Abstract
Ideal root system architecture (RSA) is important for efficient nutrient uptake and high yield in crops. We cloned and characterized a key RSA regulatory gene, GRAVITROPISM LOSS 1 (OsGLS1), in rice (Oryza sativa L.). The gls1 mutant displays an increased root growth angle, longer primary roots, more adventitious roots and greater nutrient uptake efficiency and grain yield in paddy fields. OsGLS1 is strongly expressed in the root tips of seedlings and in leaves at the flowering stage. OsGLS1 encodes a RING finger E3 ubiquitin ligase mainly localizing at the basal plasma membrane (PM) in several root cell types when phosphorylated on its Ser-30 residue. OsGLS1 interacts with, ubiquitinates and promotes the degradation of basally localized PIN-FORMED 2 (OsPIN2) via the 26S proteasome, thus establishing the typical apical PM localization of OsPIN2 and polar auxin transport, ultimately shaping RSA. This previously unidentified OsGLS1-OsPIN2 regulatory pathway will contribute to an optimal RSA for enhancing nutrient efficiency in rice and other crops.
Subject terms: Plant physiology, Plant morphogenesis, Plant transporters
Li et al. characterized the gene GRAVITROPISM LOSS 1 (OsGLS1) in rice, which regulates root system architecture (RSA) to enhance nutrient uptake and yield. OsGLS1 promotes the degradation of OsPIN2, influencing auxin transport and root growth patterns.
Introduction
Root system architecture (RSA) is important for water and nutrient uptake and for plant anchoring into the soil substrate. RSA determines the distribution of roots in the soil and integrates the length, thickness, number and growth angle of the primary root, adventitious roots, lateral roots and root hairs1. RSA exhibits great plasticity in response to the availability of water, nutrients and other environmental signals in the soil and strongly influences nutrient uptake and grain yield in crops2–4. A deeper RSA has been reported to facilitate the uptake of subsoil water, nitrogen (N) and other mobile nutrients that are more abundant in deeper soil layers, helping maintain high yield of plants experiencing drought5–7. By contrast, a shallower RSA is beneficial for the capture of phosphorus (P) and other immobile nutrients from the topsoil layer, and it can also increase yield under specific stress conditions such as high salinity in rice8–10. Thus, identifying key RSA genes and understanding their molecular mechanisms would benefit the breeding of crop varieties with an RSA better adapted to different stresses or suboptimal soil conditions.
The root gravitropic response is an important factor determining the spatial pattern of root growth, thereby affecting RSA, and consists of three steps in Arabidopsis (Arabidopsis thaliana): first, gravity sensing and signaling initiation by the root cap; second, translocation of the gravity signal to the elongation zone; and third, a root curvature response11. Different hypotheses have been proposed to explain how cells sense gravity, with the prevailing theory (the starch-statolith theory) stating that amyloplasts in root cap cells act as statoliths by sedimenting along the gravity vector12. Transduction of the sedimentation signal relies on the phosphorylation of LAZY1-LIKE (LZY) proteins, which enhances their interaction with TRANSLOCONS AT THE OUTER ENVELOPE MEMBRANE OF CHLOROPLASTS (TOC) proteins at the surface of amyloplasts, leading to enrichment of LZYs on the amyloplast surface and their polar relocation to the new basal plasma membrane (PM) in Arabidopsis roots13,14. The root curvature response following gravity sensing and signal translocation is explained by the Cholodny-Went theory, which involves the redistribution of auxin in the root tip via auxin carriers14. The phytohormone auxin plays an important role in the entire root gravitropic response, which starts with the production of amyloplasts through regulation of the expression of key starch granule biosynthesis genes, such as STARCH SYNTHASE 4 (SS4), PHOSPHOGLUCOMUTASE (PGM) and ADENOSINE DIPHOSPHATE GLUCOSE PYROPHOSPHORYLASE 1 (ADG1), in root tip cells in Arabidopsis15. Auxin transporters, such as PIN-FORMED2 (PIN2), PIN3 and PIN7, are also important in the root response to gravity. When roots are reoriented in the same plane, the amyloplasts sediment toward the new lower side of the PM in columella cells; PIN3 and PIN7 then rapidly and polarly relocalize to this side of the PM through transcytosis, driving auxin flow toward the lower side of the root tip. These changes in auxin flux affect PIN2 localization and abundance, with less PIN2 on the new upper side of the root and more on the lower side of the newly oriented root; this asymmetric PIN2 accumulation contributes to an asymmetric auxin gradient in the root elongation zone, ultimately leading to asymmetric growth and gravitropic bending along the gravity vector16. However, the mechanisms underlying the asymmetric distribution of PIN2 in plants are still unclear.
Rice (Oryza sativa), a staple food for more than half of the world’s population, has a typical fibrous root system, making it an excellent system for studying root traits in cereal crops17. Root growth angle (RGA), which defines how plant roots grow toward the gravity vector, is one of the most important determinants of RSA in soil12. Many genes have been reported to affect RGA in rice. Among them, OsPIN1 genes, OsPIN2 and AUXIN RESISTANT 1 (OsAUX1) encode auxin transporters that affect root gravitropic responses by transporting auxin between gravity-sensing columella cells and elongating epidermal cells18–20. RICE MORPHOLOGY DETERMINANT (OsRMD), encoding an actin-binding protein, affects RGA responses to low external phosphate21, and DEEPER ROOTING 1 (OsDRO1) and its homolog quantitative trait locus for SOIL SURFACE ROOTING 1 (OsqSOR1), predominantly expressed in root tips, participate in root gravitropic responses7,9. Although several genes are known to affect RGA, the underlying molecular mechanism remains unclear in rice.
To understand the mechanism underlying RGA, we previously identified mutants with defects in root gravitropism from an ethyl methanesulfonate (EMS)–mutated population of the japonica rice cultivar Xiushui 63 (XS63) and isolated the gravitropism loss 1 (gls1) mutant with shallower RSA22. Here, we cloned the causal gene mutated in gls1 and elucidated how its shapes RSA and its potential for breeding applications in improving nutrient uptake efficiency and grain yield. This work defines a hitherto unidentified signaling module for the establishment and maintenance of normal apical polar localization of OsPIN2 in rice root outer cell layers (including epidermis, exodermis and sclerenchyma cells) and provides targets for breeding crops with nutrient-efficient RSA and high yield.
Results
gls1 has a shallower root system architecture
To explore the biological role of OsGLS1 in rice growth and development, we investigated the phenotype of the gls1 mutant when grown in hydroponic culture. Compared to wild-type XS63 seedlings, 7-day-old gls1 mutant seedlings produced longer primary root, more adventitious roots, taller shoots and wider RGA (Supplementary Fig. 1a–e). Gravitropic response assays showed a loss of the root gravitropic response of gls1; however, its shoot gravitropic response was normal (Supplementary Fig. 1f, h). Furthermore, the spatial distribution of roots of 4-week-old gls1 plants was clearly larger than that of XS63 (Supplementary Fig. 1g). The dry weights of shoots and roots of 7-week-old gls1 plants were ~23.1% and ~42.3% higher than those of XS63 plants, respectively, suggesting that the OsGLS1 mutation enhances shoot and root growth (Supplementary Fig. 1i, j). To examine root architecture in soil, we reconstructed the 3D root structure of XS63 and gls1 plants using X-ray computerized tomography. We observed that the gls1 mutant had a larger RGA (92.2° ± 2.3) than XS63 (50.3° ± 4.5), and gls1 roots were more concentrated in the surface layer of the soil (Fig. 1a, b). These results suggest that OsGLS1 negatively regulates plant growth and plays an important role in controlling RSA.
Fig. 1. gls1 has a shallower root system and higher grain yield in soil.
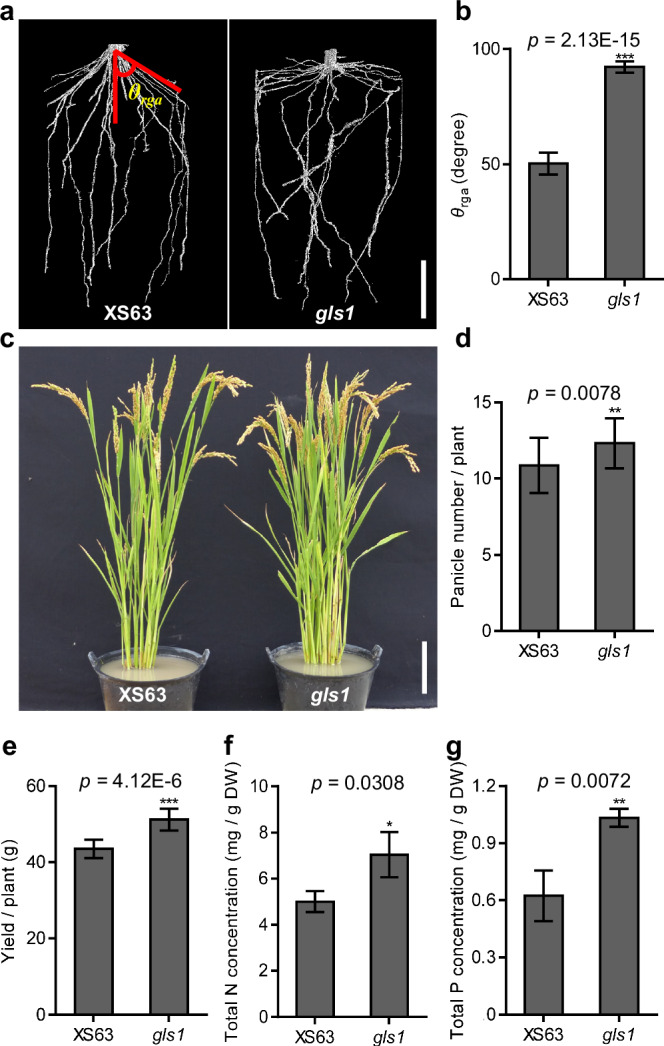
a Root system architecture of seedlings for the wild-type XS63 and the gls1 mutant grown for 21 days in 8-cm-diameter pots filled with sterilized Kettering loam, as revealed by X-ray computed tomography. b Root growth angle (θrga) of XS63 and gls1 in a measured using ImageJ. Representative photograph (c), panicle number (d), yield per plant (e), and total nitrogen (N; f) and phosphorus (P; g) concentrations in shoots of XS63 and gls1 plants at the maturation stage grown in the paddy field (means ± standard deviation (SD), n = 10 in b and e, n = 22 in d, n = 3 in f and g). Scale bars, 3 cm in a, 10 cm in c. Asterisks indicate significance differences as determined by a two-tailed Student’s t-test (*p < 0.05, **p < 0.01, ***p < 0.001).
gls1 shows increased nutrient uptake and grain yield in paddy soil
In agricultural systems, most nutrients are generally highly concentrated near the soil surface, largely from the decomposition of plant matter and the application of fertilizers or organic amendments23. Especially in paddy fields, most nutrients, such as N, P and potassium (K), are easily soluble in water and readily accumulate in the surface soil. Therefore, shallower roots would be well suited to take up nutrients from the topsoil. To test whether the shallower roots of the gls1 mutant facilitate nutrient uptake in soil, we fertilized plants grown in pots with 15N-labeled urea and compound fertilizers (Supplementary Fig. 2a). We measured the concentrations of 15N, P, K, calcium (Ca) and magnesium (Mg) in different leaves, panicles and tillers before fertilization (0 day) and at different time points after fertilization (2, 4, 6 and 8 days). The concentrations of 15N reached values 33.7%–51% higher in various tissues of the gls1 mutant than in XS63 plants (Supplementary Fig. 2b). The concentrations of P, K, Ca and Mg were also significantly higher in the panicles of gls1 plants at all time points after fertilization (Supplementary Fig. 2c–f). These results suggest that the gls1 mutant exhibits greater nutrient uptake in soil.
We also investigated the growth performance of gls1 and XS63 plants in the greenhouse and paddy field. In both growing environments, the gls1 mutant had a significantly larger RGA and shallower root distribution than XS63 (Supplementary Fig. 3a, b and 4a). The effective panicle number, panicle length and yield per plant for gls1 were about 13.4%, 5.7% and 17.6% higher than for XS63 in paddy fields, while plant height, seed setting, thousand-grain weight, grain length and grain width were not clearly affected (Fig. 1c–e and Supplementary Fig. 4b–h). Furthermore, the total N and total P concentrations were ~40.9% and ~65.8% higher in gls1 shoots, respectively, compared to those in XS63 shoots in paddy fields (Fig. 1f, g), which is consistent with the results obtained in the greenhouse (Supplementary Fig. 3c, d). We conclude that mutation of OsGLS1 improves nutrient uptake and grain yield in soil.
To determine whether OsGLS1 directly regulates nutrient uptake and/or transport, we measured nutrient concentrations in gls1 and XS63 grown in hydroponic solution containing 15N-labeled urea. The 15N concentrations in the shoot and flag leaf of gls1 were similar as those in XS63 at all time points tested (Supplementary Fig. 5a, b). Furthermore, we detected no significant differences in the P, K, iron (Fe), Ca, Mg, sodium (Na), zinc (Zn) or manganese (Mn) concentration of roots, shoots and leaves from gls1 and XS63 plants (Supplementary Fig. 5c–j). These results suggest that mutation of OsGLS1 has no effect on nutrient uptake or transport in hydroponic culture.
Cloning of OsGLS1 and complementation analysis
We previously showed that gls1 is a single recessive mutation, which we mapped to chromosome 422. To identify the causal gene, we fine-mapped gls1 using a segregating F2 population (1400 plants with the mutant phenotype) derived from a cross between gls1 and Kasalath (indica). We narrowed down the location of the candidate gene to a 104-kb region between markers RM16253 and RM16260 on the short arm of chromosome 4 (Supplementary Fig. 6a). Genome sequencing of gls1 and XS63 revealed a single insertion of a G (1538 bp downstream from the start codon) in the second exon of LOC_Os04g01160 in gls1 (Supplementary Fig. 6b), leading to a premature stop codon. To test whether the larger root architecture of gls1 is caused by the mutation of LOC_Os04g01160 (hereafter OsGLS1), we generated a complementation construct comprising the OsGLS1 promoter driving the OsGLS1 coding sequence cloned in-frame and upstream of the GREEN FLUORESCENT PROTEIN (GFP) sequence, which we then introduced into the gls1 mutant (gls1 GLS1pro:GLS1-GFP). We obtained several transgenic lines with similar RGA and other root traits to XS63, indicating that OsGLS1 is the causal gene mutated in the gls1 mutant and that the GLS1-GFP fusion is functional (Supplementary Fig. 6c, d).
For an independent confirmation of the identity of OsGLS1, we generated three additional gls1 alleles, named gls1-2, gls1-3 and gls1-4, in the Heijing 2 background (HJ2, a japonica rice variety with short growth period) using clustered regularly interspaced short palindromic repeat (CRISPR)–CRISPR-associated nuclease 9 (Cas9)-mediated gene editing. The gls1-2, gls1-3 and gls1-4 mutants have a 4-bp deletion, 5-bp deletion and 1-bp insertion in the second exon of OsGLS1, respectively, resulting in frameshifts and premature stop codons in OsGLS1 (Supplementary Fig. 7a, b). All three mutants displayed significantly larger RGA compared to that of HJ2 seedlings (Supplementary Fig. 7c, d). These results confirm that the abnormal RSA seen in gls1 is caused by mutation of OsGLS1.
OsGLS1 is polarly localized at the plasma membrane of root outer cell layers
We investigated the expression pattern of OsGLS1 by generating transgenic lines expressing the β-GLUCURONIDASE (GUS) reporter gene driven by the OsGLS1 promoter (GLS1pro:GUS) in the HJ2 background. GUS staining of the GLS1pro:GUS lines revealed that OsGLS1 is expressed in all tissues tested, with higher expression in root tips and stem bases of 7-day-old seedlings. Longitudinal and cross sections of the primary root elongation zone showed that OsGLS1 is mainly expressed in the epidermis, exodermis and sclerenchyma cells of rice roots (Supplementary Fig. 8a–l). Moreover, RT-qPCR analysis showed that OsGLS1 is ubiquitously expressed in different tissues, with higher expression in the stem bases and root tips (0–1 cm from root tips) at the seedling stage (7 days old) and in flag leaves at the flowering stage (50 days old) (Supplementary Fig. 8m).
We investigated the subcellular localization of OsGLS1 using the gls1-2 GLS1pro:GLS1-GFP complementation lines, revealing that OsGLS1 is polarly localized at the basal PM of cells in root outer cell layers. To counterstain the PM, we used FM4-64 staining, a lipophilic styryl dye (Fig. 2a, b). We further quantified the fluorescence intensity of GLS1-GFP and FM4-64 at different PM sites, including upper, middle, lower and basal PM of root epidermal cells, and found that the ratios of GLS1-GFP to FM4-64 of lower and basal PM were significantly higher than that of upper and middle PM, indicating an higher protein abundance of OsGLS1 at the lower and basal PM (Supplementary Fig. 9b, c). In addition, we detected a weak GLS1-GFP signal in the cytoplasm (Supplementary Fig. 9a, b). In situ immunostaining with an anti-GFP antibody showed that OsGLS1-GFP mainly localizes to the basal PM in root epidermis, exodermis and sclerenchyma cells, with a relatively weak signal in stele cells (Supplementary Fig. 9d). Altogether, these results indicate that OsGLS1 is a polarly localized protein that mainly localizes to the basal PM of root outer cell layers.
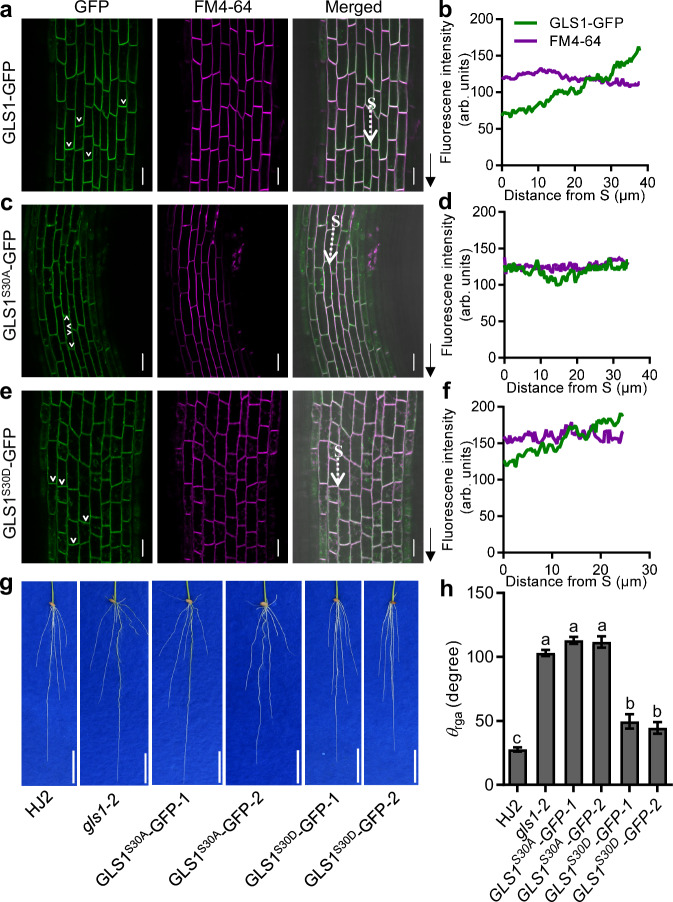
Fig. 2. The polar localization of OsGLS1 at the plasma membrane is required for its effect on root growth angle.
Localization of various GLS1-GFP variants (green fluorescence) and FM4-64 staining (magenta fluorescence) in cells of the primary root cells from gls1-2 GLS1pro:GLS1-GFP (GLS1-GFP; a), gls1-2 GLS1pro:GLS1S30A-GFP (GLS1S30A-GFP; c) and gls1-2 GLS1pro:GLS1S30D-GFP (GLS1S30D-GFP; e) transgenic lines. Quantitative analysis of the fluorescence intensity of GFP and FM4-64 along the white dashed arrow in a (b), c (d) and e (f). S indicates the starting point of fluorescence intensity measurement. White arrowheads mark the polarity of GLS1-GFP; black arrows indicate the direction of root tips in a, c and e; arb. units, arbitrary units. Root phenotypes (g) and root growth angle (θrga; h) of the wild-type HJ2, the gls1-2 mutant and GLS1S30A-GFP and GLS1S30D-GFP transgenic lines grown on germination paper for 7 days after germination. Scale bars, 20 μm in a, c, e and 3 cm in g. In h, data represent means ± SD (n = 12); different letters indicate significant differences (p < 0.01; one-way ANOVA with Tukey’s honestly significant difference test).
Phosphorylation of OsGLS1 at Ser-30 is important to its polar localization
To determine which region of OsGLS1 is required for its polar localization, we produced transgenic plants harboring constructs encoding different truncations of OsGLS1 fused to GFP under the control of the cauliflower mosaic virus (CaMV) 35S or the OsGLS1 promoter (Supplementary Fig. 10a). GLS11–170-GFP (amino acids 1–170 of OsGLS1 fused to GFP) showed polar localization at the PM of root epidermal cells like full-length GLS1-GFP. By contrast, GLS1171–690-GFP and GLS1120–690-GFP showed non-polar localization at the PM and also accumulated in the nuclei of root epidermal cells. The GLS180–690-GFP and GLS140–690-GFP fusions did not localize to the PM of root epidermal cells (Supplementary Fig. 10b–g), suggesting that the N-terminal 39 amino acids are essential for the polar localization of OsGLS1 at the PM.
Phosphorylation was previously shown to be an important mechanism affecting polar localization to the PM24. Accordingly, we looked for phosphorylated residues in OsGLS1-GFP immunoprecipitated by using an anti-GFP antibody–conjugated to beads using protein extracts from the roots of 7-day-old gls1-2 GLS1pro:GLS1-GFP seedlings by liquid chromatography–tandem mass spectrometry analysis. We identified three phosphorylation sites (Ser-30, Thr-644 and Thr-654) in immunoprecipitated OsGLS1 (Supplementary Fig. 11). To determine whether these phosphorylation sites contribute to the polar localization of OsGLS1 at the PM, we transformed the gls1-2 mutant with the vector GLS1pro:GLS13D-GFP (Ser-30, Thr-644 and Thr-654 replaced with Asp) or GLS1pro:GLS13A-GFP (Ser-30, Thr-644 and Thr-654 replaced with Ala) and asked whether they complemented the mutant phenotype (Supplementary Fig. 12a). Indeed, we observed an almost complete rescue of RGA in gls1-2 GLS1pro:GLS13D-GFP seedlings, to the same level as in HJ2, while the RGA of gls1-2 GLS1pro:GLS13A-GFP seedlings was similar to that of gls1-2 (Supplementary Fig. 12b,c). Furthermore, GLS13D-GFP, but not GLS13A-GFP, was polarly localized to the basal PM, similar to GLS1-GFP (Supplementary Fig. 12d). These results suggest that phosphorylation at these three residues is important for the polar localization of OsGLS1.
The N-terminal 39 amino acids of OsGLS1 are important for its polar localization at the PM and include the Ser-30 residue. An alignment of OsGLS1 and homologs from other plant species indicated that Ser-30 is conserved in most OsGLS1-like proteins (Supplementary Fig. 13), suggesting that the phosphorylation of Ser-30 is important for the polar localization of OsGLS1. To test this hypothesis, we generated gls1-2 GLS1pro:GLS1S30A-GFP (encoding OsGLS1 with Ser-30 replaced with Ala) and gls1-2 GLS1pro:GLS1S30D-GFP (Ser-30 residue replaced with Asp) transgenic lines. For each construct, we selected two representative independent transgenic lines with similar OsGLS1 transcript levels for phenotyping (Supplementary Fig. 12a). We observed that GLS1S30D-GFP, but not GLS1S30A-GFP, was polarly localized to the basal PM (Fig. 2c–f). Furthermore, the RGA of gls1-2 GLS1pro:GLS1S30D-GFP seedlings was rescued to levels close to HJ2, whereas the GLS1pro:GLS1S30A-GFP construct did not rescue the mutant phenotype (Fig. 2g, h). These results indicate that the phosphorylation of Ser-30 is essential for the polar localization of OsGLS1 to the PM.
OsGLS1 and OsPIN2 show genetic interaction
We previously reported that OsPIN2 is an important gene regulating RSA in rice19. OsPIN2 is mainly expressed in root epidermis, exodermis and sclerenchyma cells, and pin2 mutants have a shallower RSA19. The similar expression pattern of OsGLS1 and OsPIN2 and their mutant phenotypes suggested a possible genetic relationship. We therefore generated the pin2 gls1 double mutant by crossing gls1-2 to the pin2 mutant in the HJ2 background. Phenotypic analysis of 7-day-old seedlings and 4-week-old plants showed that the adventitious root number, primary root length and root hair length of the pin2 gls1 double mutant were similar to those of the pin2 mutant but significantly lower than in the gls1-2 mutant. The RGA of the pin2 gls1 double mutant was similar to that of the gls1-2 mutant and significantly larger than that of the pin2 mutant (Supplementary Figs. 14 and 15). These results suggest that OsGLS1 and OsPIN2 are involved in root development through the same genetic pathway.
OsGLS1 regulates the abundance and subcellular localization of OsPIN2
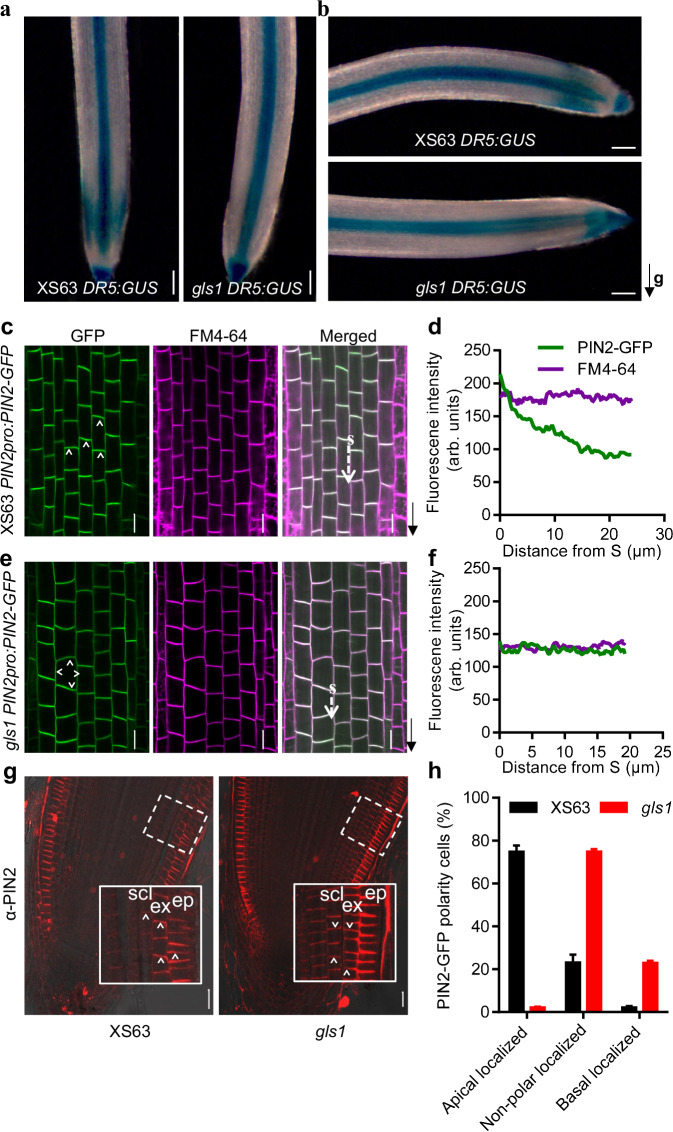
Auxin is important in root growth and development. An RT-qPCR analysis detected no clear difference in the expression levels of auxin biosynthesis–related OsYUCCA genes between the roots of wild-type XS63 and gls1 (Supplementary Fig. 16a). To investigate whether auxin distribution was affected in gls1, we transformed the DR5:VENUS and DR5:GUS reporter constructs (with VENUS or GUS driven by a synthetic auxin-responsive promoter DR5) into XS63 and crossed the resulting lines to gls1 to introduce the reporter lines in the gls1 background. In roots grown vertically, we observed lower auxin signaling output in gls1 root outer cell layers based on VENUS fluorescence and GUS staining, whereas the auxin signaling output was higher in the stele cells and the root cap cells compared to those in XS63 roots; auxin signaling output was similar on both sides of root outer cell layers in gls1 and XS63 (Fig. 3a and Supplementary Fig. 17a, b). Following a rotation of 90° to bring the roots into a horizontal orientation, we observed more intense GUS staining on the lower side of the elongation zone in XS63 roots, but not in gls1 roots (Fig. 3b), suggesting that normal auxin signaling output is affected in gls1. These observations suggest that OsGLS1 might be involved in auxin distribution.
Fig. 3. Mutation of OsGLS1 affects auxin distribution and the polar localization of OsPIN2.
GUS staining in the root tips of 7-day-old DR5:GUS seedlings in the XS63 and gls1 backgrounds growing vertically (a) and 2 h after reorientation by 90°. The black arrow indicates the direction of gravity (g; b). Localization of PIN2-GFP (green fluorescence) and FM4-64 (magenta fluorescence) in the primary root tip from 7-day-old seedlings of PIN2pro:PIN2-GFP in XS63 (c) and gls1 PIN2pro:PIN2-GFP (e). The black arrows indicate the direction of root tips. Quantitative analysis of fluorescence intensity along the white dashed arrows in c (d) and in e (f). S refers the starting point of fluorescence intensity measurement. g Immunostaining of OsPIN2 in the root tips of 5-day-old XS63 (left) and gls1 (right) seedlings using an antibody against OsPIN2 (anti-PIN2; red fluorescence). ep, epidermis; ex, exodermis; scl, sclerenchyma. The white arrowheads indicate PIN2 polarity at the plasma membrane. h Percentage of cells with apical, non-polar or basal OsPIN2 localization in epidermis, exodermis and sclerenchyma cells of XS63 and gls1 as revealed by immunostaining (means ± SD; n = 4). Scale bars, 100 μm in a and b, 20 μm in c, e and g.
An RT-qPCR analysis established that the expression levels of the auxin transport–related OsAUX and OsPIN genes are comparable between gls1 and XS63 (Supplementary Fig. 16b, c). To test whether OsGLS1 might modulate the abundance of auxin transporters, we generated AUX1pro:AUX1-GFP and PIN2pro:PIN2-GFP transgenic lines in XS63 and crossed them to gls1 to obtain gls1 AUX1pro:AUX1-GFP and gls1 PIN2pro:PIN2-GFP reporter lines. We observed no difference in the localization or intensity of AUX1-GFP in root epidermis cells of XS63 and gls1 (Supplementary Fig. 18a). Similarly, in situ immunostaining with an anti-OsPIN1a/b antibody revealed a similar basal PM localization and abundance for OsPIN1a and OsPIN1b in cortex and stele cells in XS63 and gls1 roots (Supplementary Fig. 18b). These results suggest that mutation of OsGLS1 does not affect the normal localization or abundance of OsAUX1, OsPIN1a or OsPIN1b in root tips. By contrast, PIN2-GFP protein abundance was significantly higher in the roots of gls1 PIN2pro:PIN2-GFP seedlings compared to that in PIN2pro:PIN2-GFP in XS63 (Supplementary Fig. 19). Yet although PIN2-GFP always localized to the apical PM in root epidermis cells of XS63, it showed no polar localization to the PM of root epidermis cells in gls1 (Fig. 3c–f). In situ immunostaining of OsPIN2 in longitudinal sections of XS63 and gls1 root tips using an anti-OsPIN2 antibody confirmed the polar localization of OsPIN2 to the apical PM in most root epidermis, exodermis and sclerenchyma cells in the meristem and elongation zones of XS63 (about 74.8%). Notably, OsPIN2 was evenly distributed between the apical and basal PM in most (about 74.9%) root outer cell layers in gls1. OsPIN2 showed polar localization to the basal PM in root cortex cells of both gls1 and XS63 (Fig. 3g, h and Supplementary Fig. 20). These results suggest that OsGLS1 is important for the normal abundance and polar localization of OsPIN2 to the PM in root epidermis, exodermis and sclerenchyma cells.
OsGLS1 physically interacts with OsPIN2
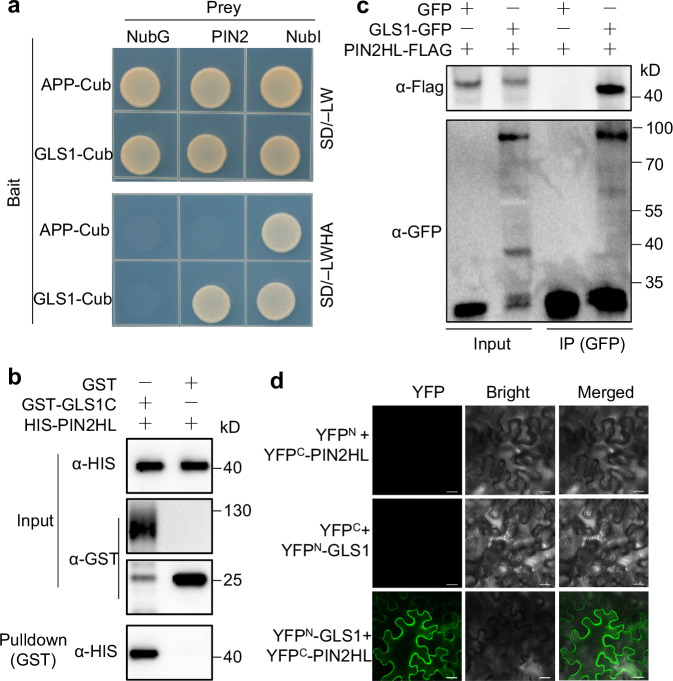
We asked whether OsGLS1 might interact directly with OsPIN2 by performing a split-ubiquitin yeast two-hybrid assay. Indeed, OsGLS1 interacts with OsPIN2 in yeast cells (Fig. 4a). Additional assays with truncated variants of both proteins mapped the interaction interface to the C-terminal region of OsGLS1 containing the VWA domain, rather than its N-terminal region containing the RING domain; similarly, the OsPIN2 central hydrophilic loop (OsPIN2HL) was important for the interaction, but the N- or C-terminal region of OsPIN2 were not (Supplementary Fig. 21a–c). In vitro pull-down assays using recombinant purified glutathione S-transferase (GST)-tagged OsGLS1 C-terminal region (amino acids 40–690, named GST-GLS1C) and His-tagged OsPIN2HL (amino acids 161–459, named HIS-PIN2HL) confirmed that OsGLS1C interacts with OsPIN2HL (Fig. 4b). Co-immunoprecipitation (Co-IP) assays using rice protoplasts transfected with the constructs GLS1-GFP and PIN2HL-FLAG showed that OsGLS1 immunoprecipitates OsPIN2HL following immunoprecipitation with an anti-GFP antibody (Fig. 4c). In addition, bimolecular fluorescence complementation (BiFC) confirmed that OsGLS1 interacts with OsPIN2HL at the PM of plant cells (Fig. 4d and Supplementary Fig. 22). These results demonstrate that OsGLS1 physically interacts with OsPIN2, both in vitro and in vivo.
Fig. 4. OsGLS1 physically interacts with OsPIN2.
a Split-ubiquitin yeast two-hybrid analysis. Cub, C-terminal half of ubiquitin (the bait APP-Cub interacts with NubI but not with NubG, which served as positive and negative controls, respectively); NubI and NubG, wild-type and mutated N-terminal fragments of ubiquitin, respectively. Yeast cells were grown on synthetic defined (SD) medium lacking leucine and tryptophan (SD/–LW) or lacking leucine, tryptophan, histidine and adenine (SD/–LWHA). b Pull-down assay. Recombinant GST-GLS1C, OsGLS1C (aa 40–690) fused to a GST tag. HIS-PIN2HL, OsPIN2HL (aa 161–459) fused to a HIS tag. After co-incubating GST-GLS1C with HIS-PIN2HL, proteins were pulled down with glutathione-Superflow resin and detected using anti-HIS antibody. c Co-immunoprecipitation (Co-IP) assays. 35S:OsPIN2HL-FLAG was co-transfected with 35S:OsGLS1-GFP or 35S:GFP in rice protoplasts. Equal amounts of total proteins were immunoprecipitated with anti-GFP antibody–conjugated beads and detected using anti-GFP and anti-Flag antibodies. d BiFC assays. OsGLS1 and OsPIN2HL were fused to the C-terminal (YFPC) and N-terminal (YFPN) halves of YFP, respectively. Combinations of constructs encoding YFPN or YFPC fused to the corresponding OsGLS1 and OsPIN2HL were used as negative controls. Fluorescence was observed using a confocal microscope. Scale bars, 20 μm.
OsGLS1 directly ubiquitinates and degrades OsPIN2
OsGLS1 contains a RING domain, which usually functions as an E3 ubiquitin ligase. To test whether OsGLS1 possesses E3 ubiquitin ligase activity, we produced a truncated OsGLS1 variant (amino acids 1–240) containing the RING domain fused to GST (GST-GLS1N). In the presence of E1, E2 and ubiquitin-Flag, GST-GLS1N self-ubiquitinated (Supplementary Fig. 23a). However, mutating Cys-127 to Ser (C127S) or His-145 to Tyr (H145Y) in the RING domain of OsGLS1 greatly decreased the E3 ubiquitin ligase activity, especially C127S (Supplementary Fig. 23a). These results suggest that OsGLS1 has E3 ubiquitin ligase activity.
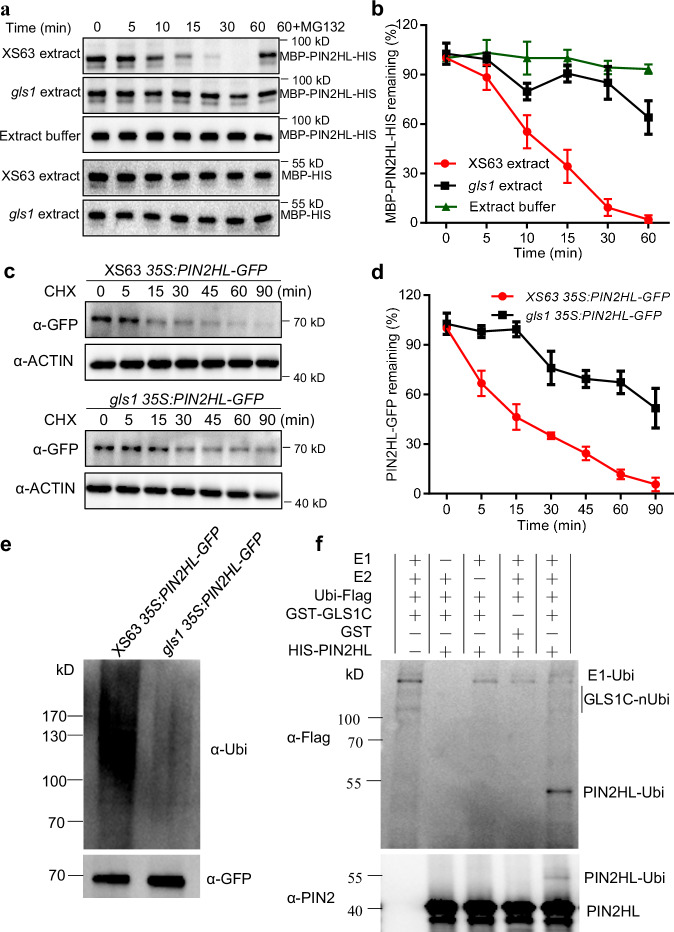
As OsPIN2 interacted with OsGLS1 and the abundance and localization of OsPIN2 was altered in gls1, we speculated that OsGLS1 might regulate the stability of OsPIN2 by mediating its ubiquitination and degradation. To test this hypothesis, we performed cell-free degradation assays using recombinant maltose-binding protein (MBP)-tagged PIN2HL-HIS with protein extracts from the roots of XS63 or gls1 seedlings. We observed the degradation of MBP-PIN2HL-HIS, but not MBP-HIS, within 30 min of incubation with protein extract from XS63, whereas MBP-PIN2HL-HIS was not fully degraded even after 60 min of incubation with protein extracts from gls1 or with extraction buffer (Fig. 5a, b). Importantly, the degradation of MBP-PIN2HL-HIS in XS63 extracts was blocked by addition of MG132, an inhibitor of the 26S proteasome, suggesting the involvement of the 26S proteasome in OsPIN2 degradation (Fig. 5a).
Fig. 5. OsGLS1 directly ubiquitinates and degrades OsPIN2 in vitro and in vivo.
OsPIN2HL degradation in cell-free degradation assays. Total proteins extracted from the roots of XS63 or gls1 seedlings were incubated with recombinant MBP-PIN2HL-HIS alone or with MG132 for the indicated times; MBP-PIN2HL-HIS was detected using anti-MBP antibody (a); the band signal intensity was quantified (b). MBP-HIS was used as a control. c, d Regulation of OsPIN2HL stability by OsGLS1 in vivo. Total proteins were extracted from the roots of 10-day-old seedlings from 35S:PIN2HL-GFP in XS63 and gls1 35S:PIN2HL-GFP transgenic lines treated with 100 μM cycloheximide (CHX) for the indicated times. PIN2HL-GFP was detected using anti-GFP antibody (c) and band signal intensity quantified (d). OsACTIN1 was used as a loading control. In b and d, data represent means ± SD (n = 3). e In vivo ubiquitination assay of OsGLS1 on OsPIN2HL. The roots of 10-day-old seedlings from 35S:PIN2HL-GFP in XS63 and gls1 35S:PIN2HL-GFP transgenic lines treated with 50 μM MG132 for 8 h were sampled for protein extraction. PIN2HL-GFP was immunoprecipitated using anti-GFP antibody-conjugated beads. Ubiquitinated PIN2HL-GFP was detected using an anti-ubiquitin antibody (α-Ubi). f Ubiquitination of OsPIN2HL by OsGLS1 in vitro. Recombinant HIS-PIN2HL was incubated with GST-GLS1C alone or in the presence of E1, E2 and ubiquitin-Flag (Ubi-Flag). Ubiquitinated HIS-PIN2HL was detected using anti-Flag and anti-PIN2 antibodies.
To further investigate whether OsGLS1 degrades OsPIN2 in rice roots, we generated transgenic plants harboring a 35S:PIN2HL-GFP transgene in the XS63 and gls1 backgrounds. Immunoblot analysis showed that PIN2HL-GFP is degraded more rapidly in XS63 than in gls1 after treatment with cycloheximide (CHX), a protein synthesis inhibitor; ~50% of PIN2HL-GFP was degraded within 15 min of CHX treatment and was completely degraded by 90 min in XS63, while PIN2HL-GFP levels remained constant for the first 15 min after CHX addition, with ~50% of PIN2HL-GFP being degraded after 90 min in gls1 (Fig. 5c, d). Additionally, the abundance of the PM localized OsPIN2 was significantly decreased when OsGLS1 was co-expressed in the rice protoplasts (Supplementary Fig. 23b). These results indicate that OsGLS1 modulates the degradation of OsPIN2 in rice.
To investigate whether OsGLS1 facilitates OsPIN2 degradation directly by ubiquitination, we examined the levels of ubiquitinated PIN2HL in PIN2HL-GFP in XS63 and gls1 PIN2HL-GFP transgenic plants treated with MG132. We observed much lower levels of ubiquitinated PIN2HL-GFP in gls1 than in XS63 (Fig. 5e). An in vitro ubiquitination assay showed that recombinant PIN2HL is ubiquitinated in the presence of GST-GLS1C and ubiquitin-Flag, E1 or E2, but not ubiquitinated in the absence of GST-GLS1C (Fig. 5f). Together, these results demonstrate that OsGLS1 directly ubiquitinates and degrades OsPIN2.
Discussion
RGA is an important factor determining the distribution of roots in soil, but the mechanisms behind its establishment have remained unclear. Here, we identified a key RGA regulatory gene, OsGLS1, and elucidated its molecular mechanism in regulating RGA in rice (Fig. 6). OsGLS1 phosphorylated on Ser-30 is polarly localized to the basal PM of root outer cell layers. OsGLS1 directly ubiquitinates and degrades OsPIN2 at the basal PM, leading to the polar accumulation of OsPIN2 at the apical PM and thus differential auxin distribution, which shapes a normal RSA. These results reveal a key target for genetic improvement of RSA for nutrient uptake and yield in crops.
Fig. 6. A working model for the regulation of root system architecture in rice by OsGLS1–OsPIN2.
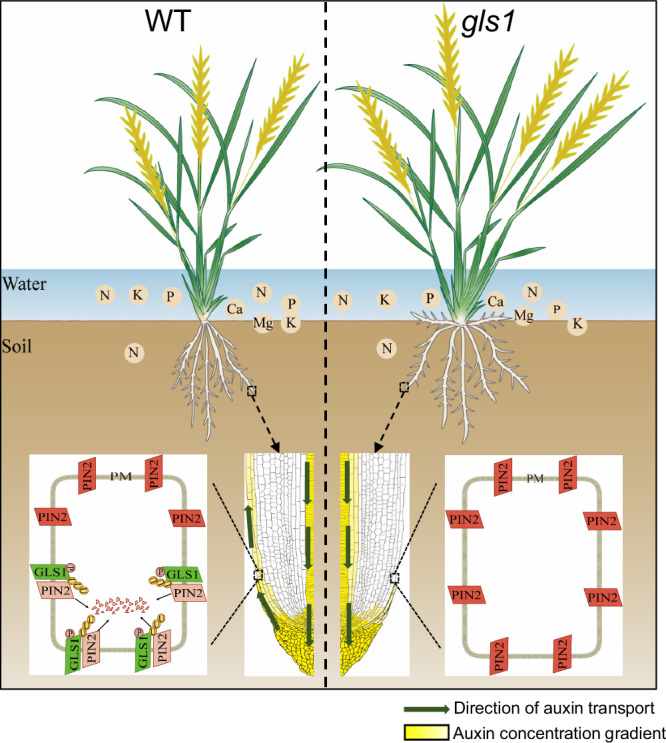
In the wild type (WT, left), OsGLS1 is mainly localized to the basal side of the plasma membrane (PM) in root epidermal cells, a process dependent on OsGLS1 Ser-30 phosphorylation. There, OsGLS1 directly ubiquitinates and degrades OsPIN2, subsequently leading to the apical PM localization of OsPIN2 to maintain normal auxin gradient distribution and root system architecture (RSA). In the gls1 mutant (right), loss of OsGLS1 function leads to a non-polar localization for OsPIN2 at the PM of root epidermal cells, resulting in abnormal auxin gradient distribution and shallower RSA, which facilitates nutrient uptake from water and topsoil in the paddy field.
OsGLS1 is an important target gene to achieve an ideal root system architecture
Root morphological traits have long been a key target of breeders for crop improvement25. Several genes are reported to be involved in RGA7,9,10,26,27; however, how the RGA is set up is still not clear. In this study, the original gls1 mutant and gene-edited gls1 alleles showed a shallower RSA with larger RGA, longer primary roots, more adventitious roots and greater root dry weight compared to the wild type (Supplementary Fig. 1), identifying OsGLS1 as a negative regulator of RSA. We observed higher nutrient uptake and grain yield in gls1 compared to the wild type under paddy soil conditions (Fig. 1 and Supplementary Figs. 2–4), indicating that the shallower and larger RSA of gls1 contribute to higher nutrient uptake and yield in paddy soil. Meanwhile, the shoot dry weight of gls1 mutant is significantly higher than that of the wild type after 5 weeks grown in solution culture, suggesting that the OsGLS1 may directly control shoot growth in rice. These results suggest that OsGLS1 is an important target gene for genetic improvement of RSA. The gls1 mutant has a very shallow root architecture that may not be suitable for drought conditions; therefore, a flooding condition in the whole growth period is required for the optimal growth and yield of gls1 in field. Therefore, downregulating OsGLS1 expression by editing its promoter or producing a weak allele may help produce a line with suitable RGA for nutrient-efficiency, drought tolerance and high yield for use in the field in future.
OsGLS1 is a key regulator for the polar localization of OsPIN2
Plant growth and development and responses to environmental conditions are largely controlled by auxin gradient distribution that is mainly generated by polarly localized PIN proteins28. OsPIN2 and its Arabidopsis homolog PIN2 play important roles in root gravitropic response and RGA through their function in establishing auxin distribution from the root cap to epidermis cells19,29. However, how PIN2 is polarly localized is still not clear. The apical polar localization of PINs in Arabidopsis was previously thought to depend on their phosphorylation, as unphosphorylated PIN1 localized to the basal PM of root epidermal cells in the pinoid (pid) mutant, and overexpression of PID or downregulation of PROTEIN PHOSPHATASE 2 A (PP2A) genes led to increased PIN (PIN1, PIN2 and PIN4) phosphorylation and consequently to a basal-to-apical shift in their localization30–32. However, further studies suggested that phosphorylation was insufficient to direct PIN1 polarity, as phosphorylated PIN1 was detected on all sides of the PM in different cell types of PID overexpression seedlings28,33. Treatment with the proteasome inhibitor MG132 resulted in apparent deficiencies in PIN2 endocytosis to the vacuole and the stability of ubiquitinated PIN2, suggesting an important role for the proteasome-ubiquitin system in regulating PIN2 endocytosis and stability29,34. The E3 ligases RING DOMAIN LIGASE 1 (RGLG1) and RGLG2 were suggested to target PIN2, as PIN2 abundance increased and its ubiquitination levels were lower in the roots of the rglg1 rglg2 double mutant35. However, a recent study showed that the abundance of PIN1 and PIN2 was comparable in the roots of rglg1 rglg2 and the wild type36, suggesting that RGLG1 and RGLG2 do not mediate PINs ubiquitination or stability. Additionally, another E3 ligase, CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), was reported to affect PIN2 stability during root gravitropism37. However, whether COP1 directly ubiquitinates PIN2 is unknown.
In this study, we demonstrated that OsGLS1 modulates the polar localization of OsPIN2 to the PM through directly degrading the pool of OsPIN2 that localizes to the basal PM via the 26S proteasome degradation pathway, based on the following lines of evidence: (1) OsGLS1 is highly expressed in root epidermis, exodermis and sclerenchyma cells (Supplementary Fig. 8j–l and Supplementary Fig. 9). There is no significant difference in the polarity orientation of OsGLS1 among the epidermis, exodermis and sclerenchyma cells, however, OsGLS1 accumulated more in epidermis cells than that in exodermis and sclerenchyma cells in root tip (Supplementary Fig. 9), same as OsPIN2 (Fig. 3g); (2) the gls1 mutant displayed a pin2-like mutant phenotype with a shallower RSA (Fig. 1a, b and Supplementary Fig. 1a, e, g); (3) OsPIN2 protein was more abundant and did not polarly localize in the PM of gls1 mutant roots, in contrast to wild-type roots (Fig. 3c–h and Supplementary Figs. 19 and 20); (4) OsGLS1 directly interacts with OsPIN2 (Fig. 4); and (5) OsPIN2 ubiquitination is lower in gls1 mutant roots compared to the wild type, and OsGLS1 directly mediates the ubiquitination and degradation of OsPIN2 in vitro and in vivo but is blocked by application of MG132 (Fig. 5). Our results provide solid genetic and cytological evidence of the important role of OsGLS1-dependent OsPIN2 ubiquitination and degradation in controlling OsPIN2 polar localization and, thus, root gravitropism. However, whether the ubiquitination or degradation of OsPIN2 is correlated with its phosphorylation is worth further study.
The RGA of the gls1 pin2 double mutant was similar to that of the gls1 single mutant but was significantly larger than that of the pin2 single mutant (Supplementary Figs. 14a,e and 15a,d), suggesting that other components contribute to OsGLS1-mediated RGA besides OsPIN2. The rice genome contains 12 OsPIN19, 27 OsABCB38 and five OsAUX genes39. Most of these genes are involved in auxin transport and rice root development, e.g., OsABCB1440, OsPIN1a–OsPIN1d18, OsPIN219, OsPIN10a41, OsAUX142, OsAUX339 and OsAUX443. The expression level of these genes was not affected in gls1 roots (Supplementary Fig. 16b,c), and OsGLS1 did not interact with OsAUX1, OsABCB1, OsABCB6 or OsABCB14 in Y2H assay (Supplementary Fig. 21d, e), although the possibility of OsGLS1 interaction with these proteins is not fully explored. Whether the localization and abundance of the auxin transporters (aside from OsPIN2, OsPIN1a, OsPIN1b and OsAUX1) are regulated by OsGLS1 needs further investigation. Notably, OsGLS1 was previously reported as SOIL-SURFACE ROOTING 1 (OsSOR1, also named MAOHUZI 2 (OsMHZ2)) involved in ethylene-mediated inhibition of root growth through targeting INDOLE-3-ACETIC ACID INDUCIBLE 26 (OsIAA26)44. However, neither OsIAA26 overexpression lines nor the iaa26 mutant showed defects in root gravitropism44, suggesting that OsGLS1-regulated RGA is not mediated by OsIAA26.
OsGLS1 phosphorylated on its Ser-30 residue localizes to the basal PM of root outer cell layers (Fig. 2). There are three OsGLS1 homologous genes (WAVY GROWTH 3 (WAV3), WAV3 HOMOLOG 1 (WAVH1) and WAVH2) in Arabidopsis, and their protein products also localize to the basal PM of root cells45,46. Although the wavh1 and wavh2 mutants show no root growth defects, the wav3 mutant has an enhanced root response to gravity; however, the wavh1 wavh2 wav3 triple mutant exhibits defects in root gravitropism45. Konstantinova et al. reported that AtPIN2 is basal polarity in root epidermis cells of wavh1 wavh2 wav3 triple mutants, but is apical polarity in wild type. Furthermore, the ubiquitin ligase activity of WAV3/WAVHs is not directly involved in affecting PIN2 polarity in Arabidopsis46. However, the molecular mechanism of AtWAV3/WAVHs impact on PIN polarity is unclear. In this study, our results show that the polarity of OsPIN2 is lost in gls1 mutant rather than a reversed polarity compared to the wild type (Fig. 3). We proved that OsGLS1 localized more on basal PM which interacts directly with, ubiquitinates and promotes the degradation of basally localized OsPIN2 (Figs. 4, 5; Supplementary Fig. 23b), thus establishing the apical PM localization of OsPIN2 in rice (Fig. 6), a mechanism different from Arabidopsis.
Methods
Plant materials and growth conditions
The japonica rice (Oryza sativa) cultivars ‘Xiushui63’ (XS63) and ‘Heijing2’ (HJ2) were used in this study. The gls1 and pin2 single mutants were generated by ethyl methanesulfonate treatment of XS63 and HJ2 seeds, respectively19,22. The gls1-2, gls1-3 and gls1-4 single mutants in the HJ2 background were generated by CRISPR/Cas9-mediated gene editing. The gls1-2 single mutant was crossed to the pin2 single mutant to produce the gls1 pin2 double mutant. The transgenic lines GLS1pro:GLS1-GFP, GLS1pro:GLS1S30A-GFP (encoding a variant of GLS1 with Ser-30 replaced with Ala), GLS1pro:GLS1S30D-GFP (Ser-30 replaced with Asp), GLS1pro:GLS13A-GFP (Ser-30, Thr-644 and Thr-654 replaced with Ala), GLS1pro:GLS13D-GFP (Ser-30, Thr-644 and Thr-654 replaced with Asp), GLS1pro:GLS140–690-GFP (encoding a variant of GLS1 lacking the N-terminal 39 amino acids (aa)), GLS1pro:GLS180–690-GFP (lacking the N-terminal 79 aa), GLS1pro:GLS1120–690-GFP (lacking the N-terminal 119 aa), 35S:GLS11-170-GFP (containing the N-terminal 170 aa), 35S:GLS1171-690-GFP (containing the C-terminal 520 aa), AUX1pro:AUX1-GFP, PIN2pro:PIN2-GFP, 35S:PIN2HL-GFP and 35S:PIN2HL-FLAG were generated by transformation of the related constructs into XS63 or HJ2. Some transgenic lines were also crossed to the gls1 mutant to produce the related transgenic lines in the gls1 mutant background.
For hydroponic culture, seedlings were grown in full-strength Kimura nutrient solution in a greenhouse under a 12-h-light (30 °C)/12-h-dark (22 °C) photoperiod, ~300 μmol m−2 s−1 photons, and ~60% humidity. The pH of the nutrient solution was adjusted to 5.5 before use, and the solution was replaced every 7 days. For field experiments, XS63 and gls1 were cultured in Hangzhou (30° N, 120° E) in the summer and Sanya (18° N, 109° E) in the winter seasons and were maintained watered throughout their growth period. X-ray computerized tomography imaging was performed as described previously19. Nicotiana benthamiana plants were cultivated in growth chambers (25 °C, 14 h light/10 h dark photoperiod, ~200 μmol m−2 s−1 photons, and ~60% humidity).
Gene cloning and complementation tests
For the map-based cloning, gls1 was crossed to the indica rice cultivar Kasalath to create a segregating F2 population from which 1400 plants with larger root growth angle were selected for mapping. The OsGLS1 gene was located in a 104-kb region between markers RM16253 and RM16260 on the short arm of chromosome 4 using simple sequence repeat markers. OsGLS1 was then cloned based on genome resequencing using the MutMap method. Briefly, genomic DNA was extracted from XS63, gls1 and F2 seedlings with the mutant phenotype using a DNeasy Plant Mini Kit (QIAGEN, 9012-90-2); genome sequencing was performed for each sample with a mean coverage of 30× using an Illumina HiSeq2500 sequencer. The putative mutations in the candidate genes were confirmed by Sanger sequencing of PCR products. The primers used for background identification are listed in Supplementary Table 1.
Construction of vectors and generation of transgenic plants
To generate the GLS1pro:GUS construct, a 2950-bp fragment upstream of the OsGLS1 translation start codon was amplified from XS63 genomic DNA using Phanta Max Super-Fidelity DNA Polymerase (Vazyme, P505-d1) and then cloned into the pBI101.3-GUS vector using a ClonExpress II One Step Cloning Kit (Vazyme, C112-02).
To complement the gls1 mutant, an ~5.0-kb genomic DNA fragment containing the promoter sequence (2950 bp of sequence upstream from the ATG) and the entire coding sequence (2067 bp) of OsGLS1 without the stop codon were amplified from XS63 genomic DNA and cDNA, respectively, using the same DNA polymerase as above, and then ligated into the pCAMBIA1300-sGFP vector using the same cloning kit, yielding the clone designated GLS1pro:GLS1-GFP.
The truncated and mutated variants of OsGLS1 were amplified from GLS1pro:GLS1-GFP using the same DNA polymerase as above and then ligated into the vector pCAMBIA1300-sGFP or modified pCAMBIA1300-sGFP with the cauliflower mosaic virus (CaMV) 35S promoter using a ClonExpress MultiS One Step Cloning Kit (Vazyme, C113-02). The vectors containing the truncated OsGLS1 variants were named GLS1pro:GLS140–690-GFP, GLS1pro:GLS180–690-GFP, GLS1pro:GLS1120–690-GFP, 35S:GLS11-170-GFP and 35S:GLS1171-690-GFP. The vectors containing the mutated OsGLS1 variants were named GLS1pro:GLS130A-GFP, GLS1pro:GLS130D-GFP, GLS1pro:GLS13A-GFP and GLS1pro:GLS13D-GFP.
The AUX1pro:AUX1-GFP and PIN2pro:PIN2-GFP expression vectors were constructed as described before42,47. To generate the 35S:PIN2HL-GFP and 35S:PIN2HL-FLAG constructs, the 1056-bp coding sequence of PIN2HL was amplified from XS63 root cDNA and cloned in-frame with the sequence encoding GFP or the FLAG tag in the modified pCAMBIA1300-sGFP or pTCK303 vector, respectively. The CRISPR/Cas9-OsGLS1 vector was generated as described previously18. All primers used above are listed in Supplementary Table 1. After confirmation by Sanger sequencing, all constructs mentioned above were transformed into XS63 or HJ2 via Agrobacterium (Agrobacterium tumefaciens) (strain EHA105)-mediated transformation of callus induced from mature embryos as described previously17.
Immunostaining
Samples from XS63, gls1 and gls1 GLS1pro:GLS1-GFP transgenic plants were subjected to immunostaining as described previously17. Briefly, the root tips of 5-day-old seedlings were fixed in paraformaldehyde solution (4% (w/v) paraformaldehyde, 60 mM sucrose in phosphate-buffered saline (PBS), pH 7.4) for 2 h at room temperature under vacuum infiltration (0.08 Mpa). After three washes with PBS, the fixed samples were embedded in 5% (w/v) agar and sectioned to ~60-μm thickness with a vibratome (VT1000 S, Leica, Germany). The sections were placed in a clean Petri dish and incubated for 2 h at 30 °C in PBS containing 0.1% (w/v) pectinase and 1.5% (w/v) driselase, then for 2 h at 30 °C in PBS containing 0.3% (v/v) Triton X-100. The samples were washed three times with PBS and blocked with PBS containing 5% (w/v) bovine serum albumin (BSA) at 30 °C for 2 h. After addition of primary antibodies against PIN1a/b (customized by Abclonal, 1/1500), PIN2 (customized by Abclonal, 1/1500) or GFP (Invitrogen, A-11122, 1/2500), the samples were incubated at 30 °C for 2 h. The samples were washed three times with PBS, blocked with 5% (w/v) BSA in PBS, exposed to secondary antibody against rabbit IgG conjugated to Alexa Fluor 555 (Invitrogen, A-21428, 1/3000) for 2 h at 30 °C, washed five times in PBS and examined under a confocal laser scanning microscope (LSM710, Zeiss) at excitation and emission wavelengths of 555 and 565 nm, respectively.
Subcellular localization analysis
Protoplast transformation was performed as described previously48. Briefly, stems of 20 two-week-old rice seedlings were cut into 0.5–1 mm strips and digested with 10 ml enzyme solution (0.6 M mannitol, 10 mM MES at PH5.7, 1.5% cellulose [w/v], 0.75% macerozyme [w/v], 0.1% BSA [w/v], 1 mM CaCl2) for 4–5 h in the dark with gentle shaking at 28 °C. Then added 10 ml of W5 medium (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES at pH 5.7) with further shaking for 10 min, and filtered through 70 μm nylon mesh and centrifuged at 1500 rpm for 4 min to collect the protoplasts. Protoplasts were observed under a 63× objective on a Zeiss Axiovert LSM 710 laser scanning microscope. Fluorescence was detected at 493 to 542 nm for GFP and 578 to 625 nm for mCherry.
Yeast two-hybrid assay
Yeast two-hybrid (Y2H) assays were performed following the instructions provided with the DUAL membrane Pairwise Interaction kit (Dualsystems Biotechnology, Switzerland). For the split-ubiquitin Y2H assay, the full-length OsGLS1 coding sequence without the stop codon was amplified using the same DNA polymerase as above and cloned in-frame with the sequence encoding the C-terminal half of ubiquitin (Cub) and the DNA-binding domain of the synthetic LexA-VP16 transcription factor in the pDHB1 vector using the same cloning kit. The full-length OsPIN2, OsAUX1, OsABCB1, OsABCB14 or OsABCB16 coding sequence without the stop codon was amplified and fused in-frame to the N-terminal half of ubiquitin (NubG) in the pPR3-STE vector using the same cloning kit. For the standard Y2H assays, the full-length or truncated OsGLS1 (GLS1, GLS1N and GLS1C) coding sequences were amplified using the same DNA polymerase and cloned into the pGBKT7 vector, and the coding sequences of the truncated OsPIN2 variants (PIN2N, PIN2HL and PIN2C) were amplified using the same DNA polymerase and cloned into the pGADT7 vector using the same cloning kit. The NMY51 or AH109 yeast strain was transformed with appropriate “bait” and “prey” plasmids according to the yeast transformation protocol (Weidi Biotechnology). Positive transformants were selected on synthetic defined (SD) medium lacking tryptophan (Trp) and leucine (Leu) for 3 days at 30 °C; positive colonies were spotted on SD medium lacking Trp, Leu, histidine (His) and adenine (Ade) before growth for 4 days at 30 °C. All primers used above are listed in Supplementary Table 1.
Bimolecular fluorescence complementation (BiFC) assay
The OsGLS1 and OsPIN2HL coding sequences were amplified using the same DNA polymerase as above and cloned into the pCB301-based vectors and in-frame with either the sequence encoding the C-terminal or N-terminal half of yellow fluorescent protein (YFP) using the same cloning kit. The resulting constructs were expressed in the rice protoplasts or leaves of Nicotiana benthamiana plants by Agrobacterium-mediated infiltration as described previously48. After 2 days, YFP fluorescent signals in the infiltrated leaves were monitored using a confocal laser scanning microscope (LSM710, Zeiss) at excitation and emission wavelengths of 488 and 500 nm, respectively. All primers used above were listed in Supplementary Table 1.
Preparation of recombinant proteins
To prepare the recombinant proteins, various expression vectors were constructed. To construct the GST-GLS1N and GST-GLS1C plasmids, the fragments of GLS1N and GLS1C, respectively, were amplified from the OsGLS1 coding sequence and inserted into the pGEX-4T-1 vector. To construct the HIS-PIN2HL and HIS-PIN2HL-MBP plasmids, the PIN2HL fragment was amplified from the OsPIN2 coding sequence and inserted into pET-28a and into a modified pET-28a in which the sequence encoding the MBP tag was cloned into pET-28a at the NdeI and EcoRI restriction sites to add the MBP tag. The GST-GLS1N and GST-GLS1C constructs were introduced into the Escherichia coli strain Transetta, while HIS-PIN2HL and HIS-PIN2HL-MBP were introduced into E. coli strain BL21. E. coli cells were grown to an OD at 600 nm of ~0.6. Recombinant protein production was then induced with the addition of 0.2 mM isopropyl-β-D-thiogalactopyranoside for 18 h at 16 °C with gentle shaking. Fusion proteins were purified using glutathione Sepharose 4B (GE Healthcare, 17-0756-05) or Ni-NTA Agarose Resin (Roche) according to the manufacturers’ instructions. All primers used above are listed in Supplementary Table 1.
Pull-down assay
The purified fusion proteins GST-GLS1C and HIS-PIN2HL or GST and HIS-PIN2HL were incubated with glutathione Sepharose 4B in PBS containing 1 mM phenylmethylsulfonyl fluoride (PMSF) for 3 h at 4 °C. The beads were washed five times with PBS and suspended in 60 μl elution buffer (50 mM Tris-HCl, pH 7.5, 10 mM glutathione). After a brief centrifugation (12,000 g, 4 °C, 10 min), the supernatant was collected and subjected to immunoblot analysis. Anti-His (Abclonal, AE003) and anti-GST antibodies (TransGen Biotechnology, HT601) were diluted 1:5,000. A rabbit anti-mouse IgG secondary antibody conjugated to peroxidase (Sigma-Aldrich, A9044) was diluted 1:10,000.
Co-immunoprecipitation (Co-IP) assay
The roots of 7-day-old transgenic seedlings harboring 35S:PIN2HL-Flag and GLS1pro:GLS1-GFP or 35S:GFP were obtained by crossing 35S:PIN2HL-Flag to GLS1pro:GLS1-GFP or 35S:GFP. The indicated samples were harvested, ground in liquid nitrogen and resuspended in Pierce IP buffer (Thermo Scientific, 87787) with freshly added 1 mM PMSF, 20 μM MG132 and protease inhibitor cocktail (Roche, 4693132001). Filtered protein extracts were centrifuged at 20,000 g, 4 °C for 10 min, and the resulting supernatant was incubated with anti-GFP magnetic beads (Chromotek, gtm-20) for 2 h at 4 °C. Beads were washed five times with washing buffer (50 mM Tris-HCl, pH 7.5, 250 mM NaCl, 0.2% (v/v) Triton X-100, 1 mM PMSF and protease inhibitor cocktail (Roche, 4693132001)). Following SDS-PAGE and transfer to nitrocellulose, GFP, GLS1-GFP and PIN2HL-FLAG were detected by immunoblotting with tag-specific antibodies. The anti-GFP (Sigma-Aldrich, G1544) and anti-FLAG antibodies (Sigma-Aldrich, F1804) were diluted 1:5000. The secondary antibody, rabbit anti-mouse IgG antibody conjugated to peroxidase (Sigma-Aldrich, AP160P) or goat-anti-rabbit IgG antibody conjugated to peroxidase (Sigma-Aldrich, 12–348) was diluted 1:10,000.
In vitro ubiquitination assay
Assays were performed as described previously with little modification44. In brief, each reaction was carried out in a 30-μl mixture containing 0.5 μg human E1 (Enzo, BML-UW9410-0050), 1.5 μg E2 UbcH5b (Enzo, BML-UW9060-0100), 2.5 μg ubiquitin-Flag (Sigma-Aldrich, U5382), 1 μg recombinant protein of interest and ubiquitination buffer (50 mM Tris-HCl, pH 7.4, 2 mM ATP, 5 mM MgCl2, 2 mM DTT, 40 μM ZnSO4 and 0.2 U inorganic pyrophosphatase). The reaction mixtures were incubated for 1 h at 37 °C with agitation in a thermomixer and the reactions stopped by addition of SDS-PAGE sample buffer. The samples were heated to 65 °C for 10 min and separated by electrophoresis on 10% (w/v) SDS-PAGE gels followed by immunoblotting using mouse anti-FLAG (Sigma-Aldrich, F1804) and rabbit anti-PIN2 antibodies (customized by Abclonal).
Cell-free degradation assay
The roots of 7-day-old XS63 and gls1 seedlings were harvested and ground into a fine powder in liquid nitrogen. Total proteins were extracted in degradation buffer containing 25 mM Tris-HCl, pH 7.5, 10 mM NaCl, 10 mM MgCl2, 4 mM PMSF, 5 mM DTT and 10 mM ATP as previously described49. The supernatant was collected by two centrifugation steps at 12,000 g for 10 min at 4 °C, and protein concentration was determined with a protein assay kit (BIO-RAD, BIO-000001). The concentration of total proteins was adjusted to equal amounts in degradation buffer for each assay. One hundred nanograms of recombinant protein was incubated in 100 μl protein extracts (equivalent to 100 μg total proteins) for the individual assays. The extracts were incubated at 28 °C, and samples were taken at the indicated intervals to determine HIS-PIN2HL-MBP or HIS-MBP abundance by immunoblotting. Quantitative analysis of immunoblots was performed using Quantity Tools of Image Lab software (Bio-Rad). The dilution for the mouse anti-MBP antibody (Sigma-Aldrich, A4213) was 1:5000.
In Vivo degradation assay
10-day-old XS63 35S:PIN2HL-GFP and gls1 35S:PIN2HL-GFP transgenic plants grown in hydroponic culture were treated with 100 μM CHX for 5, 15, 30, 45, 60 and 90 min. Proteins were extracted from roots using extraction buffer containing 25 mM Tris, pH 7.5, 1 mM EDTA, 150 mM NaCl, 1% (v/v) Triton X-100, and 5% (v/v) glycerol plus 1 mM PMSF, 20 mM MG132, and 1× protease inhibitor cocktail (Roche, 4693132001). Anti-GFP antibody (Sigma-Aldrich, G1544) was used to detect PIN2HL-GFP fusion proteins, and anti-Actin antibody (Sigma-Aldrich, A3853) was used as a loading control.
RNA extraction, reverse transcription and RT-qPCR
Total RNA was extracted from different plant tissues using TRIzol reagent (Invitrogen, 15596026). First-strand cDNA was synthesized from 2 μg total RNA using HiScript II Reverse Transcriptase (Vazyme, R201-02). qPCR was performed using SYBR® Green I Master Mix (Roche, 4913850001) on a LightCycler® 480 II Real Time PCR system (Roche) according to the manufacturer’s instructions. The specificity of the reactions was verified by melting-curve analysis. Relative RNA levels for each gene were calculated from cycle threshold (CT) values according to the ΔCT method (Applied Biosystems, http://www.appliedbiosystems.com). The qPCR program included a denaturation step (3 min, 95 °C) and 45 cycles for an amplification and detection step (10 s, 95 °C denaturation; 10 s, 60 °C primer annealing; 20 s, 72 °C amplification), followed by a melting-curve program (denaturation at 95 °C; cooling and holding at 65 °C for 60 s and heating at a speed of 0.1 °C/s to 95 °C). At the end, a cooling step of 5 min at 40 °C was performed. OsACTIN, OsUBQ5, OseEF-1a and OsGAPDH were used as internal controls for RT-qPCR. The primer sequences are listed in Supplementary Table 2.
Shoot gravistimulation and measurement
Rice seeds were surface sterilized using 75% (v/v) ethanol for 2 min and 30% (v/v) bleach for 30 min with gentle shaking and washed three times with sterile double-distilled water. The seeds were allowed to germinate on plates filled with 0.5% (w/v) agarose for 3 days in the dark at 30 °C and then rotated 90° (i.e., from their original vertical orientation to horizontal), and curvature was measured after 6 h. Digital images were taken from at least 20 seedlings at each time point, and their shoot gravitropic curvature was analyzed using ImageJ software (http://rsb.info.nih.gov/ij/).
Measurements of 15N and other nutrients
For 15N absorption experiment under soil culture, rice seedlings of the wild type XS63 and gls1 mutant plants were grown in polyvinyl chloride columns (20-cm diameter × 40-cm height) containing sieved soil (<2 mm) and supplied with 0.66 g 15N-labeled urea and 0.48 g mixed fertilizers (fertilized separately at tillering and grain filling stage). Tissues were sampled at different time points after the second fertilization. For 15N absorption experiment under solution culture, 4-week-old rice plants grown in hydroponic culture were grown in solution without N for 1 week, and the plants were then transferred to a full-strength nutrient solution containing 0.25 mM 15N-labeled urea for different time before harvest. For 15N measurements, about 10 mg powder of each sample was analyzed using a MAT253-Flash EA1112-MS system (Thermo Scientific). The concentrations of other nutrient elements in plants were measured by ICP analysis as previously described50. About 10 mg of dried sample was digested with HNO3:H2O2 (5:1; v/v) using microwave digestion. After cooling, the digested sample was diluted to 50 ml with distilled water. The ion concentrations in the solution were measured using an ICP-OES instrument (Optima 8000, Perkin Elmer). The entire experiment was repeated twice and each time with three replicates.
Statistical analysis and reproducibility
The experiments performed in this study were repeated at least two times, and similar results were obtained each time. All results were analyzed and plotted in GraphPad Prism v.9 (www.graphpad.com/features). SPSS software was used for statistical analysis. Significant differences between two sets of data were determined by unpaired two-tailed Student’s t-test (*p < 0.05, **p < 0.01 and ***p < 0.001); differences among more than two sets of data were analyzed by one-way analysis of variance (ANOVA) followed by Tukey’s honestly significant difference test (p < 0.01).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2021YFF1000400), the Natural Science Foundation of Zhejiang Province, China (LZ24C150001; LQ22C020001), the National Natural Science Foundation of China (32372806), the Fundamental Research Funds for the Central Universities (226-2024-00102), and the Ministry of Education and Bureau of Foreign Experts of China (B14027).
Author contributions
C.M. and Y.Li designed the research. Y.Li, M.R., Y.W., L.W., K.Z., H.G., M.L., Y.Liu, J.Z., and J.X. performed the experiments. Y.Li, M.R., X.M., Z.W., C.L., and S.Z. analyzed the data. Y.Li, and C.M. wrote the article.
Peer review
Peer review information
Nature Communications thanks Angus Murphy, and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The authors declare that all data supporting the findings of this study are available within the manuscript and the Supplementary files. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yong Li, Meiyan Ren.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55324-5.
References
- 1.Khan, M. A., Gemenet, D. C. & Villordon, A. Root system architecture and abiotic stress tolerance: current knowledge in root and tuber crops. Front. Plant Sci.7, 1584–1597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shahzad, Z. & Amtmann, A. Food for thought: how nutrients regulate root system architecture. Curr. Opin. Plant. Biol.39, 80–87 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rogers, E. D. & Benfey, P. N. Regulation of plant root system architecture: implications for crop advancement. Curr. Opin. Biotech.32, 93–98 (2015). [DOI] [PubMed] [Google Scholar]
- 4.de Dorlodot, S. et al. Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci.12, 474–481 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Wasson, A. et al. Traits and selection strategies to improve root systems and water uptake in water-limited wheat crops. J. Exp. Bot.63, 3485–3498 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Lynch, J. Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Ann. Bot.112, 347–357 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uga, Y. et al. Control of root system architecture by DEEPERROOTING 1 increases rice yield under drought conditions. Nat. Genet.45, 1097–1102 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Lynch, J. P. Root phenes for enhanced soil exploration and phosphorus acquisition: tools for future crops. Plant Physiol.156, 1041–1049 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kitomi, Y. et al. Root angle modifications by the DRO1 homolog improve rice yields in saline paddy fields. Proc. Natl Acad. Sci. USA117, 21242–21250 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oo, A. Z. et al. Synergy between a shallow root system with a DRO1 homologue and localized P application improves P uptake of lowland rice. Sci. Rep.11, 9484 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su, S. H., Gibbs, N. M., Jancewicz, A. L. & Masson, P. H. Molecular mechanisms of root gravitropism. Curr. Biol.27, 964–972 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Kirschner, G. K. et al. ENHANCED GRAVITROPISM 2 encodes a STERILE ALPHA MOTIF-containing protein that controls root growth angle in barley and wheat. Proc. Natl Acad. Sci. USA118, e2101526118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishimura, T. et al. Cell polarity linked to gravity sensing is generated by LZY translocation from statoliths to the plasma membrane. Science381, 1006–1010 (2023). [DOI] [PubMed] [Google Scholar]
- 14.Chen, J. Y. et al. Amyloplast sedimentation repolarizes LAZYs to achieve gravity sensing in plants. Cell186, 1–15 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang, Y. Z. et al. Auxin-mediated statolith production for root gravitropism. New Phytol.224, 761–774 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Roychoudhry, S. et al. Antigravitropic PIN polarization maintains non-vertical growth in lateral roots. Nat. Plants9, 1500–1513 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu, J. S. et al. CRD 1, an Xpo1 domain protein, regulates mi RNA accumulation and crown root development in rice. Plant J.100, 328–342 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Li, Y. et al. Functional divergence of PIN1 paralogous genes in rice. Plant Cell Physiol.60, 2720–2732 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Wang, L. L. et al. LARGE ROOT ANGLE1, encoding OsPIN2, is involved in root system architecture in rice. J. Exp. Bot.69, 385–397 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giri, J. et al. Rice auxin influx carrier OsAUX1 facilitates root hair elongation in response to low external phosphate. Nat. Commun.9, 1408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, G. et al. Rice actin binding protein RMD controls crown root angle in response to external phosphate. Nat. Commun.9, 2346 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi, J. H., Hao, X., Wu, Z. C. & Wu, P. A new genetic factor for root gravitropism in rice (Oryza sativa L.). J. Zhejiang Univ.-Sc. B10, 777–783 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorup-Kristensen, K. et al. Digging deeper for agricultural resources, the value of deep rooting. Trends Plant Sci.25, 406–417 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Huang, F. et al. Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell22, 1129–1142 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tracy, S. R. et al. Crop improvement from phenotyping roots: highlights reveal expanding opportunities. Trends Plant Sci.25, 105–118 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Gamuyao, R. et al. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature488, 535–539 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Huang, Y. et al. Improving rice nitrogen-use efficiency by modulating a novel monouniquitination machinery for optimal root plasticity response to nitrogen. Nat. Plants9, 1902–1914 (2023). [DOI] [PubMed] [Google Scholar]
- 28.Barbosa, I. C. R., Hammes, U. Z. & Schwechheimer, C. Activation and polarity control of PIN-FORMED auxin transporters by phosphorylation. Trends Plant Sci.23, 523–538 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Abas, L. et al. Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. cell biol.8, 249–256 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Friml, J. et al. A PINOID-dependent binary switch in apical-basal polar PIN targeting directs auxin efflux. Science306, 862–865 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Dhonukshe, P. et al. Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS (N/S) motifs to direct apical PIN recycling. Development137, 3245–3255 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Michniewicz, M. et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell130, 1044–1056 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Weller, B. et al. Dynamic PIN-FORMED auxin efflux carrier phosphorylation at the plasma membrane controls auxin efflux-dependent growth. Proc. Natl Acad. Sci. USA114, 887–896 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laxmi, A., Pan, J., Morsy, M. & Chen, R. Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana. PloS one3, e1510 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leitner, J. et al. Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Proc. Natl Acad. Sci. USA109, 8322–8327 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Retzer, K. et al. Endosomally localized RGLG-Type E3 RING-Finger ligases modulate sorting of ubiquitylation-mimic PIN2. Int. J. Mol. Sci.23, 6767 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sassi, M. et al. COP1 mediates the coordination of root and shoot growth by light through modulation of PIN1- and PIN2-dependent auxin transport in Arabidopsis. Development139, 3402–3412 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Verrier, P. et al. Plant ABC proteins–a unified nomenclature and updated inventory. Trends Plant Sci.13, 151–159 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Wang, M. et al. The auxin influx carrier, OsAUX3, regulates rice root development and responses to aluminium stress. Plant Cell Environ.42, 1125–1138 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Xu, Y. et al. OsABCB14 functions in auxin transport and iron homeostasis in rice (Oryza sativa L.). Plant J.79, 106–117 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Zhang, Q. et al. The putative auxin efflux carrier OsPIN3t is involved in the drought stress response and drought tolerance. Plant J.72, 805–816 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Yu, C. et al. The auxin transporter, OsAUX1, is involved in primary root and root hair elongation and in Cd stress responses in rice (Oryza sativa L.). Plant J.83, 818–830 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Ye, R. et al. Primary root and root hair development regulation by OsAUX4 and its participation in the phosphate starvation response. J. Integr. Plant Biol.63, 1555–1567 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Chen, H. et al. E3 ubiquitin ligase SOR1 regulates ethylene response in rice root by modulating stability of Aux/IAA protein. Proc. Natl Acad. Sci. USA115, 4513–4518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakai, T. et al. The wavy growth 3 E3 ligase family controls the gravitropic response in Arabidopsis roots. Plant J.70, 303–314 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Konstantinova, N. et al. WAVY GROWTH Arabidopsis E3 ubiquitin ligases affect apical PIN sorting decisions. Nat. Commun.13, 5147 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, S. et al. VLN2 regulates plant architecture by affecting microfilament dynamics and polar auxin transport in rice. Plant Cell27, 2829–2845 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang, Z. et al. PROTEIN PHOSPHATASE95 regulates phosphate homeostasis by affecting phosphate transporter trafficking in rice. Plant Cell32, 740–757 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, F. et al. CASEIN KINASE2-dependent phosphorylation of PHOSPHATE2 fine-tunes phosphate homeostasis in rice. Plant Physiol.183, 250–262 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He, Q. et al. OsbHLH6 interacts with OsSPX4 and regulates the phosphate starvation response in rice. Plant J.105, 649–667 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the manuscript and the Supplementary files. Source data are provided with this paper.