demonstrating that KIF18A knockdown and inhibition can have robust anti-tumor activity in vivo, paving the way for therapeutic translation. In this study, the insensitivity of CINLow tumors to VLS-1272 was not evaluated with in vivo studies needed to confirm translation of the CIN dependency. However, published data with other KIF18A inhibitors36 demonstrated differential CIN sensitivity to KIF18A inhibition in vivo.
Previous efforts to therapeutically target mitotic kinesins in cancer has had limited success, in part due to their ability to interfere with mitosis in proliferating normal cells as well as tumor cells14. In this study, we demonstrate that inhibition of KIF18A is selective for chromosomally unstable tumor cells while sparing proliferating non-transformed and primary cells. Treatment with VLS-1272 resulted in dose-dependent tumor growth inhibition without body weight loss (an indicator of toxicity). Furthermore, hematological lineages in vivo are largely unaffected by KIF18A inhibition, in contrast to anti-mitotics which can result in adverse effects such as neutropenia14. These findings are consistent with those observed with KIF18A loss of function in normal cells. While KIF18A loss impacts chromosome alignment in both normal cells and tumor, normal cell progress through mitosis, while CINHigh cells cannot18,33,59.
The anti-proliferative impact of VLS-1272 requires activation of the SAC. Cells insensitive to VLS-1272, such as normal and CINLow cells, may have mechanisms that can satisfy the SAC despite KIF18A inhibition, allowing mitotic progression, whereas CINHigh tumor cells trigger the SAC upon KIF18A inhibition resulting in mitotic arrest. Furthermore, cells with high levels of chromosome instability often have mitotic aberrations, such as lagging chromosomes60, that need to be corrected, which likely result in mitotic delays61–63. Thus, the chromosome alignment function of KIF18A is more crucial in cells that need to compensate for more frequent mitotic abnormalities. While VLS-1272 alters KIF18A localization in CINLow cells, chromosome congression is not altered. Evaluation of kinetochore protein phosphorylation to measure kinetochore-microtubule attachment status could provide additional insight as to how CINLow cells are able to achieve full chromosome congression in the absence of KIF18A function. The selectivity of KIF18A inhibition for CINHigh cells versus normal cells distinguishes VLS-1272 from clinically tested anti-mitotic drugs and suggests that KIF18A inhibitors might be powerful anti-cancer agents with a favorable therapeutic index.
Chromosome instability is prevalent in many (estimated >80%) solid tumor types, including cancers with high unmet medical need such as lung, ovarian, and stomach cancer, suggesting that KIF18A inhibitors may have broad clinical applicability64–66. CIN can drive metastatic progression through cytosolic DNA activation of a cGAS/STING/non-canonical NF-κB pathway9. A recent comprehensive clinical assessment of metastatic tumors demonstrated a high correlation of CIN with metastatic burden in multiple tumor types67. Therefore, KIF18A inhibitors may represent a promising treatment capable of combating both primary tumors and metastatic disease even in settings where patients are refractory to conventional therapies.
Many known standard-of-care therapies promote chromosome instability on their own through different mechanisms68–70. Thus, in tumors with lower levels of CIN, combinations with these agents may promote CIN and enhance their responsiveness to KIF18A inhibition. In addition, CIN can drive drug resistance through clonal heterogeneity that carries drug resistant mutations as well as copy number and pathway alterations8,71–74. Killing CINHigh tumor cells with a KIF18A inhibitor could delay drug resistance by limiting cancer cell adaptation and plasticity, thus reducing the rate by which drug-resistant phenotypes arise. Furthermore, the lack of impact of VLS-1272 on T cell proliferation warrants exploration of combination with immune checkpoint inhibitors, which are increasingly being used as standard-of-care in multiple tumor types. Future studies are aimed at uncovering the full clinical potential of KIF18A inhibition.
Methods
Ethics statement
Tumor studies involving animals were approved by Volastra Therapeutics and the Institutional Animal Care and Use Committee (IACUC) of WuXi AppTec following the guidance of the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). Mice were kept in individual ventilation cages at constant temperature (20–26 °C) and humidity (40–70%) with a 12 h light-dark cycle. When the mean tumor volume exceeded 2000 mm3, all animals in the given group were euthanized. This maximal mean tumor volume was not exceeded in this study. PBMCs were purified from healthy donors with written informed consent for blood withdrawal. The blood donation protocol was approved by the Institutional Review Board at the Shanghai Liquan Hospital.
Computational identification of genes essential in chromosomally unstable cells
Copy number profiles for cancer cell lines were obtained from DepMap (The Cancer Dependency Map)23. Whole exome sequencing based segment files were downloaded from Version 2020 Q2 release. Segment files were analyzed for computing FGA (fraction genome altered) for all cell lines. Copy Number Segments with absolute log ratio >=0.2 were marked as altered. The sum of the segment sizes for all segments marked altered were then normalized to the total genome size to obtain the fraction of the genome altered.
DEPMAP cell lines were classified into CINHigh, CINMed and CINLow groups, with CINHigh being the top FGA quartile and CINLow the bottom FGA quartile. DEMETER2-based gene effect scores from an RNAi Screen across 712 cell lines75 were used to assess essentiality. Essential genes were defined as those with a gene effect score < -1 in the RNAi screen. To identify genes that were essential in CINHigh cell lines at the pan-cancer level, the odds ratio from a Fisher’s Exact test was used to associate the CIN level of cell lines with essentiality scores for each gene in a systematic genome-wide analysis. Further, p values obtained from the Fisher’s exact test for all genes were then adjusted for false discovery using the Benjamini-Hochberg test. Genes with FDR < 0.25 were defined as ones with the most significant association between CIN and gene essentiality. All statistical analysis was performed in R statistical software.
Cell lines
CAL51 (DSMZ ACC-302), JIMT-1 (AddexBio C0006005), MDA-MB-231 (ATCC HTB-26), and hTERT RPE-1 (ATCC CRL-4000) cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco) supplemented with 10% Fetal Bovine Serum (FBS) (Gibco) and 100 units/ml penicillin and 100 μg/ml streptomycin (Thermo Fisher Scientific). MCF10A (ATCC CRL-10317) cells were grown in DMEM/F12 medium supplemented with 5% horse serum, epidermal growth factor (20 ng/ml), insulin (10 µg/ml), hydrocortisone (0.5 mg/ml) (Lonza), cholera toxin (100 ng/ml) (Sigma Aldrich), 100 units/ml penicillin, and 100 μg/ml streptomycin (Thermo Fisher Scientific). HCC1806 (ATCC CRL-2335) and HCC15 (DSMZ ACC-496) cells were grown in RPMI 1640 Medium (Gibco) supplemented with 10% FBS (Gibco) and 100 units/ml penicillin and 100 μg/ml streptomycin (Thermo Fisher Scientific). NIH:OVCAR-3 (ATCC HTB-161)) cells were grown in RPMI 1640 Medium (Gibco) supplemented with 20% FBS (Gibco) and 100 units/ml penicillin and 100 μg/ml streptomycin (Thermo Fisher Scientific). HT-55 (ECACC 85061105) cells were grown in EMEM (EBSS) supplemented with 2 mM glutamine, 1% non-essential amino acids (NEAA), and 20% FBS. All cell lines tested negative for mycoplasma contamination using MycoAlert Mycoplasma Detection Kit (Lonza LT07-318). No commonly misidentified cell lines were used in the study. Information on cell line authentication by STR was provided by the commercial sources for each cell line.
Generation of cells expressing KIF2A or KIF2B
MDA-MB-231 cells were engineered to stably express Cas9 by lentiviral transduction of the pRCE2B-EFS-Cas9-2a-Blast vector (Cellecta). Cells were selected in blasticidin (EMD Millipore 203351) and a Cas9-expressing clone was isolated. Next, KIF2A or KIF2B were introduced by lentiviral transduction of the pLVX-IRES-Neo vector (Takara) containing eGFP-tagged human KIF2A or KIF2B. Cells were selected in 500 μg/mL G418 (Sigma-Aldrich G8168) and sorted for GFP positivity by FACS.
KIF18A CRISPR knockout and proliferation assay
MDA-MB-231-Cas9-KIF2A or MDA-MB-231-Cas9-KIF2B cells were transfected twice with non-targeting sgRNA (Synthego scrambled sgRNA #1) or pooled sgRNAs targeting KIF18A (Synthego Gene Knockout Kit v2 sequences 5’- AAACUGACUUCUUCUUGUUU, 5’- AGCAGCUGGAUUUCAUAAAG, 5’- AUCAACAAUGUCUGUCACUG) using RNAiMAX (Thermo 13778-075), according to the manufacturer’s protocol. Cells were cultured in complete medium and on day 6 post-transfection they were harvested, counted, and equal cell numbers were plated in 96 well plates. Proliferation was assessed at the indicated time points using CellTiter-Glo 2.0 reagent (Promega G9243), according to the manufacturer’s protocol.
KIF18A siRNA knockdown
KIF18A siRNA studies were performed using Silencer Select siRNA (Thermo Fisher Scientific s37882) and Lipofectamine RNAiMAX (Thermo Fisher Scientific 13778-075), according to the manufacturer’s protocol. Cells were transfected overnight before being plated for proliferation assays and Western blotting. Proliferation and knockdown were assessed 72 h after replating.
Cell proliferation and viability assays
Cells were plated for 7-day viability studies in 96 well plates 24 h before indicated treatments. Following indicated compound treatments, the plates were equilibrated to room temperature 30 min before the addition of CellTiter-Glo (CTG) reagent (Promega (G9241)). Plates were rocked at room temperature for 10 min covered from light. The luminescence was then recorded on a plate reader (PerkinElmer) and IC50s were calculated using curve-fitting in Graphpad Prism software.
To assess cell death, cells were plated 24 h before compound administration. Enumeration of dead cells was performed by trypan blue (Thermo Fisher 15250061) staining and cell counting on a Countess 3 automated cell counter (Thermo Fisher) three days after treating with VLS-1272. Kinetic dead cell assessment was completed by imaging cells on an Incucyte live-cell analysis instrument (Sartorius), where cells were dosed with VLS-1272 and 5 ug/mL propidium iodide (PI) (Thermo Fisher P1304MP). Images were taken every 6 h at 10x magnification. PI-positive (dead cells) were counted using Incucyte software (Sartorius) and plotted in Prism (GraphPad).
Inducible KIF18A shRNA cell line
JIMT-1 cells were transduced with commercially available SMARTvector inducible shRNA lentiviral particles containing shRNA constructs targeting KIF18A (Cat. V3SH7670-225957177 and V3SH7670-228478740) or a non-targeting control (Horizon Discovery Cat. VSC6572) using Lipofectamine3000 (Thermo Fisher L3000015) according to manufacturer’s specifications. Cells were selected in 2 μg/mL puromycin (Invivogen ant-pr-1) to maintain expression of the inducible shRNA constructs and maintained in culture media as listed above. Inducible knockdown of KIF18A was achieved by treating cells with 0, 0.1, or 1 µg/mL doxycycline (Sigma-Aldrich D9891-5G) for 72 h.
In vivo doxycycline-inducible KIF18A knockdown in JIMT- 1 tumor xenografts
The JIMT-1 control shRNA and JIMT-1 KIF18A shRNA tumor cell lines were maintained in DMEM supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 2 ug/mL puromycin and 1% Anti-Anti. The cells growing in exponential growth phase were harvested and counted for tumor inoculation. 6–8-week-old female SCID Beige mice (Beijing Vital River Laboratory Animal Technology Co., Ltd.) were inoculated subcutaneously at the right flank with 10 × 106 JIMT-1 control shRNA or JIMT-1 KIF18A shRNA cells in 0.2 mL of PBS mixed with Matrigel (50:50). Animals were grouped for treatment 7 days after tumor inoculation when the average tumor volume was between 99 and 103 mm3. Doxycycline (Takara Bio Inc. 631311) was dissolved in saline at a stock concentration of 5 mg/mL and was used for an initial bolus of doxycycline that was administered at a dose of 50 mpk i.p. at the start of the experiment. Normal and 625 ppm doxycycline chow were given and replenished daily, and the daily consumption was recorded. Tumor sizes were measured twice per week in two dimensions using a caliper, and the volumes were expressed in mm3 using the formula: V = 0.5(a x b2) where a and b are the long and short diameters of the tumor, respectively.
For western blot analysis of KIF18A expression from tumors, animals (4 per group) were euthanized, and tumors were collected 4 days post initiation of doxycycline treatment. A sample of the excised tumors were cut up to about 30–100 mg. Tumor tissues samples were placed in a microcentrifuge tube containing stainless steel beads (Qiagen 69989) and RIPA Buffer (Sigma R0278) containing 1% Protease Inhibitor Cocktail (Roche 69989) and 1% Phosphatase Inhibitor Cocktail 2 Sigma (P5726). The tissue was ground using a TissueLyser (Shanghai Jingxin (JXFSTPRP-CL)) at 50 Hz for 5 min. Tissue lysates were kept on ice for 30 min and vortexed every 5 min. Samples were centrifuged at 21,130 x g at 4 °C for 10 min and the supernatants were transferred to a new 1.5 mL microcentrifuge tube kept on ice. Protein concentrations were measured with a BCA Protein Assay Kit (Thermo Scientific 26619). Samples were diluted to the same final concentration (2 μg/μL) using RIPA buffer plus 4x LDS Sample Buffer (Thermo Scientific NP0007) and 10x Sample Reducing Agent Thermo Scientific (NP0009). Samples were heated at 100 °C for 10 min and separated using SDS-PAGE and immunoblotted. Blots were blocked in Odyssey blocking buffer (LI-COR 927-60001). Membranes were probed with rabbit anti-KIF18A (1:500, Fortis Lifesciences A301-080A) and rabbit anti-GAPDH (1:2000, Cell Signaling Technologies 5174). Westerns were blotted with IRDye® 800CW Goat anti-Rabbit IgG Secondary Antibody (1:10000, LI-COR 926-32211) and imaged on an Odyssey CLx infrared imaging system (LI-COR).
Western blotting for cell culture
Cells in culture were lysed in RIPA lysis and extraction buffer (Thermo Fisher Scientific 89901) supplemented with protease/phosphatase inhibitor Cocktail (Cell Signaling Technologies 5872S) at 4 °C for 30 min. Samples were spun at 18k x g for 20 min at 4 °C to remove the protein supernatant from the insoluble fraction. The concentration of protein lysates was measured using the Precision Red Advanced Protein Assay kit (Cytoskeleton, Inc. ADV02-A). A 30 µg sample of lysate was loaded and separated using SDS-PAGE and immunoblotting. Blots were blocked and blotted with antibodies diluted in Intercept TBS protein-free blocking buffer (LI-COR 927-80001). Antibodies used in these studies were rabbit anti-KIF18A (1:500, Fortis Lifesciences A301-080A), rabbit anti-GAPDH (1:2500, Abcam ab9485), rabbit anti-Caspase-3 (1:1000, Cell Signaling Technology 9662) and rabbit anti-Vinculin (1:3000, Abcam ab129002). Westerns were blotted with goat anti-rabbit IgG (H + L) antibody conjugated to IRDye 680RD (1:20000, LI-COR 926-68071) and imaged on an Odyssey CLx infrared imaging system (LI-COR).
T cell proliferation assay
Fresh PBMCs were prepared from 2 healthy human donors. PBMCs were resuspended with EasySep Buffer (STEMCELL Technologies, Inc.) at a density of 5 × 107 cells/ml, and T cells were isolated using the EasySep Human T Cell Isolation Kit (STEMCELL Technologies, Inc.). Resuspended T cells in RPMI1640 Complete Medium (RPMI1640 supplemented with 10% HI-FBS, 1% P/S and 55 µM 2-mercaptomethanol) were plated at a density of 1 × 105 cells per well. Serial dilutions of VLS-1272 were added, and Dynabeads human T cell activator CD3/CD28 beads (Thermo Fisher Scientific 11161D) were added to each well at a bead-to-cell ratio of 1:6. Plates were placed in the incubator for 72 h before CTG readout.
In vitro immunofluorescence assays
Cells were plated 24 h before treatment in a 96 well plate (Perkin Elmer 6055300). Cells were treated with compound for 48 h before being fixed in cold methanol for 15 min. After permeabilization with 0.1% Triton X-100/PBS for 10 min, samples were blocked with 5% BSA/PBS prior to incubation with primary antibodies. Primary and secondary antibodies were diluted to the desired concentrations in 5% BSA/PBS and each were incubated at room temp for 1 h in between washes. Primary antibodies used in these studies included mouse anti-α-tubulin clone DM1A (1:2000, Millipore Sigma T6199), rat anti-phospho Histone H3 (Ser10) clone 6G8B7 (1:2000, Millipore Sigma (05-636-I)), rabbit anti-KIF18A (1:500, Fortis Lifesciences (A301-080A)). Secondary antibodies used in these studies included goat anti-mouse IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (1:2000 (IF), Thermo Fisher Scientific (A11001)), Goat anti-Rat IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 (1:2000 (IF), Thermo Fisher Scientific (A11077)), Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 (1:2000 (IF), Thermo Fisher Scientific (A21244)). Cells were also stained with 1 ug/mL DAPI (Sigma Aldrich) to stain the DNA. Images were acquired using an Operetta CLS High Content Analysis System and images and cellular features were identified and analyzed using Harmony High-Content Imaging and Analysis Software (PerkinElmer). Plates were scanned with a 10x objective (NA 0.3, WD 5.2 mm) to identify and count mitotic cells positive for pHH3 staining versus the total number of identified nuclei. Z-stacks of mitotic cells (pHH3-positive) were then rescanned with a 63x water immersion objective (NA 1.15, WD 0.6 mm) to collect images for quantitative analysis. The maximum projection of the recorded Z-stacks was used for representative images and for quantitative analyses. Using the Harmony software, quantification of KIF18A mis-localization was measured by determining the ratio of KIF18A staining that overlapped with α-tubulin versus DAPI-stained DNA. For determination of chromosome congression, the area of DAPI-stained DNA in the pHH3-positive cells was measured and quantified.
Micronuclei assay
Cells were cultured for 3 days and fixed prior to reaching confluence. Cells were stained with 4’,6-Diamidine-2’-phenylindole dihydrochloride (DAPI) and images were acquired using a 20x water immersion objective on the Operetta CLS high-content imaging system (Perkin Elmer). Micronuclei were quantified with Harmony software (Perkin Elmer).
Cell cycle profiling
Cells were plated 24 h before treatment in a 6 well plate. Cells were treated with compound for 48 h before trypsinization and fixation by standard methods through the addition of 500 μL ice cold methanol, added dropwise with vortexing. Cells were washed with PBS and stained using FxCycle PI/RNase Staining Solution (Thermo Fisher F10797). DNA content was measured on a Northern Lights instrument (Cytek). The entire singlet population was analyzed with a gate that was created to capture the population of cells positively stained with the FxCycle PI/RNase Staining Solution (Supplementary Fig. 1). Data analysis of the cell cycle profile was analyzed in FloJo using the Dean-Jett-Fox model.
Synthesis of VLS-1272
The synthetic scheme of VLS-1272 is shown in Supplementary Fig. 8.
All reagents were purchased from commercial sources and used without purification. NMR spectra were obtained on a Bruker AMX-400 instrument. Chemical shifts are reported in parts per million (ppm, δ) downfield from tetramethylsilane. Mass spectra were obtained with ESI from a Shimazdu LCMS-2020 system.
Synthesis of methyl 3-(N-(tert-butyl)sulfamoyl)benzoate (2) To a solution of 2-methylpropan-2-amine (0.27 mL, 2.6 mmol) in 5 mL of CH2Cl2 were added iPr2NEt (1.3 mL, 7.7 mmol) and methyl 3-chlorosulfonylbenzoate (0.90 g, 3.8 mmol). The mixture was stirred at 20 °C for 2 h, concentrated, combined with 20 mL of H2O, and was extracted with EtOAc (2 ×10 mL). The combined extracts were washed with brine (10 mL), dried over anhydrous Na2SO4, filtered, and concentrated methyl methyl 3-(N-(tert-butyl)sulfamoyl)benzoate (2, 1.0 g, ~96% crude) as yellow oil used without purification.
Synthesis of 3-(N-(tert-butyl)sulfamoyl)benzoic acid (3)40. To a solution of 2 (0.90 g, 3.3 mmol) in 6 mL of THF and 2 mL of H2O was added LiOH monohydrate (0.70 g, 17 mmol). The mixture was stirred at 20 °C for 2 h, concentrated to about 2 mL, and the pH was adjusted to 3 by the addition of 4 M HCl. The mixture was combined with 10 mL of H2O and extracted with EtOAc (2 × 10 mL). The extracts were combined, washed with 10 mL of brine, dried over anhydrous Na2SO4, filtered, and concentrated to provide 3-(N-(tert-butyl)sulfamoyl)benzoic acid (3, 0.60 g, 70% yield, 99% purity) as a white solid. 1H NMR (400 MHz, DMSO-d6) δ ppm 13.51 (br s, 1H), 8.38 (s, 1H), 8.11–8.13 (m, 1H), 8.04–8.06 (m, 1H), 7.68–7.70 (m, 2H), 1.09 (s, 9H).
Synthesis of ethyl spiro[2.5]octane-6-carboxylate (5) To a 0 °C solution of 1.5 L of 1 M Et2Zn in hexanes with 0.90 L of CH2Cl2 was added dropwise a mixture of TFA (0.17 kg, 1.50 mol) and 300 mL of CH2Cl2 over 2.5 h. The mixture was stirred at 0 °C for 1 h, and a mixture of CH2I2 (0.12 L, 1.5 mol) in 300 mL of CH2Cl2 was added slowly at 0 °C. After stirring at 0 °C for 1 h, ethyl 4-methylenecyclohexanecarboxylate (0.1 kg, 0.54 mmol, 90% purity) in 300 mL of CH2Cl2 was added slowly at 0 °C. The mixture was stirred at 20 °C for 2 h, the reaction was poured into sat 2 L of NH4Cl and the resulting mixture was extracted with PE (800 mL). The organic phase was washed with 100 mL of brine, dried over anhydrous Na2SO4, filtered, concentrated, and purified by flash chromatography (5% EtOAc in petroleum ether) to provide compound ethyl spiro[2.5]octane-6-carboxylate (5, 0.10 kg, 87% yield) as a colorless oil. ESI MS calcd for C11H15NO4S, 257.07; found, 256.0 [M-H]-. 1H NMR (400 MHz, CDCl3) δ ppm 4.11–4.16 (m, 2H), 2.29-2.32 (m, 1H), 1.88-1.91 (m, 2H), 1.64–1.68 (m, 4H), 1.24-1.28 (t, 3H), 0.97-0.98 (m, 2H), 0.21-0.28 (m, 4H).
Synthesis of spiro[2.5]octan-6-ylmethanol (6) To a suspension of LiAlH4 (63 g, 1.7 mol) in 2.1 L of THF was added 5 (0.30 kg, 1.7 mol) in 300 mL of THF at 0 °C. The mixture was stirred at 0 °C for 1 h, then 62.4 mL of H2O was added slowly to maintain the temperature between 5 and 10 °C. A 15% aqueous solution of NaOH (62.4 mL), and an additional 187 mL of H2O were added. The mixture was filtered the filtrate was concentrated to provide compound spiro[2.5]octan-6-ylmethanol (6, 0.80 kg, 86% yield) as a colorless oil. 1H NMR (400 MHz, CDCl3) δ ppm 3.49 (d, J = 2.4 Hz, 2H), 1.70–1.76 (m, 4H), 1.49–1.51 (m, 1H), 1.14–1.15 (m, 2H), 0.88–0.91 (m, 2H), 0.17–0.27 (m, 4H).
Synthesis of spiro[2.5]octane-6-carbaldehyde (7)76 To a solution of 6 (0.20 kg, 1.4 mol) in 2 L of CH2Cl2 was added pyridinium chlorochromate (0.46 kg, 2.1 mol). The mixture was stirred at 20 °C for 1 h. Silica gel (0.50 kg) was added and the mixture was filter through a short column of Celite and silica gel, washing with CH2Cl2 to afford 2.5 L of brown solution of spiro[2.5]octane-6-carbaldehyde (7, assumed 1.4 mol, ~0.56 M).
Synthesis of 5”-bromodispiro[cyclopropane-1,1’-cyclohexane-4’,3”-indoline] (9) To a 0 °C solution of 7 (0.20 kg, 1.5 mmol) in 2.3 L of CH2Cl2 was slowly added 4-bromophenyl)hydrazine hydrochloride (0.26 kg, 1.2 mol). The mixture was stirred at 0 °C for 30 min, and 0.41 kg of TFA (3.6 mol) was added, and then the mixture was stirred at 20 °C for 12 h. To the mixture was added 1.1 kg of TFA (9.2 mol) and Et3SiH (0.54 kg, 4.6 mmol) at 0 °C. After stirring at 20 °C for 2 h, the mixture was concentrated and combined with 1.5 L of EtOAc and 1.5 L of H2O, the pH was adjusted to 7 with saturated NaHCO3, and the layers were separated. The organic phase was washed with 1 L of brine, dried over Na2SO4, filtered, and concentrated. The residue was triturated with 1 L of petroleum ether for 30 min to provide 0.11 kg of 5”-bromodispiro- [cyclopropane-1,1’-cyclohexane-4’,3”-indoline] (9). The mother liquor was concentrated and purified by flash chromatography (5-33% EtOAc in petroleum ether) to provide an additional 33 g of 9. Total yield: 0.14 kg, 31% yield as a gray solid. ESI MS calcd for C15H18BrN, 291.06/293.06; found, 292.1/294.1 [M + H]+. 1H NMR (400 MHz, CDCl3) δ 7.17 (d, J = 2.00 Hz, 1H), 7.14-7.08 (m, 1H), 6.52–6.48 (m, 1H), 3.51–3.44 (m, 2H), 2.17–1.95 (m, 2H), 1.77–1.71 (m, 4H), 1.00–0.87 (m, 2H), 0.42–0.23 (m, 4H).
Synthesis of 3-(5”-bromodispiro[cyclopropane-1,1’-cyclohexane-4’,3”-indoline]-1”-carbonyl)-N-(tert-butyl)benzenesulfonamide (10) To a solution of 3 (3.0 g, 12 mmol) in 35 mL of CH2Cl2 was added HATU (9.0 g, 24 mmol) and iPr2NEt (7.5 mL, 43 mmol). The mixture was stirred at 0 °C for 15 min, and then 9 (3.7 g, 13 mmol) was added. The resulting mixture was stirred at 20 °C for 12 h, combined with 50 mL of EtOAc (50.0 mL) and washed with H2O (40.0 mL x 3) and 40 mL of brine, dried over Na2SO4, filtered, and concentrated to give 7.0 g of black solid, which was triturated with 28 mL of iPrOH (28.0 mL) at 20 °C for 45 min. The suspension was filtered to provide 3-(5”-bromodispiro[cyclopropane-1,1’-cyclohexane-4’,3”-indoline]-1”-carbonyl)-N-(tert-butyl)benzenesulfonamide (10, 4.2 g, 79% purity, 53% yield) as a brown solid. ESI MS calcd for C26H31BrN2O3S, 530.12/532.12; found, 531.2/533.2 [M + H]+. 1H NMR (400 MHz, DMSO-d6) δ ppm 8.04 (s, 1H), 7.99 (d, J = 7.2 Hz 2H), 7.85 (d, J = 7.2 Hz, 1H), 7.73–7.69 (t, J = 7.2 Hz, 1H), 7.67 (s, 1H), 7.51 (s, 1H), 7.41 (br s, 1H), 3.90 (s, 2H), 1.77–1.74 (m, 2H), 1.65–1.62 (m, 4H), 1.11 (s, 9H), 0.84–0.81 (br d, J = 12.8 Hz, 2H), 0.26 (m, 4H).
Synthesis of N-(tert-butyl)-3-(5”-(methylsulfonamido)dispiro[cyclopropane-1,1’-cyclohexane-4’,3”-indoline]-1”-carbonyl)benzenesulfonamide (11) A mixture of 15 g of 10 (28 mmol), 4.3 g of CuI (22 mmol), 8 g of methanesulfonamide (84 mmol), 3.2 g of N1,N2-dimethylcyclohexane-1,2-diamine (22 mmol), and 18 g of K3PO4 (84 mmol) in 150 mL of degassed DMF was stirred at 160 °C for 3 h under an N2 atmosphere. The reaction mixture was poured in 1 L of H2O and extracted twice with 0.60 L of EtOAc. The extracts were combined, washed with 0.10 L of brine, dried over anhydrous Na2SO4, filtered, and concentrated. The residue was stirred in 0.15 L of MeOH at 50 C for 2 h, filtered, and the filter cake was washed twice with 10 mL of MeOH and dried under vacuum to provide N-(tert-butyl)-3-(5”-(methylsulfonamido)dispiro[cyclopropane-1,1’-cyclohexane-4’,3”-indoline]-1”-carbonyl)benzenesulfonamide (VLS-1272), 9.1 g, 57% yield, 99.1% purity (Supplementary Fig. 9) as an off-white solid. ESI HR-MS calcd for C27H35N3O5S2, 545.2018; found, 546.2100 [M + H]+. 1H NMR (400 MHz, DMSO-d6) δ ppm 9.58 (br s, 1H), 8.11–7.91 (m, 3H), 7.88–7.67 (m, 1H), 7.74–7.59 (m, 2H), 7.14 (d, J = 1.88 Hz, 2H) 3.88 (br d, J = 1.63 Hz, 2H), 2.94 (s, 3H), 1.74–1.49 (m, 6H), 1.11 (s, 9H), 0.94–0.80 (m, 2H), 0.34–0.18 (m, 4H). 13C NMR (100 MHz, DMSO-d6) δ ppm 11.4, 11.6, 17.7, 29.7, 31.3, 35.3, 44.0, 53.4, 59.9, 115.6, 117.4, 120.1, 124.7, 127.8, 130.0, 130.5, 134.7, 137.4, 138.1, 142.3, 144.6, 166.4. The 1H NMR and 13C NMR traces are shown in Supplementary Fig. 10-12.
Confirmation of structure of VLS-1272 by single-crystal X-ray crystallography
12 mg VLS-1272 was dissolved in 1 mL CH2Cl2/methanol (1:1) and kept in a half sealed 4 mL vial. The solution evaporates slowly at rt. Crystals were observed on the second day (Supplementary Fig. 13).
Empirical formula: C27H35N3O5S; Formula weight545.70; Temperature: 300.6(9) K; Crystal system: triclinic; Space group: P-1; a: 9.71930(10) Å; b: 11.0906(2) Å; c 13.0749(2) Å; α: 97.4860(10) °; β: 94.1990(10) °; γ: 100.0080(10) °; Volume: 1369.46(4) Å3; Z: 2; ρcalcg: 1.323 cm3; μ: 2.107 mm−1; F(000): 580.0; Crystal size: 0.2 × 0.2 × 0.05 mm3; Radiation: CuKα (λ = 1.54184); 2Θ range for data collection: 6.852 to 133.202°; Index ranges: −11 ≤ h ≤ 11, −13 ≤ k ≤ 13, −12 ≤ l ≤ 15; Reflections collected: 23964; Independent reflections: 4833 [Rint = 0.0500, Rsigma = 0.0321]; Data/restraints/parameter: 4833/40/342; Goodness-of-fit on F2: 1.074; Final R indexes [I > = 2σ (I)]: R1 = 0.0456, wR2 = 0.1219; Final R indexes [all data]: R1 = 0.0499, wR2 = 0.1260.
Preparation of orally formulated VLS-1272
To prepare spray-dried dispersion of VLS-1272 (VLS-1272-SDD), VLS-1272 (5.0 g) was dissolved in 500 mL of 80/20 CH2Cl2 and MeOH. HPMC AS-MG (15 g, Shin-Etsu) was added and stirred until dissolved. Spray drying was conducted using a mini spray dryer (Buchi B-290) with the following parameters: high performance cyclone, inlet temperature 79–83 °C, outlet temperature 55–58 °C, pump 10%, N2 flow rate 40 min, aspirator 100%. The resulting product was placed in a vacuum drying oven at RT overnight, then at 40 °C for 6 h to provide 17.1 g (86% yield) of VLS-1272-SDD (26.6% API, 73.4% HPMC AS-MG). The product was confirmed amorphous by XRPD and packed in portions in sealed foil bags at 4 °C and warmed to room-temperature before opening for use.
Biochemistry assays supporting development of VLS-1272
The sequence for KIF18A protein (amino acid residues 1-374) and GFP-tagged KIF18A (amino acid residues 1-480) was cloned into the pFastBac1-His-linker-TEV backbone and expressed in a baculovirus expression system. Protein was purified using analytical columns on an HPLC for biochemical testing.
Biochemical assays used for compound screening were run in a reaction buffer consisting of 80 mM PIPES (pH 6.9), 1 mM MgCl2, 75 mM KCl, 1 mM EGTA, 1 mM DTT, 0.01% BSA, 0.005% Tween-20, and 10 µM Taxol. Compounds or DMSO were dispensed into 384-well plates with an Echo liquid handler (Beckman). KIF18A (aa 1-374) diluted to 4 nM in the reaction buffer was added and pre-incubated with compounds or DMSO for 30 min at room temperature. A substrate mix containing 0.2 mg/mL microtubules and 2 mM ATP diluted in reaction buffer was added 1:1 with enzyme mix. Reactions were carried out for 60 min at 28 °C. ADP-Glo Max R1 was added 1:1 to the reaction mixture and incubated at room temperature for 4 h. ADP-Glo Max R2 was then added 1:1 to the reaction mixture and incubated at room temperature for 1 h before reading the luminescence on a plate reader. For side-by-side comparison of VLS-1272 and AM-1882, reactions were performed with 1.25 nM KIF18A and 50 uM ATP for 30 min. AM-1882 was purchased from MedChemExpress (KIF18A-IN-1; Cat. No.: HY-145034). For ATP-dependence assays, the concentration of enzyme varied with ATP concentration, with 0.25 nM KIF18A used with 10, 33, and 100 uM ATP, 1 nM KIF18A with 330 nM ATP and 2 nM KIF18A with 1000 nM ATP. IC50 values were calculated using curve-fitting in GraphPad Prism software. For global progress curve analysis of time/dose-dependent inhibition of KIF18A, lower enzyme concentration (0.25 nM final) was used to slow down ATP depletion overnight to maintain steady state enzyme kinetics. ATP concentrations used with mouse KIF18A, KIF19, KIF18B, KIF11 and KIFC1 are indicated in Table 1.
Docking of VLS-1272 and AM-1882 in KIF18A homology model
The published homology model40 was prepared in the Molecular Operating Environment (MOE) software, Version 2022.02 (Chemical Computing Group ULC, 2022) using the QuikPrep protocol with default settings. VLS-1272 and AM-1882 were docked into the site defined by the ligand in the homology model using the Triangle Matcher method for initial placement. The top 30 poses by the London dG score were refined by the Induced Fit method. The top five poses by the GBVI/WSA dG score were collected and analyzed. Figures were prepared in the PyMOL Molecular Graphics System, Version 2.5.2, Schrödinger, LLC.
Polymerized microtubules for gliding motility assay
X-Rhodamine labeled polymerized microtubules were prepared by diluting lyophilized X-Rhodamine labeled tubulin (Cytoskeleton, TL620M, porcine) to a final concentration of 2 mg/mL in 1X BRB80 (80 mM K-PIPES pH 6.8, 1 mM EGTA, 1 mM MgCl2) with 2 mM GTP. The labeled tubulin was then combined with 4X the volume of 5 mg/mL unlabeled tubulin (Cytoskeleton, T240, porcine) diluted in 1X BRB80 with 2 mM GTP. The combined labeled and unlabeled tubulin was clarified by spinning in a tabletop Optima TLX ultracentrifuge with a TLA-100 rotor (Beckman) for 10 min, 95,000 RPM, at 4 °C. The subsequent supernatant was incubated at 37 °C for 20 min. Taxol was added to a final concentration of 200 μM, and the solution was incubated for an additional 20 min at 37 °C. The subsequent polymerized microtubules were kept protected from light prior to use and diluted to a final concentration of 25 μg/mL.
Gliding motility assay
Flow cell chambers were assembled using a 25 × 75 mm glass slide (Thermo Scientific, 3011-002), a 24 × 60 mm coverslip (Deckgläser), and two pieces of double-sided tape which resulted in a ∼20 μl chamber. 50 μL of 10% Anti-GFP (3E6)-Mouse Antibody (Life Tech, A-11120) diluted in 1X BRB80 was incubated in the flow cell chamber for ten minutes. 330 nM purified KIF18A (1-480) His-cleaved GFP protein diluted in 50 μL Flow Cell Buffer (FCB; 1X BRB80, 100 mM KCl, 3 mM MgATP, 1 μM taxol, 1 mM DTT, 0.01% BSA and 1X Oxygen Scavenger Mix (OSM; 0.045 mg/mL catalase, 0.066 mg/mL glucose oxidase, and 5.8 mg/mL glucose)) was added and incubated at room temperature for five minutes. The chamber was washed with 50 μL Wash Buffer (WB; 1× BRB80, 300 mM KCl, 3 mM MgATP, and 0.01% BSA) and then incubated with 50 μL 1% Pluronic F-127 (Sigma, p2443) for 1 minute to passivate the surface of the chamber. The chamber was then washed with 50 μL of FCB. 50 μL of 25 μg/mL X-Rhodamine labeled taxol stabilized microtubules diluted in FCB or 50 μL of 25 μg/mL X-Rhodamine labeled taxol stabilized microtubules combined with 25 nM VLS-1272 diluted in FCB was added to the flow cell chamber and incubated for five minutes prior to imaging. Microtubule gliding was imaged with HiLo microscopy on a Ti2E inverted microscope (Nikon Instruments) with LUN-F laser launch and iLas2 TIRF/photostimulation controller driven by NIS Elements with 100X Apo TIRF (1.49 numerical aperture) objective lens collecting images every 3 s for a duration of 3 min. Images were captured using an iXon Life 897 EMCCD camera with flow chambers maintained in an Okolab environmental chamber at 37 °C. To quantify microtubule gliding velocity, the tips of microtubules were tracked using MTrackJ in ImageJ/Fiji. The total microtubule displacement distance was then divided by the duration of the track to determine gliding velocity. Mean and standard deviation are reported for three replicates. The total number of tracks analyzed was 249 KIF18A 1-480 without inhibitor and 177 for KIF18A 1-480 with VLS-1272.
Profiling of VLS-1272 across a cell line panel
To determine the effect of VLS-1272 on viability in a large cell line panel, cells were plated 24 h before administration of different doses of VLS-1272 with a top dose of 3 µM. Cells were treated for 168 h before performing CTG assays as described previously.
For each of the 198 cell lines treated with VLS-1272, area under the curve (AUC) was calculated from viability assessments after 7-day treatment. AUC = 1 indicates no effect on viability, and AUC = 0 indicates complete inhibition of proliferation.
Comparison of the VLS-1272 profile to the profile of each gene in the DepMap dataset
Spearman’s rho was used to compare the AUC profile of VLS-1272 to each gene knockdown effect profile in the DepMap RNAi dataset. P-values were subsequently adjusted for multiple testing using the Benjamini-Hochberg method. This process was repeated to compare the AUC profile of VLS-1272 to each gene knockout effect profile in the DepMap CRISPR dataset. For these analyses the ‘CRISPR_(DepMap_Public_23Q2+Score, Chronos).csv’ and ‘RNAi_(Achilles+DRIVE+Marcotte,_DEMETER2).csv’ files were obtained from DEPMAP (V23Q2 release)23–28, and only genes with at least 20 cell lines to compare were included. A median of 114 cell lines for each of 16,789 genes from the RNAi dataset and a median of 192 cell lines for each of 17,928 genes from the CRISPR dataset were used.
Comparison of the VLS-1272 profile to the profile of anti-mitotic drugs in the PRISM dataset
AUC values for a curated list of 21 anti-mitotic compounds profiled in 128 cell lines that were also present in the VLS-1272 panel were obtained from the ‘secondary-screen-dose-response-curve-parameters.csv’ file from DEPMAP PRISM55. In some instances, data was not readily available for all 21 anti-mitotic compounds in all of the 128 cell lines. As suggested by PRISM, screen data from ‘MTS010’ was preferentially used when available. Spearman’s rho was used to compare the response in each cell line for all pairwise combinations of the 22 compounds using complete observations only. As a control, Spearman’s rho was also used to compare response to each compound with the KIF18A values from the CRISPR and RNAi DepMap files mentioned above23–28.
In vivo PK and CBC studies
For single dose PK studies, 6–8-week-old male CD-1 mice (Jihui Laboratory Animal Co. Ltd.) were administered 1 mg/kg VLS-1272 intravenously into the tail vein, or 30 mg/kg VLS-1272-SDD (vehicle: 5% TPGS (Tocofersolan (MCE cat #109605)) and 0.5% Methylcellulose (400cp) (Aladdin, cat #K2005343) in ddH2O) via oral gavage. Blood was collected at the indicated timepoints and drug concentrations in the plasma were measured.
For multi-day dosing of non-tumor bearing mice, female 6–8-week-old SCID Beige mice (Jihui Laboratory Animal Co. Ltd.) were dosed twice daily by oral gavage with 30 mg/kg or 60 mg/kg of VLS-1272-SDD. Blood was collected and hematological lineages were analyzed by complete blood count using Sysmex XN-1000 Hematology Analyzer.
Tumor growth inhibition studies
HCC15 xenograft efficacy studies were initiated by culturing the cells in RPMI-1640 medium supplemented with 10% fetal bovine serum and 1% Anti-Anti at 37 °C in an atmosphere of 5% CO2 in air. OVCAR-3 xenograft efficacy studies were initiated by culturing the cells in RPMI-1640 medium supplemented with 20% fetal bovine serum, 0.01 mg/mL bovine insulin and 1% Anti-Anti at 37 °C in an atmosphere of 5% CO2 in air. The tumor cells were routinely subcultured twice weekly and were harvested while growing in an exponential growth phase and counted for tumor inoculation. HCC15 and OVCAR-3 tumor cells (10 × 106 cells) were each mixed in 0.2 mL of PBS with Matrigel (at a 50:50 ratio) and were inoculated subcutaneously on the right flank of each mouse. In the HCC15 study, 6–8-week-old female SCID Beige mice (Beijing Vital River Laboratory Animal Technology Co., Ltd.) were used. In the OVCAR-3 study, 6–8-week-old female BALB/c nude mice (Beijing Vital River Laboratory Animal Technology Co., Ltd.) were used. When the average HCC15 tumor volume reached about 111 mm3, the animals were randomized, and treatment was initiated. For the HCC15 study, nutrient gel and sunflower seeds were provided to all groups as indicated in the Source Data for Supplementary Fig. 7a. When the average OVCAR-3 tumor volume reached about 173 mm3, the animals were randomly grouped, and treatment was initiated. Vehicle (5% TPGS (Tocofersolan MCE cat #109605) and 0.5% Methylcellulose (400cp) (Aladdin, cat #K2005343) in ddH2O) or VLS-1272-SDD suspended in vehicle were dosed by oral gavage. Tumor sizes were measured twice per week in two dimensions using a caliper. Tumor volume is expressed in mm3 using the formula: V = 0.5 a x b2 where a and b are the long and short diameters of the tumor, respectively.
Tumor dissection/preparation
Tumors were transferred to centrifuge tubes containing formalin. Tissue was processed and embedded according to standard methods using a tissue processor (Leica ASP300S) and a tissue embedding center (Leico EG 1150H + C), respectively. Sections of 4 μm thickness were made using a manual rotary microtome (Leica RM2235) for IHC and H&E.
pHH3 staining of tumor sections
Automated IHC and H&E were performed with the BondRx platform. IHC for pHH3 was run using Bond Epitope Retrieval Solution 1 (Leica (AR9961)) for 20 min at 100 °C. pHH3 primary antibody dilution of 1:3200 (Millipore (06-750) was used for staining. H&E was run using Bond Polymer Refine Detection (Leica DS9800), hematoxylin (BASO BA-4041) and eosin (BASO BA-4022) for 5 min each. All slides were scanned on an Aperio Versa (Leica) at 20x magnification.
KIF18A immunofluorescence of in vivo samples
KIF18A co-immunofluorescence with α -tubulin in HCC15 tumor xenografts was performed on paraffin cross-sections through end-stage tumors (day 35–40 of twice daily dosing beginning when tumors reached 150 mm in diameter). Tissue sections were dewaxed with Xylenes (Fisher) and rehydrated through a graded ethanol series. Heat-mediated antigen retrieval was performed with pH 6 sodium citrate buffer (Sigma-Aldrich, C9999), prior to washing of sections in running tap water and 1X phosphate buffered saline (PBS, Fisher). Sections were permeabilized with 0.3% Triton X-100 (Sigma) and blocked with 2% bovine serum albumin (BSA, Sigma-Aldrich, A9647) and 5% goat serum in 0.3% Triton X-100/PBS. Primary antibodies for KIF18A (Sigma-Aldrich, HPA039484, 1:250) and α-tubulin (Cell Signaling Technologies, 3873S, clone: [DM1A], 1:1000) were applied overnight in a humidified chamber at 4 °C. Following PBS washes, sections were incubated with secondary antibodies for 2 h at room temperature in a humidified chamber using Alexa Fluor-568 goat anti-rabbit IgG (Invitrogen, A11011) and Alexa Fluor-488 goat anti-mouse (Invitrogen, A11001) at 1:400. After PBS washes and nuclear counter-staining with DAPI (0.4 mg/ml in PBS), sections were mounted with Prolong Gold Anti-fade (Invitrogen, P36934). Images were captured using a Zeiss 880 Laser Scanning Confocal Airyscan microscope.
IF quantification was performed on confocal Z-slice snapshots captured using Bitplane Imaris imaging software. Following channel separation with FIJI software, Intermodes auto-thresholds were applied to the α-tubulin and DAPI-highlighted regions. The generated masks were overlayed with the KIF18A fluorescence to quantify the ratio of KIF18A fluorescence co-localized with α-tubulin over KIF18A co-localized with DAPI (FIJI). For each study, data was collected from 5 mice per treatment group from 6-7 fields per tumor. Chromosomal congression was quantified from the surface area of DAPI signal captured from the same confocal Z-slice snapshots (Bitplane Imaris, FIJI).
Statistics & reproducibility
No statistical method was used to predetermine sample size. No data were excluded from the analyses. Randomization was performed in grouping tumor-bearing mice into vehicle and compound treatment groups. Investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank Aliasghar Mortazi and Ethan Benjamin for computational analysis, Song Chen for flow cytometry assistance, and our contract research partners for their contributions: Yuzhou Xu and Ning Zheng for biochemistry at Shanghai ChemPartner Co., Ltd.; Yingjia Zhang, Xiling Wang and Chaoran Chen for cell biology at Shanghai ChemPartner Co., Ltd., and Yao (Sonia) Shuai for in vivo pharmacology at WuXi AppTec Co., Ltd. J.S. is supported by grant NIH R35 GM144133 and K.A.Q. is supported by grant NSF GRF 1842491.
Author contributions
Data acquisition and analysis by A.F.P., R.S., M.C.C., T.Y.F., M.D.A., A.C., R.L., N.B., K.A.Q. and A.R.R. Computational analysis by M.J., A.V., R.S.D. and D.A.C. Experimental design, data analysis and study supervision by A.F.P., R.Z., M.D.A., J.S., C.A-A., S.E.B., R.S.D., S.D., D.A.C. and C.H.E. Conceptualization and project design by R.Z., S.M.S., D.A.C. and C.H.E. Manuscript preparation and revisions by A.F.P., D.A.C. and C.H.E.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The data that support the findings of this study are provided within the paper, Supplementary Information, Supplementary Data, and Source Data files. DEPMAP cancer cell line data is from public sources (https://depmap.org/portal/), with analyses using the versions or files specified in the Methods. The published human KIF18A crystal structure (3LRE [10.2210/pdb3lre/pdb]) was used for molecular docking. Source data are provided with this paper.
Competing interests
A.F.P., R.Z., M.J., R.S., M.C.C., A.V., T.Y.F., M.D.A., A.C., R.L., N.B., C.A-A., S.E.B., R.S.D., S.D., S.M.S., D.A.C. and C.H.E. are current or former employees of Volastra Therapeutics and may own equity in Volastra Therapeutics. J.S. received funding from Volastra Therapeutics. D.A.C. is an inventor on a patent (WO/2023/028564) filed by Volastra Therapeutics describing KIF18A inhibitors including VLS-1272. D.A.C., S.M.S. and S.D. are inventors on a patent (WO/2024/178255) filed by Volastra Therapeutics describing formulations of KIF18A inhibitors including VLS-1272.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55300-z.
References
- 1.McGranahan, N., Burrell, R. A., Endesfelder, D., Novelli, M. R. & Swanton, C. Cancer chromosomal instability: therapeutic and diagnostic challenges. Embo Rep.13, 528–538 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakhoum, S. F., Danilova, O. V., Kaur, P., Levy, N. B. & Compton, D. A. Chromosomal Instability Substantiates Poor Prognosis in Patients with Diffuse Large B-cell Lymphoma. Clin. Cancer Res.17, 7704–7711 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi, C.-M. et al. Chromosomal instability is a risk factor for poor prognosis of adenocarcinoma of the lung: Fluorescence in situ hybridization analysis of paraffin-embedded tissue from Korean patients. Lung Cancer64, 66–70 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Carter, S. L., Eklund, A. C., Kohane, I. S., Harris, L. N. & Szallasi, Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet38, 1043–1048 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Walther, A., Houlston, R. & Tomlinson, I. Association between chromosomal instability and prognosis in colorectal cancer: a meta-analysis. Gut57, 941 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Sansregret, L., Vanhaesebroeck, B. & Swanton, C. Determinants and clinical implications of chromosomal instability in cancer. Nat. Rev. Clin. Oncol.15, 139–150 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Lee, A. J. X. et al. Chromosomal Instability Confers Intrinsic Multidrug Resistance. Cancer Res71, 1858–1870 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lukow, D. A. et al. Chromosomal instability accelerates the evolution of resistance to anti-cancer therapies. Dev. Cell10.1016/j.devcel.2021.07.009 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bakhoum, S. F. et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature553, 467–472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz, G. K. et al. Phase I study of barasertib (AZD1152), a selective inhibitor of Aurora B kinase, in patients with advanced solid tumors. Invest N. Drug31, 370–380 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Manfredi, M. G. et al. Characterization of Alisertib (MLN8237), an Investigational Small-Molecule Inhibitor of Aurora A Kinase Using Novel In Vivo Pharmacodynamic Assays. Clin. Cancer Res.17, 7614–7624 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Olmos, D. et al. Phase I Study of GSK461364, a Specific and Competitive Polo-like Kinase 1 Inhibitor, in Patients with Advanced Solid Malignancies. Clin. Cancer Res.17, 3420–3430 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Wengner, A. M. et al. Novel Mps1 Kinase Inhibitors with Potent Antitumor Activity. Mol. Cancer Ther.15, 583–592 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Yan, V. C. et al. Why Great Mitotic Inhibitors Make Poor Cancer Drugs. Trends Cancer6, 924–941 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayr, M. I., Storch, M., Howard, J. & Mayer, T. U. A Non-Motor Microtubule Binding Site Is Essential for the High Processivity and Mitotic Function of Kinesin-8 Kif18A. Plos One6, e27471 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stumpff, J., Wagenbach, M., Franck, A., Asbury, C. L. & Wordeman, L. Kif18A and Chromokinesins Confine Centromere Movements via Microtubule Growth Suppression and Spatial Control of Kinetochore Tension. Dev. Cell22, 1017–1029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stumpff, J., Dassow, G., von, Wagenbach, M., Asbury, C. & Wordeman, L. The Kinesin-8 Motor Kif18A Suppresses Kinetochore Movements to Control Mitotic Chromosome Alignment. Dev. Cell14, 252–262 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marquis, C. et al. Chromosomally unstable tumor cells specifically require KIF18A for proliferation. Nat. Commun.12, 1213 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen-Sharir, Y. et al. Aneuploidy renders cancer cells vulnerable to mitotic checkpoint inhibition. Nature 1–6 10.1038/s41586-020-03114-6 (2021). [DOI] [PMC free article] [PubMed]
- 20.Quinton, R. J. et al. Whole-genome doubling confers unique genetic vulnerabilities on tumour cells. Nature 1–6 10.1038/s41586-020-03133-3 (2021). [DOI] [PMC free article] [PubMed]
- 21.Liu, X. et al. Germinal Cell Aplasia in Kif18a Mutant Male Mice Due to Impaired Chromosome Congression and Dysregulated BubR1 and CENP-E. Genes Cancer1, 26–39 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reinholdt, L. G., Munroe, R. J., Kamdar, S. & Schimenti, J. C. The mouse gcd2 mutation causes primordial germ cell depletion. Mech. Dev.123, 559–569 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Cancer Dependency Map. https://depmap.org/portal/depmap/.
- 24.Meyers, R. M. et al. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat. Genet49, 1779–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dempster, J. M. et al. Extracting Biological Insights from the Project Achilles Genome-Scale CRISPR Screens in Cancer Cell Lines. Biorxiv 720243 10.1101/720243 (2019).
- 26.Dempster, J. M. et al. Chronos: a cell population dynamics model of CRISPR experiments that improves inference of gene fitness effects. Genome Biol.22, 343 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pacini, C. et al. Integrated cross-study datasets of genetic dependencies in cancer. Nat. Commun.12, 1661 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghandi, M. et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu, Z. et al. Deep learning predicts chromosomal instability from histopathology images. Iscience24, 102394 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bakhoum, S. F. et al. Numerical chromosomal instability mediates susceptibility to radiation treatment. Nat. Commun.6, 5990 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakhoum, S. F., Thompson, S. L., Manning, A. L. & Compton, D. A. Genome stability is ensured by temporal control of kinetochore–microtubule dynamics. Nat. Cell Biol.11, 27–35 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laucius, C. D., Orr, B. & Compton, D. A. Chromosomal instability suppresses the growth of K-Ras-induced lung adenomas. Cell Cycle18, 1702–1713 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Czechanski, A. et al. Kif18a is specifically required for mitotic progression during germ line development. Dev. Biol.402, 253–262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang, Y. et al. Defects in chromosome congression and mitotic progression in KIF18A-deficient cells are partly mediated through impaired functions of CENP-E. Cell Cycle8, 2643–2649 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Catarinella, M., Grüner, T., Strittmatter, T., Marx, A. & Mayer, T. U. BTB‐1: A Small Molecule Inhibitor of the Mitotic Motor Protein Kif18A. Angew. Chem. Int Ed.48, 9072–9076 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Payton, M. et al. Small-molecule inhibition of kinesin KIF18A reveals a mitotic vulnerability enriched in chromosomally unstable cancers. Nat. Cancer5, 66–84 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, R. & Wong, K. High performance enzyme kinetics of turnover, activation and inhibition for translational drug discovery. Expert Opin. Drug Dis.12, 17–37 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Lu, H. & Tonge, P. J. Drug–target residence time: critical information for lead optimization. Curr. Opin. Chem. Biol.14, 467–474 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Copeland, R. A. The drug–target residence time model: a 10-year retrospective. Nat. Rev. Drug Discov.15, 87–95 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Tamayo, N. A. et al. Targeting the Mitotic Kinesin KIF18A in Chromosomally Unstable Cancers: Hit Optimization Toward an In Vivo Chemical Probe. J Med. Chem.10.1021/acs.jmedchem.1c02030 (2021). [DOI] [PubMed]
- 41.Niwa, S. et al. KIF19A Is a Microtubule-Depolymerizing Kinesin for Ciliary Length Control. Dev. Cell23, 1167–1175 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Janssen, L. M. E. et al. Loss of Kif18A Results in Spindle Assembly Checkpoint Activation at Microtubule-Attached Kinetochores. Curr. Biol.28, 2685–2696.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Zeng, X., Xu, W. K., Lok, T. M., Ma, H. T. & Poon, R. Y. C. Imbalance of the spindle-assembly checkpoint promotes spindle poison-mediated cytotoxicity with distinct kinetics. Cell Death Dis.10, 314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maan, M. et al. Tank Binding Kinase 1 modulates spindle assembly checkpoint components to regulate mitosis in breast and lung cancer cells. Biochimica Et. Biophysica Acta Bba-Mol. Cell Res1868, 118929 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gliech, C. R. et al. Weakened APC/C activity at mitotic exit drives cancer vulnerability to KIF18A inhibition. EMBO J. 1–29 10.1038/s44318-024-00031-6 (2024). [DOI] [PMC free article] [PubMed]
- 46.Lejonc, J. L., Vernant, J. P., Macquin, I. & Castaigne, A. MYOCARDIAL INFARCTION FOLLOWING VINBLASTINE TREATMENT. Lancet316, 692 (1980). [DOI] [PubMed] [Google Scholar]
- 47.Deangelis, L. M., Gnecco, C., Taylor, L. & Warrell, R. P. Evolution of neuropathy and myopathy during intensive vincristine/corticosteroid chemotherapy for non‐hodgkin’s lymphoma. Cancer67, 2241–2246 (1991). [DOI] [PubMed] [Google Scholar]
- 48.House, K. W., Simon, S. R. & Pugh, R. P. Chemotherapy‐induced myocardial infarction in a young man with hodgkin’s disease. Clin. Cardiol.15, 122–125 (1992). [DOI] [PubMed] [Google Scholar]
- 49.Rowinsky, E. K., Eisenhauer, E. A., Chaudhry, V., Arbuck, S. G. & Donehower, R. C. Clinical toxicities encountered with paclitaxel (Taxol). Semin Oncol.20, 1–15 (1993). [PubMed] [Google Scholar]
- 50.Postma, T. J., Vermorken, J. B., Liefting, A. J. M., Pinedo, H. M. & Heimans, J. J. Paclitaxel-induced neuropathy. Ann. Oncol.6, 489–494 (1995). [DOI] [PubMed] [Google Scholar]
- 51.Forsyth, P. et al. Prospective study of paclitaxel-induced peripheral neuropathy with quantitative sensory testing. J. Neuro-oncol.35, 47–53 (1997). [DOI] [PubMed] [Google Scholar]
- 52.Parmar, M. K. B. et al. Paclitaxel plus platinum-based chemotherapy versus conventional platinum-based chemotherapy in women with relapsed ovarian cancer: the ICON4/AGO-OVAR-2.2 trial. Lancet361, 2099–2106 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Bois et al. A Randomized Clinical Trial of Cisplatin/Paclitaxel Versus Carboplatin/Paclitaxel as First-Line Treatment of Ovarian Cancer. Jnci J. Natl Cancer Inst.95, 1320–1329 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Dermitzakis, E. V. et al. The impact of paclitaxel and carboplatin chemotherapy on the autonomous nervous system of patients with ovarian cancer. Bmc Neurol.16, 190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corsello, S. M. et al. Discovering the anticancer potential of non-oncology drugs by systematic viability profiling. Nat. Cancer1, 235–248 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mayr, M. I. et al. The Human Kinesin Kif18A Is a Motile Microtubule Depolymerase Essential for Chromosome Congression. Curr. Biol.17, 488–498 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Sepaniac, L. A. et al. Micronuclei in Kif18a mutant mice form stable micronuclear envelopes and do not promote tumorigenesis. J. Cell Biol.220, e202101165 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laboratories, C. R. SCID Beige Mouse Hematology. https://www.criver.com/sites/default/files/resources/FoxChaseSCID%C2%AEBeigeMouseClinicalPathologyData.pdf.
- 59.Fonseca, C. L. et al. Mitotic chromosome alignment ensures mitotic fidelity by promoting interchromosomal compaction during anaphaseAlignment ensures formation of a single nucleus. J. Cell Biol.218, 1148–1163 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson, S. L. & Compton, D. A. Examining the link between chromosomal instability and aneuploidy in human cells. J. Cell Biol.180, 665–672 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karg, T., Warecki, B. & Sullivan, W. Aurora B–mediated localized delays in nuclear envelope formation facilitate inclusion of late-segregating chromosome fragments. Mol. Biol. Cell26, 2227–2241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith, L., Plug, A. & Thayer, M. Delayed replication timing leads to delayed mitotic chromosome condensation and chromosomal instability of chromosome translocations. Proc. Natl. Acad. Sci.98, 13300–13305 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bakhoum, S. F. et al. The mitotic origin of chromosomal instability. Curr. Biol.24, R148–R149 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cimini, D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim Biophys. Acta1786, 32–40 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Ben-David, U. & Amon, A. Context is everything: aneuploidy in cancer. Nat. Rev. Genet21, 44–62 (2020). [DOI] [PubMed] [Google Scholar]
- 66.Drews, R. M. et al. A pan-cancer compendium of chromosomal instability. Nature606, 976–983 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nguyen, B. et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell185, 563-575.e11 (2022). [DOI] [PMC free article] [PubMed]
- 68.Szikriszt, B. et al. A comprehensive survey of the mutagenic impact of common cancer cytotoxics. Genome Biol.17, 99 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murugaesu, N. et al. Tracking the Genomic Evolution of Esophageal Adenocarcinoma through Neoadjuvant Chemotherapy. Cancer Discov.5, 821–831 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee, H.-S. et al. Effects of Anticancer Drugs on Chromosome Instability and New Clinical Implications for Tumor-Suppressing Therapies. Cancer Res.76, 902–911 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meyer, M. et al. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc. Natl. Acad. Sci.112, 851–856 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Piotrowska, Z. et al. Heterogeneity Underlies the Emergence of EGFRT790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third-Generation EGFR Inhibitor. Cancer Discov.5, 713–722 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eyler, C. E. et al. Single-cell lineage analysis reveals genetic and epigenetic interplay in glioblastoma drug resistance. Genome Biol.21, 174 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aissa, A. F. et al. Single-cell transcriptional changes associated with drug tolerance and response to combination therapies in cancer. Nat. Commun.12, 1628 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McFarland, J. M. et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat. Commun.9, 4610 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu, H. et al. Inhibitors of Cathepsin S. (2008).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The data that support the findings of this study are provided within the paper, Supplementary Information, Supplementary Data, and Source Data files. DEPMAP cancer cell line data is from public sources (https://depmap.org/portal/), with analyses using the versions or files specified in the Methods. The published human KIF18A crystal structure (3LRE [10.2210/pdb3lre/pdb]) was used for molecular docking. Source data are provided with this paper.