Abstract
Background
Clinical and epidemiological analyses have found an association between coronavirus disease 2019 (COVID-19) and knee osteoarthritis (KOA). Infection with COVID-19 may increase the risk of developing KOA.
Objectives
This study aimed to investigate the potential causal relationship between COVID-19 and KOA using Mendelian randomization (MR) and to explore the underlying mechanisms through a systematic bioinformatics approach.
Methods
Our investigation focused on exploring the potential causal relationship between COVID-19, acute upper respiratory tract infection (URTI) and KOA utilizing a bidirectional MR approach. Additionally, we conducted differential gene expression analysis using public datasets related to these three conditions. Subsequent analyses, including transcriptional regulation analysis, immune cell infiltration analysis, single-cell analysis, and druggability evaluation, were performed to explore potential mechanisms and prioritize therapeutic targets.
Results
The results indicate that COVID-19 has a one-way impact on KOA, while URTI does not play a causal role in this association. Ribosomal dysfunction may serve as an intermediate factor connecting COVID-19 with KOA. Specifically, COVID-19 has the potential to influence the metabolic processes of the extracellular matrix, potentially impacting the joint homeostasis. A specific group of genes (COL10A1, BGN, COL3A1, COMP, ACAN, THBS2, COL5A1, COL16A1, COL5A2) has been identified as a shared transcriptomic signature in response to KOA with COVID-19. Imatinib, Adiponectin, Myricetin, Tranexamic acid, and Chenodeoxycholic acid are potential drugs for the treatment of KOA patients with COVID-19.
Conclusions
This study uniquely combines Mendelian randomization and bioinformatics tools to explore the possibility of a causal relationship and genetic association between COVID-19 and KOA. These findings are expected to provide novel perspectives on the underlying biological mechanisms that link COVID-19 and KOA.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-024-02074-4.
Keywords: COVID-19, Knee osteoarthritis (KOA), Mendelian randomization (MR), Bioinformatics analysis, Acute upper respiratory tract infection (URTI)
Introduction
As the number of COVID-19 survivors increases, the significance of post-COVID-19 syndrome and its persistent adverse symptoms becomes apparent. Osteoarthritis (OA), an age-related degenerative disease, affects approximately 250 million individuals globally, results in the loss of joint function due to the limited capacity of cartilage regeneration [1]. Emerging evidence suggests a potential link between COVID-19 and knee osteoarthritis (KOA). Research has indicated a higher prevalence of KOA in patients with COVID-19 [2]. Patients with COVID-19 often exhibit hypocalcaemia, vitamin D deficiency, and immobility due to the disease, contributing to bone demineralization [3]. Research also indicates the impact of the COVID-19 pandemic on the physical therapy and surgical treatment of patients with KOA, leading to an increased risk of thrombosis after surgery [4–7]. However, there is currently no scientific evidence establishing a direct causal relationship between COVID-19 and KOA. Given the lack of pharmacological or cell-based therapies for OA, especially in the context of the COVID-19 pandemic, it is crucial to comprehend the potential impact of COVID-19 on KOA.
Notably, COVID-19 shares similarities with acute upper respiratory tract infection (URTI). Both illnesses are caused by viruses that can be transmitted through various routes and can lead to severe respiratory symptoms, often requiring hospitalization. Notably, COVID-19 has a longer incubation period than common influenza [8–10]. Thus, our study firstly explores the causal associations between COVID-19, URTI, and KOA by employing the Mendelian randomization (MR) method [11]. Further research is essential to gain a comprehensive understanding of the relationship between COVID-19 and KOA, particularly concerning cellular and molecular mechanisms. With recent advancements in genetic analysis techniques and bioinformatics, our study combines both methods to explore the genetic links between these diseases. Additionally, we assessed the interaction among genes with potential crosstalk to gain deeper insights into the pathophysiological processes connecting KOA with COVID-19. Overall, this research provides new insights into the relationship between COVID-19 and KOA.
Methods
Preparation of GWAS data and bidirectional MR analysis
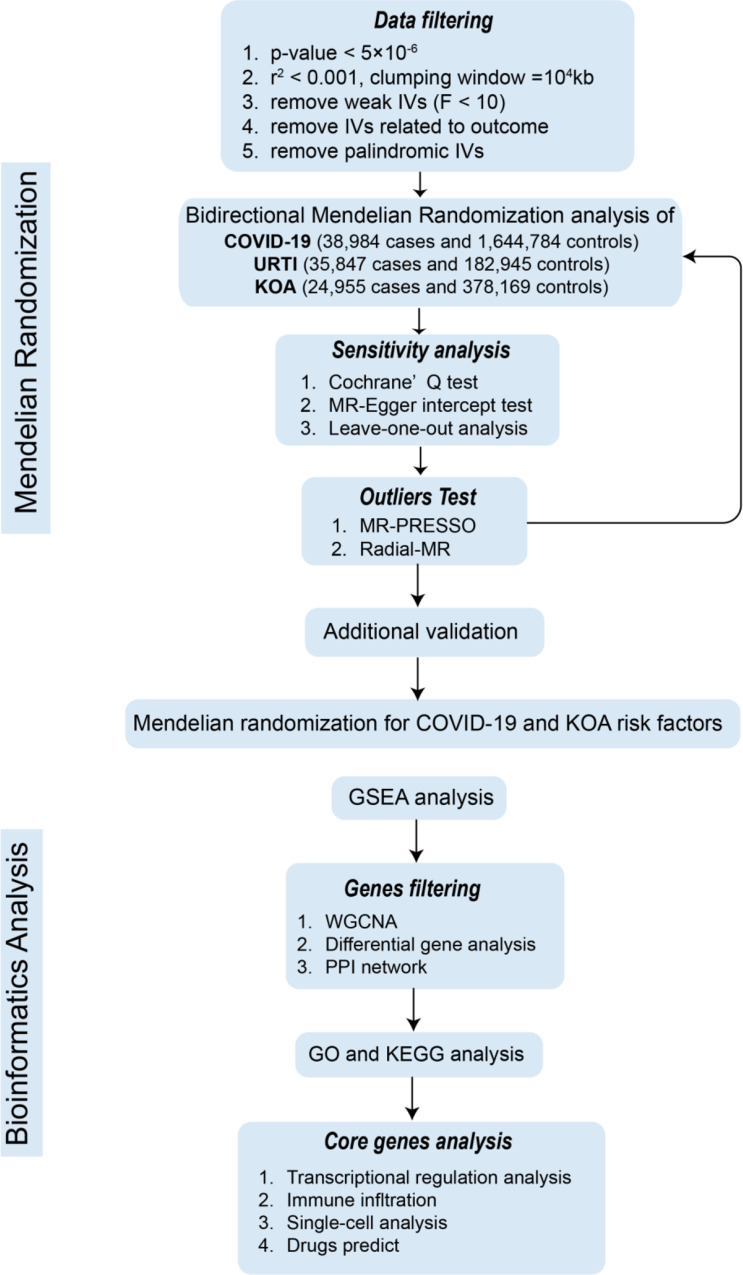
GWAS data for COVID-19, KOA, and URTI were sourced from the IEU OPEN GWAS PROJECT [12], with the following sample sizes: COVID-19 (38,984 cases and 1,644,784 controls), KOA (24,955 cases and 378,169 controls ), and URTI (35,847 cases and 182,945 controls). The KOA data came from a meta-analysis primarily involving participants aged 40–69, both male and female, while the COVID-19 and URTI studies did not focus on specific age or gender groups. A GWAS explores genotypic variations, particularly single-nucleotide polymorphisms (SNPs), throughout the entire genome to establish the association between genes and traits. To preserve the independence of SNPs associated with exposure, we implemented criteria of r2 > 0.001 and a clump window distance greater than 10,000 kb. Furthermore, a significance level of p-value < 5e-6 was established for the GWAS to ensure sufficient statistical power in the MR analysis. The effectiveness of the instrument was evaluated using the F statistic, with a threshold of greater than 10 for all SNPs incorporated in the analysis [13].
The primary analytical approach utilized in this study was the inverse variance weighted (IVW) method [14], where COVID-19 and URTI were examined as exposures, while KOA as the outcome. It is crucial to acknowledge that employing SNPs as IVs may introduce certain biases, such as horizontal pleiotropy and heterogeneity. To assess the presence of horizontal pleiotropy, we analysed the intercept of the MR Egger method, MR-PRESSO method and evaluated heterogeneity using both the IVW and MR Egger approaches. Additionally, we calculated the power statistic for the IVW estimates using an online tool, mRnd (https://shiny.cnsgenomics.com/mRnd/) [15]. A statistical power of over 80% is recommended for sufficient reliability. Figure 1 illustrates the study flow.
Fig. 1.
Work flow
Identification of DEGs from transcriptome datasets
The series matrix file data for GSE180226, GSE197143 and GSE206606 were obtained from the NCBI GEO public database. GSE180226 consists of 20 patients with COVID-19 and 3 control patients. E-MTAB-12,184 series matrix file data from the ArrayExpress database (https://www.ebi.ac.uk/biostudies/arrayexpress), which includes 12 transcriptome datasets for 6 healthy subjects and 6 patients with KOA, was also downloaded. GSE197143 and GSE206606 provide a list of DEGs related to URTI. Additional file: Table S1 contains more detailed information. To analyse the datasets, the R software packages “limma” [16] and “DESeq2” [17] were utilized. The criteria used to identify DEGs were |log FC| ≥ 1 and adjusted p-value < 0.05. Volcano plots were created to visualize differential gene clusters.
Gene set enrichment analysis of all genes
We utilized gene set enrichment analysis (GSEA) [18], which relies on predefined gene sets derived from functional annotations or previous experimental results, to rank genes according to their differential expression between two sample types. This approach helps determine whether the predefined gene set is enriched at either the top or bottom of the ranking table. To conduct our analysis, we utilized the R package “clusterProfiler” [19] and investigated the gene expression profiles of GSE180226 and E-MTAB-12,184. We created a comprehensive list of genes and ranked them in descending order based on their fold change values. The reference gene set used was “c5.ontology.all.v2023.1.Hs.symbols.gmt” from the MSigDB database [20]. Statistical significance was determined at an adjusted p-value < 0.05.
WGCNA for coexpression network construction and module identification
Weighted gene coexpression network analysis (WGCNA) is a bioinformatics method utilized to uncover patterns of gene association among various samples. It can identify genes closely related to the development of diseases through clustering and modularization. In this research, we utilized the R package “WGCNA” [21] to establish a gene coexpression network for COVID-19 and KOA individually.
GO and KEGG analyses of DEGs
The genes obtained from the intersection of the module genes identified by WGCNA and the DEGs were considered to have a significant impact on the pathogenesis of COVID-19 and KOA. Consequently, these genes were subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses [22] using the R package “clusterProfiler”. A p-value < 0.05 was used to quantify the top listed functional terms and pathways of common DEGs.
Construction of the protein‒protein interaction network
The STRING database (https://string-db.org/) was utilized to build a protein‒protein interaction (PPI) network. We employed Cytoscape software to visualize the results of coexpression network analysis. Furthermore, we utilized the GeneMANIA online tool (https://genemania.org/) to perform gene coexpression network analysis and predict gene set functions.
Analysis of transcriptional regulatory networks involving core genes
Transcription factors (TFs) play a pivotal role in regulating gene expression. We utilized the KnockTF database (https://bio.liclab.net/KnockTF/index.php) to predict potential TFs that may regulate core genes. Furthermore, the mRNA‒miRNA coregulation network was established by utilizing pertinent data obtained from the miRDB database (https://mirdb.org/). Motif prediction was carried out using the “RcisTarget” package [23] in R. This package calculates the area under the curve (AUC) for each motif-motif pair and computes the normalized enrichment score (NES) based on the distribution of AUC values. Motifs that exhibit higher NES scores are considered to be significantly associated with the research object.
Immune infiltration analysis
The CIBERSORT algorithm [24] was employed to estimate the relative abundance of immune infiltrating cells within the microenvironment. This algorithm analyses the expression matrix of immune cell subtypes using support vector regression (SVM) and deconvolution techniques. We applied the CIBERSORT algorithm to the transcriptome data and utilized the results for Spearman correlation analysis between gene expression and immune cell content. To ensure robustness, we conducted 1000 permutations. This analysis offers valuable insights into the contributions of various immune cell types to the development and progression of diseases.
Single-cell analysis associated with core genes
Single-cell data were preprocessed using the “Seurat” package [25]. Following manual quality control, we employed the “harmony” package [26] to mitigate batch effects. We utilized the UMAP method to identify spatial relationships between each pair of clusters. Subsequently, the clusters were annotated using the cellmarker2.0 database (http://bio-bigdata.hrbmu.edu.cn/CellMarker) and manually curated data.
Potential therapeutic drugs
The SPIED3 tool (http://www.spied.org.uk/cgi-bin/hgnc-spied3.cgi) was used to predict potential therapeutic drugs using the Connectivity Map 2.0 datasets [27]. This tool identifies small molecule drugs that exhibit negative correlations with the expression profiles of DEGs.
Results
Mendelian randomization (MR) analysis
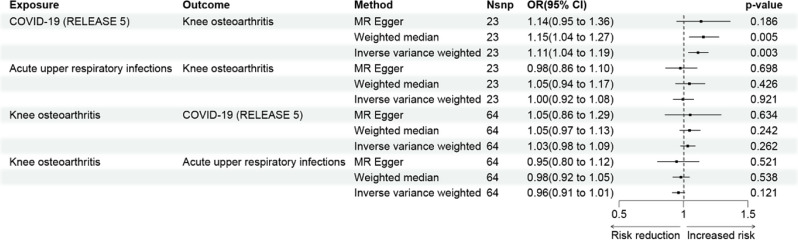
The findings from the MR analysis revealed a negative genetic effect of COVID-19 on KOA, as indicated by an odds ratio (OR) of 1.11 (95% confidence interval [CI]: 1.04–1.19, p-value = 0.003) (Fig. 2). In accordance with the Bonferroni correction, a p-value below 0.025 (0.05/2) is considered to indicate statistically significant evidence of a causal relationship. Therefore, our findings demonstrate statistical significance. However, no statistically significant association was observed between URTI and KOA (p-value > 0.025). Additional analyses, including sensitivity analysis, heterogeneity testing, and horizontal pleiotropy testing, were conducted to assess potential bias in the MR results. No significant bias was detected (Additional file: Table S2 and S3). Reverse MR analysis was also performed to examine the influence of KOA as an exposure on COVID-19 and URTI, but no statistically significant associations were found. By setting α at 5%, we achieved a significant statistical power of 0.95 (> 0.8). This indicates that the independent variables provided accurate estimates of causal effects, ensuring the reliability of the results.
Fig. 2.
Potential causal associations of COVID-19, URTI and KOA in MR analyses
Moreover, we conducted additional validation, which further supports our view that COVID-19 infection may have negative effects on the knee joint (Additional file: Table S4). After excluding outliers using Radial-MR method (Additional file: Figure S1), the next MR analysis passed the outlier test. The IVW method showed a P-value of 0.0002.
It was also hypothesized that COVID-19 could influence the risk of KOA through a KOA risk factor. To investigate this further, additional MR analysis was conducted using genome-wide association studies (GWAS) of potential KOA risk factors as outcomes. These risk factors included body mass index (BMI), osteoporosis, pain in joints, muscle or soft tissue injuries, and metabolic disorders such as type 1 and type 2 diabetes. Among these factors, only BMI showed MR evidence indicating that COVID-19 could potentially lead to higher BMI (IVW: PFDR =0.006; Additional file: Table S5).
In conclusion, these findings suggest that COVID-19 may have a negative impact on the development of osteoarthritis.
Differential gene expression analysis
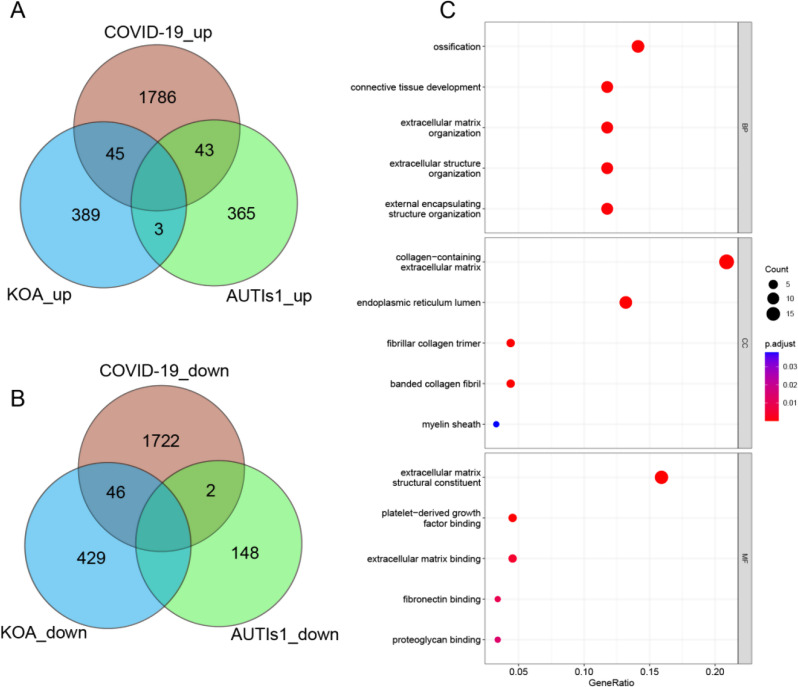
For the transcriptome analysis of COVID-19, we performed differential gene expression analysis by comparing 19 disease cohorts with 3 control cohorts (one disease sample was excluded to ensure data integrity and precision). The analysis revealed a total of 3644 DEGs, with 1874 upregulated and 1770 downregulated (Additional file: Figure S2A). For the transcriptomic analysis of KOA, we obtained the E-MTAB-12,184 series matrix data file from ArrayExpress in BioStudies. We analysed a total of 12 transcriptome datasets, encompassing 6 datasets from healthy subjects and 6 datasets from patients with KOA. This analysis led to the identification of 912 DEGs (Additional file: Figure S2B), with 437 genes exhibiting increased expression and 475 genes with decreased expression. Additionally, we selected 411 upregulated DEGs and 150 downregulated DEGs based on the same control standards utilized for URTI from GSE206606. We also identified 89 upregulated DEGs and 1 downregulated DEG from GSE197143 (Additional file: Figure S2C and S2D). Subsequently, we conducted a search to identify common DEGs between COVID-19 and KOA, excluding DEGs associated with URTI. This effort resulted in the identification of 91 DEGs (Fig. 3A and B; Additional file: Table S6).
Fig. 3.
Identification of common DEGs. A-B Venn diagram identifies coupregulated and codownregulated DEGs of KOA, COVID-19, and URTI. C GO enrichment analysis of 91 DEGs
Gene set enrichment analysis of all genes
Due to the limited information obtained from GO and KEGG enrichment analyses, we employed GSEA, a method capable of examining enrichment signals, to perform a more comprehensive analysis [18]. The GSEA results revealed that 572 items from E-MTAB-12,184 and 341 items from GSE180226 were enriched in the hallmark gene set. Subsequently, we examined the intersection of pathways and functions shared between these datasets, which led to the identification of 16 commonly inhibited items and 23 commonly activated items in the disease group compared to the healthy group (Additional file: Figure S3). These results demonstrated negative enrichment primarily in ribosome- and mRNA-related biological processes, while positive enrichment was observed in signalling receptors and extracellular matrix-related biological processes. These findings provide valuable insights into the potential interaction between COVID-19 and KOA, warranting further investigation.
GO and KEGG analyses of DEGs
We conducted functional enrichment analysis on 91 differentially expressed genes (DEGs) identified from two separate datasets. This analysis covered four main categories: biological process (BP), cellular component (CC), molecular function (MF), and KEGG pathways.
In BP analysis, the results highlighted the processes related to ossification, connective tissue development, extracellular matrix organization, and the organization of extracellular and external encapsulating structures, indicating their importance in bone development. CC analysis revealed significant enrichment of components such as the collagen-containing extracellular matrix, endoplasmic reticulum lumen, and collagen trimers. MF analysis emphasized functions related to extracellular matrix structural constituents, glycosaminoglycan binding, and sulfur compound binding (Fig. 3C). KEGG pathway analysis identified the top 7 enriched pathways, including the PI3K/AKT signalling pathway, ECM-receptor interaction, protein digestion and absorption, focal adhesion, and human papillomavirus infection. These findings suggest their potential relevance in the context of these DEGs (Additional file: Figure S4).
WGCNA construction and hub module identification
WGCNA is a bioinformatics approach that focuses on the connection between clinical features and coexpression modules, yielding comprehensive, reliable, and biologically significant study results [28]. Genes within the same module are believed to be functionally interconnected, allowing for the identification of biologically significant modules and hub genes that may be shared by COVID-19 and KOA. In our analysis of the expression profile data of COVID-19, we conducted WGCNA with a threshold parameter β set to 5 (Additional file: Figure S5A). This analysis led to the identification of eight distinct gene modules. Notably, the brown module showed the strongest positive correlation with the disease state, with a correlation coefficient of 0.79 (p-value = 1e-5), as depicted in Additional file: Figure S5B. This module contains 691 genes. Expanding our analysis to encompass the expression spectrum of KOA, we identified twelve gene modules using a threshold β value of 9 (Additional file: Figure S5C). Among these, the blue module exhibited the highest positive association with the disease state, with a correlation coefficient of 0.92 (p-value = 2e-5), as illustrated in Additional file: Figure S5D. This module encompasses 1622 genes.
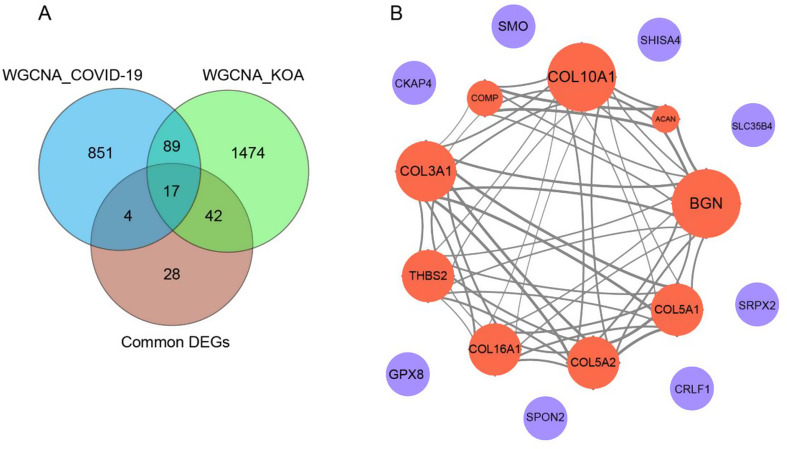
We extended our analysis by conducting an intersection analysis, comparing the brown module of COVID-19 with the blue module of KOA. This analysis revealed 106 intersecting genes. When cross-referencing this gene set with the 91 previously identified common DEGs of COVID-19 and KOA, we ultimately identified a group of 17 common genes, as shown in Fig. 4A.
Fig. 4.
A Venn diagrams identify the intersection of genes between WGCNA module genes and common DEGs. B Visualization of the protein‒protein interaction (PPI) network of the 17 genes. Red indicates upregulated DEGs and blue downregulated genes in COVID-19 and KOA
Protein‒protein interaction network construction
PPI analysis plays a vital role in uncovering potential interactions and functional associations among shared target genes. In our study, we integrated the 17 shared target genes into a PPI network using the online search tool STRING. This network consists of 9 nodes and 54 edges, with a PPI interaction score exceeding 0.4 (Fig. 4B). Notably, the nine nodes corresponding to the upregulated DEGs exhibited network interactions, whereas the downregulated DEGs did not. This suggests that the upregulated DEGs may have greater biological significance and play crucial roles in the pathological process of KOA in relation to COVID-19.
Furthermore, we used GeneMANIA to perform biological function analysis to identify genes that share properties and functions with the nine core genes (Additional file: Figure S6A). This analysis revealed 20 closely associated genes, demonstrating coexpression, physical interactions, shared protein domains, pathways, and colocalization that underlie their functional associations. The primary functions of these genes involve extracellular matrix structural constituents, collagen trimers, collagen-containing extracellular matrix, and growth factor binding.
To further investigate the molecular mechanisms linking the association between KOA and COVID-19, we conducted an analysis of GO and KEGG pathway annotations for the nine shared core genes. As anticipated, all nine genes displayed significant enrichment for the ontology terms “extracellular matrix” and “protein digestion and absorption” (Additional file: Figure S6B-E). Overall, these nine genes (COL10A1, BGN, COL3A1, COMP, ACAN, THBS2, COL5A1, COL16A1, COL5A2) were identified as the core genes for further investigation in our study.
Immune infiltration analysis
The immune microenvironment plays a pivotal role in the development of COVID-19 and KOA and has significant implications for the diagnosis, prognosis, and treatment response of patients. In this study, we investigated the relationship between the nine aforementioned genes and immune cell infiltration across two datasets. Our objective was to discern differences in immune cell presence and gene correlation between disease and healthy control groups.
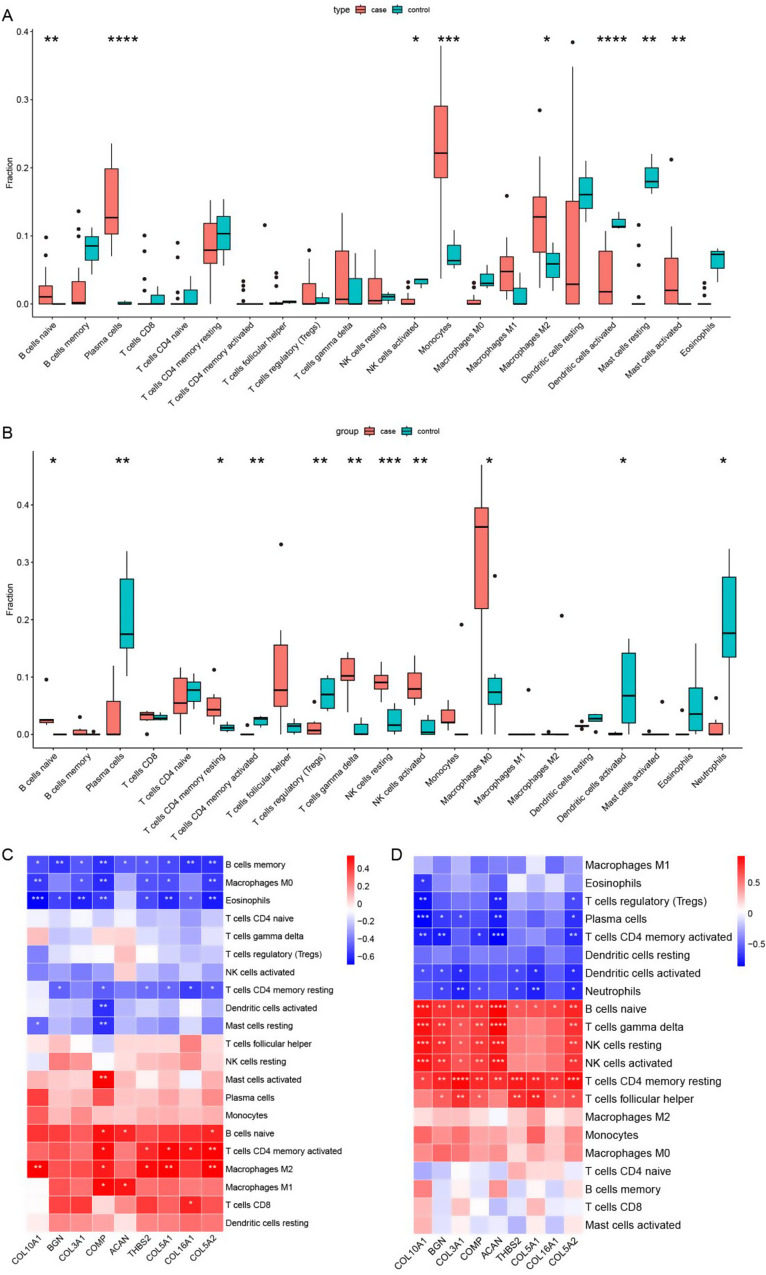
We employed the CIBERSORT algorithm to analyse the DEGs associated with COVID-19 and DEGs associated with KOA separately. In the COVID-19 dataset (GSE180226), we quantified the fractions of twenty-two distinct types of immune cells, which were visually represented using a box plot (Fig. 5A). Similarly, we illustrated the distribution of these immune cells using the DEGs obtained from the KOA dataset (E-MTAB-12184), and the results are presented in Fig. 5B. Additionally, we visually presented correlations between these 22 immune cell types and the nine core genes in the two datasets, as depicted in Fig. 5C and D.
Fig. 5.
Analysis of immune infiltration associated with COVID-19 and KOA. A-B Box plot showing the distribution of immune cells in the COVID-19 and KOA samples, A for COVID-19 and B for KOA. The round dots indicate the outliners. C-D The correlation between the nine core genes and immune cells, C for COVID-19 and D for KOA. The symbol * (A-D) represents p-value < 0.05, **p-value < 0.01, ***p-value < 0.001, **** p-value < 0.0001
We noted substantial disparities in the distribution and proportions of various immune cell types between the COVID-19 and healthy control groups. The case group had higher infiltration fractions for naive B cells, plasma cells, monocytes, M2 macrophages, and activated mast cells, while the infiltration fractions of activated NK cells, resting dendritic cells, and resting mast cells were lower compared to the control group. Similarly, when comparing KOA patients to healthy controls, we observed an increased infiltration fraction of naive B cells, resting memory CD4 T cells, resting gamma delta T cells, resting NK cells, activated NK cells, and M0 macrophages. In contrast, there was a decreased infiltration fraction of plasma cells, activated memory CD4 T cells, regulatory T cells (Tregs), activated dendritic cells, and neutrophils within the cartilage of KOA patients. Interestingly, naive B cells exhibited a significant increase in infiltration in both COVID-19 and KOA samples.
Correlation analysis between the 22 immune cell types and the nine identified core genes revealed significant positive correlations of the core genes with naive B cells, resting memory CD4 T cells, and M2 macrophages in the COVID-19 dataset. However, negative correlations were observed with memory B cells, M0 macrophages, and eosinophils. The gene COMP displayed a significant positive correlation with activated mast cells, while showing a negative correlation with activated dendritic cells and resting mast cells. Furthermore, in the KOA dataset, the same set of genes exhibited positive correlations with naive B cells, gamma delta T cells, resting NK cells, activated NK cells, resting memory CD4 T cells, and follicular helper T cells, while displaying a negative correlation with plasma cells, activated memory CD4 T cells, activated dendritic cells, and neutrophils. Additionally, the correlation analysis between immune cells and core genes demonstrated positive correlations for naive B cells and resting memory CD4 T cells in both COVID-19 and KOA samples. However, eosinophils exhibited a negative correlation with these genes in both sets of samples. These findings provide further evidence for the crucial role played by immune cells in the pathogenic processes of COVID-19 and KOA.
Transcriptional regulation analysis
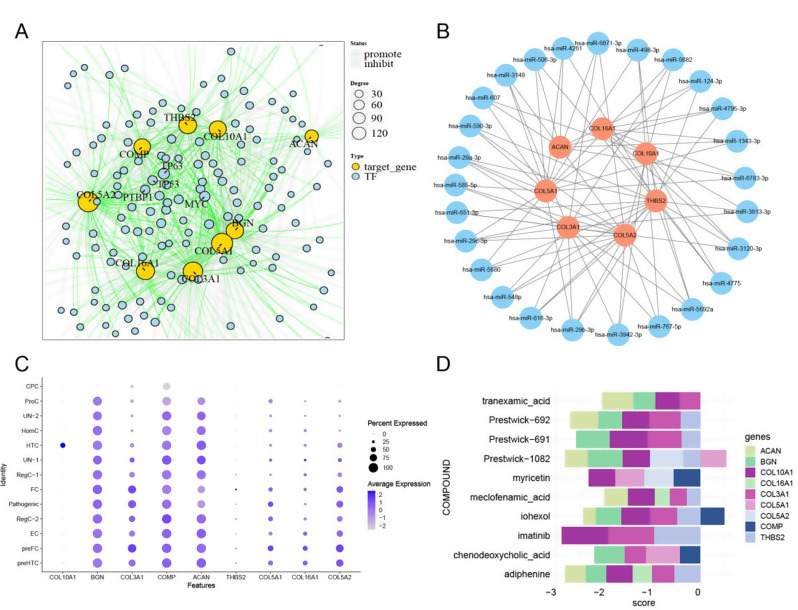
Analysis of transcriptional regulation is a critical aspect in understanding the complex interactions among integrative TFs, miRNAs, and hub genes. It provides valuable insights into the underlying biological processes involved in disease pathogenesis [29–31]. In this study, we utilized the KnockTF database to identify 223 TFs for the nine core genes. The TF-gene relationship pairs resulting from the analysis are visualized in Fig. 6A. Among these TFs, TP63 and MYC exhibited the highest degree of connectivity, with 20 and 16 connections, respectively. This suggests their potential major roles in different cell types. TP63 was found to inhibit the expression of the genes BGN, COL10A1, COL16A1, COL5A1, and COMP while promoting that of THBS2. In contrast, MYC was found to inhibit the expression of the ACAN, BGN, COL16A1, COL3A1, COL5A1 and COMP genes but promote that of THBS2.
Fig. 6.
A TF networks of core genes, the yellow line denotes promotion and the grey line inhibition. Only the nodes with degrees exceeding 10 are presented by their names. B mRNA‒miRNA networks of hub genes, with red indicating mRNA and blue miRNA. C Expression profiles of the nine core genes in different cellular subtypes of cartilage tissue. D Potential therapeutics for KOA patients with COVID-19
MicroRNAs (miRNAs) are short noncoding RNA molecules that play a crucial role in gene regulation by binding to mRNA molecules, inhibiting translation, or causing mRNA degradation of target genes [32, 33]. In our study, we further investigated whether miRNAs associated with the hub genes regulate the transcription or degradation of specific risk genes. miRNAs associated with the nine shared core genes were obtained from the miRDB database, and the miRNA-gene network was visualized using Cytoscape software. The resulting network comprises 617 nodes and 751 edges. Only nodes with degrees greater than 3 are displayed (Fig. 6B). Remarkably, hsa-miR-5692a (target mRNA: THBS2, COL10A1, COL3A1, COL5A2), hsa-miR-29b-3p (target mRNA: COL3A1, COL5A1, COL5A2, THBS2), hsa-miR-29c-3p (target mRNA: COL3A1, COL5A1, COL5A2, THBS2), hsa-miR-29a-3p (target mRNA: COL3A1, COL5A1, COL5A2, THBS2), and hsa-miR-3120-3p (target mRNA: THBS2, COL3A1, COL10A1, COL16A1) were identified as the most connected miRNAs, suggesting their potential interactive role in the two diseases.
Additionally, we performed motif TF annotation and key gene screening. The motif transfac_pro_M01583 had the highest normalized enrichment score (NES) of 6.99. Genes BGN, COL10A1, and COL3A1 were found to be enriched in this motif. The enriched motifs and their corresponding TFs are visually represented in Additional file: Figure S7.
Core gene expression in single cells
We conducted single-cell analysis using the GSE220243 dataset. The dataset was processed using the “Seurat” R package, employing the UMAP algorithm to cluster all cells into thirteen distinct classes. To accurately classify the cells, we utilized annotation data from CellMarker 2.0 and curated data from relevant research papers. The comprehensive classification of cells includes prehypertrophic chondrocyte (preHTC), prefibrocartilage chondrocyte (preFC), effector chondrocyte (EC), regulatory chondrocyte (RegC), fibrocartilage chondrocyte (FC), hypertrophic chondrocyte (HTC), homeostatic chondrocyte (HomC), proliferative chondrocyte (ProC), pathogenic chondrocyte (pathogenic), cartilage progenitor cell (CPC), and two additional unidentified categories.
To gain further insights into the intricacies of these cell types, we examined the expression levels of the nine genes across these cell types. This analysis, as depicted in Fig. 6C, provided critical information about the specific gene expression patterns within the different chondrocyte subtypes. Notably, the genes COL10A1 and THBS2 were found to be specifically expressed in HTC and FC. However, COL3A1, COL5A1 and COL5A2 showed higher expression in FC, pathogenic, preFC and preHTC. BGN, COMP, and ACAN were found to be highly expressed in almost all cell subtypes.
Small molecule drug prediction
In the final phase of our study, we conducted further investigations into potential drugs based on the nine core genes. Initially, we utilized the SPIED3 tool to predict target drugs (Fig. 6D). The input for this analysis consisted of the core genes and their corresponding LogFC values obtained from the differential gene expression analysis of the KOA dataset. The results indicated that the compound “imatinib” had the lowest score of “-2.89”, suggesting its potential effectiveness in inhibiting the expression of three genes: COL10A1 (-0.98), THBS2 (-0.96), and COL3A1 (-0.95). These findings provide valuable insights for preventing and treating KOA in the aftermath of the COVID-19 pandemic.
Discussion
In the wake of the COVID-19 pandemic, there has been a dedicated research effort to understand its underlying complications and consequences [34, 35]. KOA, characterized by articular cartilage abrasion, is an age-related degenerative disease that affects millions of people worldwide. Although KOA itself does not directly threaten patients’ lives, its chronic and progressive nature can significantly impact their quality of life and even increase mortality rates [3, 36]. Increasingly, studies are confirming the link between COVID-19 and KOA. Overall, how the impact of COVID-19 on KOA and the most effective treatment approaches remain unclear, and the relationship between COVID-19 and KOA is not fully understood. Therefore, investigating the mechanism of how COVID-19 affects KOA has crucial clinical significance for early recognition and intervention.
COVID-19 can impact KOA in one direction, but URTI does not play a causal role
In this study, bidirectional MR analysis was initially employed to explore potential causal links among three conditions: COVID-19, KOA, and URTI. The results indicated a significant association between KOA and COVID-19. However, we did not observe a significant association between URTI and KOA. Notably, despite the considerable similarities between COVID-19 and URTI, such as their viral origins and modes of transmission through droplets, airborne aerosols, or direct contact, as well as the presence of common clinical symptoms such as fever, cough, and respiratory distress, our MR analysis yielded distinct findings. These differences in causality between COVID-19 and URTI may be attributed to subtle genetic variations between the two conditions. Further investigation is essential to gain a comprehensive understanding of the underlying factors regarding how COVID-19 affects KOA.
Ribosomal dysfunction could play a mediating role in connecting COVID-19 with KOA
GSEA results revealed predominantly negative enrichment in ribosome- and mRNA-related biological processes. Ribosomes, which function as protein synthesis factories in cells, play a crucial role in infection and the human antiviral response. Studies have demonstrated that viral nonstructural proteins (NSPs) interact with various ribosomal states, disrupting mRNA translation. Notably, SARS-CoV-2 interferes with ribosome mRNA translation and employs multiple strategies, including degrading host mRNA and blocking host mRNA export, in addition to inhibiting host mRNA translation [3]. There is a growing body of experimental evidence suggesting that molecular mechanisms associated with ribosome biogenesis and activity are dysregulated in OA [37–39]. These ribosome abnormalities occur at various stages of ribosome biogenesis, from early development to maturation, resulting in preferential translation in OA [40]. Although traditional explanations for OA have centred around an imbalance between the anabolism and catabolism of joint tissue, Van and colleagues have proposed an alternative perspective. They consider OA as an acquired ribosomopathy and suggest that targeting ribosomes offers a novel approach for developing disease-modifying treatments for OA [41]. Overall, further research is warranted to explore the effects of ribosomes on KOA.
COVID-19 may influence the metabolic processes of the extracellular matrix, potentially impacting the normal development of joints
Identifying significant gene ontology and molecular pathways has enhanced our understanding of how COVID-19 increases the risk of morbidity in KOA. Our findings indicated that the 91 common DEGs exhibit strong associations with the ECM. Additionally, the GO and KEGG pathway annotations of the nine core genes shared among the DEGs support this conclusion. This suggests that the nine core genes play a crucial role in the construction and degradation of the ECM, as well as other essential biological processes.
The knee joint is a complex structure consisting of various tissues, including articular cartilage, the synovial membrane, the joint capsule, menisci, subchondral bone, infrapatellar and suprapatellar fat pads, and tensile connective tissues such as tendons and ligaments [42]. Articular cartilage comprises a dense ECM with sparsely distributed chondrocytes displaying varying morphology and potentially diverse functions [43]. The ECM constitutes a complex network of proteoglycans, collagens, water, minerals, and fibrous proteins, imparting biomechanical properties to the articular cartilage [44]. Under normal conditions, chondrocytes maintain homeostasis of articular cartilage by regulating the synthesis and degradation of ECM [45]. However, this balance is disrupted by injury, inflammation, or other stimuli, leading to chondrocyte stress, ECM degradation, and abnormal accumulation of damaged proteins. Organelles such as the endoplasmic reticulum and mitochondria may also become dysfunctional. These events culminate in the degradation of cartilage, apoptosis of chondrocytes, and dysfunction of the subchondral bone, ultimately resulting in KOA [7]. Moreover, COVID-19 induces a systemic inflammatory response and abnormal expression of ECM components [46, 47]. The ECM is regulated by SARS-CoV-2 infection and plays a crucial role in the pathogenesis of the infection. An overactive ECM can exacerbate the progression of disease and its regulation can help alleviate symptoms [48].
Additionally, several studies have reported the involvement of inflammatory signalling pathways, including the PI3K/AKT, IL-17, TNF, NF-κB, and MAPK signalling pathways, in the osteoarthritic process [49–52]. It has been observed that activation of the PI3K/AKT pathway inhibits chondrocyte apoptosis and promotes chondrocyte proliferation [53, 54]. Moreover, the ECM-receptor interaction pathway is a critical signalling and metabolic pathway in progression of KOA [55]. The PI3K/AKT signalling pathway plays a pivotal role in regulating cell proliferation and maintaining the biological characteristics of malignant cells [56]. Overall, there is a strong connection between the PI3K/AKT signalling pathway and the ECM. Research has indicated that regulation of the PI3K/AKT signalling pathway can influence degradation of the cytoplasmic matrix [57]. Thus, investigating alterations in the ECM during the pathological mechanisms of COVID-19 and KOA may contribute to a better understanding and more effective management of KOA after COVID-19.
The nine fundamental genes exhibit a robust correlation with the occurrence of knee osteoarthritis
A specific set of genes (COL10A1, BGN, COL3A1, COMP, ACAN, THBS2, COL5A1, COL16A1, COL5A2) was identified as the common transcriptomic signature in response to KOA with COVID-19. COL10A1 is specifically expressed by HTC. It plays a vital role in the deposition of other matrix molecules within the hypertrophic zone, creating an environment essential for haematopoiesis, mineralization, and modelling in endochondral ossification [58]. BGN is a small proteoglycan highly expressed in the growing skeleton and human skin, specifically in differentiating keratinocytes [59]. It belongs to the small leucine-rich proteoglycan (SLRP) family of proteins. BGN is involved in bone growth, muscle development and regeneration, and collagen fibril assembly across various tissues. Additionally, it may play a role in regulating inflammation and innate immunity. COL3A1 is found in most soft connective tissues, along with type I collagen and plays a role in regulating cortical development. Gene expression analysis has confirmed increased fibrogenesis in COVID-19 patients, as evidenced by the overexpression of genes related to collagen biosynthesis and ECM biosynthesis and degradation, including COL3A1 and COL5A1. Notably, COL3A1 is overexpressed in both COVID-19 and cases of usual interstitial pneumonia/idiopathic pulmonary fibrosis (UIP/IPF) [60]. COL3A1 is also a potential diagnostic biomarker for OA [61]. In the lungs of COVID-19 patients, there was a significant increase in key collagen and matricellular transcripts, particularly type III procollagen (COL3A1). This increase was observed to be higher than the levels seen in patients with influenza A or interstitial lung disease (ILD) [62]. COMP is a noncollagenous ECM protein that plays a structural role in cartilage by interacting with other ECM proteins, including collagens and fibronectin. It also mediates the interaction of chondrocytes with the cartilage ECM through cell surface integrin receptors [63, 64]. ACAN is a major proteoglycan in the ECM of cartilaginous tissues and plays a primary role in providing resistance to compression in cartilage. Studies have shown an increased expression of the Nos2 gene and a decreased expression of the Acan gene in cartilage tissue from rats with experimental OA compared to control animals. This indicates the activation of inflammatory and destructive processes in the tissue [65]. Various ACAN gene variants have also been linked to skeletal disorders [66]. THBS2 is an adhesive glycoprotein that mediates cell-to-cell and cell-to-matrix interactions. It may serve as a novel biomarker in end-stage OA and plays a role in ECM-receptor interactions [67–70]. As members of the collagen family of proteins, COL5A1 and COL5A2 are minor connective tissue components that are widely distributed throughout the body. They bind to various molecules and play an essential role in regulating the assembly of tissue-specific matrices [71–73]. COL16A1 is a member of the collagen family associated with fibril-forming collagens, such as type I and II collagens. It helps to maintain the integrity of the ECM and is highly expressed in fibroblasts, keratinocytes, smooth muscle, and the amnion [74].
Interestingly, these genes were upregulated and interacted with each other, while the remaining genes were downregulated and did not exhibit any distinct connections. These nine core genes, which have similar functions, may have significant implications in the onset and progression of KOA with COVID-19 when their expression is increased. They could serve as potential targets for therapeutic intervention.
The miR-29 family may participate in the regulatory process of the pathogenesis and development of KOA with COVID-19
In recent years, miRNAs have gained increasing attention. These small noncoding RNAs, typically 21–25 nucleotides long, consist of sequences that complement the 3′ UTR of their target mRNAs, leading to mRNA degradation or inhibition of mRNA translation [75]. miRNA subsets have clinical relevance as biomarkers, offering insights into the occurrence, progression, genetic links, and stages of diseases. The field of miRNA therapeutics has rapidly advanced due to the use of bioinformatic approaches to identify miRNA-binding sites, understand their associated biological pathways in target genes, and the availability of in vitro and in vivo preclinical research models [76].
In our study, we constructed an mRNA‒miRNA regulatory network, with hsa-miR-5692a, hsa-miR-29b-3p, hsa-miR-29c-3p, hsa-miR-29a-3p, and hsa-miR-3120-3p exhibiting the highest average connectivity among the nine core genes. These miRNAs are closely related to key genes, as indicated by bioinformatics analysis, suggesting their potential pivotal roles in the pathogenesis of OA. Studies have shown that the miR-29 family, a group of microRNAs, plays an early role in the development of OA. Their expression is also regulated in cartilage during the progression of OA [77]. Notably, there are multiple binding sites for miR-29a-3p and miR-29b-3p on the SARS-CoV-2 genome, and they target DEGs involved in the immune response. This suggests their potential utility as biomarkers and therapeutic agents [78, 79]. Studies have demonstrated that hsa-miR-29b-3p regulates mRNA targets involved in endothelial dysfunction and the inflammatory response in SARS-CoV-2 infection, contributing to severe lung injury and immunothrombosis [80]. Additionally, miR-29b-3p is upregulated in cartilage tissue from patients with OA and is associated with altered expression and secretion of molecules related to cartilaginous degeneration. This upregulation facilitates chondrocyte apoptosis and contributes to the progression of OA [81]. Furthermore, miR-29b-3p has been shown to directly bind to the 3′-UTR of COL1A1 and COL3A1 mRNAs, leading to their degradation or preventing translation [82]. Soluble factors derived from OA cartilage can promote the expression of miR-29b-3p in cocultured BMSCs and inhibit the expression and secretion of COL1A1 and COL3A1 genes and proteins [83]. Collectively, these studies underscore the crucial role of miRNAs in the onset and development of COVID-19 and KOA.
Naive B cells, along with the nine core genes, offer valuable entry points to explore the relationship between the two diseases
The onset and progression of COVID-19 are closely linked to immunological inflammation, which can lead to immune irregularities such as imbalanced responses, cytokine storms, and heightened neutrophil activation. This, in turn, results in the release of proinflammatory cytokines such as interleukin-1β and tumour necrosis factor-α [84–86]. Simultaneously, immune dysfunction plays a significant role in the development of KOA. Although KOA has traditionally been considered a condition affecting joint cartilage, emerging evidence suggests the involvement of the immune system. Factors such as genetics, metabolism, or mechanical stress can initially damage cartilage, leading to the release of specific autoantigens that trigger an immune response. The immune system preserves ECM health and repairs damage after injury. Disruptions in this balance are associated with various diseases [87]. Immune cells, including neutrophils, monocytes, and macrophages, remodel the ECM by producing enzymes such as serine proteases and matrix metalloproteinases (MMPs). These enzymes degrade ECM components like collagen and fibronectin, creating breakdown products that may act as autoimmunogens [88–90], worsening conditions like KOA. This immune response involves the infiltration of joint tissues by T cells, B cells, and macrophages, which leads to the release of cytokines and chemokines, as well as the activation of the complement system. This cascade results in the release of cartilage-degrading factors, such as MMPs and prostaglandin E2 (PGE2), which further exacerbate cartilage damage [91].
The findings of our study indicate a higher presence of naive B cells in the disease group compared to the control group. Naive B cells play a dual role in the context of COVID-19. They form the foundation for generating antibodies to combat SARS-CoV-2. However, in severe COVID-19 cases, naive B cells are implicated in the development of autoreactivity, possibly influenced by the inflammatory environment characteristic of the disease [92, 93]. In OA, stromal cells derived from synovitis play a crucial role in supporting the survival of B-cells. However, B-cells in OA show changes in their ability to proliferate and differentiate, although they still maintain antibody production, particularly in the synovium [94]. In summary, naive B cells appear to be involved in the development of autoreactivity, potentially due to the inflammatory environment. Their role may have implications for understanding the immunological consequences of diseases.
In summary, the role of immunity in the development of KOA is of great significance. Thorough analysis of the infiltration pattern of immune cells in KOA is crucial for enhancing patient prognosis. Our research identified eosinophils, macrophages, and naive B cells, along with nine core genes, as potential avenues for investigating the relationship between these two diseases and exploring their potential as novel therapeutic targets. However, it is important to note that the immune responses triggered by COVID-19 may exacerbate KOA to some extent. Nonetheless, additional experimental validation is necessary to confirm the correlation between specific subtypes of immune cells and their impact on KOA.
Differential expression of the nine core genes in subcellular cartilage tissue
Subsequently, we acquired a dataset consisting of single-cell samples of KOA from the GEO database, specifically for the purpose of conducting single-cell annotation analysis. Single-cell analysis is a powerful tool for detecting cellular heterogeneity and uncovering underlying mechanisms [95]. By elucidating the molecular and functional profiles of different chondrocyte subtypes and understanding their interactions with other cell types in the joint, we can significantly enhance our understanding of joint biology and OA pathology.
Among the ten subchondrocytes that have been annotated, HTC have been identified as crucial regulatory cells for bone growth. These cells are associated with blood vessel invasion and are surrounded by a calcified extracellular matrix that supports endochondral ossification [58]. Previous research has shown that COL10A1 is a reliable marker for HTC cells in articular cartilage [96], which is consistent with our analysis results.
FC is primarily found in the late stages of OA and they express a high ratio of genes associated with unfavourable OA outcomes. They also have the capacity for vascularization, implying their role in promoting OA progression [38]. THBS2 showed the highest average expression level and the largest percentage expression level among FC, indicating its potential as a biomarker for FC (Additional file: Figure S8).
Promising therapeutics
OA is a complex, systemic disease with multifactorial triggers. Current treatments, such as nonsteroidal anti-inflammatory drugs and glucocorticoids, have limitations in terms of moderate effectiveness and potential side effects with long-term use [97, 98]. Currently, pain management and joint replacement surgery are the primary approaches for severe cases of pain and joint issues. However, these methods have limitations and associated risks, especially for elderly patients. Given the complexity of OA onset, there are currently no definitive treatments [99]. This is particularly important in the context of COVID-19, and studying potential drug targets may offer benefits to patients with both conditions.
This study identified potential therapeutic drugs for the nine core genes through screening using the SPIE3 web tool, providing valuable support for future research on treatment and therapeutic interventions. We conducted a search for relevant reports on several of the anticipated medications. Imatinib (Imatinib mesylate) is a protein tyrosine kinase inhibitor that has been shown to prevent and treat rheumatoid arthritis (RA) induced by type II collagen antibody in mice [100]. Treatment with imatinib reduced the number of synovial mast cells expressing tryptase in mouse knees and attenuated murine OA [101]. Adiponectin, a circulating adipokine, has been strongly associated with various forms and stages of OA [102]. Myricetin has been demonstrated to be a potent protective molecule against OA. It reduces the synthesis of inflammatory cytokines and attenuates the progression of OA, partially by suppressing the activation of IL-1β/MAPK pathway [103]. Activation of the Nrf2/HO-1 signalling pathway with myricetin has also shown promise in attenuating ECM degradation in human chondrocytes and ameliorating murine OA [104]. Tranexamic acid has been suggested as an effective and cost-effective drug for reducing blood loss in cemented primary hip arthroplasty for OA [105]. It significantly reduced cartilage-destructive lesions and increased cartilage hypertrophy [106]. Chenodeoxycholic acid (CDCA) is another potential therapeutic agent for OA. CDCA significantly decreased cartilage degradation on the surface of the femoral condyles and mitigated pathological changes in articular cartilage and the synovial membrane. It also significantly reduced the release of matrix metalloproteinase-1 (MMP-1), matrix metalloproteinase-3 (MMP-3), interleukin-1β (IL-1β), and prostaglandin E2 (PGE2) in synovial fluid [107]. In the external prediction results, various drugs demonstrate a positive effect on KOA. However, experimental verification is necessary for many of them. Development of these drugs can help prevent and treat KOA after COVID-19.
Conclusions
Understanding the precise pathophysiological mechanism of the relationship between COVID-19 and KOA is of paramount importance. This understanding can pave the way for the development of precise and reliable biomarkers and modern therapeutic agents. Such advancements would have a significant impact on reducing the economic burden associated with joint replacement surgeries and lowering the morbidity rates of degenerative conditions.
Our study, which combines MR and bioinformatics tools, provides evidence supporting a causal effect of COVID-19 on the risk of KOA. This finding is particularly noteworthy because COVID-19, despite being classified as a respiratory infection, differs from other respiratory infections in this aspect.
We conducted a comprehensive analysis of transcriptome data for patients with COVID-19 and to identify DEGs and hub genes that are common to both diseases. Subsequent analyses, including functional enrichment, WGCNA, transcriptional regulation, and single-cell sequencing, revealed the potential copathogenesis between COVID-19 and KOA, mediated by hub genes such as COL10A1, BGN, COL3A1, COMP, ACAN, THBS2, COL5A1, COL16A1, and COL5A2.
It is important to acknowledge the limitations of this study that arise from the use of a computational biology approach. First, computational biology approaches inherently introduce bias as they rely on existing datasets, algorithms, and models. This bias can affect the ability to accurately capture and reproduce underlying genetic links. Second, the availability of standard-compliant datasets for KOA may be limited, which can impact the comprehensiveness of the analysis. Additionally, challenges in sample collection can result in a lack of experimental validation for the pathogenic role of the identified signature key genes, TFs, miRNAs, and other factors within a limited time period. Further research through randomized controlled double-blind prospective studies is necessary.
In summary, our study represents the first attempt to integrate MR and bioinformatics approaches to investigate the intricate relationship between COVID-19 and KOA. This study provides a solid theoretical foundation for future research in the field of KOA and COVID-19. This work opens up new avenues for exploring the molecular mechanisms underlying these two complex conditions and offers hope for the development of more effective prevention and treatment strategies in the future.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Abbreviations
- COVID-19
Coronavirus Disease 2019
- KOA
Knee Osteoarthritis
- UTRI
Upper Respiratory Tract Infection
- MR
Mendelian Randomization
- WGCNA
Weighted Gene Coexpression Network Analysis
- DEGs
Differentially Expressed Genes
- ECM
Extracellular Matrix
- SNPs
Single-Nucleotide Polymorphisms
- IVW
Inverse Variance Weighted Method
- IVs
Instrumental Variables
- GSEA
Gene Set Enrichment Analysis
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PPI
Protein‒Protein Interaction
- TFs
Transcription Factors
- miRNAs
MicroRNAs
Author contributions
Study supervision: J.P. Study concept and design: X.Z., J.L. and J.P. Data collection: X.Z. and J.L. MR analysis: J.L. and X.Z. Bioinformatics analysis: X.Z. Single-cell RNAseq analysis: Q. M. Drafting of the manuscript: X.Z.and J.L. Revision of the manuscript: J.P. and J.G. All authors critically reviewed and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32470699) and the Research Startup Funds of Chongqing Medical University.
Data availability
All data used for analysis were obtained from published studies and public databases. The datasets utilized for Mendelian randomization (MR) analysis can be accessed through the IEU OPEN GWAS PROJECT (https://gwas.mrcieu.ac.uk/datasets/) using the following accession numbers: ebi-a-GCST011073, finn-b-j10_UPPERINFEC, and ebi-a-GCST007090. Additionally, the datasets supporting the bioinformatics analysis discussed in this article are available in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/) under the accession numbers GSE180226, GSE197143, GSE206606, and GSE220243, as well as in the ArrayExpress database (https://www.ebi.ac.uk/biostudies/arrayexpress) under the accession number E-MTAB-12,184.
Declarations
Ethics approval and consent to participate
Not applicable. The data used for analysis were obtained from published studies and public databases, no additional ethics approval was needed.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiao Zheng and Jinhao Li contributed equally to this work.
References
- 1.Kim JR, Yoo JJ, Kim HA. Therapeutics in Osteoarthritis based on an understanding of its Molecular Pathogenesis. Int J Mol Sci. 2018;19(3). [DOI] [PMC free article] [PubMed]
- 2.Huzum B, Curpan AS, Puha B, Serban DN, Veliceasa B, Necoara RM et al. Connections between Orthopedic Conditions and Oxidative Stress: Current Perspective and the Possible Relevance of Other Factors, Such as Metabolic Implications, Antibiotic Resistance, and COVID-19. Medicina (Kaunas). 2022;58(3). [DOI] [PMC free article] [PubMed]
- 3.Lauwers M, Au M, Yuan S, Wen C. COVID-19 in Joint Ageing and Osteoarthritis: current status and perspectives. Int J Mol Sci. 2022;23(2). [DOI] [PMC free article] [PubMed]
- 4.Battista S, Dell’Isola A, Manoni M, Englund M, Palese A, Testa M. Experience of the COVID-19 pandemic as lived by patients with hip and knee osteoarthritis: an Italian qualitative study. BMJ Open. 2021;11(10):e053194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cordani C, Lazzarini SG, Del Furia MJ, Kiekens C, Arienti C, Negrini S. Arthralgia: a map of Cochrane evidence relevant to rehabilitation for people with post COVID-19 condition. Eur J Phys Rehabil Med. 2022;58(6):870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuellen G, Liesenfeld O, Kowald A, Barrantes I, Bastian M, Simm A, et al. The preventive strategy for pandemics in the elderly is to collect in advance samples & data to counteract chronic inflammation (inflammaging). Ageing Res Rev. 2020;62:101091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michael JW, Schlüter-Brust KU, Eysel P. The epidemiology, etiology, diagnosis, and treatment of osteoarthritis of the knee. Dtsch Arztebl Int. 2010;107(9):152–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 Novel Coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27(8):1133–63. [DOI] [PubMed] [Google Scholar]
- 12.Zheng H, Shi YZ, Liang JT, Lu LL, Chen M. Modifiable factors for migraine prophylaxis: a mendelian randomization analysis. Front Pharmacol. 2023;14:1010996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Z, Zheng Z, Zhang F, Wu Y, Trzaskowski M, Maier R, et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun. 2018;9(1):224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng J, Baird D, Borges MC, Bowden J, Hemani G, Haycock P, et al. Recent developments in mendelian randomization studies. Curr Epidemiol Rep. 2017;4(4):330–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in mendelian randomization studies. Int J Epidemiol. 2013;42(5):1497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Love MI, Huber W, Anders S. Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling Tumor infiltrating Immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177(7):1888–e90221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16(12):1289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams G. SPIEDw: a searchable platform-independent expression database web tool. BMC Genomics. 2013;14(1):765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi K, Bing ZT, Cao GQ, Guo L, Cao YN, Jiang HO, et al. Identify the signature genes for diagnose of uveal melanoma by weight gene co-expression network analysis. Int J Ophthalmol. 2015;8(2):269–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152(6):1237–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. [DOI] [PubMed] [Google Scholar]
- 31.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10(4):252–63. [DOI] [PubMed] [Google Scholar]
- 32.Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234(5):5451–65. [DOI] [PubMed] [Google Scholar]
- 33.Vishnoi A, Rani S. MiRNA Biogenesis and Regulation of diseases: an overview. Methods Mol Biol. 2017;1509:1–10. [DOI] [PubMed] [Google Scholar]
- 34.Parikh S, Gomez O, Davis T, Lyon Z, Corces A. Avascular necrosis as a Sequela of COVID-19: a Case Series. Cureus. 2023;15(2):e35368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soroa G, Álvarez A, Monge I, Navarro D, Roca O. Osteonecrosis and osteomyelitis of the Proximal Third of Tibia as a late Sequela of COVID-19: a Case Report. Plast Aesthet Nurs (Phila). 2022;42(4):190–6. [DOI] [PubMed] [Google Scholar]
- 36.Chao PY, Lee LY. [Impact of the COVID-19 pandemic on Health-Related Quality of Life in older adults with preoperative knee Osteoarthritis]. Hu Li Za Zhi. 2023;70(3):26–36. [DOI] [PubMed] [Google Scholar]
- 37.Chou CH, Jain V, Gibson J, Attarian DE, Haraden CA, Yohn CB, et al. Synovial cell cross-talk with cartilage plays a major role in the pathogenesis of osteoarthritis. Sci Rep. 2020;10(1):10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji Q, Zheng Y, Zhang G, Hu Y, Fan X, Hou Y, et al. Single-cell RNA-seq analysis reveals the progression of human osteoarthritis. Ann Rheum Dis. 2019;78(1):100–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei J, Hui A. Review of ribosome interactions with SARS-CoV-2 and COVID-19 mRNA vaccine. Life (Basel). 2022;12(1). [DOI] [PMC free article] [PubMed]
- 40.Hwang HS, Lee MH, Kim HA. TGF-β1-induced expression of collagen type II and ACAN is regulated by 4E-BP1, a repressor of translation. Faseb j. 2020;34(7):9531–46. [DOI] [PubMed] [Google Scholar]
- 41.van den Akker GGH, Caron MMJ, Peffers MJ, Welting TJM. Ribosome dysfunction in osteoarthritis. Curr Opin Rheumatol. 2022;34(1):61–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blalock D, Miller A, Tilley M, Wang J. Joint instability and osteoarthritis. Clin Med Insights Arthritis Musculoskelet Disorders. 2015;8:15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sebastian A, McCool JL, Hum NR, Murugesh DK, Wilson SP, Christiansen BA et al. Single-cell RNA-Seq reveals Transcriptomic Heterogeneity and Post-traumatic Osteoarthritis-Associated early molecular changes in mouse articular chondrocytes. Cells. 2021;10(6). [DOI] [PMC free article] [PubMed]
- 44.Bhosale AM, Richardson JB. Articular cartilage: structure, injuries and review of management. Br Med Bull. 2008;87:77–95. [DOI] [PubMed] [Google Scholar]
- 45.Deng ZH, Li YS, Gao X, Lei GH, Huard J. Bone morphogenetic proteins for articular cartilage regeneration. Osteoarthritis Cartilage. 2018;26(9):1153–61. [DOI] [PubMed] [Google Scholar]
- 46.Sarker H, Haimour A, Toor R, Fernandez-Patron C. The emerging role of epigenetic mechanisms in the causation of aberrant MMP activity during human pathologies and the Use of Medicinal drugs. Biomolecules. 2021;11(4). [DOI] [PMC free article] [PubMed]
- 47.Zhou Y, Horowitz JC, Naba A, Ambalavanan N, Atabai K, Balestrini J, et al. Extracellular matrix in lung development, homeostasis and disease. Matrix Biol. 2018;73:77–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang JJ, Wang CW, Liu Y, Zhang YY, Yang NB, Yu YC, et al. Role of the extracellular matrix in COVID-19. World J Clin Cases. 2023;11(1):73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balabko L, Andreev K, Burmann N, Schubert M, Mathews M, Trufa DI, et al. Increased expression of the Th17-IL-6R/pSTAT3/BATF/RorγT-axis in the tumoural region of adenocarcinoma as compared to squamous cell carcinoma of the lung. Sci Rep. 2014;4:7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han Y, Li X, Yan M, Yang M, Wang S, Pan J, et al. Oxidative damage induces apoptosis and promotes calcification in disc cartilage endplate cell through ROS/MAPK/NF-κB pathway: implications for disc degeneration. Biochem Biophys Res Commun. 2019;516(3):1026–32. [DOI] [PubMed] [Google Scholar]
- 51.Li J, Zheng J. Theaflavins prevent cartilage degeneration via AKT/FOXO3 signaling in vitro. Mol Med Rep. 2019;19(2):821–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang W, Hsu P, Zhong B, Guo S, Zhang C, Wang Y, et al. MiR-34a enhances Chondrocyte apoptosis, senescence and facilitates development of Osteoarthritis by Targeting DLL1 and regulating PI3K/AKT pathway. Cell Physiol Biochem. 2018;48(3):1304–16. [DOI] [PubMed] [Google Scholar]
- 53.Ke H, Mou X, Xia Q. Remifentanil repairs cartilage damage and reduces the degradation of cartilage matrix in post-traumatic osteoarthritis, and inhibits IL-1β-induced apoptosis of articular chondrocytes via inhibition of PI3K/AKT/NF-κB phosphorylation. Ann Transl Med. 2020;8(22):1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun K, Luo J, Guo J, Yao X, Jing X, Guo F. The PI3K/AKT/mTOR signaling pathway in osteoarthritis: a narrative review. Osteoarthritis Cartilage. 2020;28(4):400–9. [DOI] [PubMed] [Google Scholar]
- 55.Rahmati M, Nalesso G, Mobasheri A, Mozafari M. Aging and osteoarthritis: central role of the extracellular matrix. Ageing Res Rev. 2017;40:20–30. [DOI] [PubMed] [Google Scholar]
- 56.Lunardi A, Webster KA, Papa A, Padmani B, Clohessy JG, Bronson RT, et al. Role of aberrant PI3K pathway activation in gallbladder tumorigenesis. Oncotarget. 2014;5(4):894–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lv W, Wu M, Ren Y, Luo X, Hu W, Zhang Q et al. Treatment of keloids through Runx2 siRNA–induced inhibition of the PI3K/AKT signaling pathway. Mol Med Rep. 2021;23(1). [DOI] [PMC free article] [PubMed]
- 58.Gu J, Lu Y, Li F, Qiao L, Wang Q, Li N, et al. Identification and characterization of the novel Col10a1 regulatory mechanism during chondrocyte hypertrophic differentiation. Cell Death Dis. 2014;5(10):e1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Traupe H, van den Ouweland AM, van Oost BA, Vogel W, Vetter U, Warren ST, et al. Fine mapping of the human biglycan (BGN) gene within the Xq28 region employing a hybrid cell panel. Genomics. 1992;13(2):481–3. [DOI] [PubMed] [Google Scholar]
- 60.Pérez-Mies B, Caniego-Casas T, Bardi T, Carretero-Barrio I, Benito A, García-Cosío M, et al. Progression to lung fibrosis in severe COVID-19 patients: a morphological and transcriptomic study in postmortem samples. Front Med (Lausanne). 2022;9:976759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li S, Wang H, Zhang Y, Qiao R, Xia P, Kong Z, et al. COL3A1 and MMP9 serve as potential diagnostic biomarkers of Osteoarthritis and are Associated with Immune Cell Infiltration. Front Genet. 2021;12:721258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ackermann M, Kamp JC, Werlein C, Walsh CL, Stark H, Prade V, et al. The fatal trajectory of pulmonary COVID-19 is driven by lobular ischemia and fibrotic remodelling. EBioMedicine. 2022;85:104296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen FH, Thomas AO, Hecht JT, Goldring MB, Lawler J. Cartilage oligomeric matrix protein/thrombospondin 5 supports chondrocyte attachment through interaction with integrins. J Biol Chem. 2005;280(38):32655–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koelling S, Clauditz TS, Kaste M, Miosge N. Cartilage oligomeric matrix protein is involved in human limb development and in the pathogenesis of osteoarthritis. Arthritis Res Ther. 2006;8(3):R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dranitsina AS, Dvorshchenko KO, Korotkyi OH, Vovk AA, Falalyeyeva TM, Grebinyk DM, et al. Expression of Nos2 and acan genes in rat knee articular cartilage in Osteoarthritis. Cytol Genet. 2019;53(6):481–8. [Google Scholar]
- 66.Stattin EL, Lindblom K, Struglics A, Önnerfjord P, Goldblatt J, Dixit A, et al. Novel missense ACAN gene variants linked to familial osteochondritis dissecans cluster in the C-terminal globular domain of aggrecan. Sci Rep. 2022;12(1):5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alford AI, Terkhorn SP, Reddy AB, Hankenson KD. Thrombospondin-2 regulates matrix mineralization in MC3T3-E1 pre-osteoblasts. Bone. 2010;46(2):464–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kyriakides TR, Zhu YH, Smith LT, Bain SD, Yang Z, Lin MT, et al. Mice that lack thrombospondin 2 display connective tissue abnormalities that are associated with disordered collagen fibrillogenesis, an increased vascular density, and a bleeding diathesis. J Cell Biol. 1998;140(2):419–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.LaBell TL, Milewicz DJ, Disteche CM, Byers PH. Thrombospondin II: partial cDNA sequence, chromosome location, and expression of a second member of the thrombospondin gene family in humans. Genomics. 1992;12(3):421–9. [DOI] [PubMed] [Google Scholar]
- 70.Zheng L, Chen W, Xian G, Pan B, Ye Y, Gu M, et al. Identification of abnormally methylated-differentially expressed genes and pathways in osteoarthritis: a comprehensive bioinformatic study. Clin Rheumatol. 2021;40(8):3247–56. [DOI] [PubMed] [Google Scholar]
- 71.Colombi M, Dordoni C, Venturini M, Ciaccio C, Morlino S, Chiarelli N, et al. Spectrum of mucocutaneous, ocular and facial features and delineation of novel presentations in 62 classical Ehlers-Danlos syndrome patients. Clin Genet. 2017;92(6):624–31. [DOI] [PubMed] [Google Scholar]
- 72.Mak KM, Png CY, Lee DJ. Type V Collagen in Health, Disease, and fibrosis. Anat Rec (Hoboken). 2016;299(5):613–29. [DOI] [PubMed] [Google Scholar]
- 73.Willard K, Mannion S, Saunders CJ, Collins M, September AV. The interaction of polymorphisms in extracellular matrix genes and underlying miRNA motifs that modulate susceptibility to anterior cruciate ligament rupture. J Sci Med Sport. 2018;21(1):22–8. [DOI] [PubMed] [Google Scholar]
- 74.Grässel S, Bauer RJ. Collagen XVI in health and disease. Matrix Biol. 2013;32(2):64–73. [DOI] [PubMed] [Google Scholar]
- 75.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–31. [DOI] [PubMed] [Google Scholar]
- 76.Hanna J, Hossain GS, Kocerha J. The potential for microRNA therapeutics and clinical research. Front Genet. 2019;10:478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Le LT, Swingler TE, Crowe N, Vincent TL, Barter MJ, Donell ST, et al. The microRNA-29 family in cartilage homeostasis and osteoarthritis. J Mol Med (Berl). 2016;94(5):583–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Askari N, Hadizadeh M, Rashidifar M. A new insight into sex-specific non-coding RNAs and networks in response to SARS-CoV-2. Infect Genet Evol. 2022;97:105195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Le L, Swingler TE, Crowe N, Driscoll C, Vincent TL, Barter MJ, et al. The microRNA-29 family in osteoarthritis. Osteoarthr Cartil. 2014;22:S41–2. [Google Scholar]
- 80.Centa A, Fonseca AS, da Silva Ferreira SG, Azevedo MLV, de Paula CBV, Nagashima S, et al. Deregulated miRNA expression is associated with endothelial dysfunction in post-mortem lung biopsies of COVID-19 patients. Am J Physiol Lung Cell Mol Physiol. 2021;320(3):L405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen L, Li Q, Wang J, Jin S, Zheng H, Lin J, et al. MiR-29b-3p promotes chondrocyte apoptosis and facilitates the occurrence and development of osteoarthritis by targeting PGRN. J Cell Mol Med. 2017;21(12):3347–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Steele R, Mott JL, Ray RB. MBP-1 upregulates miR-29b that represses Mcl-1, collagens, and matrix-metalloproteinase-2 in prostate cancer cells. Genes Cancer. 2010;1(4):381–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mayer U, Benditz A, Grässel S. miR-29b regulates expression of collagens I and III in chondrogenically differentiating BMSC in an osteoarthritic environment. Sci Rep. 2017;7(1):13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sur S, Steele R, Isbell TS, Ray R, Ray RB. Circulatory exosomes from COVID-19 patients trigger NLRP3 inflammasome in endothelial cells. mBio. 2022;13(3):e0095122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang J, Li Q, Qiu Y, Lu H. COVID-19: imbalanced cell-mediated immune response drives to immunopathology. Emerg Microbes Infect. 2022;11(1):2393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sutherland TE, Dyer DP, Allen JE. The extracellular matrix and the immune system: a mutually dependent relationship. Sci (New York NY). 2023;379(6633):eabp8964. [DOI] [PubMed] [Google Scholar]
- 88.Jabłońska-Trypuć A, Matejczyk M, Rosochacki S. Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. J Enzyme Inhib Med Chem. 2016;31(sup1):177–83. [DOI] [PubMed] [Google Scholar]
- 89.Cawston TE, Young DA. Proteinases involved in matrix turnover during cartilage and bone breakdown. Cell Tissue Res. 2010;339(1):221–35. [DOI] [PubMed] [Google Scholar]
- 90.Kolesnikoff N, Chen CH, Samuel MS. Interrelationships between the extracellular matrix and the immune microenvironment that govern epithelial tumour progression. Clinical science (London, England: 1979). 2022;136(5):361 – 77. [DOI] [PMC free article] [PubMed]
- 91.Haseeb A, Haqqi TM. Immunopathogenesis of osteoarthritis. Clin Immunol. 2013;146(3):185–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Feldman J, Bals J, Altomare CG, St Denis K, Lam EC, Hauser BM et al. Naive human B cells engage the receptor binding domain of SARS-CoV-2, variants of concern, and related sarbecoviruses. bioRxiv. 2021. [DOI] [PMC free article] [PubMed]
- 93.Woodruff MC, Ramonell RP, Haddad NS, Anam FA, Rudolph ME, Walker TA, et al. Dysregulated naive B cells and de novo autoreactivity in severe COVID-19. Nature. 2022;611(7934):139–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xie X, Doody GM, Shuweihdi F, Conaghan PG, Ponchel F. B-cell capacity for expansion and differentiation into plasma cells are altered in osteoarthritis. Osteoarthritis Cartilage. 2023;31(9):1176–88. [DOI] [PubMed] [Google Scholar]
- 95.Lee J, Hyeon DY, Hwang D. Single-cell multiomics: technologies and data analysis methods. Exp Mol Med. 2020;52(9):1428–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shen G. The role of type X collagen in facilitating and regulating endochondral ossification of articular cartilage. Orthod Craniofac Res. 2005;8(1):11–7. [DOI] [PubMed] [Google Scholar]
- 97.Pelletier JP, Martel-Pelletier J, Rannou F, Cooper C. Efficacy and safety of oral NSAIDs and analgesics in the management of osteoarthritis: evidence from real-life setting trials and surveys. Semin Arthritis Rheum. 2016;45(4 Suppl):S22–7. [DOI] [PubMed] [Google Scholar]
- 98.Savvidou O, Milonaki M, Goumenos S, Flevas D, Papagelopoulos P, Moutsatsou P. Glucocorticoid signaling and osteoarthritis. Mol Cell Endocrinol. 2019;480:153–66. [DOI] [PubMed] [Google Scholar]
- 99.Di Francesco M, Fragassi A, Pannuzzo M, Ferreira M, Brahmachari S, Decuzzi P. Management of osteoarthritis: from drug molecules to nano/micromedicines. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2022;14(3):e1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Koyama K, Hatsushika K, Ando T, Sakuma M, Wako M, Kato R, et al. Imatinib mesylate both prevents and treats the arthritis induced by type II collagen antibody in mice. Mod Rheumatol. 2007;17(4):306–10. [DOI] [PubMed] [Google Scholar]
- 101.Wang Q, Lepus CM, Raghu H, Reber LL, Tsai MM, Wong HH et al. IgE-mediated mast cell activation promotes inflammation and cartilage destruction in osteoarthritis. Elife. 2019;8. [DOI] [PMC free article] [PubMed]
- 102.Ilia I, Nitusca D, Marian C. Adiponectin in Osteoarthritis: pathophysiology, relationship with obesity and presumptive diagnostic biomarker potential. Diagnostics (Basel). 2022;12(2). [DOI] [PMC free article] [PubMed]
- 103.Liu X, Fang H, Zeng C, Cai D. Myricetin attenuates osteoarthritis by blockade of the IL-1β/MAPK pathway. Osteoarthr Cartil. 2018;26.
- 104.Pan X, Chen T, Zhang Z, Chen X, Chen C, Chen L, et al. Activation of Nrf2/HO-1 signal with Myricetin for attenuating ECM degradation in human chondrocytes and ameliorating the murine osteoarthritis. Int Immunopharmacol. 2019;75:105742. [DOI] [PubMed] [Google Scholar]
- 105.Niskanen RO, Korkala OL. Tranexamic acid reduces blood loss in cemented hip arthroplasty: a randomized, double-blind study of 39 patients with osteoarthritis. Acta Orthop. 2005;76(6):829–32. [DOI] [PubMed] [Google Scholar]
- 106.Vignon E, Mathieu P, Bejui J, Descotes J, Hartmann D, Patricot LM, et al. Study of an inhibitor of plasminogen activator (tranexamic acid) in the treatment of experimental osteoarthritis. J Rheumatol Suppl. 1991;27:131–3. [PubMed] [Google Scholar]
- 107.Yan ZW, Dong J, Qin CH, Zhao CY, Miao LY, He CY. Therapeutic effect of Chenodeoxycholic Acid in an experimental rabbit model of Osteoarthritis. Mediators Inflamm. 2015;2015:780149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used for analysis were obtained from published studies and public databases. The datasets utilized for Mendelian randomization (MR) analysis can be accessed through the IEU OPEN GWAS PROJECT (https://gwas.mrcieu.ac.uk/datasets/) using the following accession numbers: ebi-a-GCST011073, finn-b-j10_UPPERINFEC, and ebi-a-GCST007090. Additionally, the datasets supporting the bioinformatics analysis discussed in this article are available in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/) under the accession numbers GSE180226, GSE197143, GSE206606, and GSE220243, as well as in the ArrayExpress database (https://www.ebi.ac.uk/biostudies/arrayexpress) under the accession number E-MTAB-12,184.