Abstract
Magnetic resonance (MR) images are commonly used to diagnose prolapsed lumbar intervertebral disc (PLID). However, for a computer-aided diagnostic (CAD) system, distinguishing between pathological abnormalities of PLID in MR images is a challenging and intricate task. Here, we propose a comprehensive model for the automatic detection and cropping of regions of interest (ROI) from sagittal MR images using the YOLOv8 framework to solve this challenge. We also propose weighted average ensemble (WAE) classification and segmentation models for the classification and the segmentation, respectively. YOLOv8 has good detection accuracy for both the lumbar region (mAP50 = 99.50%) and the vertebral disc (mAP50 = 99.40%). The use of ROI approaches enhances the accuracy of individual models. Specifically, the classification accuracy of the WAE classification model reaches 97.64%, while the segmentation model achieves a Dice value of 95.72%. This automatic technique would improve the diagnostic process by offering enhanced accuracy and efficiency in the assessment of PLID.
Keywords: Magnetic resonance imaging, Prolapsed lumbar intervertebral disc, YOLOv8, Weighted average ensemble, ROI
Subject terms: Engineering, Biomedical engineering, Diagnosis
Introduction
Lower back pain (LBP) has become an emerging problem in global health, impacting around 7.5% of the global population1. Currently, it is an increasingly important issue, with individuals facing significant disabilities being 77% more susceptible to LBP2. Ossification of the facets, spinal stenosis, or intervertebral disc (IVD) herniation can result in nerve compression, thereby exacerbating illness conditions associated with LBP3. A specific example of this problem is the prolapsed lumbar intervertebral disc (PLID), also known as a herniated disc, which occurs when the disc material protrudes over the normal intervertebral boundaries in the lumbar area. Because of the pressure exerted by the displacement of the spinal cord, patients experience symptoms like numbness and discomfort in the lower extremities, as well as generalized irritation. The process of disc herniation follows a series of well-defined stages. The degree of lumbar pain severity is directly correlated with the degree of nucleus displacement outside its normal boundaries throughout these stages4,5. In instances of significant severity, the level of discomfort may require the implementation of surgical measures. Consequently, an accurate herniation diagnosis is necessary for effective lower back pain therapy.
The identification of lumbar disc herniation is often dependent on the utilization of imaging modalities such as computed tomography (CT), X-rays, myelography, and magnetic resonance imaging (MRI)6,7. While CT, X-rays, and myelography utilize radiation for the purpose of medical imaging, which can potentially have adverse effects on the human body, MRI emerges as a comparatively safer alternative. When it comes to medical imaging, MRI is considered the safer option compared to CT scans, X-rays, and myelography due to the requirement of using radiation that has adverse effects on human health. As a non-invasive imaging modality, MRI utilizes radiofrequency and magnetic fields to generate accurate visual depictions of soft tissues in the spinal region, such as nerves and discs, rather than the utilization of ionizing radiation. Therefore, the safety profile and capacity to provide comprehensive imaging make MRI largely acknowledged as the most feasible technique for visualizing intervertebral disc herniation8. The utilization of a non-invasive methodology is of utmost importance within the framework of lower back pain, as it guarantees a comprehensive and secure evaluation for accurate diagnosis and appropriate therapy.
Over the past few years, substantial efforts have been devoted to developing techniques, including machine learning (ML) and deep learning (DL) techniques, for the detection and diagnosis of PLID. Schmidt et al.9 created a graphical model that employs a probabilistic approach for labeling vertebral discs in whole-spine MR images. This method is semi-automated and requires user intervention to extract features from the image manually. To solve this problem, Prisilla et al.10 proposed a fully automated YOLO model to detect lumbar intervertebral discs, which achieves detection accuracy above 90%. Several machine learning techniques have been put forth to categorize disc herniation. Hybrid models were created by Unal et al.11 to identify disc abnormalities from lumbar spine MR scans after image characteristics were manually removed. With an accuracy of 97.75%, this model was utilized to divide lumbar disc disease into two groups. Fully automated feature extraction techniques, like DL, have been presented as a solution to the issue, as the prior work required feature extraction to be done manually to train the ML model. Kubaisi et al.12 proposed a convolutional neural network (CNN) model with a transfer learning strategy to deal with the problem of insufficient training data and used the regions of interest (ROI) method, leading to the best possible results for classifying disc states into two classes. They investigated different CNN models to suggest the best one for the problem. Alsmirat et al.13 developed a computer-aided diagnostic (CAD) system using AlexNet, known as the CNN model. The researchers employed transfer learning, ROI, and data augmentation techniques to further enhance the accuracy and robustness of the model. The recognition accuracy was 91.38%, while the detection accuracy was 95.65%. Šušteršic et al.6 introduced a new deep learning strategy involving boundary box cropping through. U-Net segmentation outcomes, then classifying sagittal MR images into five categories using DiscNet. The classification accuracy of the model was 87% for the axial image and 91% for the sagittal MR image. Various segmentation models have also been put forth to diagnose disc herniation. Support vector machines (SVM) and a histogram of oriented gradients (HOG) feature extractor were used in a machine learning (ML) approach developed by Ghosh et al.14 for the identification of vertebral discs. The MR sagittal images of the spine were segmented into three different classes, such as spine, vertebrae, and intervertebral disc, using the auto-context approach. In another study, Sáenz-Gamboa et al.15 produced multiclass segmentation of the lumbar spine using an ensemble model. They ensembled different variants of U-Net models for segmenting twelve types of tissue in the sagittal plane of the spine. They achieved 76.7% mean intersection over union (mIoU) without background information. Furthermore, the BianqueNet segmentation model was introduced by Zheng et al.16 to quantify intervertebral disc degeneration (IVD) with 90.19% mIoU for the intervertebral disc. Even though there is a significant advancement in ML and DL for the classification and segmentation of disc herniation, these techniques still need expertise in interaction and a time-consuming approach due to the many image processing and semi-automated approaches for the ROI.
In this study, we have developed a comprehensive automated approach for identifying and extracting the ROI from sagittal MR images. This automated ROI extraction technique enhances classification accuracy and reduces processing power. By cropping the IVD regions, CNN models can better focus on the pathological abnormalities caused by disc herniation in MR images. Additionally, we perform both classification and segmentation of lumbar disc herniations. The automated ROI detection is executed in two stages using the YOLOv8 framework. The first stage involves identifying and cropping the lumbar area, followed by detecting the IVDs within the cropped lumbar regions. We evaluate various pre-trained Keras CNN models for image classification along with several image segmentation models, assessing their performance with and without fine-tuning using ROI images. To classify disc herniation, we use a weighted average ensemble (WAE) classification model, and for lumbar region segmentation, we employ a WAE segmentation model. Custom loss functions are used in segmentation tasks to address class imbalance issues. Since individual models often suffer from low prediction accuracy due to high bias or variance, our proposed fully automated WAE models can classify and segment MR images with high accuracy. These models are trained and tested on a private PLID dataset, delivering outstanding results. This methodology enhances the diagnostic process by providing greater efficiency and precision in evaluating lumbar disc herniation.
Materials and methods
A. Collection of dataset
In this study, the diagnostic records and spine MR images of individuals with LBP are obtained from the radiology information system at Mymensingh Medical College Hospital. These data records were obtained between March 30, 2023, and April 5, 2023, under the close surveillance and monitoring of the hospital authority and qualified health professionals. Besides, the corresponding authority of Mymensingh Medical College Hospital has permitted us to use this data for study and research by providing a "no objection certificate." The classification involved the collection of mid-sagittal T2W lumbar MR images from 286 patients diagnosed with PLID and 11 patients with no significant spinal problems. In the segmentation task, mid-sagittal T2W lumbar MR images were used from a larger number of 403 patients, including 286 with PLID, 25 with protrusion and disc bulge, 17 with spinal canal narrowing, 64 with a degenerative disc, and 11 with normal findings. Some irrelevant images were excluded during the study. These images are obtained using a Philips 1.5 T MRI scanner. The study was conducted in accordance with Mymensingh Medical College Hospital’s relevant guidelines and regulations, and the full dataset (MR images and associated medical records) used in this study was anonymized and de-identified. Ethical approval for this study was obtained from the Human Research Ethics Committee (HREC) of Mymensingh Medical College Hospital (Approval Number: 01-24012).
B. Labeling of the dataset
Initially, MicroDICOM software (Version: DICOM viewer 2023.1, https://www.microdicom.com/news/207-march-7-2023-new-version-dicom-viewer.html) was used to convert the DICOM pictures into JPG format. The APEER ANNOTATE17 web application is used to create masks for semantic segmentation, and these masks are exported as a tiff file, which is a lossless compression format. For the detection task, the Roboflow web application is used to label images18. Various imaging series of MRI, including axial and sagittal T1W, T2W, T2*W, and spectral attenuated inversion recovery (SPAIR), are used to diagnose disc herniation in the clinic19. However, this study employed T2W and T2*W MR images. Each series of MR images has multiple numbers of frames. Here, we only use a mid-sagittal image for each patient. Because the mid-sagittal slice has more picturesque features of disc herniation than other slices and the patients who have no T2W image, we prefer the T2*W image for increasing the amount of data.
C. Proposed system
The techniques proposed for machine learning have been executed in the following manner:
Data augmentation: In the current study, our engaged dataset has been augmented to minimize overfitting and increase the number of training pictures.
ROI detection: Initially, mid-sagittal MR scans were used to identify lumbar regions and intervertebral discs using the YOLOV8 model.
Bounding box-based ROI cropping: In this stage, the bounding box area found by the YOLOv8 model is cropped.
Classification of disc herniation: At this stage, the intervertebral disc, which is divided into two classes—the herniated disc and the lack of herniation—is used to classify disc herniation.
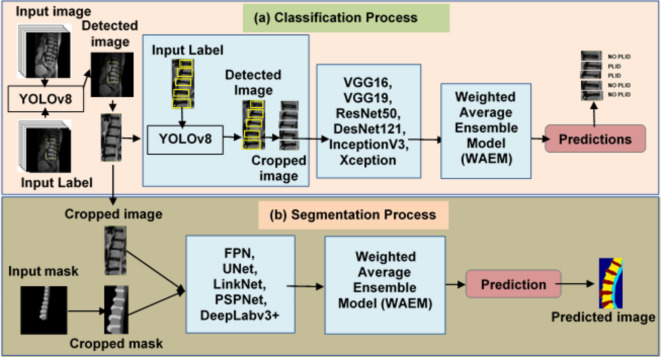
Lumbar spine segmentation: Furthermore, using sagittal T2W MR images, the lumbar regions are segmented into four unique classes: vertebral disc, vertebrae, spine, and backdrop. The overall workflow of the proposed method is shown in Fig. 1.
Fig. 1.
The overall workflow of the proposed method. (a) First, the YOLOv8 model is used to detect lumbar regions in sagittal MR images. The detected bounding box areas are then cropped. Subsequently, the IVDs are further detected using the YOLOv8 model. These IVDs are then classified into PLID and NO PLID categories using CNN models. (b) The cropped lumbar regions are then inputted into segmentation models to segment the lumbar regions into three distinct classes: vertebral disc, vertebrae, and spine.
Google Colab, a platform with a graphics processing unit (GPU) backend, was used for this research. The Intel Core i5-8257U central processor unit (CPU) running at 3.90 GHz and a Tesla K80 GPU with 10 GB of RAM made up the processing gear used in this study. Models were implemented using the Keras and TensorFlow frameworks20.
Data augmentation
Data augmentations were needed to increase the training data in machine learning21. This improves the performance and robustness of the machine-learning models21. For the detection tasks, the MR images were set to a mode of ‘black’ and had a rotation range spanning from −200 to + 200. In order to adjust brightness, the mid-sagittal MR images were adjusted to a brightness range of −25 to + 25 percent. For the classification and segmentation tasks, the following augmentation was performed:
Random rotation: The rotation parameter was set to a range of 00 to 300, utilizing the ‘nearest’ mode for image orientations.
Brightness: The brightness levels ranged from 10 to 50 percent.
Horizontal flip: A horizontal flip was performed.
Shifting: Horizontal and vertical shifts ranged from 0 to 20 percent, with ‘nearest mode’.
Transformations that had physical significance were performed. Some irrelevant transformations, such as the vertical shift, were excluded. The datasets were divided as follows: 75%, 15%, and 10% were used for training, validation, and testing, respectively. In addition, the image size used for this study is 400 × 400.
Region of interest (ROI) detection
The YOLO model has been utilized in several applications for detecting, segmenting, and classifying medical images22,23. This network functions on a single stage, in contrast to the two-stage detection mechanism of the region-based convolutional neural network (R-CNN). Singular neural networks process information more quickly than two-stage detection networks24. The sagittal MR image detection is accomplished using YOLOv8. In this study, we set epochs to 100, a batch size of 8, a learning rate of 0.02, and stochastic gradient descent (SGD) with a momentum value of 0.9.
ROI cropping using bounding box
The cropping ROI was executed to remove unnecessary data from the image. It reduces the computer’s processing power and increases the accuracy of the ML models25. ROI cropping can crop a portion of the image needed to highlight for medical purposes, such as lumbar regions. Two-stage detection using the YOLOv8 model is used to crop the lumbar regions and IVD. In the first stage, the lumbar regions are encircled by a rectangular box that is the detection output of the YOLOv8 model. The ROI of masks is cropped, corresponding to the rectangular box of their image. In the second stage, the IVDs are identified within the cropped lumbar regions, and these discs are then further cropped for use in the classification task. The dimension of a rectangular box is augmented by 10% to ensure that the necessary data associated with the ROI is addressed.
Weighted average ensemble methods
Ensembling is a methodology employed in machine learning that involves the combination of decisions derived from multiple models. This study employed a weighted average ensembling approach, wherein weight values were assigned based on the performance of the models. Finding the optimum model’s weight is the key objective of the ensembling approach. In this study, the grid search algorithm was employed to ascertain the optimal weight for the model. Grid search is a widely used method for determining the optimal combination of parameters within a predefined range.
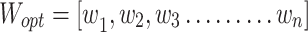 |
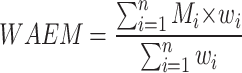 |
1 |
The optimal weights given to the models, which vary from 0 to 1, are indicated by the variable "Wopt". “WAEM” stands for the weighted average ensemble model, which generates predictions based on the weighted models, whereas "Mi" denotes the individual models. In the classification task the n value was 6 and in the segmentation task it was 8.
Disc herniation classification
In this stage, several pre-trained Keras models were employed to determine the optimal models for a WAE approach. The primary objective of classification is to categorize MR images of the spine as either suffering from herniation or not. Six different Keras models have been investigated:
Visual Geometry Group (VGG16/19)26,
Residual Networks (ResNet50)27,
InceptionV328,
Extreme Inception (Xception)29,
Densely Connected Convolutional Networks (DenseNet121)30
In earlier research, these models showed excellent performance in disc state classification12,13,30. There are two variants of the VGGNet architecture: VGG16 and VGG19. The VGG16 and VGG19 models have fully linked, pooling, and convolutional layers in their architecture. Interestingly, VGG19 contains fourteen convolution layers, whereas VGG16 has only twelve26. This is a considerable difference in the amount of convolution layers between the two. A residual block with up to 150 levels has been introduced by the ResNet architecture27. ResNet50 was selected for this study because it performed better than the previous ResNet versions. The InceptionV3 model comprises various layers, including convolution, pooling, batch normalization, dropouts, and fully connected layers28. The Xception model incorporates depth wise separable convolutions into its architecture. This convolution reduces computation time compared to classical convolution29. The accuracy of classifications may decrease as the layers in a CNN become more profound and complex, and the information from the input layers may potentially diminish with every successive layer added. DenseNet introduces a novel architecture incorporating dense blocks to establish connections between layers. The dense block comprises convolution, batch normalization, and ReLU activation layers30. This study investigated transfer learning from ImageNet with and without fine-tuning approaches for disc state classification. Utilizing the transfer learning approach minimizes the challenge caused by limited training data31. The investigation was performed as follows:
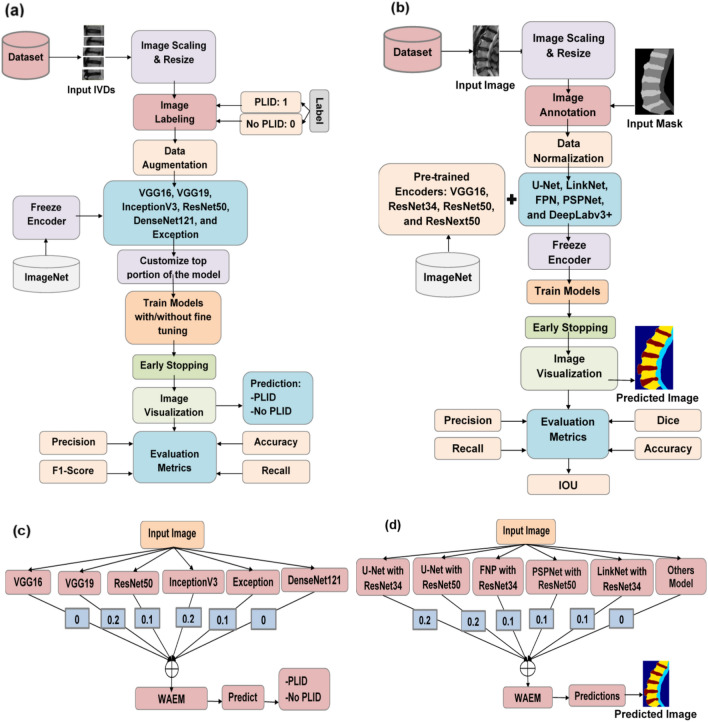
The bootstrapping approach is used to train these pre-trained models. For our classification needs, we modify the top layers of the pre-trained models while freezing their convolution layers. We refer to this process as not being fine-tuned. Some convolution layers that have been frozen earlier are unfrozen and trained on a bespoke dataset during the fine-tuning phase. Both with and without fine-tuning, ROI images are run through the models. In this study, we configure the model with 2 dense layers, 1 dropout layer with a dropout rate of 20%, epochs of 35, a batch size of 5, and use the Adam optimizer. The softmax activation function is applied, with categorical cross-entropy as the loss function. The learning rate is set to 0.001 for feature extraction and 0.0001 for the fine-tuning approach. Table S1 in the supplementary materials displays the hyperparameters used in training. The training procedure is terminated using the early stop function with patience 10 to avoid the training data overfitting. The optimization of models has been achieved by altering the hyperparameters. Various techniques are employed to address class imbalances in datasets, including data augmentation, class weighting, and algorithmic modifications. Class weighting refers to assigning reduced weights to the majority class and increased weights to the minority class during training. We assign a weight of 0.702 to the NO PLID data and a weight of 1.73 to the PLID data. The weights are set as follows: 0.1 for ResNet50 and Exception, 0.2 for VGG16 and InceptionV3, 0 for VGG16 and DenseNet121. A grid search procedure was employed to determine the optimal weighted values. Images without an ROI are also examined using these approaches. The training procedure was ended by using the early stop function to avoid overfitting the training data. Evaluation measures were computed once the models were trained. These models were weighted average ensembles, and a grid search procedure was used to identify the best-weighted values. The overall classification workflow is shown in Fig. 2a, and the ensembling process is shown in Fig. 2b.
Fig. 2.
Flowchart depicting the entire training and ensembling process. The details of (a) the training procedure for classification tasks, (b) the training procedure for segmentation tasks, (c) the ensembling procedure for classification tasks, and (d) the ensembling procedure for segmentation tasks. Once the models complete their training, they are combined using weighted average ensembling techniques.
Custom loss function
Dice loss32 measures the similarity between predicted and ground truth segmentation masks. The model tries to reduce the dice loss during training. Dice loss is especially useful for imbalanced segmentation tasks. Dice loss can be represented as,
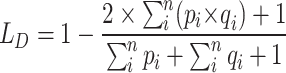 |
2 |
where n represents the number of pixels in the segmentation mask, 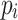 is the probability of classifying pixel “i” as belonging to the object in the predicted mask.
is the probability of classifying pixel “i” as belonging to the object in the predicted mask. 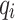 is the ground truth probability of classifying pixel “i” as belonging to the object in the ground truth mask. The focal loss33 is a loss function commonly employed in ML, with its primary purpose being to reduce the effect of class imbalance challenges. It can be represented as,
is the ground truth probability of classifying pixel “i” as belonging to the object in the ground truth mask. The focal loss33 is a loss function commonly employed in ML, with its primary purpose being to reduce the effect of class imbalance challenges. It can be represented as,
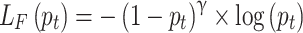 |
3 |
Here γ ˃ 0 when γ = 1, it works like a cross-entropy function, 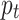 represents the predicted probability for the correct class. We used compound loss34,
represents the predicted probability for the correct class. We used compound loss34,
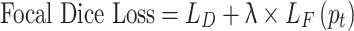 |
4 |
The trade-off between dice and focal loss is λ.
Lumbar region segmentation
Several segmentation models have been investigated after various preprocessing techniques, including ROI cropping, annotation, and resizing. In this stage, the following segmentation models were used:
U-Net35
LinkNet36
Feature Pyramid Network (FPN)37
Pyramid Scene Parsing Network (PSPNet)38
DeepLabv3 + 39
Images are categorized using the U-Net architecture by allocating a specified class to each pixel. Two components comprise the system: an encoder that extracts features from images and lowers them to a lower dimension, and a decoder that concatenates these features to produce the required output35. LinkNet refers to the network that results when addition is used instead of concatenation in U-Net36. Again, FPN refers to the network that emerges when a 1 × 1 convolution is done, and the U-Net concatenation operation is also altered to addition37. A pyramid pooling module built within the PSPNet architecture efficiently absorbs global information into the network38. These models do not use distinct layers but operate very similarly to the U-Net. An encoder and a decoder component make up the Deeplabv3 + architecture. Contextual information is extracted from the image via its atrous convolution layer39. The U-Net, LinkNet, FPN, and PSPNet architectures were used in the Segmentation Models repository on GitHub. Each of these designs had an encoder that was previously trained using ImageNet40. Pre-trained encoders from VGG19, ResNet34/50, and ResNext5041 were employed in the current study. Even though more encoders were tested, the results were insufficient to meet the desired goals. The procedures were performed as follows:
Individual models like U-Net, LinkNet, FPN, and PSPNet with pretrained encoders such as VGG16, ResNet34, ResNet50, and ResNext50, as well as DeepLabv3 + with ResNet50, are examined on private MR datasets with PLID. We set the epoch to 30, the batch size to 3, use the Adam optimizer with a learning rate of 0.003, and implement an early stop function with patience 10 to terminate training. Here we have especially used the custom loss function ‘Dice Focal Loss’ to reduce the effect of class imbalance challenges by using class weights of 0.25. Table S2 in the supplementary materials displays the hyperparameters used in the training process. After training the models, we combine them using a weighted average-ensembling approach. The weights are set as follows: 0.1 for FNP with ResNet34, LinkNet with ResNet34 and PSPNet with ResNet50, 0.2 for U-Net with ResNet34 and ResNet50, and 0 for the others model. A grid search procedure is used to determine the optimal weight values. Figure 2c, d explains the overall segmentation processes.
Evaluation metrics
A thorough understanding may not always be obtained from classification accuracy only, especially if there is an imbalance in the distribution of classes within the observed data. Rather, a better comprehension of unbalanced data is provided by accuracy, the area under the receiver operating characteristic (ROC) curve, recall, f1-score, and confusion matrix42. The models produce better results for some assessment measures. An explanation of assessment metrics is provided below.
Accuracy: it quantifies the model’s accuracy by determining the proportion of correct outcomes by the model relative to the total number of input cases. Accuracy can be represented as
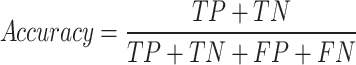 |
5 |
Specificity: the metric measures the proportion of correct negative outcomes by the model to the total number of negative instances. Specificity can be represented as
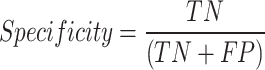 |
6 |
Precision: It measures the proportion of correct positive outcomes by the model to the total number of cases the model classified as positive. Precision can be represented as
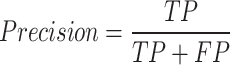 |
7 |
Recall: the metric measures the proportion of correct positive outcomes by the model to the dataset’s total number of positive cases. Recall can be represented as
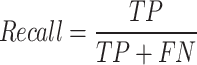 |
8 |
F-Score: it is calculated by taking the mean of the recall and precision value harmonics, providing a more balanced evaluation of the model’s overall effectiveness. F-Score can be represented as
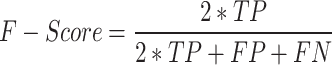 |
9 |
Mean average precision (mAP): the average precision (AP) indicates how precision varies with recall as the classification threshold is altered. mAP is a metric that calculates the average precision over multiple categories in a multi-class detection task. The number of classes is represented here by M. Formula of mAP can be written as,
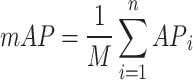 |
10 |
Dice similarity coefficient (DSC): it provides a numerical measure of how closely the predicted segmentation map (A) matches the actual segmentation map (B)43. DSC can be represented as,
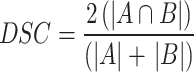 |
11 |
Intersection over union (IoU): ): it provides a quantitative measure of the amount of overlap that exists between the predicted segmentation maps (A) and the ground truth segmentation maps (B) by dividing the area of their intersection by the area of their union44. The mean Intersection over Union (mIoU) values were computed using Eq. (5) for various class labels denoted by "i”. IoU can be represented as
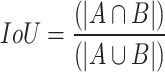 |
12 |
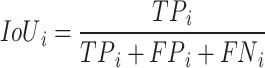 |
13 |
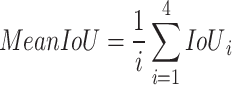 |
14 |
where FP represents the occurrence of a false positive, TP represents the occurrence of a true positive, FN represents the occurrence of a false negative, and TN represents the occurrence of a true negative.
Results and discussions
A. Detection results
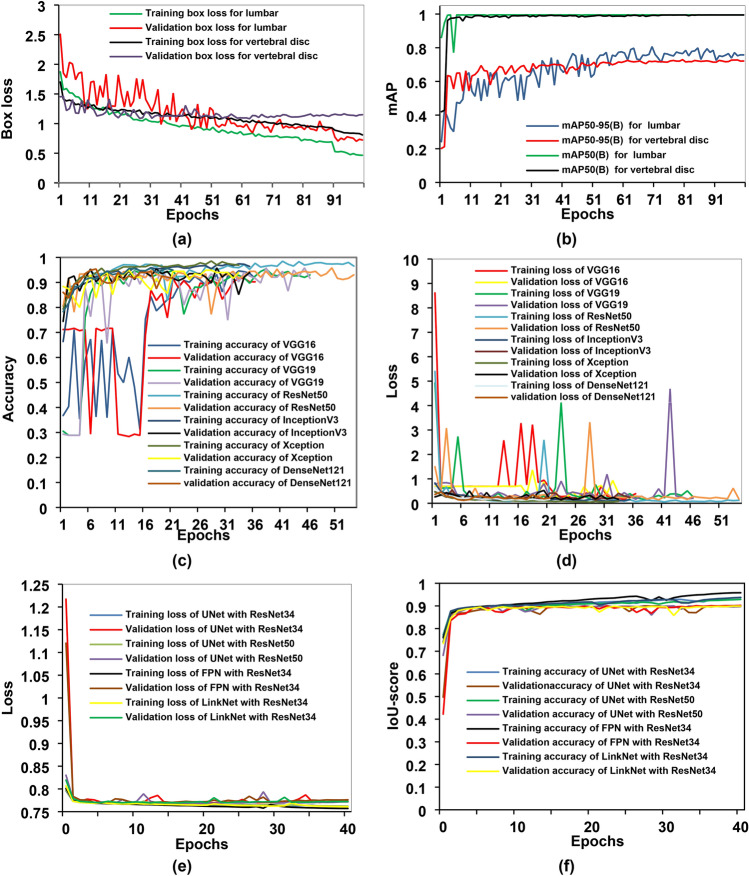
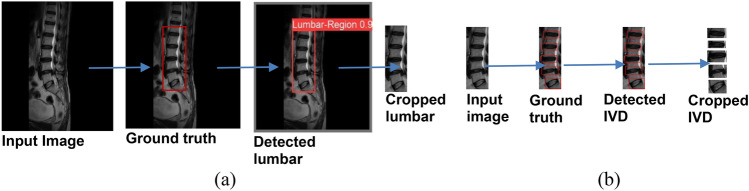
The training and validation box loss and mAP curves for sagittal MR images are shown in Fig. 3a, b. Initially, the validation curve shows some irregularities at the beginning of the training process. These fluctuations can be attributed to the variability in the spine MR images or an insufficient amount of training and validation data. Such inconsistencies in the dataset can lead to unusual patterns in the validation curve during the early stages of training. The assessments of the experiment employed mAP50 and mAP50-95 metrics to evaluate the clarity of the experimental results. For lumbar region detection, the mAP50, precision, and recall values are 99.50%, 99.9%, and 100.00%, respectively. For vertebral disc detection, the mAP50 is 99.40%, with precision at 98.70% and recall at 99.30%. The results show that the model performs remarkably well in detecting both the lumbar area and vertebral discs, with nearly perfect precision and recall metrics. The high mAP50 values in both categories imply robust performance at lower confidence thresholds. However, the drop in mAP50-95, notably in the vertebral discs, indicates that the model’s performance is less consistent at higher confidence thresholds. This is usual, as higher confidence thresholds necessitate more precise localization. Overall, the findings are extremely positive, especially given the vital importance of correct detection in medical imaging. The nearly perfect precision and recall values demonstrate the model’s reliability in real-life applications. Improvements could focus on improving performance at higher confidence thresholds, resulting in even greater accuracy across all levels of overlap. A confidence value of 0.25 was utilized during the object detection process, resulting in a 100 percent detection rate for the lumbar and intervertebral discs. Only detections with confidence scores greater than 0.25 are considered valid. The detection results for the lumbar region and IVDs are shown in Fig. 4. Initially, the YOLOv8 model was used to detect the lumbar spine region. Once detected, the model cropped the lumbar regions based on these detections, as illustrated in Fig. 4a. The cropped lumbar regions were then reintroduced into the YOLOv8 model for a second round of detection, this time focusing on identifying the lumbar IVDs. The model detected the lumbar IVDs and subsequently cropped these regions according to the new set of detections, as shown in Fig. 4b.
Fig. 3.
Training and validation metrics charts of various models. (a,b) Depict the training and validation box loss and mAP curve, respectively, for both lumbar region and IVD detection using YOLOv8. (c,d) Represent the accuracy and loss curves of different fine-tuned CNN classification models. Additionally, (e,f) show the training and validation loss and IoU score curves of different segmentation models.
Fig. 4.
The detection and cropping outcomes for both (a) the lumbar region and (b) the IVD in an MR image. The initial stage (a) shows detection and the cropping of lumbar regions. Subsequently, the second stage (b) depicts the detection and cropping of IVD following the identification of the lumbar regions.
The YOLOv8 model used here accurately identifies the lumbar regions and IVDs, providing bounding boxes for the regions of interest (ROI). After detection, YOLOv8’s outputs are used to crop and isolate the detected IVDs, providing clean and focused input data for the classification task. This cropping step significantly reduces noise and irrelevant information in the input, which improves the performance of subsequent classification models (such as CNN architectures). By providing high-quality, precisely cropped ROIs, YOLOv8 enhances the feature extraction process and ensures that the classification models can focus on the relevant features indicative of conditions like disc herniation.
The YOLOv8 model is also renowned for its exceptional speed, making it suitable for real-time applications. The other model such as Faster R-CNN is typically slower due to their more complex processing pipelines. The YOLOv8 model has demonstrated superior accuracy compared to Faster R-CNN and EfficientDet. For instance, the YOLOv8 model achieved an mAP50 of 0.62 with a GPU latency of 1.3 ms, while Faster R-CNN achieved an mAP50 of 0.41 with a GPU latency of 54 ms and EfficientDet achieved an mAP50 of 0.4745. The YOLOv8 model also excels in real-time object detection, offering superior speed and efficiency compared to Faster R-CNN, SSD, and RetinaNet46. While transformer-based models such as DETR have shown promise, YOLOv8 delivers competitive accuracy with lower computational overhead47.
B. Classification results
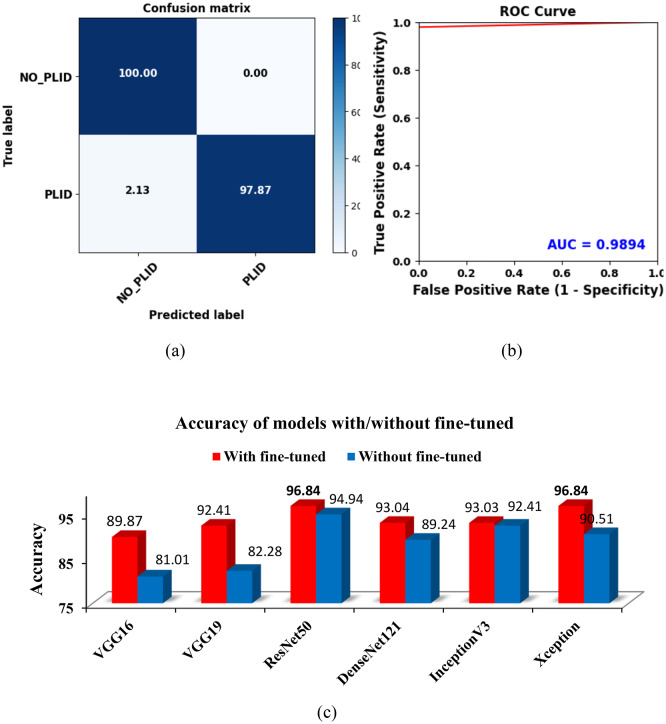
Using ROI (IVDs) images, feature extraction and fine-tuned models were examined throughout the classification phase. Figure 3c, d displays the accuracy and loss curves for the various fine-tuned CNN models. It shows that the ResNet50 model has the lowest loss and the greatest accuracy for both the training and validation phases. However, due to insufficient training and validation data, there were variations in the accuracy and loss curves. These inconsistencies underscore the impact of data quantity on the model’s performance. The class imbalance issue has emerged in this case since there are more normal instances than PLID cases. To address the issue, we weighted the majority class with a lower value during training and the minority class with a higher value. This adjustment helps the model focus more on the minority class. As a result of this weighting approach, the WAE model achieved a prediction accuracy of 100% for normal images and 97.87% for herniated disc images, as illustrated in Fig. 5a. Additionally, as depicted in Fig. 5b, the model’s AUC is a commendable 98.94%, with a true positive rate of 97.87% and a false positive rate of 0. This indicates the model’s strong performance in distinguishing between normal and herniated disc cases.
Fig. 5.
Evaluation of the WAEM’s performance in the classification task. (a) The confusion matrix of the WAEM highlights its remarkable accuracy for normal images. (b) The receiver operating characteristic (ROC) curve, where the AUC of the WAEM model demonstrates its strong ability to distinguish between different classes. (c) A graphical comparison of individual model’s accuracy with and without fine-tuned approaches, showcasing the significant performance enhancement achieved through the use of ROI and fine-tuning techniques.
We also performed the ablation studies, which assess the importance and effectiveness of each component in achieving the overall results of the proposed deep learning model. The results of the ablation study for the WAE classification and segmentation models are shown in Table 1. For the classification task, when ROI cropping is removed and fine-tuning is applied, the model achieves an accuracy of 93.10%. When ROI cropping is used but fine-tuning is not applied, the accuracy increases to 96.84%, showing a 3.74% improvement. Finally, when both ROI cropping and fine-tuning are used together, the model’s accuracy reaches 99.37%, which is an additional 2.53% improvement. This demonstrates that both ROI cropping and fine-tuning are crucial for optimizing the classification model.
Table 1.
Results of the ablation study for WAE classification and segmentation model.
| Method | ROI cropping | Fine-tuning | Custom loss function | Accuracy (%) |
|---|---|---|---|---|
| Proposed WAE classification model | ✓ | ✓ | – | 99.37 |
| Without fine-tuning | ✓ | × | – | 96.84 |
| No ROI cropping | × | ✓ | – | 93.10 |
| mIoU | ||||
|---|---|---|---|---|
| Proposed WAE segmentation model | – | – | ✓ | 91.86 |
| No custom loss function | – | – | × | 91.63 |
Based on Table 2, which presents evaluation metrics for disc state classification using various fine-tuned models on ROI (IVD) images, the WAEM has the best accuracy of any model at 99.37% and a top AUC score of 98.94%, suggesting strong performance in differentiating between different disc states. Notably, the WAEM model achieves perfect specificity and precision, indicating that it accurately detects all positive instances while eliminating false positives. Furthermore, it has a high recall of 97.87% and a remarkable F1-Score of 98.93%, exhibiting both sensitivity to disc conditions and overall balanced performance across accuracy and recall parameters. These findings highlight the efficiency of the ensemble technique, which potentially utilizes the capabilities of multiple models to improve classification accuracy and reliability in medical applications. Figure 5c depicts a graphical representation of the accuracy of the different CNN models with and without a fine-tuned method. The results clearly show that using ROI and fine-tuning approaches improves model performance significantly. Notably, the fine-tuned ResNet50 and Xception models achieved the highest accuracy (99.84%). This demonstrates the benefits of optimizing model parameters using ROI and fine-tuning procedures to improve classification accuracy. Table 3 compares the performance of the suggested WAEM model with existing work from various authors. The proposed WAEM model achieves outstanding results, with an accuracy of 99.37%, the highest among all models given. It also achieves a perfect precision of 100%, suggesting that its predictions contain no false positives. Furthermore, WAEM has a great recall of 97.87% and a remarkable F1-Score of 98.93%, demonstrating its capacity to accurately identify disc states. Other models, including VGG16, ResNet101, AlexNet, and KNN, reported lower accuracies and varying performance metrics across experiments and datasets. This comparison demonstrates WAEM’s greater accuracy and precision, indicating that it can serve as a robust classification model for disc state identification.
Table 2.
Detailed evaluation metrics for disc state classification of ROI images, comparing performance with and without fine-tuned models.
| Models | Accuracy with fine-tuning (%) | Accuracy without fine-tuning (%) | AUC (%) | Specificity (%) | Precision (%) | Recall (%) | F1-score (%) |
|---|---|---|---|---|---|---|---|
| VGG16 | 89.87 | 81.01 | 82.98 | 100 | 100 | 65.96 | 79.49 |
| VGG19 | 92.41 | 82.28 | 87.23 | 100 | 100 | 74.47 | 85.37 |
| ResNet50 | 96.84 | 94.94 | 95.91 | 98.20 | 95.65 | 93.62 | 94.62 |
| DenseNet121 | 93.04 | 89.24 | 88.91 | 99.09 | 97.37 | 78.73 | 87.06 |
| InceptionV3 | 93.03 | 92.41 | 91.98 | 94.60 | 87.50 | 89.36 | 88.42 |
| Xception | 96.84 | 90.51 | 96.52 | 97.30 | 93.75 | 95.75 | 94.74 |
| AEM | 98.10 | 93.67 | 96.81 | 100 | 100 | 93.62 | 96.70 |
| WAEM | 99.37 | 96.84 | 98.94 | 100 | 100 | 97.87 | 98.93 |
Table 3.
Comparative analysis of proposed classification models with existing approaches.
| Authors | Models | No. of class | No. of patients | Accuracy (%) | Precision (%) | Recall (%) | F1-score (%) |
|---|---|---|---|---|---|---|---|
| Proposed model | WAEM | 2 | 286 | 99.37 | 100 | 97.87 | 98.93 |
| A. kubaisi et al.12 | VGG16 | 2 | 181 | 89.01 | 90.91 | 90.91 | 90.91 |
| Q. Pan et al.53 | ResNet101 | 5 | 500 | 88.76 | – | – | – |
| M. Alsmirat et al.13 | AlexNet | 8 | 164 | 91.38 | – | – | – |
| A. Al Imran et al.54 | KNN | 2 | 310 | 85.20 | 90.00 | 88.00 | 88.99 |
C. Segmentation results
Various pre-trained encoders and models were evaluated using ROI images. Through a grid search approach, weights were assigned as follows: 0.2 to U-Net with ResNet34 and ResNet50, 0.1 to LinkNet with ResNet34, 0.1 to PSPNet with ResNet50, and 0.1 to FPN with ResNet34, while the other networks received a weight of 0. The FPN model with ResNet34 achieved the highest validation accuracy and the lowest loss. Due to differences in MR scans and the limited quantity of training data, the models showed some overfitting. Nevertheless, the results were still effective for diagnostic purposes. The insufficient data for training and validation caused some fluctuations in the loss and accuracy curves, as illustrated in Fig. 3e, f. A comparative analysis of segmentation results generated by different models is provided, where Fig. 6a shows distinct input images using PLID and Fig. 6b the ground truth segmentation. Figure 6c–g displays the anticipated segmentation outcomes of different segmentation models. It can be shown from Fig. 6g that the utilization of WAE models yielded superior outcomes in the context of segmentation. The predicted results of single models, specifically Fig. 6e, f, exhibit over-segmentation and miss-segmentation in certain areas. However, the WAEM exhibits more precise segmentation than other models. The individual models were also evaluated with encoders such as VGG19, DenseNet121, InceptionV3, MobileNetV2, and Efficientb7, but we did not get satisfactory results for the datasets, so we excluded these models from the ensembling process. Ablation studies for the segmentation task shown in Table 1, without using the custom loss function, the model achieves a mIoU of 91.63%. When the custom loss function is applied, the mIoU increases slightly to 91.86%, showing an improvement of 0.23%. The results highlight the significant role of ROI cropping and fine-tuning in enhancing classification accuracy, while the custom loss function provides a modest but important improvement in segmentation performance. Table 4 provides a comprehensive evaluation of various models with pre-trained encoders, focusing on metrics such as mIoU and Dice coefficient. The U-Net model using the ResNet34 encoder has the highest metrics, with a mIoU of 91.55% and a Dice score of 95.57%. It indicates outstanding segmentation performance, which outperforms other encoder configurations. LinkNet, although slightly behind, demonstrates competitive performance with ResNet34, achieving a mIoU of 91.27% and a Dice score of 95.42%. However, models like FPN and PSPNet exhibit a drop in performance, particularly with the VGG16 encoder, which has the lowest mIoU at 86.18% and a Dice score of 92.52%. PSPNet with ResNet50 and ResNext50 configurations also displays relatively lower performance, with mIoUs of 90.71% and 89.50%, respectively, and corresponding Dice scores. The DeepLabv3 + model, evaluated only with ResNet50, records an mIoU of 90.20% and a Dice score of 94.83%, showing strong but not leading performance. These findings demonstrate U-Net’s superior performance across multiple encoders, particularly ResNet34, emphasizing its robustness and reliability in medical image segmentation tasks. The variations in performance between models and encoder configurations highlight the importance of model architecture and encoder selection in optimizing segmentation results. Table 5 presents a comprehensive assessment of the proposed WAE model compared to previous methods. WAEM, investigated on a dataset of 403 patients, obtained a Dice coefficient of 95.74%, one of the highest previously reported. WAEM also performs exceptionally well in segmenting certain anatomical structures, with a spine IoU of 89.37%, a vertebrae IoU of 92.18%, and a vertebral disc IoU of 90.24%, for a mean IoU of 91.86%. The others performance metrics achieved by the WAE segmentation model are a Dice score of 95.74%, an accuracy of 96.22%, a recall of 95.63%, and a precision of 95.86%. These results outperformed previous models, such as Zheng et al.'s BianqueNet16, which, despite having a high Dice score of 95.51% and comparable vertebral disc IoU, lags slightly in overall mean IoU. Notably, U-Net models and other methods such as Spine-GAN and SVM show significantly lower performance, particularly in the mean IoU. The improved attention U-Net by Wang et al.48 records a high Dice score but lacks comprehensive IoU metrics.
Fig. 6.
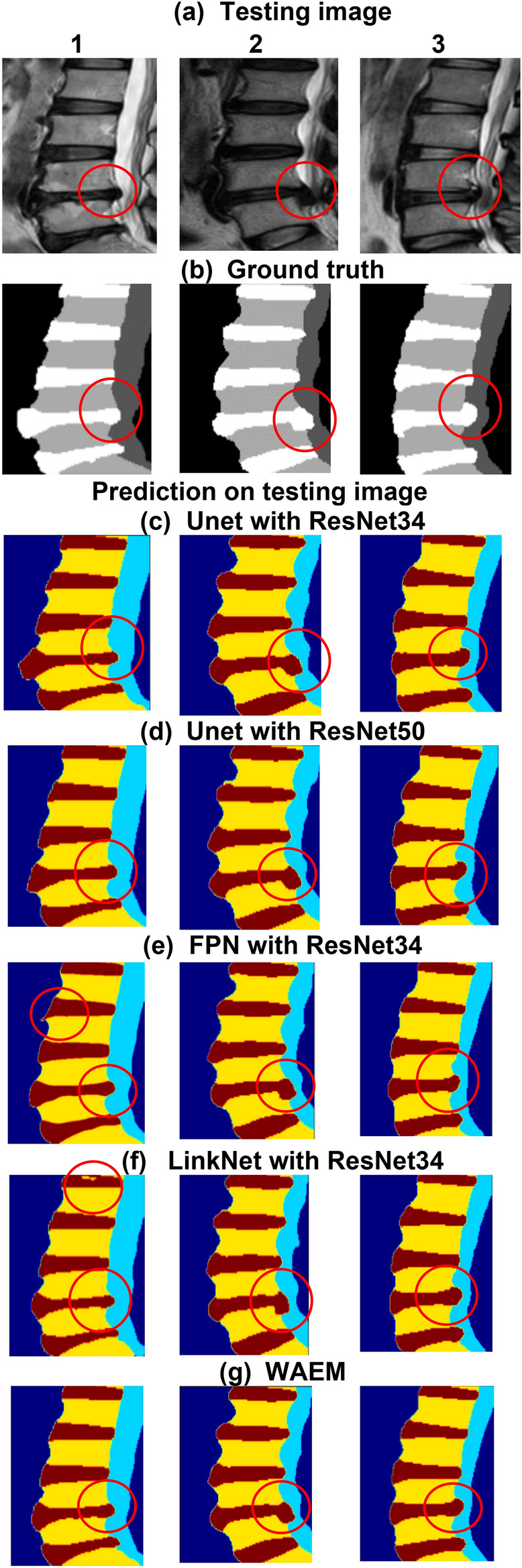
A comparison of segmentation outcomes produced by different models. (a) Showcases various input images with PLID. (b) The ground truth segmentation of the input images. (c–g) The predicted segmentation results of different segmentation models. (e,f) Represent the segmentation outcomes of single models, highlighting instances of over-segmentation and miss-segmentation in certain areas. The WAEM, represented in (g), demonstrates more precise segmentation compared to other individual models.
Table 4.
Comparative evaluation metrics for different models utilizing pre-trained encoders.
| Models | ResNet34 | ResNet50 | ResNext50 | VGG16 | ||||
|---|---|---|---|---|---|---|---|---|
| mIoU (%) | Dice (%) | mIoU (%) | Dice (%) | mIoU (%) | Dice (%) | mIoU (%) | Dice (%) | |
| U-Net | 91.55 | 95.57 | 91.20 | 95.38 | 90.90 | 95.22 | 90.83 | 95.18 |
| LinkNet | 91.27 | 95.42 | 90.11 | 94.77 | 90.83 | 95.17 | 90.64 | 95.07 |
| FPN | 90.89 | 95.21 | 89.85 | 94.63 | 89.73 | 94.56 | 86.18 | 92.52 |
| PSPNet | 90.84 | 95.18 | 90.71 | 95.11 | 89.50 | 94.43 | 90.39 | 94.93 |
| DeepLabv3 + | – | – | 90.20 | 94.83 | – | – | – | – |
Table 5.
Detailed contrast between the proposed method and previous approaches.
| Authors | Models | No. of patients | Dice (%) | Spine IoU (%) | Vertebrae IoU (%) | Vertebral disc IoU (%) | Mean IoU (%) |
|---|---|---|---|---|---|---|---|
| Proposed Model | WAEM | 403 | 95.74 | 89.37 | 92.18 | 90.24 | 91.86 |
| Zheng et al.16 | BianqueNet | 223 | 95.51 | 90.35 | 94.25 | 90.19 | 91.60 |
| J. Jairo et al.15 | U-Net | 181 | – | 75.50 | 86.00 | 88.70 | 83.40 |
| S. Wang et al.48 | Improved Attention U-Net | 180 | 95.01 | – | – | – | – |
| Z. Han et al.55 | Spine-GAN | 253 | 87.10 | 87.00 | 81.00 | 87.30 | – |
| S. Ghosh et al.14 | SVM | 212 | – | – | – | 84.00 | 84.00 |
| S. Pang et al.56 | SpineParseNet | 215 | 89.22 | – | – | – | – |
| Cheng et al.57 | MultiResU-Net | 180 | – | – | – | 77.10 | – |
Recently, Saeed et al.49 proposed the multi-scale feature fusion (MSFF) network for spine fracture segmentation using CT images. This approach involves a complex framework with six modules for spatial feature extraction, channel-wise feature enhancement, and positional focus on the region of interest. It is particularly effective in handling multi-scale features and improving segmentation border refinement. In another study, the same authors50 introduced a similar MSFF model employing multi-scale feature fusion techniques with five modules, but this approach was tailored for MRI images. In their further study51, they proposed a 3D MRU-Net, which incorporates residual blocks (MobileNetv2) and cascaded hierarchical atrous spatial pyramid pooling (CHASPP). This model facilitates multi-view feature extraction from 3D CT images. Including attention modules in the decoder enhances the model’s ability to focus on regions of interest. Furthermore, the same author52 introduced CHASPPRAU-Net for spine segmentation and 3D MRU-Net with residual blocks (MobileNetv2) for vertebra recognition. These models demonstrate a more sophisticated approach to handling 3D data while incorporating attention mechanisms. In contrast, our proposed feature extraction approach leverages pre-trained encoders such as VGG19, ResNet34/50, and ResNext50, trained on ImageNet for segmentation tasks. This approach focuses on transfer learning techniques using various architectures, including U-Net, LinkNet, FPN, PSPNet, and Deeplabv3 + . These models rely primarily on encoders for feature extraction and decoders for feature concatenation in segmentation tasks. While our approach emphasizes pre-trained 2D encoders for feature extraction, the works by Saeed et al. introduce more complex multi-scale feature fusion techniques. Their methods involve multiple modules, MobileNetv2 as an encoder (similar to our work), and CHASPP layers for feature extraction in spine-related segmentation tasks. Overall, the WAEM model establishes a new benchmark for accuracy and reliability in spine and vertebral disc segmentation, demonstrating its robustness and usefulness in clinical settings for precise anatomical segmentation.
To address the overfitting issues, we have included several measures to ensure the robustness of our model. The training and validation curves show minimal gap, indicating that overfitting is not significant. Additionally, we employed techniques such as dropout regularization, data augmentation, and an early stopping function to further mitigate overfitting. The fine-tuned pre-trained models which are trained on a large dataset were also used specifically for our work, leveraging transfer learning to improve generalization with a relatively small dataset. These efforts collectively demonstrate our commitment to reduce overfitting effectively. We acknowledge that cross-validation was not performed in this study due to time and computational constraints. However, we plan to include cross-validation in future work as part of a more comprehensive evaluation. Furthermore, although validation on independent datasets was not feasible due to a lack of access to external data, we have outlined this as a priority for future research. In the current study, to simulate independent validation and assess generalization ability, we used separate splits for training, validation, and testing, ensuring that the test set remained entirely isolated during model training. This approach allowed us to provide a robust evaluation of the model’s performance within the constraints of the available data. The accuracy of the models may degrade when applied to data from MR scanning devices of different manufacturers, due to variations in calibration and data acquisition parameters. The complexity of anatomical structures necessitates significant expertise for the manual segmentation of mid-sagittal MR images. Furthermore, categorizing intervertebral discs into specific groups is challenging, as some data points are ambiguous. For future work, it is crucial to expand the dataset, particularly with more instances of disc herniation, to improve the robustness of the models. Increasing the volume of training data can also help mitigate overfitting in detection and segmentation models.
Conclusions
We have developed an innovative approach for the accurate detection and cropping of ROI in medical imaging, specifically targeting the lumbar regions of the spine and IVDs using the YOLOv8 framework. To enhance the overall accuracy of the classification models, fine-tuned techniques are applied. To address the issue of insufficient training data, transfer learning is employed. Moreover, the problem of class imbalance is solved by assigning higher weights to the minority class and lower weights to the majority class during the training phase. The WAE classification model has demonstrated significant improvements in lumbar IVDs classification performance, achieving an accuracy of 99.37%, an F1-score of 98.93%, a precision of 100%, a recall of 97.87%, and an AUC of 98.94%. For the WAE segmentation model, we incorporated a range of pre-trained encoders, including U-Net, LinkNet, PSPNet, and FPN with VGG16, ResNet34, ResNet50, and ResNext50 encoders, as well as DeepLabv3 + with a ResNet50 encoder, all of which are pre-trained on ImageNet. This combination of models, alongside the WAE technique, has resulted in substantial performance enhancements. The WAE segmentation model achieved a mIoU of 91.86%, a Dice score of 95.74%, an accuracy of 96.22%, a recall of 95.63%, and a precision of 95.86%. With this developed automated method, PLID assessments might be done more quickly and with more precision, greatly improving the diagnosis procedure.
Supplementary Information
Acknowledgements
We are grateful to the Department of Radiology at Mymensingh Medical College Hospital.
Author contributions
Conceptualization, A. Sayed, G.M.M. Rahman; Data collection, A. Sayed, H. Ahmed, and A. Islam; Methodology, A. Sayed; Formal analysis, A. Sayed, A. Islam, H. Ahmed, and A. Hossain; Investigation and Supervision, G.M.M. Rahman; Writing original draft, A. Sayed, M. S. Islam, J. Park, A. Islam, and R. Shahrior; writing review, M. S. Islam; Editing, A. Sayed, M. S. Islam, A. Islam, J. Park and A. Hossain.
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Informed consent statement
Written informed consent from the patients is waived due to the retrospective nature of the data collection and the use of de-identified MR images, and full ethical approval has been granted.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
G. M. Mahmudur Rahman, Email: mahmud@bme.kuet.ac.bd.
Md. Sherajul Islam, Email: sheraj_kuet@eee.kuet.ac.bd.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-84301-7.
References
- 1.Wu, A. et al. Global low back pain prevalence and years lived with disability from 1990 to 2017: Estimates from the Global Burden of Disease Study 2017 (in Eng). Ann. Transl. Med.8(6), 299 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartvigsen, J. et al. What low back pain is and why we need to pay attention (in Eng). Lancet391(10137), 2356–2367 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Urban, J. P. & Roberts, S. xDegeneration of the intervertebral disc (in Eng). Arthritis Res. Ther.5(3), 120–130 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan Xin Hui Nicole, H. N. A. S. K. W. Classification of lumber spine disc herniation using machine learning methods. Orient. J. Comput. Sci. Technol.14, 01–10 (2021).
- 5.Fardon, D. F., Williams, A. L., Dohring, E. J., Murtagh, F. R., Gabriel Rothman, S. L. & Sze, G. K. Lumbar disc nomenclature: version 2.0: Recommendations of the combined task forces of the North American Spine Society, the American Society of Spine Radiology and the American Society of Neuroradiology (in Eng). Spine J.14(11), 2525–2545 (2014). [DOI] [PubMed]
- 6.Sustersic, T. et al. A deep learning model for automatic detection and classification of disc herniation in magnetic resonance images (in Eng). IEEE J. Biomed. Health Inform.26(12), 6036–6046 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Vitosevic, F., Rasulic, L. & Medenica, S. M. Morphological characteristics of the posterior cerebral circulation: An analysis based on non-invasive imaging (in Eng). Turk. Neurosurg.29(5), 625–630 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Katti, G., Ara, S. & Shireen, D. Magnetic resonance imaging (MRI) - A review. Int. J. Dent. Clin. 3, 03/31 (2011).
- 9.Schmidt, S. et al. Spine detection and labeling using a parts-based graphical model (in Eng). Inf. Process. Med. Imaging20, 122–133 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Prisilla, A. A. et al. An approach to the diagnosis of lumbar disc herniation using deep learning models (in Eng). Front. Bioeng. Biotechnol.11, 1247112 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unal, Y., Polat, K., Kocer, H. E. & Hariharan, M. Detection of abnormalities in lumbar discs from clinical lumbar MRI with hybrid models. Appl. Soft Comput.33, 65–76 (2015).
- 12.Al-kubaisi, A. & Khamiss, N. N. A transfer learning approach for lumbar spine disc state classification. Electronics11, 1. 10.3390/electronics11010085
- 13.Alsmirat, M., Al-Mnayyis, N., Al-Ayyoub, M. & Al-Mnayyis, A. Deep learning-based disk herniation computer aided diagnosis system from MRI axial scans. IEEE Access10, 32315–32323 (2022). [Google Scholar]
- 14.Ghosh, S. & Chaudhary, V. Supervised methods for detection and segmentation of tissues in clinical lumbar MRI. Comput. Med. Imag. Graph.38(7), 639–649 (2014). [DOI] [PubMed]
- 15.Sáenz-Gamboa, J. J., Domenech, J., Alonso-Manjarrés, A., Gómez, J. A. & de la Iglesia-Vayá, M. Automatic semantic segmentation of the lumbar spine: Clinical applicability in a multi-parametric and multi-center study on magnetic resonance images (in Eng). Artif. Intell. Med.140, 102559 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng, H.-D. et al. Deep learning-based high-accuracy quantitation for lumbar intervertebral disc degeneration from MRI. Nat. Commun. 13(1), 841 (2022). [DOI] [PMC free article] [PubMed]
- 17.Suchanek, A. ZEISS Arivis Cloud. https://www.apeer.com/home/ (2023).
- 18.Dwyer, J. N. A. B. Roboflow Web Application. https://app.roboflow.com/ (2020).
- 19.Jensen, E. C. Technical review, types of imaging, part 4—Magnetic resonance imaging. Anat. Rec.297(6), 973–978 (2014). [DOI] [PubMed]
- 20.Gulli, A. & Pal, S. Deep Learning with Keras 318 (Packt Publishing, 2017). [Google Scholar]
- 21.Aghnia Farda, N., Lai, J. C., Wang, P. Y., Lee, J., Liu, W. & Hsieh, I. H. Sanders classification of calcaneal fractures in CT images with deep learning and differential data augmentation techniques (in Eng). Injury52(3), 616–624 (2021). [DOI] [PubMed]
- 22.Bloice, M. D., Stocker, C. & Holzinger, A. Augmentor: An image augmentation library for machine learning. arXiv preprint arXiv:1708.04680 (2017).
- 23.Terven, J. & Cordova-Esparza, D. A comprehensive review of YOLO: From YOLOv1 to YOLOv8 and beyond. arXiv preprint arXiv:2304.00501 (2023).
- 24.Morbekar, A., Parihar, A. & Jadhav, R. Crop disease detection using YOLO. In 2020 International Conference for Emerging Technology (INCET). 1–5 (IEEE, 2020).
- 25.Yamakawa, M., Shiina, T., Nishida, N. & Kudo, M. Optimal cropping for input images used in a convolutional neural network for ultrasonic diagnosis of liver tumors. Jpn. J. Appl. Phys. 59(SK), SKKE09 (2020).
- 26.Simonyan, K. & Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014).
- 27.He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 770–778 (2016).
- 28.Xia, X., Xu, C. & Nan, B. Inception-v3 for flower classification. In International Conference on Image, Vision and Computing (ICIVC). 783–787 (IEEE, 2017).
- 29.Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 1251–1258 (2017).
- 30.Huang, G., Liu, Z., Van Der Maaten, L. & Weinberger, K. Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 4700–4708 (2017).
- 31.Kim, H. E., Cosa-Linan, A., Santhanam, N., Jannesari, M., Maros, M. E. & Ganslandt, T. Transfer learning for medical image classification: A literature review. BMC Med. Imaging22(1), 69 (2022). [DOI] [PMC free article] [PubMed]
- 32.Sudre, C. H., Li, W., Vercauteren, T., Ourselin, S. & Jorge Cardoso, M. Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: Third International Workshop, DLMIA 2017, and 7th International Workshop, ML-CDS 2017, Held in Conjunction with MICCAI 2017, Québec City, QC, Canada, September 14, Proceedings 3, 2017. 240–248 (Springer, 2017). [DOI] [PMC free article] [PubMed]
- 33.Lin, T.-Y., Goyal, P., Girshick, R., He, K. & Dollár, P. Focal loss for dense object detection. In Proceedings of the IEEE International Conference on Computer Vision. 2980–2988 (2017).
- 34.Zhu, W. et al. Anatomynet: Deep 3D squeeze-and-excitation u-nets for fast and fully automated whole-volume anatomical segmentation. BioRxiv. 392969 (2018).
- 35.Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, 2015. 234–241 (Springer, 2015).
- 36.Chaurasia, A. & Culurciello, E. Linknet: Exploiting encoder representations for efficient semantic segmentation. In IEEE Visual Communications and Image Processing (VCIP), 2017. 1–4 (IEEE, 2017).
- 37.Bodur, R., Bhattarai, B. & Kim, T.-K. A Unified Architecture of Semantic Segmentation and Hierarchical Generative Adversarial Networks for Expression Manipulation. arXiv preprint arXiv:2112.04603 (2021).
- 38.Zhao, H., Shi, J., Qi, X., Wang, X. & Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2881–2890 (2017).
- 39.Chen, L.-C., Zhu, Y., Papandreou, G., Schroff, F. & Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV). 801–818 (2018).
- 40.Iakubovskii, P. Segmentation Models. https://github.com/qubvel/segmentation_models. (2019)
- 41.Xie, S., Girshick, R., Dollár, P., Tu, Z. & He, K. Aggregated residual transformations for deep neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 1492–1500 (2017).
- 42.Hossin, M. & S. M.N. A review on evaluation metrics for data classification evaluations. Int. J. Data Min. Knowl. Manag. Process5, 01–11 (2015).
- 43.Zou, K. H. et al. Statistical validation of image segmentation quality based on a spatial overlap index (in Eng). Acad. Radiol.11(2), 178–189 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahman, M. & Wang, Y. Optimizing Intersection-Over-Union in Deep Neural Networks for Image Segmentation. 234–244 (2016).
- 45.Abramov, M. YOLOv8 vs Faster R-CNN: A Comparative Analysis. https://keylabs.ai/blog/yolov8-vs-faster-r-cnn-a-comparative-analysis/ (2024).
- 46.Sohan, M., Sai Ram, T. & Rami Reddy, C. V. A review on YOLOv8 and its advancements. In Data Intelligence and Cognitive Informatics, Singapore, 2024. 529–545 (Springer, 2024).
- 47.Deschenaux, J. How Robust are Pre-Trained Object Detection ML Models like YOLO or DETR?https://www.lakera.ai/blog/how-robust-are-pre-trained-object-detection-ml-models (2023).
- 48.Wang, S., Jiang, Z., Yang, H., Li, X. & Yang, Z. Automatic segmentation of lumbar spine MRI images based on improved attention U-Net. Comput. Intell. Neurosci.2022, 4259471 (2022). [DOI] [PMC free article] [PubMed]
- 49.Saeed, M. U., Bin, W., Sheng, J. & Mobarak Albarakati, H. An automated multi-scale feature fusion network for spine fracture segmentation using computed tomography images. J. Imag. Inform. Med. 37(5), 2216–2226 (2024). [DOI] [PMC free article] [PubMed]
- 50.Saeed, M. U., Bin, W. , Sheng, J., Albarakati, H. & Dastgir, A. MSFF: An automated multi-scale feature fusion deep learning model for spine fracture segmentation using MRI. Biomed. Signal Process. Control91, 105943 (2024).
- 51.Saeed, M. U., Bin, W., Sheng, J., Ali, G. & Dastgir, A. 3D MRU-Net: A novel mobile residual U-Net deep learning model for spine segmentation using computed tomography images. Biomed. Signal Process. Control86, 105153 (2023).
- 52.Saeed, M. U., Dikaios, N., Dastgir, A., Ali, G., Hamid, M. & Hajjej, F. An automated deep learning approach for spine segmentation and vertebrae recognition using computed tomography images. Diagnostics13(16). 10.3390/diagnostics13162658 [DOI] [PMC free article] [PubMed]
- 53.Pan, Q. et al. Automatically diagnosing disk bulge and disk herniation with lumbar magnetic resonance images by using deep convolutional neural networks: Method development study (in Eng). JMIR Med. Inform.9(5), e14755 (2021). [DOI] [PMC free article] [PubMed]
- 54.Imran, A. A., Rifat, M. R. & Mohammad, R. Enhancing the Classification Performance of Lower Back Pain Symptoms Using Genetic Algorithm-Based Feature Selection. 455–469 (2019).
- 55.Han, Z., Wei, B., Mercado, A., Leung, S. & Li, S. Spine-GAN: Semantic segmentation of multiple spinal structures (in Eng). Med. Image Anal.50, 23–35 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Pang, S. et al. SpineParseNet: Spine parsing for volumetric MR image by a two-stage segmentation framework with semantic image representation. IEEE Trans. Med. Imaging40(1), 262–273 (2021). [DOI] [PubMed] [Google Scholar]
- 57.Cheng, Y. K. et al. Automatic segmentation of specific intervertebral discs through a two-stage MultiResUNet model (in Eng). J. Clin. Med.10(20), 17 (2021). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.