Abstract
Molecular dynamics (MD) with the ReaxFF force field is used to study the structural damage to HIV capsid protein and gp120 protein mediated by reactive oxygen species (ROS). Our results show that with an increase in ROS concentration, the structures of the HIV capsid protein and gp120 protein are more severely damaged, including dehydrogenation, increase in oxygen-containing groups, helix shortening or destruction, and peptide bond breaking. In particular, we noticed that extraction of H atoms from N atoms by ROS was significantly higher than that from C atoms. There was no significant difference in the effect of ROS on dehydrogenation and shortening or breaking of the helices. In contrast, the impact of O on the increase in oxygen-containing groups and the fracture of peptide bonds in the gp120 protein is more significant than that of O3, and the effect of O3 is greater than that of ˙OH. In addition, the degree of structural damage of the gp120 protein was greater than that of the capsid protein. These detailed findings deepen our understanding of the role of ROS in regulating the structure and function of the HIV capsid protein and gp120 protein and provide valuable insights for plasma therapy for acquired immune deficiency syndrome (AIDS).
Molecular dynamics (MD) with the ReaxFF force field is used to study the structural damage to HIV capsid protein and gp120 protein mediated by reactive oxygen species (ROS).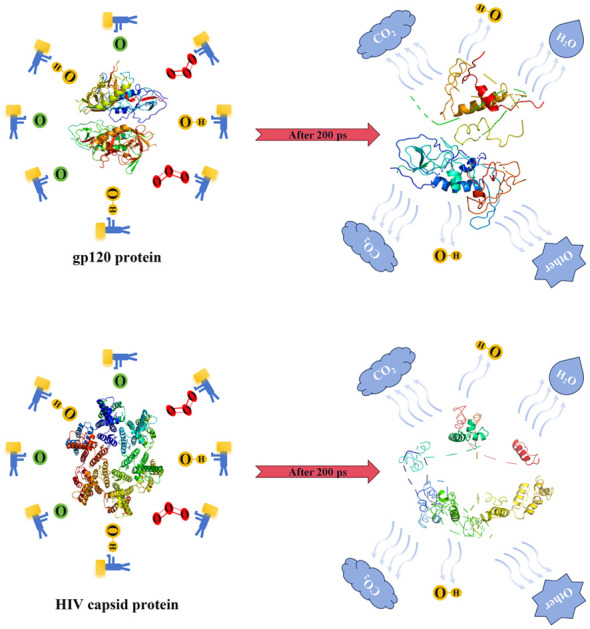
1. Introduction
Cold atmospheric plasma (CAP) is a partially ionized gas that possesses the characteristics of being generated at atmospheric pressure and operating under room-temperature/physiological-temperature conditions.1–5 It is composed of ions and free electrons, which form and produce reactive oxygen species (ROS) and reactive nitrogen species (RNS) after energetic excitation, including ground-state oxygen atoms (O), hydroxyl radicals (˙OH), ozone molecules (O3), peroxynitrite (HNO3) and nitric oxide (NO).6 Based on the above characteristics of plasma, it has important potential for application in biochemistry, materials, and physics.7–15 For example, Okada et al. treated green algae cells with plasma, reducing the activity of the green algae cells and thus reducing the fluorescence effect of chlorella.16 Dave et al. modified a catheter substrate by irradiating it under plasma and found that the treated catheter substrate successfully prevented biofilm formation.17 Guo et al. used plasma to degrade tetracycline antibiotics. After the reaction, hydroxylation, dehydroxylation, aldehyde formation, deammoniation, carbonylation, and other phenomena occurred, with the overall process being efficient and environmentally friendly, and having low energy consumption.18 Therefore, understanding the interaction between plasma and matter is of great scientific importance.
Since ROS can directly interact with organisms and be targeted,19–22 they can effectively change biological structure, activity, function, etc., such as regulating cell growth rate, inactivating viruses, and other effects that more and more researchers are paying attention to.23–31 For example, Attri et al. treated plant seeds and found that the treated seeds had enhanced antioxidant activity.32 Yang et al. used plasma to oxidize coronavirus spike proteins, destroying and inactivating their molecular structure.33
The HIV capsid protein is the key factor of the HIV structure. It is involved in the process of virus assembly, maturation, and immune escape. If the HIV capsid protein is destroyed, it can effectively inhibit HIV transmission and replication.34 For example, Link et al. designed GS-6207 to bind tightly at the conserved interface between capsid protein monomers, making the capsid protein unable to interact with other proteins, thereby inhibiting HIV replication.35 The gp120 protein is located on the surface of the virus particle. It is responsible for recognizing and binding to the CD4 receptor on the host cell’s surface, thus initiating the virus invasion process.36 The structure and function of the gp120 protein have an important influence on the infectivity and virulence of HIV.37 For example, Huang et al. used 35O22-specific antibodies to bind to the gp120 protein and gp41 protein, which significantly reduced the binding of HIV to host cells in patients.38 Featherstone et al. used SERINC5 cell protein to bind to HIV protein to change the gp120 protein conformation, and found that HIV-1 infection could be significantly inhibited.37
Reactive molecular dynamics (RMD) simulations can track molecular structure, especially the breaking and formation of chemical bonds, which is of great significance for understanding the interaction effect of ROS and RNS on biological macromolecules. Attri et al. simulated the interaction of materials such as RONS produced by plasma with Mdm2-p53.32 After oxidation, the Mdm2 protein could not inhibit the activity of tumor suppressor protein p53, resulting in high activity of anti-tumor protein p53. Ding et al. simulated the reaction of ROS-oxidized hyaluronic acid (HA) with drugs and found that the acetylamino group in HA was oxidized to an unsaturated acyl group, which led to cross-linking between HA and the drug.39
This work uses RMD simulations to investigate ROS-mediated HIV damage from the outer gp120 protein to internal capsid protein. In the second part, we introduce the parameters related to the simulation and the statistical method used in the data. In the third part, we analyze the obtained data from the primary structure of the proteins to the tertiary structure. Our research showed that ROS-mediated damage mainly involved dehydrogenation, the increase of oxygen-containing groups, and the breaking of peptide bonds. In the fourth part, we summarize our findings and provide insights into potential applications.
2. Model and methods
2.1. ReaxFF
The ReaxFF force field was proposed by Van Duin et al. in 2001 based on the theories of classical mechanics and quantum mechanics.40 The force field uses the bond order to judge the breaking of the chemical bond between two atoms and describes the change in the chemical bond in the simulation process,41 which is commonly used in RMD.41,42
The total energy of the system is divided into the bond energy (Ebond); over/under coordination energy (Eover/under); valence angle energy (Eval); penalty energy (Epen); torsion energy (Etors); conjugation energy (Econj); non-bonded van der Waals interaction (EvdWaals); and Coulomb interaction (ECoulomb); that is,Esystem = Ebond + Eover + Eunder + Eval + Epen + Etors + Econj + EvdWaals + ECoulomb
This work uses a ReaxFF force field that contains C/H/O/N/S elements and is suitable for calculating the reaction of proteins in a water environment.43 For example, it successfully simulates the reactions of glucose in aqueous solutions and the plasma degradation of aflatoxin B1.44,45
2.2. Simulation model
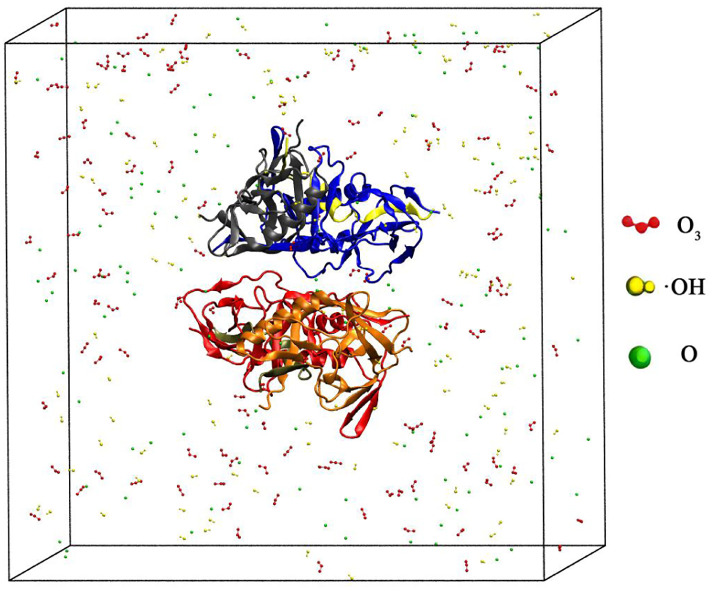
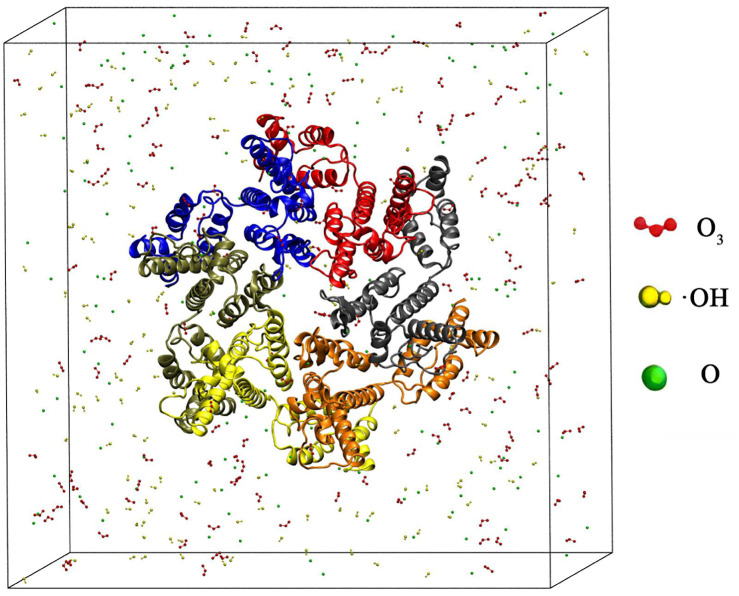
As shown in Fig. 1, a cube simulation box with periodic boundary conditions was constructed using Packmol. The gp120 protein obtained from the PDB database is 73.67 Å × 89.55 Å × 66.57 Å, labelled 3TGR and located in the center of the box, and the box length is 140 Å.46 Then we randomly put ROS particles into the box, where O 3 is red, ˙OH is yellow and O is green. Finally, we filled the box with water molecules. In Fig. 2, the capsid protein (labelled 6SLU) reaction simulator was constructed in the same way as that of the gp120 protein, where the difference was that the box length was changed to 150 Å.
Fig. 1. Schematic diagram of a simulation box where the gp120 protein is placed in the center. ROS, namely O3 (red), ˙OH (yellow), and O (green), were randomly placed in the box, where water molecules filled the box and were not shown.
Fig. 2. Schematic diagram of a simulation box where the capsid protein is placed in the center. ROS, namely O3 (red), ˙OH (yellow), and O (green), were randomly placed in the box, where water molecules filled the box and were not shown.
We used LAMMPS to simulate the ROS oxidation mechanism of the HIV capsid protein through Reaxff force fields.40–43,47 We adopted an NVT ensemble and set the time step to 0.5 fs and the temperature to 277 K, which is the optimal temperature for the survival of the gp120 protein.34,35 We found that after 140 ps, the type and quantity of products no longer changed, so the reaction could be considered complete. We still extended the reaction time to 200 ps to ensure the reaction was completely finished.33
We defined some parameters for better analysis, among which DC–H and DN–H, obtained based on VESTA,48 are the protein’s C–H and N–H bond reduction rates. IC O and DCO–NH represent the rate at which carbonyl groups increase in the protein and the rate at which peptide bonds decrease, calculated with OpenBabel by converting the structure file after reaction into a Simplified Molecular Input Line Entry Specification (SMILES) format, including the information on chemical bonds in the protein.49Dhelix represents the reduction rate of the number of amino acids constituting the helices in the protein, which was calculated using PyMOL.50 All results are the statistical average with three independent calculations.
3. Results and discussion
3.1. ROS oxidise the gp120 protein
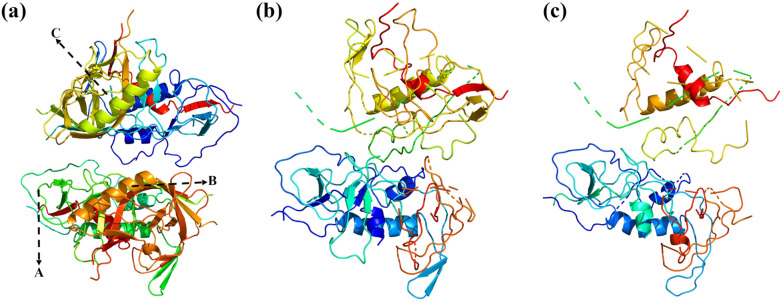
Our simulations show that ROS oxidation leads to significant changes in the primary and tertiary structure of the gp120 protein, as shown in Fig. 3. For example, the amino acid sequence at A changes from “CRNVSTVQCT” to “GRNVSTVQCT”, indicating a change in the primary structure. The helices in the secondary structure are shortened or disappear. For example, the amino acid sequence on the helix at B changes from “ESKWNHTLQKVGEEL” to “NNKTFNGTGP”, and the helix at B is shortened due to the change in the primary structure (amino acid sequence). In addition, peptide bonds are broken, such as the amino acid sequence at C changing from “RNGTYNHTG” to “RN-TYNH-TG”. In addition, it can be seen from Fig. 3 that the tertiary structure changes significantly with the reaction time.
Fig. 3. Schematic diagram of changes in the gp120 protein when oxidized by ROS with the increase in the reaction time, where (a) is the initial moment, (b) is 50 ps, and (c) is 200 ps.
The results in Table 1 show that DN–H, DC–H, IC O, Dhelix, and DCO–NH increase with the increase in CROS, which indicates that the reaction occurred more vigorously. Specifically, DN–H ranges from 3.37% to 12.22%, DC–H ranges from 0.03% to 0.40%, IC O ranges from 51.25% to 117.21%, Dhelix ranges from 50.23% to 56.46%, and DCO–NH ranges from 7.75% to 13.05%. It is worth noting that the maximum value of IC O exceeds 100%, indicating a substantial increase in the number of carbonyl groups in the gp120 protein. Based on the above phenomenon, we concluded that the ROS concentration was one of the key regulatory factors in the degree of oxidation of the gp120 protein, and the mechanism is multidimensional, which will be discussed and analyzed below.
Table 1. Statistics of ROS oxidation of the gp120 protein. CROS is the ROS concentration, while DC–H and DN–H represent the reduction rate of C–H and N–H bonds, respectively. IC O and DCO–NH indicate the rate at which the carbonyl group increases and the rate at which the peptide bond decreases, respectively, and Dhelix denotes the reduction rate of amino acids comprising the helices.
| C ROS | D N–H | D C–H | I C O | D helix | D CO–NH |
|---|---|---|---|---|---|
| 23.06 μmol ml−1 | −3.37% | −0.03% | +51.25% | −50.23% | −7.75% |
| 103.51 μmol ml−1 | −9.39% | −0.18% | +63.75% | −53.78% | −9.95% |
| 311.46 μmol ml−1 | −12.22% | −0.40% | +117.21% | −56.46% | −13.05% |
To distinguish the contribution of different ROS species to gp120 protein oxidation, we calculated the degree of oxidation of the gp120 protein at the concentrations of O, O3, and ˙OH in Table 2. DN–H, DC–H, IC O, Dhelix, and DCO–NH were positively correlated with the increase in the concentration of O, O3, and ˙OH. A horizontal comparison of O, O3, and ˙OH showed no significant difference in DN–H, Dhelix, and DCO–NH. However, for IC O, the effect of O is stronger than that of O3, and the effect of O3 is stronger than that of ˙OH. The main reason for this phenomenon is that in the aqueous environment, different ROS can be converted to each other, which makes the process more complicated.22 They can react quickly and directly with functional groups in proteins, resulting in a decrease in the number of peptide bonds and a corresponding increase in the number of carbonyl groups.51 O3 decomposes into O2 and O at a lower decomposition rate.52,53 As a result, O3 has less effect than O. ˙OH reacts with water to produce relatively low-oxidizing H2O2, which results in the impact being minimal. This suggests that although the three ROS species contribute similarly to dehydrogenation, they play different roles in peptide bond breakdown and carbonyl formation, which are key indicators of protein oxidation.54
Statistics of the oxidization of the gp120 protein by ˙OH, O3 and O with different concentrations, where C˙OH, CO3, and CO denote the concentrations of ˙OH, O3 and O, respectively.
| C O | D N–H | D C–H | I C O | D helix | D CO–NH |
|---|---|---|---|---|---|
| 7.68 μmol ml−1 | −1.64% | −0.03% | +50.00% | −50.12% | −8.07% |
| 34.48 μmol ml−1 | −5.61% | −0.13% | +93.80% | −63.17% | −12.81% |
| 103.6 μmol ml−1 | −8.02% | −0.17% | +97.36% | −63.78% | −14.11% |
| C ˙OH | D N–H | D C–H | I C O | D helix | D CO–NH |
|---|---|---|---|---|---|
| 7.68 μmol ml−1 | −0.66% | −0.08% | +56.25% | −53.41% | −8.07% |
| 34.48 μmol ml−1 | −3.52% | −0.10% | +62.50% | −55.19% | −10.47% |
| 103.6 μmol ml−1 | −7.39% | −0.23% | +68.20% | −61.20% | −13.06% |
| C O3 | D N–H | D C–H | I C O | D helix | D CO–NH |
|---|---|---|---|---|---|
| 7.68 μmol ml−1 | −2.45% | −0.02% | +41.88% | −52.41% | −9.56% |
| 34.48 μmol ml−1 | −5.59% | −0.11% | +65.50% | −52.84% | −11.27% |
| 103.6 μmol ml−1 | −8.79% | −0.19% | +82.52% | −54.78% | −13.41% |
3.2. ROS oxidise the capsid protein
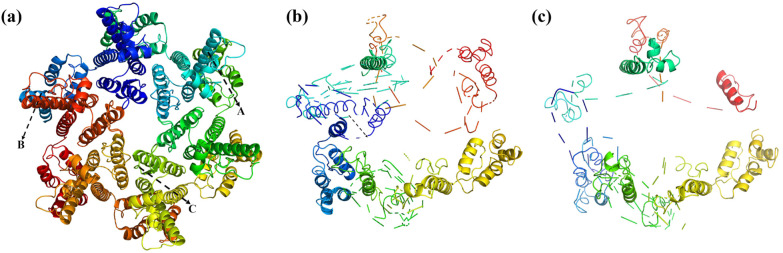
Similar to the gp120 protein, ROS-mediated oxidation of the HIV capsid protein caused dehydrogenation, an increase in oxygen-containing groups, helix shortening or elimination, and peptide bond breakage. Our simulations show that the oxidative effect of ROS causes significant changes in both the primary and tertiary structures of the HIV capsid protein, as shown in Fig. 4. For example, the amino acid sequence at A in the primary structure changes from “TINEEAAEWD” to “MVHMEAAEWD”. The helices in the secondary structure are shortened or disappear. For example, the helix at B is changed from eleven amino acids, “TLEEMMTACQG”, to seven, “EEMMTAC”. In addition, the amino acid sequence at C changes from “PIVQNIQGQM” to “PIV-NIQ-QM” with peptide bond breakage. In addition, according to the diagram, the tertiary structure changes significantly with the reaction time, which we will discuss and analyze below.
Fig. 4. Schematic diagram of the change in the capsid protein when oxidized by ROS with the increase in the reaction time, where (a) is the initial moment, (b) is 50 ps, and (c) is 200 ps.
The ROS-mediated structural damage of the HIV capsid protein mainly includes dehydrogenation, increase in oxygen-containing groups, shortening or elimination of helices, and breaking of peptide bonds. Our simulation results are shown in Table 3, with the corresponding variables in the caption. The results showed that DN–H, DC–H, IC O, Dhelix, and DCO–NH increased with the increase in CROS, indicating that the reaction was more violent and the structural changes in the HIV capsid protein were more obvious. With the increase in CROS, DN–H ranges from 3.13% to 12.48%, and DC–H ranges from 0.01% to 0.36%, indicating that ROS capture H atoms from N atoms more strongly than H atoms from C atoms. IC O ranging from 31.41% to 41.03% showed a large increase in the number of carbonyl groups, and there was a significant increase in DCO–NH, which, combined with dehydrogenation, resulted in a significant decrease in the number of helices in the HIV capsid protein.55
Table 3. Statistics of ROS oxidation of the HIV capsid protein. CROS is the ROS concentration, while DC–H and DN–H represent the reduction rate of C–H and N–H bonds, respectively. IC O and DCO–NH indicate the rate at which the carbonyl group increases and the rate at which the peptide bond decreases, respectively, and Dhelix denotes the reduction rate of amino acids comprising the helices.
| C ROS | D N–H | D C–H | I C O | D helix | D CO–NH |
|---|---|---|---|---|---|
| 23.01 μmol ml−1 | −3.13% | −0.01% | +31.41% | −51.54% | −6.14% |
| 103.28 μmol ml−1 | −8.00% | −0.07% | +34.62% | −52.12% | −7.01% |
| 309.86 μmol ml−1 | −12.48% | −0.36% | +41.03% | −55.13% | −10.04% |
Similar to the previous part, to distinguish the contribution of different ROS species to HIV capsid protein oxidation, we calculated the oxidation degree by O, O3, and ˙OH at different concentrations. The results are shown in Table 4, where CX represents the concentration of X. As the concentration increases, DN–H, DC–H, IC O, Dhelix, and DCO–NH increase in a positive correlation. Horizontal comparison of O, O3, and ˙OH showed no significant difference in DN–H, DC–H, IC O, Dhelix, and DCO–NH.
Statistics of the oxidization of the HIV capsid protein by ˙OH, O3 and O with different concentrations, where C˙OH, CO3, and CO denote the concentrations of ˙OH, O3 and O, respectively.
| C O | D N–H | D C–H | I C O | D helix | D CO–NH |
|---|---|---|---|---|---|
| 7.09 μmol ml−1 | −1.40% | −0.03% | +35.83% | −55.23% | −6.36% |
| 31.85 μmol ml−1 | −4.74% | −0.04% | +42.05% | −57.42% | −8.88% |
| 95.55 μmol ml−1 | −9.27% | −0.15% | +54.55% | −64.50% | −11.37% |
| C ˙OH | D N–H | D C–H | I C O | D helix | D CO–NH |
|---|---|---|---|---|---|
| 7.09 μmol ml−1 | −0.96% | −0.01% | +32.05% | −50.54% | −4.97% |
| 31.85 μmol ml−1 | −3.08% | −0.02% | +39.42% | −53.56% | −6.54% |
| 95.55 μmol ml−1 | −6.07% | −0.04% | +44.55% | −55.62% | −7.22% |
| C O3 | D N–H | D C–H | I C O | D helix | D CO–NH |
|---|---|---|---|---|---|
| 7.09 μmol ml−1 | −1.40% | −0.04% | +30.77% | −49.46% | −5.51% |
| 31.85 μmol ml−1 | −4.72% | −0.06% | +35.90% | −50.32% | −6.19% |
| 95.55 μmol ml−1 | −9.34% | −0.26% | +50.00% | −52.87% | −10.01% |
These results indicate that dehydrogenation is one of the main protein oxidation types, along with carbonylation, helical reduction, and peptide bond breaking. As shown in Tables 3 and 4, and Fig. 4, we observed all the phenomena of ROS-mediated oxidation of the HIV capsid protein. Dehydrogenation can promote changes in the protein’s primary structure. The increase in oxy-containing groups, such as carbonyl groups, will increase the degree of helix reduction to a certain extent, thus destroying the secondary structure of the HIV capsid protein. The breaking of peptide bonds will produce a new carbonyl group, breaking the peptide chain and affecting the tertiary structure. With the increase in ROS concentration, the oxidation degree of the HIV capsid protein increased significantly, which would inhibit the replication and transmission of HIV.
4. Conclusions
This work used MD simulations with ReaxFF force fields to investigate ROS-mediated structural damage to the gp120 protein and HIV capsid protein. Our results show that with increasing ROS concentrations, the structures of the gp120 protein and HIV capsid protein are oxidized more, especially via dehydrogenation, increasing oxygen-containing groups, shortening or breaking the helices and disrupting peptide bonds, and thus disrupting the primary, secondary, and tertiary structures of the HIV capsid and gp120 proteins. In particular, we found that ROS have a significantly higher ability to extract H atoms from N atoms than from C atoms. At the same ROS concentration, the degree of structural damage to gp 120 was greater than that to the HIV capsid protein. The effect of O on the increase in oxygen-containing groups and the fracture of peptide bonds during the oxidation of the gp120 protein is more significant than the effect of O3, and the effect of O3 is greater than that of ˙OH.
Our findings have important implications for understanding the role of ROS in regulating the structure and function of the HIV capsid protein and gp120 protein and for further elucidation of the mechanisms of primary, secondary, and tertiary structural disruption during the response. Currently, researchers are working to develop microplasma devices that are sufficiently tiny to potentially have applications in both subcutaneous and internal structures, which may broaden the scope of application, no longer limited to treating superficial tissues.56 Thus, this work can provide new insights into therapy for AIDS by utilizing ROS to destroy the proteins involved in HIV replication. However, the problem of determining the safe dose of ROS in normal cells and the effective dose to inhibit the activity of the HIV capsid protein and gp120 protein remains unresolved. The combination of experiments and simulations is particularly important, as it may provide new insights into the plasma treatment of HIV.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Author contributions
Cunjia Pan: data curation (equal); formal analysis (equal); methodology (equal); software (equal); validation (equal); writing – original draft (equal). Qiaoyue Chen: data curation (equal); formal analysis (equal); visualization (equal); writing – original draft (equal). Danfeng Liu: data curation (equal); formal analysis (equal); visualization (equal); writing – original draft (equal). Mingming Ding: conceptualization (lead); funding acquisition (lead); investigation (equal); writing – review & editing (equal). Lili Zhang: investigation (equal); supervision (equal); writing – review & editing (equal).
Conflicts of interest
There are no conflicts to declare.
Acknowledgments
This work is financially supported by the National Natural Science Foundation of China (No. 22227804).
References
- Nijdam S. Desai K. V. Park S.-J. Sun P. P. Sakai O. Lister G. Eden J. G. Plasma Sources Sci. Technol. 2022;31:123001. [Google Scholar]
- Shao T. Wang R. Zhang C. Yan P. High Volt. 2018;3:14–20. [Google Scholar]
- Bruggeman P. J. Iza F. Brandenburg R. Plasma Sources Sci. Technol. 2017;26:123002. [Google Scholar]
- Lee J. Moon H. Ku B. Lee K. Hwang C.-Y. Baek S. J. Int. J. Mol. Sci. 2020;21:4556. doi: 10.3390/ijms21124556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privat-Maldonado A. Schmidt A. Lin A. Weltmann K.-D. Wende K. Bogaerts A. Bekeschus S. Oxid. Med. Cell. Longevity. 2019;2019:9062098. doi: 10.1155/2019/9062098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts K. Hamaguchi S. Ito T. Karahashi K. Knoops H. C. M. Mackus A. J. M. Kessels W. M. M. E. Plasma Sources Sci. Technol. 2022;31:103002. [Google Scholar]
- Shanker M. A. Khanashyam A. C. Pandiselvam R. Joshi T. J. Thomas P. E. Zhang Y. Rustagi S. Bharti S. Thirumdas R. Kumar M. Kothakota A. Food Control. 2023;151:109793. [Google Scholar]
- Therriault J. Servaes S. Tissot C. Rahmouni N. Ashton N. J. Benedet A. L. Karikari T. K. Macedo A. C. Lussier F. Z. Stevenson J. Wang Y.-T. Fernandez-Arias J. Stevenson A. Socualaya K. Q. Haeger A. Nazneen T. Aumont E. Hosseini A. Rej S. Vitali P. Triana-Baltzer G. Kolb H. C. Soucy J.-P. Pascoal T. A. Gauthier S. Zetterberg H. Blennow K. Rosa-Neto P. Alzheimer’s Dementia. 2023;19:4967–4977. doi: 10.1002/alz.13026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth J.-P. Mozetic M. Nikiforov A. Oehr C. Plasma Sources Sci. Technol. 2022;31:103001. [Google Scholar]
- Adamovich I. Agarwal S. Ahedo E. Alves L. L. Baalrud S. Babaeva N. Bogaerts A. Bourdon A. Bruggeman P. J. Canal C. Choi E. H. Coulombe S. Donko Z. Graves D. B. Hamaguchi S. Hegemann D. Hori M. Kim H. H. Kroesen G. M. W. Kushner M. J. Laricchiuta A. Li X. Magin T. E. Thagard S. M. Miller V. Murphy A. B. Oehrlein G. S. Puac N. Sankaran R. M. Samukawa S. Shiratani M. Simek M. Tarasenko N. Terashima K. Thomas Jr E. Trieschmann J. Tsikata S. Turner M. M. van der Walt I. J. van de Sanden M. C. M. von Woedtke T. J. Phys. D: Appl. Phys. 2022;55:373001. [Google Scholar]
- Okyere A. Y. Rajendran S. Annor G. A. Curr. Res. Food Sci. 2022;5:451–463. doi: 10.1016/j.crfs.2022.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z. Bai Y. Bai Y. Chu Y. Gu S. Xiang X. Ding Y. Zhou X. Food Chem. 2022;377:131932. doi: 10.1016/j.foodchem.2021.131932. [DOI] [PubMed] [Google Scholar]
- Domonkos M. Ticha P. Trejbal J. Demo P. Appl. Sci. 2021;11:4809. [Google Scholar]
- Oehrlein G. S. Hamaguchi S. Plasma Sources Sci. Technol. 2018;27:023001. [Google Scholar]
- Ma C. Nikiforov A. De Geyter N. Morent R. Ostrikov K. K. Curr. Opin. Chem. Eng. 2022;36:100764. [Google Scholar]
- Okada T. Chang C.-Y. Kobayashi M. Shimizu T. Sasaki M. Kumagai S. Arch. Biochem. Biophys. 2016;605:11–18. doi: 10.1016/j.abb.2016.04.001. [DOI] [PubMed] [Google Scholar]
- Dave P. Balasubramanian C. Hans S. Patil C. Nema S. K. Plasma Chem. Plasma Process. 2022;42:815–831. [Google Scholar]
- Guo J. Zhang Y. Molecules. 2023;28:3850–3859. [Google Scholar]
- Kim S. J. Chung T. H. Sci. Rep. 2016;6:20332. doi: 10.1038/srep20332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Meo S. Reed T. T. Venditti P. Manuel Victor V. Oxid. Med. Cell. Longevity. 2016;16:1245049. doi: 10.1155/2016/1245049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D. Talbot A. Nourmohammadi N. Sherman J. H. Cheng X. Keidar M. Biointerphases. 2015;10:040801. doi: 10.1116/1.4938020. [DOI] [PubMed] [Google Scholar]
- Santosh K. C. Abolfath R. Sci. Rep. 2022;12:19853. doi: 10.1038/s41598-022-23727-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkonen A. Kuchitsu K. Phytochemistry. 2015;112:22–32. doi: 10.1016/j.phytochem.2014.09.016. [DOI] [PubMed] [Google Scholar]
- Han L. Patil S. Boehm D. Milosavljevic V. Cullen P. J. Bourke P. Appl. Environ. Microbiol. 2016;82:450–458. doi: 10.1128/AEM.02660-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postiglione A. E. Muday G. K. Front. Plant Sci. 2020;11:968. doi: 10.3389/fpls.2020.00968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik N. K. Ghimire B. Li Y. Adhikari M. Veerana M. Kaushik N. Jha N. Adhikari B. Lee S.-J. Masur K. von Woedtke T. Weltmann K.-D. Choi E. H. Biol. Chem. 2019;400:39–62. doi: 10.1515/hsz-2018-0226. [DOI] [PubMed] [Google Scholar]
- Laroussi M. Lu X. Keidar M. J. Appl. Phys. 2017;122:020901. [Google Scholar]
- Weltmann K. D. von Woedtke T. Plasma Phys. Controlled Fusion. 2017;59:014031. [Google Scholar]
- Sierla M. Waszczak C. Vahisalu T. Kangasjarvi J. Plant Physiol. 2016;171:1569–1580. doi: 10.1104/pp.16.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keidar M. Plasma Sources Sci. Technol. 2015;24:033001. doi: 10.1088/0963-0252/21/3/034006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V. Straltsova D. Medvedev S. S. Pozhvanov G. A. Sokolik A. Yurin V. J. Exp. Bot. 2014;65:1259–1270. doi: 10.1093/jxb/eru004. [DOI] [PubMed] [Google Scholar]
- Attri P. Okumura T. Koga K. Shiratani M. Wang D. Takahashi K. Takaki K. Agronomy. 2022;12:482. [Google Scholar]
- Yang Z. Xiao A. Liu D. Shi Q. Li Y. Plasma Processes Polym. 2023;20:2200242. [Google Scholar]
- Amellal B. Murphy R. Maiga A. Brucker G. Katlama C. Calvez V. Marcelin A. G. HIV Med. 2008;9:790–793. doi: 10.1111/j.1468-1293.2008.00632.x. [DOI] [PubMed] [Google Scholar]
- Link J. O. Rhee M. S. Tse W. C. Zheng J. Somoza J. R. Rowe W. Begley R. Chiu A. Mulato A. Hansen D. Singer E. Tsai L. K. Bam R. A. Chou C.-H. Canales E. Brizgys G. Zhang J. R. Li J. Graupe M. Morganelli P. Liu Q. Wu Q. Halcomb R. L. Saito R. D. Schroeder S. D. Lazerwith S. E. Bondy S. Jin D. Hung M. Novikov N. Liu X. Villasenor A. G. Cannizzaro C. E. Hu E. Y. Anderson R. L. Appleby T. C. Lu B. Mwangi J. Liclican A. Niedziela-Majka A. Papalia G. A. Wong M. H. Leavitt S. A. Xu Y. Koditek D. Stepan G. J. Yu H. Pagratis N. Clancy S. Ahmadyar S. Cai T. Z. Sellers S. Wolckenhauer S. A. Ling J. Callebaut C. Margot N. Ram R. R. Liu Y.-P. Hyland R. Sinclair G. I. Ruane P. J. Crofoot G. E. McDonald C. K. Brainard D. M. Lad L. Swaminathan S. Sundquist W. I. Sakowicz R. Chester A. E. Lee W. E. Daar E. S. Yant S. R. Cihlar T. Nature. 2020;584:614–618. doi: 10.1038/s41586-020-2443-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. Guo Y.-C. Zhang X.-L. Deng L. Sang P. Yang L.-Q. Liu S.-Q. Biochim. Biophys. Acta, Biomembr. 2020;1862:183217. doi: 10.1016/j.bbamem.2020.183217. [DOI] [PubMed] [Google Scholar]
- Featherstone A. Aiken C. J. Virol. 2020;94:594. doi: 10.1128/JVI.00594-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. Kang B. H. Pancera M. Lee J. H. Tong T. Feng Y. Imamichi H. Georgiev I. S. Chuang G.-Y. Druz A. Doria-Rose N. A. Laub L. Sliepen K. van Gils M. J. de la Pena A. T. Derking R. Klasse P.-J. Migueles S. A. Bailer R. T. Alam M. Pugach P. Haynes B. F. Wyatt R. T. Sanders R. W. Binley J. M. Ward A. B. Mascola J. R. Kwong P. D. Connors M. Nature. 2014;515:138–147. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y. Wang X. Li S. Lian-Li Li Q. Xu T. Zhao T. Zhang Y. T. Int. J. Biol. Macromol. 2023;242:124944. doi: 10.1016/j.ijbiomac.2023.124944. [DOI] [PubMed] [Google Scholar]
- van Duin A. Dasgupta S. Lorant F. Goddard W. I. J. Phys. Chem. A. 2001;105:9396–9409. [Google Scholar]
- Aktulga H. Fogarty J. Pandit S. Grama A. Parallel Comput. 2012;38:245–259. [Google Scholar]
- Senftle T. P. Hong S. Islam M. M. Kylasa S. B. Zheng Y. Shin Y. K. Junkermeier C. Engel-Herbert R. Janik M. J. Aktulga H. M. Verstraelen T. Grama A. van Duin A. C. T. npj Comput. Mater. 2016;2:15011. [Google Scholar]
- Monti S. Corozzi A. Fristrup P. Joshi K. L. Shin Y. K. Oelschlaeger P. van Duin A. C. T. Barone V. Phys. Chem. Chem. Phys. 2013;15:15062. doi: 10.1039/c3cp51931g. [DOI] [PubMed] [Google Scholar]
- Cui H. Lai R. Yuan S. Liao C. Wang A. Li G. J. Chem. Theory Comput. 2023;19:4286–4298. doi: 10.1021/acs.jctc.3c00013. [DOI] [PubMed] [Google Scholar]
- Li S. Yao X. Wang X. Tian S. Zhang Y. Innovative Food Sci. Emerging Technol. 2022;80:103101. [Google Scholar]
- Ni T. Gerard S. Zhao G. Dent K. Ning J. Zhou J. Shi J. Anderson-Daniels J. Li W. Jang S. Engelman A. N. Aiken C. Zhang P. Nat. Struct. Mol. Biol. 2020;27:855–862. doi: 10.1038/s41594-020-0467-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A. P. Aktulga H. M. Berger R. Bolintineanu D. S. Brown W. M. Crozier P. S. in’t Veld P. J. Kohlmeyer A. Moore S. G. Nguyen T. D. Shan R. Stevens M. J. Tranchida J. Trott C. Plimpton S. J. Comput. Phys. Commun. 2022;271:108171. [Google Scholar]
- Momma K. Izumi F. J. Appl. Crystallogr. 2011;44:1272–1276. [Google Scholar]
- O’Boyle N. M. Banck M. James C. A. Morley C. Vandermeersch T. Hutchison G. R. J. Cheminf. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. Abstr. Pap. Am. Chem. Soc. 2004;228:313–314. [Google Scholar]
- Akagawa M. Free Radical Res. 2021;55:307–320. doi: 10.1080/10715762.2020.1851027. [DOI] [PubMed] [Google Scholar]
- Arriaga D. K. Kang S. Thomas A. A. J. Org. Chem. 2023;88:13720–13726. doi: 10.1021/acs.joc.3c01372. [DOI] [PubMed] [Google Scholar]
- Jodzis S. Patkowski W. J. Electrost. 2017;85:43–51. [Google Scholar]
- Cabiscol E. Tamarit J. Ros J. Mass Spectrom. Rev. 2014;33:21–48. doi: 10.1002/mas.21375. [DOI] [PubMed] [Google Scholar]
- Funato Y. Miki H. Methods. 2014;65:184–189. doi: 10.1016/j.ymeth.2013.06.032. [DOI] [PubMed] [Google Scholar]
- Brany D. Dvorska D. Halasova E. Skovierova H. Int. J. Mol. Sci. 2020;21:2932. doi: 10.3390/ijms21082932. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.