Abstract
Increasing evidence points toward an essential role for complement activation in the pathogenesis of diabetic kidney disease (DKD). However, the precise molecular mechanisms remain unclear, and the pathway predominantly contributing to complement activation in DKD is of particular interest. In this study, the glomerular proteome, especially the profiles of the complement proteins, was analyzed in kidney biopsies from 40 DKD patients and 10 normal controls using laser microdissection-assisted liquid chromatography-tandem mass spectrometry (LMD-LC-MS/MS). The glomerular abundances of three proteins related to classical pathway (CP) (C1q, C1r, C1s), five proteins related to alternative pathway (AP) (CFB, CFH, CFHR1, CFHR3, CFHR5), one common protein related to CP and lectin pathway (LP) (C4), and six proteins related to terminal complement pathway (C3, C5, C6, C7, C8, C9) were significantly increased in DKD. Notably, none of the proteins unique to the lectin complement pathway, including mannose-binding lectin (MBL) and its associated proteins, were detected in DKD glomeruli. Furthermore, the glomerular complement proteins of CP and AP were positively correlated with glomerular pathological grades and proteinuria, and negatively correlated with eGFR in DKD patients. Our results highlight a critical role for complement activation of the CP and AP, rather than the LP, in DKD progression.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-84900-4.
Keywords: Complements, Diabetic kidney disease, Classical pathway, Alternative pathway, Lectin pathway, Proteomics
Subject terms: Diabetic nephropathy, Complement cascade
Introduction
Diabetic kidney disease (DKD), affecting nearly 40% of patients with diabetes, is the leading cause of end stage renal disease (ESRD) worldwide1. The global prevalence of diabetes mellitus has been rising rapidly, with an anticipated increase from 422 million in 2023 to 642 million by 2040, coupled with a continuously growing incidence of DKD. Despite advancements in anti-glycemic and anti-hypertensive therapies, DKD contributes to substantial morbidity, mortality, and economic burden in diabetic patients. Therefore, identifying novel therapeutic targets to prevent DKD progression is imperative.
DKD develops via a variety of complex pathogenic mechanisms. Novel applications of proteomic technologies have provided new insights into the role of the complement system in DKD pathogenesis. Urine proteomics of DKD patients identified up-regulated differentially expressed proteins (DEPs), including C9, C3, C4 and complement factor H (CFH), enriched in the complement cascade2,3. Plasma proteomics revealed that collectin-11, an initiator of the complement lectin pathway (LP), was elevated in patients with incipient DKD, positioned at the core “traffic link” of protein–protein interactions, and proposed as a candidate biomarker for DKD4. Moreover, DKD patients had the highest urine/plasma ratio of complement C35, which suggests that local complement activation has a more critical role than circulation-based complement activation. Proteomic analysis of proteins within the glomeruli provides direct evidence of altered proteins and pathways, offering stronger insights into the pathogenic mechanisms of DKD. However, knowledge regarding the glomerular proteome in DKD is still limited.
The molecular triggers and initial pathways for complement activation depend on the illness setting and vary among different renal diseases6. Activation of the LP by aberrantly glycated proteins in a hyperglycemic milieu was proposed to be the main link between complement and DKD7–10. However, an increasing body of evidence highlights the involvement of the complement system’s classical pathway (CP) and alternative pathway (AP) in DKD. Complement C1q was identified to bind to advanced glycation end products11,12, thereby activating the CP. Glomerular transcriptomic data of DKD patients associated with renal histopathology showed that C1q in glomeruli may accelerate DKD progression13. Bioinformatic analysis of gene expression data revealed that, in addition to C1q, complement factor B (CFB) was associated with DKD development14,15. Targeting CFB attenuates DKD both in vivo and in vitro15,16. The precise molecular mechanisms underlying complement activation in DKD have not been elucidated, and the pathway that predominantly contributes to complement activation in DKD is of particular interest.
This study aimed to identify altered protein expression profiles in the glomeruli of DKD patients using laser capture microdissection (LCM) followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS), determine the key complement proteins in DKD pathogenesis, and elucidate the potential mechanism of complement activation underlying DKD development.
Materials and methods
Human biopsy specimens
Forty patients with biopsy-proven diabetic nephropathy, who had been referred to the First Affiliated Hospital of Zhengzhou University between January 2019 and April 2020, were enrolled in this study. All the patients met the American Diabetes Association diagnostic criteria for type 2 diabetes (T2D). The diagnosis and classification of diabetic nephropathy were based on the criteria proposed by the Renal Pathology Society in 201017. Patients with coincident nondiabetic renal disease or inadequate renal tissue (< 10 glomeruli) were excluded. Human kidney tissues were collected from the remaining paraffin-embedded core needle renal biopsy specimens with ethical approval. The baseline clinical and pathological data of DKD patients were collected at the time of kidney biopsy. Normal controls of kidney tissue samples from the unaffected parts of surgical nephrectomies (taken at least 2 cm away from the cancer site) were obtained from 10 age- and sex-matched patients. The control group had normal basic laboratory parameters (e.g., eGFR, SCr, hematuria, and proteinuria) and no history of diabetes. Renal biopsies from six patients with arterionephrosclerosis and no history of diabetes were included as disease controls. Each specimen was routinely examined under light and immunofluorescence microscopies. This study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Research Committee of the First Affiliated Hospital of Zhengzhou University. All participants gave written informed consent.
Glomerular isolation by laser-capture microdissection
First, 8-µm-thick sections of formalin-fixed and paraffin-embedded renal tissue were cut onto Leica membrane glass slides (Leica NO.11600288, Leica Microsystems, Mannheim, Germany), de-paraffinized and stained with hematoxylin. The glomeruli were identified and microdissected with laser capture techniques (Leica LMD7, Leica Microsystems, Mannheim, Germany) with a total area of approximately 500,000 μm2 per patient. Microdissected tissue was collected into 200-µL Eppendorf tubes containing 20 µL Tris/EDTA/0.002% Zwittergent 3–16 buffer. Samples were processed by incubation at 98 °C for 90 min and then ultrasonicated in a water bath for 90 min. The proteins extracted from the microdissected tissue fragments were then digested by 0.2 µg trypsin overnight at 37 °C. After desalination using ZipTip C18 microcolumns, the peptides were prepared for liquid chromatography and tandem mass spectrometry (MS).
Proteomics by tandem MS
Peptide samples were analyzed using the EASY-nLC 1200 system coupled to a Q-Exactive HF-X Orbitrap mass spectrometer (Thermo Scientific) using a linear gradient method with an initial concentration of 4% buffer B, which was increased to 7% at 1 min, with subsequent increases to 25% (95 min), 40% (111 min), 100% (116 min), and a hold for 4 min at a flowrate of 600 nL per minute. MS spectra were acquired in data-dependent acquisition (DDA) mode, consisting of a full MS scan with an m/z range of 350–2000 and a resolution of 60,000 at 200 m/z, followed by MS/MS scans of the top 20 parent ions (intensity threshold of 27,000 counts/s; 15,000 resolution at 200 m/z).
A list of proteins based on the peptides identified by MS was generated for each patient. Protein identifications were accepted if they could be established on the basis of at least one unique peptide identified with high confidence (false discovery rate FDR < 1%). The raw MS data were processed using Sequest HT node within Proteome Discoverer (PD) software (Version 2.4; Thermo Fisher Scientific, Waltham, MA, USA). The peptide abundance was calculated using the intensity of all precursors, and the protein abundance was determined by the total intensity of all matched peptides. The protein abundance was normalized based on the total abundance of all proteins in the sample set to a constant value of 106.
Pathways and protein interaction network analysis
Protein-protein interaction (PPI) network analysis was performed using the Search Tool for the Retrieval of Interacting Genes (STRING) 10.0 database (http://string-db.org). The interaction confidence score of the PPI network analysis was set to 0.9. The PPI network was visualized using Cytoscape (Version 3.7.2; Institute for Systems Biology, Seattle, WA, USA), and the most significant module was identified using the MCODE plug-in. To explore the hub nodes in the most significant module of the PPI network, all nodes were scored and ranked using the topological analysis provided by CytoHubba. Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of the upregulated protein signatures were analyzed by clusterProfiler package (version 3.16.0) in R (version 4.0.0; University of Auckland, Auckland, New Zealand). A heat map of the DEPs was generated using pheatmap (version 1.0.12) in R.
Validation of glomerular complements by immunohistochemistry (IHC)
Paraffin-embedded sections were cut, deparaffinized, and hydrated by soaking in 100% xylene and descending sequential ethanol concentrations, followed by microwave heating in citrate buffer (pH 6.0) for 20 min to repair antigen. The sections were then incubated in 0.3% H2O2 in phosphate-buffered saline to block endogenous peroxidase activity. After washing, sections were incubated with either anti-C1q antibodies (1:200 dilution, 16889-1-AP; Proteintech), anti-C3 antibodies (1:200 dilution, ab200999; Abcam), anti-C4 antibodies (1:300 dilution, 22233-1-AP; Proteintech), anti-C5b-9 antibodies (1:400 dilution, ab55811; Abcam), anti-MBL antibodies (1:200 dilution, ab23457; Abcam), anti-MASP1 antibodies (1:400 dilution, 21837-1-AP; Proteintech), anti-CFB antibodies (1:50 dilution, ab192577; Abcam), or anti-CFH antibodies (1:50 dilution, ab170036; Abcam) overnight at 4 °C in a moist chamber. HRP-conjugated secondary antibodies (1:2000 dilution, Zhongshan Golden Bridge, Beijing, China) were incubated for 20 min at 37 °C, and an avidin-biotin-peroxidase complex (1:20 dilution, Zhongshan Golden Bridge) was applied. Color development was induced by diaminobenzidine treatment. Finally, the sections were counterstained with hematoxylin, dehydrated, sealed, and observed using light microscopy. The extent of glomerular staining for complement proteins was evaluated at ×400 magnification and scored semiquantitatively as follows: 0, no staining; 1, weak and spotty intraglomerular staining; 2, moderate and segmental intraglomerular staining; and 3, strong and diffuse intraglomerular staining. The scorer was blinded to the disease state.
Statistical analysis
Continuous variables are presented as mean ± SD or median (interquartile range). For comparisons between different groups, the Student’s t-test was used for parametric data, and the Wilcoxon rank-sum test was used for nonparametric data. Noncontinuous variables were expressed as proportions and compared using the chi-square test or Fisher’s exact test. The correlations between the abundance of each glomerular complement component and proteinuria, baseline eGFR, and renal pathology were assessed. Pearson’s correlation coefficients were used to measure the associations between normally distributed variables. Spearman’s rank-order correlation tests were performed to analyze the correlations among non-normally distributed variables or rank variables. Associations between glomerular abundances of complement proteins and the combined endpoint of ESRD and/or death were assessed using Cox regression analysis. Protein abundance data were log2-transformed to calculate the hazard ratio (HR). A P value of ˂ 0.05 was considered statistically significant. All analyses were performed using SPSS 23.0 (IBM SPSS Inc. Chicago, IL, USA).
Results
Clinical and histological characteristics
Of the 40 patients with DKD, 18 were men and 22 were women, with a mean age of 64.5 ± 6.9 years and a median diabetes duration of 10 (6,18) years at the time of biopsy. 87.5% (35/40) of the DKD patients were complicated by hypertension on admission. Upon biopsy, the patients had a mean eGFR of 36.4 ± 3.2 mL/min/1.73m2 and a mean urine protein of 5.93 ± 0.87 g/24 h. The mean level of glycated hemoglobin (HbA1c) was 9.1 ± 1.2%. Decreased levels of serum C3 were found in 4 (10.0%) patients. According to the pathological classification of diabetic nephropathy17, the patients were divided into four classes of glomerular lesions with separate scoring for interstitial and tubular involvement. Among the 40 patients, 4 were classified as class I (glomerular basement membrane thickening), 12 as class II (mesangial expansion), 12 as class III (nodular sclerosis), and 12 as class IV (advanced diabetic glomerulosclerosis). The relevant demographic, clinical, and histological information is presented in Table 1.
Table 1.
Clinical and pathologic data of patients with DKD.
| DKD (n = 40) | |
|---|---|
| Age (years), mean ± SD | 64.5 ± 6.9 |
| Gender (male/female) | 18/22 |
| Diabetes duration (years), median (IQR) | 10 (6,18) |
| History of hypertension, n (%) | 35 (87.5%) |
| Fasting plasma glucose (mg/dl), mean ± SD | 204 ± 35.2 |
| Glycated hemoglobin (HbA1c), mean ± SD | (9.1 ± 1.2) % |
| SCr (µmol/L), mean ± SD | 201.1 ± 21.2 |
| eGFR (ml/min/1.73 m2), mean ± SD | 36.4 ± 3.2 |
| Proteinuria (g/24 h), mean ± SD | 5.93 ± 0.87 |
| Hematuria, n (%) | 12 (30%) |
| Serum C3 (g/L), mean ± SD | 1.17 ± 0.21 |
| Glomerular class (I/II/III/IV), n | 4/12/12/12 |
| IFTA score (0/1/2/3), n | 0/15/6/19 |
SD, standard deviation; IQR, interquartile range; SCr, serum creatinine; eGFR, estimated glomerular filtration rate; C3, complement 3; IFTA, interstitial fibrosis and tubular atrophy.
Glomerular proteome profiles of human DKD biopsies
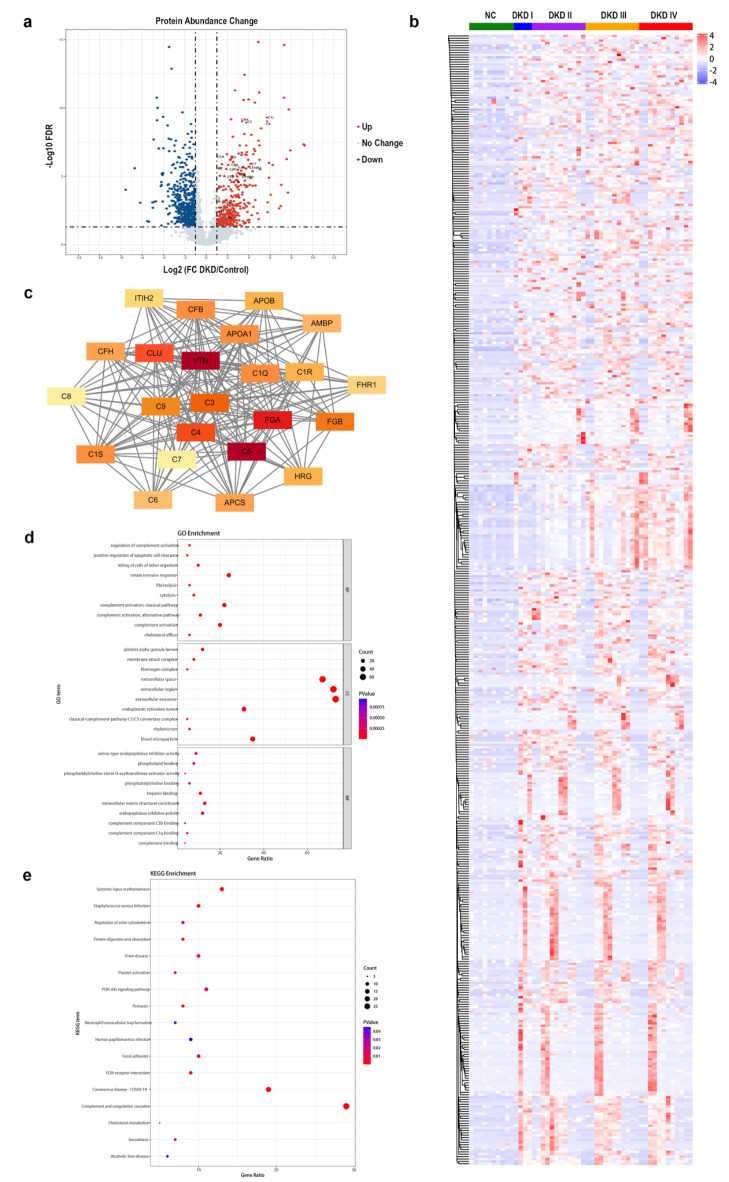
A total of 5834 proteins were identified in the dissected glomeruli of 40 DKD patients and 10 normal controls. Of these, 1246 proteins exhibited a significantly differentially expressed abundance in laser micro-dissected glomerular tissues from DKD patients with the criteria of |log2 fold change (log2 FC)|≥1 and P < 0.05, of which 475 upregulated proteins and 771 downregulated proteins were identified (Fig. 1a). The expression profiles of these 1246 proteins are shown in a hierarchical clustering heatmap (Fig. 1b). A total of 270 DEPs were subjected to the STRING database. Consequently, 23 hub proteins were identified, 19 of which were upregulated and 4 downregulated. 13 complement proteins (C1q, C1r, C1s, C3, C4, C5, C6, C7, C8, C9, CFB, CFH, and CFHR1) were included in the upregulated hub proteins (Fig. 1c).
Fig. 1.
Differential glomerular proteome profiles between DKD and normal controls. (a) Volcano plot of FCs and P values for 5834 proteins identified in the dissected glomeruli of 40 DKD patients, compared with 10 normal controls. A total of 1246 proteins exhibited a significantly differentially expressed abundance (475 upregulated proteins in red, 771 downregulated proteins in blue). (b) Heatmap representing the hierarchical clustering analyses of the proteins significantly differentially expressed in the DKD glomeruli. (c) Networks of the protein-protein interactions of DEPs in DKD glomeruli mapped using STRING. The 23 hub proteins are illustrated as nodes with differential colors, whose intensity depends on the significance of the interactions. (d) Top 10 GO terms of in the upregulated BPs, CCs, and MFs of DEPs between DKD patients and normal controls. (e) Scatterplot for the top 10 upregulated pathways in KEGG enrichment of DEPs between DKD patients and normal controls. DKD, diabetic kidney disease; NC, normal controls; FC, fold change; DEPs, differentially expressed proteins; STRING, the Search Tool for the Retrieval of Interacting Genes; GO, Gene ontology; BP, biological processes; CC, cellular component; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes.
The ClusterProfiler package was used for GO enrichment analysis to explore the potential functions of the 270 DEPs. GO classification showed that the top 5 upregulated biological processes (BPs) were as follows: innate immune response, complement activation (CP), complement activation, complement activation (AP), killing of cells of other organism (Fig. 1d). Classical-complement-pathway C3/C5 convertase complex ranked among the top 10 cellular components (CCs) terms (Fig. 1d). Moreover, complement component C1q binding, complement component C3b binding, and complement binding were on the list of the top 10 molecular functions (MFs) terms (Fig. 1d), confirming activation of complement CP and AP as the main function of DEPs. Complement and coagulation cascades were the most enriched among the top 10 upregulated pathways in KEGG analysis, which demonstrated that the DEPs may participate in the progression and development of DKD by regulating the complement cascade (Fig. 1e).
Proteomic analysis of complement proteins in DKD glomeruli
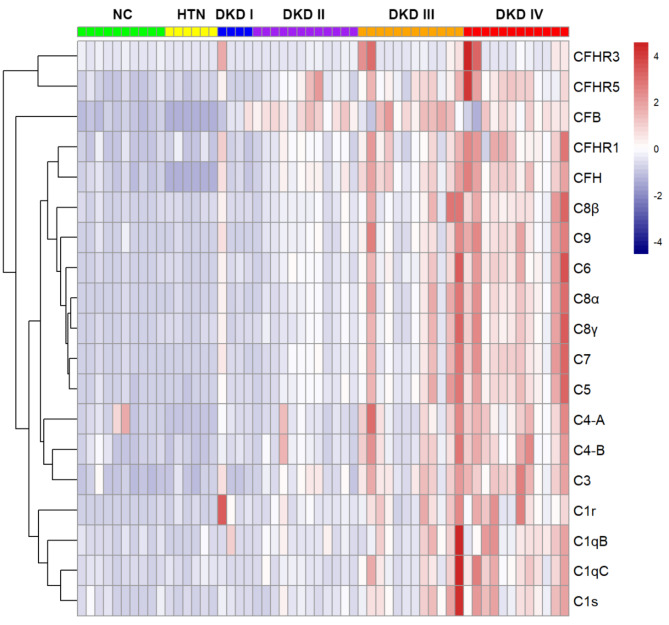
The expression profiles of complement proteins in the 40 DKD patients and 10 normal controls are shown in Table 2; Fig. 2. As the spectral analysis results indicated in the table, the glomerular abundances of three classical complement pathway proteins (C1q, C1r, C1s), five alternative complement pathway proteins (CFB, CFH, CFHR1, CFHR3, CFHR5), one classical-lectin complement pathway common protein C4 and six terminal complement pathway proteins (C3, C5, C6, C7, C8, C9) were significantly higher in DKD than those in normal controls. None of the complement proteins exhibited increased glomerular abundance in renal biopsies from patients with arterionephrosclerosis compared to normal controls (Fig. 2). C3, C9, and C4 were the top three proteins with high relative spectral abundances (total relative abundance: 15743, 11845, and 7991, respectively), and their corresponding fold-change values (DKD/NC) were 3.2, 7.2, and 4.8, respectively. Notably, proteins exclusively acting on the lectin complement pathway, including collectins (MBL, collectin-10, collectin-11), ficolins (ficolin-1, ficolin-2, ficolin-3) and proteins of the MBL-associated serine protease (MASP) family (MASP-1, MASP-2, MASP-3, MAp19, MAp44), were not detected in the glomeruli of DKD patients. The expression details of LP proteins in the glomeruli of DKD patients and controls are listed in Supplementary Table S1.
Table 2.
The complement proteins in the glomerular proteome between the DKD patients and normal controls.
| Proteins | Normalized abundance in DKD | Fold change (DKD/controls) | P-value versus control | Fold change to controls | P-value between classes | |||
|---|---|---|---|---|---|---|---|---|
| Class I DKD | Class II DKD | Class III DKD | Class IV DKD | |||||
| C1qB | 234 | 5.6 | < 0.001 | 3.0 | 2.5 | 7.1 | 8.1 | 0.041 |
| C1qC | 268 | 4.7 | < 0.001 | 1.2 | 1.6 | 6.1 | 7.4 | 0.004 |
| C1r | 79 | 69.2 | < 0.001 | 86.3 | 27.8 | 80.1 | 93.9 | 0.088 |
| C1s | 116 | 4.2 | < 0.001 | 1.3 | 1.6 | 5.8 | 6.2 | 0.004 |
| C2 | 3 | 25.6 | < 0.001 | 5.5 | 24.3 | 33.3 | 25.9 | 0.276 |
| C3 | 15,743 | 3.2 | < 0.001 | 1.6 | 2.1 | 3.6 | 4.4 | < 0.001 |
| C4-A | 490 | 2.0 | 0.011 | 0.8 | 0.9 | 2.6 | 3.0 | 0.004 |
| C4-B | 7991 | 4.8 | < 0.001 | 1.7 | 2.5 | 5.8 | 7.0 | 0.003 |
| C5 | 4789 | 10.4 | < 0.001 | 3.6 | 4.8 | 12.7 | 16.1 | 0.001 |
| C6 | 1350 | 19.1 | < 0.001 | 7.2 | 7.8 | 22.2 | 31.4 | 0.001 |
| C7 | 1428 | 14.8 | < 0.001 | 5.3 | 6.2 | 16.8 | 24.7 | 0.001 |
| C8α | 926 | 21.1 | < 0.001 | 6.0 | 8.1 | 26.0 | 34.2 | 0.001 |
| C8β | 1384 | 12.5 | < 0.001 | 4.5 | 4.8 | 16.3 | 19.1 | 0.007 |
| C8γ | 1397 | 13.7 | < 0.001 | 4.3 | 5.7 | 16.5 | 22.1 | 0.003 |
| C9 | 11,845 | 7.2 | < 0.001 | 3.0 | 3.3 | 8.0 | 11.7 | 0.001 |
| CFB | 115 | 4.5 | < 0.001 | 2.6 | 4.9 | 5.3 | 4.1 | 0.035 |
| CFI | 6 | 11.4 | < 0.001 | 11.6 | 10.5 | 14.7 | 9.0 | 0.694 |
| CFH | 1061 | 3.5 | < 0.001 | 1.7 | 2.3 | 4.1 | 4.7 | < 0.001 |
| CFHR1 | 181 | 7.4 | < 0.001 | 3.7 | 4.0 | 8.0 | 11.5 | 0.002 |
| CFHR3 | 256 | 22.0 | 0.019 | 21.9 | 1.4 | 27.8 | 36.7 | 0.327 |
| CFHR5 | 484 | 6.7 | < 0.001 | 2.5 | 5.0 | 6.9 | 9.7 | 0.032 |
C4-A and C4-B are the two isotypes of C4. CFB, complement factor B; CFI, complement factor I; CFH, complement factor H; CFHR1, complement factor H related protein 1; CFHR3, complement factor H related protein 3; CFHR5, complement factor H related protein 5;
Fig. 2.
Differentially glomerular expression profiles of complement proteins in patients with diabetic kidney disease, compared to normal controls and arterionephrosclerosis. In the heatmap, each row represents one complement protein, and each column corresponds to one patient’s dataset. Normalized relative abundances of glomerular complement proteins are depicted by a pseudo-color scale, with red indicating a higher level of protein expression, white indicating an equal level, and blue indicating a lower expression level compared with mean protein abundance of each sample. The dendrogram displays the outcome of unsupervised hierarchical clustering analysis, placing similar proteome profile values near to each other. DKD, diabetic kidney disease; NC, normal controls; HTN, hypertension; CFB, complement factor B; CFH, complement factor H; CFHR, complement factor H related protein.
Validation of complement proteins in DKD glomeruli by IHC
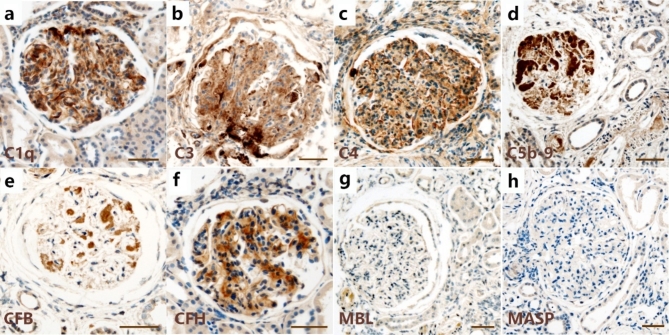
To validate the glomerular expression of complement observed by proteomic evaluation, IHC was performed on a separate cohort of human biopsies diagnosed with DKD and on normal controls for eight complement proteins in three pathways: C1q, C3, C4, C5b-9, CFB, CFH, MBL, and MASP. The characteristics of the validation cohort are shown in Supplementary Table S2. Immunohistochemical analysis verified the GBM localization of C1q, C3, C4, and C5b-9, with strong positive staining for nodular nephrosclerosis (Fig. 3a–d). The CFB also showed a GBM immunostaining pattern (Fig. 3e), whereas the CFH showed a mesangial staining pattern (Fig. 3f). Immunoreactivity intensity analysis confirmed the significantly higher expression of C1q, C3, C4, C5b-9, CFB, and CFH in the glomeruli of DKD patients than in normal controls, consistent with the quantitative proteomics results. Remarkably, neither of the two unique proteins in the LP, MBL or MASP, were detected in the glomeruli of DKD biopsies (Fig. 3g, h). The details of IHC scores for each complement protein are provided in Supplementary Table S3.
Fig. 3.
Immunohistochemistry (IHC) staining for C1q, C3, C4, C5b-9, CFB, CFH, MBL, and MASP. (a–e) Representative images of C1q, C3, C4, C5b-9, and CFB staining showing a GBM pattern of immunostaining in kidney tissues from patients with DKD. (f) Immunohistochemical diagram of CFH in kidneys with DKD depicting mesangial deposition. (g, h) Representative images of MBL and MASP staining showing no deposition was found in the glomeruli of kidneys with DKD. Scale bars represent 50 μm. CFB, complement factor B; CFH, complement factor H; MBL, mannose-binding lectin; MASP, mannose-binding lectin-associated serine protease; GBM, glomerular basement membrane; DKD, diabetic kidney disease.
Correlations between glomerular complement proteins and clinical or pathological parameters
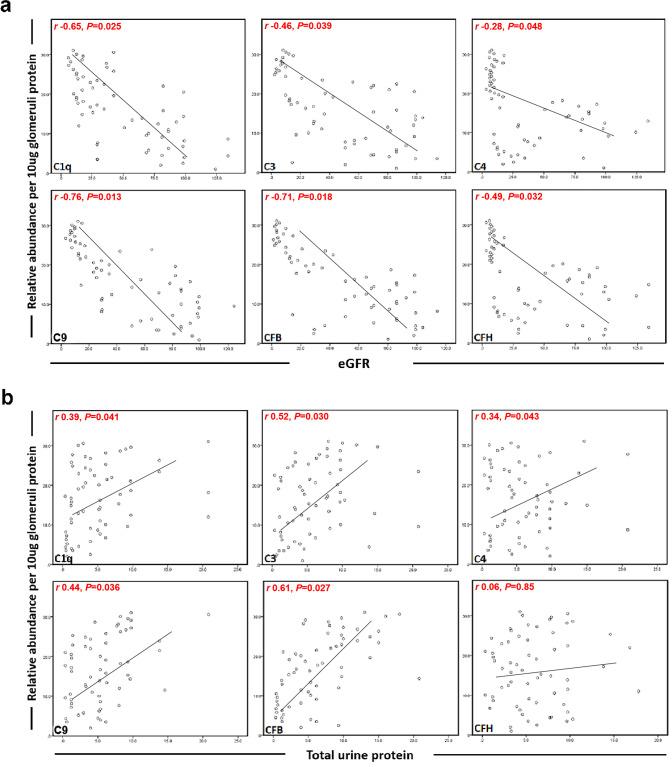
The correlations between the abundance of each glomerular complement protein and clinical parameters at the time of biopsy were assessed. The abundance of the six glomerular complement proteins (C1q, C3, C4, C9, CFB, and CFH) was significantly negatively correlated with eGFR (Pearson r= -0.65, -0.46, -0.28, -0.76, -0.71, and − 0.49, respectively). The abundance of most glomerular complement proteins (C1q, C3, C4, C9, and CFB) positively correlated with total urine protein per 24 h (Pearson r = 0.39, 0.52, 0.34, 0.44, and 0.61, respectively); however, the glomerular abundance of CFH did not correlate with total urine protein per 24 h (P = 0.85). The correlation between glomerular complement proteins and clinical markers in DKD patients is illustrated in Fig. 4.
Fig. 4.
Correlations between the abundances of individual glomerular complement proteins and baseline eGFR (a) and total urine protein per 24 h (b) in patients with DKD. Pearson correlation coefficients are indicated in the figures. CFB, complement factor B; CFH, complement factor H.
Of note, the abundances of complement proteins C1q, C1s, C3, C4, C5, C6, C7, C8, C9, CFB, CFH, CFHR1, and CFHR5 showed positive correlations with glomerular pathological grades (P < 0.001 to 0.041). However, C1r and CFHR3 were not associated with pathological grades (P = 0.088 and 0.327, respectively) (Table 2).
Correlations between glomerular complement proteins and outcomes
Follow-up data were collected from 32 patients (80.0%), with a median follow-up duration of 12 months. 18 patients (56.3%) reached the combined endpoint of progression to ESRD and/or death. Glomerular abundances of C1q, C1r, C1s, C3, C4, C5, C6, C7, C8, C9, CFH, CFHR1, CFHR3, and CFHR5 were significantly associated with the combined ESRD/death outcome (Table 3). The strongest risk factors for unfavourable outcomes were glomerular C3 (HR per doubling, 4.29; 95% confidence interval [CI], 2.00 to 9.21) and CFH (HR per doubling, 4.29; 95% CI, 1.97 to 9.35). After adjusting for serum creatinine (mg/dL), glomerular abundance of CFH remained predictive of disease progression (HR per doubling, 3.18; 95% CI, 1.49 to 6.76).
Table 3.
Association of glomerular abundance of individual complement protein with the combined outcome of ESRD and/or death.
| Protein | Unadjusted survival analysis | Multivariable analysis | ||
|---|---|---|---|---|
| Hazard ratio per doubling (95% CI) | P-value | Hazard ratio per doubling (95% CI) | P-value | |
| C1qB | 1.628 (1.142, 2.323) | 0.007 | 1.503 (1.074, 2.104) | 0.018 |
| C1qC | 2.455 (1.645, 3.664) | < 0.001 | ||
| C1r | 1.463 (1.097, 1.951) | 0.010 | 1.391 (1.045, 1.852) | 0.024 |
| C1s | 2.145 (1.462, 3.147) | < 0.001 | 1.893 (1.299, 2.760) | 0.001 |
| C3 | 4.294 (2.002, 9.210) | < 0.001 | ||
| C4-A | 2.199 (1.480, 3.269) | < 0.001 | 1.954 (1.328, 2.875) | 0.001 |
| C4-B | 2.597 (1.620, 4.164) | < 0.001 | ||
| C5 | 2.149 (1.437, 3.212) | < 0.001 | ||
| C6 | 2.064 (1.421, 2.999) | < 0.001 | ||
| C7 | 2.017 (1.388, 2.929) | < 0.001 | ||
| C8α | 1.991 (1.393, 2.845) | < 0.001 | ||
| C8β | 1.804 (1.305, 2.495) | < 0.001 | 1.601 (1.155, 2.219) | 0.005 |
| C8γ | 1.974 (1.393, 2.798) | < 0.001 | ||
| C9 | 2.267(1.478, 3.477) | < 0.001 | ||
| CFB | 0.634 (0.372, 1.078) | 0.092 | ||
| CFH | 4.290 (1.969, 9.350) | < 0.001 | 3.175 (1.493, 6.755) | 0.003 |
| CFHR1 | 2.392 (1.467, 3.902) | < 0.001 | 1.887 (1.177, 3.024) | 0.008 |
| CFHR3 | 1.182 (1.053, 1.326) | 0.004 | 1.171 (1.044, 1.314) | 0.007 |
| CFHR5 | 2.203 (1.378, 3.521) | 0.001 | 2.103 (1.290, 3.429) | 0.003 |
Protein abundance data were log2-transformed to calculate the hazard ratio (HR). C4-A and C4-B are the two isotypes of C4. CI, confidence interval; CFB, complement factor B; CFI, complement factor I; CFH, complement factor H; CFHR1, complement factor H related protein 1; CFHR3, complement factor H related protein 3; CFHR5, complement factor H related protein 5.
Discussion
In our study, complement proteins of the CP and AP were identified in the glomeruli of kidney biopsies from DKD patients using LMD-LC-MS/MS. Furthermore, the abundance of the complement proteins correlated with proteinuria, renal function, pathological grades, and outcomes. Interestingly, none of the complement proteins of the LP were detected in the glomerular proteome of DKD kidneys. Our results provide new insights into the underlying mechanisms of complement activation in diabetic glomerulopathy.
Complement activation plays an important role in the onset and progression of DKD6. Multiple complement proteins and activation products, such as C1q, MBL, Bb, C4d, iC3b, C3a, C5a, and C5b-9, were elevated in the plasma/serum and urine samples of DKD patients, indicative of activated complement pathways in DKD18–22. Similarly, the deposition of complement proteins, including C1q, C3, C4d, C3a, C5a, and C5b-9 was found in the glomeruli of DKD patients and associated with worse renal outcomes12,13,23–25. In contrast, decreased serum C3 levels were observed in 68.0% of patients in a DKD cohort and were significantly associated with more severe kidney damage and poor renal outcomes26. In DKD mice, reduced proteinuria and attenuated glomerular injury were observed following antagonism or knockout of receptors for complement activation products C3a or C5a22,24,25,27,28. These clinical and experimental findings highlight the essential role for complement activation in the pathogenesis of DKD. In our study, 13 of the 19 upregulated hub proteins in the PPI network of the DEPs belonged to the complement system. Moreover, the abundance of glomerular complement proteins was correlated with the level of proteinuria, degree of pathological lesions, and impairment of renal function. Our results support the profound involvement of the complement system in the development of DKD.
Among these three activation pathways, the LP is of particular interest in DKD. Since LP is triggered by the binding of pattern recognition proteins to specific ligands, predominantly sugars, it is reasonable to propose that LP is activated by altered glycated proteins on cell surfaces due to high glucose levels in patients with diabetes. However, the evidence for LP activation in DKD from circulation and kidney glomeruli appears contradictory. Serum levels of MBL were significantly increased in DKD patients9,10,29,30. In contrast, glomerular MBL deposits were rarely observed by IHC in autopsied diabetic cases, regardless of the presence of diabetic nephropathy12. Similarly, another study failed to detect glomerular MBL by IHC in renal biopsies from DKD patients13. Our glomerular proteomic analysis using LMD-LC-MS/MS further confirmed the absence of unique proteins of the LP, including MASPs, collectins, and ficolins, in the glomeruli of DKD patients. These contradictions prompted us to investigate the exact role of LP activation in diabetic glomerular lesions.
Moreover, the complement pathways involved may differ depending on the site of renal injury, such as the glomeruli, tubules, or tubular interstitium31. MBL and MASP-1 in the tubular interstitium were closely related to the degree of tubular interstitial damage in DKD patients, suggesting that the LP may contribute to renal tubular interstitial lesions in diabetic nephropathy32. However, our glomerular proteome findings indicated an unclear role of LP activity in glomerular injury in DKD. Further studies are needed to clarify the precise mechanisms underlying glomerular lesions in DKD.
In addition, the evidences for the relationship between MBL and DKD were mainly concerning type 1 diabetes (T1D)31. Increased levels of MBL were linked to the development of renal injury in patients with T1D9,10,29. A Danish study reported associations between high MBL-expressing MBL2 genotypes and the risk of arising DKD in T1D patients33. MBL deficiency attenuated renal changes in a streptozotocin-induced mouse model of T1D8. In contrast, the correlation between MBL and DKD in T2D is less well-defined31. The serum/plasma levels of MBL and collectin11 were significantly increased in T2D patients with incipient and overt nephropathy4,30,34. Further research is needed to explore the role of LP in the development of DKD among patients with T2D.
In our study, complement proteins of the other two pathways, i.e., CP and AP, rather than LP, were identified in the glomeruli of DKD. In addition, increased glomerular abundances of C1q and C4d were correlated with more severe kidney damage and unfavorable renal outcomes, consistent with previous studies13,23,35,36, and highlighting the essential role of CP activation in the progression of DKD. However, the underlying mechanisms how CP is activated in DKD remain unclear. Conventionally, the immune complex mediates the initiation of complement CP through the binding of C1q to the Fc regions of antibodies in an immune complex. Interestingly, C1q has been identified de novo as a binding protein for advanced glycation end products11, providing a possible explanation for triggering CP in diabetes. A better understanding of the activation of complement CP in the diabetic state will depend on a deeper recognition of C1q in future investigations.
The role of complement AP activation in the pathogenesis of DKD has gained increasing attention. Bioinformation analysis revealed that the differentially expressed genes were enriched in alternative complement pathway in podocytes from DKD patients15. Our study showed that complement AP proteins, including CFB, CFH, CFHR1, CFHR3, and CFHR5, were significantly increased in the glomeruli of DKD patients. Furthermore, AP activation was identified as the third most upregulated biological process in the GO analysis of the glomerular proteome in DKD. High glucose is proposed to induce the glycation of complement regulators, leading to dysfunction of their regulatory ability, followed by overactivation of the complement AP6,37. However, whether the overactivation of complement AP in the setting of diabetes is due to glycation-induced impairment of complement regulators needs further exploration.
In conclusion, this was the first study to investigate the glomerular proteome in human kidney biopsies from DKD patients. Activation of complement CP and AP, but not the LP, was identified in the glomeruli of DKD patients and correlated with the severity of glomerular injury. Our results address the critical roles of the CP and AP in DKD development. Because complement activation was not attenuated by conventional DKD treatments, our findings highlight the need for further research to better understand the mechanism of complement activation in the pathogenesis of DKD and the potential use of complement blockade in the disease.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Special thanks to Dr. Qingwei Wang and Wenyan Wu for their help with the collection of normal kidney tissues. This study was supported by a grant from the National Natural Science Foundation of China (82270766), and a grant from the Key Scientific and Technological Project of the Ministry of Health of China (SBGJ202102126).
Author contributions
G.L.X. was responsible for study design. Y.Y. and Y.Z. conducted the study, analyzed the data, and wrote the manuscript. Y.L. participated in conducting the study and analyzing the data. X.J.Z, K.H., D.D.K., and T.T. contributed to discussion and reviewed and edited the manuscript. J.H.Y. and Z.P.X. assisted in the data analysis. L.L. and R.M.H. were responsible for data collection. W.M.X. and W.Y. contributed to the revision of the manuscript.
Data availability
All the data in this study are available from the corresponding author on reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yang Yang and Ying Zhang contributed equally to this work
References
- 1.Sagoo, M. K. & Gnudi, L. Diabetic nephropathy: An overview. Methods Mol. Biol. 3–7, (2067). 10.1007/978-1-4939-9841-8_1 (2020). [DOI] [PubMed]
- 2.Fan, G. et al. Urine proteomics identifies biomarkers for diabetic kidney disease at different stages. Clin. Proteom.18, 32. 10.1186/s12014-021-09338-6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao, L. et al. Urinary complement proteins and risk of end-stage renal disease: Quantitative urinary proteomics in patients with type 2 diabetes and biopsy-proven diabetic nephropathy. J. Endocrinol. Invest.44, 2709–2723. 10.1007/s40618-021-01596-3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu, H., Deng, S., Zheng, M. & Hu, K. iTRAQ plasma proteomics analysis for candidate biomarkers of type 2 incipient diabetic nephropathy. Clin. Proteom.16, 33. 10.1186/s12014-019-9253-1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Araumi, A. et al. Urinary and plasma proteomics to discover biomarkers for diagnosing between diabetic nephropathy and minimal change nephrotic syndrome or membranous nephropathy. Biochem. Biophys. Rep.27, 101102. 10.1016/j.bbrep.2021.101102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petr, V. & Thurman, J. M. The role of complement in kidney disease. Nat. Rev. Nephrol.19, 771–787. 10.1038/s41581-023-00766-1 (2023). [DOI] [PubMed] [Google Scholar]
- 7.Fortpied, J., Vertommen, D. & Van Schaftingen, E. Binding of mannose-binding lectin to fructosamines: A potential link between hyperglycaemia and complement activation in diabetes. Diabetes Metab. Res. Rev.26, 254–260. 10.1002/dmrr.1079 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Ostergaard, J. A. et al. Diabetes-induced changes in mannan-binding lectin levels and complement activation in a mouse model of type 1 diabetes. Scand. J. Immunol.77, 187–194. 10.1111/sji.12027 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Saraheimo, M. et al. Increased levels of mannan-binding lectin in type 1 diabetic patients with incipient and overt nephropathy. Diabetologia48, 198–202. 10.1007/s00125-004-1594-1 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Hansen, T. K. et al. Association between mannose-binding lectin, high-sensitivity C-reactive protein and the progression of diabetic nephropathy in type 1 diabetes. Diabetologia53, 1517–1524. 10.1007/s00125-010-1742-8 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Chikazawa, M. et al. Identification of C1q as a binding protein for advanced glycation end products. Biochemistry55, 435–446. 10.1021/acs.biochem.5b00777 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Bus, P. et al. Complement activation in patients with diabetic nephropathy. Kidney Int. Rep.3, 302–313. 10.1016/j.ekir.2017.10.005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiao, Y. et al. Activation of complement C1q and C3 in glomeruli might accelerate the progression of diabetic nephropathy: Evidence from transcriptomic data and renal histopathology. J. Endocrinol. Invest.13, 839–849. 10.1111/jdi.13739 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu, B. et al. Investigation of the mechanism of complement system in diabetic nephropathy via bioinformatics analysis. J. Diabetes Res.2021 (5546199). 10.1155/2021/5546199 (2021). [DOI] [PMC free article] [PubMed]
- 15.Lu, Q. et al. Complement factor B in high glucose-induced podocyte injury and diabetic kidney disease. JCI Insight. 6, e147716. 10.1172/jci.insight.147716 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun, Z. J., Chang, D. Y., Chen, M. & Zhao, M. H. Deficiency of CFB attenuates renal tubulointerstitial damage by inhibiting ceramide synthesis in diabetic kidney disease. JCI Insight. 7, e156748. 10.1172/jci.insight.156748 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tervaert, T. W. et al. Pathologic classification of diabetic nephropathy. J. Am. Soc. Nephrol.21, 556–563. 10.1681/ASN.2010010010 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Morita, Y. et al. Complement activation products in the urine from proteinuric patients. J. Am. Soc. Nephrol.11, 700–707. 10.1681/ASN.V114700 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Li, X. Q., Chang, D. Y., Chen, M. & Zhao, M. H. Complement activation in patients with diabetic nephropathy. Diabetes Metab.45, 248–253. 10.1016/j.diabet.2018.04.001 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Pelletier, K. et al. Clinical value of complement activation biomarkers in overt diabetic nephropathy. Kidney Int. Rep.4, 797–805. 10.1016/j.ekir.2019.03.004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng, J. M. et al. Pathological significance of urinary complement activation in diabetic nephropathy: A full view from the development of the disease. J. Diabetes Investig. 10, 738–744. 10.1111/jdi.12934 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan, S. M. et al. Complement C5a induces renal Injury in diabetic kidney disease by disrupting mitochondrial metabolic agility. Diabetes69, 83–98. 10.2337/db19-0043 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Duan, S. et al. Association of glomerular complement C4c deposition with the progression of diabetic kidney disease in patients with type 2 diabetes. Front. Immunol.11, 2073. 10.3389/fimmu.2020.02073 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yiu, W. H. et al. Complement C5a inhibition moderates lipid metabolism and reduces tubulointerstitial fibrosis in diabetic nephropathy. Nephrol. Dial Transpl.33, 1323–1332. 10.1093/ndt/gfx336 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Li, X. Q., Chang, D. Y., Chen, M. & Zhao, M. H. Deficiency of C3a receptor attenuates the development of diabetic nephropathy. BMJ Open. Diabetes Res. Care. 7, e000817. 10.1136/bmjdrc-2019-000817 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, J. et al. Implication of decreased serum complement 3 in patients with diabetic nephropathy. Acta Diabetol.55, 31–39. 10.1007/s00592-017-1060-4 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Morigi, M. et al. C3a receptor blockade protects podocytes from injury in diabetic nephropathy. JCI Insight. 5, e131849. 10.1172/jci.insight.131849 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, L. et al. Complement C5 activation promotes type 2 diabetic kidney disease via activating STAT3 pathway and disrupting the gut-kidney axis. J. Cell. Mol. Med.25, 960–974. 10.1111/jcmm.16157 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaunisto, M. A. et al. Elevated MBL concentrations are not an indication of association between the MBL2 gene and type 1 diabetes or diabetic nephropathy. Diabetes58, 1710–1714. 10.2337/db08-1495 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan, L. Z., Tong, Q. & Xu, J. Elevated serum levels of mannose-binding lectin and diabetic nephropathy in type 2 diabetes. PloS One. 10, e0119699. 10.1371/journal.pone.0119699 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan, S. M., Snelson, M., Ostergaard, J. A. & Coughlan, M. T. The complement pathway: New insights into immunometabolic signaling in diabetic kidney disease. Antioxid. Redox Signal.37, 781–801. 10.1089/ars.2021.0125 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng, J. M. et al. Lectin-induced renal local complement activation is involved in tubular interstitial injury in diabetic nephropathy. Clin. Chim. Acta. 482, 65–73. 10.1016/j.cca.2018.03.033 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Hansen, T. K. et al. Association between mannose-binding lectin and vascular complications in type 1 diabetes. Diabetes53, 1570–1576. 10.2337/diabetes.53.6.1570 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Hansen, T. K. et al. Mannose-binding lectin and mortality in type 2 diabetes. Arch. Intern. Med.166, 2007–2013. 10.1001/archinte.166.18.2007 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Tang, X. et al. The clinical impact of glomerular immunoglobulin M deposition in patients with type 2 diabetic nephropathy. Am. J. Med. Sci.356, 365–373. 10.1016/j.amjms.2018.07.001 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Sun, Z. J. et al. Complement deposition on renal histopathology of patients with diabetic nephropathy. Diabetes Metab.45, 363–368. 10.1016/j.diabet.2018.08.011 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Flyvbjerg, A. The role of the complement system in diabetic nephropathy. Nat. rev. Nephrol.13, 311–318. 10.1038/nrneph.2017.31 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data in this study are available from the corresponding author on reasonable request.