Abstract
Radiotherapy aims to achieve a high tumor control probability while minimizing damage to normal tissues. Personalizing radiotherapy treatments for individual patients, therefore, depends on integrating physical treatment planning with predictive models of tumor control and normal tissue complications. Predictive models could be improved using a wide range of rich data sources, including tumor and normal tissue genomics, radiomics, and dosiomics. Deep learning will drive improvements in classifying normal tissue tolerance, predicting intra-treatment tumor changes, tracking accumulated dose distributions, and quantifying the tumor response to radiotherapy based on imaging. Mechanistic patient-specific computer simulations (‘digital twins’) could also be used to guide adaptive radiotherapy. Overall, we are entering an era where improved modeling methods will allow the use of newly available data sources to better guide radiotherapy treatments.
Radiotherapy aims to optimize the ratio between tumor eradication probability and normal tissue toxicity probabilities. This requires selecting the best dose distribution and the best dose-fractionation prescription. Treatment personalization has been limited to date and focuses on mainly physical factors that constrain the possible dose distribution. However, genuinely personalizing a course of radiotherapy for any patient ultimately depends on integrating the physical aspects of treatment planning with quantitative models of predicted disease control and normal tissue complication risk. Tumors vary enormously concerning genomic alterations, vascular supply, cell density, transcriptomic profiles, hypoxia, glucose consumption, cellular radiation repair capacity, heterogeneity, and many other factors.1 Invasion into surrounding tissues is also a highly variable process.2–4 In addition, local control may depend on multiple host factors, including immune-related variables, such as tumor invasive lymphocytes (TILS).5,6 Hence, individual tumors can be said to have individual dose-response curves that are steeper than population dose-response curves.7 As discussed below, multidimensional information could further define cancer subtypes with steeper dose-response characteristics compared to the population average.
Figure 1 shows an example of dose-response curves for tumor control probability (TCP) and multiple toxicity endpoints, estimated for an early-stage non-small lung cancer treatment plan delivered at MSK.8–10 Most of these curves are relatively shallow, making a truly personalized optimization of radiotherapy for this patient difficult. Nonetheless, there are many reasons to believe that predictions willl improve. Cancer imaging relevant to radiotherapy response has become multimodal and quantitative and can be deployed during a course of RT. In-room imaging technologies and AI autosegmentation tools allow us to image and adapt radiotherapy routinely. The ability to sample or image many disease characteristics, including how they may change over the course of radiotherapy,11 and to integrate all this information, together with biological predictors, into dose-response prediction models, promises a new era of clinical and research progress in radiotherapy. The challenge is to answer how all this information should be integrated into dose-response and physical-response (e.g., tumor regression) predictions. Which data elements or features are predictive, nonredundant, and practical to collect and routinely analyze? What is the optimal data integration and prediction method: through statistical models such as nomograms, comprehensive AI models, or minimal-complexity mechanistic mathematical models?
Figure 1.
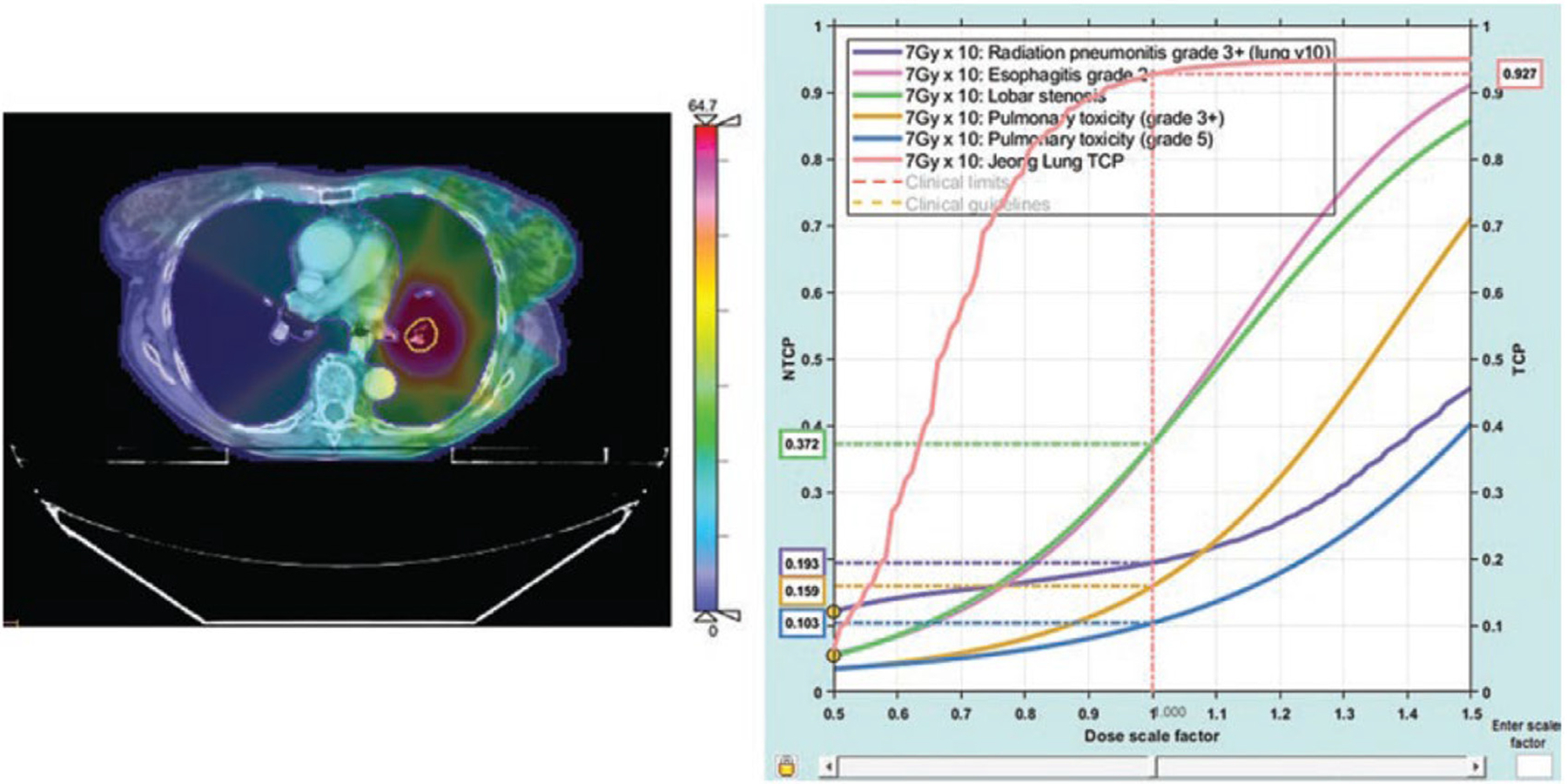
Lung cancer radiotherapy patients have individualized dose-response curves. Dose-response predictions, for tumor eradication and normal tissue risks, calculated for an early-stage nonsmall lung cancer treatment plan. Left: A planning CT scan overlaid with a dose colorwash. Right: the left-most curve shows the probability of local control, which is high for this dose and fractionation prescription (93%). The risk of toxicities (pneumonitis, esophagitis, bronchial stenosis) are shown. All curves intersect a 1.0 scale factor at the prescription fraction size. Data science could drive the development of predictive models that account for underlying biology and radiobiology, providing an improved basis for treatment decisions [reprinted with permission from10]).
The need to accelerate efforts to model cancer and cancer treatments mathematically is now well-recognized. In a recent (2021) NIH-DOE series of workshops on advancing computational approaches for predictive oncology in the clinical and research domains of radiation oncology,12 one key question identified was “How can we develop computational models to simulate how radiation kills a cell, affects a group of cells, transforms a tumor, and impacts a patient— and how does underlying patient-specific heterogeneity affect this process at multiple scales?” An important conclusion of the workshop was that patient-specific computer simulation “…provides a paradigm shift in oncology because it will enable data-driven simulation models to be integrated into routine clinical care—a key step toward the goals of precision oncology: to uniquely and continuously tailor treatment to each patient over time.”
We are entering an era where new or improved informatics tools and analysis methods allow us to use many rich data sources within reach to better guide cancer research and treatments. Models of individualized disease response and normal tissue toxicity dose-response curves could be integrated directly into the planning optimization process.13 New deep-learning tools in development will be able to track and monitor accumulated doses14–17 and even predict tumor regression patterns over the course of treatment,18 providing an improved basis for decision-making and plan adaptations. In this article, we briefly review a selection of information sources and model-building approaches for physically and biologically optimized radiotherapy. These include new opportunities based on real-world data, multidimensional imaging, radiomics and deep learning analyses, tumor and normal tissue genomics, monitoring longitudinal tumor changes, normal tissue complication modeling, and tumor control modeling. We emphasize the opportunities to advance radiation oncology via data science approaches, but this is not to diminish the critical role of hypothesis-driven preclinical studies and prospective clinical trials. Some important topics of relevance to data science, including blood based assays of tumor cells or circulating tumor DNA,19 are only mentioned briefly.
Missing Real World Data (RWD) in radiation oncology.
Predictive model building typically starts with retrospective institutional datasets. However, the lack of well-annotated real-world datasets is a major obstacle to advancement in radiation oncology.20 While data on patient characteristics, imaging, pathology, genomics, and treatments can theoretically be integrated, this is rarely done in practice and is not well-supported by current informatics systems. Our experience is that the reliability (i.e., correctness) of data collected during clinical workflows correlates with clinical usefulness.21,22 This is yet another incentive to develop more effective clinical workflow tools in radiation oncology, a win-win proposition.23
New informatics tools and AI large language models (LLMs) could help create more complete “patient data stories” by automatically and reliably extracting information from unstructured health record narratives, inferring context, and linking data elements.24–30 This could increase RWD’s availability to improve research and clinical care.31,32 Data standards, shared ontologies, and increasingly standardized electronic health record systems support the feasibility of large-scale RWD datasets.33,34 Patient data stories can be made comparable and computable using graphs.35 Graph-based analyses could probe numerous real-world issues, such as differences in outcomes reported between different classes of healthcare systems36 or possibly the impact of an initially incorrect cancer diagnosis. However, fully representing the patient story computationally, even for the application of clinical guidelines,37 is an unsolved problem. The scale and presence of errors in clinical datasets also represents methodological obstacles. Using a new generation of AI tools to monitor narrative dictation in real time to flag ambiguities or to identify missing data elements could increase the efficiency and reliability of RWD collection. For clinical data mining, an “hypothesis laundering” approach could leverage machine learning on large “dirty” RWD datasets to generate a relevant hypothesis, which could then be rigorously tested on a smaller, curated “clean” dataset. This would balance the benefits of analyzing large datasets with the need for rigorous hypothesis testing. Creating comprehensive, integrated RWD datasets and computable patient stories has implications beyond specific medical fields and could potentially improve the applicability of medical guidelines and algorithms more broadly. Capturing comprehensive clinical data that is structured and computable could enable a new era of true learning health systems.20
Connecting radiobiology and genomics.
Tumor and germline genomics have shown significant potential for improving patient-specific dose-response predictions. Torres-Roca and colleagues developed the Radiosensitivity Index (RSI) based on tumor mRNA expression profiles for 10 key genes and demonstrated impressive predictive power for local control and survival endpoints across multiple cancer types.38–42 This has been further refined into the genomically adjusted radiation dose (GARD) metric by Scott et al.43 These results firmly establish the relevance of gene expression patterns in determining clinical radiation response. Increasingly comprehensive multi-omic screens of radiosensitivity have recently become available.44 Manem and colleagues have recently developed multigene mRNA radiosensitivity predictors using multiple preclinical datasets.45,46
The genomics of normal tissue radiation response (often called radiogenomics) is also a promising target for continued progress. Single gene validation studies have generally not been positive, probably because effect sizes from individual alleles are typically small,47 although the XRCC1 single-nucleotide polymorphism (SNP) appears to be important.48 Efforts to build multigene predictive models by the REQUITE genome-wide association studies (GWAS) consortium have shown promising performance for multiple endpoints.49,50 In addition, several recent studies have shown that machine-learning approaches applied to germline GWAS can classify toxicity risk based on multi-SNP models.51–55 In particular, the novel preconditioned random forest regression (PRFR) algorithm developed by Jung Hun Oh and colleagues54 has successfully predicted risks for multiple postradiotherapy endpoints, including late rectal bleeding,54 erectile dysfunction,54 urinary toxicity,56 hematuria,52 and radiation-induced second breast cancers.57 Post-hoc network analyses of the models can identify relevant interacting genes and biological pathways contributing to toxicity risk. For normal tissue toxicity, tissue-specific genes and pathways involved in maintenance or function are often identified as important, rather than DNA damage repair pathways. The REQUITE consortium has also shown that an AI sparse autoencoder approach is effective for multi-SNP toxicity prediction.58 These results together indicate the potential of machine learning and AI methods applied to GWAS to produce genomic profile risk models predictive enough for clinical use. A critical future obstacle will be making this area of research beneficial to all patients, not just those in the largest ethnic and racial groups.59 The decreasing cost of obtaining patient germline profiles makes this approach feasible for clinical implementation. However, continued multi-institutional collaborations will be required to produce clinically useful and validated tools.
Quantitative imaging biomarkers can help refine patient-specific dose-response curves.
Many baseline imaging parameters are relevant to radiotherapy response.60–62 Quantitative imaging has shown significant potential in predicting and assessing radiotherapy response through various biomarkers and techniques.63 We mention a few salient results: FDG-PET imaging predictors often correlate with poor outcomes. For example, higher SUVmax values correlate with more aggressive disease and radioresistance in head and neck cancers and NSCLC.64 A modeling analysis of published data for oropharyngeal cancer estimated that tumors with abovemedian SUVmax values required about 20% higher radiation dose to achieve the same local control rates as those patients with below-median values.62 MRI apparent diffusion coefficient (ADC) imaging has been shown to correlate with local control for multiple tumor sites.65–67 Lower diffusion relates to greater cellular density and worse local control.68 Pretreatment hypoxia, imaged, for example, using PET F18-labeled misonidazole (F-MISO)69,70 has been correlated with poor outcomes in many tumor sites.71 Hypoxia has also been associated with an immune-cold microenvironment in colorectal tumors.72 It should be emphasized that all these methods of interrogating the tumor are closely linked. Figure 2 illustrates this by relating image-based metrics extracted for an H&N cancer cohort.73 Closely correlated metrics are directly connected in the graph. It may be surprising to see a strong correlation between, for example, MRI-ADC values and PET-FDG mean standard uptake values in the lesion. Understanding these correlations could be a key to defining actionable yet practical clinical biomarkers. Quantitative imaging measures of residual disease may also be coupled with serum molecular measures of disease under development.74,75
Figure 2.
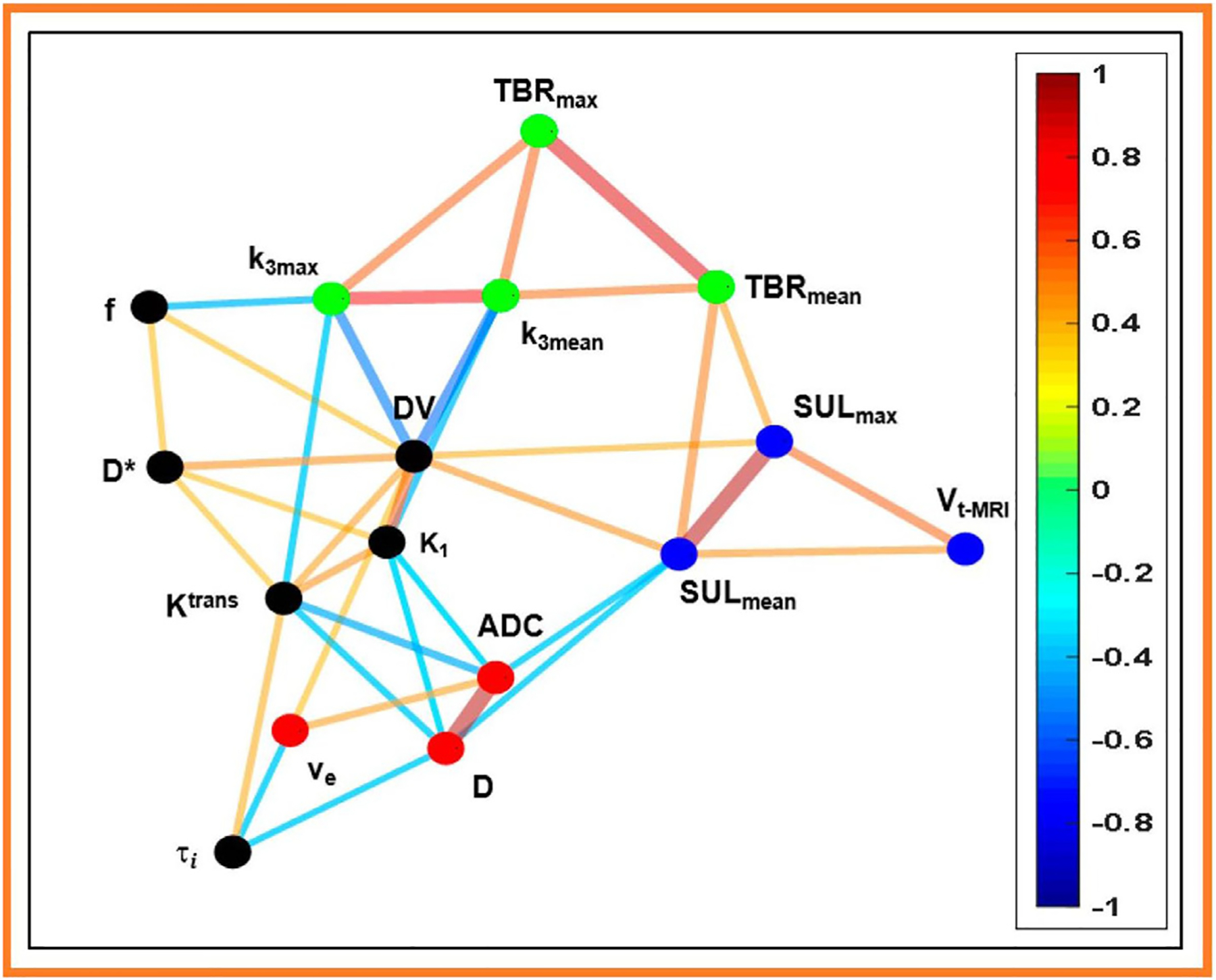
A relatedness graph of quantitative metrics derived from images for H&N cancer patients (N=23), as reported by Paudyal et al.73 Image types include FDG-PET/CT (SUL [lean body mass corrected SUV], FMSIO-PET/CT (K1 [perfusion], k3 [hypoxia], TBR [hypoxia tumor blood ratio], and DV [distribution volume]), DW-MRI (ADC [apparent diffusion coefficient]), IVIM (D [true diffusion coefficient], D* [pseudo diffusion coefficient,] and f [perfusion fraction]), and FXR DCE-MRI (Ktrans [volume transfer constant], ve [volume fraction of extravascular and extracellular space], and τi [intracellular water mean lifetime]). A spin-glass model was used to derive closely-related communities, indicated by node color. Edge color refers to Spearman’s rank correlation coefficient, ρ. (Used with permission).
Extracting more information from images: radiomics.
Extracting more detailed imaging features, called ‘radiomics,’ provides potentially useful inputs for predictive models.76–78 Recent reviews summarize evidence that radiomic models can provide histopathological and prognostically relevant information for many disease sites and subtypes.79–93 Radiomic features could be tested for inclusion in patient-specific dose-response models. For H&N tumors, Mukherjee et al.94 showed that radiomics models can predict tumor grade, perineural invasion, lymphovascular invasion, extracapsular spread, and HPV status. van Dijk et al.95 recently reported on an internally- and externally-validated model that used CT features and machine learning to identify H&N cancer classes of widely varying local control and overall survival levels. In some cases, the multi-modal bivariate distributions of image features may be important, as shown by Vallières et al.96 in predicting metastatic spread from sarcoma tumors using co-located FDG-PET and MRI features. Important problems still persist with radiomics methods, however, including understanding the inter-relationship of unusual features with more standard prognostic metrics such as tumor volume.97
Predicting tumor phenotypes with deep learning.
More recently, artificial intelligence (AI) methods have often been used to further probe the predictive value of image characteristics.98,99 Deep learning AI image analysis tools essentially start with simple features and build up many combined features of different characteristics and scales.100 The learning process emphasizes features meaningful to the selected learning task, which may be an outcome endpoint. A recent review concluded that deep-learning-based models outperformed radiomic feature modeling in 65% of reported comparisons.101 However, low-dimensional radiomic models are more explainable, and understanding what key imaging features drive classification in individual cases may be critical to clinical adoption and to advancing cancer science.
Computer science advances in algorithms and techniques are being rapidly translated into medical imaging applications, and in particular, the impact of deep learning models on predicting outcomes based on clinical images is accelerating. For example, the recent publication of De Blase et al.102 showed that multiple standard clinical images could be combined, in this case including CT and FDG-PET images, as well as gross disease planning contours, within deep learning frameworks to predict multiple endpoints, including the risk of local failure and distant progression. Ma et al.103 in work from the same group, showed that a hybrid CNN vision transformer architecture could be used to develop a robust model to predict multiple endpoints, including local failure, distant progression, and overall survival. Patients in the low-risk groups (about 40%) for local-regional control and distant progression had few events during followup. HPV status was captured and accounted for separately from the deep learning predictions in the final prediction.
Another recent approach to using deep learning was published by Jiang et al.,104 who showed that multitask learning for gastric cancers could improve both performance and the interpretability of deep learning predictive models. Learned endpoints included the response to chemotherapy and immunotherapy and the tumor microenvironment. Significant splits in survival curve predictions may reflect distinct biology or radiobiology. It should be emphasized that validated image classification signatures could be applied to scans for surgical patients and correlated with underlying genomic and pathological signatures from surgical specimens to identify cancer subtypes. The correlation of tumor genomics with radiological phenotype is often called ‘radiogenomics,’ which is often confused with using the same term to mean radiosensitivity genomics. Shui et al. recently reviewed the substantial progress in relating specific radiomic features to clinically meaningful genetic tumor subtypes.105 Qian et al.106 have recently reviewed efforts to correlate lung cancer radiomics with underlying genetic or pathology markers. Deep learning approaches for cancer subtype classification and survival prediction using radiological imaging data show promise for providing biological insights, even without full integration with genomic data.107 For example, a study on lung adenocarcinoma patients used deep learning and radiomics networks based on CT imaging to predict histologic subtypes.108 The ability to predict outcomes from imaging alone could potentially guide treatment decisions and provide prognostic information noninvasively. Even without full data integration, deep learning overall survival classifications could be compared with genomic processing of tumor surgical specimens (mRNA, methylomics, copy number variations, spatial transcriptomics, etc.), potentially extending the ability of imaging to ‘see biology.’ The noninvasive nature of imaging could allow for longitudinal monitoring of tumor evolution and treatment response in ways not possible with tissue sampling.
While radiomic and deep learning quantitative imaging correlates show promise, challenges remain in harmonizing data across different scanners and protocols to reduce bias and improve clinical applicability.109 Ongoing research aims to overcome these obstacles and enable the widespread use of quantitative imaging in predicting and assessing radiotherapy response.
Capturing the longitudinal phenotypes of cancer.
The concept of a temporal disease trajectory in cancer and the potential for improved tracking of tumor growth rates represents a significant opportunity in oncology. We use the term temporal disease trajectory to refer to the progression of cancer over time, including its initiation, growth, response to treatment, and potential recurrence. Understanding this temporal and spatial trajectory is likely helpful for several reasons. In particular, a better understanding of the disease trajectory can help predict patient outcomes and adjust treatment strategies accordingly. The rate of growth can be an indicator of tumor aggressiveness, helping to guide treatment intensity. Studying temporal trajectories across many patients can reveal new insights into cancer biology, such as subtype and treatment efficacy. New streams of temporal information are also critical to leveraging the “digital twin” approach to guiding cancer care.110,111
Monitoring tumor growth rates before, during, and after treatment would be a paradigm-shifting practice that offers many benefits. This could provide a rapid assessment of whether a chosen therapy is effective, allowing real-time adaptations to treatment plans based on tumor response and possibly identifying the development of treatment resistance before it becomes clinically apparent. Detailed growth rate data can also drive new biological investigations, for example, by correlating growth rates with genomic data to identify key drivers of tumor progression and exploring how the tumor microenvironment (e.g., hypoxia, cellular density) influences growth rates and treatment response. As one example, Attalah et al. reported that the growth rate of lung tumors before RT is a strong negative predictor of both local control and overall survival.112 They later validated this finding in a separate cohort.113 They concluded that “SGR [tumor specific growth rate] can be used in conjunction with other well-known predictive factors to formulate a practical predictive model to identify subgroups of the patient at higher risk of recurrence after SBRT.”
Changes in imaging parameters during radiotherapy have also been shown to correlate with outcomes. For example, for nonsmall cell lung cancer (NSCLC) patients, a more substantial reduction in SUVmax observed over the course of radiotherapy correlated with better outcomes.114 As discussed further below, the resolution of PET-detected hypoxia early in treatment is associated with better outcomes.115 Intratreatment changes in MRI apparent diffusion coefficient (ADC) values, related to tumor cell density, also predict outcomes.66,116 Trada et al. have shown that, in head and neck cancer, a combination of increasing ADC and decreasing FDG-PET metabolic tumor volume correlates with treatment success.117 Intratreatment changes in cfDNA may also help guide adaptations.118 For NSCLC patients treated with stereotactic body radiotherapy, Residual FDG-PET at 12 weeks post-RT is well-correlated with local failure.119
Changes in imaging features during therapy, often called delta-radiomics, have been extracted to predict response. Delta-radiomics features have been shown to be predictive in multiple settings, beyond baseline values, for example: H&N local progression 3 months post-RT based on cone-beam CTs,120 local control for NSCLC based on weekly CTs,121 pancreatic cancer pathological response based on per-fraction CTs,122 and NSCLC response to immunotherapy post-RT using contrast CTs taken after 2 to 4 treatment cycles.123 Regarding normal tissue toxicity, in RT for H&N cancer, weekly CT delta-radiomics have also been used by van Dijk et al. to improve the early prediction of xerostomia.124 Thor et al. showed that early expansion of the esophagus during RT for NSCLC as determined on weekly MRIs was correlated with the later development of severe acute esophagitis.125
Longitudinal tracking of metastases.
For patients who develop metastases, quantifying the time course could be impactful. For example, Hsu et al.126,127 recently developed a deep learning methodology and software system for automatically tracking the appearance and location of brain metastases across multiple longitudinal MRI scans. Patients vary widely in terms of temporal and spatial patterns in the appearance of brain metastases, e.g., widespread metastases appearing a few months after treatment vs. an isolated metastasis appearing years later. Understanding how such factors should guide treatment management requires routine quantitative tracking in space and time. Tracking brain metastases’ temporal/spatial appearance and correlating with underlying primary and metastatic biology may lead to valuable scientific and clinical insights. AI tools for monitoring the volume change of tumors can be more reliable than by-eye RECIST size marking.128 New and future AI methods will likely be able to track tumor changes in detail, even giving information about how a tumor grows or loses mass and the variability of mass density changes across the tumor volume. Patient-specific histograms of tumor mass gains or losses per unit time could provide a new window into disease prognosis and treatment response.
Data integration challenges.
Standard nomograms typically use regression modeling to combine quantified risk levels from multiple data elements. However, the high-dimensional information in genomics, radiological imaging, and pathology features must arguably be related. Hence, more effective integration of multiple data types requires building topological cross-links that are currently primarily unknown. For several reasons, multimodal data integration in oncology offers significant potential for improving research, diagnosis, and clinical management. Different diagnostic modalities provide complementary insights that are crucial for comprehensive treatment management. For example, integrating imaging, histopathology, genomics, and clinical data can offer a more complete picture of a patient’s condition than any single modality alone.129,130 Combining data from multiple modalities can potentially increase the accuracy of prognostic models. Integrated analyses can potentially reveal overlaps or shared dependencies between different data types, providing a deeper understanding of disease mechanisms and patient outcomes.130 Multimodal data integration may be more effective at identifying important cancer subtypes or host interactions that are not apparent when examining single data types in isolation.131 In addition to graph-based integration methods, AI-driven data fusion strategies include early fusion (combining raw data or features at the input level), late fusion (aggregating predictions from separate unimodal models), and intermediate fusion (iteratively learning improved feature representations under a multimodal context).130 Disparate systems of different kinds and scales must be integrated for optimal predictive power. Consequently, multiscale modeling coupled with machine learning is also an important data integration and modeling methodology.132,133
AI methods can be used to convert one type of data into another type (imperfectly), potentially establishing important co-dependencies. For example, a deep learning model can use standard pathology slides to estimate molecular marker densities usually only available with more advanced immunofluorescence methods.134 The problem of integrating high dimensional data of multiple data types into a joint network is an active area of investigation. Jaume et al.135 presented a transformer model (named SurvPath) that can learn the relationships between whole slide pathology features and bulk transcriptomics. Outside of AI, high-dimensional biomedical data is typically represented in a connected graph format, with links indicating a strong correlation or known chemical reactions. Some networks can be built from known interactions (such as protein interaction networks), whereas network topologies for other data types need to be constructed, for example for radiomic features. This can be done, for example, by analyzing the correlations of radiomic features derived from relevant image collections.136 A subject sample can be represented as a normalized distribution in such a network. Optimal mass transport theory is a powerful method for quantifying the closeness of any two samples. Multiple data types can be represented as multigraphs with some inter-modality cross-links. Using such methods, meaningful comparisons can be made on high-dimensional spaces with a modest number of subject samples. As an example of multimodal non-AI data fusion, Pouryahya et al.131 integrated breast cancer multiomics (mRNA, copy number variations, and methylomics) on a graph network representing known gene interactions. Unsupervised optimal mass transport distances were used to cluster distinct subtypes, resulting in identifying a biologically distinct and particularly lethal subtype.
The heart of radiotherapy is the tissue-registered dose distribution. Hence, developing improved Normal Tissue Complication Probability (NTCP) models is crucial for advancing radiotherapy treatment planning and optimizing patient outcomes. One key is assembling appropriately large multi-institutional datasets. In addition to the QUANTEC papers,137 which are now more than a decade old, many of the important issues are discussed in the comprehensive book edited by Rancati and Fiorino.138 Despite extensive research into NTCP models, clinical practice still heavily relies on dose-volume threshold constraints. While these constraints are practical, they greatly oversimplify the complex relationship between the dose distribution and tissue response. Validated NTCP models have the potential to provide more nuanced and relevant information about treatment outcomes compared to standard dose-volume constraint guidelines.139,140 As noted above, many NTCP risk curves are relatively shallow, meaning that complications risk changes gradually with dose. Converting these curves into binary dose-volume constraints can lead to overemphasizing specific thresholds and losing important context about the continuous nature of risk.137
A significant challenge in developing robust NTCP models is the limited variance in dose-distribution data from institutions that adhere to consistent dose-volume constraints. This lack of variability in potentially predictive dose-volume metrics makes it difficult to derive comprehensive NTCP models that accurately reflect the full range of the dose-response relationship. Unsurprisingly, NTCP models that combine multi-institutional and multiprotocol data can demonstrate altered tolerance relationships141. There is an opportunity to create multi-institutional treatment plan dose-distribution databases, which could provide a broader range of dose-distribution data, allowing for the development of more robust and generalizable NTCP models.
While privacy issues are an obstacle for multi-institutional databases, advanced techniques for mapping dose-distributions to representative patient anatomies offer a potential solution. Point-wise correlations with outcomes, called ‘voxel based analysis,’ can provide essential insights into toxicity predictors.142–146 These methods could allow institutions to contribute anonymized dose-distribution data while maintaining patient privacy.
Most NTCP model building is low-dimensional, using only a few fitting parameters to conserve statistical power. One effective method for building low-dimensional and interpretable dose volume predictive models is to use ensemble model methods, for example, by combining (‘bagging’) models constructed on bootstrap data replicates. Thor et al.147 used this approach to effectively model the overall survival impact of irradiating cardiopulmonary structures in the RTOG 0617 clinical trial data.
Spatial NTCP models aim to geometrically represent the 3D dose distribution, allowing for a more comprehensive analysis of dose patterns and their impact on normal tissue complications.148 Analogous to genomics and radiomics, ‘dosiomics’ is a term used to describe the more general extraction and modeling of dose features to predict outcomes.149 Multiple recent efforts have been made to improve toxicity predictions using deep learning based primarily on the dose distribution.150 Men et al.151 used a deep learning approach to predict xerostomia following H&N RT based on the dose distributions of RTOG trial 0522. Humbert-Vidan et al.152 recently reported that deep learning could beat a random forest approach for predicting mandibular osteoradionecrosis. Reber et al.153 recently investigated machine learning and deep learning approaches to predicting osteoradionecrosis from dose-distribution data. Machine learning on extracted features had superior performance compared to deep learning. Other work has demonstrated that deep learning can be useful in classifying the risk of xerostomia in head and neck treatment plans151.
A recent exciting trend is the combination of tissue image features, whether extracted by radiomics or deep learning, with dose distribution features to predict toxicity risk. El Naqa et al.154 recently showed in a pioneering deep learning analysis that liver MRI images could be used to classify liver function and feed into a more traditional NTCP model of liver toxicity that accounts for dose-volume characteristics. Bin et al.155 used lung ventilation images in addition to the dose distributions as inputs to deep learning to improve the prediction of post-RT pneumonitis. An exciting aspect of their study was that they weighted the dose distribution by estimated ventilation values to create ‘functional dose’ maps. Zhang et al.156 recently constructed a nomogram that combined a lung radiomics score with a dosiomics score and other clinical variables to predict pneumonitis.
Predicting the tumor dose-response curve.
Tumor Control Probability (TCP) modeling has evolved significantly to incorporate more realistic and mechanistic factors that impact tumor response to radiation therapy. We do not consider radiopharmaceutical therapy here, which has a set of distinct issues.157 As noted above, many TCP curves are relatively shallow (changes of 1–2% absolute increase in TCP per 1% increase in dose are typical.158) This shallowness is presumably due to a mix of previously unquantified factors that impact response. The mathematical goal of TCP modeling is to produce models that agree with clinical data and have steep dose-response curves. The scientific goal is to create models with a valid mechanistic basis so they can be extrapolated beyond the current data, even if just hypothetically. Here, we summarize some key factors in TCP modeling. The cellular access to oxygen or lack thereof (hypoxia) is nonuniform on both the microscale and the macroscale in human tumors.159,160 Hypoxia is known to correlate with poor clinical outcomes, as noted above.161 Incorporating hypoxia into TCP modeling is critical because hypoxic tumor cells are more radioresistant162,163 and also because hypoxia negatively affects other important factors, particularly reducing cellular proliferation.164,165 Not all tumor cells are stem cells capable of regrowing a tumor. Variation in stem cell fraction is also important and likely impacts the dose needed for control, similar to what is shown in preclinical studies.166 This is not currently accounted for in human TCP models,166 although spatial variability of tumor stem cell density could be taken into account.167,168 Tumors are also known to be heterogenous in clonal lineage. Cellular radiosensitivity measured using in vitro cell survival experiments based on repeat biopsy samples has demonstrated very high variability (coefficients of variation of 30–40% being typical169). Early bioeffect models sought to compare values of “biologically equivalent dose” (BED), rather than predicting the resulting TCP directly. This was done primarily using the Poisson distribution, based on in vitro experiments, to calculate the probability of zero clonogens surviving after radiotherapy. Models further incorporated the linear-quadratic (LQ) cell survival model, which accounts for both single-particle-track DNA lethal damage (linear in fraction dose) and multiple-track lethal interactions (quadratic in fraction dose). Cellular proliferation itself is also a vital resistance mechanism, as mentioned above. Proliferation can increase during a course of RT due to cell death and decreased resource competition, mimicking a reversal of Gompertzian (i.e., slowing) growth curves. This can reduce overall treatment efficacy, especially for longer treatment courses.170 For this reason, a time factor has often been added to the LQ framework to reflect a reduction in local control for treatments of increasing overall treatment time past an assumed ‘kickoff time.’171 This applies for tumors that proliferate significantly during conventionally fractionated radiotherapy (i.e., most tumors, excluding very indolent tumors such as prostate).171 The LQ model of tumor control has been used to analyze many radiotherapy datasets,172–174 and has been extended, for example, to account for the kinetics of repair processes.175
Going beyond these simple bioeffect equations, some models attempt a more mechanistic approach. One critical test of any mechanistic TCP model is whether it can recapitulate clinical dose-response relationships with reasonable parameters. Unfortunately, validating biological mechanisms is almost always a challenge due to a lack of adequate test data. As one potential approach to capturing the dynamic response to radiotherapy, Jeong et al. have published a flexible tumor radiobiological simulation model based on established radiobiological principles as well as the novel concept of an “energy budget,” representing the fact that, in a tumor, the competition for locally available oxygen invariably limits the fraction of cells that can progress through the cell cycle.176,177 The model recapitulates many critical aspects of known tumor biology and radiobiology, including doses required to achieve local control, tumor volume regression during RT, high cell loss factors, volume doubling times much longer than cell cycle times, and the increase in the dose required as overall treatment time increases past 3–4 weeks. The usefulness of the model was demonstrated in a fit to all published RT protocol results for early-stage non-small cell lung cancer178 (see Figure 3). In a further test, the same model could fit all the early-stage NSCLC radiotherapy outcome data from the Japanese carbon ion beam therapy results.179 As shown by Crispin-Ortuzar, the model is also consistent with previously reported image-based predictors of radiotherapy response.180 It is an open question whether this level of tumor dynamics is always needed as opposed to approaches with simplified dynamics.181 One interesting prediction of this mechanistic model is that radiotherapy may be more effective if delivered in a ‘primer shot’ approach, by giving a single fraction of 5–8 Gy followed by a break, to allow tumor reoxygenation, of 10 days to 2 weeks.182
Figure 3.
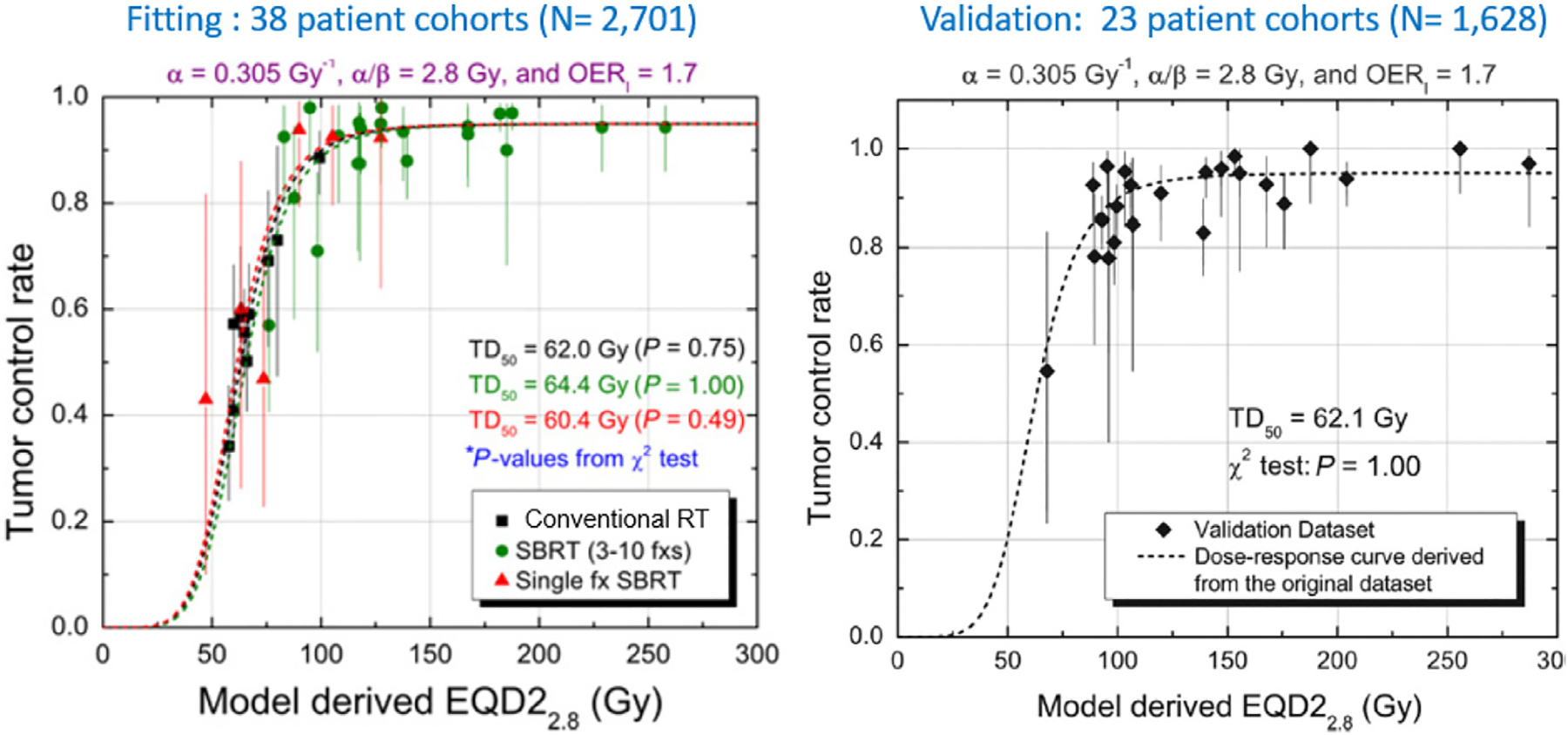
The Jeong model fit to all analyzable Stage I (localized) NSCLC cohorts as reported in Jeong et al.178 Left: 4-parameter model fit to 38 cohorts (published by 2010 and before, plus two internal datasets); Right: Validation test of fitted model to separate cohorts (published 2009–2014.) This mechanistic model is the first model of any kind shown to well-fit RT response across all fractionation regimes, from single fraction delivery to 2 Gy per weekday. The model identifies the point of dose intensity beyond which response flattens, regardless of fractionation (reprinted with permission.)
Shrinkage of NSCLC tumors during RT has been positively correlated with improved outcomes.183,184 Future developments of this or other TCP models could incorporate patient-specific imaging (e.g., FDG-PET, FMISO-PET hypoxia imaging, DWI-MRI) at baseline and changes measured during treatment using cone-beam CT (CBCT) or other modalities.185–187 Compartment modeling is just one possible approach.
A different approach to tumor response modeling emphasizes using partial differential equations to model the physical image of a tumor including the invasive edge.2,186,188–190 This can be linked with image-based machine learning as well. This approach has been used to predict tumor response to radiotherapy, for example in brain and breast tumors.191 An advantage of this approach is that it leverages the behavior of the tumor shape for learning.
TCP models could incorporate any number of features representing different characteristics of the problem.188,192,193 The impact of an immune contribution to dose-response may be considerable. For example, recurrence risk following post-op radiotherapy in the SweBCG91RT trial was reduced when tumor samples contained tumor-infiltrating lymphocytes (TILs).194 Immune ‘hot’ tumors could be identifiable partly through radiomics or deep learning models.195,196 The best modeling approach depends on the clinical use case, the disease site, histology, and the treatment modality. Preclinical studies are also an underused potential source of modeling data.
Local control is undoubtedly also a function of dose to subclinical disease, which has been shown to have an exceptionally shallow dose response.197 Unexplained failures to control early-stage NSCLC tumors, even at high doses, may be related to hard-to-image tumor cells in the peri-tumoral region, as indicated by the results of Davey’s model.198,199 In prostate cancer, the impact of daily kV image-guidance on outcomes has been shown to be related to the predicted likelihood of extracapsular extension.200 Bortfeld and colleagues have recently been studying how the nonimageable component of disease could be modeled and probabilistically estimated.201,202 Unkelbach and colleagues have been using models to estimate the probabilistic likelihood of lymph node spread in head and neck patients.203
The impact on TCP of low-dose regions inside or, more commonly, on the edge of a tumor is relatively unknown, although calculations can be made under assumptions.197 The ICRU paradigm (updated in ICRU 83204) rather dogmatically assumes that the proper guidance in RT is not to allow any drop in the prescription dose near the edge of a tumor. In the presence of localization uncertainty, subclinical disease uncertainty, and unavoidable dose-rolloff, this ‘paranoid target volume’ approach implies that large margins of normal tissue must also be irradiated to high doses. This already seems at odds with the recognition that RT is a balance between disease eradication and normal tissue toxicity.205,206 Fortunately, the stereotactic prescription approach that allows for more target volume dose heterogeneity and is more comfortable with dose gradients at the edge of a tumor seems to have superseded or at least supplemented the ICRU 83 guidance. Continuing improvements in pretreatment and on-table imaging also support this transition. The ICRU approach does not optimally manage how localization uncertainties are handled.207 A critical knowledge gap could be filled by inter-institutional analyses of the impact of target dose heterogeneity (e.g., GTV and PTV dose heterogeneity) on TCP. We hope these brief comments clarify that personalized TCP models could become much more refined and comprehensive while retaining scientific interpretability. Nonetheless, deep learning approaches directly on target dose distributions may lead to improvements in predictive power. For example, Dudas et al.208 recently used deep learning to estimate the risk of local failure for NSCLC treatments. Analysis showed that the dose periphery was the most predictive region (see Fig. 4). This may be related to peri-tumoral sub-clinical extension, as predicted by Davey’s model, discussed above.
Figure 4.
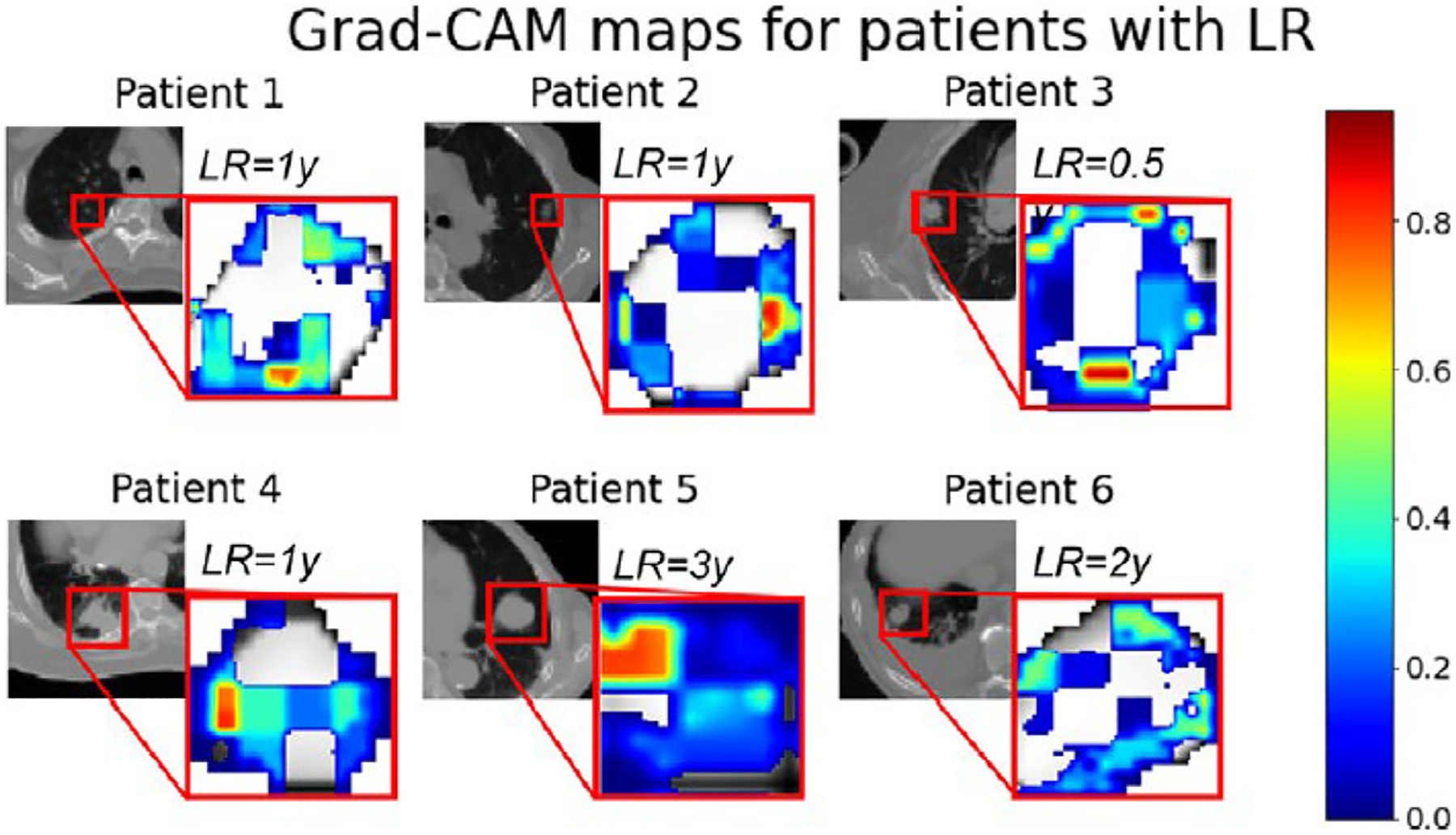
In a deep learning model published by Dudas et al.208 to predict the likelihood of local failure, attention analysis shows that the dose near the target volume edge was critical. (reprinted with permission.)
In summary, there is an urgent need to identify tumor subtypes and other radio-response factors to provide steeper dose-response relationships to guide treatment planning and personalized prescriptions. Validated predictive models, although difficult to build and validate, could utilize widely available data sources to have a positive impact on the treatment process of millions of cancer patients worldwide. Advances in radiation oncology modeling could connect radiobiology with current themes in cancer biology, support the rational use of multimodality imaging and integration with genomics, and give clinicians much more precise and personalized insights into prescription and planning tradeoffs. Predicting and monitoring early tumor response to treatment, informed by digital twin tumor simulations, could guide adaptive tumor prescription changes. How much effort will be required to perform these studies, and what is the potential payoff? Figure 5 shows our speculative schematic summarizing many of the topics reviewed here, ordered by the estimated level of resources required along the x-axis and the estimated upper ceiling of potential significance along the y-axis. We invite you to mark up the image based on your own estimations of what efforts would be most impactful. While many studies may be pursued by single institutions, multi-institutional studies, and even cooperative group studies will typically be required for themes to reach their full translational significance. Many under-developed aspects of radiation oncology data science could drive simultaneous improvements in disease control, toxicity avoidance, and efficient patient management.
Figure 5.
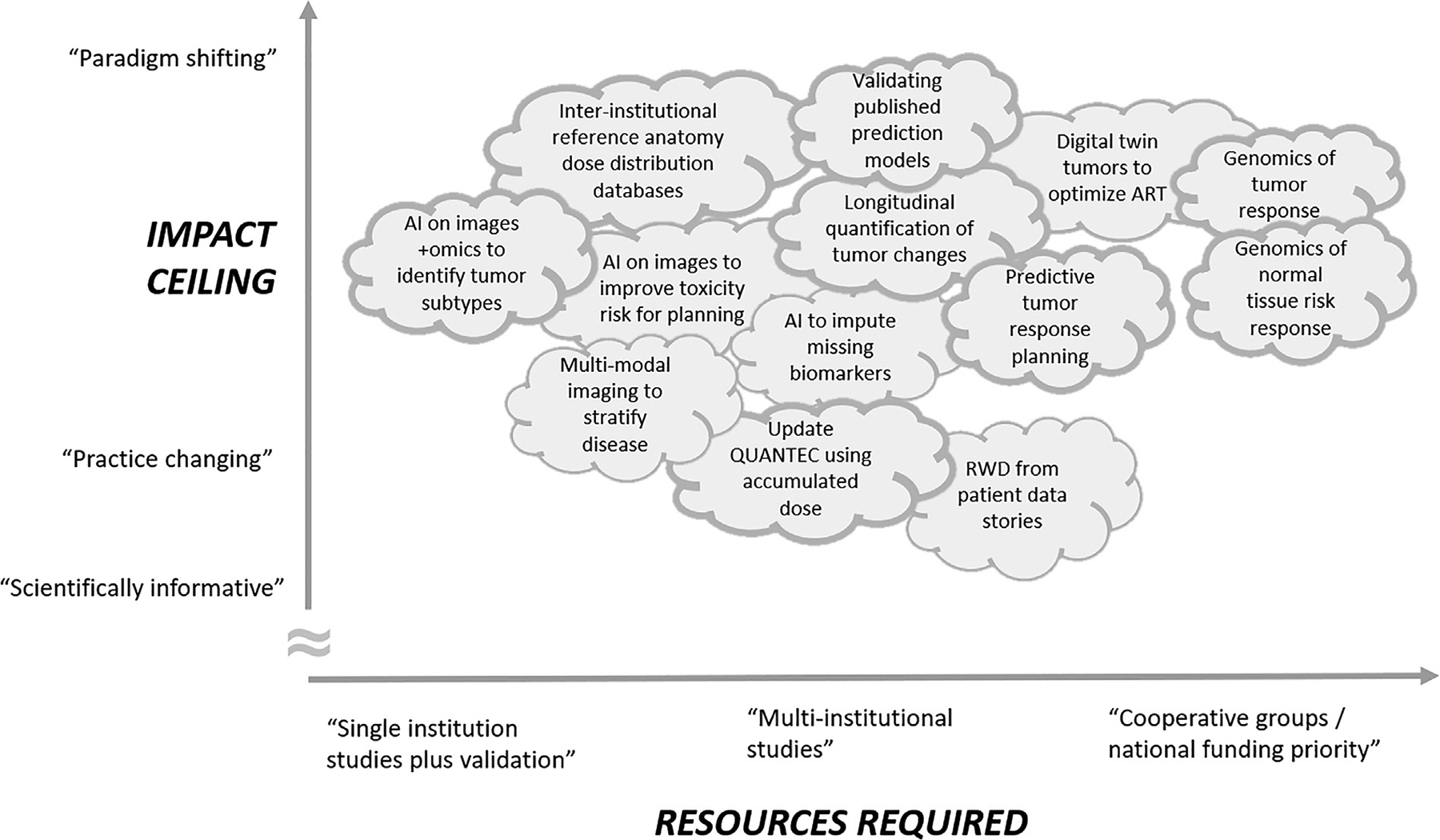
To stimulate thinking, we estimate the payoff versus resources required for developing and implementing modeling approaches we have discussed. We invite the reader to ‘correct’ the diagram.
Acknowledgments
The author wishes to acknowledge numerous discussions on these topics with colleagues at MSK and elsewhere, including Amita Shukla-Dave, Jung Hun Oh, Harini Veeraraghavan, Andrew Jackson, Maria Thor, John Humm, Aditya Apte, Jeho Jeong, Mireia Crispin-Ortuzar, Zeno Guow, Jan-Jakob Sonke, and Philippe Lambin.
References
- 1.Marusyk A, Polyak K: Tumor heterogeneity: causes and consequences. Biochim Biophys Acta 1805(1):105–117, 2010. 10.1016/j.bbcan.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen L, Painter K, Surulescu C, Zhigun A: Mathematical models for cell migration: a non-local perspective. Philos Trans R Soc Lond B Biol Sci 375, 2020:20190379. 10.1098/rstb.2019.0379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pandya P, Orgaz JL, Sanz-Moreno V: Modes of invasion during tumour dissemination. Mol Oncol 11(1):5–27, 2017. 10.1002/1878-0261.12019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carey SP, D’Alfonso TM, Shin SJ, Reinhart-King CA: Mechanobiology of tumor invasion: engineering meets oncology. Crit Rev Oncol Hematol 83 (2):170–183, 2012. 10.1016/j.critrevonc.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddy AV, Hill CS, Sehgal S, et al. : High neutrophil-to-lymphocyte ratio following stereotactic body radiation therapy is associated with poor clinical outcomes in patients with borderline resectable and locally advanced pancreatic cancer. J Gastrointest Oncol 13(1):368–379, 2022. 10.21037/jgo-21-513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Celebi F, Agacayak F, Ozturk A, et al. : Usefulness of imaging findings in predicting tumor-infiltrating lymphocytes in patients with breast cancer. Eur Radiol 30(4):2049–2057, 2020. 10.1007/s00330-019-06516-x [DOI] [PubMed] [Google Scholar]
- 7.Zagars GK, Schultheiss TE, Peters LJ: Inter-tumor heterogeneity and radiation dose-control curves. Radiother Oncol 8(4):353–361, 1987. 10.1016/s0167-8140(87)80186-x [DOI] [PubMed] [Google Scholar]
- 8.Iyer A, Apte AP, Bendau E, et al. : ROE (Radiotherapy Outcomes Estimator): An open-source tool for optimizing radiotherapy prescriptions. Comput Methods Programs Biomed 242, 2023:107833. 10.1016/j.cmpb.2023.107833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen I, Iyer A, Thor M, et al. : Simulating the Potential of Model-Based Individualized Prescriptions for Ultracentral Lung Tumors. Advances in Radiation Oncology 8, 2023(6):101285. 10.1016/j.adro.2023.101285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deasy JO, Jeong J, Thor M, et al. : Optimizing Lung Cancer Radiotherapy Treatments Using Personalized Dose-Response Curves. Springer; Berlin Heidelberg, 1–24, 2022. 10.1007/174_2022_307 [DOI] [Google Scholar]
- 11.Bylicky MA, Shankavaram U, John-Aryankalayil M, et al. : Multiomic-based molecular landscape of FaDu xenograft tumors in mice after a combinatorial treatment with radiation and an HSP90 inhibitor identifies adaptation-induced targets of resistance and therapeutic intervention. Mol Cancer Ther 23:577–588, 2024. 10.1158/1535-7163.MCT-23-0796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchsbaum JC, Jaffray DA, Ba D, Borkon LL, Chalk C. Predictive Radiation Oncology−A New NCI−DOE Scientific Space and Community. Published online 2022. https://meridian.allenpress.com/radiation-research/article-abstract/197/4/434/477175 [DOI] [PMC free article] [PubMed]
- 13.Modiri A, Stick LB, Rice SR, et al. : Individualized estimates of overall survival in radiation therapy plan optimization—A concept study. Med Phys 45(11):5332–5342, 2018. https://aapm.onlinelibrary.wiley.com/doi/abs/10.1002/mp.13211?casa_token=0kd1DGcwdZoAAAAA:xIuMnM9OhDMpsJzZhFklOkHb6HkNab9HcfEOvCt-1kQ_bun-yc9oKby3h-jwJUcOYFwil−Ku75h9BecoA [DOI] [PubMed] [Google Scholar]
- 14.McCulloch MM, Cazoulat G, Anderson BM, et al. : Improving GI Toxicity Models Through Deep Learning-Based Segmentation and Biomechanical Model-Based Dose Accumulation. Int J Radiat Oncol Biol Phys 111(3): S45–S46, 2021. 10.1016/j.ijrobp.2021.07.124 [DOI] [Google Scholar]
- 15.Smolders A, Lomax A, Weber DC, Albertini F: Deep learning based uncertainty prediction of deformable image registration for contour propagation and dose accumulation in online adaptive radiotherapy. Phys Med Biol 68, 2023(24):245027. 10.1088/1361-6560/ad0282 [DOI] [PubMed] [Google Scholar]
- 16.McCulloch MM, Cazoulat G, Svensson S, et al. : Leveraging deep learning-based segmentation and contours-driven deformable registration for dose accumulation in abdominal structures. Front Oncol 12, 2022:1015608. 10.3389/fonc.2022.1015608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alam S, Veeraraghavan H, Tringale K, et al. : Inter- and intrafraction motion assessment and accumulated dose quantification of upper gastrointestinal organs during magnetic resonance-guided ablative radiation therapy of pancreas patients. Phys Imaging Radiat Oncol 21:5461, 2022. 10.1016/j.phro.2022.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, Rimner A, Hu YC, et al. : Toward predicting the evolution of lung tumors during radiotherapy observed on a longitudinal MR imaging study via a deep learning algorithm. Med Phys 4699–4707, 2019. 10.1002/mp.13765. Published online August 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen SA, Liu MC, Aleshin A: Practical recommendations for using ctDNA in clinical decision making. Nature 619(7969):259–268, 2023. 10.1038/s41586-023-06225-y [DOI] [PubMed] [Google Scholar]
- 20.McNutt TR, Benedict SH, Low DA, et al. : Using Big Data Analytics to Advance Precision Radiation Oncology. Int J Radiat Oncol Biol Phys 101(2):285–291, 2018. 10.1016/j.ijrobp.2018.02.028 [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Symons J, Agapow P, et al. : Best practices in the real-world data life cycle. PLOS Digit Health 1, 2022(1):e0000003. 10.1371/journal.pdig.0000003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El Naqa I Perspectives on making big data analytics work for oncology. Methods 111:32–44, 2016. 10.1016/j.ymeth.2016.08.010 [DOI] [PubMed] [Google Scholar]
- 23.Deasy JO, Deasy JO, Stetson PD: A platform for continuous learning in oncology. Nature Cancer 2(7):675–676, 2021. 10.1038/s43018-021-00239-z [DOI] [PubMed] [Google Scholar]
- 24.Chen S, Guevara M, Ramirez N, et al. : Natural language processing to automatically extract the presence and severity of esophagitis in notes of patients undergoing radiotherapy. JCO Clin Cancer Inform 7, 2023:e2300048. 10.1200/CCI.23.00048 [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Yu Z, Guo Y, Bian J, Wu Y. Clinical Relation Extraction Using Transformer-based Models. arXiv [csCL]. 2021. arXiv preprint arXiv:2107.08957. http://arxiv.org/abs/2107.08957 [Google Scholar]
- 26.Chen A, Paredes D, Yu Z, et al. Identifying Symptoms of Delirium from Clinical Narratives Using Natural Language Processing. arXiv [csCL]. 2023. arXiv preprint arXiv:2304.00111. http://arxiv.org/abs/2304.00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munzir SI, Hier DB, Oommen C, Carrithers MD. A Large Language Model Outperforms Other Computational Approaches to the High-Throughput Phenotyping of Physician Notes. arXiv [csAI]. 2024. arXiv preprint arXiv:2406.14757. http://arxiv.org/abs/2406.14757 [Google Scholar]
- 28.Alsentzer E, Rasmussen MJ, Fontoura R, et al. : Zero-shot interpretable phenotyping of postpartum hemorrhage using large language models. NPJ Digit Med 6(1):212, 2023. 10.1038/s41746-023-00957-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Wang H, Yerebakan HZ, Shinagawa Y, Luo Y. FHIR-GPT Enhances Health Interoperability with Large Language Models. medRxiv. 2024:2023.10.17.23297028. NEJM AI, AIcs2300301. doi: 10.1101/2023.10.17.23297028 [DOI] [Google Scholar]
- 30.Bitterman DS, Goldner E, Finan S, et al. : An end-to-end natural language processing system for automatically extracting radiotherapy events from clinical texts: NLP to extract radiotherapy events from text. Int J Radiat Oncol Biol Phys 2023. 10.1016/j.ijrobp.2023.03.055. Published online March 27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.National Academies of Sciences, Engineering, and Medicine, Health and Medicine Division, Board on Health Sciences Policy: Barriers and Disincentives to the Use of Real-World Evidence and Real-World Data. National Academies Press; (US: ), 2019.. Accessed June 28, 2024 https://www.ncbi.nlm.nih.gov/books/NBK540112/ [Google Scholar]
- 32.Liu F, Panagiotakos D: Real-world data: a brief review of the methods, applications, challenges and opportunities. BMC Med Res Methodol 22(1):287, 2022. 10.1186/s12874-022-01768-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morin O, Valli eres M, Braunstein S, et al. : An artificial intelligence framework integrating longitudinal electronic health records with real-world data enables continuous pan-cancer prognostication. Nature Cancer 2(7):709–722, 2021. 10.1038/s43018-021-00236-2 [DOI] [PubMed] [Google Scholar]
- 34.Mayo CS, Feng MU, Brock KK, et al. : Operational Ontology for Oncology (O3): A professional society-based, multistakeholder, consensusdriven informatics standard supporting clinical and research use of real-world data from patients treated for cancer. Int J Radiat Oncol Biol Phys 117(3):533–550, 2023. https://www.sciencedirect.com/science/article/pii/S0360301623005254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrodt J, Dudchenko A, Knaup-Gregori P, Ganzinger M: Graph-Representation of Patient Data: a Systematic Literature Review. J Med Syst 44(4):86, 2020. 10.1007/s10916-020-1538-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfister DG, Rubin DM, Elkin EB, et al. : Risk Adjusting Survival Outcomes in Hospitals That Treat Patients With Cancer Without Information on Cancer Stage. JAMA Oncol 1(9):1303–1310, 2015. 10.1001/jamaoncol.2015.3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott P, Heigl M, McCay C, et al. : Modelling clinical narrative as computable knowledge: The NICE computable implementation guidance project. Learn Health Syst 7(4):e10394, 2023. 10.1002/lrh2.10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shridhar R, Strom T, Springett GM, et al. : Radiosensensitivity Index shows promise for predicting outcomes with adjuvant radiation in resected pancreatic cancer patients. Int J Radiat Oncol Biol Phys 90(1): S174, 2014. https://www.researchgate.net/profile/Ravi-Shridhar/publication/280277705_Radiosensensitivity_Index_Shows_Promise_for_Predicting_Outcomes_With_Adjuvant_Radiation_in_Resected_Pancreatic_Cancer_Patients/links/5bf6e0c592851ced67d0d3e1/Radiosensensitivity-Index-Shows-Promise-for-Predicting-Outcomes-With-Adjuvant-Radiation-in-Resected-Pancreatic-Cancer-Patients.pdf [Google Scholar]
- 39.Eschrich S, Zhang H, Zhao H, et al. : Systems Biology Modeling of the Radiation Sensitivity Network: A Biomarker Discovery Platform. International Journal of Radiation Oncology*Biology*Physics 75(2):497–505, 2009. 10.1016/j.ijrobp.2009.05.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torres-Roca JF, Eschrich S, Zhao H, et al. : Prediction of radiation sensitivity using a gene expression classifier. Cancer Res 65(16):7169–7176, 2005. 10.1158/0008-5472.CAN-05-0656 [DOI] [PubMed] [Google Scholar]
- 41.Eschrich SA, Fulp WJ, Pawitan Y, et al. : Validation of a radiosensitivity molecular signature in breast cancer. Clin Cancer Res 18(18):5134–5143, 2012. 10.1158/1078-0432.CCR-12-0891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strom T, Hoffe SE, Fulp W, et al. : Radiosensitivity index predicts for survival with adjuvant radiation in resectable pancreatic cancer. Radiother Oncol 117(1):159–164, 2015. 10.1016/j.radonc.2015.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott JG, Berglund A, Schell MJ, et al. : A genome-based model for adjusting radiotherapy dose (GARD): a retrospective, cohort-based study. Lancet Oncol 18(2):202–211, 2017. 10.1016/S1470-2045(16)30648-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yard BD, Adams DJ, Chie EK, et al. : A genetic basis for the variation in the vulnerability of cancer to DNA damage. Nat Commun 7:11428, 2016. 10.1038/ncomms11428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kolnohuz A, Ebrahimpour L, Yolchuyeva S, Manem VSK: Gene expression signature predicts radiation sensitivity in cell lines using the integral of dose−response curve. BMC Cancer 24(1):2, 2024. 10.1186/s12885-023-11634-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manem VSK: Development and validation of genomic predictors of radiation sensitivity using preclinical data. BMC Cancer 21(1):937, 2021. 10.1186/s12885-021-08652-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chesmore K, Bartlett J, Williams SM: The ubiquity of pleiotropy in human disease. Hum Genet 137(1):39–44, 2018. 10.1007/s00439-017-1854-z [DOI] [PubMed] [Google Scholar]
- 48.Gong L, Luo M, Sun R, Qiu L, Chen C, Luo Z: Significant Association Between XRCC1 Expression and Its rs25487 Polymorphism and Radiotherapy-Related Cancer Prognosis. Front Oncol 11, 2021:654784. 10.3389/fonc.2021.654784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Franco NR, Massi MC, Ieva F, et al. : Development of a method for generating SNP interaction-aware polygenic risk scores for radiotherapy toxicity. Radiother Oncol 159:241–248, 2021. 10.1016/j.radonc.2021.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Weijst L, Azria D, Berkovic P, et al. : The correlation between pre-treatment symptoms, acute and late toxicity and patient-reported health-related quality of life in non-small cell lung cancer patients: Results of the REQUITE study. Radiother Oncol 176:127–137, 2022. 10.1016/j.radonc.2022.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hosoya N, Miyagawa K: Implications of the germline variants of DNA damage response genes detected by cancer precision medicine for radiological risk communication and cancer therapy decisions. J Radiat Res 62(Supplement_1):i44–i52, 2021. 10.1093/jrr/rrab009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oh JH, Lee S, Thor M, et al. : Predicting the germline dependence of hematuria risk in prostate cancer radiotherapy patients. Radiother Oncol 185, 2023:109723. 10.1016/j.radonc.2023.109723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee S, Kerns S, Ostrer H, Rosenstein B, Deasy JO: Machine Learning on a Genome-Wide Association Study to Predict Late Genitourinary Toxicity Following Prostate Radiotherapy. 2018. Int. J. Radiat. Oncol. Biol. Phys 101(1):128–135, 2018. https://www.sciencedirect.com/science/article/pii/S0360301618301263/pdf?md5=8e216860cc3ec57a8fd94a863a086a64&pid=1-s2.0-S0360301618301263-main.pdf&_valck=1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oh JH, Kerns S, Ostrer H, Powell SN, Rosenstein B, Deasy JO: Computational methods using genome-wide association studies to predict radiotherapy complications and to identify correlative molecular processes. Sci Rep 7:43381, 2017. 10.1038/srep43381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deichaite I, Hopper A, Krockenberger L, et al. : Germline genetic biomarkers to stratify patients for personalized radiation treatment. J Transl Med 20(1):360, 2022. 10.1186/s12967-022-03561-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee S, Kerns S, Ostrer H, Rosenstein B, Deasy JO, Oh JH: Machine Learning on a Genome-wide Association Study to Predict Late Genitourinary Toxicity After Prostate Radiation Therapy. Int J Radiat Oncol Biol Phys 101(1):128–135, 2018. 10.1016/j.ijrobp.2018.01.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee S, Liang X, Woods M, et al. : Machine learning on genome-wide association studies to predict the risk of radiation-associated contralateral breast cancer in the WECARE Study. PLoS One 15, 2020(2): e0226157. 10.1371/journal.pone.0226157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Massi MC, Gasperoni F, Ieva F, et al. : A Deep Learning Approach Validates Genetic Risk Factors for Late Toxicity After Prostate Cancer Radiotherapy in a REQUITE Multi-National Cohort. Front Oncol 10, 2020:541281. 10.3389/fonc.2020.541281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abdelkarem OAI, Choudhury A, Burnet NG, Summersgill HR, West CML: Effect of Race and Ethnicity on Risk of Radiotherapy Toxicity and Implications for Radiogenomics. Clin Oncol 34(10):653–669, 2022. 10.1016/j.clon.2022.03.013 [DOI] [PubMed] [Google Scholar]
- 60.Ling CC, Humm J, Larson S, et al. : Towards multidimensional radiotherapy (MD-CRT): biological imaging and biological conformality. Int J Radiat Oncol Biol Phys 47(3):551–560, 2000. 10.1016/s0360-3016(00)00467-3 [DOI] [PubMed] [Google Scholar]
- 61.Rafat M, Ali R, Graves EE: Imaging radiation response in tumor and normal tissue. Am J Nucl Med Mol Imaging 5(4):317–332, 2015. https://www.ncbi.nlm.nih.gov/pubmed/26269771 [PMC free article] [PubMed] [Google Scholar]
- 62.Jeong J, Setton JS, Lee NY, Oh JH, Deasy JO: Estimate of the impact of FDG-avidity on the dose required for head and neck radiotherapy local control. Radiother Oncol 111(3):340–347, 2014. 10.1016/j.radonc.2014.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O’Connor JPB, Aboagye EO, Adams JE, et al. : Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol 14(3):169–186, 2017. 10.1038/nrclinonc.2016.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu J, Dong M, Sun X, Li W, Xing L, Yu J: Prognostic value of 18F-FDG PET/CT in surgical non-small cell lung cancer: A meta-analysis. PLoS One 11, 2016(1):e0146195. 10.1371/journal.pone.0146195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hatakenaka M, Nakamura K, Yabuuchi H, et al. : Pretreatment Apparent Diffusion Coefficient of the Primary Lesion Correlates With Local Failure in Head-and-Neck Cancer Treated With Chemoradiotherapy or Radiotherapy. International Journal of Radiation Oncology*Biology*Physics 81(2):339–345, 2011. 10.1016/j.ijrobp.2010.05.051 [DOI] [PubMed] [Google Scholar]
- 66.Lo CH, Huang WY, Hsiang CW, et al. : Prognostic Significance of Apparent Diffusion Coefficient in Hepatocellular Carcinoma Patients treated with Stereotactic Ablative Radiotherapy. Sci Rep 9(1):14157, 2019. 10.1038/s41598-019-50503-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gladwish A, Milosevic M, Fyles A, et al. : Association of Apparent Diffusion Coefficient with Disease Recurrence in Patients with Locally Advanced Cervical Cancer Treated with Radical Chemotherapy and Radiation Therapy. Radiology 279(1):158–166, 2016. 10.1148/radiol.2015150400 [DOI] [PubMed] [Google Scholar]
- 68.Casares-Magaz O, van der Heide UA, Rørvik J, Steenbergen P, Muren LP: A tumour control probability model for radiotherapy of prostate cancer using magnetic resonance imaging-based apparent diffusion coefficient maps. Radiother Oncol 119(1):111–116, 2016. 10.1016/j.radonc.2016.02.030 [DOI] [PubMed] [Google Scholar]
- 69.Xu Z, Li XF, Zou H, Sun X, Shen B: 18F-Fluoromisonidazole in tumor hypoxia imaging. Oncotarget 8(55):94969–94979, 2017. 10.18632/oncotarget.21662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Horsman MR, Mortensen LS, Petersen JB, Busk M, Overgaard J: Imaging hypoxia to improve radiotherapy outcome. Nat Rev Clin Oncol 9:674, 2012. 10.1038/nrclinonc.2012.171 [DOI] [PubMed] [Google Scholar]
- 71.Busk M, Overgaard J, Horsman MR: Imaging of Tumor Hypoxia for Radiotherapy: Current Status and Future Directions. Semin Nucl Med 50(6):562–583, 2020. 10.1053/j.semnuclmed.2020.05.003 [DOI] [PubMed] [Google Scholar]
- 72.Craig SG, Humphries MP, Alderdice M, et al. : Immune status is prognostic for poor survival in colorectal cancer patients and is associated with tumour hypoxia. Br J Cancer 123(8):1280–1288, 2020. 10.1038/s41416-020-0985-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paudyal R, Grkovski M, Oh JH, et al. : Application of Community Detection Algorithm to Investigate the Correlation between Imaging Biomarkers of Tumor Metabolism, Hypoxia, Cellularity, and Perfusion for Precision Radiotherapy in Head and Neck Squamous Cell Carcinomas. Cancers 13(15):3908, 2021. 10.3390/cancers13153908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Semenkovich NP, Szymanski JJ, Earland N, Chauhan PS, Pellini B, Chaudhuri AA: Genomic approaches to cancer and minimal residual disease detection using circulating tumor DNA. J Immunother Cancer 11, 2023(6):e006284. 10.1136/jitc-2022-006284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moser T, Heitzer E: Surpassing sensitivity limits in liquid biopsy. Science 383(6680):260–261, 2024. 10.1126/science.adn1886 [DOI] [PubMed] [Google Scholar]
- 76.Huang EP, O’Connor JPB, McShane LM, et al. : Criteria for the translation of radiomics into clinically useful tests. Nat Rev Clin Oncol 20 (2):69–82, 2023. 10.1038/s41571-022-00707-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lambin P, Rios-Velazquez E, Leijenaar R, et al. : Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48(4):441–446, 2012. 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lambin P, Leijenaar RTH, Deist TM, et al. : Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14 (12):749–762, 2017. 10.1038/nrclinonc.2017.141 [DOI] [PubMed] [Google Scholar]
- 79.Wang Q, Xu J, Wang A, et al. : Systematic review of machine learning-based radiomics approach for predicting microsatellite instability status in colorectal cancer. Radiol Med 128(2):136–148, 2023. 10.1007/s11547-023-01593-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu D, Yan Y, Jiang M, et al. : Predictive value of radiomics-based machine learning for the disease-free survival in breast cancer: a systematic review and meta-analysis. Front Oncol 13, 2023:1173090. 10.3389/fonc.2023.1173090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huynh LM, Hwang Y, Taylor O, Baine MJ: The Use of MRI-Derived Radiomic Models in Prostate Cancer Risk Stratification: A Critical Review of Contemporary Literature. Diagnostics (Basel) 13(6):1128, 2023. 10.3390/diagnostics13061128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chaddad A, Tan G, Liang X, et al. : Advancements in MRI-Based Radiomics and Artificial Intelligence for Prostate Cancer: A Comprehensive Review and Future Prospects. Cancers 15(15):3839, 2023. 10.3390/cancers15153839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Atmakuru A, Chakraborty S, Faust O, et al. : Deep learning in radiology for lung cancer diagnostics: A systematic review of classification, segmentation, and predictive modeling techniques. Expert Syst Appl 255, 2024:124665. 10.1016/j.eswa.2024.124665 [DOI] [Google Scholar]
- 84.Wu L, Lou X, Kong N, Xu M, Gao C: Can quantitative peritumoral CT radiomics features predict the prognosis of patients with non-small cell lung cancer? A systematic review. Eur Radiol 33(3):2105–2117, 2023. 10.1007/s00330-022-09174-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shehata M, Abouelkheir RT, Gayhart M, et al. : Role of AI and Radiomic Markers in Early Diagnosis of Renal Cancer and Clinical Outcome Prediction: A Brief Review. Cancers 15(10):2835, 2023. 10.3390/cancers15102835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Adusumilli P, Ravikumar N, Hall G, Swift S, Orsi N, Scarsbrook A: Radiomics in the evaluation of ovarian masses a systematic review. Insights Imaging 14(1):165, 2023. 10.1186/s13244-023-01500-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Di Donato V, Kontopantelis E, Cuccu I, et al. : Magnetic resonance imaging-radiomics in endometrial cancer: a systematic review and meta-analysis. Int J Gynecol Cancer 33(7):1070–1076, 2023. 10.1136/ijgc-2023-004313 [DOI] [PubMed] [Google Scholar]
- 88.Tortora M, Gemini L, Scaravilli A, et al. : Radiomics Applications in Head and Neck Tumor Imaging: A Narrative Review. Cancers 15, 2023(4). 10.3390/cancers15041174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jia LL, Zhao JX, Zhao LP, Tian JH, Huang G: Current status and quality of radiomic studies for predicting KRAS mutations in colorectal cancer patients: A systematic review and metaanalysis. Eur J Radiol 158, 2023:110640. 10.1016/j.ejrad.2022.110640 [DOI] [PubMed] [Google Scholar]
- 90.Yuan E, Chen Y, Song B: Quality of radiomics for predicting microvascular invasion in hepatocellular carcinoma: a systematic review. Eur Radiol 33(5):3467–3477, 2023. 10.1007/s00330-023-09414-5 [DOI] [PubMed] [Google Scholar]
- 91.Wu T, Gao C, Lou X, Wu J, Xu M, Wu L: Predictive value of radiomic features extracted from primary lung adenocarcinoma in forecasting thoracic lymph node metastasis: a systematic review and meta-analysis. BMC Pulm Med 24(1):246, 2024. 10.1186/s12890-024-03020-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Philip MM, Welch A, McKiddie F, Nath M: A systematic review and meta-analysis of predictive and prognostic models for outcome prediction using positron emission tomography radiomics in head and neck squamous cell carcinoma patients. Cancer Med 12(15):16181–16194, 2023. 10.1002/cam4.6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Elsayed B, Alksas A, Shehata M, et al. : Exploring Neoadjuvant Chemotherapy, Predictive Models, Radiomic, and Pathological Markers in Breast Cancer: A Comprehensive Review. Cancers 15(21):5288, 2023. 10.3390/cancers15215288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mukherjee P, Cintra M, Huang C, et al. : CT-based Radiomic Signatures for Predicting Histopathologic Features in Head and Neck Squamous Cell Carcinoma. Radiol Imaging Cancer 2, 2020(3):e190039. 10.1148/rycan.2020190039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van Dijk LV, Mohamed A Sr, Ahmed S, et al. : Head and neck cancer predictive risk estimator to determine control and therapeutic outcomes of radiotherapy (HNC-PREDICTOR): development, international multi-institutional validation, and web implementation of clinicready model-based risk stratification for head and neck cancer. Eur J Cancer 178:150–161, 2023. 10.1016/j.ejca.2022.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Valli eres M, Freeman CR, Skamene SR, El Naqa I: A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol 60(14):5471–5496, 2015. 10.1088/0031-9155/60/14/5471 [DOI] [PubMed] [Google Scholar]
- 97.Welch ML, McIntosh C, Haibe-Kains B, et al. : Vulnerabilities of radiomic signature development: The need for safeguards. Radiother Oncol 130:2–9, 2019. 10.1016/j.radonc.2018.10.027 [DOI] [PubMed] [Google Scholar]
- 98.Hosny A, Aerts HJ, Mak RH: Handcrafted versus deep learning radiomics for prediction of cancer therapy response. The Lancet Digital Health 1(3):e106–e107, 2019. 10.1016/S2589-7500(19)30062-7 [DOI] [PubMed] [Google Scholar]
- 99.Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL: Artificial intelligence in radiology. Nat Rev Cancer 18(8):500–510, 2018. 10.1038/s41568-018-0016-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Raschka S: Machine Learning Q and AI: 30 Essential Questions and Answers on Machine Learning and AI. No Starch Press, 2024.. San Francisco: https://www.amazon.com/Machine-Learning-AI-Essential-Questions/dp/1718503768/ref=sr_1_1?crid=33S6LDRHYXFU0&dib=eyJ2IjoiMSJ9.1pcW4FOhUJRVjHov__NPKWIwgVg8rzTeTdM5KuIg−yRBV0U4IFbeghXcGyEVPcrHWI1nmpG4qT4PSnQ09ZV8z-QyDZZeVj3I0oj7_xf0TAohyLk-CEodxFSoAO5o_mTe-qhS0jYa5oU6vKnWwPONkwPYOIlHOCFjfuqEIsiUnxuq7q0psfFnuj82PDO53xsMtTnkLueThqdtn6OuwFCEp3bI8_Ty8uYCT5sh7yRX3Ok.wAXs1taifzg0cQJkYGTzvqx_6E33TXMAZm9j23moN_M&dib_tag=se&keywords=sebastian+raschka&qid=1713884537&sprefix=sebastian+rachka%2Caps%2C59&sr=8-1 [Google Scholar]
- 101.Demircioğlu A: Are deep models in radiomics performing better than generic models? A systematic review. Eur Radiol Exp 7(1):11, 2023. 10.1186/s41747-023-00325-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.De Biase A, Ma B, Guo J, et al. : Deep learning-based outcome prediction using PET/CT and automatically predicted probability maps of primary tumor in patients with oropharyngeal cancer. Comput Methods Programs Biomed 244, 2024:107939. 10.1016/j.cmpb.2023.107939 [DOI] [PubMed] [Google Scholar]
- 103.Ma B, Guo J, De Biase A, et al. : PET/CT based transformer model for multi-outcome prediction in oropharyngeal cancer. Radiother Oncol 197, 2024:110368. 10.1016/j.radonc.2024.110368 [DOI] [PubMed] [Google Scholar]
- 104.Jiang Y, Zhang Z, Wang W, et al. : Biology-guided deep learning predicts prognosis and cancer immunotherapy response. Nat Commun 14(1):5135, 2023. 10.1038/s41467-023-40890-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shui L, Ren H, Yang X, et al. : The Era of Radiogenomics in Precision Medicine: An Emerging Approach to Support Diagnosis, Treatment Decisions, and Prognostication in Oncology. Front Oncol 10, 2020:570465. 10.3389/fonc.2020.570465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Qian L, Wu T, Kong S, et al. : Could the underlying biological basis of prognostic radiomics and deep learning signatures be explored in patients with lung cancer? A systematic review. Eur J Radiol 171, 2024:111314. 10.1016/j.ejrad.2024.111314 [DOI] [PubMed] [Google Scholar]
- 107.Kim H, Goo JM, Lee KH, Kim YT, Park CM: Preoperative CT-based Deep Learning Model for Predicting Disease-Free Survival in Patients with Lung Adenocarcinomas. Radiology 296(1):216–224, 2020. 10.1148/radiol.2020192764 [DOI] [PubMed] [Google Scholar]
- 108.Wang C, Shao J, Lv J, et al. : Deep learning for predicting subtype classification and survival of lung adenocarcinoma on computed tomography. Transl Oncol 14, 2021(8):101141. 10.1016/j.tranon.2021.101141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gurney-Champion OJ, Mahmood F, van Schie M, et al. : Quantitative imaging for radiotherapy purposes. Radiother Oncol 146:66–75, 2020. 10.1016/j.radonc.2020.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.National Academies of Sciences, Engineering, and Medicine: Opportunities and Challenges for Digital Twins in Biomedical Research: Proceedings of a Workshop−in Brief. Washington, DC: The National Academies Press, 2023.. 10.17226/26922 Accessed August 14, 2024 [DOI] [PubMed] [Google Scholar]
- 111.of Engineering NA, Science C, Board T, et al. Opportunities and Challenges for Digital Twins in Biomedical Research. Published online 2023. Washington, DC. https://www.ncbi.nlm.nih.gov/books/NBK592664 [Google Scholar]
- 112.Atallah S, Cho BCJ, Allibhai Z, et al. : Impact of Pretreatment Tumor Growth Rate on Outcome of Early-Stage Lung Cancer Treated With Stereotactic Body Radiation Therapy. International Journal of Radiation Oncology*Biology*Physics 89(3):532–538, 2014. 10.1016/j.ijrobp.2014.03.003 [DOI] [PubMed] [Google Scholar]
- 113.Atallah S, Le LW, Bezjak A, MacRae R, Hope AJ, Pantarotto J: Validating impact of pretreatment tumor growth rate on outcome of early-stage lung cancer treated with stereotactic body radiation therapy. Thorac Cancer 12(2):201–209, 2021. 10.1111/1759-7714.13744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kong FMS, Frey KA, Quint LE, et al. : A Pilot Study of [18F]Fluorodeoxyglucose Positron Emission Tomography Scans During and After Radiation-Based Therapy in Patients With Non−Small-Cell Lung Cancer. J Clin Orthod 25(21):3116–3123, 2007. 10.1200/JCO.2006.10.3747 [DOI] [PubMed] [Google Scholar]
- 115.Löck S, Perrin R, Seidlitz A, et al. : Residual tumour hypoxia in head-and-neck cancer patients undergoing primary radiochemotherapy, final results of a prospective trial on repeat FMISO-PET imaging. Radiother Oncol 124(3):533–540, 2017. 10.1016/j.radonc.2017.08.010 [DOI] [PubMed] [Google Scholar]
- 116.King AD, Chow KK, Yu KH, et al. : Head and neck squamous cell carcinoma: diagnostic performance of diffusion-weighted MR imaging for the prediction of treatment response. Radiology 266(2):531–538, 2013. 10.1148/radiol.12120167 [DOI] [PubMed] [Google Scholar]
- 117.Trada Y, Keall P, Jameson M, et al. : Changes in serial multiparametric MRI and FDG-PET/CT functional imaging during radiation therapy can predict treatment response in patients with head and neck cancer. Eur Radiol 33(12):8788–8799, 2023. 10.1007/s00330-023-09843-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sanz-Garcia E, Zhao E, Bratman SV, Siu LL: Monitoring and adapting cancer treatment using circulating tumor DNA kinetics: Current research, opportunities, and challenges. Sci Adv 8(4):eabi8618, 2022. 10.1126/sciadv.abi8618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bollineni VR, Widder J, Pruim J, Langendijk JA, Wiegman EM: Residual 18F-FDG-PET uptake 12 weeks after stereotactic ablative radiotherapy for stage I non-small-cell lung cancer predicts local control. Int J Radiat Oncol Biol Phys 83(4):e551–e555, 2012. 10.1016/j.ijrobp.2012.01.012 [DOI] [PubMed] [Google Scholar]
- 120.Sellami S, Bourbonne V, Hatt M, et al. : Predicting response to radiotherapy of head and neck squamous cell carcinoma using radiomics from cone-beam CT images. Acta Oncol 61(1):73–80, 2022. 10.1080/0284186X.2021.1983207 [DOI] [PubMed] [Google Scholar]
- 121.Fave X, Zhang L, Yang J, et al. : Delta-radiomics features for the prediction of patient outcomes in non−small cell lung cancer. Sci Rep 7 (1):588, 2017. 10.1038/s41598-017-00665-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nasief H, Zheng C, Schott D, et al. : A machine learning based delta-radiomics process for early prediction of treatment response of pancreatic cancer. NPJ Precis Oncol 3:25, 2019. 10.1038/s41698-019-0096-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cousin F, Louis T, Dheur S, et al. : Radiomics and Delta-Radiomics Signatures to Predict Response and Survival in Patients with Non-Small-Cell Lung Cancer Treated with Immune Checkpoint Inhibitors. Cancers 15(7):1968, 2023. 10.3390/cancers15071968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.van Dijk LV, Langendijk JA, Zhai TT, et al. : Delta-radiomics features during radiotherapy improve the prediction of late xerostomia. Sci Rep 9(1):12483, 2019. 10.1038/s41598-019-48184-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Alam SR, Zhang P, Zhang SY, et al. : Early Prediction of Acute Esophagitis for Adaptive Radiation Therapy. Int J Radiat Oncol Biol Phys 110 (3):883–892, 2021. 10.1016/j.ijrobp.2021.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hsu DG, Ballangrud ÅM, Shamseddine A, et al. : Automatic segmentation of brain metastases using T1 magnetic resonance and computed tomography images. Phys Med Biol 66, 2021(17):175014. 10.1088/1361-6560/ac1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hsu DG, Ballangrud Å, Prezelski K, et al. : Automatically tracking brain metastases after stereotactic radiosurgery. Physics and Imaging in Radiation Oncology 27, 2023:100452. 10.1016/j.phro.2023.100452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dercle L, Zhao B, Gönen M, et al. : Early Readout on Overall Survival of Patients With Melanoma Treated With Immunotherapy Using a Novel Imaging Analysis. JAMA Oncol 8(3):385–392, 2022. 10.1001/jamaoncol.2021.6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Boehm KM, Aherne EA, Ellenson L, et al. : Multimodal data integration using machine learning improves risk stratification of high-grade serous ovarian cancer. Nat Cancer 3(6):723–733, 2022. 10.1038/s43018-022-00388-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lipkova J, Chen RJ, Chen B, et al. : Artificial intelligence for multimodal data integration in oncology. Cancer Cell 40(10):1095–1110, 2022. 10.1016/j.ccell.2022.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pouryahya M, Oh JH, Javanmard P, et al. aWCluster: A novel integrative network-based clustering of multiomics for subtype analysis of cancer data. IEEE/ACM Trans Comput Biol Bioinform. Published online 2020. https://ieeexplore.ieee.org/abstract/document/9266088/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rockne RC, Hawkins-Daarud A, Swanson KR, et al. : The 2019 mathematical oncology roadmap. Phys Biol 16, 2019(4):041005. 10.1088/1478-3975/ab1a09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Alber M, Tepole AB, Cannon WR, et al. : Integrating machine learning and multiscale modeling—perspectives, challenges, and opportunities in the biological, biomedical, and behavioral sciences. npj Digital Medicine 2(1):1–11, 2019. 10.1038/s41746-019-0193-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ghahremani P, Li Y, Kaufman A, et al. : Deep Learning-Inferred Multiplex ImmunoFluorescence for Immunohistochemical Image Quantification. Nat Mach Intell 4(4):401–412, 2022. 10.1038/s42256-022-00471-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jaume G, Vaidya A, Chen RJ, Williamson DFK, Liang PP, Mahmood F: Modeling dense multimodal interactions between biological pathways and histology for survival prediction. In: In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2024. p. 11579–11590, 2024. https://openaccess.thecvf.com/content/CVPR2024/papers/Jaume_Modeling_Dense_Multimodal_Interactions_Between_Biological_Pathways_and_Histology_for_CVPR_2024_paper.pdf [Google Scholar]
- 136.Oh JH, Apte AP, Katsoulakis E, et al. : Reproducibility of radiomic features using network analysis and its application in Wasserstein k-means clustering. J Med Imaging (Bellingham) 8, 2021(3):031904. 10.1117/1.JMI.8.3.031904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Marks LB, Yorke ED, Jackson A, et al. : Use of Normal Tissue Complication Probability Models in the Clinic. Int J Radiat Oncol Biol Phys 76 (3, Supplement):S10–S19, 2010. 10.1016/j.ijrobp.2009.07.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rancati T, Fiorino C: Modelling Radiotherapy Side Effects: Practical Applications for Planning Optimisation. CRC Press, 2019.. Boca Raton: https://play.google.com/store/books/details?id=WCGeDwAAQBAJ [Google Scholar]
- 139.Deasy JO, Mayo CS, Orton CG: Treatment planning evaluation and optimization should be biologically and not dose/volume based. Med Phys 42(6):2753–2756, 2015. 10.1118/1.4916670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Allen Li X, Alber M, Deasy JO, Jackson A: The use and QA of biologically related models for treatment planning: Short report of the TG166 of the therapy physics committee of the AAPM. Medical 39(3):1386–1409, 2012 [DOI] [PubMed] [Google Scholar]
- 141.Thor M, Jackson A, Zelefsky MJ, et al. : Inter-institutional analysis demonstrates the importance of lower than previously anticipated dose regions to prevent late rectal bleeding following prostate radiotherapy. Radiother Oncol 127(1):88–95, 2018. 10.1016/j.radonc.2018.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Palma G, Monti S, Pacelli R, et al. : Radiation Pneumonitis in Thoracic Cancer Patients: Multi-Center Voxel-Based Analysis. Cancers 13 (14):3553, 2021. 10.3390/cancers13143553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Palma G, Monti S, Conson M, Pacelli R, Cella L: Normal tissue complication probability (NTCP) models for modern radiation therapy. Semin Oncol 46(3):210–218, 2019. 10.1053/j.seminoncol.2019.07.006 [DOI] [PubMed] [Google Scholar]
- 144.Palma G, Monti S, Cella L: Voxel-based analysis in radiation oncology: A methodological cookbook. Phys Med 69:192–204, 2020. 10.1016/j.ejmp.2019.12.013 [DOI] [PubMed] [Google Scholar]
- 145.Beasley W, Thor M, McWilliam A, et al. : Image-based Data Mining to Probe Dosimetric Correlates of Radiation-induced Trismus. Int J Radiat Oncol Biol Phys 102(4):1330–1338, 2018. 10.1016/j.ijrobp.2018.05.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Craddock M, Nestle U, Koenig J, et al. : Cardiac Function Modifies the Impact of Heart Base Dose on Survival: A Voxel-Wise Analysis of Patients With Lung Cancer From the PET-Plan Trial. J Thorac Oncol 18(1):57–66, 2023. 10.1016/j.jtho.2022.09.004 [DOI] [PubMed] [Google Scholar]
- 147.Thor M, Deasy JO, Hu C, et al. : Modeling the impact of cardio-pulmonary irradiation on overall survival in NRG Oncology trial RTOG 0617. Clin Cancer Res 26(17):4643–4650, 2020. 10.1158/1078-0432.CCR-19-2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ebert MA, Gulliford S, Acosta O, et al. : Spatial descriptions of radiotherapy dose: normal tissue complication models and statistical associations. Phys Med Biol 66(12):12TR01, 2021. 10.1088/1361-6560/ac0681 [DOI] [PubMed] [Google Scholar]
- 149.Gabry s HS, Buettner F, Sterzing F, Hauswald H, Bangert M: Design and selection of machine learning methods using radiomics and dosiomics for NTCP modeling of xerostomia. Front Oncol 8:35, 2018. https://www.frontiersin.org/articles/10.3389/fonc.2018.00035/abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Appelt AL, Elhaminia B, Gooya A, Gilbert A, Nix M: Deep Learning for Radiotherapy Outcome Prediction Using Dose Data A Review. Clin Oncol 34(2):e87–e96, 2022. 10.1016/j.clon.2021.12.002 [DOI] [PubMed] [Google Scholar]
- 151.Men K, Geng H, Zhong H, Fan Y, Lin A, Xiao Y: A Deep Learning Model for Predicting Xerostomia Due to Radiation Therapy for Head and Neck Squamous Cell Carcinoma in the RTOG 0522 Clinical Trial. Int J Radiat Oncol Biol Phys 105(2):440–447, 2019. 10.1016/j.ijrobp.2019.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Humbert-Vidan L, Patel V, Andlauer R, King AP, Guerrero Urbano T: Prediction of Mandibular ORN Incidence from 3D Radiation Dose Distribution Maps Using Deep Learning. Applications of Medical Artificial Intelligence. Springer Nature; Switzerland, 49–58, 2022. 10.1007/978-3-031-17721-7_6 [DOI] [Google Scholar]
- 153.Reber B, Van Dijk L, Anderson B, et al. : Comparison of Machine-Learning and Deep-Learning Methods for the Prediction of Osteoradionecrosis Resulting From Head and Neck Cancer Radiation Therapy. Adv Radiat Oncol 8, 2023(4):101163. 10.1016/j.adro.2022.101163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.El Naqa I, Johansson A, Owen D, et al. : Modeling of Normal Tissue Complications Using Imaging and Biomarkers After Radiation Therapy for Hepatocellular Carcinoma. Int J Radiat Oncol Biol Phys 100 (2):335–343, 2018. 10.1016/j.ijrobp.2017.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bin L, Yuan T, Zhaohui S, et al. : A deep learning-based dual-omics prediction model for radiation pneumonitis. Med Phys 48(10):6247–6256, 2021. 10.1002/mp.15079 [DOI] [PubMed] [Google Scholar]
- 156.Zhang Z, Wang Z, Yan M, et al. : Radiomics and Dosiomics Signature From Whole Lung Predicts Radiation Pneumonitis: A Model Development Study With Prospective External Validation and Decision-curve Analysis. International Journal of Radiation Oncology*Biology*Physics 115(3):746–758, 2023. 10.1016/j.ijrobp.2022.08.047 [DOI] [PubMed] [Google Scholar]
- 157.Mellhammar E, Dahlbom M, Vilhelmsson-Timmermand O, Strand SE: Tumor Control Probability and Small-Scale Monte Carlo Dosimetry: Effects of Heterogenous Intratumoral Activity Distribution in Radiopharmaceutical Therapy. J Nucl Med 64(10):1632–1637, 2023. 10.2967/jnumed.123.265523 [DOI] [PubMed] [Google Scholar]
- 158.Okunieff P, Morgan D, Niemierko A, Suit HD: Radiation dose-response of human tumors. Int J Radiat Oncol Biol Phys 32(4):1227–1237, 1995. 10.1016/0360-3016(94)00475-z [DOI] [PubMed] [Google Scholar]
- 159.Hill RP, Bristow RG, Fyles A, Koritzinsky M, Milosevic M, Wouters BG: Hypoxia and Predicting Radiation Response. Semin Radiat Oncol 25 (4):260–272, 2015. 10.1016/j.semradonc.2015.05.004 [DOI] [PubMed] [Google Scholar]
- 160.Toma-Dasu I, Dasu A: Modelling tumour oxygenation, reoxygenation and implications on treatment outcome. Comput Math Methods Med 2013(1):141087. 10.1155/2013/141087. 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Milosevic M, Warde P, Ménard C, et al. : Tumor hypoxia predicts biochemical failure following radiotherapy for clinically localized prostate cancer. Clin Cancer Res 18(7):2108–2114, 2012. 10.1158/1078-0432.CCR-11-2711 [DOI] [PubMed] [Google Scholar]
- 162.Carlson DJ, Stewart RD, Semenenko VA: Effects of oxygen on intrinsic radiation sensitivity: A test of the relationship between aerobic and hypoxic linear-quadratic (LQ) model parameters. Med Phys 33(9):3105–3115, 2006. 10.1118/1.2229427 [DOI] [PubMed] [Google Scholar]
- 163.Bristow RG, Hill RP: Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer 8(3):180–192, 2008. 10.1038/nrc2344 [DOI] [PubMed] [Google Scholar]
- 164.Riffle S, Hegde RS: Modeling tumor cell adaptations to hypoxia in multicellular tumor spheroids. J Exp Clin Cancer Res 36(1):102, 2017. 10.1186/s13046-017-0570-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Riffle S, Pandey RN, Albert M, Hegde RS: Linking hypoxia, DNA damage and proliferation in multicellular tumor spheroids. BMC Cancer 17(1):338, 2017. 10.1186/s12885-017-3319-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Baumann M, Krause M, Hill R: Exploring the role of cancer stem cells in radioresistance. Nat Rev Cancer 8(7):545–554, 2008. 10.1038/nrc2419 [DOI] [PubMed] [Google Scholar]
- 167.Radonic S, Besserer J, Meier V, Bley CR, Schneider U: A Novel Analytical Population Tumor Control Probability Model Includes Cell Density and Volume Variations: Application to Canine Brain Tumor. Int J Radiat Oncol Biol Phys 110(5):1530–1537, 2021. 10.1016/j.ijrobp.2021.03.021 [DOI] [PubMed] [Google Scholar]
- 168.Wu HJ, Temko D, Maliga Z, et al. : Spatial intra-tumor heterogeneity is associated with survival of lung adenocarcinoma patients. Cell Genomics 2, 2022(8):100165. 10.1016/j.xgen.2022.100165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.West CM, Davidson SE, Roberts SA, Hunter RD: Intrinsic radiosensitivity and prediction of patient response to radiotherapy for carcinoma of the cervix. Br J Cancer 68(4):819–823, 1993. https://www.ncbi.nlm.nih.gov/pubmed/8398714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Withers HR, Taylor JM, Maciejewski B: The hazard of accelerated tumor clonogen repopulation during radiotherapy. Acta Oncol 27 (2):131–146, 1988. https://www.ncbi.nlm.nih.gov/pubmed/3390344 [DOI] [PubMed] [Google Scholar]
- 171.Fowler JF, Lindstrom MJ: Loss of local control with prolongation in radiotherapy. Int J Radiat Oncol Biol Phys 23(2):457–467, 1992. 10.1016/0360-3016(92)90768-d [DOI] [PubMed] [Google Scholar]
- 172.Vogelius IR, Bentzen SM: Dose response and fractionation sensitivity of prostate cancer after external beam radiotherapy: A meta-analysis of randomized trials. Int J Radiat Oncol Biol Phys 100(4):858–865, 2018 [DOI] [PubMed] [Google Scholar]
- 173.Joiner MC, Bentzen SM: Fractionation: the linear-quadratic approach. Basic Clinical Radiobiology. CRC press, 112–124, 2018. https://www.taylorfrancis.com/chapters/edit/10.1201/9780429490606-9/fractionation-linear-quadratic-approach-michael-joiner-s%C3%B8ren-bentzen [Google Scholar]
- 174.Shuryak I, Carlson DJ, Brown JM, Brenner DJ: High-dose and fractionation effects in stereotactic radiation therapy: Analysis of tumor control data from 2965 patients. Radiother Oncol 115(3):327–334, 2015. 10.1016/j.radonc.2015.05.013 [DOI] [PubMed] [Google Scholar]
- 175.Zaider M, Minerbo GN: Tumour control probability: a formulation applicable to any temporal protocol of dose delivery. Phys Med Biol 45(2):279–293, 2000. https://www.ncbi.nlm.nih.gov/pubmed/10701504 [DOI] [PubMed] [Google Scholar]
- 176.Jeong J, Deasy JO: Modeling the relationship between fluorodeoxyglucose uptake and tumor radioresistance as a function of the tumor microenvironment. Comput Math Methods Med 2014, 2014:847162. 10.1155/2014/847162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Jeong J, Shoghi KI, Deasy JO: Modelling the interplay between hypoxia and proliferation in radiotherapy tumour response. Phys Med Biol 58(14):4897–4919, 2013. 10.1088/0031-9155/58/14/4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Jeong J, Oh JH, Sonke JJ, et al. : Modeling the Cellular Response of Lung Cancer to Radiation Therapy for a Broad Range of Fractionation Schedules. Clin Cancer Res 23(18):5469–5479, 2017. 10.1158/1078-0432.CCR-16-3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Jeong J, Taasti VT, Jackson A, Deasy JO: The Relative Biological Effectiveness of Carbon Ion Radiation Therapy for Early Stage Lung Cancer. Radiother Oncol 2020. 10.1016/j.radonc.2020.09.027. Published online September 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Crispin-Ortuzar M, Jeong J, Fontanella AN, Deasy JO: A radiobiological model of radiotherapy response and its correlation with prognostic imaging variables. Phys Med Biol 62(7):2658–2674, 2017. 10.1088/1361-6560/aa5d42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zahid MU, Mohsin N, Mohamed ASR, et al. : Forecasting Individual Patient Response to Radiation Therapy in Head and Neck Cancer With a Dynamic Carrying Capacity Model. Int J Radiat Oncol Biol Phys 111(3):693–704, 2021. 10.1016/j.ijrobp.2021.05.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gouw ZAR, Jeong J, Rimner A, et al. : Primer shot” fractionation with an early treatment break is theoretically superior to consecutive weekday fractionation schemes for early-stage non-small cell lung cancer. Radiother Oncol 190, 2024:110006. 10.1016/j.radonc.2023.110006 [DOI] [PubMed] [Google Scholar]
- 183.Jabbour SK, Kim S, Haider SA, et al. : Reduction in Tumor Volume by Cone Beam Computed Tomography Predicts Overall Survival in Non-Small Cell Lung Cancer Treated With Chemoradiation Therapy. Int J Radiat Oncol Biol Phys 92(3):627–633, 2015. 10.1016/j.ijrobp.2015.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wald P, Mo X, Barney C, et al. : Prognostic Value of Primary Tumor Volume Changes on kV-CBCT during Definitive Chemoradiotherapy for Stage III Non−Small Cell Lung Cancer. J Thorac Oncol 12 (12):1779–1787, 2017. 10.1016/j.jtho.2017.08.010 [DOI] [PubMed] [Google Scholar]
- 185.Gérard M, Corroyer-Dulmont A, Lesueur P, et al. : Hypoxia Imaging and Adaptive Radiotherapy: A State-of-the-Art Approach in the Management of Glioma. Front Med 6:117, 2019. 10.3389/fmed.2019.00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wu C, Lorenzo G, Hormuth DA 2nd, et al. : Integrating mechanism-based modeling with biomedical imaging to build practical digital twins for clinical oncology. Biophys Rev 3, 2022(2):021304. 10.1063/5.0086789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hormuth DA II, Farhat M, Christenson C, et al. : Opportunities for improving brain cancer treatment outcomes through imaging-based mathematical modeling of the delivery of radiotherapy and immunotherapy. Adv Drug Deliv Rev 187, 2022:114367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jackson PR, Juliano J, Hawkins-Daarud A, Rockne RC, Swanson KR: Patient-Specific Mathematical Neuro-Oncology: Using a Simple Proliferation and Invasion Tumor Model to Inform Clinical Practice. Bull Math Biol 77(5):846–856, 2015. 10.1007/s11538-015-0067-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Rockne R, Alvord EC Jr, Rockhill JK, Swanson KR: A mathematical model for brain tumor response to radiation therapy. J Math Biol 58 (4–5):561–578, 2009. 10.1007/s00285-008-0219-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Jacobs J, Rockne RC, Hawkins-Daarud AJ, et al. : Improved model prediction of glioma growth utilizing tissue-specific boundary effects. Math Biosci 312:59–66, 2019. 10.1016/j.mbs.2019.04.004 [DOI] [PubMed] [Google Scholar]
- 191.Rahman MM, Feng Y, Yankeelov TE, Oden JT: A fully coupled space−time multiscale modeling framework for predicting tumor growth. Comput Methods Appl Mech Eng 320:261–286, 2017. 10.1016/j.cma.2017.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Enderling H, Alfonso JCL, Moros E, Caudell JJ, Harrison LB: Integrating Mathematical Modeling into the Roadmap for Personalized Adaptive Radiation Therapy. Trends Cancer Res 5(8):467–474, 2019. 10.1016/j.trecan.2019.06.006 [DOI] [PubMed] [Google Scholar]
- 193.Leder K, Pitter K, LaPlant Q, et al. : Mathematical modeling of PDGF-driven glioblastoma reveals optimized radiation dosing schedules. Cell 156(3):603–616, 2014. 10.1016/j.cell.2013.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Stenmark Tullberg A, Puttonen HAJ, Sjöström M, et al. : Immune Infiltrate in the Primary Tumor Predicts Effect of Adjuvant Radiotherapy in Breast Cancer; Results from the Randomized SweBCG91RT Trial. Clin Cancer Res 27(3):749–758, 2021. 10.1158/1078-0432.CCR-20-3299 [DOI] [PubMed] [Google Scholar]
- 195.Tong H, Sun J, Fang J, et al. : A Machine Learning Model Based on PET/CT Radiomics and Clinical Characteristics Predicts Tumor Immune Profiles in Non-Small Cell Lung Cancer: A Retrospective Multicohort Study. Front Oncol 11, 2021:603882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Masson-Grehaigne C, Lafon M, Palussiére J, et al. : Enhancing Immunotherapy Response Prediction in Metastatic Lung Adenocarcinoma: Leveraging Shallow and Deep Learning with CT-Based Radiomics across Single and Multiple Tumor Sites. Cancers 16(13):2491, 2024. 10.3390/cancers16132491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Withers HR, Peters LJ, Taylor JM: Dose-response relationship for radiation therapy of subclinical disease. Int J Radiat Oncol Biol Phys 31 (2):353–359, 1995. 10.1016/0360-3016(94)00354-N [DOI] [PubMed] [Google Scholar]
- 198.Davey A, Thor M, van Herk M, et al. : PD-0673 Validation of the interaction between peritumour density and dose for local relapse in lung SABR. Radiother Oncol 170:S608–S610, 2022. 10.1016/s0167-8140(22)02920-6 [DOI] [Google Scholar]
- 199.Davey A, Thor M, van Herk M, et al. : Predicting cancer relapse following lung stereotactic radiotherapy: an external validation study using real-world evidence. Front Oncol 13, 2023:1156389. 10.3389/fonc.2023.1156389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Munck Af Rosenschold P, Zelefsky MJ, Apte AP, et al. : Image-guided radiotherapy reduces the risk of under-dosing high-risk prostate cancer extra-capsular disease and improves biochemical control. Radiat Oncol 13(1):64, 2018. 10.1186/s13014-018-0978-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Buti G, Ajdari A, Chen YL, Bridge CP, Sharp GC, Bortfeld T: Integrating muscle fiber orientation from visible human data into radiotherapy target volumes. Phys Med Biol 69, 2024(14):145006. 10.1088/1361-6560/ad5d50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bortfeld T, Shusharina N, Craft D: Probabilistic definition of the clinical target volume—implications for tumor control probability modeling and optimization. Phys Med Biol 66(1):01NT01, 2021. 10.1088/1361-6560/abcad8 [DOI] [PubMed] [Google Scholar]
- 203.Ludwig R, Schubert AD, Barbatei D, et al. : Modelling the lymphatic metastatic progression pathways of OPSCC from multi-institutional datasets. Sci Rep 14(1):15750, 2024. 10.1038/s41598024-66012-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Gregoire V, Mackie TR: Dose prescription, reporting and recording in intensity-modulated radiation therapy: a digest of the ICRU Report 83. Imaging Med 3(3):367–373, 2011. 10.2217/iim.11.22 [DOI] [PubMed] [Google Scholar]
- 205.Witte MG, Sonke JJ, Siebers J, Deasy JO, van Herk M: Beyond the margin recipe: the probability of correct target dosage and tumor control in the presence of a dose limiting structure. Phys Med Biol 62 (19):7874–7888, 2017. 10.1088/1361-6560/aa87fe [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Deasy JO, Jeong J: Radiobiological Principles for Adaptive Radiotherapy. Adaptive Radiation Therapy. Boca Raton, FL: CRC Press, Taylor & Francis Group, 3–18, 2011. https://www.taylorfrancis.com/chapters/edit/10.1201/b10517-8/radiobiological-principles-adaptive-radiotherapy-joseph-deasy-jeho-jeong [Google Scholar]
- 207.Shepherd A, James SS, Rengan R: The Practicality of ICRU and Considerations for Future ICRU Definitions. Semin Radiat Oncol 28 (3):201–206, 2018. 10.1016/j.semradonc.2018.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Dudas D, Saghand PG, Dilling TJ, Perez BA, Rosenberg SA, El Naqa I: Deep Learning-Guided Dosimetry for Mitigating Local Failure of Patients With Non-Small Cell Lung Cancer Receiving Stereotactic Body Radiation Therapy. Int J Radiat Oncol Biol Phys 119(3):990–1000, 2024. 10.1016/j.ijrobp.2023.11.059 [DOI] [PubMed] [Google Scholar]


