Abstract
The T-cell receptor (TCR)/CD3 complex plays an essential role in the immune response and is a key player in cancer immunotherapies. There are two classes of TCR/CD3 complexes, defined by their TCR chain usage (αβ or γδ). Recently reported structures have revealed the organization of the αβ TCR/CD3 complex, but similar studies regarding the γδ TCR/CD3 complex have lagged behind. Here, we report cryoelectron microscopy (cryoEM) structural analysis of two γδ TCRs, G115 (Vγ9 Vδ2) and 9C2 (Vγ5 Vδ1), in complex with CD3 subunits. Our results show that the overall subunit organization of the γδ TCR/CD3 complexes is similar to αβ TCRs. However, both γδ TCRs display highly mobile extracellular domains (ECDs), unlike αβ TCRs, which have TCR ECDs that are rigidly coupled to its transmembrane (TM) domains. We corroborate this finding in cells by demonstrating that a γδ T-cell specific antibody can bind a site that would be inaccessible in the more rigid αβ TCR/CD3 complex. Furthermore, we observed that the Vγ5 Vδ1 complex forms a TCR γ5 chain-mediated dimeric species whereby two TCR/CD3 complexes are assembled. Collectively, these data shed light on γδ TCR/CD3 complex formation and may aid the design of γδ TCR-based therapies.
Subject terms: Cryoelectron microscopy, Membrane proteins, Antigen processing and presentation, Immunotherapy
γδTCRs detect and initiate immune responses to various antigens. Here, Hoque et al. report cryoEM structures of two γδTCRs bound by Fabs, revealing their assembly with CD3 signaling components and clonotype-dependent propensity for dimerization.
Introduction
T-cells are specialized immune cells that protect the body against pathogenic threats by recognizing foreign antigens via their T-cell receptor (TCR). T-cell subsets are broadly classified based on their TCR chain usage, αβ or γδ1. In humans, γδ T-cells are further subdivided by their δ-chain usage. In the peripheral blood, the most abundant γδ T-cell population utilizes the semi-invariant γ9δ2 TCR chain pairing2–5. In some peripheral tissues, like the liver, intestines, and the dermis of the skin, δ1-chain expressing T-cells are enriched5. Unlike αβ TCRs that almost always recognize peptide-MHC antigens, γδ TCRs can detect MHC-like antigens as well as ligands that structurally diverge from the MHC fold6. While many of the ligands recognized by γδ T-cells remain unknown, the γδ TCR is thought to recognize molecules that are upregulated during cellular stress, such as lipid species in the context of CD1, and phosphoantigen accumulation sensed by butyrophilins6–11. pMHC-independent target cell recognition makes γδ T-cells attractive for cell therapies as there may be a lower risk of graft-versus-host disease12–14.
On the T-cell surface, TCR chain heterodimers (TCRαβ or TCRγδ) associate with three CD3 homo- and hetero-dimers (CD3ζζ, CD3εγ, CD3εδ in humans)15,16 to form heterooctameric TCR/CD3 complexes. The TCR chains direct ligand specificity by binding directly to antigen, but unlike other receptors, lack intracellular signaling domains. On the other hand, the CD3 chains do not play a role in ligand recognition but contain ITAM sites that are phosphorylated upon ligand engagement17–19. Thus, in a functional TCR/CD3 complex the TCR chains guide specific ligand engagement, while the CD3 chains drive the cellular response.
From a structural perspective, the αβ TCR heterodimer extracellular domains (ECDs) and their complexes with pMHC have been studied extensively, yielding important insights into specific ligand recognition20–22. More recent studies of the entire αβ TCR/CD3 complex, both with and without ligand, have provided additional insight into the organization of the TCR/CD3 complex23–25. These studies demonstrated that the αβ TCR ECD, which is responsible for pMHC binding, is rigidly coupled to the remainder of the signaling complex via extensive contacts between TCR constant regions and CD3 subunits. Structural studies of the γδ TCR have lagged behind that of the αβ TCR, and have almost exclusively been limited to structures of TCR ECDs and their complexes with ligands26–28. It has been proposed that the γδ TCR/CD3 complex may adopt an alternative organization relative to the αβ TCR/CD3, due to differences in glycosylation patterns of CD3δ and the proximity of TCR and CD3 chains as determined by crosslinking experiments29–32. However, recently reported structures of γδ TCR/CD3 complexes have called these claims into question28.
Here, we present cryoEM structures of two γδ TCR clones, G115 (Vγ9 Vδ2) and 9C2 (Vγ5 Vδ1) in complex with CD3 and in the presence of Fab fragments of antibodies targeting CD3 or TCRδ33,34. Our results show that the overall subunit organization of the γδ TCR/CD3 is analogous to the αβ TCR/CD3 complex, with a conserved TM domain architecture driving assembly. However, we find that the γδ TCR ECD is highly mobile due to lack of stabilizing interactions with the CD3 ECDs. This differs notably from the αβ TCR ECD, which has a relatively rigid association with the CD3 ECD that restricts its movement. Surprisingly, we observe a Vγ5-mediated dimeric species for the 9C2 TCR. Collectively, our results elucidate the organization of γδ TCR/CD3 complexes and provide potential insights into the unique structural properties γδ TCRs that may underpin their specialized roles in T-cell mediated immunity.
Results
Purification of two full length γδ TCR/CD3 complexes
We chose two γδ TCR clones, G115 and 9C2, for which crystal structures of the ECDs have been solved previously, for structural analysis as full-length complexes with CD326,27. The G115 γδ TCR heterodimer clone utilizes a Vγ9 Vδ2 chain pairing and binds butyrophilin molecules upon the accumulation of intracellular phosphoantigens35–37. The 9C2 γδ TCR heterodimer utilizes a Vγ5 Vδ1 chain pairing and directly recognizes lipid antigens presented in the context of CD1 molecules27 (Supplementary Fig. 1a). Both TCR clones were produced utilizing the same constant regions for the TCR γ-chain (TRGC1), as well as the TCR δ-chain (TRDC). We adapted a previously described scheme for producing detergent-solubilized αβ TCR/CD3 complexes24 and purified both γδ TCR/CD3 complexes as singular monodisperse peaks with similar elution volumes via size exclusion chromatography (Supplementary Fig. 1b).
CryoEM structure of the flexible G115 (Vγ9 Vδ2) TCR/CD3 complex
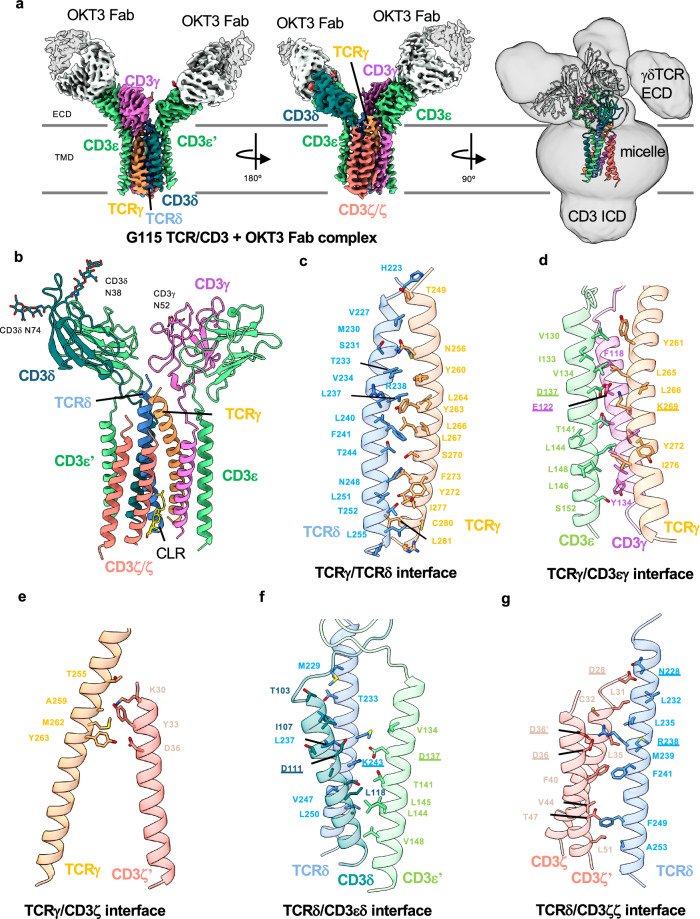
We began by using single particle cryoEM to characterize the G115 γδ TCR/CD3 complex (henceforth referred to as G115 TCR), which uses Vγ9 and Vδ2 in its TCR chains. To aid in cryoEM image processing by increasing the molecular mass of the complex, we pre-incubated the sample with purified Fabs of the commercially available CD3ε-binding antibody OKT334,38. The OKT3 monoclonal antibody is widely used as a T-cell activator39 and was the first clinically approved monoclonal antibody40. We determined a cryoEM structure of the OKT3 Fab-bound G115 TCR to 3.3 Å resolution (Fig. 1; Supplementary Fig. 2a, b; Table 1). The cryoEM map showed density for the TM helices of the γδ TCR, the ECD and TM domains of the CD3ε/δ/γ/ζ chains, as well as two copies of the OKT3 Fab bound to the CD3ε subunits in the CD3εδ and CD3εγ dimers, respectively (Fig. 1a and Supplementary Fig. 2c). Side chain densities were clearly resolved in most of these regions, allowing confident model building. N-linked glycans were modeled for residues N38 and N74 on the CD3δ chains, as well as N52 on the CD3γ chain (Fig. 1b). In the TM region, three densities corresponding to lipids positioned between the TCRγ, TCRδ, CD3ζ, and CD3γ chains were apparent (Supplementary Fig. 3a). We tentatively assigned cholesterol (CLR) to the interior lipid density in the lower half of the TM domain (Fig. 1b). In addition, we observed a density that corresponds to an S-palmitoyl moiety at TCRγ residue C279 (Supplementary Fig. 2c). This residue has been predicted to be a high confidence target for palmitoylation by the SwissPalm server41. However, the functional significance of this post translational modification is unknown.
Fig. 1. CryoEM reconstruction and modeling of the G115 (Vγ9 Vδ2) TCR/CD3 complex.
a Left and middle: two views of a 3.27 Å resolution map of the G115 TCR/CD3 complex bound by OKT3 fab. Right: cryoEM map was Gaussian filtered to 2.5 σ and the contour level was reduced to allow visualization of a weak γδ TCR ECD density. b Structure of the G115 TCR/CD3 complex. Interfaces between TCRγ/TCRδ (c), TCRγ/CD3εγ (d), TCRγ/CD3ζ (e), TCRδ/CD3εδ (f), and TCRδ/CD3ζζ (g). Chains are shown in cartoon representation and color-coded as indicated and interfacial residues are shown as sticks. Underlined residues highlight conserved electrostatic interactions between αβ and γδ TCR-CD3 interaction networks.
Table 1.
CryoEM data, structure refinement, and validation
| G115 TCR/CD3/ OKT3 Fab | G115 TCR ECD/ Fab 1 | 9C2 TCR ECD/ Fab 2 | 9C2 TCR ECD/ Fab 3 | |
|---|---|---|---|---|
| Data collection and processing | ||||
| Magnification | 105,000 | 105,000 | 105,000 | 105,000 |
| Voltage (kV) | 300 | 300 | 300 | 300 |
| Electron exposure (e–/Å2) | ~50 | ~40 | ~40 | ~40 |
| Defocus range | −1.0 to −2.2 | −1.0 to -2.2 | −1.0 to -2.2 | −1.0 to −2.2 |
| Pixel size (Å) | 0.839 | 0.839 | 0.839 | 0.839 |
| # of Movies | 15,639 | 8,599 | 8,037 | 5,854 |
| Initial number of particles | 7.2 M | 10.0 M | 5.1 Ma | 3.1 M |
| Particles selected after 2D classification | 1.7 M | 250 K | 64 K | 46 K |
| Final selected particles | 290,758 | 156,210 | 31,918 | 29,351 |
| Symmetry imposed | C1 | C1 | C2 | C2 |
| Map resolution (Å) | 3.27 | 3.21 | 3.45 | 3.46 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 |
| Refinement | ||||
| Initial Model used | 8ES7 | 8DFW | 4LFH | 4LFH |
| Model composition | ||||
| Non-hydrogen atoms | 8397 | 5222 | 10,480 | 10,522 |
| Protein residues | 1054 | 660 | 1306 | 1310 |
| Ligands | 7 | 4 | 12 | 10 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.002 | 0.003 | 0.002 | 0.003 |
| Bond angles (°) | 0.685 | 0.605 | 0.599 | 0.596 |
| Validation | ||||
| MolProbity score | 2.04 | 1.99 | 2.36 | 2.55 |
| Rotamer outliers (%) | 3.38 | 0.00 | 4.18 | 6.41 |
| Clash score | 6.55 | 12.25 | 9.75 | 11.43 |
| Ramachandran plot | ||||
| Favored (%) | 95.92 | 94.17 | 94.57 | 94.74 |
| Allowed (%) | 4.08 | 5.83 | 5.43 | 5.26 |
| Disallowed (%) | 0.00 | 0.00 | 0.00 | 0.00 |
| Deposition ID | ||||
| PDB | 9CQ4 | 9CQ7 | 9CQ8 | 9CQL |
| EMDB | 45,808 | 45,810 | 45,811 | 45,814 |
aCombined particle counts from 2D template based and Topaz particle picking. Duplicate particles were removed after 2D classification.
Surprisingly, our reconstruction lacked clear density corresponding to the ECD of γδ TCR chains, suggesting it is highly mobile relative to the rest of the complex and thus not resolved upon averaging. Notably, the individual particles that contributed to our final reconstruction appear to show signal corresponding to the γδ TCR-chain ECDs, indicating the complex is intact on the cryoEM grid, and the absence of signal for the TCR ECD in the reconstruction is not due to proteolytic cleavage or degradation of the detergent-solubilized complex (Supplementary Fig. 2d). Indeed, when we applied a Gaussian filter and lowered the contour level of our high-resolution map, we see a lobe of density that likely corresponds to the flexible ECDs of the γδ TCR chains in the TCR/CD3 complex map (Fig. 1a, right panel). The highly mobile nature of the γδ TCR ECDs in the context of the TCR/CD3 complex contrasts starkly with αβ TCR ECDs, which are more rigidly coupled to the TM region and thereby well-resolved in cryoEM structures24,25,42.
Structural organization of the G115 TCR/CD3 complex
Excepting the mobile and poorly resolved ECD of the TCR chains, our structure of the G115 TCR/CD3 shows that the TCRγδ chains assemble with CD3 chains in a similar manner to the αβTCR complex. The TCRγ and TCRδ TM helices are adjacent to each other and positioned along the center of the assembly with the three CD3 heterodimers on the perimeter (Fig. 1b). Sterol-shaped lipid densities, which occupy a crevice in between TCRδ, CD3δ, and CD3ζ chains, appear to play a role in stabilizing the TM domain architecture. Interestingly, sterol coordination at this site has been shown to be involved in αβ TCR activation43.
The interface between the TCRγ and TCRδ TM helices is stabilized primarily by hydrophobic interactions between the helices, as well as potential hydrogen bonds between S231 (TCRδ)/N256 (TCRγ) and T252 (TCRδ)/Y272 (TCRγ) (Fig. 1c). TCRγ also has a significant interface with the TM helices of CD3εγ heterodimer wherein electrostatic interaction between K269 of TCRγ and two acidic residues, E122 (CD3γ) and D137 (CD3ε) near the midpoint of the membrane appears to be critical (Fig. 1d). TCRγ also makes several contacts with the extracellular half of one of the CD3ζ TM helices (Fig. 1e). Analogous to the TCRγ/CD3εγ interface, TCRδ contacts both chains of the CD3δε heterodimer, including via a bifurcated salt bridge between K243 (TCRδ) and D137 (CD3ε)/D111 (CD3δ) (Fig. 1f). Finally, TCRδ uses R238 to coordinate electrostatically with the D36 residues of both protomers of the CD3ζζ homodimer (Fig. 1g). Notably, most of the electrostatic interactions mentioned above are conserved in the αβTCR/CD3 TM domain assembly (Supplementary Fig. 4b, c)42.
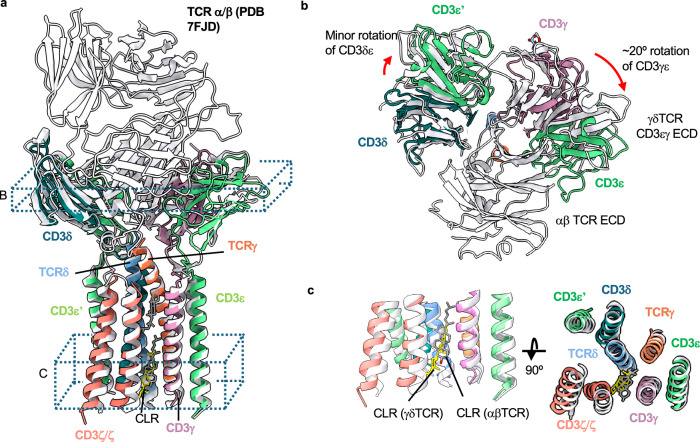
While the overall subunit organization is conserved between γδ TCR/CD3 and αβ TCR/CD3 complexes, TCR-based structural alignment of our OKT3 Fab-bound model to a published αβ TCR/CD3 complex (PDB ID: 7FJD) revealed some noteworthy differences (Fig. 2a). This includes a ~ 20° rotation in the CD3εγ heterodimer toward the region where the TCR ECD would be in a rigidly coupled complex (Fig. 2b). This rotation opens a gap between the CD3εγ and CD3ε’δ heterodimer ECDs, which contact each other in αβ TCR/CD3. We did not observe any substantial differences in the TM domain organizations of αβ and γδTCRs, consistent with the conserved interactions at the subunit interfaces and overall conserved positioning of sterol-shaped lipid (Fig. 2c).
Fig. 2. Structural comparison of the G115 TCR/CD3 complex to an αβ TCR/CD3 complex.
a The G115 TCR/CD3 complex was aligned to an αβ TCR/CD3 complex (PDB:7FJD) via the TCR γ and β chains using matchmaker command in ChimeraX. G115 TCR chains are colored as indicated, while the entire αβ TCR/CD3 complex is colored white. b, c Cross sectional and top-down views are depicted from regions indicated in a. The rotation angles shown in b were estimated between the centroids of each domain (CD3εδ or CD3εγ ECDs, comparing γδ and αβ TCRs) calculated in ChimeraX.
CryoEM structures of mobile γδ TCR ECDs
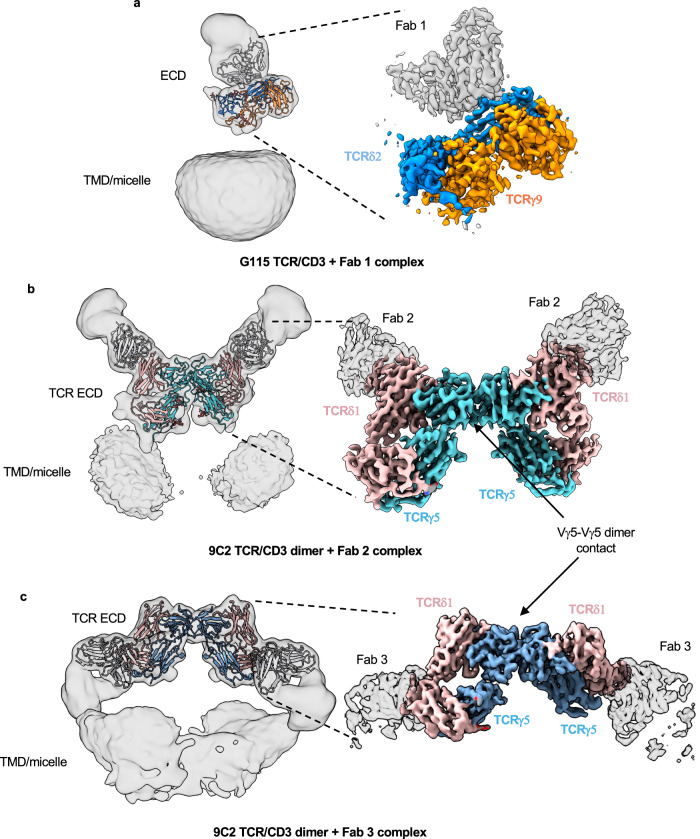
As described above, we could only resolve weak density for the G115 TCR ECD in our OKT3 Fab complex, presumably due to TCR ECD flexibility relative to the remainder of the complex. To aid in visualization of the γδ TCR ECD by cryoEM, we made a complex of G115 TCR with the Fab fragment of a previously described TCR Vδ2-chain binder33 (Fab 1). Unlike the OKT3-bound sample, 2D class averages for the G115 TCR/Fab 1 complex displayed well resolved features for the TCR ECD bound by Fab 1 and faint signal for the micelle/TM domain (Supplementary Fig. 5a). We ultimately obtained a 3.2 Å resolution map in which the TCR ECD and Fab 1 V region were sufficiently resolved to permit model building (Fig. 3a; Supplementary Fig. 5a–c; Table 1). We note the appearance of a weak micelle-like density in our G115 TCR/Fab 1 map upon applying a Gaussian filter, further supporting the notion that the TCR ECD is flexible relative to the membrane bound portion of the molecule (Fig. 3a, left panel). Alignment of the γδTCR ECD to the αβTCR ECD reveals substantial structural differences (Supplementary Fig. 4a). In the αβ TCR/CD3 complex, the TCRα DE loop makes contacts with CD3δ, while TCRβ makes contacts with CD3ε (Supplementary Fig. 4a). The flexibility of the γδ TCR ECD can be rationalized by lack of sequence conservation between the γδ and αβ TCR ECD constant regions (Supplementary Fig. 4b, c), which apparently eliminates most of the important TCR/CD3 ECD contacts observed in αβ TCR. Nonetheless, our structure of G115 TCR ECD/Fab 1 complex demonstrates that the TCR ECD remains structurally intact in our detergent-solubilized cryoEM sample despite its lack of rigid association with the remainder of the complex.
Fig. 3. CryoEM structures of γδ TCR ECDs bound by anti-TCRVδ Fabs.
a CryoEM map of G115 TCR bound by Fab 1. b CryoEM map of 9C2 TCR bound by Fab 2. c CryoEM map of 9C2 TCR bound by Fab 3. Densities are color coded based on the built atomic model. Each left panel shows Gaussian filtered maps (2.5 σ) at low threshold with fitted atomic models to enable visualization of the TMD and micelle densities. Locally refined (a, b) or higher threshold sharpened (c) maps shown on the right display higher resolution features for the TCR ECD and bound Fab V domains.
We wanted to test whether the flexibility we observed for the TCR ECD was specific to the G115 TCR or a generalized phenomenon amongst γδ TCRs using different V genes. To this end, we sought to determine the structure of the detergent-solubilized 9C2 TCR/CD3 complex (henceforth referred to as 9C2 TCR) which utilized Vγ5 Vδ1. To aid in TCR ECD reconstruction for the 9C2 TCR, we separately utilized the Fab fragments of two distinct TCR Vδ1-chain binding antibodies (Fabs 2 and 3)33.
We first collected cryoEM data of the 9C2 TCR bound by Fab 2. Like the G115 TCR ECD, the 9C2 TCR showed high resolution features for the Fab-bound TCR ECD (Supplementary Fig. 6a). However, in stark contrast to the monomeric G115 TCR, our 2D class averages of 9C2 TCR showed dimeric species (Supplementary Fig. 6a). We generated a 3.5 Å resolution C2-symmetric map of this dimeric species in which side chain densities for most residues were observable for the TCR ECDs and Fab 2 variable regions (Fig. 3b, right panel; Supplementary Fig. 6; Table 1). Applying a Gaussian filter revealed two weak micelle densities situated beneath each protomer of the dimeric TCR ECD/Fab 2 complex (Fig. 3b, left panel). The dimer interface is in a germline encoded region of TCR Vγ5 (Fig. 3b, right panel). We expand on this interface in the following section.
We next determined a 3.5 Å resolution cryoEM structure of the 9C2 TCR bound to a Fab 3 (Fig. 3c, right panel; Supplementary Fig. 7; Table 1). In our structure, Fab 3 binds TCR δ1 at the linker connecting the variable and constant regions, employing a binding angle distinct from Fab 2. Like the 9C2/Fab 2 complex, we observed a Vγ5-chain mediated dimer species in the Fab 3 complex structure, with two micelle densities positioned beneath the TCR chain ECDs (Fig. 3c, left panel). In addition, we observed a small population of particles (~10 K) where the dimeric γδ TCR is embedded in one micelle density (Supplementary Fig. 7d). Unfortunately, we were unable to reconstruct a high-resolution map of this set of particles, which may have been more representative of a γδ TCR dimer constrained in cis at the cell membrane than our current dimer structures, where the TM domains of each TCR/CD3 complex are contained within separate micelles. Taken together, these data indicate that γδ TCR ECDs are flexible across different clonotypes and reveal the presence of a Vγ5-mediated dimeric species of detergent-solubilized 9C2 TCR/CD3 complex.
Structural analysis of the 9C2 TCR ECD dimer interface
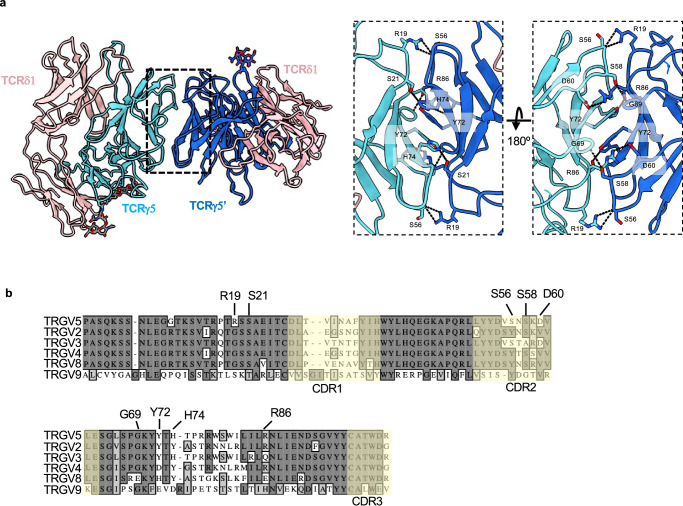
Using the higher resolution 9C2 TCR/Fab 2 structure, we analyzed the residues that are involved in the Vγ5-mediated dimeric interface (Fig. 4a). R19 of each Vγ5 protomer makes a hydrogen bond with the backbone nitrogen of S56 at the periphery of the interface. The core of this interface is mediated by a π-π interaction between Y72 in each protomer. Additionally, hydrogen bonds were observed between side chains of residues D60/R86 and H74/S21, while S58 makes a hydrogen bond with the backbone nitrogen of G69. To assess the sequence conservation of Vγ5 residues at the dimer interface, we performed a sequence alignment of TCR γ-chain variable regions (TRGV) (Fig. 4b). Overall, the variable region is well conserved throughout all the sequences we analyzed apart from TRGV9, which is exemplified by the G115 TCR that only showed monomeric species in this study. We find that the R19/S56 interaction is likely only found in the TRGV5-utilizing TCRs, as the arginine in position 19 is a glycine in TRGV2/3/4/8. Likewise, the D60/R86 electrostatic interaction is not conserved in other TCRγ chains; TRGV3, the only other TRGV in which D60 is conserved, has a glutamine at position 86. Y72, which makes an aromatic interaction at the core of this dimeric interface, is conserved in TRGV2 and TRGV3 but replaced with charged residues in TRGV4, V8, and V9. Overall, the residues mediating Vγ5 dimerization are not well conserved, but we cannot rule out the possibility of different modes of dimerization used by other TCRγ chains.
Fig. 4. Vγ5 Vδ1 TCR dimer interface and its sequence conservation.
a Structure of the 9C2 TCR ECD (Fab 2 complex used, Fab models hidden for clarity) with each TCRγ protomer colored a different shade of blue. Analysis of the TCRVγ5-mediated dimer interface with interacting residues shown as sticks and dashed lines representing putative hydrogen bonds. Note: TCRγ Y72 of protomer 1 forms a π-π interaction with Y72 of protomer 2. b Sequence alignment of various germline encoded TCR γ-chain variable regions. Residues that have been identified in the dimerization interface are labeled. CDRs are indicated with a faint yellow tint.
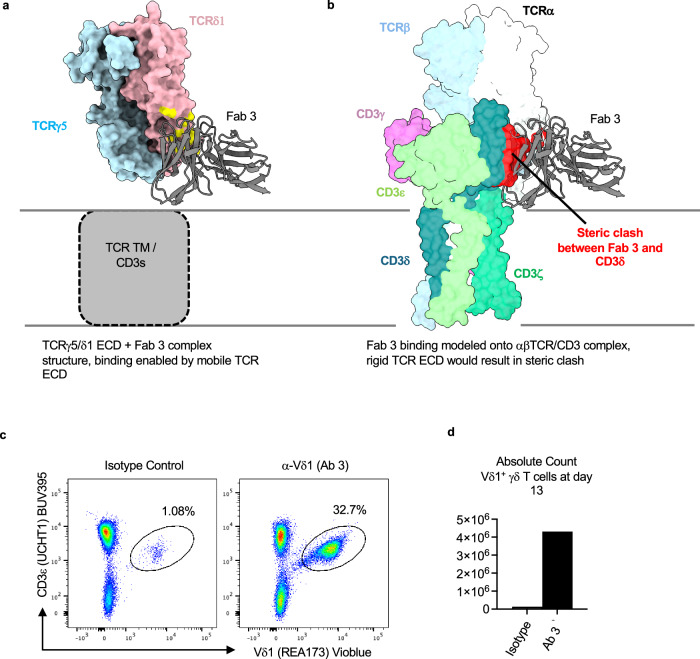
Fab 3 binds a TCRδ1 epitope that would be obscured in the αβ TCR-CD3 complex
We noted that Fabs 2 and 3 bind the 9C2 TCRδ1 chain using distinctly different mechanisms (Fig. 3b, c). While Fab 2 approaches the TCRδ V region from the opposite side of the weak micelle densities (Fig. 3b, left panel), Fab 3 binds using an angle such that its C region is positioned adjacent to the micelles (Fig. 3c, left panel). Considering this unique binding mode that apparently brings Fab 3 in potential overlap with the membrane plane, we surmised that the binding of Fab 3 to the 9C2 γδ TCR/CD3 complex is only possible due to the lack of rigid coupling between the γδ TCR ECD and the rest of the complex. Indeed, structural alignment of our γδ TCR ECD/Fab 3 complex to αβ TCR/CD3 complex indicated that the region where Fab 3 would bind onto TCRα is occluded by the CD3δ ECD of the CD3εδ heterodimer (Fig. 5b). To confirm that Fab 3 can bind a Vδ1 γδ TCR/CD3 complex expressed on cell surface, we tested the ability of the parental IgG of Fab 3 (Ab 3) to expand Vδ1 γδ T cells from human donor PBMCs. Plate-bound Ab 3 indeed selectively expanded Vδ1 γδ T-cells after 13 days of culture (Fig. 5c, d). This data supports our structural observations by showing that the γδ TCR ECD is highly flexible in a functional cellular environment, as the Fab 3 epitope would be masked in the more rigid αβ TCR/CD3 complex.
Fig. 5. Fab 3 binding region is masked in the rigid αβ TCR/CD3 complex.
a Depiction of the Fab 3 binding site in a Fab 3 bound 9C2 γδTCR ECD structure. The Fab variable region is shown as cartoon, whereas the TCR ECD is shown as surface. The Fab 3 epitope is highlighted in yellow. b Bound Fab 3 was modeled onto the αβ TCR from PDB ID 8ES7 through alignment of the TCR β-chain to TCRγ. The Fab variable region is shown as a cartoon, whereas the TCR ECD is shown as surface view. Apparent steric clash between Fab 3 and CD3δ is highlighted in red. Expansion of Vδ1 γδ T-cells from human donor PBMCs following culture with Ab 3. T-cell purity (c, gated on live, CD45+ cells), and counts (d) were assessed on day 13. N = 1 for flow cytometry experiments.
Discussion
We utilized cryoEM to determine the structures of two clonotypic γδ TCR/CD3 complexes bound by the Fab fragments of antibodies directed against CD3 or TCR δ chains. We find some overall structural similarities between the αβ and γδ TCR/CD3 complexes (Fig. 2). Both TCRs utilize the same stoichiometry in their CD3 chain usage (Fig. 1), retain a similar transmembrane domain architecture (Fig. 2), and bind a lipid resembling cholesterol at analogous locations (Fig. 2c)24,25,42. In addition, our results show that the γδ TCR/CD3 complex differs in two major ways from αβ TCR/CD3. First, the extracellular domain of the γδ TCR heterodimer is flexible relative to the membrane embedded portion of the molecule and the CD3 ECD heterodimers (Figs. 1, 3, 5), due to absence of the extensive interactions between the γδ TCR constant regions and the CD3 ECDs that are present in αβTCRs (Supplementary Fig. 4). This observation also implies that interactions between the TM helices44 are sufficient to drive the assembly of the TCR/CD3 complex, and that the ECD contacts observed in αβ TCRs may help enforce rigidity while not necessarily contributing to the complex stability. Secondly, for the 9C2 (Vγ5 Vδ1) TCR, we observed a surprising dimeric species mediated by the Vγ5 chain (Fig. 3b, c).
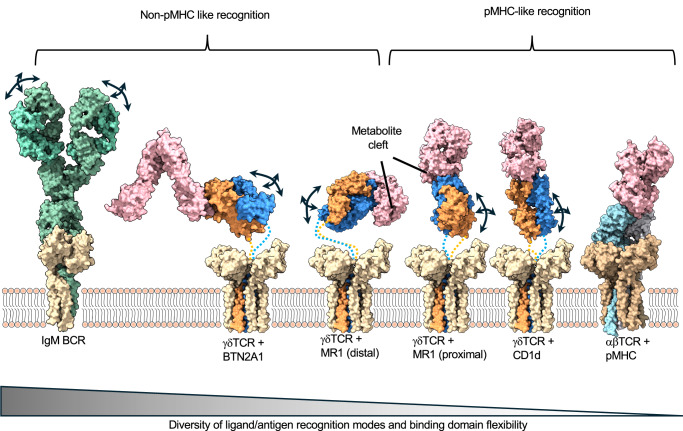
The flexibly coupled ECD of γδ TCR/CD3 may allow for binding to a more diverse range of antigens than αβ TCRs, which are restricted to pMHCs and related molecules (Fig. 6). In the case of the αβ TCR/pMHC-I interaction, the TCR typically engages its ligand in a head-to-head orientation using an overall conserved docking polarity that places the TCR Vα and Vβ over the α2 and α1 helix of MHC-I, respectively45. This docking polarity has been demonstrated to be critical for productive TCR signaling and coreceptor localization with CD3ζ46. As such, a rigidly coupled αβ TCR ECD would be required to effectively transmit the docking polarity at the TCR/pMHC interface to the transmembrane and cytosolic side of the TCR/CD3/coreceptor complex, where signaling occurs. However, for γδ TCR, CD4/CD8 co-receptors are not required47, and there is not only a wider range of ligands that can be engaged but the mode of engagement can vary. For example, the 9C2 TCR binds CD1d/lipid complexes in a classical MHC-like interaction27, while the G115 TCR recognizes BTN2A1 in a side-on orientation that leaves the CDRs largely uninvolved37. This difference in ligand engagement geometry is further highlighted when considering the metabolite presenting MHC-like molecule MR1. Although αβ TCR engages MR1 in MHC-like fashion by docking on top of the metabolite-binding cleft48, γδ TCRs displays diversity in its engagement of MR1; G83 (Vγ8 Vδ3) TCR binds the side of the metabolite binding cleft in MR1 independent of antigen49 and the G7 (Vγ9 Vδ1) TCR clone binds distal from the metabolite binding cleft50. The latter non-canonical γδ TCR/MR1 interaction presumably requires a flexible γδ TCR ECD, as observed in our cryoEM data, to contact the distal portion of the MR1 molecule.
Fig. 6. γδ TCR ECD flexibility correlates with the diversity of antigen recognition by the γδ TCR.
Immune receptor models are organized by their ligand binding geometries. αβ TCR/CD3/pMHC complex (rightmost) is characterized as having a rigid ECD and conserved pMHC-docking mode. BCR (left most, IgM receptor shown) has highly mobile antigen binding (Fab) regions and unlimited ways of engaging antigens. γδTCRs (hypothetical composite models of TM/CD3 and ECD/ligand complexes shown, with flexible linkers depicted as dotted lines) have a αβTCR-like architecture but use their mobile ECDs to engage ligands in diverse ways. Arrows represent flexibility in ligand/antigen recognition domains. PDB ID 7XQ8 was used for the IgM BCR. PDB ID 8DFW was used for γδ TCR ECD/BTN2A1 complex. PDB ID 6MWR was used for the γδ TCR ECD/MR1 complex (cleft distal). PDB ID 7LLI was used for γδ TCR ECD/MR1 complex (cleft proximal). PDB ID 4LHU was used for γδ TCR ECD/CD1d complex. PDB ID 8ES8 was used for αβ TCR/CD3/pMHC complex.
In addition to diverse ligand recognition, the flexibility of γδ TCRs may explain the differential glycosylation patterns observed on CD3δ in αβ and γδ TCR/CD3 complexes29. A highly flexible γδTCR ECD may allow increased access for glycosyltransferase enzymes to the surface of CD3δ, resulting in the addition of more complex N-linked glycans in the case of γδTCR/CD3. Conversely, the rigid αβ TCR ECD may serve to limit the activity of glycosyltransferase enzymes on CD3δ by steric hindrance.
The differences in rigidity of γδ and αβ TCRs may also shed light on their mechanisms of activation. A rigid TCR ECD may explain why the αβ TCR has been shown to be mechanosensitive while the γδ TCR has not51, as force transduction across the receptor would likely be more efficient when the domains are rigidly coupled. Consistent with this notion, a chimeric TCR that contains γδ variable domains but αβ constant domains is mechanosensitive51, while a chimeric TCR with a γδ constant domains and αβ variable domains resulted in impaired function52. Another theory of αβ TCR activation is that of conformational change, in which the receptor allosterically transmits pMHC engagement from the TCR ECD to the CD3 chains53,54. Notably, conformational changes have not been observed in the αβ TCR/CD3 upon pMHC engagement in cryoEM structures using the detergent-solubilized receptor24,25. Nonetheless, for γδ TCR, the lack of contacts between the TCR ECD and the rest of the receptor would seemingly exclude a role for an allosteric conformational change induced by ligand binding in activation. A third model for TCR activation is kinetic-segregation, whereby the tight intermembrane spacing enforced by the immune synapse results in exclusion of proteins with large ECDs, including the inhibitory phosphatases CD45 and CD14855. The flexible ECD of the γδ TCR is compatible with the kinetic-segregation model, as the limited length of the connecting peptides would produce synaptic spacings in the same range as αβ TCRs. Indeed, a recent report showed that γδ TCR triggering displays properties that conform to the kinetic-segregation model56. Taken together, these lines of evidence suggest that the activation of γδ TCRs, by virtue of their flexible TCR ECDs, involves distinct molecular mechanisms from αβ TCRs, though the general principles at the synapse level are likely similar.
We report that the 9C2 TCR/CD3 complex displays Vγ5- mediated dimerization (Figs. 3, 4), while our cryoEM data for G115 (Vγ9 Vδ1) TCR/CD3 complex only showed monomeric species (Fig. 3a). Of note, this mode of dimerization is also present as crystal contacts in x-ray structures of 9C2, both alone and in complex with CD1d antigen (Supplementary Fig. 8)27. This suggests that dimerization may be promoted by high local concentration of the TCR ECD and does not require the full-length complex. The relevance of this dimer for γδ TCR function needs further study, but we speculate that this interface may act to induce local clustering of TCRs on the cell surface57,58 that could result in a more potent T-cell response.
During the preparation of our manuscript, two other groups independently published structures of γδ TCR/CD3 complexes28,59. Xin et al. characterized the same two γδ TCR clones that we describe here, while Gully et al. characterized the MR1 reactive G83 (Vγ8 Vδ3) γδ TCR. The structural findings of Xin et al. and Gully et al. are largely consistent with those described here (Supplementary Fig. 9). Both groups report that the γδ TCR ECDs are highly flexible relative to the CD3 and TM domains, while Xin et al. report a γ5-chain mediated dimeric species for the 9C2 TCR. The three structures display a very similar TM domain organization (Supplementary Fig. 9a–d). Our dimeric Fab 2 bound 9C2 TCR ECD model is also very similar to the corresponding structure generated by Xin et al. (PDB: 8JBV), especially in the Vγ5-Vγ5 dimer interface with many of the residues positioned in a nearly identical manner (Supplementary Fig. 9e–g). Xin et al. demonstrated via FRET that Vγ5 residues D60, Y72, and R86 are important for dimerization of the 9C2 TCR in cells, which agrees with our structures.
An interesting finding reported by Xin et al. is their medium resolution reconstruction of a full-length dimerized 9C2 TCR/CD3 with the two sets of TM domains embedded within a single micelle; we were unable to obtain a similar structure because only a minor population of our particles contained two TCR/CD3s embedded within the same micelle (Supplementary Fig. 7d). Aligning our dimeric 9C2 TCR ECD/Fab 3 structure to the full-length 9C2 TCR structure obtained by Xin et al (PDB ID: 8JCB) revealed a minor clash between the Fab and CD3γ in one of the subunits (Supplementary Fig. 9h). However, this clash is not likely to perturb binding of the antibody because of the mobility of the γδ TCR ECD.
Additionally, Xin et al. report that the dimeric 9C2 TCR/CD3 complex has a left-shifted SEC elution volume relative to that of the G115 TCR/CD3 complex, while we observed that the elution volumes of 9C2 and G115 are essentially unchanged, indicating the 9C2 TCR dimers were generated upon sample concentration or vitrification. The basis for the subtle differences in dimerization behavior observed in our experiments and those of Xin et al. remain unclear. Nonetheless, these three studies reinforce the unique structural features of γδ TCR/CD3 complexes and allow for new routes of investigation for an enigmatic subset of T-cells.
Methods
Construct design
TCR/CD3 construct designs were adapted from previous approaches (Supplementary Fig. 1)24. Briefly, TCR and CD3 chain DNA constructs were codon-optimized and synthesized by GenScript. The full-length G115 and 9C2 TCR constructs were comprised of the δ-chain followed by the γ-chain with an intervening furin cleavage sequence and a P2A cleavage site. The CD3 construct was designed as previously described in ref.24.
γδ TCR/CD3 expression
γδTCR/CD3 complexes were expressed using BacMam-mediated viral transduction in HEK293F cells (ThermoFisher cat# R79007). BacMam virus for each construct was produced in ExpiSf9 cells maintained in ExpiSf CD media. P1 viral stocks were concentrated by centrifugation at 72,500 x g in a Ti45 rotor for one hour at 4 °C. The viral stocks were then resuspended in 2% FBS/Freestyle 293 media. HEK293F cells were then transduced with a 1:1 mixture of G115 TCR/CD3 viruses or 9C2 TCR/CD3 viruses (as shown in Supplementary Fig. 1a) and incubated at 37 °C. 12–16 h post transduction, 10 mM Sodium Butyrate was added to the culture. 48 h after transduction, Cells were harvested by centrifugation, washed with ice cold PBS + Protease inhibitor, and stored at −80 °C for downstream applications.
γδ TCR/CD3 isolation and purification
HEK293F cells were thawed and resuspended in buffer containing 20 mM HEPES pH 8.0, 150 mM NaCl, 1% Glyco-diosgenin (GDN), and EDTA-free cOmplete protease inhibitors (Roche). The mixture was stirred for 1 h at 4 °C and then clarified by centrifugation at 30,000 x g for 20 min. The lysate was then added to GFP nanobody-coupled Sepharose resin pre-equilibrated with SEC buffer (20 mM HEPES pH 8.0, 150 mM NaCl, 0.01% GDN) and rotated at 4 °C for 1.5 h. The resin was then collected in a gravity column and washed with SEC buffer several times. The washed resin was then collected and PreScission protease and 0.5 mM DTT were added and incubated overnight at 4 °C to liberate the TCR/CD3 complex from the Sepharose beads. This mixture was then added to a gravity column, the flow through was collected, concentrated using a 100 kDa MWCO Amicon Ultra centrifugal filter, and injected into a Superose 6 Increase 10/300 GL column. Peak fractions were collected, concentrated using a 100 kDa MWCO filter, and either frozen at −80 °C or prepared directly for cryoEM analyses.
Fab and γδTCR/CD3 complex sample preparation
OKT3 Fab was cleaved from OKT3 IgG antibody using IdeS enzyme and standard protocols. Anti-TCRVδ chain Fabs 1, 2, and 3 were cleaved from IgGs using Pierce Mouse IgG1 Fab and F(ab′)2 Preparation Kit (ThermoFisher) using protocols provided by the manufacturer. Fc domains were removed from the samples using CaptureSelect multispecies Fc resin (ThermoFisher). Fabs were further purified by SEC, concentrated and stored at −80 °C prior to use.
G115 TCR/CD3 + OKT3 Fab complex was generated by mixing the two components at a 1:1.2 molar ratio. G115 TCR/CD3 + Fab 1, 9C2 TCR/CD3 + Fab 2, and 9C2 TCR/CD3 + Fab 3 complexes were generated by mixing the full-length receptor and Fab at 1:0.6 molar ratio. An internal CD3-binding Fab was added to each of the latter three samples, but we were unsuccessful in obtaining high resolution reconstructions of the complex between CD3 and this Fab.
CryoEM grid preparation and data collection
UltrAuFoil 1.2/1.3 grids (Quantifoil), freshly plasma cleaned in a Solarus II (Gatan) using a H2/O2 gas mixture, were used for each sample. In the case of the G115 TCR/CD3 + OKT3 Fab complex, 0.01% fluorinated octyl-maltoside (FOM) was added immediately before freezing to help overcome preferred orientation60. Samples were plunge frozen (blot time 7–15 s, blot force 0) in liquid ethane cooled by liquid nitrogen in a Vitrobot Mark IV operated at 4 °C and 100% humidity.
Grids were loaded into a Titan Krios G3i electron microscope equipped with a BioQuantum K3. Images were collected in counting mode at a magnification of 105kx, yielding a pixel size of 0.839 Å. For data collection, a defocus range of −1.0 to −2.2 µm was used, the energy filter was inserted with a width of 20 eV, and the 100 µm objective aperture was inserted. Each movie was dose fractionated into 46 frames over a 1.6 s exposure and had a total dose of ~40 or ~50 e-/Å2.
CryoEM data processing
All datasets were preprocessed in a similar manner. Briefly, movies were imported into cryoSPARC v4.361 and preprocessed using Patch Motion Correction and Patch CTF Estimation. Micrographs were curated using a 3.5 Å CTF cutoff, except the Fab 3-bound 9C2 complex which utilized a 4.0 Å cutoff. 2D class averages generated from blob-picking random subsets of micrographs were used as templates to pick particles from the respective datasets. TOPAZ picking62 was also used for the Fab 2-bound 9C2 TCR/CD3 complex. Further processing steps are described below:
OKT3 bound G115 TCR/CD3 complex: 7.2 M particles from template-based picking were subjected to two rounds of 2D classification, only keeping particles contributing to class averages with clear features of the micelle-embedded receptor complex, yielding 1.7 million particles. Three successive cycles of ab initio and heterogenous refinement were done, with the best class selected to proceed to the next round. This resulted in a clean class of 290 K particles. Non-uniform refinement63 was then performed yielding a map with a nominal resolution of 3.27 Å.
Fab 1 bound G115 TCR/CD3 complex: 10.0 M particles from template-based picking were subjected to multiple rounds of 2D classification, keeping class averages clearly showing the TCR ECD/Fab 1 complex, yielding 250 K particles. Two successive cycles of ab initio and heterogenous refinement were done, with the best class selected to proceed to the next round, resulting in a final subset of 156 K particles. Non-uniform refinement was then performed yielding a map with a nominal resolution of 3.58 Å. Local refinement using a mask focusing on the TCR ECD and the Fab V domain yielded a map with a nominal resolution of 3.21 Å.
Fab 2 bound 9C2 TCR/CD3 complex: Multiple rounds of 2D classification were conducted separately on 2D template-picked (2.5 M) and TOPAZ-picked (2.8 M, using a model generated from clean template picks from this dataset) particles, selecting class averages showing clear features of the dimerized TCR ECD/Fab 2 complex. Particles after 2D classification from 2D template picking and TOPAZ picking were combined and duplicates removed, resulting in a stack of 64 K particles that were subjected to ab initio reconstruction with 3 classes. 32 K particles from the best ab initio class were refined with C2 symmetry using non-uniform refinement, yielding a map with a resolution of 3.41 Å. Local refinement using a mask focusing on the TCR ECD and the Fab V domain yielded a C2-symmetric 3.45 Å resolution map with improved features.
Fab 3 bound 9C2 TCR/CD3 complex: 3.1 M particles from template picking were subjected to multiple rounds of 2D classification, keeping class averages showing dimerized TCR/Fab 3 complex with two micelle densities present. ~46 K particles from this process were then subjected to ab initio refinement (3 classes). The best class (29 K particles) was selected and nonuniform refinement was performed with C2 symmetry yielding a map with a nominal resolution of 3.46 Å. We also observed a rare population of particles that displayed 2D class averages showing dimerized 9C2 ECD contained within one micelle density; however, we were unable to resolve a high-resolution map of these particles.
Model building and refinement
The model of the G115 TCR/CD3 complex was built using a published structure of αβ TCR/CD3 complex (PDB ID: 8ES7) as an initial model. The TM domains of the αβ TCR chains were deleted and replaced with an AF264 prediction of the TCR γ- and δ-chains. The model of the G115 TCR ECD was built using a published crystal structure of the G115 TCR ECD bound to BTN2A1 (PDB ID: 8DFW) as an initial model. The 9C2 TCR model building used a published ECD crystal structure (PDB ID: 4LFH) as an initial model. All Fabs were initially modeled using AF2 predictions. The models were iteratively built manually in COOT 0.9.8.94 EL65 and real-space refined in PHENIX 1.21.166. All refinements were performed with secondary structure, Ramachandran, and geometry restraints turned on. The structures were validated with Molprobity as implemented in PHENIX. All structural figures were generated in ChimeraX67 or Pymol 2.5.468.
Structural alignments and interaction network determination
H-bond and contact residue networks were determined by selecting the chains of interest and performing either the “Find H-bond” or “Contacts” functions in ChimeraX, respectively. For all structural alignments, the “matchmaker” function was utilized in ChimeraX.
Acquisition of peripheral blood mononuclear cells (PBMCs)
Human PBMCs for validating the activity of the agonistic anti-Vδ1 antibody were obtained from consenting volunteers via a commercial vendor, AllCells. More detailed information on AllCells’ donor procurement process and range of donor characteristics/criteria can be found on AllCells website: https://allcells.com/. Sex or gender analysis was not carried out since the PBMCs were used in in vitro studies and are isolated from their original biological context. Thus, sex and/or gender does not apply to the fundamental biological process studied in this case.
Expansion of Vδ1 expressing γδ T-cells
Human Vδ1+ γδ T cells were expanded by incubating healthy donor PBMCs with plate-bound agonistic α-Vδ1(Ab 3) or Isotype antibody control in media containing recombinant human IL-2. At day 7, cell cultures were transferred to new plates without agonistic α-Vδ antibody and continued to culture in media containing IL-2. Final T cell counts, and purity were assessed on day 13 by AOPI labeling (Nexcelom) and flow cytometry using detection antibodies for human CD45 (clone HI30), human CD3 (clone UCHT1), and human Vδ1 (clone REA173).
Multisequence alignment and annotation of TCR chains
TRDC, TRAC, TRGC1/2, TRBC1/2, and TRGV protein sequences were obtained from UniProt and aligned via MUSCLE69.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Yi Zhou, Erich Joachim Goebel, and Audrey Baeyens for their invaluable input, Nam Nguyen for assistance with cell culture and protein expression, and the Regeneron cloud/HPC teams for supporting cryoEM data storage and processing. This project was funded by Regeneron Pharmaceuticals.
Author contributions
M.H., K.S., T.Z., L.L.M., M.C.F., E.S., W.C.O., and J.C.L. conceptualized the studies. M.H., K.S., L.L.M., T.R., J.J., expressed and purified proteins. M.H. and KS prepared samples for cryoEM, acquired, and built the atomic models with contributions from MCF. JBG conducted the γδ T-cell expansion and flow cytometry studies. M.C.F., E.S., W.C.O., and J.C.L. supervised the overall project. M.H., K.S., and T.Z. drafted the manuscript with contributions from M.C.F., E.S., W.C.O., and J.C.L. The manuscript was finalized by all authors.
Peer review
Peer review information
Nature Communications thanks Thomas Herrmann, Chen Sun and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials or have been deposited in the following databases: The cryoEM maps and coordinates generated in this study have been deposited at the PDB and EMDB with the following accession codes: 9CQ4, EMD-45808 (G115 TCR/CD3 complex bound by OKT3 Fab); 9CQ7, EMD-45810 (G115 TCR ECD bound by Fab 1); 9CQ8, EMD-45811 (dimeric 9C2 TCR ECD bound by Fab 2); 9CQL and EMD-45814 (dimeric 9C2 TCR ECD bound by Fab 3), respectively. Regeneron materials described here may be made available to qualified, academic, noncommercial researchers through a material transfer agreement upon request at https://regeneron.envisionpharma.com/vt_regeneron/. For questions about how Regeneron shares materials, use the email address preclinical.collaborations@regeneron.com.
Competing interests
All authors are employees of Regeneron Pharmaceuticals and own options and/or stock. JCL and WCO are officers of Regeneron Pharmaceuticals.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mohammed Hoque, Email: mohammed.hoque@regeneron.com.
Tong Zhang, Email: tong.zhang@regeneron.com.
Kei Saotome, Email: kei.saotome@regeneron.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55467-5.
References
- 1.Sun, L., Su, Y., Jiao, A., Wang, X. & Zhang, B. T cells in health and disease. Signal Transduct. Target. Ther.8, 235 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parker, C. M. et al. Evidence for extrathymic changes in the T cell receptor gamma/delta repertoire. J. Exp. Med.171, 1597–1612 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groh, V. et al. Human lymphocytes bearing T cell receptor gamma/delta are phenotypically diverse and evenly distributed throughout the lymphoid system. J. Exp. Med.169, 1277–1294 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Triebel, F. et al. A novel human V delta gene expressed predominantly in the Ti gamma A fraction of gamma/delta+ peripheral lymphocytes. Eur. J. Immunol.18, 2021–2027 (1988). [DOI] [PubMed] [Google Scholar]
- 5.Ribeiro, S. T., Ribot, J. C. & Silva-Santos, B. Five layers of receptor signalling in γδ T cell differentiation and activation. Front. Immunol.6 (2015). 10.3389/fimmu.2015.00015 [DOI] [PMC free article] [PubMed]
- 6.Deseke, M. & Prinz, I. Ligand recognition by the γδ TCR and discrimination between homeostasis and stress conditions. Cell. Mol. Immunol.17, 914–924 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morita, C. T., Jin, C., Sarikonda, G. & Wang, H. Nonpeptide antigens, presentation mechanisms, and immunological memory of human Vγ2Vδ2 T cells: discriminating friend from foe through the recognition of prenyl pyrophosphate antigens. Immunol. Rev.215, 59–76 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Marlin, R. et al. Sensing of cell stress by human γδ TCR-dependent recognition of annexin A2. Proc. Natl. Acad. Sci. USA114, 3163–3168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan, L. et al. Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate Vγ9Vδ2 T cells. Nature621, 840–848 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrmann, T. & Karunakaran, M. M. Phosphoantigen recognition by Vγ9Vδ2 T cells. Eur. J. Immunol. 2451068 (2024). [DOI] [PubMed]
- 11.Harly, C. et al. Key implication of CD277/butyrophilin-3 (BTN3A) in cellular stress sensing by a major human γδ T-cell subset. Blood120, 2269–2279 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cela, M. et al. Gamma delta T lymphocyte regeneration after T lymphocyte-depleted bone marrow transplantation from mismatched family members or matched unrelated donors. Bone Marrow Transplant.17, 243–247 (1996). [PubMed] [Google Scholar]
- 13.Yabe, M. et al. Transition of T cell receptor gamma/delta expressing double negative (CD4-/CD8-) lymphocytes after allogeneic bone marrow transplantation. Bone Marrow Transplant.14, 741–746 (1994). [PubMed] [Google Scholar]
- 14.Godder, K. et al. Long term disease-free survival in acute leukemia patients recovering with increased γδ T cells after partially mismatched related donor bone marrow transplantation. Bone Marrow Transplant.39, 751–757 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Kuhns, M. S., Davis, M. M. & Garcia, K. C. Deconstructing the form and function of the TCR/CD3 complex. Immunity24, 133–139 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Call, M. E., Pyrdol, J. & Wucherpfennig, K. W. Stoichiometry of the T‐cell receptor–CD3 complex and key intermediates assembled in the endoplasmic reticulum. EMBO J.23, 2348–2357 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun, Z.-Y. J., Kim, K. S., Wagner, G. & Reinherz, E. L. Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3ϵγ heterodimer. Cell105, 913–923 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Bettini, M. L. et al. Cutting Edge: CD3 ITAM Diversity Is Required for Optimal TCR Signaling and Thymocyte Development. J. Immunol.199, 1555–1560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Love, P. E. & Hayes, S. M. ITAM-mediated signaling by the T-cell antigen receptor. Cold Spring Harb. Perspect. Biol.2, a002485 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kass, I., Buckle, A. M. & Borg, N. A. Understanding the structural dynamics of TCR-pMHC interactions. Trends Immunol.35, 604–612 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Sewell, A. K. Why must T cells be cross-reactive? Nat. Rev. Immunol.12, 669–677 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mariuzza, R. A., Agnihotri, P. & Orban, J. The structural basis of T-cell receptor (TCR) activation: An enduring enigma. J. Biol. Chem.295, 914–925 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong, D. et al. Structural basis of assembly of the human T cell receptor–CD3 complex. Nature573, 546–552 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Saotome, K. et al. Structural analysis of cancer-relevant TCR-CD3 and peptide-MHC complexes by cryoEM. Nat. Commun.14, 2401 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sušac, L. et al. Structure of a fully assembled tumor-specific T cell receptor ligated by pMHC. Cell185, 3201–3213.e3219 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allison, T. J., Winter, C. C., Fournié, J.-J., Bonneville, M. & Garboczi, D. N. Structure of a human γδ T-cell antigen receptor. Nature411, 820–824 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Uldrich, A. P. et al. CD1d-lipid antigen recognition by the γδ TCR. Nat. Immunol.14, 1137–1145 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Xin, W. et al. Structures of human γδ T cell receptor-CD3 complex. Nature (2024). 10.1038/s41586-024-07439-4 [DOI] [PMC free article] [PubMed]
- 29.Krangel, M. S. et al. T3 glycoprotein is functional although structurally distinct on human T-cell receptor gamma T lymphocytes. Proc. Natl Acad. Sci. USA84, 3817–3821 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alarcon, B. et al. The T-cell receptor gamma chain-CD3 complex: implication in the cytotoxic activity of a CD3+ CD4- CD8- human natural killer clone. Proc. Natl Acad. Sci. USA84, 3861–3865 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Neerven, J., Coligan, J. E. & Koning, F. Structural comparison of alpha/beta and gamma/delta T cell receptor-CD3 complexes reveals identical subunit interactions but distinct cross-linking patterns of T cell receptor chains. Eur. J. Immunol.20, 2105–2111 (1990). [DOI] [PubMed] [Google Scholar]
- 32.Morath, A. & Schamel, W. W. αβ and γδ T cell receptors: Similar but different. J. Leukoc. Biol.107, 1045–1055 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Aya Jakobovits, O. F., Andy An-deh Lin, Marianne Theresa Santaguida, Radhika Chetan Desai, Yifeng Frank Jing, Daulet Kadyl Satpayev, Yan Li. Methods for selective expansion of gamma delta t-cell populations and compositions thereof. United States patent (2017).
- 34.Norman, D. J. Mechanisms of action and overview of OKT3. Ther. Drug Monit.17, 615–620 (1995). [DOI] [PubMed] [Google Scholar]
- 35.Willcox, C. R. et al. Phosphoantigen sensing combines TCR-dependent recognition of the BTN3A IgV domain and germline interaction with BTN2A1. Cell Rep.42, 112321 (2023). [DOI] [PubMed] [Google Scholar]
- 36.Mamedov, M. R. et al. CRISPR screens decode cancer cell pathways that trigger γδ T cell detection. Nature621, 188–195 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fulford, T. S. et al. Vγ9Vδ2 T cells recognize butyrophilin 2A1 and 3A1 heteromers. Nat. Immunol.25, 1355–1366 (2024). [DOI] [PubMed] [Google Scholar]
- 38.Kjer-Nielsen, L. et al. Crystal structure of the human T cell receptor CD3εγ heterodimer complexed to the therapeutic mAb OKT3. Proc. Natl Acad. Sci.101, 7675–7680 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwab, R., Crow, M. K., Russo, C. & Weksler, M. E. Requirements for T cell activation by OKT3 monoclonal antibody: role of modulation of T3 molecules and interleukin 1. J. Immunol.135, 1714–1718 (1985). [PubMed] [Google Scholar]
- 40.Smith, S. L. Ten years of Orthoclone OKT3 (muromonab-CD3): a review. J. Transpl. Coord.6, 109–119 (1996). quiz 120-101. [DOI] [PubMed] [Google Scholar]
- 41.Blanc, M. et al. SwissPalm: Protein Palmitoylation database [version 1; peer review: 3 approved]. F1000Res.4 (2015). 10.12688/f1000research.6464.1 [DOI] [PMC free article] [PubMed]
- 42.Dong, D. et al. Structural basis of assembly of the human T cell receptor-CD3 complex. Nature573, 546–552 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Chen, Y. et al. Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility. Mol. Cell82, 1278–1287.e1275 (2022). [DOI] [PubMed] [Google Scholar]
- 44.Call, M. E., Pyrdol, J., Wiedmann, M. & Wucherpfennig, K. W. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell111, 967–979 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rossjohn, J. et al. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol.33, 169–200 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Zareie, P. et al. Canonical T cell receptor docking on peptide-MHC is essential for T cell signaling. Science372 (2021). 10.1126/science.abe9124 [DOI] [PubMed]
- 47.Witherden, D. A., Johnson, M. D. & Havran, W. L. Coreceptors and Their Ligands in Epithelial γδ T Cell Biology. Front. Immunol.9 (2018). 10.3389/fimmu.2018.00731 [DOI] [PMC free article] [PubMed]
- 48.Eckle, S. B. G. et al. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J. Exp. Med.211, 1585–1600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rice, M. T. et al. Recognition of the antigen-presenting molecule MR1 by a Vδ3+ γδ T cell receptor. Proc. Natl. Acad. Sci.118, e2110288118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Nours, J. et al. A class of γδ T cell receptors recognize the underside of the antigen-presenting molecule MR1. Science366, 1522–1527 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Mallis, R. J. et al. Molecular design of the γδT cell receptor ectodomain encodes biologically fit ligand recognition in the absence of mechanosensing. Proc. Natl Acad. Sci.118, e2023050118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tao, C. et al. γδTCR immunoglobulin constant region domain exchange in human αβTCRs improves TCR pairing without altering TCR gene-modified T cell function. Mol. Med Rep.15, 1555–1564 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dopfer, E. P. et al. The CD3 conformational change in the gammadelta T cell receptor is not triggered by antigens but can be enforced to enhance tumor killing. Cell Rep.7, 1704–1715 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Gil, D., Schamel, W. W., Montoya, M., Sanchez-Madrid, F. & Alarcon, B. Recruitment of Nck by CD3 epsilon reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell109, 901–912 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Davis, S. J. & van der Merwe, P. A. The kinetic-segregation model: TCR triggering and beyond. Nat. Immunol.7, 803–809 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Li, F. et al. Ligand-induced segregation from large cell-surface phosphatases is a critical step in γδ TCR triggering. Cell Rep.43 (2024). 10.1016/j.celrep.2024.114761 [DOI] [PMC free article] [PubMed]
- 57.Purtic, B., Pitcher, L. A., van Oers, N. S. C. & Wülfing, C. T cell receptor (TCR) clustering in the immunological synapse integrates TCR and costimulatory signaling in selected T cells. Proc. Natl Acad. Sci.102, 2904–2909 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Minguet, S., Swamy, M., Alarcón, B., Luescher, I. F. & Schamel, W. W. A. Full Activation of the T Cell Receptor Requires Both Clustering and Conformational Changes at CD3. Immunity26, 43–54 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Gully, B. S. et al. Structure of a fully assembled γδ T cell antigen receptor. Nature (2024). 10.1038/s41586-024-07920-0 [DOI] [PMC free article] [PubMed]
- 60.Vénien-Bryan, C. & Fernandes, C. A. H. Overview of Membrane Protein Sample Preparation for Single-Particle Cryo-Electron Microscopy Analysis. Int. J. Mol. Sci.24 (2023). 10.3390/ijms241914785 [DOI] [PMC free article] [PubMed]
- 61.Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods16, 1153–1160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods17, 1214–1221 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr D. Biol. Crystallogr.66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D. Struct. Biol.75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pettersen, E. F. et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci.30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.The PyMOL Molecular Graphics System, Version 2.5 Schrödinger, LLC.
- 69.Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials or have been deposited in the following databases: The cryoEM maps and coordinates generated in this study have been deposited at the PDB and EMDB with the following accession codes: 9CQ4, EMD-45808 (G115 TCR/CD3 complex bound by OKT3 Fab); 9CQ7, EMD-45810 (G115 TCR ECD bound by Fab 1); 9CQ8, EMD-45811 (dimeric 9C2 TCR ECD bound by Fab 2); 9CQL and EMD-45814 (dimeric 9C2 TCR ECD bound by Fab 3), respectively. Regeneron materials described here may be made available to qualified, academic, noncommercial researchers through a material transfer agreement upon request at https://regeneron.envisionpharma.com/vt_regeneron/. For questions about how Regeneron shares materials, use the email address preclinical.collaborations@regeneron.com.