Abstract
Background
Renal cell cancer (RCC) is the most common and highly malignant subtype of kidney cancer. Mesenchymal stromal cells (MSCs) are components of tumor microenvironment (TME) that influence RCC progression. The impact of RCC-secreted small non-coding RNAs (sncRNAs) on TME is largely underexplored. Here, we comprehensively analysed the composition of exosomal sncRNAs secreted by RCC cells to identify those that influence MSCs.
Methods
Exosomal sncRNAs secreted by RCC cells and normal kidney cells were analyzed using RNAseq, followed by qPCR validation. MSCs were treated by conditioned media (CM) derived from RCC cells and transfected with piRNA, followed by the analysis of proliferation, viability, migration and immunocytochemical detection of piRNA. Expression of MSCs genes was evaluated using microarray and qPCR. TCGA data were analyzed to explore the expression of sncRNAs in RCC tumors.
Results
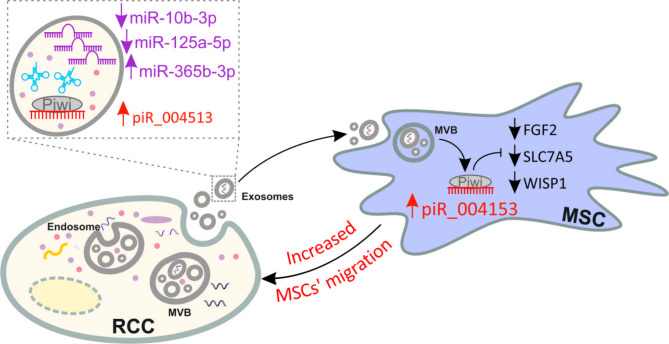
RNAseq revealed 40 miRNAs, 71 tRNAs and four piRNAs that were consistently secreted by RCC cells. qPCR validation using five independent RCC cell lines confirmed that expressions of miR-10b-3p and miR-125a-5p were suppressed, while miR-365b-3p was upregulated in exosomes from RCC cells when compared with normal kidney proximal tubules. The expression of miR-10b-3p and miR-125a-5p was decreased, whereas the expression of miR-365b-3p was increased in RCC tumors and correlated with poor survival of patients. Expressions of tRNA-Glu, tRNA-Gly, and tRNA-Val were the most increased, while tRNA-Gln, tRNA-Leu, and tRNA-Lys were top decreased in RCC exosomes when compared with normal kidney cells. Moreover, hsa_piR_004153, hsa_piR_016735, hsa_piR_019521, and hsa_piR_020365 were consistently upregulated in RCC exosomes. piR_004153 (DQ575660.1; aliases: hsa_piRNA_18299, piR-43772, piR-hsa-5938) was the most highly expressed in exosomes from RCC cells when compared with normal kidney cells. Treatment of MSCs with RCC CM resulted in upregulation of piR_004153 expression. Transfection of MSCs with piR_004153 stimulated their migration and viability, and altered expression of 35 genes, including downregulation of FGF2, SLC7A5, and WISP1. Immunocytochemistry confirmed the nuclear localization of piR_004153 transfected in MSCs.
Conclusion
RCC cells secrete multiple sncRNAs, including piR_004153 which targets MSCs, alters expression of FGF2, SLC7A5, and WISP1, and stimulates their motility and viability. To our knowledge, this is the first study showing that cancer-derived piRNA can enhance MSC migration.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-024-02001-1.
Keywords: Renal cell cancer, RCC, miRNA, tRNA, piR_004153, piRNA, Piwi-interacting RNA, Mesenchymal stromal cells, MSC
Background
Tumor cells release extracellular vesicles (EVs) that contain multiple types of molecules, including small non-coding RNAs (sncRNAs) such as miRNAs, piRNAs, or tRNAs [1]. Despite multiple studies analyzing the sncRNA composition of EVs released by cancer cells [2, 3], the functional significance of EVs components remains underexplored.
While the role of miRNAs in cancer development and progression has been investigated in many studies, including ours [4–6], the functions of piRNAs remain largely elusive. piRNAs (PIWI-interacting RNAs) are small (21–35 nt), non-coding RNAs that regulate gene expression using different mechanisms. Their primary function is the epigenetic silencing of genes in the nucleus. They can also act like miRNAs, by binding to target transcripts to trigger their degradation or inhibit their translation in the cytoplasm. In contrast to miRNAs, they do not interact with AGO proteins and are generated from single-strand precursors in a DICER-independent mechanism [7]. Remarkably, the regulatory potential of piRNAs is much bigger than miRNAs: the human genome encodes more than 8 x 106 piRNAs compared with ~ 2 x 103 miRNA precursors [7]. The expression of piRNAs is dysregulated in cancers, leading to aberrant expression of their target genes, including oncogenes and tumor suppressors. Thereby, piRNAs can regulate apparently all cancer-related processes, including proliferation, migration, invasion, apoptosis, and senescence [8]. Recent studies revealed that the role of transfer RNAs (tRNAs) goes far beyond the simple transfer of amino acids during translation. tRNAs actively influence the carcinogenic process by affecting the translation of oncogenes and tumor suppressors [9]. Furthermore, tRNAs are the source of multiple sncRNAs, including tRNA-derived fragments (tRFs) and tRNA-derived small RNAs (tsRNAs). Some of these tRNA derivatives may act similarly to miRNAs or piRNAs, thereby contributing to metastatic progression [10].
Renal cell cancer (RCC) is the most common type of kidney cancer, causing more than 175,000 deaths annually worldwide. The prognosis for RCC patients highly depends on the stage of the disease. Patients with localized RCC can be successfully treated by surgical removal of the tumor, resulting in 91–93% 5-year survival for stage I RCC. Unfortunately, approximately 30% of RCC patients are diagnosed with metastatic disease. In such cases, the prognosis drops dramatically, resulting in only 12–32% 5-year survival for stage IV RCC patients [6, 11]. Patients with metastatic RCC are treated with targeted therapies involving multikinase inhibitors (sorafenib, sunitinib, pazopanib, and axitinib), inhibitors of the mTOR1 complex (temsirolimus and everolimus), and immune checkpoint inhibitors. Unfortunately, even these therapies do not guarantee success. The recently introduced immunomodulating drugs prolong median overall survival by only 6 months when compared with everolimus [12].
Mesenchymal stem cells (mesenchymal stromal cells, MSCs) are multipotent cells that reside in different tissues. Their primary niche is the bone marrow, from which they can be recruited to the sites of injury and participate in healing processes. These MSCs’ activities result from their ability to differentiate into various cell lineages, including osteoblasts, chondrocytes, and adipocytes, as well as to secrete factors such as VEGF and HGF [13, 14]. The idea of MSC homing to tumor tissue was inspired by the concept that “tumors are wounds that do not heal” [15] and was further validated by multiple studies that demonstrated their ability to stimulate cancer progression [16, 17]. Specifically, MSCs can promote tumor progression by activating proliferation, migration, invasiveness, stemness, epithelial-mesenchymal transition (EMT), and chemoresistance [16]. They also suppress immune responses, facilitating tumor growth, metastatic spread and supporting drug-resistant tumor dormancy [18]. MSCs contribute to the progression of RCC, by stimulating cancerous proliferation, migration, and tumor growth [19, 20]. However, the exact mechanisms by which MSCs could be attracted to RCC tumors are largely unknown.
In our recent study, we found that conditioned media from RCC cells derived from advanced tumors stimulate the motility of MSCs. AREG (amphiregulin), FN1 (fibronectin), and DPP4 were identified as modulators of MSC migration [21]. Here, we hypothesized that MSCs’ migration could also be modulated by small non-coding RNAs released by RCC cells. To verify this hypothesis, we analyzed exosomal sncRNAs secreted by RCC cells and found that exosomes derived from advanced RCC tumors secrete piRNA hsa_piR_004153 that targets MSCs to induce transcriptional reprograming and stimulate their migration towards RCC cells. To the best of our knowledge, this is the first study showing that MSCs can be targeted by extracellular piRNAs.
Methods
Cell lines
RPTEC/TERT1 (CRL-4031; hTERT-immortalized epithelial cell line isolated from the proximal tubules), Caki-1 (HTB-46; ccRCC skin metastasis), 786-O (CRL-1932; ccRCC primary tumor), A498 (HTB-44; ccRCC primary tumor) cell lines were obtained from ATCC (American Type Culture Collection), while KIJ265T (ccRCC IV stage primary tumor) and KIJ308T (ccRCC II stage primary tumor) cell lines were a kind gift from Doctor John A. Copland and the Mayo Foundation for Medical Education and Research. The cell lines were cultured as reported [22].
MSCs isolation and characterization
MSCs were isolated from bone marrow (BM) obtained during standard orthopedic surgeries under the approval of the Local Bioethics Committee (Approval no. KB/115/2016) and with the written informed consent of patients. Isolation and characterization of BM-MSCs were performed as recently described [21]. Data confirming MSCs identity are shown in Supplementary Figure S1. MSCs were cultured as described previously [21].
Collection of RCC CM and isolation of RNA for RNAseq purposes
The isolation of exosomal RNA was performed using 10 ml of CM. The collected CM was centrifuged at 250xg for 15 min, and the supernatant was used for the isolation of exosomes using Cell Culture Media Exosome Purification and RNA Isolation Mini Kit (Norgen Biotek, Thorold, Canada; Cat. 60700) that utilize the all-in-one procedure for the purification of exosomes and the subsequent isolation of RNA. All procedures were done in accordance with the manufacturer’s instructions. Briefly, the CM was transferred into a new tube, following the addition of ExoC Buffer and Slurry E. After incubation (5 min, RT) and centrifugation (2000xg for 5 min), the supernatant was discarded. The exosomes were isolated from pellet ExoR Buffer and Mini Filter Spin Column. Next, the eluted exosomes were used for RNA isolation as indicated in the manufacturer’s instructions. The RNA elution was concentrated to 20 µL by using RNA Clean-Up and Concentration MicroElute Kit (Norgen Biotek, Cat. 61000).
RNAseq analysis
RNAseq was performed under conditions provided in Supplementary Methods file S1. Data were normalized using a trimmed mean of M-values (TMM) [23]. TMM normalized counts were used for differential expression (DE) analysis with the Benjamini-Hochberg procedure used to adjust false discovery rate (FDR) [24]. The criteria for significant DE were log fold change of ≥ 1 or ≤ -1 at p-value and FDR of ≤ 0.05. The analysis was done using EdgeR: 3.26.7. RNAseq data were deposited in NCBI/GEO database (acc. no. GSE256016).
Isolation of RNA from MSCs
RNA was isolated using the GeneMATRIX Universal RNA/miRNA Purification Kit (EURX, Gdansk, Poland) following the manufacturer’s protocol.
Reverse transcription and qPCR of miRNA and piRNA
To analyze miRNA expression, cDNA was synthesized using the miRCURY LNA Universal cDNA Synthesis kit (Qiagen, Inc.). Subsequently, qPCR was performed using specific LNA primers for each miRNA (miR-10b-3p: YP00205909; miR-17-5p: YP02119304; miR-21-5p: YP00204230; miR-193a-5p: YP00204665; miR-125a-5p: YP00204339; miR-365a-3p: YP00204622, miR-455-3p: YP00204035 and let-7i-5p: YP00204394). Primers for RT and Real-time reactions for measuring piRNA molecule expression were designed using software: http://www.srnaprimerdb.com/DesignResult. Sequences of primers are given in Supplementary Table S1. The reverse transcription reaction was performed using the RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) and a mixture of reverse transcription primers for each piRNA whose expression was studied. Real-time PCR was performed using Roche reagents. The expressions of the tested piRNAs: piR_004153, piR_016521, piR_019735, and piR_020365 were normalized to the expression of the reference piR_014620.
Reverse transcription and qPCR of tRNA
The exosomal RNA samples were pretreated by demethylation followed by RNA precipitation and then reverse transcription was performed by using the rtStar™ tRNA - optimized First-Strand cDNA Synthesis Kit (Arraystar, Cat#: AS-FS-004, Rockville, MD, USA). The manufacturer’s protocol of RNA demethylation was modified by reducing the volume of the reaction mixture by 30%. Then, RNA precipitation was performed by using UltraPure™ Phenol: Chloroform: Isoamyl Alcohol (25:24:1, v/v) (Invitrogen, Cat. No.:15593031, Carlsbad, CA, USA), Chloroform (Avantor, Cat. No.: PA-06-234431116) and Isopropanol (Chempur, cat. No.: 363-117515002-1 L). The precipitation protocol was modified by doubling the volume of the precipitation mixtures, extending the incubation time from 10 to 20 min, and extending the centrifugation time from 10 to 15 min. cDNA synthesis was performed according to the manufacturer’s protocol. Then, cDNA was 2-fold diluted in nuclease-free water and qPCR reactions were performed using rtStar™ Pre-designed Human tRNA qPCR Primer Pair (cat#: AS-NR-001-1-M Rockville, MD, USA) and the Arraystar SYBR® Green Real-Time qPCR Master Mix (Arraystar, Cat#: AS-MR-005-5, Rockville, MD, USA) according to manufacturer’s protocol. The following qPCR conditions were used: initial denaturation at 95 °C for 10 min and 40 cycles of 95 °C for 10 s, 60 °C for 1 min, followed by melting curve analysis (95 °C for 5 min, 65 °C for 1 min; continuous reading of fluorescence from 65 to 97 °C with 0.11 °C/sec ramp rate and five acquisitions per °C). The primers used in the study are provided in Supplementary Table S1. The expression of tRNAs was normalized to the expression of stably expressed molecule His-GTG-1 tRNA and calculated using the 2 − ΔCq method [25, 26].
Treatment of MSCs with CM
MSC cells were treated with CM from RPTEC/TERT1, Caki-1 or KIJ265T cells as described previously [21].
Transfection of MSCs with piRNA
Based on the sequence of the piR_004153 molecule from the piRNAdb database [27], https://www.pirnadb.org/information/pirna/hsa-piR-5748, a synthetic piRNA molecule was synthesized (Genomed; 5’-UCCCUGGUGGUCUAGUGGUUAGGAUUCGGCAC-3’BIOTIN) and transfected into MSC cells using Lipofectamine 2000 (Thermo Fisher) with 45 pmol of piRNA or negative control (unspecific synthetic molecule; 5’-UGCUUUGCACGGUAACGCCUGUUUU-3’BIOTIN) per well in a 12-well plate. RNA was isolated 48 h after transfection, while protein isolation and functional cell assays were done 72 h after transfection.
Transfection and immunocytochemical detection of biotinylated piRNA
Biotinylated piRNA was purchased from Genomed and used for transfection of ccRCC cells with Lipofectamine 2000. Cells were grown on glass coverslips, rinsed with PBS, and fixed with 4% paraformaldehyde for 8 min at room temperature. They were then permeabilized with 0.25% TritonX-100 for 3 min, followed by blocking non-specific binding sites with 2% BSA (bovine serum albumin) in TBST (Tris-buffered saline with Tween) for 1 h. Afterwards, the coverslips were incubated with a specific secondary antibody labeled with Cy-3 dye (Monoclonal Anti-Biotin-Cy3; Sigma #C5585), diluted in 2% BSA in TBST (1:100), for 1 h in the dark. To stain the nuclei, cells were treated with DAPI (4′,6-diamidino-2-phenylindole) for 5 min. Finally, the cells were examined using a laser scanning confocal microscope (LSM 800, AxioObserver Z.1; Zeiss, Oberkochen, Germany) with ZEN 3.7 software (Zeiss).
Migration
Migration was measured using a classical wound-healing assay as well as a transwell assay. The latter was performed using CytoSelect™ 96-Well Cell Migration Assay (8 μm, Fluorometric Format) (Cell Biolabs, Inc., San Diego, CA, USA; # CBA-106) following the instructions provided by the manufacturer. For the wound-healing assay, MSCs were placed at a density of 5 × 10^4 cells per well on a 12-well plate. After 24 h, the cells underwent transfection using piRNA molecules. After 48 h post-transfection a wound was made by scraping the surface with a sterile 200 µl pipette tip, and a first image of the wound was taken using a Zeiss Axio Observer.D1 microscope. After an additional 24-hour incubation period, a second image was captured to evaluate the healing of the wound and the migration of MSCs.
Viability
Cell viability was measured by staining cells with trypan blue, according to a previously described protocol [28].
Proliferation
The proliferation was measured using the Cell Proliferation ELISA, BrdU (colorimetric) kit (Roche Diagnostics, GmbH), following the instructions provided by the manufacturer.
Microarray analysis
Microarray analysis of MSC transfected with piR_004153 was performed as previously described [4]. Microarray data were deposited at NCBI GEO (GSE263824).
Bioinformatic analysis
Data on miRNA expression in RCC tumors and correlation with RCC patients’ survival were retrieved from ENCORI/starBase [29]. miRNA target genes were predicted using miRnet 2.0 [30]. GO (Gene ontology) analysis was performed using ShinyGO 0.80 [31]. tRF molecules (that regulate gene expression, in a mechanism similar to miRNAs) that could be cut out from analyzed tRNAs were predicted using the OncotRF bioinformatics program (http://bioinformatics.zju.edu.cn/OncotRF/; [32]. Only those tRF that are highly expressed in KIRC and have the potential impact on the survival of ccRCC patients were selected for further analysis. These analyses were also performed using OncotRF.
Statistical analysis
The analyzed data were retrieved from at least three independent biological experiments. Statistical analysis was performed using a t-test or ANOVA followed by Dunnett’s multiple comparison test, p < 0.05 was considered statistically significant.
Results
miRNA secreted by RCC cells are enriched in the regulators of key oncogenic pathways
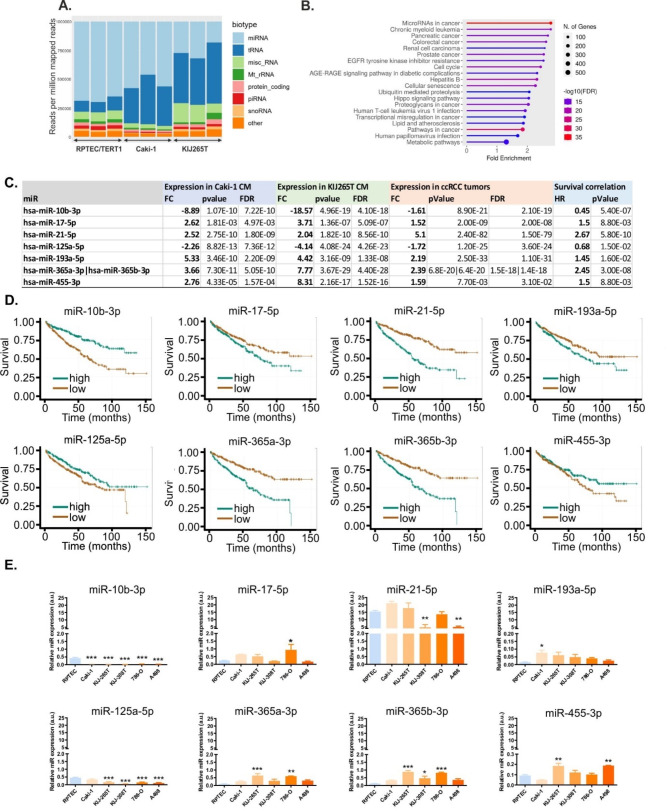
RNAseq of RCC CM revealed multiple sncRNA biotypes, with miRNAs and tRNAs representing the largest sncRNA fractions (Fig. 1A).
Fig. 1.
miRNAs and tRNAs are the largest fractions of sncRNAs secreted by RCC cells. (A) Representation of sncRNA biotypes. (B) Top enriched KEGG pathways among the genes predicted as miRNA targets. (C) microRNAs selected for validation. The table shows the results of RNAseq (miRNA expression in Caki-1 and KIJ265T CM), miRNA expression in RCC tumors (n = 517 RCC tumors, n = 71 non-tumorous kidney samples), and correlation of survival of RCC patients (n = 517). TCGA/KIRC data were retrieved via ENCORI. (D) Kaplan-Meyer plots showing correlation of miRNA expression with survival of RCC patients. (E) qPCR validation of RNAseq data. The plots show expression of microRNAs in CM isolated from RCC cell lines (Caki-1, KIJ265T, KIJ308T, 786-O, and A481) and non-tumorous control cell line RPTEC/TERT1. Note different scales for miR-193-5p and miR-455. Statistical analysis: One-way ANOVA with Dunnett’s multiple comparisons posttest. * p < 0.05, ** p < 0.01, *** p < 0.001. n = 3 independent biological experiments
When compared with CM from control cells, CM isolated from Caki-1 and KIJ265T showed 83 and 107 altered miRNAs, respectively (Supplementary Table S2). There were 40 consistently altered miRNAs in CM from Caki-1 and KIJ265T cells, including 17 upregulated and 23 downregulated miRNAs when compared with RPTEC/TERT1 cell line (Supplementary Table S2). miRnet predicted > 12,000 experimentally verified gene targets of the identified exosomal miRNAs (Supplementary Table S3). GO analysis of the predicted gene targets showed that top enriched KEGG pathways including ‘microRNAs in cancer’ and various cancers (e.g. chronic myeloid leukemia, pancreatic cancer, colorectal cancer, renal cell carcinoma or prostate cancer), as well as ‘EGFR tyrosine kinase inhibitor resistance’ and ‘metabolic pathways’ (Fig. 1B).
The expression of genes encoding aberrantly secreted miRNAs is disturbed in RCC tumors
To find the potential clinical relevance of 40 miRNAs that were commonly altered in CM from both RCC cell lines, we analyzed their expression in RCC tumors (Supplementary Table S2). The expression of 31 of these miRNAs was disturbed in RCC tumors when compared with control tissues, while the altered expression of 17 miRNAs correlated with poor survival of patients. Based on the concordance between the direction of expression changes in RCC tumors and CM from RCC cell lines, as well as the association with poor survival of patients, 8 miRNAs were selected for further validation (Supplementary Table S4, Fig. 1C and D). qPCR validation in the CM isolated from five RCC cell lines (Caki-1, KIJ265T, KIJ308T, 786-O, and A498) and the non-cancerous cell line RPTEC/TERT1 confirmed uniformly suppressed expression of miR-10b-3p in CM from all analyzed RCC cell lines, while miR-125a-5p was suppressed in CM from four analyzed cell lines (Fig. 1E). The other miRNAs showed variable patterns of expression. miR-17-5p was statistically significantly increased in CM from the 786-O cell line. Following RNAseq data, miR-193a-5p tended to be upregulated in all CM although statistical significance was reached only for Caki-1. miR-365a-3p was statistically significantly upregulated in CM from KIJ265T and 786-O, while miR-365b-3p was increased in CM from KIJ265T, KIJ308T, and 786-O. miR-455-3p was statistically significantly upregulated in CM from KIJ265T and A498. miR-21-5p was the most abundant miR in all analyzed CM; however, its expression was suppressed in CM from KIJ308T and A498 cells (Fig. 1E). The predicted targets of three miRNAs (hsa-miR-10b-3p, hsa-miR-125a-5p, hsa-miR-365b-3p) with uniformly altered expression in CM from at least three cell lines included those enriched in KEGG cancer-related pathways as well as in the regulation of protein transport, programmed cell death and metabolism (Supplementary Table S5, Supplementary Table S6, Supplementary Table S7, Supplementary Figure S2).
tRNAs are the components of RCC non-coding secretome
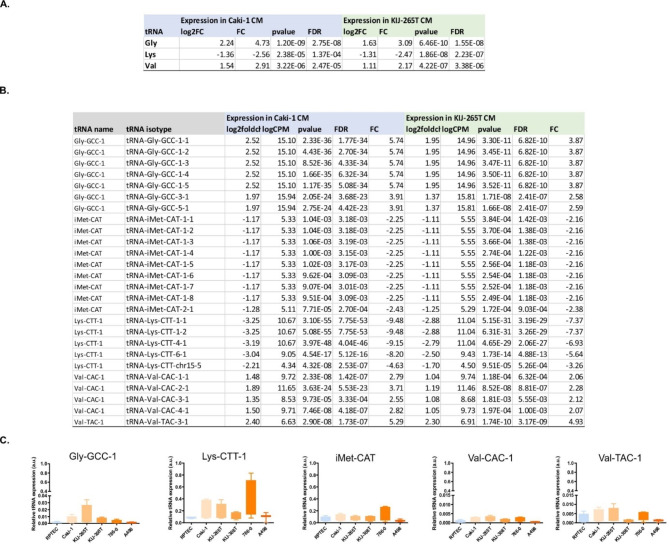
RNAseq revealed that tRNAs coding for Gly and Val were consistently up-regulated in CM from Caki-1 and KIJ256T, while tRNAs coding for Lys were concomitantly down-regulated in CM from both cell lines (│FC│≥2, FDR < 0.05) (Fig. 2A and B). This is consistent with previous data showing that tRNAVal is one of the three top altered tRNA types across multiple cancer types [33]. Regarding the specific tRNA isotypes, there were 109 altered tRNAs in CM from Caki-1 cells and 123 tRNAs differentially secreted by KIJ265T cells. Of note, 71 tRNAs were consistently altered in CM from both cell lines, including 47 tRNAs that were up-regulated and 24 tRNAs that were down-regulated (│FC│≥2, FDR < 0.05) (Supplementary Table S8). The most upregulated anticodons included TCC, GCC, and TAC, while top downregulated anticodons included CTG, TAA, and CTT.
Fig. 2.
tRNAs secreted by RCC cells. A. tRNAs altered in CM from Caki-1 and KIJ265T cell lines when compared with control RPTEC/TERT1 cells, classified by the transferred amino acid. The table shows results of RNAseq. B. Specification of tRNA isotypes with consistently altered expression in CM Caki-1 and CM KIJ265T (RNAseq data). C. qPCR validation of RNAseq results. Statistical analysis: One-way ANOVA with Dunnett’s multiple comparisons posttest. Data from 3-to-4 independent biological experiments
Using OncotRF we identified tRNAs that could serve as a source for tRFs (Supplementary Table S9). The expression of many of those tRFs was altered in RCC tumors and correlated with poor prognosis for patients (Supplementary Table S9). Based on the results of OncotRF analysis and the criteria described in Methods, five tRNAs were selected for qPCR validation (Fig. 2C). This analysis revealed a trend for upregulation of Gly-GCC, Val-CAC, and Val-TAC, no changes for ciMet-CAT, as well as suppression of Lys-CTT; however, the data were not statistically significant (Fig. 2C).
piRNAs are aberrantly secreted by RCC cells
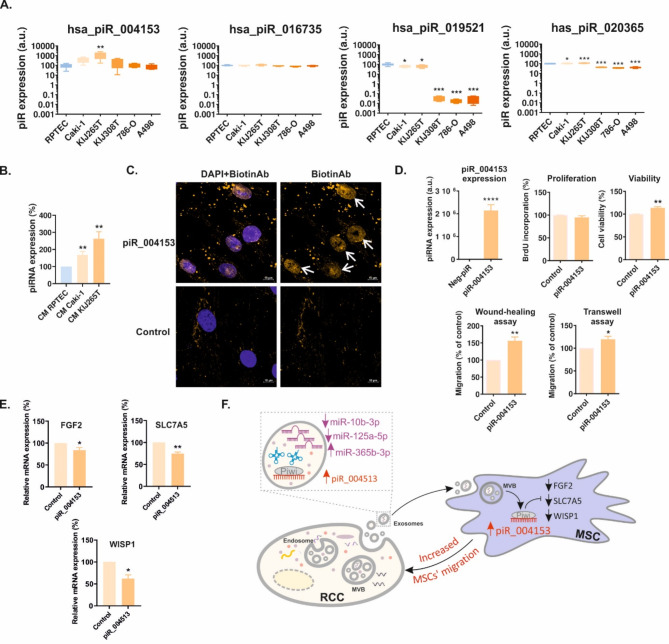
RNAseq revealed that there were 13 piRNAs of which levels were statistically significantly altered in CM from Caki-1 cells and 19 piRNAs that were statistically significantly altered in CM from KIJ265T cells when compared with control cell line RPTEC/TERT1 (Supplementary Table S10). CM from both analyzed RCC cell lines shared four piRNAs (has_piR_004153, has_piR_016735, hsa_piR_019521, and has_piR_020365) of which levels were consistently upregulated when compared with CM derived from RPTEC/TERT1 cells (Supplementary Table S10). qPCR validation showed that among all analyzed piRNAs, hsa_piR_004153 was the most highly expressed in CM from RCC cells (Fig. 3A). piR_004153 was statistically significantly upregulated in CM from KIJ265T cells and tended to be increased in CM from Caki-1 when compared with control RPTEC/TERT1 cell line. hsa_piR_019521 expression was decreased in CM from all analyzed RCC cell lines, while hsa_piR_020365 was increased in CM from Caki-1 and KIJ265T cells and suppressed in CM from the residual RCC cell lines. Hsa_piR_016735 level was not changed in any of the analyzed CM (Fig. 3A).
Fig. 3.
piR_004153 secreted by RCC cells stimulates migration of MSCs. (A) qPCR validation of expression of four piRNAs selected based on RNAseq. The plots show the results of qPCR analysis in CM derived from RCC cell lines. (B) piR_004153 expression is increased in MSCs treated with CM from RCC cells. The plot shows the results of qPCR analysis. (C) Immunocytochemical visualization of MSCs transfected with biotin-labelled piR_004153 or control scrambled oligonucleotide. The cells were stained with antibodies against biotin (yellow), nucleus was visualized using DAPI. White arrows show the nuclear localization of biotinylated piR_004153. Yellow dots are presumably artefactual precipitates of anti-biotin antibody. (D) The effects of piR_004153 on the functioning of MSCs. The plots show piR_004153 expression following its transfection in MSCs, the effect of piR_004153 on MSCs proliferation (BrdU assay), cell viability (Trypan-blue assay), and migration (wound-healing and transwell migration assay). (E) piR_004153 alters expression of MSCs genes. The plots show results of qPCR analysis of MSCs transfected with piR_004513. (F) The scheme showing non-coding RCC secretome and the impact of piR_004513 on MSCs gene expression and migration. All data were retrieved from at least 3 independent biological experiments. Statistical analysis was performed using ANOVA with Dunnett’s Multiple Comparison Test (A, B) or t-test (D, E). * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001
piR_004153 secreted by RCC cells is taken-up by MSCs to stimulate their migration and viability
In our previous study, we showed that CM derived from the most advanced RCC tumors (Caki-1, KIJ265T) stimulated the migration of MSCs [21]. Given that hsa_piR_004153 was the most highly expressed piRNA in CM from RCC cells (Fig. 3A), we reasoned that it could exert its effect on MSCs. Treatment of MSCs with RCC CM resulted in an increased expression of piR_004153 (Fig. 3B), while transfection of MSCs with biotinylated piR_004153 led to its nuclear localization as revealed by immunocytochemistry (Fig. 3C). Transfection of MSCs with piR_004153 stimulated migration and moderately increased viability of MSCs, without affecting proliferation (Fig. 3D). These results indicated that piR_004153 secreted by RCC cells can be taken up by MSCs to enhance their motility and viability.
To further analyze the influence of piR_004153 on MSC, we performed microarray analysis of MSCs transfected with piR_001453. This analysis revealed altered expression of 35 genes of which 6 were selected for validation (Supplementary Table S11). qPCR confirmed that piR_004153 decreased expression of FGF2, SLC7A5, and WISP1 in MSCs (Fig. 3E). These data show that RCC cells from advanced tumors secrete piR_004153 that targets MSC, reprograms their transcriptome, and stimulates motility.
Discussion
In this study we show that renal cancer cells secrete multiple exosomal sncRNAs, including miRNAs, piRNAs and tRNAs. Among them, we identified piRNA piR_004153, which is highly secreted by cells derived from advanced RCC tumors and taken-up by MSCs to stimulate their migration towards RCC cells and reprogram their transcriptome which might increase their tumorigenic potential. To our knowledge, this is the first study showing that cancer-derived piRNA can affect MSCs migration.
Despite multiple studies on extracellular sncRNAs in RCC, their function is still largely unknown. sncRNAs can be detected in serum/plasma of RCC patients and are considered potential biomarkers [34]. Most studies on the role of exosomal sncRNAs in RCC focus on miRNAs [35–37]. We found several miRNAs with consistently altered exosomal secretion in RCC cells. Their role in RCC is largely unknown. miR-125a-5p was previously described as an exosomal miRNA in RCC and regulated by branched-chain keto-acid dehydrogenase kinase (BCKDK) [38]. To our knowledge, there are no published data on exosomal miR-10b-3p and miR-365b-3p in RCC, thus their role in the pathology of renal cancer requires further elucidation.
The clinical significance of extracellular tRNAs and tRNA fragments (tRFs) comes from their diagnostic and regulatory potential. tRNAs and tRFs are released by multiple cell types and are detectable in human biofluids [39]. Surprisingly, the data on extracellular tRNAs and tRFs in RCC are scarce. 5’tRNA4-Val-AAC expression was decreased in RCC tumors and inversely correlated with tumor stage and grade. However, no changes in 5’tRNA4-Val-AAC level were found in patients’ sera [40]. Expressions of 5’-tRNA-Arg-CCT, 5’-tRNA-Glu-CTC, 5’-tRNA-Leu-CAG, and 5’-tRNA-Lys-TTT were decreased in RCC tumors, while 5’-tRNA-Lys-TTT inversely correlated with ISUP grades. 5’-tRNA-Arg-CCT, 5’-tRNA-Glu-CTC and 5’-tRNA-Lys-TTT levels were also decreased in sera of RCC patients [41]. Our RNAseq analysis showed consistently altered expression of 71 extracellular tRNAs in CM from both analyzed RCC cell lines. However, qPCR analysis did not validate these data. Thus, the significance of extracellular tRNAs in RCC requires further analysis.
The data on piRNA in RCC are scarce and limited to fewer than 10 studies [42–46], none of which addressed the exact role of exosomal piRNAs in RCC. Regarding piR_004153, it was reported that its expression is reduced in tumor tissue and serum of colorectal cancer patients [47], upregulated in plasma EVs of smokers [48], and plasma of patients with Parkinson’s disease [49]. These data may possibly suggest that increased extracellular piR_004153 concentrations may be commonly associated with these pathologies. Since inflammation is a common feature of smoking status [50], Parkinson’s disease [51], and renal cancer [52], this provides an interesting hypothesis that piR_004153 may be involved in inflammatory processes. MSCs migrate to the sites of inflammation, including tumors which are often referred to as “wounds that do not heal”. The possible role of piR_004153 in inflammation requires further exploration in clinical settings.
In our study, treatment of MSCs with CM collected from RCC cells increased intracellular piR_004153 levels, while transfection of MSCs with piR_004153 induced changes in MSCs transcriptome. Specifically, piR_004153 strongly decreased the expression of WISP1 and SLC7A5, and moderately attenuated expression of FGF2. WISP1 (WNT1 Inducible Signaling Pathway Protein 1) is a well-known regulator of MSCs proliferation and differentiation [53, 54]. Interestingly, WISP1 silencing in BM-MSCs induces proapoptotic signaling [55], which suggests that piR_004153-induced viability of MSCs is probably not mediated by WISP1. To our knowledge, there are no studies addressing the role of endogenously expressed WISP1 in MSCs migration, and this should be further experimentally explored. Regarding other cell types, WISP1 was shown to either promote or stimulate migration [56]. In contrast, much more is known regarding the significance of WISP1 expressed by MSCs in the context of cancer. MSCs-derived fibroblasts (MSC-DF) stimulate the progression of melanoma in a WISP1-dependent manner [57]. Specifically, low WISP1 expression in MSC-DF is required to promote metastatic progression of melanoma [57]. These data coincide with the fact that BM-MSCs are the key source of cancer-associated fibroblasts, the crucial tumor-promoting components of TME [57]. Further, WISP1 expression is decreased in BM-MSCs derived from patients with myelodysplastic syndrome, a heterogeneous group of hematopoietic malignancies with increased risk of developing acute myeloid leukemia [58]. Altogether, these data suggest that decreased WISP1 expression is associated with pro-cancerous MSCs activities. We found that piR_004153 reduced the expression of SLC7A5 (solute carrier family 7 member 5), a regulator of MSCs osteogenic differentiation [59]. This is in line with studies showing that cancer cells reprogram BM-MSC differentiation to facilitate malignant progression. For instance, cells of acute myeloid leukemia (AML) inhibit osteogenic differentiation and promote adipogenic commitment of BM-MSCs [60]. T-cell acute lymphoblastic leukemia EVs inhibit osteogenic differentiation of MSCs [61]. It was also suggested that osteogenic-resistant MSCs contribute to the development of head and neck cancer [62]. SLC7A5 in cancer plays mainly oncogenic role and stimulates proliferation, migration and invasiveness [63]. However, its role in non-cancerous cells is more complex. For instance, selective disruption of SLC7A5 gene in intestine epithelium leads to dedifferentiation of Paneth cells, induction of stemness, increased proliferation and migration of crypt base stem cells [64]. The role of SLC7A5 in the regulation of MSCs migration and viability requires further experimental studies. piR_004153 also suppressed the expression of FGF2 (fibroblast growth factor 2) (Fig. 3E); however, its role in MSCs is less well understood. BM-MSCs transduced with an FGF2-expressing vector differentiate into tenocytes in a MAPK (mitogen-activated protein kinase)-dependent manner [65] and support vascular regeneration [66]. Interestingly, treatment of MSCs with extracellular FGF2 stimulates their migration [67]; however the role of endogenously expressed FGF2 in MSCs motility and viability have not been explored. Treatment of MSCs with piR_004153 resulted in minimal changes in expression of FGF2 which suggests that this gene rather does not mediate piR_004513 effects in MSCs. The role of MSCs-expressed FGF2 in the cancer context requires elucidation.
The data on the role of piRNA in MSCs is limited. It is known that BM-MSCs secrete exosomes that contain piRNAs [68]. Moreover, the profiles of non-coding RNAs, including piRNAs undergo rapid changes during BM-MSC differentiation towards osteo- and chondrogenic lineages, as well as loss of BM-MSCs adherence [69, 70]. BM-MSCs secrete exosomes that contain piRNAs [68]. Regarding the specific piRNA effects on MSCs functioning, Liu et al. showed that piRNA-36741 promotes the osteogenic differentiation of BM-MSC and attenuates ovariectomy-induced osteoporosis in mice by inducing BMP2 (Bone morphogenetic protein 2) expression [71]. None of those studies addressed MSCs targeting by external exosomes released by cancer cells.
Conclusion
We show that RCC cells secrete multiple exosomal sncRNAs, including miRNAs, piRNAs, and tRNAs, that have the potential to influence TME. Indeed, we demonstrate that MSC motility and viability are stimulated by piR_004153 secreted by RCC cells (Fig. 3F). Future studies are needed to explore the mechanisms behind altered secretion of piRNAs by RCC, as well as exact mechanisms of piRNA uptake by MSCs. Considering previous studies showing that MSCs can stimulate RCC progression [19, 20], interference with piRNA-mediated homing of MSC to RCC tumors may possibly open new options for renal cancer treatment.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: Supplementary Methods file S1. RNAseq conditions. Supplementary Table S1. Primers used for qPCR analysis of piRNA and tRNA
Supplementary Material 2: Supplementary Table S2. miRNAs aberrantly expressed in exosomes isolated from conditioned media of Caki-1 and KIJ265T cells. The table shows results of RNAseq. Volcano plots are provided alongside the data
Supplementary Material 3: Supplementary Table S3. Genes predicted as targets of 40 miRNAs of which expression was consistently altered in CM from Caki-1 and KIJ265T cells. Prediction was performed using miRnet (https://www.mirnet.ca/miRNet/home.xhtml)
Supplementary Material 4: Supplementary Table S4. microRNAs with concordant changes of expression in exosomes and tumor tissues of RCC patients
Supplementary Material 5: Supplementary Table S5. Genes predicted as targets of 3 validated miRNAs (hsa-miR-10b-3p, hsa-miR-125a-5p, hsa-miR-365b-3p), consistently altered in CM from at least three cell RCC cell lines. Prediction was performed using miRnet (https://www.mirnet.ca/miRNet/home.xhtml)
Supplementary Material 6: Supplementary Table S6. Enriched KEGG pathways among the genes predicted as targets of 3 validated miRNAs (hsa-miR-10b-3p, hsa-miR-125a-5p, hsa-miR-365b-3p), consistently altered in CM from at least three cell RCC cell lines. The table shows results of analysis performed using ShinyGO 0.80 (http://bioinformatics.sdstate.edu/go/)
Supplementary Material 7: Supplementary Table S7. Enriched biological processes among the genes predicted as targets of 3 validated miRNAs (hsa-miR-10b-3p, hsa-miR-125a-5p, hsa-miR-365b-3p), consistently altered in CM from at least three cell RCC cell lines. The table shows results of analysis performed using ShinyGO 0.80 (http://bioinformatics.sdstate.edu/go/)
Supplementary Material 8: Supplementary Table S8. tRNAs aberrantly expressed in exosomes isolated from conditioned media of Caki-1 and KIJ265T cells. The table shows results of RNAseq. Volcano plots are provided alongside the data
Supplementary Material 9: Supplementary Table S9. The expression of tRFs is disturbed in RCC tumors. tRFs were predicted as derived from tRNA of which altered expression was found in exosomes isolated from RCC cell lines when compared with normal kidney cells. The analysis was performed using OncotRF, using data from TCGA N = 544 for RCC tumors, N = 72 for normal kidney tissue samples
Supplementary Material 10: Supplementary Table S10. piRNAs aberrantly expressed in exosomes isolated from conditioned media of Caki-1 and KIJ265T cells. The table shows results of RNAseq. Volcano plots are provided alongside the data
Supplementary Material 11: Supplementary Table S11. piR_004153 alters expression of MSCs genes. The table shows results of microarray analysis of RNA isolated from MSCs transfected with piR_004513 mimic or non-targeting scrambled oligonucleotide, cultured in five independent cell culture wells per each group. Volcano plot is provided alongside the data
Acknowledgements
The authors acknowledge a kind gift of KIJ265T and KIJ308T cell lines by Doctor John A. Copland and Mayo Foundation of Medical Education and Research.
Abbreviations
- AREG
Amphiregulin
- ATCC
American Type Culture Collection
- BM
Bone marrow
- BMP2
Bone morphogenetic protein 2
- BSA
Bovine serum albumin
- CM
Conditioned media
- FGF2
Fibroblast growth factor 2
- FN1
Fibronectin
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MAPK
Mitogen-activated protein kinase
- MSCs
Mesenchymal stem/stromal cells
- RCC
Renal cell cancer
- SLC7A5
Solute carrier family 7 member 5
- sncRNAs
Small non-coding RNAs
- TBST
Tris-buffered saline with Tween
- tRFs
tRNA-derived fragments
- WISP1
WNT1 Inducible Signaling Pathway Protein 1
Author contributions
Conceptualization: AP-W. Methodology: JB, MG, PP, KH, BS-R, UJ, HK, WZ-W, AB (Anna Burdzinska), MK. Validation: JB, MG, PP, KH, AB (Alex Białas) BR, AA-O, JŻ-K. Formal analysis: JB, PP, KH, BS-R, UJ, HK, AB (Anna Burdzinska), MK, LP, AP-W. Investigation: JB, MG, PP, KH, BS-R, UJ, AB (Alex Białas), HK, RI-N, BR, AA-O, JŻ-K, WZ-W, AB (Anna Burdzinska), MK, AP-W. Resources: UJ, MK, AB, LP, AP-W. Data Curation: JB, UJ, HK, MK, AP-W. Writing - Original Draft: AP-W, JB, PP. Writing - Review & Editing: JB, MG, PP, KH, BS-R, UJ, AB (Alex Białas), HK, RI-N, BR, AA-O, JŻ-K, WZ-W, AB (Anna Burdzinska), MK, LP, AP-W. Visualization: JB, AP-W. Supervision: UJ, MK, LP, AP-W. Project administration: JB, AP-W. Funding acquisition: AP-W.
Funding
The study was financially supported by National Science Centre, Poland (grant no. 2018/29/B/NZ5/01211) to AP-W.
Data availability
RNAseq and microarray data are available in NCBI/GEO database (acc. no. GSE256016 and GSE263824, respectively). The residual data generated or analyzed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
The study was performed in accordance with the Declaration of Helsinki and under the approval of the Local Bioethics Committee at the Medical University of Warsaw (Approval no. KB/115/2016) and written informed consent of patients.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Joanna Bogusławska, Email: joanna.boguslawska@cmkp.edu.pl.
Agnieszka Piekiełko-Witkowska, Email: apiekielko@cmkp.edu.pl.
References
- 1.O’Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol Cell Biol. 2020;21(10):585–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sadovska L, Zayakin P, Eglitis K, Endzelins E, Radovica-Spalvina I, Avotina E, et al. Comprehensive characterization of RNA cargo of extracellular vesicles in breast cancer patients undergoing neoadjuvant chemotherapy. Front Oncol. 2022;12:1005812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu QW, He Y, Xu WW. Molecular functions and therapeutic applications of exosomal noncoding RNAs in cancer. Exp Mol Med. 2022;54(3):216–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boguslawska J, Poplawski P, Alseekh S, Koblowska M, Iwanicka-Nowicka R, Rybicka B et al. MicroRNA-Mediated metabolic reprograming in Renal Cancer. Cancers (Basel). 2019;11(12). [DOI] [PMC free article] [PubMed]
- 5.Boguslawska J, Rodzik K, Poplawski P, Kedzierska H, Rybicka B, Sokol E, et al. TGF-beta1 targets a microRNA network that regulates cellular adhesion and migration in renal cancer. Cancer Lett. 2018;412:155–69. [DOI] [PubMed] [Google Scholar]
- 6.Boguslawska J, Kryst P, Poletajew S, Piekielko-Witkowska A. TGF-beta and microRNA interplay in Genitourinary Cancers. Cells. 2019;8(12). [DOI] [PMC free article] [PubMed]
- 7.Mokarram P, Niknam M, Sadeghdoust M, Aligolighasemabadi F, Siri M, Dastghaib S, et al. PIWI interacting RNAs perspectives: a new avenues in future cancer investigations. Bioengineered. 2021;12(2):10401–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su JF, Concilla A, Zhang DZ, Zhao F, Shen FF, Zhang H, et al. PIWI-interacting RNAs: Mitochondria-based biogenesis and functions in cancer. Genes Dis. 2021;8(5):603–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinzaru AM, Tavazoie SF. Transfer RNAs as dynamic and critical regulators of cancer progression. Nat Rev Cancer. 2023;23(11):746–61. [DOI] [PubMed] [Google Scholar]
- 10.Huang SQ, Sun B, Xiong ZP, Shu Y, Zhou HH, Zhang W, et al. The dysregulation of tRNAs and tRNA derivatives in cancer. J Exp Clin Cancer Res. 2018;37(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Padala SA, Barsouk A, Thandra KC, Saginala K, Mohammed A, Vakiti A, et al. Epidemiology of renal cell carcinoma. World J Oncol. 2020;11(3):79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powles T, Staehler M, Ljungberg B, Bensalah K, Canfield SE, Dabestani S, et al. Updated EAU guidelines for Clear Cell Renal Cancer patients who fail VEGF targeted therapy. Eur Urol. 2016;69(1):4–6. [DOI] [PubMed] [Google Scholar]
- 13.Li JH, Fan WS, Wang MM, Wang YH, Ren ZG. Effects of mesenchymal stem cells on solid tumor metastasis in experimental cancer models: a systematic review and meta-analysis. J Transl Med. 2018;16(1):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Chen QH, Liu AR, Xu XP, Han JB, Qiu HB. Synergism of MSC-secreted HGF and VEGF in stabilising endothelial barrier function upon lipopolysaccharide stimulation via the Rac1 pathway. Stem Cell Res Ther. 2015;6:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315(26):1650–9. [DOI] [PubMed] [Google Scholar]
- 16.Lazennec G, Lam PY. Recent discoveries concerning the tumor - mesenchymal stem cell interactions. Biochim Biophys Acta. 2016;1866(2):290–9. [DOI] [PubMed] [Google Scholar]
- 17.Galie M, Konstantinidou G, Peroni D, Scambi I, Marchini C, Lisi V, et al. Mesenchymal stem cells share molecular signature with mesenchymal tumor cells and favor early tumor growth in syngeneic mice. Oncogene. 2008;27(18):2542–51. [DOI] [PubMed] [Google Scholar]
- 18.Bartosh TJ, Ullah M, Zeitouni S, Beaver J, Prockop DJ. Cancer cells enter dormancy after cannibalizing mesenchymal stem/stromal cells (MSCs). Proc Natl Acad Sci U S A. 2016;113(42):E6447–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du T, Ju G, Wu S, Cheng Z, Cheng J, Zou X, et al. Microvesicles derived from human Wharton’s jelly mesenchymal stem cells promote human renal cancer cell growth and aggressiveness through induction of hepatocyte growth factor. PLoS ONE. 2014;9(5):e96836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsiao WC, Sung SY, Liao CH, Wu HC, Hsieh CL. Vitamin D3-inducible mesenchymal stem cell-based delivery of conditionally replicating adenoviruses effectively targets renal cell carcinoma and inhibits tumor growth. Mol Pharm. 2012;9(5):1396–408. [DOI] [PubMed] [Google Scholar]
- 21.Poplawski P, Zarychta-Wisniewska W, Burdzinska A, Boguslawska J, Adamiok-Ostrowska A, Hanusek K, et al. Renal cancer secretome induces migration of mesenchymal stromal cells. Stem Cell Res Ther. 2023;14(1):200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poplawski P, Alseekh S, Jankowska U, Skupien-Rabian B, Iwanicka-Nowicka R, Kossowska H, et al. Coordinated reprogramming of renal cancer transcriptome, metabolome and secretome associates with immune tumor infiltration. Cancer Cell Int. 2023;23(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the false Discovery Rate - a practical and powerful Approach to multiple testing. J R Stat Soc B. 1995;57(1):289–300. [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
- 26.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–8. [DOI] [PubMed] [Google Scholar]
- 27.Piuco R, Galante PAF, piRNAdb. A piwi-interacting RNA database. bioRxiv. 2021:2021.09.21.461238.
- 28.Strober W. Trypan Blue Exclusion Test of cell viability. Curr Protoc Immunol. 2015;111:A3. B 1-A3 B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang L, Zhou G, Soufan O, Xia J. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020;48(W1):W244–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36(8):2628–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao D, Sun X, Zhou L, Amanullah M, Pan X, Liu Y, et al. OncotRF: an online resource for exploration of tRNA-derived fragments in human cancers. RNA Biol. 2020;17(8):1081–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Ye Y, Gong J, Ruan H, Liu CJ, Xiang Y, et al. Global analysis of tRNA and translation factor expression reveals a dynamic landscape of translational regulation in human cancers. Commun Biol. 2018;1:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barth DA, Drula R, Ott L, Fabris L, Slaby O, Calin GA, et al. Circulating non-coding RNAs in renal cell carcinoma-pathogenesis and potential implications as clinical biomarkers. Front Cell Dev Biol. 2020;8:828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sequeira JP, Constancio V, Lobo J, Henrique R, Jeronimo C. Unveiling the World of circulating and exosomal microRNAs in renal cell carcinoma. Cancers (Basel). 2021;13:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butz H, Nofech-Mozes R, Ding Q, Khella HWZ, Szabo PM, Jewett M, et al. Exosomal MicroRNAs are diagnostic biomarkers and can mediate cell-cell communication in renal cell carcinoma. Eur Urol Focus. 2016;2(2):210–8. [DOI] [PubMed] [Google Scholar]
- 37.Boussios S, Devo P, Goodall ICA, Sirlantzis K, Ghose A, Shinde SD et al. Exosomes in the diagnosis and treatment of renal cell Cancer. Int J Mol Sci. 2023;24(18). [DOI] [PMC free article] [PubMed]
- 38.Yang K, Xu C, Sun H, Xuan Z, Liu Y, Li J, et al. Branched-chain keto-acid dehydrogenase kinase regulates vascular permeability and angiogenesis to facilitate tumor metastasis in renal cell carcinoma. Cancer Sci. 2023;114(11):4270–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torres AG, Marti E. Toward an understanding of extracellular tRNA Biology. Front Mol Biosci. 2021;8:662620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nientiedt M, Deng M, Schmidt D, Perner S, Muller SC, Ellinger J. Identification of aberrant tRNA-halves expression patterns in clear cell renal cell carcinoma. Sci Rep. 2016;6:37158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao C, Tolkach Y, Schmidt D, Kristiansen G, Muller SC, Ellinger J. 5’-tRNA halves are dysregulated in Clear Cell Renal Cell Carcinoma. J Urol. 2018;199(2):378–83. [DOI] [PubMed] [Google Scholar]
- 42.Zhang W, Zheng Z, Wang K, Mao W, Li X, Wang G, et al. piRNA-1742 promotes renal cell carcinoma malignancy by regulating USP8 stability through binding to hnRNPU and thereby inhibiting MUC12 ubiquitination. Exp Mol Med. 2023;55(6):1258–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Du X, Li H, Xie X, Shi L, Wu F, Li G, et al. piRNA-31115 promotes Cell Proliferation and Invasion via PI3K/AKT pathway in Clear Cell Renal Carcinoma. Dis Markers. 2021;2021:6915329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez VD, Vucic EA, Thu KL, Hubaux R, Enfield KS, Pikor LA, et al. Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Sci Rep. 2015;5:10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Busch J, Ralla B, Jung M, Wotschofsky Z, Trujillo-Arribas E, Schwabe P, et al. Piwi-interacting RNAs as novel prognostic markers in clear cell renal cell carcinomas. J Exp Clin Cancer Res. 2015;34(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Wu X, Gao H, Jin JM, Li AX, Kim YS, et al. Piwi-Interacting RNAs (piRNAs) are Dysregulated in Renal Cell Carcinoma and Associated with Tumor Metastasis and Cancer-Specific Survival. Mol Med. 2015;21(1):381–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qu A, Wang W, Yang Y, Zhang X, Dong Y, Zheng G, et al. A serum piRNA signature as promising non-invasive diagnostic and prognostic biomarkers for colorectal cancer. Cancer Manag Res. 2019;11:3703–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sundar IK, Li D, Rahman I. Small RNA-sequence analysis of plasma-derived extracellular vesicle miRNAs in smokers and patients with chronic obstructive pulmonary disease as circulating biomarkers. J Extracell Vesicles. 2019;8(1):1684816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aguilar MA, Ebanks S, Markus H, Lewis MM, Midya V, Vrana K, et al. Neuronally enriched microvesicle RNAs are differentially expressed in the serums of Parkinson’s patients. Front Neurosci. 2023;17:1145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Al Rifai M, DeFilippis AP, McEvoy JW, Hall ME, Acien AN, Jones MR, et al. The relationship between smoking intensity and subclinical cardiovascular injury: the multi-ethnic study of atherosclerosis (MESA). Atherosclerosis. 2017;258:119–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pajares M, A IR, Manda G, Bosca L, Cuadrado A. Inflammation in Parkinson’s Disease: Mechanisms and Therapeutic Implications. Cells. 2020;9(7). [DOI] [PMC free article] [PubMed]
- 52.Kruk L, Mamtimin M, Braun A, Anders HJ, Andrassy J, Gudermann T et al. Inflammatory networks in renal cell carcinoma. Cancers (Basel). 2023;15(8). [DOI] [PMC free article] [PubMed]
- 53.Cernea M, Tang W, Guan H, Yang K. Wisp1 mediates Bmp3-stimulated mesenchymal stem cell proliferation. J Mol Endocrinol. 2016;56(1):39–46. [DOI] [PubMed] [Google Scholar]
- 54.Shimomura T, Yoshida Y, Sakabe T, Ishii K, Gonda K, Murai R, et al. Hepatic differentiation of human bone marrow-derived UE7T-13 cells: effects of cytokines and CCN family gene expression. Hepatol Res. 2007;37(12):1068–79. [DOI] [PubMed] [Google Scholar]
- 55.Schlegelmilch K, Keller A, Zehe V, Hondke S, Schilling T, Jakob F, et al. WISP 1 is an important survival factor in human mesenchymal stromal cells. Gene. 2014;551(2):243–54. [DOI] [PubMed] [Google Scholar]
- 56.Singh K, Oladipupo SS. An overview of CCN4 (WISP1) role in human diseases. J Transl Med. 2024;22(1):601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shao H, Cai L, Moller M, Issac B, Zhang L, Owyong M, et al. Notch1-WISP-1 axis determines the regulatory role of mesenchymal stem cell-derived stromal fibroblasts in melanoma metastasis. Oncotarget. 2016;7(48):79262–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Falconi G, Fabiani E, Fianchi L, Criscuolo M, Raffaelli CS, Bellesi S, et al. Impairment of PI3K/AKT and WNT/beta-catenin pathways in bone marrow mesenchymal stem cells isolated from patients with myelodysplastic syndromes. Exp Hematol. 2016;44(1):75–83. e1-4. [DOI] [PubMed] [Google Scholar]
- 59.Ni F, Zhang T, Xiao W, Dong H, Gao J, Liu Y, et al. IL-18-Mediated SLC7A5 overexpression enhances osteogenic differentiation of human bone marrow mesenchymal stem cells via the c-MYC pathway. Front Cell Dev Biol. 2021;9:748831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H, Wang Y, Yang F, Feng S, Chang K, Yu X, et al. Clonal MDS/AML cells with enhanced TWIST1 expression reprogram the differentiation of bone marrow MSCs. Redox Biol. 2023;67:102900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuan T, Shi C, Xu W, Yang HL, Xia B, Tian C. Extracellular vesicles derived from T-cell acute lymphoblastic leukemia inhibit osteogenic differentiation of bone marrow mesenchymal stem cells via miR-34a-5p. Endocr J. 2021;68(10):1197–208. [DOI] [PubMed] [Google Scholar]
- 62.Stucky A, Gao L, Sun L, Li SC, Chen X, Park TH, et al. Evidence for AJUBA-catenin-CDH4-linked differentiation resistance of mesenchymal stem cells implies tumorigenesis and progression of head and neck squamous cell carcinoma: a single-cell transcriptome approach. Blood Genom. 2021;5(1):29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kanai Y. Amino acid transporter LAT1 (SLC7A5) as a molecular target for cancer diagnosis and therapeutics. Pharmacol Ther. 2022;230:107964. [DOI] [PubMed] [Google Scholar]
- 64.Bao L, Fu L, Su Y, Chen Z, Peng Z, Sun L, et al. Amino acid transporter SLC7A5 regulates cell proliferation and secretary cell differentiation and distribution in the mouse intestine. Int J Biol Sci. 2024;20(6):2187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cai TY, Zhu W, Chen XS, Zhou SY, Jia LS, Sun YQ. Fibroblast growth factor 2 induces mesenchymal stem cells to differentiate into tenocytes through the MAPK pathway. Mol Med Rep. 2013;8(5):1323–8. [DOI] [PubMed] [Google Scholar]
- 66.Zhang F, Peng WX, Wang L, Zhang J, Dong WT, Wu JH, et al. Role of FGF-2 transfected bone marrow mesenchymal stem cells in Engineered Bone tissue for repair of avascular necrosis of femoral head in rabbits. Cell Physiol Biochem. 2018;48(2):773–84. [DOI] [PubMed] [Google Scholar]
- 67.Awan B, Turkov D, Schumacher C, Jacobo A, McEnerney A, Ramsey A, et al. FGF2 induces Migration of Human Bone Marrow Stromal cells by increasing core fucosylations on N-Glycans of Integrins. Stem Cell Rep. 2018;11(2):325–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang A, Liu J, Zhuang X, Yu S, Zhu S, Liu Y, et al. Identification and comparison of piRNA expression profiles of Exosomes Derived from Human Stem cells from the apical papilla and bone marrow mesenchymal stem cells. Stem Cells Dev. 2020;29(8):511–20. [DOI] [PubMed] [Google Scholar]
- 69.Della Bella E, Stoddart MJ. Cell detachment rapidly induces changes in noncoding RNA expression in human mesenchymal stromal cells. Biotechniques. 2019;67(6):286–93. [DOI] [PubMed] [Google Scholar]
- 70.Della Bella E, Menzel U, Basoli V, Tourbier C, Alini M, Stoddart MJ. Differential Regulation of circRNA, miRNA, and piRNA during early osteogenic and chondrogenic differentiation of human mesenchymal stromal cells. Cells. 2020;9(2). [DOI] [PMC free article] [PubMed]
- 71.Liu J, Chen M, Ma L, Dang X, Du G. piRNA-36741 regulates BMP2-mediated osteoblast differentiation via METTL3 controlled m6A modification. Aging. 2021;13(19):23361–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Supplementary Methods file S1. RNAseq conditions. Supplementary Table S1. Primers used for qPCR analysis of piRNA and tRNA
Supplementary Material 2: Supplementary Table S2. miRNAs aberrantly expressed in exosomes isolated from conditioned media of Caki-1 and KIJ265T cells. The table shows results of RNAseq. Volcano plots are provided alongside the data
Supplementary Material 3: Supplementary Table S3. Genes predicted as targets of 40 miRNAs of which expression was consistently altered in CM from Caki-1 and KIJ265T cells. Prediction was performed using miRnet (https://www.mirnet.ca/miRNet/home.xhtml)
Supplementary Material 4: Supplementary Table S4. microRNAs with concordant changes of expression in exosomes and tumor tissues of RCC patients
Supplementary Material 5: Supplementary Table S5. Genes predicted as targets of 3 validated miRNAs (hsa-miR-10b-3p, hsa-miR-125a-5p, hsa-miR-365b-3p), consistently altered in CM from at least three cell RCC cell lines. Prediction was performed using miRnet (https://www.mirnet.ca/miRNet/home.xhtml)
Supplementary Material 6: Supplementary Table S6. Enriched KEGG pathways among the genes predicted as targets of 3 validated miRNAs (hsa-miR-10b-3p, hsa-miR-125a-5p, hsa-miR-365b-3p), consistently altered in CM from at least three cell RCC cell lines. The table shows results of analysis performed using ShinyGO 0.80 (http://bioinformatics.sdstate.edu/go/)
Supplementary Material 7: Supplementary Table S7. Enriched biological processes among the genes predicted as targets of 3 validated miRNAs (hsa-miR-10b-3p, hsa-miR-125a-5p, hsa-miR-365b-3p), consistently altered in CM from at least three cell RCC cell lines. The table shows results of analysis performed using ShinyGO 0.80 (http://bioinformatics.sdstate.edu/go/)
Supplementary Material 8: Supplementary Table S8. tRNAs aberrantly expressed in exosomes isolated from conditioned media of Caki-1 and KIJ265T cells. The table shows results of RNAseq. Volcano plots are provided alongside the data
Supplementary Material 9: Supplementary Table S9. The expression of tRFs is disturbed in RCC tumors. tRFs were predicted as derived from tRNA of which altered expression was found in exosomes isolated from RCC cell lines when compared with normal kidney cells. The analysis was performed using OncotRF, using data from TCGA N = 544 for RCC tumors, N = 72 for normal kidney tissue samples
Supplementary Material 10: Supplementary Table S10. piRNAs aberrantly expressed in exosomes isolated from conditioned media of Caki-1 and KIJ265T cells. The table shows results of RNAseq. Volcano plots are provided alongside the data
Supplementary Material 11: Supplementary Table S11. piR_004153 alters expression of MSCs genes. The table shows results of microarray analysis of RNA isolated from MSCs transfected with piR_004513 mimic or non-targeting scrambled oligonucleotide, cultured in five independent cell culture wells per each group. Volcano plot is provided alongside the data
Data Availability Statement
RNAseq and microarray data are available in NCBI/GEO database (acc. no. GSE256016 and GSE263824, respectively). The residual data generated or analyzed during this study are included in this published article and its supplementary information files.