Abstract
Computational methods for predicting protein function are of great significance in understanding biological mechanisms and treating complex diseases. However, existing computational approaches of protein function prediction lack interpretability, making it difficult to understand the relations between protein structures and functions. In this study, we propose a deep learning-based solution, named DPFunc, for accurate protein function prediction with domain-guided structure information. DPFunc can detect significant regions in protein structures and accurately predict corresponding functions under the guidance of domain information. It outperforms current state-of-the-art methods and achieves a significant improvement over existing structure-based methods. Detailed analyses demonstrate that the guidance of domain information contributes to DPFunc for protein function prediction, enabling our method to detect key residues or regions in protein structures, which are closely related to their functions. In summary, DPFunc serves as an effective tool for large-scale protein function prediction, which pushes the border of protein understanding in biological systems.
Subject terms: Machine learning, Gene ontology, Protein function predictions, Sequence annotation, Protein structure predictions
DPFunc, a deep-learning-based tool, uses protein domain information to accurately predict protein functions and detect corresponding regions in protein structures. DPFunc serves as an effective tool for large-scale protein function prediction.
Introduction
Proteins are fundamental units that perform functions to accomplish various life activities1,2. The individual proteins after mutations, for example, are necessary to verify that the specific functions are retained3–5. While traditional wet-lab experiments have long been the gold standard for accurately determining protein functions6, their time-consuming and costly have spurred the development of automated protein function prediction methods. Till now, less than 1% of protein sequences are annotated by Gene Ontology (GO) terms7, which can be divided into three ontologies: molecular functions (MF), cellular components (CC), and biological process (BP)8,9. Consequently, developing computational methods for automated protein function prediction is crucial for bridging the widening gap between the number of known annotations and protein sequences generated by high-throughput technology10,11, which benefits biologists in discovering proteins of interest and serve as a guide for protein virtual screening and protein design12,13.
Traditional computational methods have long relied on homology similarity, inferring protein function based on known proteins and applying that knowledge to proteins of interest6,14. More recently, several machine learning and deep learning classifiers15–19 have been proposed to learn the latent relationships between protein sequences and functions, surpassing the performance of traditional homology-based methods. Propelled by high-throughput technologies, a vast amount of biological data have been produced, including gene expression20, biomedical text21, protein-protein interaction (PPI)22, and homology relationship23. These data provide new perspectives for protein function prediction. The Critical Assessment of Functional Annotation (CAFA) competition24,25 has proven the advancements in incorporating these biological data. Notably, most of these methods rely on specific features generated from additional information (for example, PPI26 or gene expression27) beyond the protein sequences themselves. Thus, although these methods achieve impressive performance, they struggle to be applied to large-scale protein datasets or those predicted from only sequenced genomes, as there is no guarantee that additional information is available for all target proteins28. Above all, it still remains a big challenge to predict protein function only based on protein sequences18,29–31.
It is widely known that protein sequences fold into three-dimensional structures and further determine specific functions32. In the early days, constrained by limited experimental protein structures, almost all sequence-based computational methods predict functions based on motifs of sequences, like DeepGOPlus16 and TALE33. With the development of deep learning, computational methods, such as ESMFold34, AlphaFold235, and AlphaFold336, can predict high-accuracy protein structures from sequences, addressing the limitations of existing sequence-based methods for protein function prediction. Now, several methods that leverage both sequences and structures have been proposed, such as DeepFRI37 and GAT-GO38. These methods construct protein contact maps based on the 3D coordinates of amino acids from protein structures39–42, then adopt different Graph Neural Networks (GNNs)43,44 to extract protein-level features. These structure-based methods have achieved some progress compared to previous methods. However, existing structure-based approaches ignore the importance of different amino acids and directly average all amino acid features as protein-level features, failing to effectively discover the relationships between functions and important domains in the structure. In fact, proteins consist of many specific domains45–48, which are closely related to both their structures and functions49. It has been shown from previous studies19,26 that it is valuable to detect domains in sequences for protein function prediction.
To address these limitations, we introduce a deep learning-based method, DPFunc, which integrates domain-guided structure information for accurate protein function prediction. The core idea is to leverage domain information within protein sequences to guide the model toward learning the functional relevance of amino acids in their corresponding structures, highlighting structure regions that are closely associated with functions. More specifically, DPFunc first extracts residue-level features from a pre-trained protein language model and then employs graph neural networks to propagate features between residues. Simultaneously, it scans the sequences and generates domains, converting them into dense representations through embedding layers. Inspired by the transformer architecture, DPFunc introduces an attention mechanism that learns whole structures and predicts functions under the guidance of corresponding domain information. With this architecture, our model is able to capture functionally crucial domains within protein structures. Comprehensive evaluations and analyses reveal that DPFunc outperforms existing state-of-the-art methods. Further exploration also demonstrates its ability to detect key motifs or residues in protein structures that exhibit strong functional correlations. In summary, DPFunc offers a more efficient way to unravel the relationships between protein structures and functions compared to existing structure-based methods. It provides researchers with important sites in the structure that may be highly relevant to functions. Moreover, DPFunc also holds the potential for widespread application across large-scale protein sequence datasets, since it can directly obtain features only from protein sequences.
Results
Overview of DPFunc
DPFunc is a deep learning-based method for protein function prediction using domain-guided structure information. The overall architecture of DPFunc is shown in Fig. 1. It consists of three modules: (1) a residue-level feature learning module based on a pre-trained protein language model and graph neural networks for propagating features between residues through protein structures which can be the native structures from the PDB database50 or the predicted structures51,52 by AlphaFold235. (2) a protein-level feature learning module for extracting the whole structure features from residue-level features guided by domain information from sequences. (3) a protein function prediction module for annotating functions to proteins based on protein-level features.
Fig. 1. Model architectures of DPFunc.
a The overview of DPFunc. It mainly consists of three aspects, including the domain information via scanning protein sequences, the residue features generated from the pre-trained protein language model, and the structure graphs constructed based on the predicted or native structures. Based on these features, a residue feature learning module and a protein feature learning module are designed to learn the residue representations and significance of residues in the structure, which are used to predict functions subsequently. b The details of the residue feature learning module. It utilizes GCN layers and residual operation to update residue features based on the pre-trained features and structure graphs. c The details of the protein feature learning module. Inspired by self-attention, it takes domain information and residue representations as input, and calculates the importance of different residues in structures to generate protein features.
The residue-level feature learning module takes protein sequences and structures as input. Based on protein sequences, it first generates the initial features for each residue from the pre-trained protein language model (ESM-1b)53. Simultaneously, it constructs contact maps based on corresponding protein structures. Subsequently, these contact maps and residue-level features can be considered as graphs and corresponding node features, which are further fed into several GCN layers to update and learn the final residue-level features. Additionally, inspired by ResNet54, this module also utilizes a residual learning framework in GCNs. The protein-level feature learning module holds the key component for transforming residue-level insights into a comprehensive representation of the entire protein structure. It first uses InterProScan55 to scan the target protein sequences, compares them to background databases, and detects the domains contained in the sequences, each of which is represented by a unique entry. Since these domains are functional units responsible for specific functions, in this module, they serve as a guide to discovering significant residues in the sequences with the residue-level features generated by the first module. Specifically, these domain entries are fed into an embedding layer to generate domain-level dense representations that capture their unique characteristics, and then summed as protein-level domain information. To assess the importance of different residues, inspired by the transformer architecture, an attention mechanism is introduced to interweave the protein-level domain features and residue-level features, which detects the importance of each residue. Subsequently, protein-level features can be obtained by weighted summation of the residue-level features and their corresponding importance scores. Then, the protein function prediction module combines the protein-level features and initial residue-level features to annotate functions to proteins through several fully connected layers. Finally, the prediction results are processed by a common post-processing procedure to ensure consistency with the structures of GO terms. These modules are integrated as an automatic function prediction framework. The details of each module can be found in “Methods".
DPFunc outperforms existing state-of-the-art methods
To evaluate the performance of DPFunc, we first compare our method to three baseline methods only based on sequences (i.e., Naive16, Blast6, and DeepGO15) and two structure-based methods (i.e., DeepFRI37 and GAT-GO38). To make a fair comparison, we use the same dataset used in previous studies37,38. The dataset contains the PDB structures validated by experiments and corresponding confirmed functions (see “Methods" for dataset details). We adopt two commonly used metrics in CAFA: Fmax and AUPR (see “Methods" for details). Fmax is the maximum F-measure, which is the harmonic mean of paired precision and recall. A higher Fmax indicates better performance. AUPR is the area under the precision-recall curve with different cut-off thresholds. Again, a larger AUPR value signifies superior model performance.
The result is illustrated in Table 1. Without the post-processing procedure, DPFuncw/o post outperforms other methods in MF, CC, and BP. And the post-processing procedure further enhances the performance improvements. Specifically, when compared to GAT-GO, DPFuncw/o post achieves an increased Fmax of 8%, 5%, and 8% in MF, CC, and BP, respectively. With the post-processing procedure, these improvements become even more significant, reaching 16%, 27%, and 23%, respectively. Similar trends are observed when considering AUPR. DPFuncw/o post consistently achieves the highest performance, improving AUPR by at least 7%, 23%, and 42% in MF, CC, and BP, respectively. After considering the post-processing procedure, the performance of our model is further improved by even 8%, 26%, and 19%, respectively. We further test the effect of different sequence identities on the performance of these methods. As illustrated in Supplementary Fig. 1, DPFunc achieves better performance in all cases with different sequence identity cut-offs. It is noteworthy that although GAT-GO also uses protein structures and the same residue-level features generated from ESM-1b, our model outperforms it. This finding indicates that domain information contained in protein sequences provides valuable insights for protein function prediction. Meanwhile, the post-processing procedure makes the predictions more logical and facilitates improving the performance of models.
Table 1.
Comparison on the PDB dataset in terms of Fmax and AUPR
| Method | MF | CC | BP | |||
|---|---|---|---|---|---|---|
| Fmax | AUPR | Fmax | AUPR | Fmax | AUPR | |
| Naïve* | 0.156 | 0.075 | 0.318 | 0.158 | 0.244 | 0.131 |
| BLAST* | 0.498 | 0.120 | 0.398 | 0.163 | 0.400 | 0.120 |
| DeepGO* | 0.359 | 0.368 | 0.420 | 0.302 | 0.295 | 0.210 |
| DeepFRI* | 0.542 | 0.313 | 0.424 | 0.193 | 0.425 | 0.159 |
| GAT-GO* | 0.633 | 0.660 | 0.547 | 0.479 | 0.492 | 0.381 |
| DPFuncw/o post | 0.681 | 0.701 | 0.571 | 0.593 | 0.531 | 0.540 |
| DPFunc | 0.731 | 0.766 | 0.689 | 0.738 | 0.606 | 0.639 |
*The performance of these methods are taken from the original paper.
Best performance among all methods for each metric is shown in bold.
To enable a more comprehensive comparison with other methods6,14–16,26,29,33, we construct a large-scale dataset. Following the CAFA challenge, we partition it into training, validation, and test sets based on distinct time stamps (see “Methods" for details). Unlike the previously utilized PDB dataset, this large-scale dataset encompasses more proteins and corresponding additional information, such as PPI and GO structures, making it possible to compare our methods with other state-of-the-art (SOTA) methods. Specifically, we compare our method against two baseline methods (BlastKNN6 and Diamond14), three sequence-based methods (DeepGOCNN16, TALE33, and ATGO33), two PPI network-based methods (DeepGO15 and DeepGraphGO26), and three composite methods that integrate the results of baseline methods and their original predictions (DeepGOPlus16, TALE+33, ATGO+29). Moreover, we choose two additional web-servers as competitors, NetGO3.056 and COFACTOR57,58, where NetGO3.0 is the current state-of-the-art method in the CAFA24,25 challenge and COFACTOR is an effective structure-based tool for predicting protein functions as a component of I-TASSER-MTD59 in the CASP60 challenge.
Table 2 shows the predictive performance of various methods for five repetitions of the experiment. Notably, to ensure a fair comparison, the post-processing procedure is applied to all methods. This standardization potentially benefits those that do not inherently incorporate such processing. Despite this, DPFunc consistently outperforms all other methods in terms of Fmax and AUPR, exhibiting particularly significant improvements in AUPR. Specifically, DPFunc exhibits AUPR improvements of at least 9.6%, 9.3%, and 8.8% for MF, CC, and BP, respectively. Similar conclusions can be drawn from Supplementary Table 1. DPFunc surpasses the other two web-servers, NetGO3.0 and COFACTOR, in the vast majority of cases, except for Fmax in BP. These comparison results further prove the ability of DPFunc in protein function prediction. To evaluate the performance of DPFunc more comprehensively, based on the results in Table 2, we choose four approaches (BlastKNN, ATGO, DeepGraphGO, and ATGO+) as the representative methods for baseline methods, sequence-based methods, PPI network-based methods, and composite methods, respectively.
Table 2.
Comparison on the large-scale dataset in terms of Fmax and AUPR
| Ontology | Methods | Fmax | p value | AUPR | p value |
|---|---|---|---|---|---|
| MF | Diamond | 0.592(-) | – | 0.387(-) | – |
| BlastKNN | 0.616(-) | – | 0.484(-) | – | |
| DeepGO | 0.301( ± 5.47e-03) | 8.40e-04 | 0.204( ± 8.21e-03) | 5.65e-04 | |
| DeepGOCNN | 0.396( ± 5.73e-04) | 3.70e-05 | 0.326( ± 4.38e-04) | 4.90e-06 | |
| TALE | 0.260( ± 2.44e-05) | 1.25e-08 | 0.158( ± 1.96e-05) | 2.57e-09 | |
| ATGO | 0.454( ± 1.25e-05) | 1.55e-07 | 0.442( ± 4.37e-06) | 4.93e-08 | |
| DeepGraphGO | 0.562( ± 8.00e-05) | 6.83e-05 | 0.533( ± 1.28e-04) | 1.37e-05 | |
| DeepGOPlus | 0.589( ± 2.13e-06) | 6.22e-06 | 0.548( ± 6.26e-05) | 1.85e-05 | |
| TALE+ | 0.602( ± 6.00e-06) | 1.74e-05 | 0.543( ± 6.89e-06) | 1.83e-06 | |
| ATGO+ | 0.622( ± 6.56e-07) | 2.80e-04 | 0.599( ± 3.86e-07) | 1.63e-06 | |
| DPFunc | 0.635( ± 3.24e-06) | – | 0.658( ± 9.22e-06) | – | |
| CC | Diamond | 0.573(-) | – | 0.283(-) | – |
| BlastKNN | 0.596(-) | – | 0.384(-) | – | |
| DeepGO | 0.574( ± 4.78e-05) | 5.71e-05 | 0.580( ± 6.34e-05) | 2.01e-05 | |
| DeepGOCNN | 0.573( ± 2.45e-04) | 6.33e-04 | 0.567( ± 2.26e-04) | 1.45e-04 | |
| TALE | 0.548( ± 1.75e-05) | 2.68e-06 | 0.510( ± 3.23e-04) | 3.62e-05 | |
| ATGO | 0.602( ± 2.76e-06) | 3.15e-06 | 0.596( ± 7.35e-07) | 3.46e-07 | |
| DeepGraphGO | 0.634( ± 4.32e-07) | 1.01e-04 | 0.590( ± 7.60e-06) | 1.61e-06 | |
| DeepGOPlus | 0.626( ± 1.44e-05) | 3.06e-04 | 0.618( ± 3.89e-05) | 4.21e-05 | |
| TALE+ | 0.608( ± 8.61e-07) | 4.99e-06 | 0.591( ± 8.34e-05) | 3.68e-05 | |
| ATGO+ | 0.633( ± 3.06e-06) | 1.12e-04 | 0.636( ± 2.13e-07) | 3.79e-06 | |
| DPFunc | 0.657( ± 7.44e-06) | – | 0.695( ± 9.18e-06) | – | |
| BP | Diamond | 0.429(-) | – | 0.197(-) | – |
| BlastKNN | 0.445(-) | – | 0.258(-) | – | |
| DeepGO | 0.328( ± 9.89e-05) | 1.05e-05 | 0.260( ± 8.05e-05) | 1.99e-05 | |
| DeepGOCNN | 0.323( ± 3.35e-04) | 1.09e-04 | 0.254( ± 3.81e-04) | 5.83e-05 | |
| TALE | 0.253( ± 2.23e-05) | 1.56e-07 | 0.152( ± 4.14e-05) | 1.67e-07 | |
| ATGO | 0.396( ± 8.64e-07) | 5.29e-07 | 0.341( ± 3.32e-07) | 2.98e-07 | |
| DeepGraphGO | 0.432( ± 2.30e-06) | 1.38e-05 | 0.389( ± 6.14e-06) | 1.70e-05 | |
| DeepGOPlus | 0.438( ± 9.94e-06) | 1.58e-04 | 0.365( ± 1.28e-05) | 1.65e-05 | |
| TALE+ | 0.427( ± 4.77e-06) | 1.63e-05 | 0.327( ± 8.03e-06) | 1.04e-06 | |
| ATGO+ | 0.456( ± 4.29e-07) | 2.06e-04 | 0.399( ± 2.76e-07) | 9.41e-06 | |
| DPFunc | 0.466( ± 2.21e-06) | – | 0.434( ± 7.17e-06) | – |
The values of Fmax and AUPR in the table are the mean and standard deviation of the results of five times repeated experiments. P values are two-tailed Student’s t-test between DPFunc and the corresponding compared methods. Best performance among all methods for each metric is shown in bold.
DPFunc exhibits the ability to learn protein features and infer the GO terms more effectively, even for unseen proteins with low sequence identities to known proteins. To verify this capability, we construct several protein sets from the test set, each with a distinct sequence identity threshold relative to training proteins. The results are shown in Fig. 2a, DPFunc consistently outperforms other methods in nearly all cases, except for the 50% threshold in BP, where it demonstrates comparable performance to ATGO+. Notably, the improvements of DPFunc are still stable as the identity threshold increases. This advantage is more pronounced in CC, where the rankings of ATGO+ and DeepGraphGO change with identities. This result persists when compared to all other SOTA methods (see Supplementary Fig. 2).
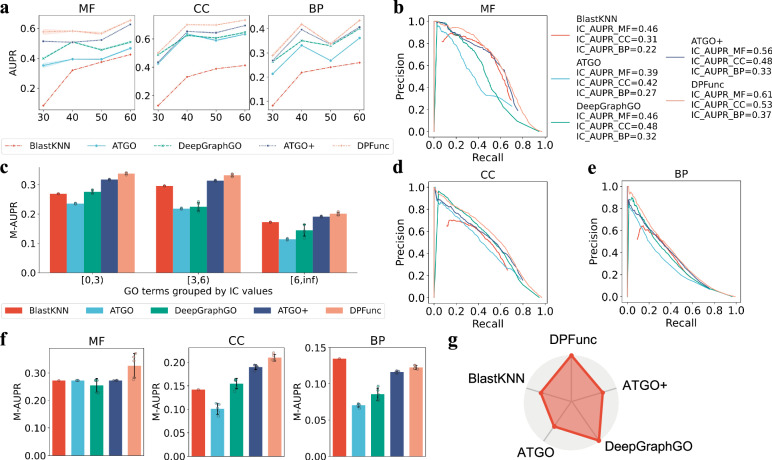
Fig. 2. Detailed analyses of model performance.
a The performance comparison of DPFunc and other representative methods on difficult protein sets with different sequence similarities to training proteins, where the data from five repeated experiments are presented as mean value +/- standard errors. b, d, e The IC weighted PR curve of DPFunc and other representative methods on MF, CC and BP, respectively. c The performance evaluation of DPFunc and other representative methods on rare GO terms with different IC values, where GO terms with higher IC values are more informative and valuable. The experiment is repeated five times for each method on the test data, reducing the effects from the random factor. The data are presented as mean value +/- standard deviation. f The performance of DPFunc and other representative methods on GO terms with deeper depths, where the distances between GO terms and root node (MF/CC/BP) are larger than 8, 6, and 8, respectively. The experiment is repeated five times for each method on the test data, reducing the effects from the random factor. The data are presented as mean value +/- standard deviation. g The coverage of predicted functions from DPFunc and other representative methods. DPFunc can predict all known functions while others can only predict parts of functions.
Beyond its overall performance, DPFunc excels in predicting informative GO terms characterized by high IC values. These terms present a greater challenge due to their few occurrences and limited training samples. As illustrated in Fig. 2c, DPFunc consistently outperforms the other methods when predicting GO terms with fewer samples, and the improvement remains for more specific GO terms (IC≥3). Notably, some methods, such as TALE, fail to accurately predict these informative GO terms (see Supplementary Fig. 3). Additionally, Fig. 2b,d-e show the performance in terms of IC-weighted AUPR (see “Methods" for details), which is different from AUPR and considers the informative of GO terms. It can be obtained that DPFunc surpasses the other methods, indicating the great potential of DPFunc in predicting informative functions. The detailed data can be obtained from Supplementary Table 2.
As functions form a loosely hierarchical structure and are related, functions with deeper depths are more specific and predicting these types of functions is more meaningful. Figure 2f shows the performance of these methods on GO terms with deeper nodes (depths > = 8 in MF and BP, depths > = 6 in CC since the maximum is 7 in CC), which evaluates the performance on each selected GO term and means their AUPR values as the final metric. DPFunc still achieves the best performance, except for being slightly weaker than BlastKNN in BP. Notably, although ATGO+ achieves comparable scores, it can only predict parts of known functions (66.3%), as shown in Fig. 2g and Supplementary Table 3. Above all, DPFunc demonstrates a distinct advantage over SOTA methods, particularly in its ability to handle unseen proteins with low sequence identity, informative GO terms with high IC values, and specific GO terms with deeper nodes.
Domain information improves the performance of DPFunc
To unequivocally demonstrate the pivotal role of domain information in DPFunc, we replace the domain attention block with a mean pooling layer, a commonly used strategy in previous studies, i.e., DeepFRI and GAT-GO. As illustrated in Fig. 3a, b, after adding the guidance of domain information, DPFunc exhibits substantial improvements in both Fmax and AUPR, compared to DPFunc w/o domain in MF, CC, and BP.
Fig. 3. The analyses of the role of domain information.
a,b The comprehensive comparison of DPFunc and DPFunc w/o domain in terms of Fmax and AUPR. c The performance on each function. AUPR values are calculated separately for each GO term (remove the perfect predicted GO terms which are shown in Fig. 3d). The median is represented by the centerline of the boxplot, while the first and third quartiles are indicated by the bounds of the box. The whiskers represent the 0.8 interquartile range (IQR). Specifically, there are 424 MF GO terms, 457 CC GO terms and 3283 BP GO terms for DPFunc. And there are 460 MF GO terms, 472 CC GO terms and 3343 BP GO terms for DPFunc w/o domain. Two-side paired t-tests are conducted on the overall performance of these two models and the resulting P values are annotated at the top of the boxes. d The number of perfect predicted GO terms. e The performance of top-k predicted functions of each protein. Since there are 8, 10, and 30 GO terms per protein on average in MF, BP, and CC, different ranges of k are selected (4-8 for MF, 7-11 for CC, and 21-33 for BP, respectively). f The performance of top-k predicted functions of each protein, where k is exactly set as 5, 9, 24 for MF, CC, and BP, respectively.
To provide a comprehensive evaluation of the differences between these two models, we test the prediction results for each individual GO term, as shown in Fig. 3c, d. Figure 3d shows the number of perfect predicted GO terms (AUPR=1), it can be observed that DPFunc with domain annotations achieves better performance. As for the other GO terms, Fig. 3c shows that DPFunc, armed with domain insights, achieves a remarkable median AUPR improvement of 12.0%, 14.7%, and 16.3% for MF, CC, and BP, respectively. These results unequivocally substantiate the unparalleled value of incorporating domain information for protein function prediction.
Moreover, to ensure model reliability, we focus on evaluating predictions with high confidence scores. Specifically, we assess the results with the top k prediction scores of these two models, where k is determined by the average number of GO terms per protein (approximately ~ 7 for MF, ~ 11 for CC, and ~ 30 for BP). As shown in Fig. 3e, it can be observed that DPFunc achieves better performance after incorporating the domain information, demonstrating mean F-measure improvements exceeding 1.6% ~ 3.1% for MF, 1.9% ~ 3.3% for CC, and 5.5% ~ 6.7% for BP. Similar conclusions can be drawn from Fig. 3f, which shows the distribution of predictions over specific k values (5 for MF, 9 for CC, and 24 for BP). In summary, our model can predict protein functions more accurately when incorporating domain information. The improvements are more striking in CC and BP.
DPFunc effectively distinguishes structure motifs and sequence identities
Since protein structures are closely related to their functions, in this section, we focus on evaluating the ability of DPFunc to discern structural motifs and their associated functions. To evaluate this ability, we first select protein pairs with low sequence similarities, and assess the similarities of their structure features using the widely adopted TM-score, a metric commonly employed in structure prediction. As illustrated in Fig. 4a, DPFunc, under the guidance of domain knowledge, demonstrates a remarkable ability to distinguish between these protein pairs, exhibiting a higher correlation with structure similarities (TM-score). In contrast, in the absence of domain information, the model struggles to differentiate structure features, resulting in consistently high structure similarities exceeding 88%, and failing to capture the nuances of dissimilar structures.
Fig. 4. The performance of DeepDugest on structure motifs learning.
a The correlations between the learned structure feature similarities and structure similarities on protein pairs with low sequence similarities. b, c The constructed structure graphs of two proteins, P0C617 and Q8NGY0, where orange points represent the edges between residues. d The functions of these two proteins (P0C617 and Q8NGY0) and corresponding related domains. e The structure alignment results of P0C617 and Q8NGY0. f, g The views of attention maps of P0C617 and Q8NGY0, where red points represent the key residues and regions detected by DPFunc. h The structure alignment results between 5JZV-A, 3WG8-A and 5Z9R-A. Dark colors in each protein represent residues that are aligned and light colors represent residues that are not aligned. i The performance of DPFunc and S2F on 47 proteins from bacteria (Bacillus subtilis, BACSU). The coordinates of each scatter indicate the F-measure values of these two methods on one protein. j The PR curve and AUPR values of DPFunc and S2F on BACSU proteins.
To further illustrate the potential of DPFunc in detecting similar structural motifs, even in the absence of sequence similarity, we conduct two case studies: P0C617 and Q8NGY0, two pivotal plasma membrane proteins that separate the cell from its external environment7. Despite their dissimilar sequences, these proteins share strikingly similar structures and same functions of maintaining plasma membrane integrity (GO:0005886). The details are shown as Fig. 4d-e. These two proteins have dissimilar sequences but similar structures to perform the same functions. DPFunc first extracts the same domain information via scanning their sequences, and these domain properties are all related to membrane functions, which are all validated in the UniProt database7. Then, Fig. 4b, c shows similar contact maps generated by their structures, and Fig. 4f, g shows similar attention maps, indicating that domain-guided insight empowers DPFunc to learn similar features from similar structures. These findings demonstrate the ability of DPFunc to capture structural resemblance and accurately predict functions, even when faced with disparate sequences, underscoring its exceptional potential in protein function prediction.
Additionally, there also exist scenarios where proteins with high sequence identities have different structures and functions. It is necessary for models to distinct these proteins and corresponding functions. Consequently, we present three proteins here to evaluate the capability of DPFunc in this scenario (PDB ID: 5JZV-A [https://www.rcsb.org/structure/5JZV], 3WG8-A [https://www.rcsb.org/structure/3WG8], 5Z9R-A [https://www.rcsb.org/structure/5Z9R], see Fig. 4h). As illustrated in Supplementary Table 6 and Supplementary Table 7, these proteins have high sequence identities but different functions. For instance, the sequence identity between 5JZV-A [https://www.rcsb.org/structure/5JZV] and 3WG8-A [https://www.rcsb.org/structure/3WG8] is 87.8% but they have only 5 common functions. For these proteins, DPFunc predicts their functions with 100% accuracy, as shown in Supplementary Fig. 4, which demonstrates the ability of our model on proteins with high sequence identities but distinct structures.
DPFunc holds the potential for annotating bacteria
With more and more protein sequences being detected, many sequenced organisms have been discovered while their functions are unknown, especially bacteria and viruses61. Accurately annotating their functions is critical to understanding the role of corresponding proteins and their association with disease61. In general, these proteins from the sequenced organisms lack other information, such as protein-protein interactions and gene expressions, representing challenges for existing computational approaches that rely on multi-type biological knowledge18. Consequently, it is meaningful to annotate these proteins from sequences62,63.
In this study, to further explore the performance of our methods, we re-divide the dataset, select a specific type of bacteria, Bacillus subtilis64, as the test data, and remove all associated species data from the training data (the details can be obtained from the Supplementary Table 4). Additionally, S2F18 is chosen as a representative method, which is proposed for annotating the sequenced organisms based on network propagation. Figure 4i, j illustrate the performance of these two methods. From Fig. 4i, it can be obtained that DPFunc gets better performance over the vast majority of proteins, with weaker performance than S2F on only 3 out of 47 proteins. It is worth noting that S2F gets nearly 0 F-measure on the majority of proteins, while DPFunc achieves significant improvements on the same proteins. Additionally, Fig. 4j illustrates the PR curve of these two methods, which demonstrates that DPFunc has a great improvement in terms of AUPR, proving the potential of DPFunc for annotating bacteria.
DPFunc effectively detects significant active sites for enzyme functions
DPFunc can also detect significant residues in proteins that are highly correlated with functions (see “Methods" for details). For instance, in enzyme reactions, the catalytic process is carried out by specific active residues65–67. In this section, we provide several cases to show the capabilities of DPFunc in active site detection. Specifically, Q9M1Y0 and Q8S929 are two cysteine proteases involved in both proteolytic activation and delipidation of ATG8 family proteins68. Previous literature68 indicates that the two proteins both have three active sites: 173-th, 368-th, and 370-th residues for Q9M1Y0, and 170-th, 364-th, and 366-th residues for Q8S929. Figure 5a shows the details of the prediction results of Q9M1Y0 and Q8S929. The red positions represent the key residues detected by DPFunc (CYS-170, PRO-305 for Q8S929, and CYS-173, PRO-369 for Q9M1Y0) and the known validated residues from previous literatures (CYS-170, ASP-364, HIS-366 for Q8S929, and CYC-173, ASP-368, HIS-370 for Q9M1Y0). Obviously, DPFunc not only accurately predicts their functions, but also highlights significant sites that overlap with known active sites. Notably, DPFunc exhibits a remarkable ability to identify potential functional hotspots for closely spaced active sites. For example, in the case of Q8S929, where the 364-th and 366-th residues are active sites, DPFunc identifies the intermediate site (the 365-th residue) as the potential functional hotspot. This remarkable ability is attributed to the power of graph neural networks, which can aggregate the information from two neighboring active sites.
Fig. 5. Key residues detected by DPFunc.
a The details detected by DPFunc on two important cysteine proteases. The red positions shown in the structures are the key residues detected by DPFunc (CYS-170, PRO-305 for Q8S929, and CYS-173, PRO-369 for Q9M1Y0). The three red residues in the detailed graphs are the active sites that have been validated (CYS-170, ASP-364, HIS-366 for Q8S929, and CYC-173, ASP-368, HIS-370 for Q9M1Y0). These residues play significant roles in autophagy and perform the functions (mediating both proteolytic activation and delipidation of ATG8 family proteins). b The red positions shown in the structures are the validated active sites of four Arabidopsis thaliana proteins (HIS-147 for Q93ZR6/WSD1, HIS-163 for Q9M3B1/WSD6, HIS-135 for Q94CK0/WSD7 and HIS-144 for Q5KS41/WSD11), performing the same functions, involving in cuticular wax biosynthesis.
Furthermore, we find four proteins from the same species (Arabidopsis thaliana) with the same functions (Wax ester synthase/diacylglycerol acyltransferase), including Q93ZR6 (WSD1) [https://www.uniprot.org/uniprotkb/Q93ZR6/entry], Q9M3B1 (WSD6) [https://www.uniprot.org/uniprotkb/Q9M3B1/entry], Q94CK0 (WSD7) [https://www.uniprot.org/uniprotkb/Q94CK0/entry] and Q5KS41 (WSD11) [https://www.uniprot.org/uniprotkb/Q5KS41/entry]. All of these proteins are significant enzymes and involved in cuticular wax biosynthesis69–72, as shown in Fig. 5(b). It is worth noting that each of these four proteins has a known active site, where HIS-147 for WSD169,70, HIS-163 for WSD670, HIS-135 for WSD770, and HIS-144 for WSD1171,72. Moreover, we use Clustal Omega73 to align these sequences. As illustrated in Supplementary Fig. 5, the four positions are aligned as expected, which further support the co-evolutionary conservation of these residues. As for these proteins, DPFunc detects all of these active sites accurately.
Additionally, we compare our method with another SOTA method74 in the field of functional site prediction. As illustrated in Supplementary Table 8, we test the two approaches on three common proteins that appear in both our study and ref. 74. The known active sites can be obtained from M-CSA database75. The results in Supplementary Fig. 6 show that our method achieves comparable performance with the method proposed in ref. 74, further supporting the effect of DPFunc on active site detection. Notably, although DPFunc detects significant active sites effectively, finding active sites in disordered regions remains a challenge that may be further explored in future models (see Supplementary Fig. 7).
Discussion
As existing protein function prediction approaches cannot extract structure features effectively and ignore the significance of different residues, in this study, we develop a deep learning-based method, called DPFunc. It incorporates domain-guided structure information to identify critical regions within protein structures, enabling accurate prediction of functions based on the latent structure motifs and key residues. Comprehensive comparisons with other state-of-the-art deep learning methods, particularly current structure-based approaches, demonstrate the advantages of our proposed approach. Notably, DPFunc also outperforms other methods on rare functions, specific functions and difficult proteins that have low sequence similarities to known proteins. Furthermore, the role of domain information in DPFunc is crucial. Under the guidance of domain information, DPFunc can predict protein functions more accurately, as further validated by analyzing the performance of top-k predicted results and individual GO terms.
Moreover, DPFunc demonstrates an impressive ability to distinguish proteins between dissimilar structures. On the other hand, several cases prove that DPFunc can learn similar structure motifs even when their sequences have low identity. Meanwhile, for those proteins from the sequenced organisms, DPFunc also shows the improvements compared to other methods. Additionally, for the functions that are performed by specific residues, DPFunc can detect key residues or regions, thus providing interpretability between the key residues and the corresponding functions, as well as enabling the discovery of potential key residues for new proteins.
DPFunc only uses protein sequences as input. Specifically, it generates domain information by scanning sequences, extracts residue features through a pre-trained protein language model, and constructs structure graphs based on the predicted structures. Importantly, all of these inputs can be obtained from the protein sequences. Consequently, DPFunc can be applied to large-scale proteins that only have sequences. In the future, we will consider introducing more structure-related biological knowledge to help models learn the relationships between structure motifs and functions. Furthermore, it is essential to consider incorporating the relationships among different protein functions. Since proteins perform functions in cellular context, their functions are dynamically transformed with the environment. How to accurately predict the dynamic functions is another challenge to be addressed in the future76.
Methods
Datasets
We use two datasets to evaluate the performance of our method. The first dataset is collected from DeepFRI37, named PDB dataset, which is a non-redundant set by clustering all PDB chains at 95% sequence identity and has also been used in previous studies such as GAT-GO38. In this dataset, the structures of proteins are obtained from the Protein Data Bank (PDB)50, which are all validated by experiment. The statistic information of proteins is illustrated in Table 3. Specifically, the original PDB dataset is split into training, validation and testing sets by cd-hit77 with 40% sequence identity, contains 36,408 proteins and 2748 GO terms, including 488 (17.8%) MF GO terms, 320 (11.6%) CC GO terms, and 1940 (70.6%) BP GO terms.
Table 3.
The statistic information of two datasets
| Dataset | MF | CC | BP | ||||
|---|---|---|---|---|---|---|---|
| PDB dataset | Train | 24837 | (80.2%) | 11162 | (70.4%) | 23386 | (79.5%) |
| Valid | 2746 | (8.9%) | 1296 | (8.2%) | 2624 | (8.9%) | |
| Test | 3399 | (10.9%) | 3400 | (21.4%) | 3400 | (11.6%) | |
| All | 30982 | (100%) | 15858 | (100%) | 29410 | (100%) | |
| CAFA dataset | Train | 31463 | (96.7%) | 42467 | (96.4%) | 47333 | (96.3%) |
| Valid | 682 | (2.1%) | 711 | (1.6%) | 767 | (1.6%) | |
| Test | 401 | (1.2%) | 877 | (2.0%) | 1039 | (2.1%) | |
| All | 32546 | (100%) | 44055 | (100%) | 49139 | (100%) |
Another dataset is collected from the UniProt and Gene Ontology database. Following the CAFA challenge, we collect the protein sequences and their corresponding functions from the UniProt database, and split them into three subsets based on timestamps, named the CAFA dataset: training data, validation data, and test data. Specifically, the training data contains the proteins released before 2020-05, the validation data encompasses the proteins released between 2020-05 and 2021-04, and the test data includes the proteins between 2021-05 and 2022-04 (published in 2022-12). Then, we collect their corresponding predicted structures from the AlphaFold database51,52. For several proteins whose structures could not be downloaded directly from the database, we use AlphaFold2 to predict their structures locally. Finally, as shown in Table 3, there are 59,397 proteins and 28,252 GO terms, including 6086 (21.5%) MF GO terms, 2492 (8.8%) CC GO terms, and 19,674 (69.6%) BP GO terms.
Residue-level feature learning module
To learn residue-level features, DPFunc first constructs graphs based on protein structures. Specifically, for a target protein pi, its residues are considered as nodes, and two residues are connected if the distance of their corresponding Cα atoms is less than 10 Å. Based on this rule, we can construct the corresponding structure graph A ∈ {0, 1}l∗l, where l represents the sequence length. Then, DPFunc utilizes the existing pre-trained protein large language model (ESM-1b) to generate the residue embeddings as node features, denoted as , where d represents the dimension of node features. Subsequently, as illustrated in Fig. 1(b), DPFunc uses two GCN layers to propagate features between residues through structure graphs. Inspired by ResNet54, DPFunc also adds a residual operation to GCN layers as follows:
| 1 |
| 2 |
where represents the residue features as the input of k-th GCN layer (k = 1, …, n and X1 = X), denotes the original graph with self-loops of each node, is the corresponding diagonal degree matrix of , is the learnable parameters of k-th GCN layer, and ReLU is a rectified linear unit function, as shown in Equation (2).
After processing several GCN layers, we can obtain the updated residue features Xn+1 from the last GCN layer, denoted as Xfinal.
Protein-level feature learning module
Traditional structure-based methods typically treat every residue equally and calculate protein features as follows:
| 3 |
where Xfinal[i] represents the features of i-th residue. In contrast, DPFunc uses domain information as guidance to find the important residues in protein structures. Specifically, DPFunc first scans the protein sequences and generates corresponding domain properties by InterProScan. Then, it employs one-hot encoding to represent these domain properties and feeds them into an embedding layer to obtain their dense representations:
| 4 |
where IPR ∈ {0, 1}1*m indicates the one-hot encoding of m domain properties, is an embedding layer, and are two linear layers, respectively.
Then, inspired by the architecture of the transformer encoder, we propose an attention mechanism based on both residue features and domain information, as illustrated in Fig. 1c. After obtaining domain embeddings H and residue features X final, DPFunc learns the latent correlations between domains and residues:
| 5 |
| 6 |
| 7 |
| 8 |
| 9 |
where represents the latent representations for attention head i, indicates the corresponding importance of residues, and Watt−iVi is the pooling results based on attention head i. Finally, the results from different attention heads are concatenated and processed as the final protein features through several feed-forward layers, denoted as . Additionally, the residual operation is also employed here to prevent the loss of features.
Function prediction, postprocessing procedure and significant residues detection
Finally, DPFunc integrates these two modules and predicts protein functions. Specifically, it utilizes the initial residue features and protein features to annotate functions:
| 10 |
| 11 |
where indicates the predicted scores of c GO terms, and MLP is the multilayer perceptron, composed of several linear layers and ReLU. It is important to note that GO terms exhibit a loosely hierarchical structure, such as ’is-a’ and ’part-of’. If a ’child’ GO term is annotated, its ’parent’ GO terms must be annotated. To ensure consistency with these hierarchical relationships, we introduce a common post-processing procedure:
| 12 |
where represents the j-th child GO terms of GO term i. Additionally, this post-processing procedure is only applied to the final predicted results, thus avoiding reducing the computational efficiency during the training process.
Additionally, once our model is trained, it can detect significant residues from the structures based on the attention mechanism. This process is illustrated in Supplementary Table 5. Firstly, the attention scores of residues can be obtained from the trained model, denoted as Watt−i in Section 4.3. Then, for each head i, these attention scores are sorted in descending order and the gaps between neighbors are calculated. Furthermore, inspired by CLEAN78, the average value of these gaps is set as the cut-off and the residues are selected to the candidate set from a higher score to a lower score until the gap between residues is larger than the cut-off. Moreover, considering the qualities of protein structures predicted by AlphaFold2, pLDDT35,36 is further used to filter the candidate sites, where higher pLDDT represents higher confidence. The residues in the candidate set with pLDDT lower than 50 are removed.
Model training and Parameter setting
As protein function prediction is an imbalanced multi-label classification problem, DPFunc utilizes focal loss as the loss function. Focal loss is specifically designed to address the challenges posed by imbalanced datasets and has shown improved performance compared to traditional binary cross-entropy loss (BCELoss):
| 13 |
| 14 |
where represents the predicted score of GO term c and y = 0, 1 denotes the corresponding true label. γ is a hyper-parameter that controls the focal scores for positive and negative labels. It can be obtained that FocalLoss is equal to BCELoss when γ is set as 0.
During the training process, we set all the dimensions of hidden layers as 1280, which are the same as the dimensions of the features generated by ESM-1b. The number of attention heads is set as 4, and the γ of focal loss is 2, which is a common default choice. We use AdamW as the optimizer with a learning rate lr = 1e − 4 and set the batch size as 64. To ensure stable performance, the models of the last three epochs are used to predict results, which are then averaged as the final results. All experiments of the DPFunc are carried out using one NVIDIA Tesla V100s GPU card with 32 GB of memory. On the CAFA dataset, our model took around 12 training epochs for 2 hours in MF, 5 hours in BP, and 2 hours in CC. On the smaller PDB dataset, 8 training epochs of our model in MF, BP, and CC all took no more than half an hour.
Evaluation metrics
In this study, five metrics are used to evaluate the performance of models and the similarity of structures, including Fmax, AUPR, M-AUPR, IC_AUPR and TM-score. Fmax is the maximum value of the harmonic average of precision and recall:
| 15 |
| 16 |
| 17 |
where f(t) is the number of proteins that predict at least one function with confidence ≥t. AUPR is the area under the precision-recall curve, and M-AUPR is the average of all AUPR values calculated for each label separately. As illustrated in Equation 17-19, IC_AUPR is different from AUPR, which considers the information content (IC) of GO terms and is calculated by the weighted precision (icPR) and recall (icRc).
| 18 |
| 19 |
| 20 |
where IC(GOj) reflects the occurrence of GO term j when its ancestors are annotated. TM-score measures the structure similarity of two proteins as follows:
| 21 |
where Psource and Ptarget are two proteins, and Psource is aligned to Ptarget. Ltarget is the sequence length of Ptarget. Lalignment is the number of paired residues and di is the distance between the i-th paired residues. is a normalized parameter. Overall, higher values of Fmax, AUPR, and M-AUPR represent better performance for protein function prediction. Similarly, a higher TM-score represents a greater degree of structure similarity between two proteins.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This work is supported in part by the National Natural Science Foundation of China under Grant (No.62225209 to M.L.), Hunan Postgraduate Research and Innovation Project (CX20240018 to W.W.), and the High Performance Computing Center of Central South University.
Author contributions
M.L. supervised the research. W.W., M.Z. and M.L. conceived the initial idea. W.W. and Y.S. collected and preprocessed the data and W.W. designed the model. W.W. and Y.S. performed the benchmark, W.W, M.Z. and M.L. designed case studies. W.F. participated in discussions. All authors wrote the manuscript and approved the final version.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The protein sequences and their functions used in this study are available in the Uniprot database [https://www.uniprot.org/uniprotkb/]. The gene ontology file can be obtained from the Gene Ontology knowledgebase [https://geneontology.org/]. The processed data for training and testing models are available in https://github.com/CSUBioGroup/DPFunc. Other data for Figs. 2–4 are provided as a Source Data file. The proteins mentioned in our cases, including P0C617, Q8NGY0, Q9M1Y0, Q8S929, Q93ZR6, Q9M3B1, Q94CK0 and Q5KS41 are available in the Uniprot repository (https://www.uniprot.org/) under their accession codes. The protein structures of 5JZV-A [https://www.rcsb.org/structure/5JZV], 3WG8-A [https://www.rcsb.org/structure/3WG8] and 5Z9R-A [https://www.rcsb.org/structure/5Z9R] are available in the PDB repository (https://www.rcsb.org/) under their accession codes. Source data are provided with this paper.
Code availability
The source codes of DPFunc are available on GitHub at https://github.com/CSUBioGroup/DPFunc, which has also been deposited in the Zenodo under accession code https://zenodo.org/records/13843028. Data are analyzed using Numpy v1.24.(https://github.com/numpy/numpy), sklearn v1.3.0 (https://scikit-learn.org/stable/), scipy v1.10.1 (https://www.scipy.org/) and Matplotlib v3.2.2 (https://matplotlib.org/). Structures are visualized by Pymol v2.5.7 (https://www.pymol.org/). Blastp v2.12.0+ https://blast.ncbi.nlm.nih.gov/doc/blast-help/downloadblastdata.htmlis used for calculating sequence identities. TM-align v2022/04/12 https://zhanggroup.org/TM-align/is used for protein structure similarity calculation.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54816-8.
References
- 1.Eisenberg, D., Marcotte, E. M., Xenarios, I. & Yeates, T. O. Protein function in the post-genomic era. Nature405, 823–826 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Ouzounis, C. A., Coulson, R. M., Enright, A. J., Kunin, V. & Pereira-Leal, J. B. Classification schemes for protein structure and function. Nat. Rev. Genet.4, 508–519 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Ng, P. C. & Henikoff, S. Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genom. Hum. Genet.7, 61–80 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Soskine, M. & Tawfik, D. S. Mutational effects and the evolution of new protein functions. Nat. Rev. Genet.11, 572–582 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Leveson-Gower, R. B., Mayer, C. & Roelfes, G. The importance of catalytic promiscuity for enzyme design and evolution. Nat. Rev. Chem.3, 687–705 (2019). [Google Scholar]
- 6.Radivojac, P. et al. A large-scale evaluation of computational protein function prediction. Nat. Methods10, 221–227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Consortium, U. Uniprot: a worldwide hub of protein knowledge. Nucleic Acids Res.47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Consortium, G. O. The gene ontology (go) database and informatics resource. Nucleic Acids Res.32, D258–D261 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Consortium, G. O. The gene ontology resource: 20 years and still going strong. Nucleic Acids Res.47, D330–D338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urzúa-Traslaviña, C. G. et al. Improving gene function predictions using independent transcriptional components. Nat. Commun.12, 1464 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark, W. T. & Radivojac, P. Analysis of protein function and its prediction from amino acid sequence. Proteins: Struct. Funct. Bioinforma.79, 2086–2096 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Madani, A. et al. Large language models generate functional protein sequences across diverse families. Nat. Biotechnol.41, 1099–1106 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sumida, K. H. et al. Improving protein expression, stability, and function with proteinmpnn. J. Am. Chem. Soc.146, 2054–2061 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using diamond. Nat. Methods12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Kulmanov, M., Khan, M. A. & Hoehndorf, R. Deepgo: predicting protein functions from sequence and interactions using a deep ontology-aware classifier. Bioinformatics34, 660–668 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulmanov, M. & Hoehndorf, R. Deepgoplus: improved protein function prediction from sequence. Bioinformatics36, 422–429 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lv, Z., Ao, C. & Zou, Q. Protein function prediction: from traditional classifier to deep learning. Proteomics19, 1900119 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Torres, M., Yang, H., Romero, A. E. & Paccanaro, A. Protein function prediction for newly sequenced organisms. Nat. Mach. Intell.3, 1050–1060 (2021). [Google Scholar]
- 19.Ibtehaz, N., Kagaya, Y. & Kihara, D. Domain-pfp allows protein function prediction using function-aware domain embedding representations. Commun. Biol.6, 1103 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scherf, U. et al. A gene expression database for the molecular pharmacology of cancer. Nat. Genet.24, 236–244 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Hunter, L. & Cohen, K. B. Biomedical language processing: what’s beyond pubmed? Mol. cell21, 589–594 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk, D. et al. The string database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic acids Res.51, D638–D646 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huerta-Cepas, J. et al. eggnog 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic acids Res.47, D309–D314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang, Y. et al. An expanded evaluation of protein function prediction methods shows an improvement in accuracy. Genome Biol.17, 1–19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou, N. et al. The cafa challenge reports improved protein function prediction and new functional annotations for hundreds of genes through experimental screens. Genome Biol.20, 1–23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.You, R., Yao, S., Mamitsuka, H. & Zhu, S. Deepgraphgo: graph neural network for large-scale, multispecies protein function prediction. Bioinformatics37, i262–i271 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu, Y.-H. et al. Tripletgo: integrating transcript expression profiles with protein homology inferences for gene function prediction. Genom. Proteom. Bioinforma.20, 1013–1027 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barot, M., Gligorijević, V., Cho, K. & Bonneau, R. Netquilt: deep multispecies network-based protein function prediction using homology-informed network similarity. Bioinformatics37, 2414–2422 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu, Y.-H., Zhang, C., Yu, D.-J. & Zhang, Y. Integrating unsupervised language model with triplet neural networks for protein gene ontology prediction. PLOS Comput. Biol.18, e1010793 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loewenstein, Y. et al. Protein function annotation by homology-based inference. Genome Biol.10, 1–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Juncker, A. S. et al. Sequence-based feature prediction and annotation of proteins. Genome Biol.10, 1–6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerstein, M. How representative are the known structures of the proteins in a complete genome? a comprehensive structural census. Fold. Des.3, 497–512 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Cao, Y. & Shen, Y. Tale: Transformer-based protein function annotation with joint sequence–label embedding. Bioinformatics37, 2825–2833 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science379, 1123–1130 (2023). [DOI] [PubMed] [Google Scholar]
- 35.Jumper, J. et al. Highly accurate protein structure prediction with alphafold. Nature596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abramson, J. et al. Accurate structure prediction of biomolecular interactions with alphafold 3. Nature 1–3 (2024). [DOI] [PMC free article] [PubMed]
- 37.Gligorijević, V. et al. Structure-based protein function prediction using graph convolutional networks. Nat. Commun.12, 3168 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai, B. & Xu, J. Accurate protein function prediction via graph attention networks with predicted structure information. Brief. Bioinforma.23, bbab502 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowie, J. U., Lüthy, R. & Eisenberg, D. A method to identify protein sequences that fold into a known three-dimensional structure. Science253, 164–170 (1991). [DOI] [PubMed] [Google Scholar]
- 40.Ouzounis, C., Sander, C., Scharf, M. & Schneider, R. Prediction of protein structure by evaluation of sequence-structure fitness: aligning sequences to contact profiles derived from three-dimensional structures. J. Mol. Biol.232, 805–825 (1993). [DOI] [PubMed] [Google Scholar]
- 41.Marks, D. S., Hopf, T. A. & Sander, C. Protein structure prediction from sequence variation. Nat. Biotechnol.30, 1072–1080 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shin, J.-E. et al. Protein design and variant prediction using autoregressive generative models. Nat. Commun.12, 2403 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kipf, T. N. & Welling, M. Semi-supervised classification with graph convolutional networks. In International Conference on Learning Representations. https://openreview.net/forum?id=SJU4ayYgl (2017).
- 44.Veličković, P. et al. Graph attention networks. In International Conference on Learning Representations. https://openreview.net/forum?id=rJXMpikCZ (2018).
- 45.Hunter, S. et al. Interpro: the integrative protein signature database. Nucleic acids Res.37, D211–D215 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paysan-Lafosse, T. et al. Interpro in 2022. Nucleic acids Res.51, D418–D427 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caetano-Anollés, G. & Caetano-Anollés, D. An evolutionarily structured universe of protein architecture. Genome Res.13, 1563–1571 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu, L. et al. Grammar of protein domain architectures. Proc. Natl Acad. Sci.116, 3636–3645 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doerks, T., Copley, R. R., Schultz, J., Ponting, C. P. & Bork, P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res.12, 47–56 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burley, S. K. et al. Protein data bank (pdb): the single global macromolecular structure archive. Protein crystallography: methods and protocols 627–641 (2017). [DOI] [PMC free article] [PubMed]
- 51.Varadi, M. et al. Alphafold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic acids Res.50, D439–D444 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varadi, M. et al. Alphafold protein structure database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Res.52, D368–D375 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rives, A. et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl Acad. Sci.118, e2016239118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition, 770–778 (2016).
- 55.Jones, P. et al. Interproscan 5: genome-scale protein function classification. Bioinformatics30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, S., You, R., Liu, Y., Xiong, Y. & Zhu, S. Netgo 3.0: protein language model improves large-scale functional annotations. Genom. Proteom. Bioinforma.21, 349–358 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, C., Freddolino, P. L. & Zhang, Y. Cofactor: improved protein function prediction by combining structure, sequence and protein–protein interaction information. Nucleic acids Res.45, W291–W299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roy, A., Yang, J. & Zhang, Y. Cofactor: an accurate comparative algorithm for structure-based protein function annotation. Nucleic acids Res.40, W471–W477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou, X. et al. I-tasser-mtd: a deep-learning-based platform for multi-domain protein structure and function prediction. Nat. Protoc.17, 2326–2353 (2022). [DOI] [PubMed] [Google Scholar]
- 60.Kryshtafovych, A., Schwede, T., Topf, M., Fidelis, K. & Moult, J. Critical assessment of methods of protein structure prediction (casp)-round xiv. Proteins: Struct. Funct. Bioinforma.89, 1607–1617 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flamholz, Z. N., Biller, S. J. & Kelly, L. Large language models improve annotation of prokaryotic viral proteins. Nature Microbiology 1–13 (2024). [DOI] [PMC free article] [PubMed]
- 62.Whisstock, J. C. & Lesk, A. M. Prediction of protein function from protein sequence and structure. Q. Rev. Biophys.36, 307–340 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Wang, W. et al. A comprehensive computational benchmark for evaluating deep learning-based protein function prediction approaches. Brief. Bioinforma.25, bbae050 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kunst, F. et al. The complete genome sequence of the gram-positive bacterium bacillus subtilis. Nature390, 249–256 (1997). [DOI] [PubMed] [Google Scholar]
- 65.Todd, A. E., Orengo, C. A. & Thornton, J. M. Plasticity of enzyme active sites. Trends Biochem. Sci.27, 419–426 (2002). [DOI] [PubMed] [Google Scholar]
- 66.del Sol, A., Fujihashi, H., Amoros, D. & Nussinov, R. Residue centrality, functionally important residues, and active site shape: analysis of enzyme and non-enzyme families. Protein Sci.15, 2120–2128 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klinman, J. P. Dynamically achieved active site precision in enzyme catalysis. Acc. Chem. Res.48, 449–456 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoshimoto, K. et al. Processing of atg8s, ubiquitin-like proteins, and their deconjugation by atg4s are essential for plant autophagy. Plant Cell16, 2967–2983 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li, F. et al. Identification of the wax ester synthase/acyl-coenzyme a: diacylglycerol acyltransferase wsd1 required for stem wax ester biosynthesis in arabidopsis. Plant Physiol.148, 97–107 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patwari, P. et al. Surface wax esters contribute to drought tolerance in arabidopsis. Plant J.98, 727–744 (2019). [DOI] [PubMed] [Google Scholar]
- 71.Takeda, S. et al. Physical interaction of floral organs controls petal morphogenesis in arabidopsis. Plant Physiol.161, 1242–1250 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takeda, S., Iwasaki, A., Tatematsu, K. & Okada, K. The half-size abc transporter folded petals 2/abcg13 is involved in petal elongation through narrow spaces in arabidopsis thaliana floral buds. Plants3, 348–358 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol.7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cagiada, M. et al. Discovering functionally important sites in proteins. Nat. Commun.14, 4175 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ribeiro, A. J. M. et al. Mechanism and catalytic site atlas (m-csa): a database of enzyme reaction mechanisms and active sites. Nucleic acids Res.46, D618–D623 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jeffery, C. J. Protein moonlighting: what is it, and why is it important? Philos. Trans. R. Soc. B: Biol. Sci.373, 20160523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. Cd-hit: accelerated for clustering the next-generation sequencing data. Bioinformatics28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu, T. et al. Enzyme function prediction using contrastive learning. Science379, 1358–1363 (2023). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The protein sequences and their functions used in this study are available in the Uniprot database [https://www.uniprot.org/uniprotkb/]. The gene ontology file can be obtained from the Gene Ontology knowledgebase [https://geneontology.org/]. The processed data for training and testing models are available in https://github.com/CSUBioGroup/DPFunc. Other data for Figs. 2–4 are provided as a Source Data file. The proteins mentioned in our cases, including P0C617, Q8NGY0, Q9M1Y0, Q8S929, Q93ZR6, Q9M3B1, Q94CK0 and Q5KS41 are available in the Uniprot repository (https://www.uniprot.org/) under their accession codes. The protein structures of 5JZV-A [https://www.rcsb.org/structure/5JZV], 3WG8-A [https://www.rcsb.org/structure/3WG8] and 5Z9R-A [https://www.rcsb.org/structure/5Z9R] are available in the PDB repository (https://www.rcsb.org/) under their accession codes. Source data are provided with this paper.
The source codes of DPFunc are available on GitHub at https://github.com/CSUBioGroup/DPFunc, which has also been deposited in the Zenodo under accession code https://zenodo.org/records/13843028. Data are analyzed using Numpy v1.24.(https://github.com/numpy/numpy), sklearn v1.3.0 (https://scikit-learn.org/stable/), scipy v1.10.1 (https://www.scipy.org/) and Matplotlib v3.2.2 (https://matplotlib.org/). Structures are visualized by Pymol v2.5.7 (https://www.pymol.org/). Blastp v2.12.0+ https://blast.ncbi.nlm.nih.gov/doc/blast-help/downloadblastdata.htmlis used for calculating sequence identities. TM-align v2022/04/12 https://zhanggroup.org/TM-align/is used for protein structure similarity calculation.