Abstract
Background
Virus-induced gene silencing (VIGS) is a rapid and powerful method for gene functional analysis in plants that pose challenges in stable transformation. Numerous VIGS systems based on Agrobacterium infiltration has been widely developed for tender tissues of various plant species, yet none is available for recalcitrant perennial woody plants with firmly lignified capsules, such as tea oil camellia. Therefore, there is an urgent need for an efficient, robust, and cost-effective VIGS system for recalcitrant tissues.
Results
Herein, we initiated the Tobacco rattle virus (TRV)-elicited VIGS in Camellia drupifera capsules with an orthogonal analysis including three factors: silencing target, virus inoculation approach, and capsule developmental stage. To facilitate observation and statistical analysis, two genes predominantly involved in pericarp pigmentation were selected for silencing efficiency: CdCRY1 (coding for a key photoreceptor affecting light-responsive perceivable anthocyanin accumulation in exocarps) and CdLAC15 (coding for an oxidase catalyzing the oxidative polymerization of proanthocyanidins in mesocarps, resulting in unperceivable red-hued mesocarps). The infiltration efficiency achieved for each gene was ~ 93.94% by pericarp cutting immersion. The optimal VIGS effect for each gene was observed at early (~ 69.80% for CdCRY1) and mid stages (~ 90.91% for CdLAC15) of capsule development.
Conclusions
Using our optimized VIGS system, CdCRY1 and CdLAC15 expression was successfully knocked down in Camellia drupifera pericarps, leading to fading phenotypes in exocarps and mesocarps, respectively. The established VIGS system may facilitate functional genomic studies in tea oil camellia and other recalcitrant fruits of woody plants.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13007-024-01320-1.
Keywords: Virus-induced gene silencing, Camellia drupifera, Pericarp pigmentation, Infiltration, CdCRY1, CdLAC15
Introduction
Camellia drupifera is a lesser-known species within the Camellia genus, primarily native to Southeast Asia, including southern China. Unlike its more famous relatives, such as Camellia sinensis (tea) and Camellia japonica, C. drupifera is valued not only for its ornamental beauty but also for its hardiness and adaptability to various soil types [1]. Additionally, its seeds are a source of a high-quality edible oils, rich in unsaturated fatty acids, and used for cooking and in cosmetics [2]. Currently, multi-omics approaches have identified several key candidate genes involved in important agronomic traits. However, the lack of an efficient genetic transformation system hinders in vivo gene function analysis in C. drupifera and related species [3, 4].
Virus-induced gene silencing (VIGS) is a widely used tool in plant functional genomics that allows for rapid, transient knockdown of gene expression. By exploiting the plant’s RNA interference (RNAi) machinery, VIGS uses recombinant viruses carrying a fragment (200–500 bp) of the target gene to trigger the degradation of homologous mRNA, thereby silencing the gene [5, 6]. This technique has been extensively applied to various plant species, enabling researchers to study gene functions involved in diverse biological processes [7, 8].
VIGS is particularly advantageous due to its speed, cost-effectiveness, and ability to silence genes in specific tissues without requiring stable transformation. Furthermore, recent advances, including the development of new viral vectors and delivery methods tailored for less amenable species, have improved its efficiency and broadened its application [9–11]. Moreover, the adaptation of VIGS for high-throughput screens has expanded its use in large-scale functional genomics studies, accelerating the discovery of genes involved in key agronomic traits [12, 13]. Nonetheless, challenges remain when optimizing VIGS for certain plant species, particularly in achieving consistent gene silencing efficiency in recalcitrant tissues and species. For instance, some plant tissues, such as woody, lignified organs, are more resistant to viral infection or exhibit lower silencing levels, complicating the study of gene function [14].
To overcome these limitations, we developed a TRV-elicited VIGS procedure in C. drupifera capsules through four infiltration approaches, including peduncle injection, direct pericarp injection, pericarp cutting immersion, and fruit-bearing shoot infusion, at five capsule developmental stages. To facilitate rapid assessment and statistical analysis, here we selected two genes predominantly involved in pericarp pigmentation for their silencing efficiency. CdCRY1, encoding a key photoreceptor cryptochrome, is speculated to affect light-responsive perceivable anthocyanin (ACN) accumulation in exocarps of C. drupifera var. ‘Hongpi’ [15, 16]; CdLAC15, encoding a laccase belonging to the family of multi-copper oxidases, is thought to be closely associated with the oxidative polymerization of proanthocyanidins (PAs) and lignins, thereby producing unperceivable red hued mesocarps in C. drupifera var. ‘Hongrou’ [17, 18]. Finally, the successful development of VIGS system in C. drupifera will significantly enhance functional genomic research in tea oil camellia, and moreover, in unyielding fruits of other recalcitrant ligneous plants.
Methods
Plant material
Camellia drupifera var. ‘Hongpi’ and C. drupifera var. ‘Hongrou’ were harvested from 20-year-old trees at Fengji Oil Tea Base, Boluo County, Huizhou City, Guangdong Province, China (E 114° 34′, N 23° 23′). Located in the south-central region of Lingnan, this site has a subtropical monsoon climate with abundant sunlight and rainfall. The average maximum and minimum temperatures are 30.0 °C and 20.0 °C, respectively. The soil is predominantly yellow loam, and the surrounding mountainous landscape, with its rich vegetation, provides ideal conditions for C. drupifera cultivation (Supplementary Fig. S1). Samples were collected in 2024, at 279 days post-pollination.
Multiple sequence comparison and phylogenetic analysis
Hidden Markov model profiles for the laccase gene family (PF07731) and photolyases/cryptochromes (PF12546) were downloaded from the Pfam database (http://pfam.xfam.org/). The HMMER 3.0 program was then used to search the transcriptome’s protein sequences for candidate CRY and LAC genes. After the initial screening, gene family members were validated using SMART (https://smart.embl-heidelberg.de/) to confirm the presence of C. drupifera CRY/LAC family members. The validated sequences were aligned using MUSCLE (v5.1, UK) for multiple sequence alignment. Finally, a maximum likelihood phylogenetic tree was built with 1000 bootstrap replicates using IQ-TREE (v2.2.0, Australia) [19]. The confidence threshold for each node was set at > 50%. The conserved domain was analyzed through the PfamScan online program (https://www.ebi.ac.uk/Tools/pfa/pfamscan/) [20].
Total RNA extraction and cDNA synthesis
Total RNA was extracted from C. drupifera at 279 days post-pollination (DAP) using the RNAprep Pure Cell/Bacteria Kit (Tiangen, China). First-strand cDNA was synthesized by reverse transcription according to the manufacturer’s instructions (Yeasen, China). Full-length cDNA was amplified from the C. drupifera cDNA library using high-fidelity DNA polymerase (Yeasen, China). Silencing vectors included pNC-TRV2, a modified version of pTRV2, and its green fluorescent protein variant, pNC-TRV2-GFP, both provided by Dr. Yan Pu from the Chinese Academy of Tropical Agricultural Sciences (Supplementary Fig. S2). Based on the CDS sequences of CdCRY1 and CdLAC15, we screened for suitable 200–300 bp cleavage sites using the Camellia sinensis shuchazao V2 CDS database and the SGN VIGS Tool (https://vigs.solgenomics.net/) [21]. The selected regions were subjected to homologous family analysis to ensure specificity, and the sequences were submitted to NCBI for comparison. Only sequences with high similarity to the target genes and < 40% similarity to other genes were selected for constructing VIGS vectors. Specific primers were designed using Primer3web (v4.1.0, https://primer3.ut.ee/; Supplementary Table S1). The PCR reaction was set up using C. drupifera cDNA as a template, with the following components: 2 µL cDNA, 2.5 µL forward primer (CdCRY1/CdLAC15-VF), 2.5 µL reverse primer (CdCRY1/CdLAC15-VR), and 25 µL 2 × Hieff® Robust PCR Master Mix (Yeasen, China), to a total volume of 50 µL. The PCR program included an initial denaturation at 98 °C for 4 min, followed by 30 cycles of 98 °C for 10 s, 59 °C for 15 s, and 72 °C for 20 s, with a final extension at 72 °C for 5 min. After recovering the amplification products from the gel, we evaluated their purity and concentration, ligated them following Nimble Clonig (NC Biotech, China) instructions, and transferred them to Escherichia coli DH5α competent cells (CAT#: DL1001, Shanghai Vidi Biotech Co., Ltd.). Positive colonies were sent to Bioengineering (Shanghai) Ltd. for sequencing to verify that the sequence matched the target.
Agrobacteria fluids preparation
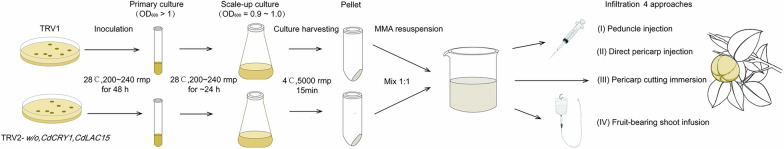
The correctly sequenced recombinant plasmids, TRV2-w/o TRV2-CdCRY1, TRV2-CdLAC15, and TRV1, were used to transform Agrobacterium. The culture was incubated at 28 °C for 2 days. Single plaques were selected and cultured for another two days in 4 mL of YEB medium containing 25 μg/mL kanamycin and 50 μg/mL rifampicin (both from Yeasen, China), at 28 °C with shaking at 200–240 rpm. The homogeneous agrobacteria solution was then transferred to 50 mL of YEB medium containing 25 μL rifampicin (50 mg/mL), 25 μL kanamycin (100 mg/mL), 5 mL MES (pH 5.6, 0.2 M), and 5 μL acetosyringone (0.1 M). The culture was diluted at a 1:20 ratio and incubated. The incubation continued at 28 °C with shaking at 200–240 rpm for 24 h. When the OD600 reached 0.9–1.0, the culture was centrifuged at 5000 rpm for 15 min to remove the supernatant. The control (TRV1 + TRV2-w/o) and experimental groups (TRV1 + TRV2-CdCRY1/TRV2-CdLAC15) were prepared by mixing agrobacteria fluids at a 1:1 volume ratio. The fluids were re-suspended in infiltration buffer (10 mM MES, 10 mM MgCl2, 200 μM acetosyringone) and adjusted OD600 to 0.8. The fluids were then left to stand for 2–3 h at room temperature before the infiltration (Fig. 1). At 279 DAP, C. drupifera capsules were collected from different experimental groups for further analysis.
Fig. 1.
Simplified flow chart of agrobacteria preparation and infiltration into C. drupifera capsules by four approaches. TRV2-w/o vector was used as control
VIGS assay
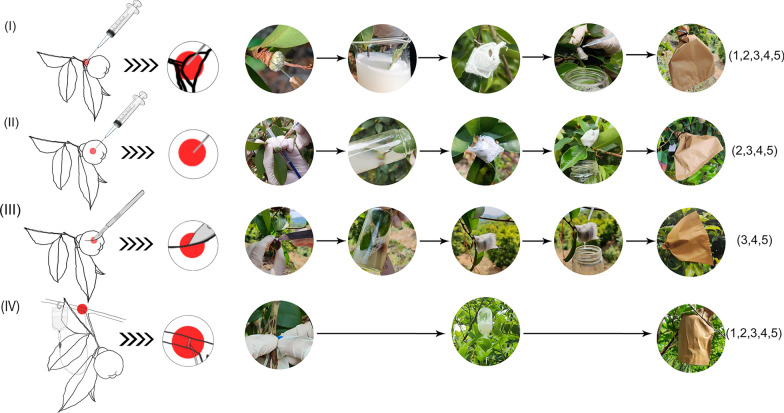
C. drupifera capsules require approximately 332 days to get fully mature after pollination. During this time, the experiment was biologically replicated across three zones (labeled X, Y, and Z, Supplementary Fig. S3). In each zone, 16 uniformly growing trees were selected for infiltration (Each gene was infiltrated into a tree using four different approaches, producing five time points per approach, with 25 to 35 capsules per time point). Four infiltration approaches were as follows (Fig. 2): (I) Peduncle injection: disposable syringes (1 mL with a 0.55 mm needle, Shandong Weigao Group Medical Polymer Material Co., Ltd., China) were used to inject 2 mL of infiltration liquid into the peduncle to a depth of 5–8 mm (As shown in the red area of Fig. 2). The penduncles were then immersed in the Agrobacterium suspension for 30 s, wrapped by gauze, and placed in a light-proof bag (outside yellow inside black shading rate 100%; Jingdong Hanli Gardening and Agricultural Specialty Store, China) for 24 h. After that, bags were removed for full light condition. (II) Direct pericarp injection: 10 mL Agrobacterium suspension was evenly injected at 5 different positions around the middle section of the capsule to a depth of 1–2 cm by syringes. The capsules were then immersed in the Agrobacterium suspension for 30 s and wrapped in gauze to ensure moisture retention. Similarly, the capsules were subjected to normal condition after 24 h dark treatment. (III) Pericarp cutting immersion: a histoculture knife (Jingdong Cherry Sage Industrial Store, China) was used to make 3–5 evenly distributed incisions, each 1 cm long and 5 mm deep, on the pericarps. After that, the capsules were also immersed for 30 s, wrapped in gauze, and maintained moist through drip irrigation followed by 24 h dark treatment. (IV) Fruit-bearing shoot infusion: 500 mL Agrobacterium suspension was transmitted through an infusion bag to the mother branch with more than 10 capsules. The pipe-puncture needle (0.51 mm, Jingdongkanglu Health Care Store, China) was inserted about 1 cm deep into the transmission layer of the tree, 20–30 cm away from the capsules, and the infusion was carried out at a rate of 100–150 drops per minute. Dark treatment was lifted until liquids drained.
Fig. 2.
Different infiltrating approaches to the capsules of C. drupifera at five different developmental stages. (I) Peduncle injection. (II) Direct pericarp injection. (III) Pericarp cutting immersion. (IV) Fruit-bearing shoot infusion. Five developmental stages indicated by numbers in parentheses: 1. 64 DAP, 2. 95 DAP, 3. 126 DAP, 4. 156 DAP, 5. 187 DAP
Phenotypic observation and gene expression analysis
To ensure study accuracy, the mesocarp and exocarp of C. drupifera were collected based on different classifications. The collected samples were observed under a laser inverted microscope (ECLIPSE Ti2-U, Nikon, Japan), and effective infestation was calculated based on the samples that emitted a green fluorescence signal (Supplementary Fig. S4). Effectively infested samples were divided into two groups: one rapidly frozen in liquid nitrogen and transferred to the laboratory for storage at − 80 °C and the other was retrieved immediately for photographic documentation and physiological experiments. Total RNA was extracted from each group and reverse transcribed into cDNA. qRT-PCR was conducted using primers specific to the VIGS silencing region, with CdPPIL as the internal reference gene. (Supplementary Table S1). Real-time quantitative PCR was performed using SYBR Green Premix ProTaqPCR reagent (Yeasen Biologicals, China) and a CFX96 system (BIO RED CFX Connect, USA). The PCR reaction mixture was 20 μL, containing 10 μL TB Green® Premix Ex Taq™ II (Tli RNaseH Plus, 2x), 0.4 μL of each primer, 1 μL of template cDNA, and 8.2 μL of double-distilled water (ddH2O). The PCR program began with an initial denaturation at 95 °C for 30 s, followed by 30 cycles of 95 °C for 5 s and 60 °C for 30 s. Each sample was analyzed in triplicate. The silencing efficiency of pTRV2-CdCRY1/CdLAC15 in injected capsules was calculated using the 2−ΔΔCt method [22].
Anthocyanin and proanthocyanidin analysis
The anthocyanin extraction starts by grinding 0.1 g of capsules rind into a fine powder using liquid nitrogen. The powder was then suspended in a 15 mL polypropylene tube with 5 mL of 80% methanol and 0.2% HCl, followed by vortexing for 30 s. Then, the mixture was sonicated at 10 °C for 15 min. After sonication, the samples were incubated for 24 h in the darkness, followed by centrifugation at 5000 rpm for 10 min. The supernatant was then transferred to a fresh tube. The anthocyanin content was assessed by measuring the extract’s absorbance at 530 nm [23].
Total soluble and insoluble PA were analyzed according to the protocol outlined by Chen et al. [24]. Briefly, ground mesocarps (0.5 g) were extracted with a 5 mL mixture of 70% acetone solution and 0.1% ascorbic acid. Thereafter, the mixture was vortexed and sonicated for 1 h at room temperature. Subsequent extraction with chloroform and hexane separated the supernatants from the insoluble PA fractions. The soluble PA content was determined spectrophotometrically after reacting with DMACA reagent (0.2% w/v DMACA [Yeasen, China] in methanol-3 N HCI) at 640 nm using a Waters 1525 Binary HPLC pump equipped with 2998 PDA and 2424 ELS detectors (Waters Corporation, Milford, MA, USA), with (+)-catechin as a standard. The insoluble PA content was determined by treating the acetone-free residue with butanol-HCl. Then, the absorbance values at 550 nm were converted into insoluble PA levels using a standard procyanidin B1 curve.
Statistical data analysis
Data analysis and plotting were performed using the R software packages tidyverse (v2.0.0), stats (v4.4.1), and ggplot2 (v3.5.1) in Excel 2019 (Microsoft Office, USA) and RStudio (v2022.02.2, Inc., USA). Significance analyses of differences between treatments were performed using two-tailed Student’s t-test, one-way analysis of variance (ANOVA), and Tukey’s multiple comparison test using SPSS software (v18.0, BM, USA). Image stitching was performed with Adobe Illustrator 2022 (Adobe Systems Incorporated, USA). Statistical significance was assessed between the experimental groups, with different letter labels indicating significant differences at the p < 0.05 level.
Results
Capsule traits of C. drupifera for observation: flavonoid-based pigmentation in pericarps
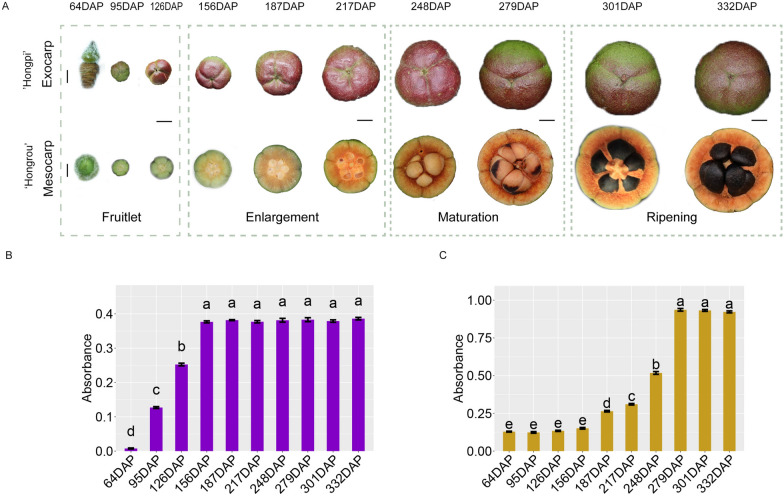
To facilitate the rapid tracking and assessment of the effects of VIGS in C. drupifera capsules, we selected two varieties with red-hued pericarps: ‘Hongpi’ with a perceivable red exocarps and ‘Hongrou’ with an unperceivable red mesocarps (Fig. 3A). In our previous metabolomic analyses, ACNs and insoluble PAs stood out among other compounds, contributing to the red coloration of exocarps and mesocarps, respectively (unpublished data). As shown in Fig. 3B, ACNs phototropically accumulated in exocarps early in capsule development (~ 95 DAP); meanwhile, insoluble PAs only began to deposit in the mesocarps at mid-development stage (~ 187 DAP; Fig. 3C).
Fig. 3.
Flavonoid-based pigmentation in C. drupifera ‘Hongpi’ exocarps and ‘Hongrou’ mesocarps throughout capsule development. A Capsule phenotype from the day after pollination to maturation (322 days) Scale bar (64 DAP) = 300 µm; Scale bar (other developmental stages) = 1 cm; The development of C. drupifera capsules occurs in four stages: Fruitlet, Enlargement, Maturation, and Ripening. The Fruitlet stage spans from 64 to 126 days after pollination (DAP), followed by the Enlargement stage from 156 to 217 DAP. The Maturation stage spans from 248 to 279 DAP, and finally, the Ripening stage occurs between 301 and 332 DAP. B Variations of ACN content at 10 different developmental stages of ‘Hongpi’ exocarps. C Variations of insoluble PA content at 10 different developmental stages of ‘Hongrou’ mesocarps
Characteristics of CdCRY1 and CdLAC15 in C. drupifera for VIGS targeting
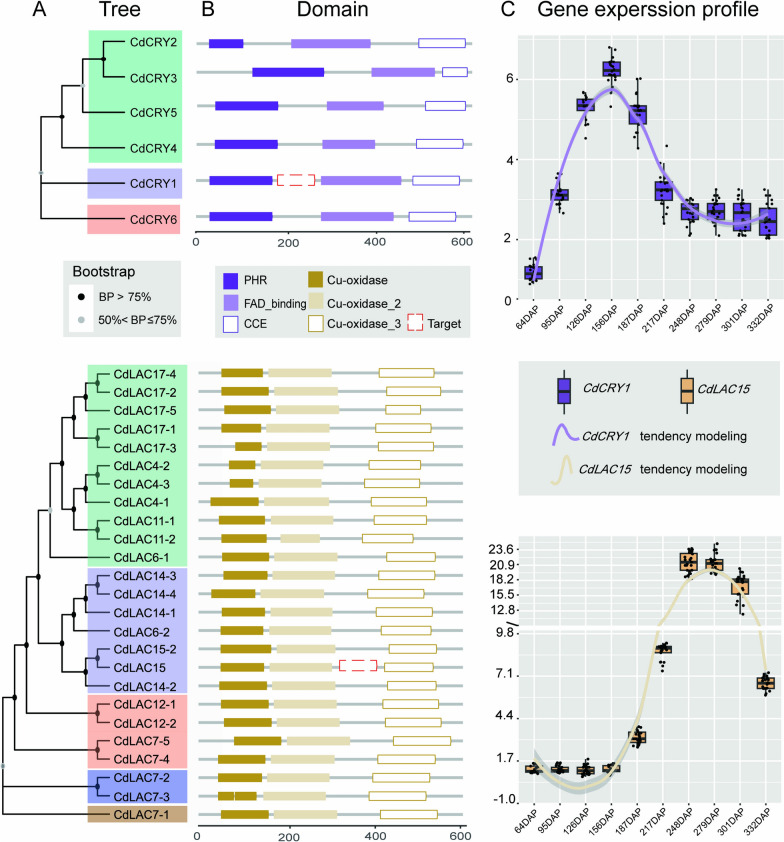
CdCRY1, a member of the cryptochrome gene family, was chosen for its consistent upregulation throughout all capsule developmental stages, as indicated by comparative transcriptome data (unpublished). Therefore, CdCRY1 is a suitable indicator for the light-responsive red exocarp trait in C. drupifera. Transcriptomic analysis identified six CdCRY homologs, all of which share three conserved domains: PHR, FAD_binding, and CCE (Fig. 4A and B). The expression profile of CdCRY1 aligns with ACN content in the exocarps in ‘Hongpi,’ suggesting its key role in the phototropic accumulation of ACNs (Fig. 4C).
Fig. 4.
Phylogenetic analyses of two target genes (CdCRY1 and CdLAC15) involved in percarp pigmentation and their expression profiles at different capsule developmental stages. A Maximum likelihood trees of 6 CdCRY homologs (upper) and 25 CdLAC homologs (lower) bootstrap values > 50% are shown on each clade. B Conserved domains shared by CdCRYs (upper) and CdLACs (lower) generated by PfamScan; a specific fragment of each gene (indicated by red dashed rectangles) was selected to construct recombinant TRV2. C Expression profiles of CdCRYs (upper) and CdLACs (lower) showing the relative expression level of each gene at 10 different developmental stages of capsules
In ‘Hongrou,’ 25 CdLACs were detected, including three typical Cu_oxidase domain-encoding laccases (Fig. 4A and B). CdLAC15, an ortholog of AtTT10 in Arabidopsis, significantly correlated with insoluble PA content in mesocarps, as evidenced by integrated metabolome and transcriptome analyses (unpublished data). CdLAC15 expression began to increase at the mid-development stage and peaked between 250 and 280 DAP, consistent with the deposition of insoluble PAs in the mesocarps of ‘Hongrou’ (Fig. 4C). A specific fragment of each gene (~ 300 bp) was selected to construct a recombinant TRV2 for precise targeting (Fig. 4B).
Optimization of agro-infiltrating approaches to C. drupifera capsules at different developmental stages
The transformation of recalcitrant capsules remains a challenge, either stably or transiently, for their firmly lignified pericarps, which greatly hamper in vivo gene functional studies in tea oil camellia. We designed four different infiltrating approaches to capsules at five developmental stages (64–187 DAP). To facilitate detection of infiltrating efficiency, we used the GFP fused TRV2 (see “Methods”). Peduncle injection (I) and fruit-bearing shoot infusion (IV) were applied at all five stages, while direct pericarp injection (II) and pericarp cutting immersion (III) could not be used at very early stages. Direct pericarp injection (II) This method was only feasible starting at 95 DAP and later. At earlier stages, the fruits were too small and delicate to be effectively infiltrated using this technique. (III) This method was applicable only from 126 DAP onwards, when the fruits had sufficiently developed to be harvested and subjected to this treatment. (Fig. 2). After eight months of follow-up, we found none of the above approaches were applicable to capsules at very early developmental stages (before 126 DAP) as they were extremely vulnerable to mechanical stress during virus inoculation. In contrast, we achieved effective infiltration with pericarp cutting immersion (III) and fruit-bearing shoot infusion (IV) at the latter three developmental stages, achieving a maximum of 100% efficiency (Supplementary Fig. 1). Remarkably, despite achieving up to 100% infiltrating efficiency in the inoculation zone of fruit-bearing shoot (IV), only 3.33% of the attached capsules were silenced to some extent. This discrepancy may be attributed to decreased viral-based vector mobility during distal transmission (Table 1).
Table 1.
Optimization of VIGS procedure with orthogonal analysis
| Gene | Infiltration approach | capsules developmental phase | Infiltration efficiency (%) | VIGS efficiency (%) |
|---|---|---|---|---|
| CdCRY1 | I | 1 | 6.67 ± 0.64 | 6.67 ± 5.77 |
| CdCRY1 | I | 2 | 6.11 ± 1.18 | 6.11 ± 5.36 |
| CdCRY1 | I | 3 | 50.34 ± 5.06 | 21.97 ± 7.50 |
| CdCRY1 | I | 4 | 50.00 ± 4.55 | 22.12 ± 6.82 |
| CdCRY1 | I | 5 | 56.67 ± 12.41 | 18.48 ± 8.64 |
| CdCRY1 | II | 1 | NA | NA |
| CdCRY1 | II | 2 | 15.60 ± 3.92 | 8.10 ± 3.03 |
| CdCRY1 | II | 3 | 62.78 ± 6.31 | 52.78 ± 11.10 |
| CdCRY1 | II | 4 | 86.97 ± 15.38 | 47.31 ± 9.88 |
| CdCRY1 | II | 5 | 83.33 ± 11.55 | 36.67 ± 11.55 |
| CdCRY1 | III | 1 | NA | NA |
| CdCRY1 | III | 2 | NA | NA |
| CdCRY1 | III | 3 | 83.57 ± 4.70 | 69.80 ± 3.04 |
| CdCRY1 | III | 4 | 93.64 ± 5.53 | 43.30 ± 2.90 |
| CdCRY1 | III | 5 | 90.00 ± 10.00 | 36.67 ± 5.77 |
| CdCRY1 | IV | 1 | 7.87 ± 6.85 | 0 |
| CdCRY1 | IV | 2 | 8.84 ± 6.96 | 0 |
| CdCRY1 | IV | 3 | 18.61 ± 1.77 | 7.87 ± 0.98 |
| CdCRY1 | IV | 4 | 24.09 ± 3.72 | 0 |
| CdCRY1 | IV | 5 | 22.12 ± 7.71 | 0 |
| CdLAC15 | I | 2 | 7.79 ± 3.67 | 7.79 ± 7.23 |
| CdLAC15 | I | 3 | 26.67 ± 2.89 | 9.72 ± 8.67 |
| CdLAC15 | I | 4 | 38.97 ± 15.44 | 18.77 ± 8.50 |
| CdLAC15 | I | 5 | 44.44 ± 10.64 | 23.74 ± 6.12 |
| CdLAC15 | II | 1 | NA | NA |
| CdLAC15 | II | 2 | 12.96 ± 8.02 | 5.56 ± 4.81 |
| CdLAC15 | II | 3 | 66.28 ± 11.06 | 33.53 ± 9.13 |
| CdLAC15 | II | 4 | 84.63 ± 8.36 | 72.41 ± 15.42 |
| CdLAC15 | II | 5 | 79.80 ± 17.32 | 76.67 ± 15.28 |
| CdLAC15 | III | 1 | NA | NA |
| CdLAC15 | III | 2 | NA | NA |
| CdLAC15 | III | 3 | 69.63 ± 16.68 | 48.52 ± 7.88 |
| CdLAC15 | III | 4 | 90.3 ± 0.52 | 87.27 ± 4.72 |
| CdLAC15 | III | 5 | 93.94 ± 5.25 | 90.91 ± 5.25 |
| CdLAC15 | IV | 1 | 6.55 ± 6.27 | 0 |
| CdLAC15 | IV | 2 | 6.11 ± 5.36 | 0 |
| CdLAC15 | IV | 3 | 9.81 ± 1.40 | 0 |
| CdLAC15 | IV | 4 | 10.83 ± 5.30 | 6.11 ± 1.18 |
| CdLAC15 | IV | 5 | 14.14 ± 7.07 | 6.73 ± 1.43 |
|
‘Hongpi’ TRV2-w/o |
I | 1 | 5.81 ± 0.54 | 0 |
|
‘Hongpi’ TRV2-w/o |
I | 3 | 54.88 ± 0.58 | 0 |
|
‘Hongpi’ TRV2-w/o |
I | 4 | 58.89 ± 8.39 | 0 |
|
‘Hongpi’ TRV2-w/o |
I | 5 | 46.63 ± 2.96 | 0 |
|
‘Hongpi’ TRV2-w/o |
II | 1 | NA | NA |
|
‘Hongpi’ TRV2-w/o |
II | 2 | 7.50 ± 1.77 | 0 |
|
‘Hongpi’ TRV2-w/o |
II | 3 | 72.96 ± 14.67 | 0 |
|
‘Hongpi’ TRV2-w/o |
II | 4 | 63.03 ± 14.70 | 0 |
|
‘Hongpi’ TRV2-w/o |
II | 5 | 74.81 ± 7.14 | 0 |
|
‘Hongpi’ TRV2-w/o |
III | 1 | NA | NA |
|
‘Hongpi’ TRV2-w/o |
III | 2 | NA | NA |
|
‘Hongpi’ TRV2-w/o |
III | 4 | 93.33 ± 11.55 | 0 |
|
‘Hongpi’ TRV2-w/o |
III | 5 | 84.07 ± 4.49 | 0 |
|
‘Hongpi’ TRV2-w/o |
IV | 1 | 5.16 ± 4.51 | 0 |
|
‘Hongpi’ TRV2-w/o |
IV | 2 | 5.34 ± 4.64 | 0 |
|
‘Hongpi’ TRV2-w/o |
IV | 3 | 10.44 ± 1.17 | 0 |
|
‘Hongpi’ TRV2-w/o |
IV | 4 | 13.89 ± 7.35 | 0 |
|
‘Hongpi’ TRV2-w/o |
IV | 5 | 15.15 ± 6.43 | 0 |
|
‘Honrou’ TRV2-w/o |
I | 1 | 0 ± 0 | 0 |
|
‘Honrou’ TRV2-w/o |
I | 2 | 0 ± 0 | 0 |
|
‘Honrou’ TRV2-w/o |
I | 3 | 6.94 ± 2.95 | 0 |
|
‘Honrou’ TRV2-w/o |
I | 4 | 58.67 ± 7.14 | 0 |
|
‘Honrou’ TRV2-w/o |
II | 1 | NA | NA |
|
‘Honrou’ TRV2-w/o |
II | 2 | 7.87 ± 6.85 | 0 |
|
‘Honrou’ TRV2-w/o |
II | 3 | 73.06 ± 13.79 | 0 |
|
‘Honrou’ TRV2-w/o |
II | 4 | 79.29 ± 11.79 | 0 |
|
‘Honrou’ TRV2-w/o |
II | 5 | 58.78 ± 4.45 | 0 |
|
‘Honrou’ TRV2-w/o |
III | 1 | NA | NA |
|
‘Honrou’ TRV2-w/o |
III | 2 | NA | NA |
|
‘Honrou’ TRV2-w/o |
III | 3 | 83.64 ± 11.82 | 0 |
|
‘Honrou’ TRV2-w/o |
III | 4 | 84.07 ± 4.49 | 0 |
|
‘Honrou’ TRV2-w/o |
III | 5 | 85.86 ± 17.23 | 0 |
|
‘Honrou’ TRV2-w/o |
IV | 1 | 7.50 ± 6.61 | 0 |
|
‘Honrou’ TRV2-w/o |
IV | 3 | 14.54 ± 4.78 | 0 |
|
‘Honrou’ TRV2-w/o |
IV | 4 | 17.19 ± 8.99 | 0 |
|
‘Honrou’ TRV2-w/o |
IV | 5 | 23.45 ± 3.65 | 0 |
| Mean 1 | 3.34 | 0.00 | 15.07 | 12.00 |
| Mean 2 | 4.74 | 4.45 | 36.22 | 47.04 |
| Mean 3 | 38.11 | 22.94 | 49.92 | 75.57 |
| Mean 4 | 28.18 | 46.14 | 1.57 | 2.57 |
| Mean 5 | 22.96 | 49.51 | ||
| Range | 34.77 | 49.51 | 48.35 | 73.00 |
| Optimum level | III | 3/5 | ||
| Principal order | III > II > I > IV | CdCRY1:III:3 | CdLAC15:III:5 |
Infiltration efficiency, number of capsules exhibiting a positive fluorescent signal within each group/total number of capsules subjected to infiltration; VlGS efficiency, number of capsules exhibiting target gene knockdown/total number of capsules subjected to infiltration; NA, Not available
Variable silencing efficiency of CdCRY1 and CdLAC15 in orthogonal combinations
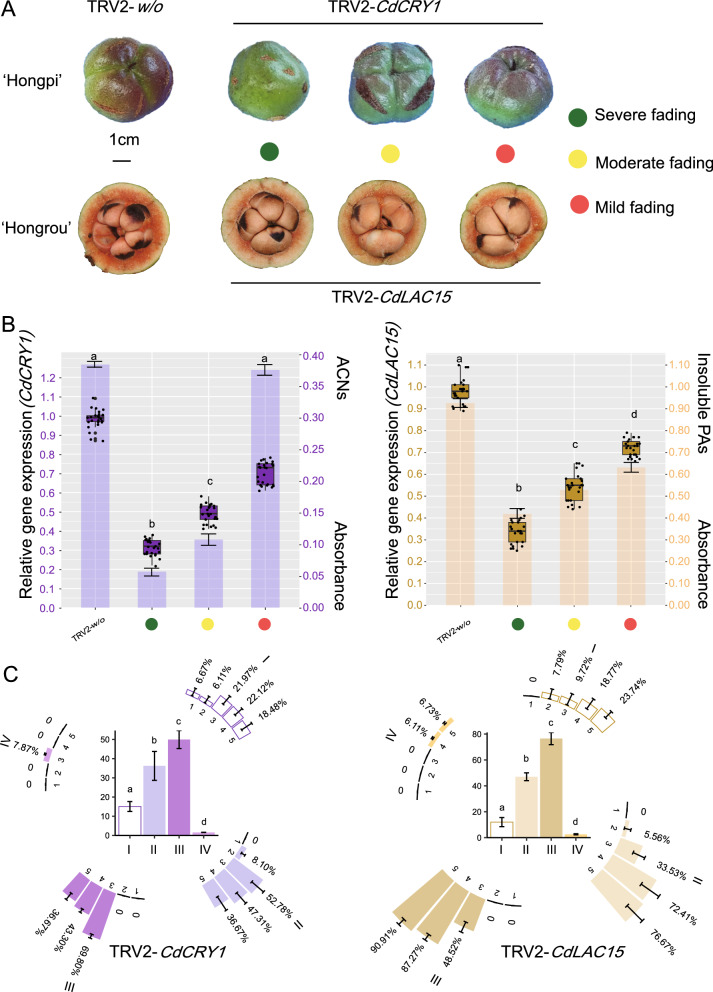
At 279 DAP, we harvested all infiltrated capsules. A total of 720 capsules were collected (306 for TRV2-w/o, 213 for TRV2-CdCRY1 and 201 for TRV2-CdLAC15) from three forest zones where the 48 C. drupifera trees were subjected to infiltration. To simplify statistical analysis for demonstrating VIGS efficiency, capsules were classified into three groups per gene based on pericarp morphology (i.e., mild, moderate, and severe fading; Fig. 5A). Accordingly, the relative expression level of each gene was quantified by qRT-PCR using TRV2-w/o as reference (see “Methods”). Down to 21%–33% of regular expression levels were detected in the optimum silencing groups presenting severe fading exocarps or mesocarps; in contrast, the lowest silencing effect was found in mild fading pericarps, retaining approx. 70%–80% expression for each gene (Fig. 5B). In comparison to the effect of CdLAC15 on the insoluble PA content of mesocarps, the ACNs content of exocarps was more sensitive to variations in CdCRY1 expression level (Fig. 5B).
Fig. 5.
Variable silencing efficiency of CdCRY1 and CdLAC15. A Three main groups of pericarp morphology for TRV2-CdCRY1 (upper) and TRV2-CdLAC15 (lower) at 279 DAP. B Boxplot diagram of CdCRY1 (left) and CdLAC15 (right) showing the relative expression level in three groups. Histogram analysis of ACNs (left) and insoluble PAs (right) in three groups. C Orthogonal analyses with three factors illustrating the effects of TRV2-CdCRY1 (left) and TRV2-CdLAC15 (right) across five different developmental stages through four different infiltrating approaches to capsules
To further evaluate VIGS efficiency in each gene, we performed an orthogonal analysis. As shown in Fig. 5C, the optimum stage to implement VIGS in pericarps, correlating with the expression profile of target gene, was immediately before and after the initial activation of genes; otherwise, we gained the less satisfactory efficiency for both CdCRY1 and CdLAC15. Moreover, pericarp cutting immersion (III) was the most effective agro-infiltrating method for the capsules VIGS in C. drupifera (Fig. 5C and Table 1).
Discussion
In this study, we successfully developed and optimized a VIGS system for transient gene silencing in the lignified pericarps of C. drupifera capsules, a woody perennial species recalcitrant to existing gene silencing methods. Our results demonstrate that this system is a valuable tool for functional genomic studies in recalcitrant tissues of woody plants like C. drupifera.
The silencing efficiency observed for the two targeted genes, CdCRY1 and CdLAC15, reflects the robustness of this VIGS system for studying gene function in capsules. The fading phenotypes observed in exocarps and mesocarps following gene knockdown support that CdCRY1 is involved in light-responsive ACNs accumulation in the exocarp, while CdLAC15 plays a role in PAs polymerization in the mesocarp (Fig. 4). These findings not only provide insights into the molecular mechanisms of pericarp pigmentation in C. drupifera, but also highlight the utility of this VIGS system for gene functional studies in woody plants with firmly lignified tissues.
Comparison with previous VIGS systems
Previous studies have shown that VIGS is an effective tool for gene silencing in herbaceous plants and some soft tissues of woody plants, but its application in lignified tissues, especially in perennial woody species like tea oil camellia C. drupifera remained limited [25]. Our system adapting TRV-based VIGS to C. drupifera overcomes previous challenges associated with tissue recalcitrance, achieving high silencing efficiency. Silencing efficiencies of up to 69.80% for CdCRY1 and 90.91% for CdLAC15 demonstrate the applicability of the system, particularly when infiltrating capsules at fruitlet stage and enlargement stage (Fig. 4 and Table 1). Moreover, our orthogonal analysis revealed that the pericarp cutting immersion method (III) was the most effective approach, surpassing traditional infiltrating methods in recalcitrant tissues (Fig. 3 and Table 1) [26].
Implications for functional genomics in woody plants
This newly optimized VIGS system has broad implications beyond the study of C. drupifera. Woody plants, particularly those with lignified tissues, pose significant challenges for stable transformation, limiting the functional validation of candidate genes identified by multi-omics approaches [27, 28]. The successful silencing of CdCRY1 and CdLAC15 in C. drupifera supports the potential of this TRV-based system for use in other woody species that are recalcitrant to transformation (Fig. 5 and Table 1). The ability to knock down genes in specific tissues and at different developmental stages offers a powerful and flexible tool for investigating traits such as fruit development, disease resistance, and secondary metabolism in other ligneous plants [29, 30].
Limitations and future directions
While this VIGS system provides a robust method for gene silencing in tea oil Camellia, several limitations remain. First, mechanical stress associated with agro-infiltration, particularly at early capsule developmental stages, caused tissue damage that decreased infiltration success. Refining delivery methods, such as using milder mechanical techniques or improving viral vector stability, could enhance efficiency, particularly in vulnerable tissues.
Additionally, although our system successfully silenced of two key genes involved in pigmentation, it remains unclear whether similar results can be achieved for genes involved in more complex metabolic pathways or those expressed at lower levels. Future research should focus on extending this method to a broader range of target genes and other organs beyond the pericarp. Moreover, combining VIGS with emerging technologies such as CRISPR/Cas-based systems could improve the precision of gene manipulation strategies for recalcitrant woody plants.
Conclusions
This study developed a novel, efficient, and cost-effective VIGS system for C. drupifera, offering an invaluable tool for functional genomics research in recalcitrant tissues. By successfully silencing CdCRY1 and CdLAC15, we demonstrated the capacity of this system in addressing challenges posed by lignified plant tissues. These findings pave the way for broader application of VIGS in woody plants, potentially advancing genetic research in other economically important species that are difficult to transform. Future research should focus on refining infiltration methods and extending the system’s application to additional genes and tissues, further enhancing its versatility in plant genomics.
Supplementary Information
Supplementary Material 2. Fig. S1. For this study, we selected Camellia drupifera. The selected trees, aged 20 years, were free from pests and diseases and exhibited vigorous growth and healthy development.
Supplementary Material 3. Fig. S2. Construction of pNC-TRV2-CdCRY1 and pNC-TRV2-CdLAC15 vectors in C. drupifera. These vectors were derived from the pTRV2 vector, with the NC cloning frame inserted at the cloning site of the original vector. The primers used were TRV2F: ggcggttcttgtgtgtgtcaac and TRV2R: caagatcagtcgagaatgtc.
Supplementary Material 4. Fig. S3. The experimental site was located at E 114°34′, N 23°23′. Three test zones, represented by X, Y, and Z circles. In each zone, 16 trees with similar growth conditions were selected for infiltration; 4 trees per gene for each infiltrating approach)
Supplementary Material 5. Fig. S4. C. drupifera capsules with negative GFP signalsand positive GFP signals. Scale bar = 50 µm.
Acknowledgements
We thank Fengji Oil Tea Base (Huizhou City, Guangdong Province, China) for providing plant materials. Special thanks to Dr. Pu Yan of the Chinese Academy of Tropical Agricultural Sciences (CAATS) for his help with the experimental strains.
Abbreviations
- ACNs
Anthocyanins
- AS
Acetosyringone
- CCE
Cryptochrome C-terminal
- CdCRY1
Cryptochrome1
- CdLAC15
Laccase15
- CdPPIL
Peptidyl-prolyl cis–trans isomerase
- Cu-oxidase
Multicopper oxidase
- Cu-oxidase_2
Multicopper oxidase 2
- Cu-oxidase_3
Multicopper oxidase 3
- DAP
Data after pollination
- DMACA
0.2% W/v DMACA in methanol-3 N HCI
- FAD_binding
Photolyase/cryptochrome alpha/beta domain profile
- HMM
Hidden markov model
- HPLC
High Performance Liquid Chromatography
- TRV
Tobacco rattle virus
- MES
2-Morpholinoethanesulphonic acid
- MgCl2
Magnesium chloride
- PAs
Proanthocyanidins
- PHR
Photolyase/cryptochrome alpha/beta domain profile
- VIGS
Virus-induced gene silencing
- YEB
Agrobacterium rhizogenes liquid medium
- TRV2-w/o
TRV2-without gene
Author contributions
B. Z. and Y. L. designed the project and revised the manuscript. H. S. conducted the experiments, including the VIGS optimization and gene silencing assays. H. C. contributed to the analysis and interpretation of the data. W. L. and S. H. assisted in sample preparation and molecular biology experiments. B. L. and W. X. performed bioinformatics analyses related to the identification of target genes. Y. S.,Y. L.and Y. G. supervised the project and provided critical revisions to the manuscript. H. S. and H. C. wrote the manuscript with input from all authors. All authors read and approved the final manuscript.
Funding
This research was funded by the Guangdong Provincial Forestry Science and Technology Innovation Project (2024KJCX004); Guangdong Basic and Applied Basic Research Foundation (2024A1515010363); Guangdong Provincial Forestry Science and Technology Innovation Project (2023KJCX006).
Availability of data and materials
The raw transcriptome data in this study came from the NCBI Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra), accession number PRJNA870661 [31]. The reference genome used in this study is publicly available from GitHub: https://github.com/Hengfu-yin/CON_genome_data or Zenodo: https://zenodo.org/record/5768785 for public access [32].
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hongjian Shen, Huajie Chen and Weimeng Li contributed equally to this work.
Contributor Information
Yong Quan Li, Email: yongquanli@zhku.edu.cn.
Bipei Zhang, Email: zhangbipei@zhku.edu.cn.
References
- 1.Le VS, Curry AS, Truong QC, Luong VD, Nguyen TL. Camelliaflosculora: a new species of Camellia section Thea series sinenses (Theaceae) from Vietnam. Brittonia. 2021;73:220–8. [Google Scholar]
- 2.Zhang F, Li Z, Zhou J, Gu Y, Tan X. Comparative study on fruit development and oil synthesis in two cultivars of Camelliaoleifera. BMC Plant Biol. 2021;21:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Purkayastha A, Dasgupta I. Virus-induced gene silencing: a versatile tool for discovery of gene functions in plants. Plant Physiol Biochem. 2009;47:967–76. [DOI] [PubMed] [Google Scholar]
- 4.Liu L, Qu J, Wang C, Liu M, Zhang C, Zhang X, et al. An efficient genetic transformation system mediated by Rhizobiumrhizogenes in fruit trees based on the transgenic hairy root to shoot conversion. Plant Biotechnol J. 2024;22:2093–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachan S, Dinesh-Kumar SP. Tobacco rattle virus (TRV)-based virus-induced gene silencing. Methods Mol Biol. 2012;894:83–92. [DOI] [PubMed] [Google Scholar]
- 6.Lange M, Yellina AL, Orashakova S, Becker A. Virus-induced gene silencing (VIGS) in plants: an overview of target species and the virus-derived vector systems. Methods Mol Biol. 2013;975:1–14. [DOI] [PubMed] [Google Scholar]
- 7.Senthil-Kumar M, Mysore KS. New dimensions for VIGS in plant functional genomics. Trends Plant Sci. 2011;16:656–65. [DOI] [PubMed] [Google Scholar]
- 8.Unver T, Budak H. Virus-induced gene silencing, a post transcriptional gene silencing method. Int J Plant Genomics. 2009;2009: 198680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Niu N, Li S, Liu Y, Xue C, Wang H, et al. Virus-induced gene silencing (VIGS) in Chinese jujube. Plants (Basel). 2023;12:2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Zhang D, Xie K, Wang Y, Liao Q, Hong Y, et al. Efficient and high-throughput pseudorecombinant-chimeric cucumber mosaic virus-based VIGS in maize. Plant Physiol. 2021;187:2865–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becker A, Lange M. VIGS—genomics goes functional. Trends Plant Sci. 2010;15:1–4. [DOI] [PubMed] [Google Scholar]
- 12.Zulfiqar S, Farooq MA, Zhao T, Wang P, Tabusam J, Wang Y, et al. Virus-induced gene silencing (VIGS): a powerful tool for crop improvement and its advancement towards epigenetics. Int J Mol Sci. 2023;24:5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panwar V, Kanyuka K. Virus-induced gene silencing in wheat and related monocot species. Methods Mol Biol. 2022;2408:95–107. [DOI] [PubMed] [Google Scholar]
- 14.Peng K, Xue C, Huang X. Enhancing virus-induced gene silencing efficiency in tea plants (Camelliasinensis L.) and the functional analysis of CsPDS. Sci Hortic. 2024;337: 113585. [Google Scholar]
- 15.Liu S, Zhang L, Gao L, Chen Z, Bie Y, Zhao Q, et al. Differential photoregulation of the nuclear and cytoplasmic CRY1 in Arabidopsis. New Phytol. 2022;234:1332–46. [DOI] [PubMed] [Google Scholar]
- 16.Mao Z, Wei X, Li L, Xu P, Zhang J, Wang W, et al. Arabidopsis cryptochrome 1 controls photomorphogenesis through regulation of H2A.Z deposition. Plant Cell. 2021;33:1961–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiao J, Zheng H, Zhou X, Huang Y, Niu Q, Ke L, et al. The functions of laccase gene GhLAC15 in fiber colouration and development in brown-colored cotton. Physiol Plant. 2024;176: e14415. [DOI] [PubMed] [Google Scholar]
- 18.Wei J, Zhang X, Zhong R, Liu B, Zhang X, Fang F, et al. Laccase-mediated flavonoid polymerization leads to the pericarp browning of litchi fruit. J Agric Food Chem. 2021;69:15218–30. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD. HMMER web server: 2018 update. Nucleic Acids Res. 2018;46:W200–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez-Pozo N, Rosli HG, Martin GB, Mueller LA. The SGN VIGS tool: user-friendly software to design virus-induced gene silencing (VIGS) constructs for functional genomics. Mol Plant. 2015;8:486–8. [DOI] [PubMed] [Google Scholar]
- 22.Kj L, Td S. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 23.Wang P, Ma G, Zhang L, Li Y, Fu Z, Kan X, et al. A sucrose-induced MYB (SIMYB) transcription factor promoting proanthocyanidin accumulation in the tea plant (Camelliasinensis). J Agric Food Chem. 2019;67:1418–28. [DOI] [PubMed] [Google Scholar]
- 24.Chen W, Zheng Q, Li J, Liu Y, Xu L, Zhang Q, et al. DkMYB14 is a bifunctional transcription factor that regulates the accumulation of proanthocyanidin in persimmon fruit. Plant J. 2021;106:1708–27. [DOI] [PubMed] [Google Scholar]
- 25.Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 2004;39:734–46. [DOI] [PubMed] [Google Scholar]
- 26.Liu Q, Xu K, Yi L, Hou Y, Li D, Hu H, et al. A rapid, simple, and highly efficient method for VIGS and in vitro-inoculation of plant virus by INABS applied to crops that develop axillary buds and can survive from cuttings. BMC Plant Biol. 2021;21:545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bélanger JG, Copley TR, Hoyos-Villegas V, Charron JB, O’Donoughue L. A comprehensive review of in planta stable transformation strategies. Plant Methods. 2024;20:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galeano E, Vasconcelos TS, Vidal M, Mejia-Guerra MK, Carrer H. Large-scale transcriptional profiling of lignified tissues in Tectonagrandis. BMC Plant Biol. 2015;15:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie L, Zhang Q, Sun D, Yang W, Hu J, Niu L, et al. Virus-induced gene silencing in the perennial woody Paeoniaostii. PeerJ. 2019;7: e7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xingqi Y, Zijia L, Yu L, Jixin Z, Yusheng Z, Dongdong L. Construction and optimization of the TRV-mediated VIGS system in Areca catechu embryoids. Sci Hortic. 2024;338: 113621. [Google Scholar]
- 31.Li Y, Liao B, Wang Y, Luo H, Wang S, Li C, et al. Transcriptome and metabolome analyses provide insights into the relevance of pericarp thickness variations in Camelliadrupifera and Camelliaoleifera. Front Plant Sci. 2022;13:1016475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin P, Wang K, Wang Y, Hu Z, Yan C, Huang H, et al. The genome of oil-Camellia and population genomics analysis provide insights into seed oil domestication. Genome Biol. 2022;23:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 2. Fig. S1. For this study, we selected Camellia drupifera. The selected trees, aged 20 years, were free from pests and diseases and exhibited vigorous growth and healthy development.
Supplementary Material 3. Fig. S2. Construction of pNC-TRV2-CdCRY1 and pNC-TRV2-CdLAC15 vectors in C. drupifera. These vectors were derived from the pTRV2 vector, with the NC cloning frame inserted at the cloning site of the original vector. The primers used were TRV2F: ggcggttcttgtgtgtgtcaac and TRV2R: caagatcagtcgagaatgtc.
Supplementary Material 4. Fig. S3. The experimental site was located at E 114°34′, N 23°23′. Three test zones, represented by X, Y, and Z circles. In each zone, 16 trees with similar growth conditions were selected for infiltration; 4 trees per gene for each infiltrating approach)
Supplementary Material 5. Fig. S4. C. drupifera capsules with negative GFP signalsand positive GFP signals. Scale bar = 50 µm.
Data Availability Statement
The raw transcriptome data in this study came from the NCBI Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra), accession number PRJNA870661 [31]. The reference genome used in this study is publicly available from GitHub: https://github.com/Hengfu-yin/CON_genome_data or Zenodo: https://zenodo.org/record/5768785 for public access [32].